- Department of Biology, Institute for Genetics, University of Cologne, Cologne, Germany
LeuO is a conserved and pleiotropic transcription regulator, antagonist of the nucleoid-associated silencer protein H-NS, and important for pathogenicity and multidrug resistance in Enterobacteriaceae. Regulation of transcription of the leuO gene is complex. It is silenced by H-NS and its paralog StpA, and it is autoregulated. In addition, in Escherichia coli leuO is antagonistically regulated by the heterodimeric transcription regulator BglJ-RcsB and by LeuO. BglJ-RcsB activates leuO, while LeuO inhibits activation by BglJ-RcsB. Furthermore, LeuO activates expression of bglJ, which is likewise H-NS repressed. Mutual activation of leuO and bglJ resembles a double-positive feedback network, which theoretically can result in bi-stability and heterogeneity, or be maintained in a stable OFF or ON states by an additional signal. Here we performed quantitative and single-cell expression analyses to address the antagonistic regulation and feedback control of leuO transcription by BglJ-RcsB and LeuO using a leuO promoter mVenus reporter fusion and finely tunable bglJ and leuO expression plasmids. The data revealed uniform regulation of leuO expression in the population that correlates with the relative cellular concentration of BglJ and LeuO. The data are in agreement with a straightforward model of antagonistic regulation of leuO expression by the two regulators, LeuO and BglJ-RcsB, by independent mechanisms. Further, the data suggest that at standard laboratory growth conditions feedback regulation of leuO is of minor relevance and that silencing of leuO and bglJ by H-NS (and StpA) keeps these loci in the OFF state.
Introduction
LeuO is a conserved and pleiotropic LysR-type transcription factor that has been best characterized in Escherichia coli and Salmonella enterica. LeuO functions both as activator and as repressor, and is presumably a tetramer, similar to other LysR-type regulators (Maddocks and Oyston, 2008; Guadarrama et al., 2014). LeuO is a master regulator with more than 100 target loci, and supposedly an important H-NS antagonist, since many LeuO-activated loci are H-NS repressed (Ueguchi et al., 1998; Chen et al., 2003; Chen and Wu, 2005; De la Cruz et al., 2007; Stoebel et al., 2008; Stratmann et al., 2008, 2012; Shimada et al., 2011; Dillon et al., 2012; Ishihama et al., 2016). In addition, genomics data revealed a significant overlap of co-regulation by LeuO and H-NS both in E. coli and in S. enterica, where 78 and 40%, respectively, of the LeuO targets are H-NS bound (Shimada et al., 2011; Dillon et al., 2012; Ishihama et al., 2016). H-NS represses transcription by formation of extended complexes on the DNA (Dillon and Dorman, 2010; Landick et al., 2015; Winardhi et al., 2015). For activation of H-NS repressed loci by LeuO several mechanisms have been proposed including alteration of the repressing H-NS nucleoprotein-complex, the prevention of spreading of the H-NS complex, and competition with H-NS for DNA binding (Chen and Wu, 2005; Shimada et al., 2011; Dillon et al., 2012). The biological role of LeuO is pleiotropic. LeuO is relevant for pathogenicity in S. enterica, for biofilm formation in Vibrio cholerae and E. coli, as well as the acid stress response and multidrug efflux in E. coli (Stoebel et al., 2008; Shimada et al., 2009, 2011; Dillon et al., 2012). Further, LeuO activates expression of the H-NS repressed genes coding for the CRISPR/Cas immunity system in E. coli and S. enterica (Pul et al., 2010; Westra et al., 2010; Medina-Aparicio et al., 2011).
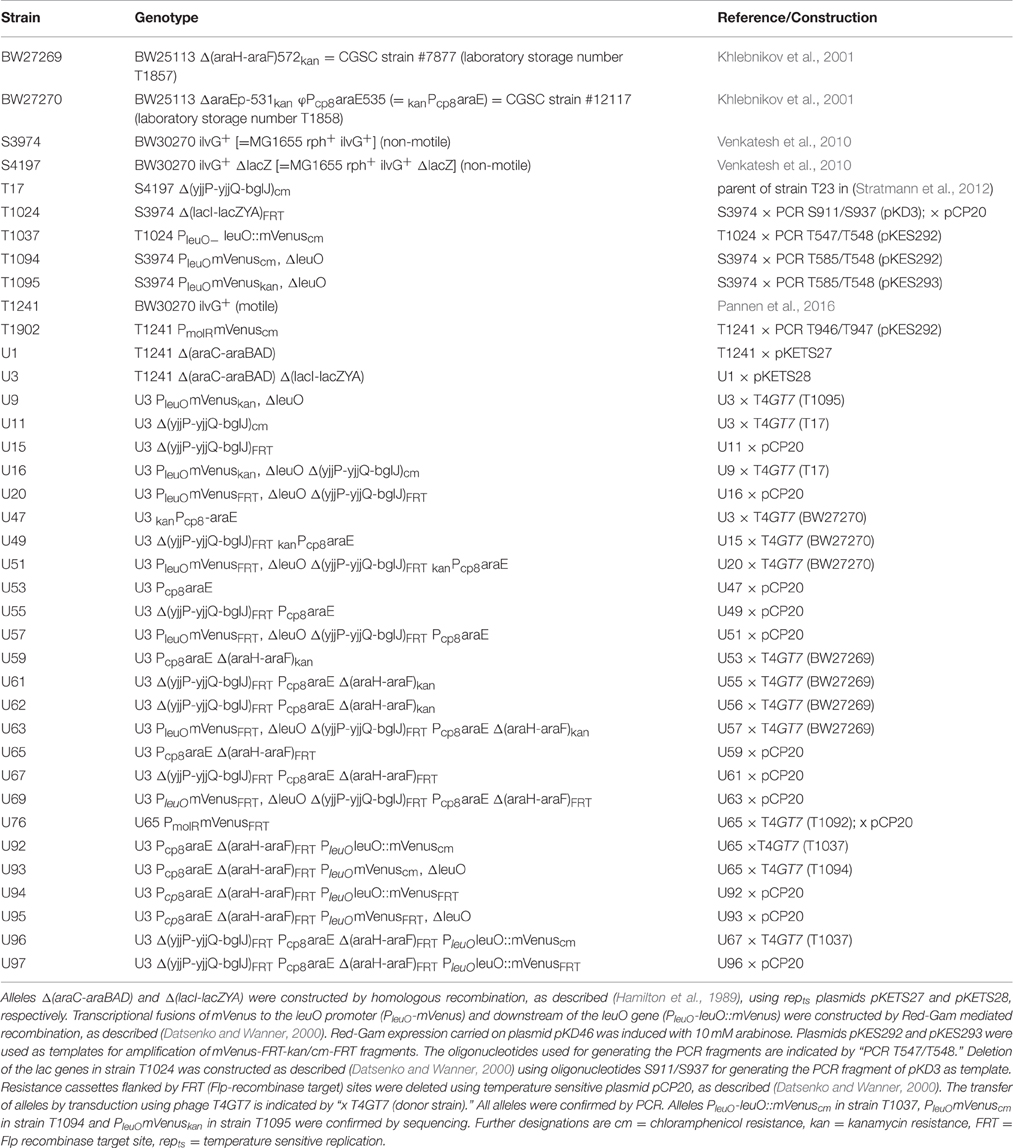
In accordance with the pleiotropic role of LeuO, transcription of leuO is tightly controlled. Under laboratory conditions the leuO gene is repressed by H-NS and by the H-NS paralog StpA, and thus the leuO gene is silent in E. coli and S. enterica (Klauck et al., 1997; Chen et al., 2001). Moderate upregulation of leuO expression was observed in stationary phase and under amino acid starvation (Fang and Wu, 1998; Fang et al., 2000; Majumder et al., 2001; Shimada et al., 2011; Dillon et al., 2012). In addition, positive autoregulation by LeuO and transcriptional coupling of leuO expression to expression of neighboring genes by DNA supercoiling has been reported (Fang and Wu, 1998; Chen et al., 2003). Furthermore, in E. coli leuO is activated by the heterodimeric transcription regulator BglJ-RcsB (Stratmann et al., 2012). Activation of leuO by BglJ-RcsB is inhibited by LeuO, and LeuO represses leuO transcription in hns and in hns stpA mutants (Figure 1A). Thus, LeuO is also a negative autoregulator (Stratmann et al., 2012). The leuO gene is preceded by at least two promoters (P1 and P2) which are repressed by H-NS and StpA and negatively autoregulated by LeuO in hns stpA mutants; the P2 promoter is activated by BglJ-RcsB (Stratmann et al., 2012). BglJ-RcsB is a heterodimer that activates transcription of various loci in E. coli (Venkatesh et al., 2010; Stratmann et al., 2012; Salscheider et al., 2014). BglJ-RcsB consists of RcsB, the response regulator of the Rcs two-component phosphorelay system (Majdalani and Gottesman, 2005), and BglJ, which has initially been found as an activator of the bgl operon (Giel et al., 1996). Further, BglJ-RcsB is active independent of phosphorylation of RcsB by the Rcs phosphorelay (Venkatesh et al., 2010; Stratmann et al., 2012; Pannen et al., 2016).
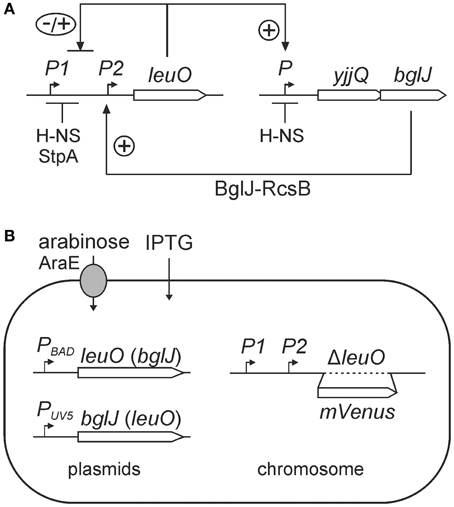
Figure 1. (A) Regulation of leuO by interlocked double-positive and negative feedback loops. Transcription of leuO is repressed by H-NS and StpA, and is activated by the BglJ-RcsB heterodimer. LeuO activates transcription of the yjjQ-bglJ operon that is also repressed by H-NS. Mutual positive regulation represents a double-positive feedback loop. In addition, LeuO inhibits activation of the leuO promoter P2 by BglJ-RcsB resembling a negative feedback. (B) Experimental system for analyzing regulation of leuO transcription by BglJ-RcsB and LeuO. To monitor leuO transcription a PleuO mVenus fusion was constructed by replacement of the native leuO gene with mVenus. The chromosomal copy of bglJ was deleted (allele Δ[yjjP-yjjQ-bglJ]) to avoid feedback regulation via LeuO. BglJ and LeuO were provided by two sets of compatible plasmids that are pKES303 (PBAD leuO, p15A-ori) and pKETS26 (PUV5 bglJ, pSC-ori) or plasmid pKES302 (PBAD bglJ) and pKETS25 (PUV5 leuO). Expression of bglJ and leuO, respectively, was induced with gradually increasing concentrations of the inducers arabinose and IPTG, respectively. To avoid feedback regulation by arabinose the strain background is Δ(araC araBAD) ΔaraH-F, Pcp8 araE resulting in constitutive expression of the arabinose transporter AraE. In addition, the lac genes were deleted, allele Δ(lacI-lacZYA), for enabling gradual induction by IPTG.
Intriguingly, activation of leuO by BglJ-RcsB is one element of a presumptive double-positive feedback loop, since LeuO in turn activates expression of the yjjQ-bglJ operon that is likewise H-NS repressed (Stratmann et al., 2008). This double-positive feedback loop is interlocked with a negative feedback loop which is based on negative autoregulation by LeuO (Figure 1). Such a network motif can function like a switch that is stable both in the OFF as well as in the ON state. Often an external signal locks such feedback loops in one state. Further, bi-stability resulting in population heterogeneity and oscillation can be based on interlocked positive and negative feedback loops (Angeli et al., 2004; Alon, 2007; Shoval and Alon, 2010).
In this study we addressed the antagonistic regulation of leuO transcription by BglJ-RcsB and LeuO, which is presumably a crucial element in the complex control of leuO expression. For quantitative and single-cell expression analysis, we established a reporter fusion of the leuO promoter region (PleuO) to mVenus and expressed bglJ and leuO in trans using tightly controlled and gradually inducible plasmidic expression systems. Expression analyses of the PleuO mVenus reporter at steady state growth conditions revealed uniform expression. The level of leuO expression correlates with the relative cellular concentration of BglJ and LeuO. The data are in agreement with a straightforward model of antagonistic regulation by the two regulators that act independently of each other.
Results
Experimental System for Analyzing Regulation of leuO Expression by BglJ and LeuO
The regulation of leuO transcription by BglJ-RcsB and LeuO is an important element in the control of the LeuO master regulator. To address regulation of leuO transcription that is directed by at least two promoters (PleuO) in dependence of the concentrations of BglJ and LeuO, a suitable experimental system was established. First, the mVenus reporter gene (coding for the yellow fluorescent protein mVenus) was fused to the leuO promoter-regulatory region by replacement of the leuO gene resulting in allele PleuO mVenus, ΔleuO (Figure 1B). Second, BglJ and LeuO were ectopically expressed from two different sets of plasmids. In one plasmid set, bglJ was expressed under control of the IPTG-inducible lacUV5 promoter (PUV5) using low-copy plasmid pKETS26 (pSC origin of replication), and leuO was expressed under control of the arabinose-inducible PBAD promoter using the low to medium copy plasmid pKES303 (pBAD30-derived, p15A origin of replication). In the other plasmid set, bglJ was expressed under control of the PBAD promoter (pKES302, p15A-ori) and leuO under control of IPTG-inducible Ptac promoter (pKEHB27, pSC-ori). The genes encoding the AraC and the LacI regulators, respectively, are also carried on these plasmids. Additionally, the yjjQ-bglJ operon was deleted resulting in allele Δ(yjjP-yjjQ-bglJ) to ensure that only plasmid-encoded BglJ is present in the cell. Note that RcsB is not limiting for activation of leuO and other loci by BglJ-RcsB (Salscheider et al., 2014; Pannen et al., 2016). Third, to allow controlled and finely tunable expression of bglJ and leuO directed by the arabinose-inducible PBAD promoter and the IPTG-inducible PUV5 and Ptac promoters, respectively, additional mutations and modifications were introduced into the reporter strain (Figure 1B). The PUV5 promoter is gradually induced over a range of inducer concentrations (IPTG) when the lactose permease gene lacY is deleted (Jensen et al., 1993). Therefore, the lacZYA operon and the lacI gene were deleted in the reporter strain resulting in allele Δ(lacI-lacZYA) (Table 1). Likewise, the arabinose regulon was modified to ensure a gradual induction of the PBAD promoter with arabinose, as described before (Khlebnikov et al., 2001; Kogenaru and Tans, 2014). Briefly, the PBAD promoter is known to have a stochastic behavior when induced with arabinose. This stochastic behavior is caused by the araE and araFGH genes encoding the arabinose transporters, because induction of the transporter genes by arabinose leads to a higher arabinose uptake and thus positive feedback (Siegele and Hu, 1997; Megerle et al., 2008). In addition, a negative feedback caused by fermentation of intracellular arabinose through the AraBAD enzymes leads to a non-gradual induction (Siegele and Hu, 1997). To avoid the negative and positive feedback, the araC gene and the araBAD and araFGH operons were deleted. Further, the low affinity arabinose transporter araE was put under the control of constitutive promoter Pcp8, as described (Khlebnikov et al., 2001; Kogenaru and Tans, 2014). The genotype of the resulting reporter strain U69 is PleuO mVenus ΔleuO Δ(yjjP-yjjQ-bglJ) φ(ΔaraEp Pcp8 araE) Δ(araH-F) Δ(araC-araBAD) Δ(lacI-lacZYA) (Table 1). Using this strain the expression level of PleuO mVenus was measured by flow-cytometry to quantify the cellular fluorescence in the population. Further, to ensure steady state conditions, cultures were grown in nutrient-poor tryptone medium. In this medium cultures that were inoculated from fresh overnight cultures to OD600 of 0.05 reached an OD600 of about 0.7–1 after 5 h of growth.
Regulation of leuO Promoter by BglJ–RcsB and by LeuO
First, activation of the PleuO mVenus fusion by BglJ-RcsB was tested. To this end, the reporter strain U69 was transformed with low-copy plasmid pKETS26 carrying bglJ under control of the IPTG-inducible PUV5 promoter (PUV5 bglJ, pSC-ori), and with plasmid pKES302 carrying bglJ under control of the arabinose-inducible PBAD promoter (PBAD bglJ, p15A-ori), respectively (Figure 2). Expression of bglJ was either not induced or induced by gradually increasing inducer concentrations. The analysis of PleuO mVenus expression by flow-cytometry revealed that gradual induction of PBAD bglJ expression (plasmid pKES302) with 2 μM–50 μM arabinose resulted in full activation of PleuO mVenus even at the very low arabinose concentration of 2 μM (Figures 2B,C). Induction of PBAD bglJ with 100 μM arabinose or higher concentrations caused growth defects. However, induction of PUV5 bglJ with IPTG concentration ranging from 10 μM to 100 μM led to a gradual increase in expression of PleuO mVenus and this increase was uniform in the population (Figures 2B,D). The presence of the PUV5 bglJ or the PBAD bglJ plasmids per se did not cause a significant increase in expression of PleuO mVenus (Figures 2B–D). Likewise, IPTG or arabinose induction of transformants of the empty vectors pBAD30 and pKETS24, respectively, had no effect (Figure 2B). Taken together these data confirm activation of leuO transcription by BglJ-RcsB, they suggest that low cellular levels of BglJ are sufficient for activation, and that the PUV5 bglJ plasmid is suitable for gradual induction of bglJ, while the PBAD bglJ plasmid is not suitable.
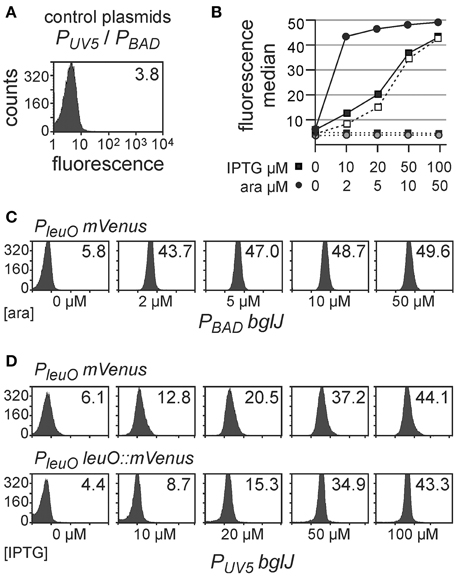
Figure 2. Activation of leuO transcription by BglJ. Expression of PleuO mVenus (in strain U69) and PleuO leuO::mVenus (strain U97) transcriptional fusions was determined by flow cytometry in absence and presence of the transcriptional activator BglJ, which was provided by plasmids. Expression was analyzed after 5 h of growth in tryptone medium without and with indicated inducer concentrations at an optical density OD600 of approximately 0.7–1. (A) Fluorescence intensity directed by PleuO mVenus in individual cells of transformants of strain U69 with the empty vectors pKETS24 (PUV5 in pSC-ori) and pBAD30 (PBAD in p15A-ori). Yellow fluorescence (X-axis) is given in arbitrary units and the Y-axis gives the number of cells that were counted. The median of the fluorescence intensity is given in the upper right corner of the graph. (B) Plot of the median fluorescence values that are shown in (C) (solid line with filled dots) and (D) (solid line with filled squares PleuO mVenus and dashed line with open squares PleuO leuO::mVenus). In addition, median fluorescence values of transformants of vector controls are shown (pKETS24, PUV5 as dotted line and filled squares, and pBAD30, PBAD dotted line with gray dots). (C) Fluorescence intensity of transformants of strain U69 with plasmids pKES302 (PBAD bglJ in p15A-ori) and pKETS24 (PUV5 in pSC-ori). The arabinose concentration used for induction of bglJ expression is given underneath the panels. (D) Fluorescence intensity of transformants of strain U69 (PleuO mVenus) with plasmids pKETS26 (PUV5 bglJ in pSC-ori) and pBAD30 (PBAD in p15A-ori), as well as of strain U97 (PleuO leuO::mVenus). The IPTG concentration used for induction of bglJ expression is given underneath the panels. Shown are representative data.
Second, autoregulation of PleuO mVenus by LeuO was analyzed using the leuO providing plasmids PUV5 leuO (pKETS25, pSC-ori) and Ptac leuO (pKEHB27, pSC-ori) which carry leuO under control of the IPTG-inducible PUV5 and Ptac promoters, respectively. In addition, a PBAD leuO plasmid (pKES303, p15A-ori) was used. The promoter PUV5 (carrying the UV5 mutation in the—10 box and the lacL8 mutation in the CRP-binding site) is ~10 times weaker than the Ptac promoter (Lanzer and Bujard, 1988), while the tightly regulated PBAD leuO plasmid presumably directs similar levels of LeuO as the Ptac leuO plasmid considering that the PBAD promoter is approximately 3 fold weaker than Ptac and that the copy number of the PBAD plasmid (pKES303, p15A-ori) is ~3-fold higher than the copy number of the pSC-derived Ptac plasmid (Guzman et al., 1995). Flow cytometry revealed a slight increase in PleuO mVenus expression at low levels of induction of plasmidic leuO (Figure 3). The data seem in agreement with weak positive autoregulation that was reported previously (Fang and Wu, 1998; Chen et al., 2003), but are statistically not significant (student's t-test, P-value > 0.05).
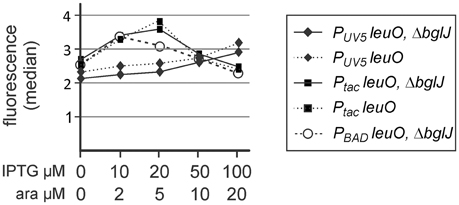
Figure 3. Autoregulation of leuO transcription. Fluorescence expression levels directed by the PleuO mVenus fusion were determined by flow cytometry. The PleuO mVenus reporter strains U69 carrying a deletion of bglJ (ΔyjjP-yjjQ-bglJ) and its isogenic wild-type bglJ+ derivative U95 were transformed with plasmids pKETS25 (pSC-ori) that carries leuO under control of PUV5, pKEHB27 (p15A-ori) that carries leuO under control of Ptac, and pKES303 (p15A-ori) that carries leuO under control of PBAD. The fluorescence median is plotted against the inducer concentration. Expression was analyzed by flow cytometry after 5 h of growth in trypton medium, IPTG, and arabinose were added at the indicated concentrations. Statistical analysis suggests that the difference in the expression level is not significant (P > 0.05).
Antagonistic Regulation of the leuO Promoter by BglJ–RcsB and by LeuO
Next we addressed antagonistic regulation of PleuO mVenus by BglJ-RcsB and by LeuO. To this end, the PleuO mVenus reporter strain U69 was transformed with the two sets of leuO and bglJ expressing plasmids. First we analyzed antagonistic regulation of leuO transcription using the plasmid set, in which bglJ is expressed under control of the PBAD promoter (PBAD bglJ, pKES302) and leuO is expressed under control of the Ptac promoter (Ptac leuO, pKEHB27). Induction of bglJ expression with 2 μM–50 μM arabinose caused full activation of PleuO mVenus (Figure 4), irrespective of the arabinose concentration, as shown above (Figure 2). Simultaneous induction of leuO by IPTG strongly reduced BglJ-RcsB-mediated activation of PleuO mVenus, but even full induction of plasmidic leuO expression with 200 μM IPTG did not completely abrogate BglJ-RcsB-mediated activation (Figure 4). These results indicate that the level of BglJ provided by the PBAD bglJ plasmid is above a threshold up to which LeuO can fully inhibit BglJ-RcsB activation. Since the PBAD bglJ plasmid does not allow gradual activation, this plasmid set does not seem suitable for gradual induction of both regulators.
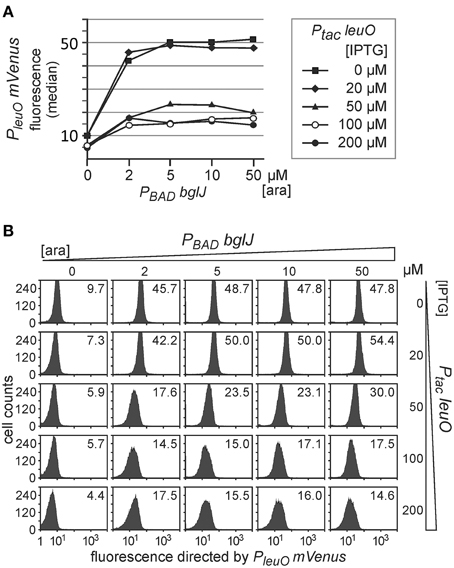
Figure 4. Antagonistic regulation of PleuO mVenus transcription by BglJ-RcsB and LeuO. Fluorescence of transformants of PleuO mVenus strain U69 with plasmids pKES302 (PBAD bglJ) and pKEHB27 (Ptac leuO) was monitored by flow cytometry. (A) The median fluorescence is plotted against the arabinose concentration used for induction of bglJ. Each line graph represents the set of data obtained of cultures grown with the specified IPTG concentration used for induction of leuO. (B) Flow cytometry data of cultures grown with increasing arabinose (rows) and IPTG (columns) concentration. Plotted in each panel are the cell counts against the fluorescence intensity. The fluorescence distribution in each panel is in agreement with uniform expression within the population. The fluorescence median that is plotted in (A) is given within each panel. Cultures were inoculated from overnight cultures to an OD600 of 0.05 and grown for 5 h in 10 ml tryptone medium containing ampicillin, chloramphenicol, as well as IPTG and arabinose at the indicated concentrations.
Second, we analyzed antagonistic regulation of PleuO mVenus using the reverse set of plasmids that includes PUV5 bglJ (pKETS26) and PBAD leuO (pKES303) (Figure 5). With this set of plasmids expression levels of BglJ are lower and gradual induction of bglJ by IPTG resulted in a gradual increase in activation of the PleuO mVenus fusion by BglJ-RcsB (Figure 5, compare with data in Figure 2). Simultaneous gradual induction of plasmidic PBAD leuO with arabinose and of PUV5 bglJ with IPTG led to a uniform decrease of expression of PleuO mVenus in the whole population as compared to level of activation by BglJ-RcsB alone (Figure 5). Induction of leuO with an arabinose concentration of 50 μM was sufficient to completely abrogate activation by BglJ-RcsB (bottom right panel, Figure 5B). A plot of the median values of the flow cytometry results visualizes the gradual effects (Figure 5A).
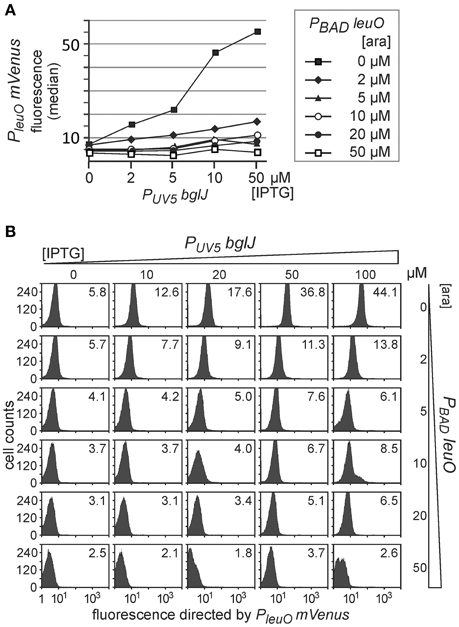
Figure 5. Antagonistic regulation of PleuO mVenus transcription by BglJ and LeuO. Transformants of PleuO mVenus strain U69 with plasmids pKES303 (PUV5 bglJ) and pKEHB28 (PBAD leuO) were grown for 5 h in tryptone medium containing arabinose and IPTG at the indicated concentrations. Fluorescence was monitored by flow cytometry. (A) The median fluorescence is plotted against the IPTG concentration that was used for induction of bglJ. Each line graph represents the set of median fluorescence data that was obtained when plasmidic leuO was induced with the indicated arabinose concentrations. (B) Flow cytometry data of cultures grown with increasing IPTG (rows) and arabinose (columns) concentration (presentation of data as in Figure 4).
Taken together, the data confirm that LeuO counteracts activation of the leuO promoter by BglJ-RcsB. Further, the data show that antagonistic regulation of the leuO promoters by LeuO and by BglJ-RcsB depends on the relative concentration of BglJ and LeuO, and the data indicate that BglJ-RcsB-mediated activation of PleuO mVenus is inhibited by LeuO only if BglJ levels are rather low. The experimental data shown in Figure 5 were used to describe PleuO activity in dependence of the concentration of BglJ and LeuO by a thermodynamic model based on Michaelis-Menten kinetics. In this model it was assumed that BglJ and LeuO regulate PleuO independently of each other. Fitting of the function to the experimental data was significant (P-value < 0.001) (function plotted in Figure 6).
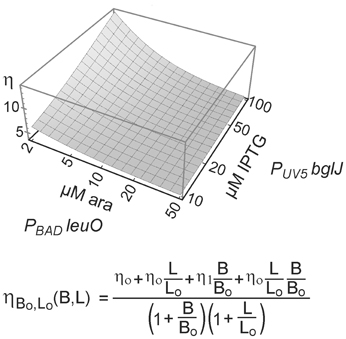
Figure 6. Modeling of antagonistic regulation of leuO transcription by BglJ-RcsB and LeuO. To describe the transcription rate directed by PleuO in dependence of the concentration of BglJ and LeuO, a thermodynamic model based on Michaelis-Menten kinetics was used. In this model it was assumed that BglJ and LeuO bind and regulate leuO transcription independently of each other. Median fluorescence values of flow cytometry data (Figure 5) were fitted to the function (bottom) describing leuO promoter activity in dependence of promoter occupancy by BglJ and LeuO. Fitting of the parameters to the experimental data by nonlinear regression according to (Fox and Weisberg, 2011) yielded P-values < 0.001. The data were plotted with Mathematica (Wolfram Research) using logarithmic scales for induction of plasmidic leuO with arabinose (ara) and of plasmidic bglJ with IPTG.
Analysis of Feedback Regulation of leuO via yjjQ–bglj and by LeuO
Next we addressed the relevance of the presumptive double-positive feedback regulation of leuO and bglJ by including the native gene of one of these two players, while providing the other one by the expression plasmid. In particular, we analyzed whether presence of the native yjjQ-bglJ operon that is activated by LeuO results in enhanced PleuO mVenus expression, when LeuO is provided in trans. Second, we tested whether the presence of native leuO might affect activation of PleuO by BglJ-RcsB.
For determining whether activation of the H-NS repressed yjjQ-bglJ operon by LeuO may yield sufficient BglJ protein for activation of PleuO we compared PleuO mVenus expression in (yjjQ-bglJ)+ strain U95 with expression in the isogenic Δ(yjjQ-bglJ) strain U69 (Figure 3). The data revealed no difference between wild-type yjjQ-bglJ+ strain U95 and Δ(yjjQ-bglJ) strain U69 suggesting that activation of yjjQ-bglJ by LeuO is either too low to provide sufficient levels of BglJ for activation of PleuO mVenus or that LeuO interferes with activation by BglJ-RcsB. Second, we analyzed whether the presence of native leuO may affect activation of the leuO promoter by BglJ-RcsB. For this analysis the leuO gene was retained at its native locus and the fluorescence reporter gene mVenus was inserted downstream of leuO (as a transcriptional fusion) resulting in allele PleuO leuO::mVenus in strain U97. Transformants of this strain with bglJ carrying plasmid pKETS26 (PUV5 bglJ, pSC-ori), were grown with IPTG concentrations ranging from 10 μM to 200 μM and PleuO leuO::mVenus expression was determined by flow cytometry. Comparison of the data obtained of PleuO leuO::mVenus with the data obtained for PleuO mVenus (ΔleuO) revealed no significant difference (Figures 2B,D). These data indicate that induction of the native leuO gene by BglJ does not provide sufficient LeuO to antagonize BglJ-RcsB-mediated activation of leuO.
Furthermore, we analyzed whether LeuO inhibits BglJ-RcsB-mediated activation of leuO transcription indirectly by downregulating BglJ-RcsB activity rather than by inhibiting activation of the leuO P2 promoter by BglJ-RcsB. To this end, activation of another BglJ-RcsB-activated promoter, the molR promoter (Salscheider et al., 2014), was analyzed in absence and presence of LeuO. BglJ was provided by PUV5 bglJ plasmid pKETS26, and LeuO was provided by PBAD leuO plasmid pKES303. As control, transformants with the empty vectors were analyzed in parallel. Activity of the molR promoter was determined using a PmolR mVenus reporter fusion. The expression analyses demonstrate that LeuO neither does affect activation of PmolR by BglJ-RcsB nor does LeuO-mediated activation of the native yjjQ-bglJ operon present in strain U76 lead indirectly to activation of PmolR (Figure 7). We note that induction of the PBAD leuO with 50 μM arabinose resulted in slower growth to OD600 = 0.6 after 5 h as compared to OD600 = 1 which may explain the 1.5-fold reduce in basal expression of PmolR mVENUS in transformants of PBAD leuO plasmid pKES303 and control plasmid PUV5 pKETS24 (Figure 7).
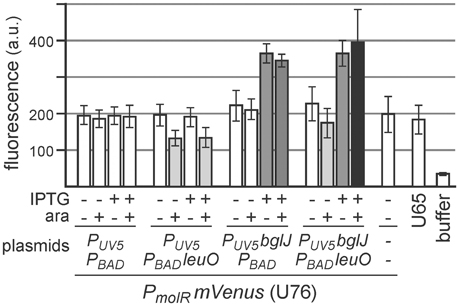
Figure 7. Activation of the molR promoter (PmolR) by BglJ-RcsB is not affected by LeuO. For determining activation of PmolR by BglJ-RcsB strain U76 was used that carries a replacement of the molR coding region by mVenus. Transformants of U76 with plasmids carrying PUV5 bglJ (pKETS26) and Ptac leuO (pKES303) as well as control plasmids (pKETS24 and pBAD30) were grown in tryptone medium for 5 h. For induction (+) IPTG (100 μM) and arabinose (50 μM) were added. When harvested, the cultures had an OD600 of approximately 1, while induction of leuO resulted in slower growth to OD600 of approximately 0.6. Yellow fluorescence of three biological replicates was determined and expression levels are given in arbitrary units (a. u.).
Discussion
In E. coli transcription of leuO is directed by at least two promoters, P1 and P2, which are repressed by H-NS and StpA. The P2 promoter requires activation by BglJ-RcsB, while LeuO inhibits activation of P2 by BglJ-RcsB. In addition, LeuO represses the leuO promoters in hns stpA mutants. Thus, leuO is antagonistically regulated by BglJ-RcsB and LeuO. The characterization of leuO transcription using a leuO promoter-mVenus reporter fusion revealed that the antagonistic regulation of leuO transcription by LeuO and by BglJ-RcsB correlates to the relative cellular amounts of these regulators. The experimental data are in agreement with a theoretical model according to which LeuO and BglJ-RcsB regulate transcription independently. Further, data indicate that double-positive feedback regulation of leuO and bglJ is of minor relevance, at least at the laboratory steady state conditions tested, since deletion of leuO and bglJ, respectively, had no significant effect on the regulation of the leuO promoter reporter fusion by LeuO and BglJ-RcsB.
Activation of the leuO P2 promoter by the BglJ-RcsB heterodimer does not occur under standard lab conditions due to H-NS-mediated repression of the yjjQ-bglJ operon (Stratmann et al., 2008, 2012). To address the antagonistic regulation of leuO transcription by BglJ-RcsB and LeuO, we tested low to medium copy plasmids for gradual induction of bglJ under control of the PUV5 and PBAD promoter, respectively. The data show that rather low amounts of BglJ are sufficient for full activation of the leuO P2 promoter (Figures 2, 4, 5). Gradual activation of leuO by BglJ-RcsB was observed only upon gradual induction of bglJ provided by the low-copy PUV5 bglJ plasmid, while bglJ expression levels directed by the PBAD bglJ plasmid turned out to be too high even when induced with just 2 μM arabinose, while induction with 100 μM arabinose caused growth defects. Likewise, we addressed autoregulation of leuO transcription by gradual induction of leuO carrying plasmids, which carry leuO under control of the PUV5, Ptac, and PBAD promoter, respectively. The data (Figure 3) indicate that positive autoregulation of leuO that was reported previously (Fang and Wu, 1998; Chen et al., 2003; Stratmann et al., 2012) is negligible at steady state growth conditions.
Further experiments, with simultaneous gradual induction of bglJ and leuO revealed that the activity of the leuO promoter correlates with the relative BglJ and LeuO concentrations (Figure 5). Interestingly, no switch-like response was observed. This might be plausible, because the distance of the LeuO DNA-binding sites to the BglJ-RcsB DNA-binding site is more than 100 bp (Stratmann et al., 2012), and LeuO and BglJ-RcsB presumably can bind simultaneously. Therefore, the LeuO-mediated inhibition of activation by BglJ-RcsB is putatively not caused by competition for binding, but by another mechanism, as for example inhibition of RNA polymerase binding to leuO promoter P2 or inhibition of transcription initiation at P2 by LeuO. Such a mechanism of repression is supported by in vitro DNA binding analyses, which revealed that LeuO inhibits open complex formation by RNA polymerase at sites mapping next to leuO promoter P1 and reduces open complex formation by RNA polymerase at sites close to P2 (Stratmann et al., 2012). A thermodynamic model based on Michaelis-Menten kinetics (Figure 6) supports the interpretation that antagonistic regulation by BglJ-RcsB and LeuO is mediated by independent mechanisms.
Previous data suggested that LeuO is controlled by interlocked double-positive and negative feedback control, because LeuO activates expression of the H-NS repressed yjjQ-bglJ operon (Stratmann et al., 2008). In the present study we analyzed whether activation of bglJ by LeuO may indirectly also turn on transcription of PleuO mVenus (Figure 3) or PmolR mVenus as another BglJ-RcsB target (Figure 7), which was not the case indicating that activation of the native yjjQ-bglJ operon by LeuO does not yield sufficient BglJ. Likewise, expression analyses of an mVenus fusion downstream of the leuO coding region yielded the same results as the PleuO mVenus reporter indicating that LeuO levels, when expressed from its native locus, remain too low to antagonize BglJ-RcsB. Taken together, double-positive feedback regulation of the leuO and yjjQ-bglJ loci is not relevant, at least at laboratory conditions, since the presence of the native leuO gene had no effect on BglJ-RcsB mediated activation of leuO that was triggered by plasmidic bglJ. Likewise the presence of native bglJ had no influence. Thus, the data suggest that repression of leuO by H-NS and StpA and of yjjQ-bglJ by H-NS dominates regulation of these loci and keeps them in the OFF state.
Materials and Methods
Strains, Media, and Plasmids
Bacterial cultures of E. coli K-12 were grown in LB (10 g/l Bacto Tryptone, 5 g/l Bacto Yeast Extract, 5 g/l NaCl) or tryptone (10 g/l Bacto Tryptone, 5 g/l NaCl) media. Antibiotics were added with concentrations of 50 μg/ml ampicillin, 15 μg/ml chloramphenicol, and 25 μg/ml kanamycin. Strains, listed in Table 1, were constructed by transduction using phage T4GT7, by Red-Gam mediated gene deletion or gene replacement, and by homologous recombination, as described (Wilson et al., 1979; Hamilton et al., 1989; Datsenko and Wanner, 2000). Plasmids and their construction are listed in Table 2 and oligonucleotides are listed in Table 3. Standard molecular techniques, such as cloning, PCR, culture growth and induction of plasmid-provided genes, were performed according to standard protocols (Ausubel et al., 2005).
Flow Cytometry and Fluorescence Assay
For expression analyses by flow cytometry cultures of transformants were inoculated from fresh overnight cultures to an OD600 of 0.05 and grown for 5 h at 37°C in 10 ml tryptone medium containing antibiotics for selection of the plasmids. The cultures were diluted to OD600 of 0.1 and kept on ice prior to analysis by flow cytometry. Flow cytometry was performed on a BD FACScalibur flow cytometer using CellQuest software (BD Biosciences, Franklin Lakes, NJ, USA). For each sample, 50,000 events were measured at a rate between 500 and 1000 events per second. The experiments were repeated at least twice and representative sets of data are shown.
Fluorescence directed by the PmolR mVenus fusion was determined by Fluorescence spectroscopy using a CLARIOstar plate reader (BMG LABTECH, Germany). Briefly, cultures were grown as for flow cytometry and the fluorescence of cells equivalent to 1.5 OD600 was measured using yellow fluorescent proteins specific excitation (495–515 nm) and detection (540–620 nm) channels. The average obtained of three biological replicates was calculated and the standard deviation is less than 25%.
Theoretical Model
To describe the transcription rate directed by PleuO in dependence of the concentration of BglJ and LeuO, a thermodynamic model based on Michaelis-Menten kinetics was used. In this model it was assumed that BglJ and LeuO regulate PleuO independently of each other. The binding probabilities were defined as B/(Bo+B) and L/(Lo+L), where B represents the concentration of BglJ in the cell, B0 the BglJ concentration at which the promoter is half occupied, L represents the concentration of LeuO and L0 the LeuO concentration at which the promoter is half occupied. Since LeuO acts as a repressor and BglJ as an activator of the leuO promoter four different states with a different expression rate were described. The basal expression level directed by PleuO in absence of BglJ and LeuO was defined as η0. In presence of LeuO and absence of BglJ, expression remains at a basal level defined as η0. However, in presence of BglJ but absence of LeuO, the expression level is higher which is defined as η1. When BglJ and LeuO are bound at the same time, the expression rate is defined as η0, because high levels of LeuO inhibit activation by BglJ, when BglJ is provided by the low-copy PUV5 bglJ plasmid. Taking these four different states into account the expression rate of leuO in dependence of LeuO and BglJ concentration was described as
The function was fitted to the median expression values determined by flow cytometry (PUV5 bglJ, and PBAD leuO, Figure 5) using non-linear regression according to (Fox and Weisberg, 2011), which yielded a high fitting significance (P-value < 0.001).
Author Contributions
HB contributed to the design of the work, acquired the data, and together with KS interpreted the data and drafted the work. KS conceived the project, contributed to the design of the work, and drafted the work.
Funding
Funding was obtained by the Deutsche Forschungsgemeinschaft through grant SCHN 371/10-2.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Thomas Stratmann and Robin Schwarzer for construction of plasmids. We are grateful for discussions and help with theoretical modeling to Prof. Johannes Berg and Alexander Klassmann, University of Cologne.
References
Alon, U. (2007). Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8, 450–461. doi: 10.1038/nrg2102
Amann, E., Ochs, B., and Abel, K.-J. (1988). Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69, 301–315. doi: 10.1016/0378-1119(88)90440-4
Angeli, D., Ferrell, J. E. Jr., and Sontag, E. D. (2004). Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc. Natl. Acad. Sci. U.S.A. 101, 1822–1827. doi: 10.1073/pnas.0308265100
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., et al. (2005). Current Protocols in Molecular Biology. New York, NY: John Wiley & Sons, Inc.
Chen, C. C., Fang, M., Majumder, A., and Wu, H. Y. (2001). A 72-base pair AT-rich DNA sequence element functions as a bacterial gene silencer. J. Biol. Chem. 276, 9478–9485. doi: 10.1074/jbc.M010501200
Chen, C. C., Ghole, M., Majumder, A., Wang, Z., Chandana, S., and Wu, H. Y. (2003). LeuO-mediated transcriptional derepression. J. Biol. Chem. 278, 38094–38103. doi: 10.1074/jbc.M300461200
Chen, C. C., and Wu, H. Y. (2005). LeuO protein delimits the transcriptionally active and repressive domains on the bacterial chromosome. J. Biol. Chem. 280, 15111–15121. doi: 10.1074/jbc.M414544200
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-A
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De la Cruz, M. A., Fernández-Mora, M., Guadarrama, C., Flores-Valdez, M. A., Bustamante, V. H., Vazquez, A., et al. (2007). LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol. Microbiol. 66, 727–743. doi: 10.1111/j.1365-2958.2007.05958.x
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Dillon, S. C., Espinosa, E., Hokamp, K., Ussery, D. W., Casadesús, J., and Dorman, C. J. (2012). LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 85, 1072–1089. doi: 10.1111/j.1365-2958.2012.08162.x
Dole, S., Kühn, S., and Schnetz, K. (2002). Post-transcriptional enhancement of Escherichia coli bgl operon silencing by limitation of BglG-mediated antitermination at low transcription rates Mol. Microbiol. 43, 217–226. doi: 10.1046/j.1365-2958.2002.02734.x
Fang, M., Majumder, A., Tsai, K. J., and Wu, H. Y. (2000). ppGpp-dependent leuO expression in bacteria under stress. Biochem. Biophys. Res. Commun. 276, 64–70. doi: 10.1006/bbrc.2000.3440
Fang, M., and Wu, H. Y. (1998). A promoter relay mechanism for sequential gene activation. J. Bacteriol. 180, 626–633.
Fox, J., and Weisberg, S. (2011). “Appendix: nonlinear regression and nonlinear least squares in R,” in An R Companion to Applied Regression. 2nd Edn. Available online at: http://socserv.socsci.mcmaster.ca/jfox/Books/Companion/appendix.html: SAGE Publications.
Giel, M., Desnoyer, M., and Lopilato, J. (1996). A mutation in a new gene, bglJ, activates the bgl operon in Escherichia coli K-12. Genetics 143, 627–635.
Guadarrama, C., Medrano-López, A., Oropeza, R., Hernández-Lucas, I., and Calva, E. (2014). The Salmonella enterica serovar typhi leuo global regulator forms tetramers: residues involved in oligomerization, dna binding, and transcriptional regulation. J. Bacteriol. 196, 2143–2154. doi: 10.1128/JB.01484-14
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130.
Hamilton, C. M., Aldea, M., Washburn, B. K., Babitzke, P., and Kushner, S. R. (1989). New method for generating deletions and gene replacements in Escherichia coli J. Bacteriol. 171, 4617–4622.
Ishihama, A., Shimada, T., and Yamazaki, Y. (2016). Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors. Nucleic Acids Res. 44, 2058–2074. doi: 10.1093/nar/gkw051
Jensen, P. R., Westerhoff, H. V., and Michelsen, O. (1993). The use of lac-type promoters in control analysis. Eur. J. Biochem. 211, 181–191. doi: 10.1111/j.1432-1033.1993.tb19885.x
Khlebnikov, A., Datsenko, K. A., Skaug, T., Wanner, B. L., and Keasling, J. D. (2001). Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147, 3241–3247. doi: 10.1099/00221287-147-12-3241
Klauck, E., Böhringer, J., and Hengge-Aronis, R. (1997). The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25, 559–569. doi: 10.1046/j.1365-2958.1997.4911852.x
Kogenaru, M., and Tans, S. J. (2014). An improved Escherichia coli strain to host gene regulatory networks involving both the AraC and LacI inducible transcription factors. J. Biol. Eng. 8, 1–5. doi: 10.1186/1754-1611-8-2
Landick, R., Wade, J. T., and Grainger, D. C. (2015). H-NS and RNA polymerase: a love–hate relationship? Curr. Opin. Microbiol. 24, 53–59. doi: 10.1016/j.mib.2015.01.009
Lanzer, M., and Bujard, H. (1988). Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. U.S.A. 85, 8973–8977.
Maddocks, S. E., and Oyston, P. C. F. (2008). Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154, 3609–3623. doi: 10.1099/mic.0.2008/022772-0
Majdalani, N., and Gottesman, S. (2005). The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59, 379–405. doi: 10.1146/annurev.micro.59.050405.101230
Majumder, A., Fang, M., Tsai, K. J., Ueguchi, C., Mizuno, T., and Wu, H. Y. (2001). LeuO expression in response to starvation for branched-chain amino acids. J. Biol. Chem. 276, 19046–19051. doi: 10.1074/jbc.M100945200
Medina-Aparicio, L., Rebollar-Flores, J. E., Gallego-Hernández, A. L., Vázquez, A., Olvera, L., Gutiérrez-Ríos, R. M., et al. (2011). The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica Serovar Typhi. J. Bacteriol. 193, 2396–2407. doi: 10.1128/JB.01480-10
Megerle, J. A., Fritz, G., Gerland, U., Jung, K., and Rädler, J. O. (2008). Timing and dynamics of single cell gene expression in the arabinose utilization system. Biophys. J. 95, 2103–2115. doi: 10.1529/biophysj.107.127191
Olins, P. O., and Rangwala, S. H. (1989). A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in Escherichia coli. J. Biol. Chem. 264, 16973–16976.
Pannen, D., Fabisch, M., Gausling, L., and Schnetz, K. (2016). Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J. Biol. Chem. 291, 2357–2370. doi: 10.1074/jbc.M115.696815
Pul, U., Wurm, R., Arslan, Z., Geissen, R., Hofmann, N., and Wagner, R. (2010). Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 75, 1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x
Salscheider, S. L., Jahn, A., and Schnetz, K. (2014). Transcriptional regulation by BglJ–RcsB, a pleiotropic heteromeric activator in Escherichia coli. Nucleic Acids Res. 42, 2999–3008. doi: 10.1093/nar/gkt1298
Shimada, T., Bridier, A., Briandet, R., and Ishihama, A. (2011). Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Mol. Microbiol. 82, 378–397. doi: 10.1111/j.1365-2958.2011.07818.x
Shimada, T., Yamamoto, K., and Ishihama, A. (2009). Involvement of leucine-response transcription factor leuo in regulation of the genes for sulfa-drug efflux. J. Bacteriol. 191, 4562–4571. doi: 10.1128/JB.00108-09
Shoval, O., and Alon, U. (2010). SnapShot: network motifs. Cell 143, 326.e321. doi: 10.1016/j.cell.2010.09.050
Siegele, D. A., and Hu, J. C. (1997). Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. U.S.A. 94, 8168–8172.
Stoebel, D. M., Free, A., and Dorman, C. J. (2008). Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154, 2533–2545. doi: 10.1099/mic.0.2008/020693-0
Stratmann, T., Madhusudan, S., and Schnetz, K. (2008). Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO. J. Bacteriol. 190, 926–935. doi: 10.1128/JB.01447-07
Stratmann, T., Pul, Ü., Wurm, R., Wagner, R., and Schnetz, K. (2012). RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants. Mol. Microbiol. 83, 1109–1123. doi: 10.1111/j.1365-2958.2012.07993.x
Ueguchi, C., Ohta, T., Seto, C., Suzuki, T., and Mizuno, T. (1998). The leuO gene-product has a latent ability to relieve the bgl silencing in Escherichia coli J. Bacteriol. 180, 190–193.
Venkatesh, G. R., Kembou Koungni, F. C., Paukner, A., Stratmann, T., Blissenbach, B., and Schnetz, K. (2010). BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J. Bacteriol. 192, 6456–6464. doi: 10.1128/JB.00807-10
Westra, E. R., Pul, U., Heidrich, N., Jore, M. M., Lundgren, M., Stratmann, T., et al. (2010). H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77, 1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x
Wilson, G. G., Young, K. Y. K., Edlin, G. J., and Konigsberg, W. (1979). High-frequency generalised transduction by bacteriophage T4. Nature 280, 80–82. doi: 10.1038/280080a0
Keywords: transcription regulator, nucleoid-associated protein, H-NS, H-NS antagonist, feedback regulation
Citation: Breddermann H and Schnetz K (2016) Correlation of Antagonistic Regulation of leuO Transcription with the Cellular Levels of BglJ-RcsB and LeuO in Escherichia coli. Front. Cell. Infect. Microbiol. 6:106. doi: 10.3389/fcimb.2016.00106
Received: 21 July 2016; Accepted: 02 September 2016;
Published: 16 September 2016.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, USAReviewed by:
Edmundo Calva, National Autonomous University of Mexico, MexicoFrancisco Ramos-Morales, University of Seville, Spain
Copyright © 2016 Breddermann and Schnetz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karin Schnetz, c2NobmV0ekB1bmkta29lbG4uZGU=
 Hannes Breddermann
Hannes Breddermann Karin Schnetz
Karin Schnetz