- 1Oral Biology, School of Dentistry, Meharry Medical College, Nashville, TN, USA
- 2Department of Physiology, Meharry Medical College, Nashville, TN, USA
- 3Fisk University, Nashville, TN, USA
- 4Department of Biochemistry and Cancer Biology, Meharry Medical College, Nashville, TN, USA
Atherosclerosis, a chronic inflammatory disease of the blood vessels, is one of the most common causes of morbidity and mortality world-wide. Involvement of Porphyromonas gingivalis in atherosclerosis is supported by observations from epidemiological, clinical, immunological, and molecular studies. Previously we reported that P. gingivalis vesicles have a much higher invasive efficiency than their originating cells. Here, we further compare the role of P. gingivalis cells and their vesicles in expression of chemoattractant proteins including CXCL1, CXCL2, and CXCL8, and adhesive molecules such as E-selectin in human umbilical vein endothelial cells (HUVECs). Both P. gingivalis 33277 cells and vesicles were able to up-regulate expression of these molecules, while the vesicles acted as more potent inducers of the inflammatory response associated with the development of atherosclerosis, consequently resulting in significant monocyte adhesion to a monolayer of HUVECs. Interestingly, we found that elevated expression of CXCL8 and E-selectin in endothelial cells induced by P. gingivalis correlated with the invasive ability of P. gingivalis cells and vesicles. Non-invasive bacterial cells and vesicles had no effect on expression of these genes. This study highlights the potential risk of P. gingivalis cells and vesicles in initiation of atherosclerosis and provides a potential target for the development of novel therapeutics against bacteria-associated atherosclerosis.
Introduction
Porphyromonas gingivalis, a Gram-negative bacterium, is associated with chronic periodontitis and with several systemic diseases including atherosclerosis (Lamont and Jenkinson, 1998; Ximénez-Fyvie et al., 2000; Hajishengallis et al., 2012; Hussain et al., 2015). Recently, a keystone pathogen hypothesis regarding the pathogenesis of periodontitis has been proposed, suggesting that the presence of P. gingivalis in the oral cavity, even at low-abundance, is capable of disturbing host-microbial homeostasis and inducing periodontitis (Hajishengallis et al., 2011, 2012). Previous studies demonstrated that P. gingivalis disrupts tissue homeostasis through manipulation of innate immunity, including complement and proinflammatory cytokines (Hajishengallis and Lamont, 2014; Hajishengallis, 2015).
P. gingivalis vesicles originate from outer membrane blebbing and contain mostly outer membrane components including lipopolysaccharides and outer membrane proteins (Xie, 2015) and exhibit the primary features of this organism. In fact, we recently demonstrated that P. gingivalis vesicles exhibit much higher invasive efficiency than their originating bacterial cells, although it appears that both invasive processes involve a clathrin-mediated endocytic machinery (Ho et al., 2015, 2016). Previous studies suggested that the effect of P. gingivalis vesicles on the human immune response system is a complicated matter and not always consistent with those induced by P. gingivalis cells. Animal studies have demonstrated that P. gingivalis vesicles with strong immunogenicity were able to elicit P. gingivalis-specific IgG and IgA in the serum of intranasal vaccinated mice as well as IgA in saliva, whereas whole cells did not (Nakao et al., 2011; Bai et al., 2015). It was suggested that the increased antigenicity found in vesicles might result from the more concentrated immune-dominant determinants on the vesicles compared to P. gingivalis cell surfaces. In addition, P. gingivalis vesicles appeared to repress immune responses induced by IFN-γ. Expression of several genes involved in IFN-γ signal transduction, including genes encoding class II transactivator, Jak1, and Jak2, proteins required for expression of MHC class II molecules, were down-regulated in vascular endothelial cells in the presence of P. gingivalis vesicles (Srisatjaluk et al., 2002). Since MHC class II molecules are essential for antigen presentation, it is likely that inhibition of their expression facilitates P. gingivalis' escape from immune surveillance.
In the study presented here, built on our previous work on comparison of host cell invasion efficiencies of P. gingivalis cells and their vesicles, we further determined the ability of P. gingivalis to induce innate immune responses in human umbilical vein endothelial cells (HUVECs). We found that after exposure to P. gingivalis cells or vesicles, HUVECs selectively expressed inflammatory mediators including IL8 and endothelial-leukocyte adhesion molecules such as E-selectin, which resulted in monocyte adhesion to HUVECs. These findings represent insight into the molecule mechanisms of P. gingivalis associated-atherogenesis.
Materials and Methods
Bacterial Strains and Vesicle Preparation and Quantification
P. gingivalis 33277 was grown from frozen stocks in TSB (trypticase soy broth) or on TSB blood agar plates supplemented with yeast extract (1 mg/mL), hemin (5 μg /mL), and menadione (1 μg/mL), and incubated at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). P. gingivalis vesicles were prepared as previously described (Furuta et al., 2009). Briefly, P. gingivalis was grown to the late exponential phase and growth media were collected by centrifugation at 10,000 × g for 15 min at 4°C and filtered through 0.22-μm-pore-size filters (Cell Treat, MA, USA) to remove residual bacteria. Vesicles were collected by ultracentrifugation at 126,000 × g for 2 h at 4°C and resuspended in phosphate-buffered saline (PBS) containing 10% glycerol.
Quantitation of P. gingivalis Vesicles
Since quantifying vesicles by their protein or lipid content in weight represents the most common way to normalize data (Kulp and Kuehn, 2010), we quantitated OMVs using both protein and lipid assays. Proteins and lipids were extracted from vesicles in PBS using a BugBuster® Protein Extraction Reagent (Novagen, MA, USA). The quantity of OMV lipid was assessed using the fluorescent lipophilic dye FM4-64 as described (Macdonald and Kuehn, 2013), and was quantitated by titration of P. gingivalis lipopolysaccharide (LPS-PG, InvivoGen, San Diego, California). Protein concentrations were determined with a Bio-Rad Protein Assay Kit (Bio-Rad, CA, USA). Results revealed that 1 × 106 P. gingivalis cells is equivalent to 100 ng protein or 3.6 μg lipid of vesicles. Thus, for in vitro infection experiments, 1 × 105 host cells were exposed to 1 × 106 P. gingivalis cells or vesicles with 100 ng protein.
Treatment of Endothelial Cells with P. gingivalis 33277 and Its Vesicles
Umbilical vein endothelial cells (HUVECs) from American Type Culture Collection (ATCC, VA, USA), were cultured in specific media, according to the manufacturer's instructions. Prior to treatment, HUVECs (1 × 105) were seeded and grown overnight in poly-L-lysine coated plates (CellTreat) at 37°C, 5% CO2, and then exposed to P. gingivalis 33277 (1 × 106) or its vesicles (100 ng). The cytotoxicity of treatments was evaluated with a Pierce LDH Cytotoxicity Assay Kit (Thermo Scientific, MA, USA). There was no cytotoxicity detected under our experimental conditions.
PCR Array
After exposure to P. gingivalis 33277 cells or their vesicles, the HUVECs were immediately transferred to TRIzol (Thermo Scientific) to release RNA. The total RNA was purified using an RNeasy Mini Kit (QIAGEN, CA, USA). A total of 500 ng RNA was reverse-transcribed to cDNA with an RT2 First strand Kit (QIAGEN) and suspended with dd-H2O in a final solution of 111 μL. cDNAs were then subjected to a Human Antibacterial Response RT2 Profiler PCR Array (QIAGEN) that includes a set of optimized real-time PCR primer assays on 96-well-plates. Real-time PCR was run on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). Data were analyzed using a RT2 Profiler PCR Array Data Analysis (version 3.5, QIAGEN). RNA from three independent cultures was analyzed.
Determination of P. gingivalis Invasive Ability Using Confocal Microscopy
HUVECs, after exposed to P. gingivalis cells or their vesicles, were fixed with 2% formaldehyde in a Vascular Cell Basal Medium (ATCC) at room temperature for 10 min after treatments, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with 10% horse serum in PBS for 1 h. The cells were then immunostained with polyclonal antibodies of P. gingivalis 33277, and anti-E-selectin monoclonal IgG (Santa Cruz Biotechnology, Texas, USA), followed by donkey anti-rabbit IgG conjugated Alex Fluor 546, chicken anti-mouse IgG conjugated Alex Fluor 488, or donkey anti-mouse IgG conjugated to Alex Fluor 546 (Thermo Scientific). Nuclei were stained with DAPI (Thermo Scientific). Confocal images were acquired using a Nikon A1R confocal microscope, and bacterial invasion abilities were determined by measuring intracellular florescence intensity in 10 random areas (5.6 × 5.6 μm) with an imaging software NIS-Elements AR 4.20.
RNA Isolation and qPCR
HUVECs treated with bacterial cells or vesicles were harvested by centrifugation and homogenized in Trizol Reagent (Thermo Scientific). The RNA in the supernatant was then purified using an RNeasy mini spin column (QIAGEN). RNA samples were digested on the column with RNase-free DNase. RT-PCR analysis was performed by using a QuantiTect SYBR Green RT-PCR Kit (QIAGEN) on the iCycler MyiQ™ Real-Time PCR detection system (Bio-Rad) according to the manufacturer's instructions. Primers are listed in Table S1. Amplification reactions consisted of a reverse transcription cycle at 50°C for 30 min, an initial activation at 95°C for 15 min, and 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. The expression levels of the investigated genes for the test sample were determined relative to the untreated calibrator sample by using the comparative cycle threshold (ΔCT) method. The ΔCT were calculated by subtracting the average CT-value of the test sample from the average CT-value of the calibrator sample, and were then used to calculate the ratio between the two by assuming 100% amplification efficiency. By loading the same amount of total RNA for any comparable samples, the ΔCT represents the difference on gene expression between the samples.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA was performed using a Human IL-8 Single Analyte ELISArray Kit (QIAGEN), according to the manufacturer's instruction. HUVECs were exposed to P. gingivalis 33277 cells or vesicles for 0, 2, 6, and 36 h. The culture media of HUVECs were collected and subjected to ELISA analysis. Concentration of IL-8 was determined using the standard curve of IL-8.
Monocyte Adhesion to HUVEC
HUVECs (5 × 104/well) were seeded on 24-well plates and cultured for 2 days to reach a confluent monolayer. HUVECs were then co-cultured with P. gingivalis 33277 cells (5 × 106) or vesicles (250 ng) for 20 h, and washed with phosphate-buffered saline (PBS) to remove unbound bacterial cells or vesicles. THP-1 cells were labeled with the fluorescent dye calcein AM (10 μM; Thermo Scientific) for 1 h, washed, and resuspended in RPMI medium containing 10% FBS. The labeled THP-1 cells (4 × 105) were incubated with HUVEC monolayers for 2 h. After being washed with RPMI medium containing 10% FBS, adhesion of THP-1 cells to HUVEC was visualized under a Nikon TE2000-E immunofluorescence microscope. Fluorescence images were analyzed by imaging software NIS-Elements AR 4.20 to measure florescence intensity in 10 random areas (5.6 × 5.6 μm).
Statistical Analyses
A student's t-test was used to determine statistical significance of the differences in expression level of inflammatory genes in HUVECs in the presence or absence of P. gingivalis cells or vesicles. P < 0.05 was considered significant. Values are shown ±SD unless stated otherwise.
Results
Differential Expression of Inflammatory Genes in HUVECs in Response to P. gingivalis Stimulation
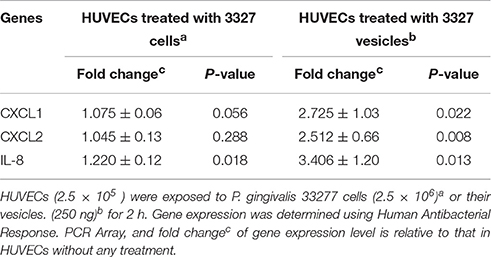
Innate immune responses in HUVECs induced by P. gingivalis cells or vesicles were first examined using a PCR array that includes 84 key genes involved in innate immune response. The relative expression level of these genes in the HUVECs exposed to either P. gingivalis cells or vesicles was determined by comparing to those observed in HUVECs without any treatment. Three genes encoding CXCL1, CXCL2, and IL-8 were found to be the most highly expressed genes in HUVECs treated with vesicles derived from P. gingivalis 33277 for 2 h (Table 1). The gene encoding IL-8 was the only one, out of 84 inflammatory genes, that was slightly up-regulated in HUVECs exposed to 33277 cells.
To assess the reliability of the PCR array results, we conducted qRT-PCR to measure mRNA levels for key genes involved in inflammatory responses individually using the identical total RNA samples used in the PCR array but different sets of primers (Table S1). Five inflammation related genes, including cxcl1, cxcl2, cxcl8 (IL-8), tlr-2, and tlr-4, were tested. Ratios of the transcripts from untreated HUVECs to HUVECs treated with 33277 cells or vesicles, determined by the PCR array or by qRT-PCR of individual genes, were in a good concordance. Expression of cxcl1, cxcl2, and IL8 were up-regulated ~2–3 fold in the vesicle-treated HUVECs compared to those in untreated cells (Figures 1A–C). Significant increase in the mRNA level of these genes was also observed in HUVECs treated with 33277 cells but at a lower degree. Up-regulated toll like receptor 4 was found in HUVECs treated with 33277 cells or vesicles, while expression of toll like receptor 2 was not altered in these cells (Figures 1D,E). Heat treatment of 33277 cells or vesicles abolished their activity to induce expression of cxcl1, cxcl2, and IL8, suggesting involvement of protein molecules. Other P. gingivalis strains including W83, the fimA mutant (FAE), and the gingipain triple mutant (KDP128, rgpA−, rgpB−, and kgp−) were also examined for their role in the induction. There was no significant alteration in expression of these inflammatory genes in the HUVECs treated with P. gingivalis cells or vesicles, except for the vesicles derived from the fimA mutant (FAE). It should be pointed out that although the fimA mutant loses its invasive activity, its vesicles are able to invade host cells without the FimA protein (Mantri et al., 2015). These data imply that the invasive ability of P. gingivalis is required for modulation of inflammatory gene expression. This is based on the observation that W83 cells and vesicles, the fimA mutant cells, the gingipain mutant cells and vesicles, and heat treated 33277 cells and vesicles showed little invasive activity (Figure 2) and were not able to significantly upregulate expression of the inflammatory genes tested. To confirm this assumption, HUVECs were pretreated with dynasore (30 μM), a potent inhibitor of P. gingivalis invasion (Ho et al., 2016). As expected, P. gingivalis 33277 cells and vesicles no long elicited expression of IL-8 in HUVECs (Figure 3).
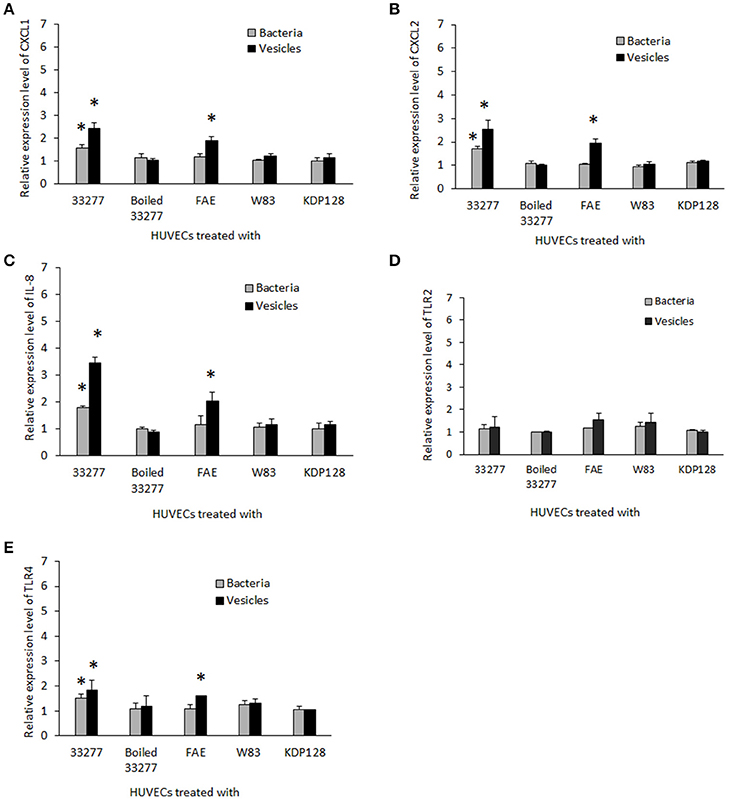
Figure 1. Expression of inflammatory gene s in human endothelial cells in response to P. gingivalis infection. HUVECs were infected with different P. gingivalis strains including wild type 33277, its isogenic fimA mutant (FAE), the isogenic ginginpain mutant (KDP128, rgpA−, rgpB−, and kgp−), heat treated 33277 and W83 strains, as well as the vesicles derived from these strains for 2h. mRNA levels of cxcl-1 (A), cxcl2 (B), il-8 (C), tlr2 (D), and tlr4 (E) were determined in the infected HUVECs using qPCR. Expression level of each gene was normalized with that of Glyceraldehyde 3-phosphate dehydrogenase gene (gapdh). Each bar represents means of fold change with standard deviation of three biological replicates relative to that found in PBS treated HUVECs (1 unit). Asterisks indicate statistical significance of expression level in HUVECs treated and untreated with P. gingivalis (P < 0.05; t-test).
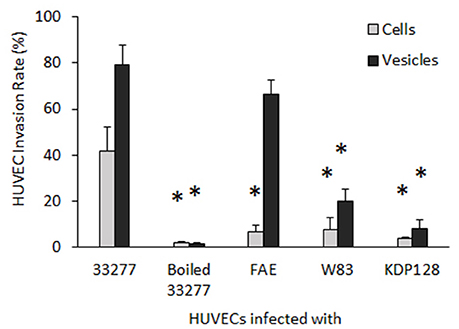
Figure 2. Invasive activity of P. gingivalis cells and vesicles into HUVECs. After exposed to P. gingivalis cells or vesicles, HUVECs were immune strained with P. gingivalis antibodies and analyzed under a confocal microscope. The number of HUVECs carrying intercellular P. gingivalis cells or vesicles (infection rate) was determined by counting the infected HUVECs in 30 random areas. Each bar represents the percentage of HUVECs with intercellular cells or vesicles. The SEs are indicated (n = 3). An asterisk indicates the statistical significance of invasive rates between P. gingivalis 33277 cells or vesicles and cells and vesicles of other P. gingivalis strains (P < 0.05; t-test).
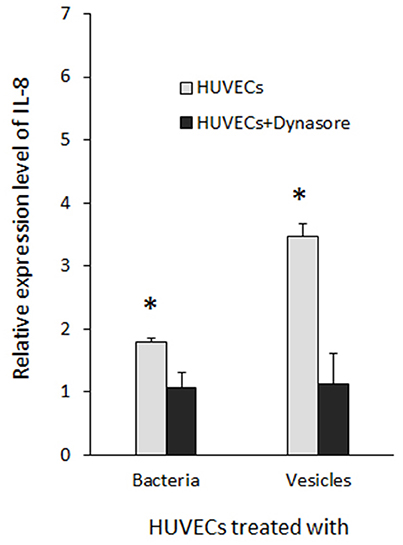
Figure 3. Inhibition of P. gingivalis induced IL-8 expression by dynasore. HUVECs were treated with dynasore (30 μM) prior to be exposed to P. gingivalis 33277 cells or vesicles. Expression level of IL-8 was determined using qRT-PCR and normalized with that of gapdh. Each bar represents means of fold change relatively to that found in HUVECs without P. gingivalis treatment (1 unit). Asterisks indicate the statistical significance of expression level in HUVECs treated and untreated with P. gingivalis (P < 0.05; t-test).
We then determined IL-8 level in the culture supernatant of HUVECs exposed to P. gingivalis 33277 cells or vesicles. IL-8 level was 90% less in the culture supernatant in the presence of 33277 cells compared to that found in the supernatant without 33277 cells, indicating degradation of IL-8 by the bacterial cells (Figure 4A). Previously, we showed that gingipains were enriched in P. gingivalis vesicles (Mantri et al., 2015), however, a much slower degradation of IL-8 was observed in the presence of 33277 vesicles, especially at 2 or 6 h exposure. We therefore speculate that membrane-associated gingipains have limited enzymatic activity.
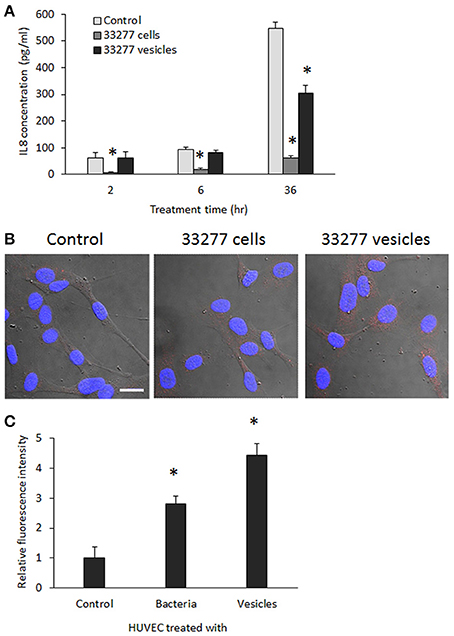
Figure 4. Determination of IL-8 level in the culture supernatants and cytoplasm of HUVECs. (A) HUVECs were exposed to P. gingivalis 33277 cells or vesicles for 2, 6, and 36 h, and culture supernatants collected. IL-1 level in the culture supernatants was measured using ELISA with a Human IL-8 Single Analyte ELISArray Kit (Qiagen). Each bar represents means of IL-8 concentration with standard deviation of three biological replicates. Asterisks indicate statistical significance of IL-8 concentration in the culture supernatants of HUVECs treated and untreated with P. gingivalis (P < 0.05; t-test). (B) Intracellular IL-8 was visualized in HUVECs using anti-E-selectin antibody and Alexa Fluor 546-conjugated secondary antibody (red) under a confocal microscope. Scale bar 20 μm. (C) Intracellular IL-8 was quantitated using an imaging software NIS-Elements AR 4.20. Asterisks indicate the statistical significance of florescence intensity in HUVECs treated and untreated with P. gingivalis (P < 0.05; t-test).
IL-8 level in the cytoplasm of HUVECs was visualized using confocal microscopy. Significantly increased expression of IL-8 was found in HUVECs treated with P. gingivalis 33277 vesicles for 18 h (Figure 4B). Quantitation of IL-8 was conducted using imaging software NIS-Elements AR 4.20 to measure florescence intensity in 10 random areas (5.6 × 5.6 μm), and 2.5- and 4-fold increase of IL-8 was observed in HUVECs treated with 33277 cells or with the vesicles compared to that in untreated HUVECs (Figure 4C), suggesting that degradation does not occur in cytoplasm. It is likely that the HUVECs treated with 33277 vesicles constitutively release more IL-8, which may maintain a stable level of IL-8 in the micro-environment, although secreted IL-8 will eventually be degraded by extracellular P. gingivalis cells and vesicles.
Induction of Adhesion Molecule Expression in HUVECs by P. gingivalis
To determine the effect of P. gingivalis infection on expression of atherosclerosis-associated proteins, we measured expression of three adhesion molecules (E-selectin, VCAM1, and ICAM-1) in HUVECs at the transcriptional level using qRT-PCR. The results showed that mRNA of E-selectin was increased about 2 and 5 fold in the HUVECs treated with P. gingivalis 33277 cells or vesicles, respectively, compared to those detected in the HUVECs without any treatment (Figure 5A). This phenomenon of up-regulated adhesion molecules appears specific to E-selectin, as there was no alteration in mRNA levels of VCAM1 and ICAM-1 in HUVECs treated P. gingivalis (data not shown). A three-fold increase in mRNA level of E-selectin was also detected in the HUVECs treated with vesicles derived from FAE (the fimA mutant) but not the FAE cells. This effect was also not observed in the HUVECs treated with P. gingivalis W83, KDP128, or heat treated 33277, nor their vesicles. Not surprisingly, treatment of HUVECs with dynasore (30 μM) blocked P. gingivalis 33277-induced up-regulation of E-selectin (Figure 5B).
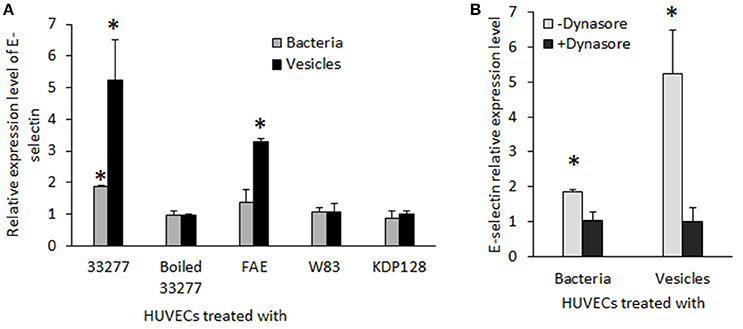
Figure 5. Expression of E-selectin in HUVECs exposed to P. gingivalis strains. (A) mRNA level of E-selectin in HUVECs in response to live P. gingivalis 33277, heat-treated 33277, the fmA mutant (FAE), the gingipain mutant (KDP128), heat treated 33277, and W83 strains and their derived vesicles was determined using qRT-PCR. (B) HUVECs were treated with dynasore (30 μM) prior to be exposed to P. gingivalis 33277 cells or vesicles. mRNA level of IL-8 was normalized with that of Glyceraldehyde 3-phosphate dehydrogenase gene (gapdh). Each bar represents means of fold change relative to that found in PBS treated HUVECs (1 unit). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations. Asterisks indicate statistical significance of expression level in HUVECs treated and untreated with P. gingivalis (P < 0.05; t-test).
To determine if expression of E-selectin protein is elevated on the surface and/or in the cytoplasm of the HUVECs exposed to 33277 cells or vesicles, we performed confocal microscopic analysis using HUVECs with or without membrane permeabilization. Consistent with mRNA level of E-selectin, E-selectin protein was also significantly enhanced in cytoplasm of HUVECs when the cells were exposed to 33277 vesicles and permeabilized before immunofluorescence staining (Figures 6A,B). Additionally, we examined the surface expression of E-selectin on the HUVECs that were not permeabilized before staining. E-selectin expression was also enhanced on the surface of the HUVECs exposed to 33277 cells and to a much greater extent when exposed to 33277 vesicles (Figure 6C).
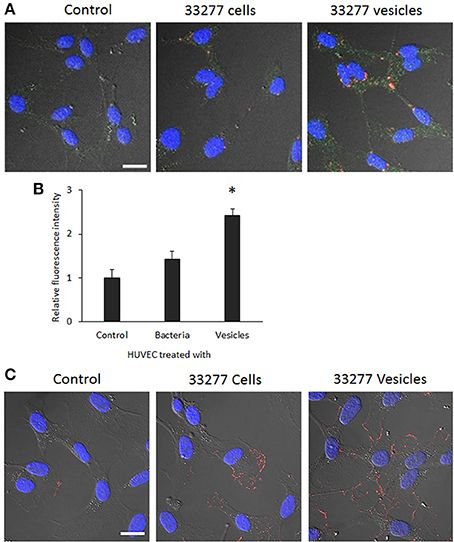
Figure 6. Visualization of E-selectin in the cytoplasm and the surface of HUVECs with confocal microscopy. (A) HUVECs were grown in a glass bottom dish for 16 h, and then infected with P. gingivalis 33277 cells or its vesicles. HUVECs were permeabilized. Internalized bacterial cells or vesicles were probed with anti-P. gingivalis polyclonal antibodies, visualized by Alexa Fluor 546-conjugated anti-rabbit IgG secondary antibody (red), E-selectin by anti-E-selectin antibody and Alexa Fluor 488-conjugated secondary antibody (green), and nucleus by DAPI (blue) Images are presented by differential interference contrast (DIC). (B) E-selectin level in the cytoplasm was determined using imaging software NIS-Elements AR 4.20. (C) Expression of E-selectin on the surface of HUVECs without permeabilization was visualized by anti-E-selectin antibody and Alexa Fluor 546-conjugated secondary antibody (red). Scale bar, 20 μm. An asterisk indicates the statistical significance of florescence intensity in HUVECs treated and untreated with P. gingivalis vesicles (P < 0.05 by t-test).
E-selectin is known as endothelial-leukocyte adhesion molecule 1, and over-expression of E-selectin on activated endothelial cells leads to initial adhesion of leukocytes to endothelium. Using an adhesion assay, we found that attachment of monocytes (THP-1) on HUVEC monolayers was significantly enhanced. Compared to the attachment of THP-1 to HUVECs, 6- and 25-fold THP-1 cells were found on HUVEC monolayers treated with either P. gingivalis 33277 cells or vesicles, respectively (Figures 7A,B). We also examined THP-1binding to HUVEC monolayers treated with dynasore or anti E-selectin antibody. As shown in Figures 7, THP-1 attachment to the monolayers was significantly reduced when dynasore (30 μM) was added to the cultural media of HUVEC monolayers during exposure of P. gingivalis vesicles, or when the monolayers were treated with anti E-selectin antibody prior addition of THP-1. These results indicate that P. gingivalis invasion and its induced E-selectin expression are key events for promoting attachment of monocytes on HUVEC monolayers.
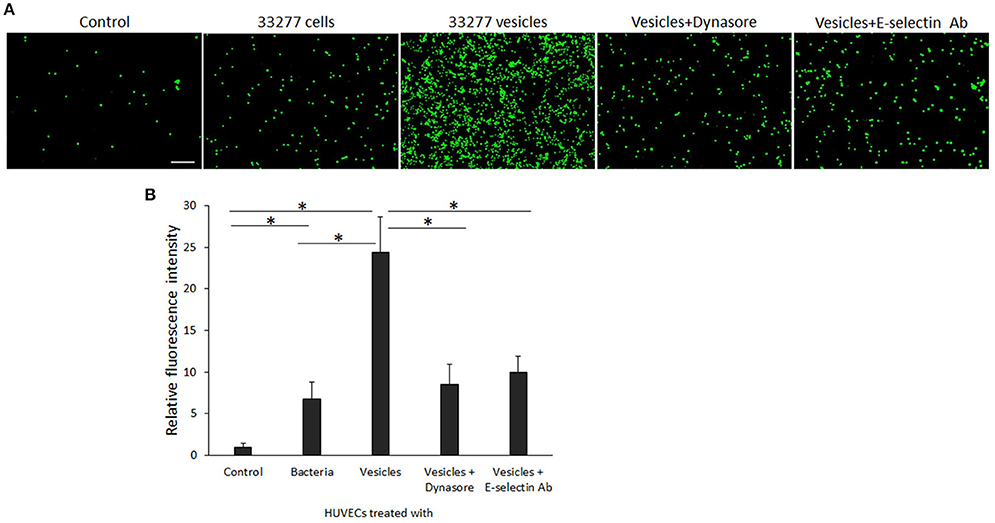
Figure 7. Attachment of monocytes to P. gingivalis-activated HUVECs. (A) The calcein AM labeled THP-1 cells (green) were incubated with P. gingivalis 33277 cell- or vesicle-activated HUVEC monolayers for 2 h. After removing the unbound THP-1 cells, THP-1 adhesion was visualized under an immunofluorescence microscope. Scale bar, 100 μm. (B) Each bar represents the mean of fluorescence intensity in 10 random areas (5.6 × 5.6 μm) of immunofluorescence images. Comparison highlighted with an asterisk indicates a significant difference between two relative levels of florescence intensity detected on HUVEC monolayers cultured under different conditions (P < 0.05 by t-test).
Discussion
An association between P. gingivalis infection and atherosclerosis has been extensively investigated in in vitro, ex vivo, and animal models, which has made this bacterium a model of atherosclerosis initiated by microorganisms (Reyes et al., 2013). Previous studies have focused on intact P. gingivalis cells, based on the discovery of proteins and DNA of this bacterium in ex vivo samples. It was speculated that P. gingivalis may enter microvasculature following tooth brushing or other dental procedures, which may lead to transient bacteremia (Kinane et al., 2005; Iwai, 2009). However, it has not been confirmed if live cells of P. gingivalis cause low-grade inflammation in the walls of arterial vessels. Based on the recent findings, including the much more efficient invasive activity of P. gingivalis vesicles and the presence of vesicle-associated major outer membrane proteins, DNA, and RNA in the vesicles (Ho et al., 2015), we propose a novel concept that vesicles serve a significant role in atherogenesis, and they likely represent a “Trojan horse” to induce infections at secondary sites, such as in the walls of vessels, more so than intact P. gingivalis cells. In agreement with this concept, data presented here demonstrate that the vesicles are also more potent inducers of inflammation responses. Expression of IL-8 and E-selectin, at both mRNA and protein levels, was enhanced in HUVECs treated with P. gingivlais cells or vesicles, and more so for those exposed to vesicles. We also observed significantly enriched E-selectin on the surfaces of HUVECs treated with P. gingivalis 33277 vesicles, as well as highly induced monocyte adhesion to these HUVECs.
Interleukin (IL)-8 is a pro-inflammatory chemokine that belongs to the CXC subfamily and is also known as CXCL8 (Bacon et al., 2002). IL-8 plays an important role in inflammation, cancer, and cardiovascular disease through cell signaling and activation (Baggiolini and Clark-Lewis, 1992; Apostolakis et al., 2009; Chen et al., 2015). P. gingivalis-induced IL-8 production has been found in neutrophils, THP-1 cells, human periodontal ligament fibroblasts, and endothelial cells (Shelburne et al., 2007; Jayaprakash et al., 2014; Zhang and Li, 2015; Damgaard et al., 2016). However, reports on expression of IL-8 in human gingival epithelial cells in response to P. gingivalis infection have been conflicting (Takeuchi et al., 2013; Fujita et al., 2014; Yee et al., 2014; Savitri et al., 2015), and it was suggested that the different epithelial cell types used in those studies affect results. When comparing the abilities of P. gingivalis 33277 cells and the vesicles purified from the culture media to induce expression of IL-8 at the transcriptional level in HUVECs, we found that the mRNA level of IL-8 was higher in HUVECs treated with vesicles than that in HUVECs treated with 33277 cells, despite identical amounts of proteins and lipids were used. Interestingly, IL-8 accumulation in the culture media of HUVECs treated with 33277 vesicles was not significantly enhanced compared to that in the culture media of untreated HUVECs, while IL-8 level was dramatically decreased in the presence of P. gingivalis 33277 cells. Degradation of IL-8 has been reported previously, and Stathopoulou et al. showed that 100% degradation of IL-8 by live P. gingivalis cells could be reached after 30 min exposure (Stathopoulou et al., 2009). It was suggested that lysine gingipain (Kgp) is likely responsible for the degradation of IL-1 in culture supernatants (Stathopoulou et al., 2009; Jayaprakash et al., 2014). We reported earlier that gingipains were selectively enriched in P. gingivalis vesicles (Mantri et al., 2015). It is likely therefore that vesicle-associated gingipains are not as efficient as gingipains secreted by live P. gingivalis cells.
The role of Selectin in atherosclerosis is known for its ability to mediate rapid on-off interaction between monocytes and endothelium, which is called rolling adhesion (McEver and Zhu, 2010; Telen, 2014). Firm adhesion of monocytes to endothelium requires the presence of cytokines such as IL-8. Gerszten et al. revealed, using videomicroscopy, that IL-8 arrested the rolling monocytes on E-selectin-expressing endothelium. This likely involves recognition of β2 leukocyte integrin and chemokine receptor CXCR2 by IL-8 (Gerszten et al., 1999). It should be pointed out that CXCR2 is a receptor for both CXCL1 and IL-8; presumably up-regulation of CXCL1 may compensate degradation of IL-8 by gingipains or have a synergistic effect in monocyte adhesion.
One striking finding of this work is that mechanisms contributing to up-regulation of IL-8 and E-selectin in HUVECs appear to involve the invasive ability of P. gingivalis cells and vesicles. Previous studies have shown an involvement of a TonB-dependent receptor (RagB), LPS, and Fimbria-dependent activation of IL-8 in primary human monocytes (Hutcherson et al., 2015), human oral keratinocytes (Luo et al., 2012), and human aortic endothelial cells (Takahashi et al., 2006). Expression of E-selectin in human endothelial cells is also up-regulated by P. gingivalis penta-acylated lipid A and fimbrial proteins (Chou et al., 2005; Reife et al., 2006; Takahashi et al., 2006). We demonstrate here that the invasive ability of P. gingivalis also contributes to elevated IL-8 and E-selectin expression in human endothelial cells, which is in agreement with an observation by Takahashi et al. that only invasive P. gingivalis strains induced production of IL-8 and E-selectin in human aortic endothelial cells (Takahashi et al., 2006). Although the mechanisms have not been clarified, our data indicate that a dynamin-mediated endocytosis is required for eliciting expression of IL-8 and E-selectin. This conclusion is based on our finding that dynasore, an inhibitor of dynamin-mediated endocytosis (Macia et al., 2006), completely blocked the ability of P. gingivalis cells and vesicles to induce IL-8, and E-selectin expression. Collectively, our findings provide evidence that the invasive P. gingivais strains, especially their vesicles, are capable of inducing the production of pro-inflammatory and adhesive molecules that are hallmarks of atherosclerosis. Therefore, identification of agents to inhibit P. gingivalis invasion is likely an efficient therapeutic approach for intervention of the bacteria-associated atherosclerosis.
Author Contributions
Conceived and designed the experiments: HX. Performed the experiments: MH and JC. Analyzed the data: ZG and HX. Contributed reagents/materials/analysis tools: JSG and HX. Wrote the paper: MH, ZG, JSG, and HX.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by NIH grants DE022428 and 025332 (HX) from NIDCR and by MD007593 and MD007586 from NIMHD. The project described was also supported by NCRR grant UL1 RR024975, which is now mediated by the NCATS (2 UL1 TR000445). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank the Meharry Office for Scientific Editing and Publications for scientific editing support (S21MD000104).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2016.00139
References
Apostolakis, S., Vogiatzi, K., Amanatidou, V., and Spandidos, D. A. (2009). Interleukin 8 and cardiovascular disease. Cardiovasc. Res. 84, 353–360. doi: 10.1093/cvr/cvp241
Bacon, K., Baggiolini, M., Broxmeyer, H., Horuk, R., Lindley, I., Mantovani, A., et al. (2002). Chemokine/chemokine receptor nomenclature. J. Interferon Cytokine Res. 22, 1067–1068. doi: 10.1089/107999002760624305
Baggiolini, M., and Clark-Lewis, I. (1992). Interleukin-8, a chemotactic and inflammatory cytokine. FEBS Lett. 307, 97–101. doi: 10.1016/0014-5793(92)80909-Z
Bai, D., Nakao, R., Ito, A., Uematsu, H., and Senpuku, H. (2015). Immunoreactive antigens recognized in serum samples from mice intranasally immunized with Porphyromonas gingivalis outer membrane vesicles. Pathog. Dis. 73:ftu006. doi: 10.1093/femspd/ftu006
Chen, W.-T., Ebelt, N. D., Stracker, T. H., Xhemalce, B., Van Den Berg, C. L., and Miller, K. M. (2015). ATM regulation of IL-8 links oxidative stress to cancer cell migration and invasion. eLife 4:e07270. doi: 10.7554/eLife.07270
Chou, H. H., Yumoto, H., Davey, M., Takahashi, Y., Miyamoto, T., Gibson, F. C. III, et al. (2005). Porphyromonas gingivalis fimbria-dependent activation of inflammatory genes in human aortic endothelial cells. Infect. Immun. 73, 5367–5378. doi: 10.1128/IAI.73.9.5367-5378.2005
Damgaard, C., Kantarci, A., Holmstrup, P., Hasturk, H., Nielsen, C. H., and Van Dyke, T. E. (2016). Porphyromonas gingivalis-induced production of reactive oxygen species, tumor necrosis factor-α, interleukin-6, CXCL8 and CCL2 by neutrophils from localized aggressive periodontitis and healthy donors: modulating actions of red blood cells and resolvin E1. J. Periodontal. Res. [Epub ahead of print]. doi: 10.1111/jre.12388
Fujita, Y., Nakayama, M., Naito, M., Yamachika, E., Inoue, T., Nakayama, K., et al. (2014). Hemoglobin receptor protein from Porphyromonas gingivalis induces interleukin-8 production in human gingival epithelial cells through stimulation of the mitogen-activated protein kinase and NF-kappaB signal transduction pathways. Infect. Immun. 82, 202–211. doi: 10.1128/IAI.01140-12
Furuta, N., Tsuda, K., Omori, H., Yoshimori, T., Yoshimura, F., and Amano, A. (2009). Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect. Immun. 77, 4187–4196. doi: 10.1128/IAI.00009-09
Gerszten, R. E., Garcia-Zepeda, E. A., Lim, Y. C., Yoshida, M., Ding, H. A., Gimbrone, M. A. Jr., et al. (1999). MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature 398, 718–723. doi: 10.1038/19546
Hajishengallis, G. (2015). Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44. doi: 10.1038/nri3785
Hajishengallis, G., Darveau, R. P., and Curtis, M. A. (2012). The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10, 717–725. doi: 10.1038/nrmicro2873
Hajishengallis, G., and Lamont, R. J. (2014). Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur. J. Immunol. 44, 328–338. doi: 10.1002/eji.201344202
Hajishengallis, G., Liang, S., Payne, M. A., Hashim, A., Jotwani, R., Eskan, M. A., et al. (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506. doi: 10.1016/j.chom.2011.10.006
Ho, M. H., Chen, C. H., Goodwin, J. S., Wang, B. Y., and Xie, H. (2015). Functional advantages of Porphyromonas gingivalis vesicles. PLoS ONE 10:e0123448. doi: 10.1371/journal.pone.0123448
Ho, M. H., Huang, L., Goodwin, J. S., Dong, X., Chen, C. H., and Xie, H. (2016). Two small molecules block oral epithelial cell invasion by Porphyromons gingivalis. PLoS ONE 11:e0149618. doi: 10.1371/journal.pone.0149618
Hussain, M., Stover, C. M., and Dupont, A. (2015). P. gingivalis in Periodontal Disease and Atherosclerosis - Scenes of Action for Antimicrobial Peptides and Complement. Front. Immunol. 6:45. doi: 10.3389/fimmu.2015.00045
Hutcherson, J. A., Bagaitkar, J., Nagano, K., Yoshimura, F., Wang, H., and Scott, D. A. (2015). Porphyromonas gingivalis RagB is a proinflammatory signal transducer and activator of transcription 4 agonist. Mol. Oral Microbiol. 30, 242–252. doi: 10.1111/omi.12089
Iwai, T. (2009). Periodontal bacteremia and various vascular diseases. J. Periodontal. Res. 44, 689–694. doi: 10.1111/j.1600-0765.2008.01165.x
Jayaprakash, K., Khalaf, H., and Bengtsson, T. (2014). Gingipains from Porphyromonas gingivalis play a significant role in induction and regulation of CXCL8 in THP-1 cells. BMC Microbiol. 14:193. doi: 10.1186/1471-2180-14-193
Kinane, D. F., Riggio, M. P., Walker, K. F., MacKenzie, D., and Shearer, B. (2005). Bacteraemia following periodontal procedures. J. Clin. Periodontol. 32, 708–713. doi: 10.1111/j.1600-051X.2005.00741.x
Kulp, A., and Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184. doi: 10.1146/annurev.micro.091208.073413
Lamont, R. J., and Jenkinson, H. F. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62, 1244–1263.
Luo, W., Wang, C. Y., and Jin, L. (2012). Baicalin downregulates Porphyromonas gingivalis lipopolysaccharide-upregulated IL-6 and IL-8 expression in human oral keratinocytes by negative regulation of TLR signaling. PLoS ONE 7:e51008. doi: 10.1371/journal.pone.0051008
Macdonald, I. A., and Kuehn, M. J. (2013). Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J. Bacteriol. 195, 2971–2981. doi: 10.1128/JB.02267-12
Macia, E., Ehrlich, M., Massol, R., Boucrot, E., Brunner, C., and Kirchhausen, T. (2006). Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10, 839–850. doi: 10.1016/j.devcel.2006.04.002
Mantri, C. K., Chen, C. H., Dong, X., Goodwin, J. S., Pratap, S., Paromov, V., et al. (2015). Fimbriae-mediated outer membrane vesicle production and invasion of Porphyromonas gingivalis. Microbiologyopen 4, 53–65. doi: 10.1002/mbo3.221
McEver, R. P., and Zhu, C. (2010). Rolling cell adhesion. Annu. Rev. Cell Dev. Biol. 26, 363–396. doi: 10.1146/annurev.cellbio.042308.113238
Nakao, R., Hasegawa, H., Ochiai, K., Takashiba, S., Ainai, A., Ohnishi, M., et al. (2011). Outer membrane vesicles of Porphyromonas gingivalis elicit a mucosal immune response. PLoS ONE 6:e26163. doi: 10.1371/journal.pone.0026163
Reife, R. A., Coats, S. R., Al-Qutub, M., Dixon, D. M., Braham, P. A., Billharz, R. J., et al. (2006). Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol. 8, 857–868. doi: 10.1111/j.1462-5822.2005.00672.x
Reyes, L., Herrera, D., Kozarov, E., Roldan, S., and Progulske-Fox, A. (2013). Periodontal bacterial invasion and infection: contribution to atherosclerotic pathology. J. Clin. Periodontol. 40(Suppl 14), S30–S50. doi: 10.1111/jcpe.12079
Savitri, I. J., Ouhara, K., Fujita, T., Kajiya, M., Miyagawa, T., Kittaka, M., et al. (2015). Irsogladine maleate inhibits Porphyromonas gingivalis-mediated expression of toll-like receptor 2 and interleukin-8 in human gingival epithelial cells. J. Periodontal. Res. 50, 486–493. doi: 10.1111/jre.12231
Shelburne, C. E., Coopamah, M. D., Sweier, D. G., An, F. Y., and Lopatin, D. E. (2007). HtpG, the Porphyromonas gingivalis HSP-90 homologue, induces the chemokine CXCL8 in human monocytic and microvascular vein endothelial cells. Cell Microbiol. 9, 1611–1619. doi: 10.1111/j.1462-5822.2007.00897.x
Srisatjaluk, R., Kotwal, G. J., Hunt, L. A., and Justus, D. E. (2002). Modulation of gamma interferon-induced major histocompatibility complex class II gene expression by Porphyromonas gingivalis membrane vesicles. Infect. Immun. 70, 1185–1192. doi: 10.1128/IAI.70.3.1185-1192.2002
Stathopoulou, P. G., Benakanakere, M. R., Galicia, J. C., and Kinane, D. F. (2009). The host cytokine response to Porphyromonas gingivalis is modified by gingipains. Oral Microbiol. Immunol. 24, 11–17. doi: 10.1111/j.1399-302X.2008.00467.x
Takahashi, Y., Davey, M., Yumoto, H., Gibson, F. C. III, and Genco, C. A. (2006). Fimbria-dependent activation of pro-inflammatory molecules in Porphyromonas gingivalis infected human aortic endothelial cells. Cell Microbiol. 8, 738–757. doi: 10.1111/j.1462-5822.2005.00661.x
Takeuchi, H., Hirano, T., Whitmore, S. E., Morisaki, I., Amano, A., and Lamont, R. J. (2013). The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-kappaB RelA/p65. PLoS Pathog. 9:e1003326. doi: 10.1371/journal.ppat.1003326
Telen, M. J. (2014). Cellular adhesion and the endothelium: E-selectin, L-selectin, and pan-selectin inhibitors. Hematol. Oncol. Clin. North Am. 28, 341–354. doi: 10.1016/j.hoc.2013.11.010
Xie, H. (2015). Biogenesis and function of Porphyromonas gingivalis outer membrane vesicles. Future Microbiol. [Epub ahead of print]. doi: 10.2217/fmb.15.63
Ximénez-Fyvie, L. A., Haffajee, A. D., and Socransky, S. S. (2000). Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27, 648–657. doi: 10.1034/j.1600-051x.2000.027009648.x
Yee, M., Kim, S., Sethi, P., Düzgünes, N., and Konopka, K. (2014). Porphyromonas gingivalis stimulates IL-6 and IL-8 secretion in GMSM-K, HSC-3 and H413 oral epithelial cells. Anaerobe 28, 62–67. doi: 10.1016/j.anaerobe.2014.05.011
Keywords: P. gingivalis, outer membrane vesicles, atherosclerosis, IL-8, E-selectin
Citation: Ho M-H, Guo Z-M, Chunga J, Goodwin JS and Xie H (2016) Characterization of Innate Immune Responses of Human Endothelial Cells Induced by Porphyromonas gingivalis and Their Derived Outer Membrane Vesicles. Front. Cell. Infect. Microbiol. 6:139. doi: 10.3389/fcimb.2016.00139
Received: 13 July 2016; Accepted: 10 October 2016;
Published: 25 October 2016.
Edited by:
Yousef Abu Kwaik, University of Louisville, USAReviewed by:
Jiri Stulik, University of Defence, Czech RepublicAshu Sharma, University at Buffalo, USA
Copyright © 2016 Ho, Guo, Chunga, Goodwin and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Xie, aHhpZUBtbWMuZWR1
 Meng-Hsuan Ho
Meng-Hsuan Ho Zhong-Mao Guo
Zhong-Mao Guo Julio Chunga
Julio Chunga J. Shawn Goodwin
J. Shawn Goodwin Hua Xie
Hua Xie