- Singapore Immunology Network, Agency for Science, Technology and Research, Singapore, Singapore
Candida albicans is responsible for ~400,000 systemic fungal infections annually, with an associated mortality rate of 46–75%. The human gastrointestinal (GI) tract represents the largest natural reservoir of Candida species and is a major source of systemic fungal infections. However, the factors that control GI colonization by Candida species are not completely understood. We hypothesized that the fungal cell wall would play an important role in determining the competitive fitness of Candida species in the mammalian GI tract. To test this hypothesis, we generated a systematic collection of isogenic C. albicans cell wall mutants and measured their fitness in the mouse GI tract via quantitative competition assays. Whereas a large variation in competitive fitness was found among mutants, no correlation was observed between GI fitness and total levels of individual cell wall components. Similar results were obtained in a set of distantly-related Candida species, suggesting that total amounts of individual cell wall components do not determine the ability of fungi to colonize the GI tract. We then subjected this collection of Candida strains and species to an extensive quantitative phenotypic profiling in search for features that might be responsible for their differences in GI fitness, but found no association with the ability to grow in GI-mimicking and stressful environments or with in vitro and in vivo virulence. The most significant association with GI fitness was found to be the strength of signaling through the Dectin-1 receptor. Using a quantitative assay to measure the amount of exposed β-glucan on the surface of fungal cells, we found this parameter, unlike total β-glucan levels, to be strongly predictive of competitive fitness in the mouse GI tract. These data suggest that fungal cell wall architecture, more so than its crude composition, critically determines the ability of fungi to colonize the mammalian GI tract. In particular, recognition of exposed β-glucan by Dectin-1 receptor appears to severely limit Candida GI fitness and hence represents a promising target to reduce fungal colonization in patients at risks of systemic candidiasis.
Introduction
The mammalian gastrointestinal (GI) tract is a complex environment hosting a large number and variety of microbes that include not only bacteria but also fungi (Qin et al., 2010; Iliev et al., 2012). While much effort has been placed in unraveling microbiota associations with host health and disease, little is known about factors that determine the ability of individual microbes, and especially fungi, to colonize the GI environment. The most frequently isolated fungi from the human GI tract belong to the Candida genus, with C. albicans consistently ranking as the most successful fungal colonizer in industrialized countries, followed by C. tropicalis, C. glabrata, C. krusei, C. parapsilosis, and C. dubliniensis (Maccallum, 2010). In addition to acting as commensals in asymptomatic individuals, Candida species are also the most important opportunistic fungal pathogens of humans (Brown et al., 2012). Candida species are able to infect the skin, mucosae and in some cases even the bloodstream of patients, especially those with a weakened immunity (Pfaller and Diekema, 2007). Clinical studies have shown that a large fraction of life-threatening systemic infections by C. albicans originate from strains residing in the patients' own GI tract (Nucci and Anaissie, 2001; Miranda et al., 2009). Therefore, understanding the factors that control the ability of Candida species to colonize the mammalian GI tract might open up opportunities for novel prevention strategies of systemic candidiasis.
Innate immunity plays a paramount role in controlling fungal infections and in limiting the systemic dissemination of Candida species from the GI tract (Koh et al., 2008). The Candida cell wall is an intricate matrix of molecular interactions between pathogen-associated molecular patterns (PAMPs) on the fungal surface and pathogen-recognition receptors (PRRs) on innate immune cells (Netea et al., 2008). These PAMP-PRR interactions are crucial not only for the recognition of fungal pathogens, but also for the initiation of appropriate immune responses (Gow and Hube, 2012). Whereas the role of the Candida cell wall composition has been extensively studied during in vitro interactions with innate immune cells and during in vivo systemic infections, little is known about its role during asymptomatic colonization of the mammalian GI tract.
The fungal cell wall is organized into a complex network of polysaccharides that are organized in layers (Figure 1A), each eliciting distinct immune responses. The innermost cell wall layer, just above the plasma membrane, is composed of chitin. It is primarily synthesized by chitin synthase 3 (encoded by CHS3) and blocks pro-inflammatory cytokine production (Bulawa et al., 1995; Mora-Montes et al., 2011). The intermediate β-glucan layer is critically dependent on (1,3)-β-glucan synthase activity (whose essential catalytic subunit is encoded by GSC1) and triggers instead strong inflammatory responses (Mio et al., 1997; Brown et al., 2003; Gow et al., 2007). This β-glucan layer is however normally masked by the less pro-inflammatory outermost mannan layer (critically dependent on PMR1 for its synthesis), which includes O-linked mannans (dependent on MNT1 and MNT2), N-linked mannans (dependent on OCH1) and phosphomannans (dependent on MNN4). This relatively ordered structure is however far from static, as the intermediate β-glucan layer can be exposed or change its conformation during infection and morphogenesis (Wheeler et al., 2008; Lowman et al., 2014). Moreover, the same cell wall layer can mediate different outcomes in different pathophysiological contexts. While Dectin-1-mediated recognition of β-glucan is for instance required for controlling systemic infections (Taylor et al., 2007) and even triggers a protective “training” of innate immunity (Quintin et al., 2012), it appears to be dispensable for controlling asymptomatic GI colonization (Vautier et al., 2012). However, the latter findings were obtained by employing a wild-type C. albicans strain, which is known to mask its β-glucan layer relatively well, hence it is currently unknown whether variation in β-glucan exposure across Candida strains and species underlies their variable ability to colonize the mammalian GI tract.
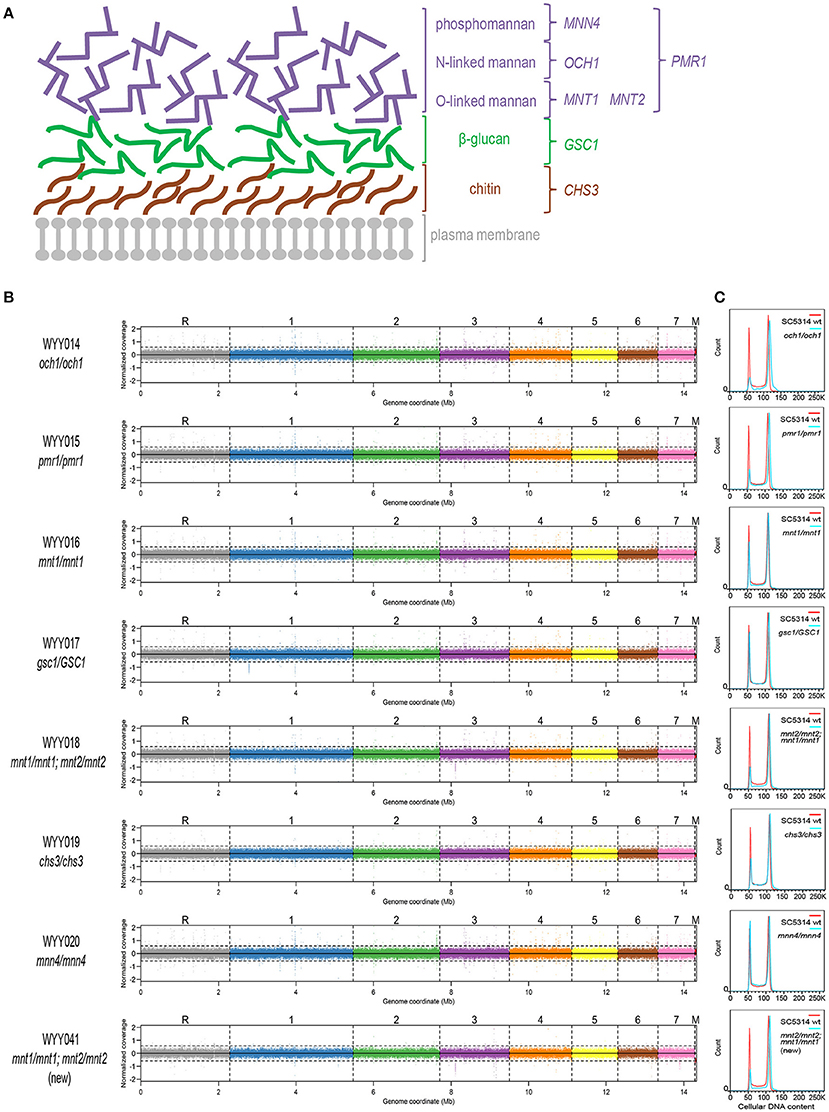
Figure 1. Euploidy and isogenicity of C. albicans cell wall mutants. (A) Schematic representation of the major cell wall components of C. albicans. The mannan (phosphomannan, N-linked mannans and O-linked mannans, in purple), β-glucan (in green) and chitin layers (in brown) are located externally to the plasma membrane (in gray). In this study, several genes involved in modifying and regulating each cell wall component were perturbed and investigated. They are listed in colors corresponding to each of the three layers. (B) The aforementioned cell wall mutants were subjected to deep sequencing and compared to the parental strain. The coverage plots, with a different color for each chromosome, illustrate lack of segmental or whole-chromosome aneuploidy (M, mitochondrial DNA). Apart from a few single-nucleotide substitutions, all sequenced mutants were perfectly isogenic to the wild-type C. albicans parental strain SC5314. (C) Cell wall mutants (blue) generated in the parental SC5314 background were analyzed for their cellular DNA content by flow cytometry and compared to that of the parental strain (red).
Here we employed a reverse genetic approach coupled with an extensive quantitative phenotypic characterization to systematically perturb the main polysaccharide layers of the C. albicans cell wall and assess their contribution in a mouse model of GI colonization. We also took advantage of the natural variation of cell wall architectures that exists within the Candida genus to validate our findings. Our data indicate that whereas most genetic perturbations in the C. albicans cell wall reduce GI fitness, several Candida species are actually fitter than C. albicans in this environment. Interestingly, the strength of signaling through the Dectin-1 receptor emerged as the strongest predictor of GI fitness across all analyzed strains and species, while no correlation was found with crude levels of any tested cell wall component. As predicted, Candida strains and species exhibiting the strongest signaling through the Dectin-1 receptor also exposed the highest amounts of β-glucan on their cell surface. Overall, these findings demonstrate a fundamental role of the Candida cell wall architecture in determining fungal colonization of the mammalian GI tract and open up new avenues for the prevention and control of opportunistic fungal infections.
Materials and Methods
Fungal Strains
C. albicans strains and other Candida species used in this study are summarized in Supplementary Table 1. Details about culture conditions and strain construction, including primers used in this study (Supplementary Table 2), can be found in Supplemental Methods. The ploidy of all generated strains was determined via a previously described quantitative flow cytometric method (Pavelka et al., 2010).
Whole-Genome Sequencing
Genomic DNA was extracted as described (Rancati et al., 2008). Sequencing libraries were prepared using Illumina sample preparation kits and indexed paired-end (PE) sequences were obtained on an Illumina HiSeq platform. All sequencing data were deposited in NCBI SRA database under accession number SRP056269. Data pre-processing, mapping and variant calling was performed in CLC Genomics Workbench, essentially as described (Liu et al., 2015). Higher level data integration and analysis was performed via customized R scripts. Detailed sequencing protocols and analysis parameters are provided in Supplemental Methods. A summary of the sequencing data and results is reported in Supplementary Tables 3, 4.
Cell Wall Composition Analysis
Logarithmically-growing C. albicans was washed three times in 50 ml of 1 mM cold phenylmethanesulfonyl fluoride (PMSF). 3 volumes of glass beads were then added to one volume of pellet and vortexed for 10–15 times at 1-min intervals to break the cells. The cell lysate was again washed three times with 50 ml cold PMSF, before addition of 3 volumes of 2% SDS to the pellet and incubation for 10 min at 100°C for two rounds. SDS was removed by washing the pellet in 1 mM PMSF and the pellet was dried overnight at 55°C before re-suspension in 1 mM PMSF to a solution of 0.01 g dry weight / ml and storage at −20°C. Total chitin was extracted and measured as described (Kapteyn et al., 1997). Total mannan were extracted from isolated cell walls by alkali treatment and further precipitated with Fehling's solution as a copper complex as described (Chaffin et al., 1998). Total glucan was extracted as described (Dijkgraaf et al., 1996) and the extracted glucan and mannan components were then measured by the Dubois method (Dubois et al., 1956) using glucose as a standard.
Mouse Experiments
Mouse experiments were conducted according to the rules and guidelines of the Agri-Food and Veterinary Authority (AVA) and the National Advisory Committee for Laboratory Animal Research (NACLAR), Singapore. The experiments were reviewed and approved by the Institutional Review Board of the Biological Resource Center, Singapore (IACUC protocol 140955). Six to ten weeks-old female C57BL/6 wild-type mice were used for all experiments. Systemic virulence was assessed after injection of 5 × 105 CFUs via the tail vein. GI competition experiments were performed essentially as described in Prieto et al. (2014). Briefly, equimolar mixtures of a test strain and a fluorescently tagged (dTomato-expressing) reference strain were intragastrically inoculated in antibiotic-treated mice (2 mg/ml streptomycin + 1500 U/ml penicillin G) and their relative frequency was monitored over time by plating stool homogenates on antibiotics-containing plates and counting fluorescent colonies under a fluorescence stereomicroscope. The relative fitness coefficient of the test strain was then obtained by linear regression according to the following formula:
where R(i) represents the ratio between the test strain and the reference strain at time i; s is the selection coefficient; γR is the growth rate of the reference strain expressed as cell divisions per hour; t represents the time points in hours and t0 the initial time point. In vitro fitness competition assays were performed essentially in the same way, except that strain mixtures were serially passaged on a daily basis in YPD medium. More details are provided in the Supplemental Methods.
Candida-Macrophage Co-cultures
J774.1 cells were plated at a density of 5 × 105 cells/well in 12-well plates for 24 h. C. albicans strains from overnight cultures were washed twice in PBS and co-cultured with the macrophages at various MOIs, ranging from 1:200 to 1:1600. Under these conditions, >99% of individual C. albicans cells were phagocytosed by the macrophages after 3 h (data not shown). Visible fungal colonies were counted after 24 h, and the percentage of phagosome-escaping cells was determined by linear regression as explained in the Supplemental Methods.
Quantitative Resistance Assays
Resistance to mCRAMP was determined on logarithmically growing cells re-suspended in sterile PBS and incubated at 37°C for 1–2 h. Cell pellets were then re-suspended in a 2-fold serial dilution of mCRAMP 1–39 (Synpeptide) and incubated at 37°C for 2 h, followed by a PBS wash, live/dead staining with 30 μM propidium iodide for 15 min and flow cytometric analysis. Half-maximal effective concentrations (EC50) were determined by non-linear regression in GraphPad Prism. The half-maximal inhibitory concentrations (IC50) of acetic and lactic acid on fungal growth were determined in vitro via turbidimetric assays as described previously (Cottier et al., 2015a) with some minor modifications (refer to Supplemental Methods). Oxidative stress resistance was determined on PBS-washed overnight cultures incubated in PBS at 37°C for 2 h. Cells were then treated with a range of tert-Butyl hydroperoxide (tBOOH) concentrations, incubated at 37°C for 1 h and subjected to live/dead analysis as described above. The NO sensitivity assay was performed following previous protocol (Chiranand et al., 2008) with some minor modifications (refer to Supplemental Methods).
High-Throughput Phenotypic Profiling
Overnight cultures of freshly revitalized C. albicans strains and Candida spp. were normalized to a final OD600 of 2 and washed once with PBS. A Freedom EVO 150 liquid-handling robot (Tecan) performed serial dilutions of the cultures and spotted 3 μl of each serial dilution onto YPD agar omnitrays representing one of a wide range of growth conditions listed in Supplementary Table 5. Each condition was tested a minimum of three independent times and growth data was acquired by a desktop scanner and analyzed using a custom R script for automated spot detection and intensity measurements followed by non-linear curve fitting across the range of serial dilutions and determination of relative growth scores as explained in Supplemental Methods.
Quantification of β-1, 3-glucan Exposure
Logarithmically-growing Candida cells were stained with anti-β-1,3-glucan monoclonal antibody (mouse IgG; Biosupplies) followed by anti-mouse IgG Alexa Fluor 488 (Molecular Probes) staining, as described previously (Wartenberg et al., 2014) with some modifications (refer to Supplemental Methods). The level of β-1,3-glucan exposure on the cell surface was then quantified on a MACSquant VYB flow cytometer (Miltenyi).
Quantification of PRR Signaling Strength
Dectin-1, Dectin-2, TLR2, and TLR4 signaling measurements were performed using a NF-kB reporter cell line (HEK-Blue™ hDectin-1b, HEK-Blue™ mDectin-2 Cells, HEK-Blue™ mTLR2 cells, HEK-Blue™ mTLR4 cells Invivogen) and its parental cell line (HEK-Blue™-null1-v or HEK-Blue™ null2 cells) as the negative control. Briefly, 1 × 105 cells/well were stimulated with 1 × 106 formaldehyde-fixed Candida cells/well and incubated in 96-well flat bottom plates for 6-20 h. Finally, NF-kB activity was quantified with the QUANTI-Blue (Invivogen) reagent accordingly to the manufacturer's instructions.
Statistical Analysis
All data were analyzed using Excel (Microsoft) and Prism (Graphpad) software. Data is reported as mean ± SD. To compare wild-type C. albicans, isogenic C. albicans cell wall mutants and Candida spp. strains fitter than or as fit as wild-type C. albicans and isogenic C. albicans cell wall mutants and Candida spp. strains less fit than wild-type C. albicans, non-parametric Mann-Whitney test was used. For all other comparisons, statistical significance was calculated using two-tailed unpaired t-tests with Welch's correction. In all analyses, p ≤ 0.05 were regarded as statistically significant.
Results
A Collection of Isogenic C. albicans Cell Wall Mutants
To assess the contribution of each cell wall component to GI tract fitness, we first generated an isogenic set of C. albicans mutants in the prototroph and otherwise wild-type reference strain SC5314, each deleted in one or more genes illustrated in Figure 1A. In particular, we generated homozygous mutants in CHS3, PMR1, MNT1, OCH1, and MNN4 using the SAT-Flipper technique, which allows inducible removal of the deletion cassette, leaving no ectopic selection marker in the genome (Sasse and Morschhauser, 2012). Since GSC1 is an essential gene, we successfully generated the corresponding heterozygous but not the homozygous mutant; on the other hand, because MNT1 was shown to be partially redundant with MNT2 (Munro et al., 2005), we also generated a double homozygous strain carrying both gene deletions in homozygosity. Using quantitative staining of cellular DNA content followed by flow cytometry, we confirmed all strains in the collection to be diploid (Figure 1B). Whole-genome sequencing further indicated absence of either whole-chromosome or segmental aneuploidy and presence of only a few point mutations, most of which were heterozygous and either non-coding or synonymous (Figure 1C, Supplementary Table 4). The mnt1/mnt1;mnt2/mnt2 strain was the only strain containing a non-synonymous mutation in homozygosity and was therefore reconstructed. The reconstructed mnt1/mnt1;mnt2/mnt2 strain was then re-sequenced and found not to carry this or other homozygous non-synonymous mutation (Supplementary Table 4). Experiments critical for this study have been repeated in parallel using both mnt1/mnt1;mnt2/mnt2 strains and found not to lead to significantly different outcomes (Supplementary Figure 1). This collection of C. albicans cell wall mutants therefore represents a carefully constructed isogenic set of strains that is well suited for quantitative phenotypic analyses without potential confounders due to undocumented genetic or karyotypic differences. Moreover, since these strains were generated without using any auxotrophic marker, they are expected to show no intrinsic deficiency in their ability to grow in nutrient-limited environments, e.g., during colonization or infection of certain host tissues, and should therefore represent an invaluable resource to study the role of the C. albicans cell wall during interaction with the host.
GI Tract Fitness of C. albicans Cell Wall Mutants
We next adopted a previously proposed in vivo competition assay aimed at comparing the ability of different C. albicans strains to colonize the mouse GI tract (Prieto et al., 2014) and extended it into a quantitative in vivo competitive fitness assay. Briefly, the original method entailed orally gavaging antibiotic-treated mice with equimolar mixtures of a test strain and a fluorescently tagged reference strain, followed by counting fluorescent and non-fluorescent colonies in plated serial dilutions of stool samples collected at specific time points from the double-colonized mice. As expected, competition of wild-type SC5314 cells against the fluorescently tagged version of the SC5314 strain led to no significant change in the relative proportion of fluorescent colonies in the mouse stool over time, indicating that presence of the fluorescence cassette was associated with no or minimal fitness cost in the mouse GI tract (Figure 2A). In contrast, our collection of isogenic cell wall mutants displayed a variety of behaviors in this assay, ranging from undetectable to significant decrease in relative frequency depending on the genotype (Figures 2B–E). In particular, while the gsc1/GSC1 and the mnn4/mnn4 mutants were essentially undistinguishable from the wild-type strain, the mnt1/mnt1 strain showed a consistent and progressive drop in relative frequency during the competition assay, reaching ~20% after ~60 h. The mnt1/mnt1; mnt2/mnt2 mutant, however, was completely outcompeted by the fluorescent strain in <40 h, suggesting that MNT1 and MNT2 are not completely redundant with each other for GI tract fitness. Moreover, while chs3/chs3 and pmr1/pmr1 mutants were outcompeted by the reference strain in a similar amount of time as the mnt1/mnt1;mnt2/mnt2 strain, the och1/och1 strain was outcompeted in ~10 h.
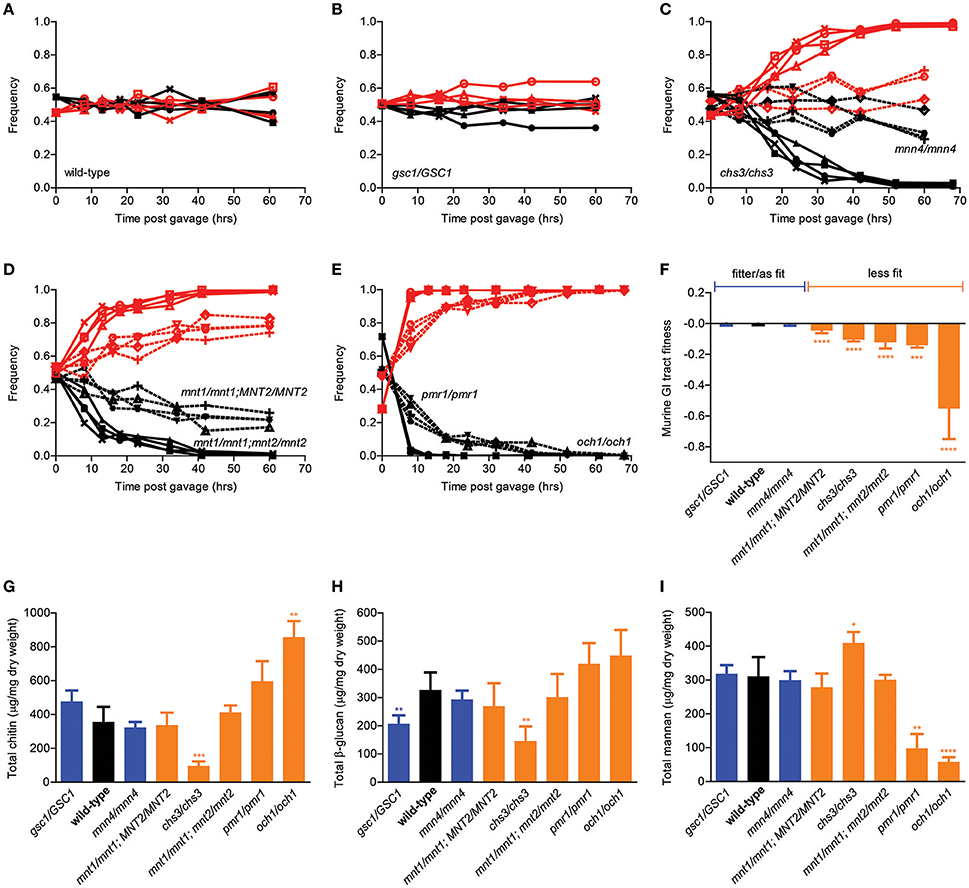
Figure 2. Murine GI tract fitness of isogenic C. albicans cell wall mutants. (A–E) Relative frequency of wild-type C. albicans (A, black line) and the isogenic cell wall mutants gsc1/GSC1 (B, black lines), mnn4/mnn4 (C, black dotted lines), chs3/chs3, (C, black solid lines), mnt1/mnt1; MNT2/MNT2 (D, black dotted lines), mnt1/mnt1; mnt2/mnt2 (D, black solid lines), pmr1/pmr1 (E, black dotted lines) and och1/och1 (E, black solid lines) in comparison to SC5314-dTomato (A–E, red lines) during an in vivo competition assay in the mouse GI tract. All experiments were initiated at a nominal ratio of 1:1 between competing strains, except for och1/och1 where the och1/och1:SC5314-dTomato ratio was 7:3. Each pair of lines represents an independent biological assay with a singly-housed mouse, with ≥4 mice per strain and ≥200 CFUs counted per time point. (F) Murine GI tract fitness of wild-type C. albicans (black) and the isogenic cell wall mutants are summarized and represented as average and standard deviation of fitted selection coefficients (see Supplemental Methods). Wild-type and mutant strains are ranked in decreasing order of murine GI tract fitness and classified into two groups: fitter than or as fit as wild-type (blue); less fit than wild-type (orange). (G–I) Cell wall polysaccharide composition of each wild-type and mutant strain in terms of total chitin (G), total β-glucan (H) and total mannan (I). Each bar graph represents the average of 3 independent biological assays. Strains are ranked and classified as in (F). Asterisks shown represent significant two-tailed p-values obtained from unpaired Welch's t-tests between wild-type and mutant strain (* < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001).
We then developed a regression method that allows us to rigorously quantitate the relative competitive fitness of each test strain using a time series of relative frequencies of fluorescent colonies as an input (see Supplemental Methods for details). By this analysis, the gsc1/GSC1 and mnn4/mnn4 strains were at least as fit as wild-type C. albicans, while all other cell wall mutants in this study displayed a wide range of fitness disadvantages (Figure 2F). In an attempt to correlate GI tract fitness with cell wall composition, we then quantified the total amounts of chitin, β-glucans and mannans in all strains but no significant association was found. In particular, among the strains with significantly reduced GI tract fitness, mnt/mnt1;MNT2/MNT2 and mnt1/mnt1;mnt2/mnt2 showed no significant alterations in crude cell wall composition; on the other hand, chs3/chs3 had significantly reduced chitin and β-glucan and significantly increased mannan levels and och1/och1 had significantly increased chitin and significantly decreased mannan levels. Overall, while genetic alterations in cell wall biosynthetic pathways clearly affect the ability of C. albicans to colonize the mouse GI tract, differential fitness in this environment cannot be attributed to relative compositional differences in the three main polysaccharide layers.
GI Tract Fitness of Candida Species
In order to extend the generality of our findings beyond C. albicans, we then repeated the in vivo competition assays in distantly related Candida species (Figures 3A–E). Interestingly, while C. krusei and C. parapsilosis were significantly less fit, C. glabrata, C. tropicalis, and C. dubliniensis were significantly fitter than C. albicans in the mouse GI tract (Figure 3F). In accordance with the cell wall mutants' data, compositional analysis of the major cell wall polysaccharides did not yield any significant association with GI tract fitness. In particular, whereas both C. glabrata and C. dubliniensis were significantly fitter than C. albicans in the mouse GI environment, the former had significantly reduced and the latter significantly increased chitin levels in comparison to C. albicans. Moreover, whereas both C. tropicalis and C. krusei showed significantly decreased β-glucan levels, the former was significantly fitter and the latter significantly less fit then C. albicans. Hence, despite the existence of a large variation in both GI tract fitness and cell wall composition within the Candida genus, these two traits appear to be largely independent from each other.
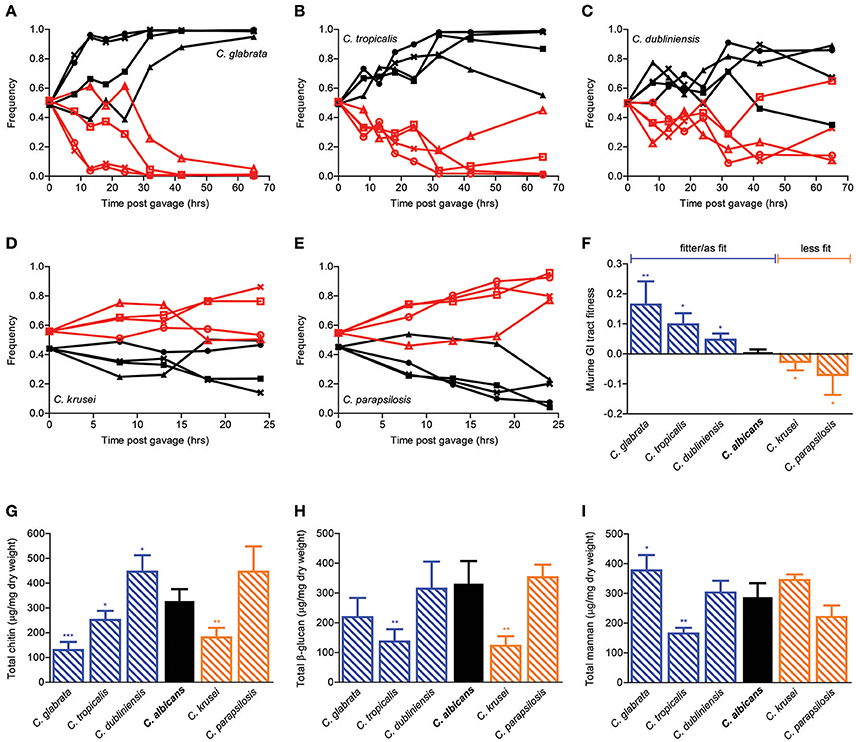
Figure 3. Murine GI tract fitness of Candida spp. strains. Relative frequency of wild-type C. glabrata (A, black), C. tropicalis (B, black), C. dubliniensis (C, black), C. krusei, (D, black), and C. parapsilosis (E, black) in comparison to SC5314-dTomato (A–E, red) during a competition assay in the mouse GI tract, represented as in Figures 2A–E. Each pair of lines represents an independent biological assay with a singly-housed mouse, with ≥4 mice per strain and ≥200 CFUs counted per time point. (F) Murine GI tract fitness of C. albicans (black) and Candida spp. (blue and orange shaded bars) represented as in Figure 2F. (G–I) Cell wall polysaccharide composition of each Candida species represented as in Figures 2G–I. Asterisks shown represent significant two-tailed p-values obtained from unpaired Welch's t-tests between wild-type and mutant strain (* < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001).
Quantitative Phenotypic Profiling Reveals No Associations with GI Tract Fitness
We next sought to identify factors underlying the variation in mouse GI tract fitness across all our C. albicans cell wall mutants and the other Candida species, employing a systematic profiling of a large number of quantitative phenotypic parameters (Supplementary Figure 2). Consistent with above findings, no significant association was found between competitive fitness in the mouse GI tract (Supplementary Figure 3A) and total chitin, mannan, β-glucan, β-1,6-glucan or β-1,3-glucan levels (Supplementary Figures 3B–F). Moreover, no association was seen between in vivo and in vitro fitness (Figure 4A), or between in vivo fitness and ability to grow under a variety of physiological or stressful environmental conditions (Supplementary Figures 3G–R), ruling out simple explanations based on overall fitness effects in some of the tested strains. Secretion of the mouse cathelicidin-related antimicrobial peptide (mCRAMP) in the GI lumen was recently shown to play an important role in controlling C. albicans colonization in the mouse GI tract (Fan et al., 2015); however no significant correlation between mouse GI tract fitness and mCRAMP resistance was observed (Figure 4B). Microbiota-derived weak organic acids, such as acetic and lactic acid, were shown to be fungistatic and proposed to contribute to colonization resistance against C. albicans in the mouse GI tract (Cottier et al., 2015b), however Candida strains and species displaying the highest competitive fitness in the mouse GI tract did not show a significant increase in acetic acid or lactic acid resistance (Figures 4C,D). An important feature of gut commensal microbes is their ability to survive in the presence of bile (Strati et al., 2016), but GI fitness was uncorrelated to resistance to ox bile or bile salts (Figures 4E,F). A trade-off between gut commensalism and systemic virulence was recently proposed (Pande et al., 2013), but no association was found between GI tract fitness and ability to kill mice during systemic infections, as all tested cell wall mutants and Candida species were significantly less virulent that wild-type C. albicans (Figure 4G). Moreover, with the only exception of the och1 mutant, the reduced systemic virulence exhibited by the C. albicans cell wall mutants did not associate with the ability to survive in phagosome-like conditions (low pH, high ROS, high NO) (Figures 4H–J). Indeed, none of the cell wall mutants displayed obvious hyphal morphogenesis defects (Supplementary Figure 4) and their ability to escape from macrophages after phagocytosis associated neither with systemic virulence nor with GI tract fitness (Figure 4K).
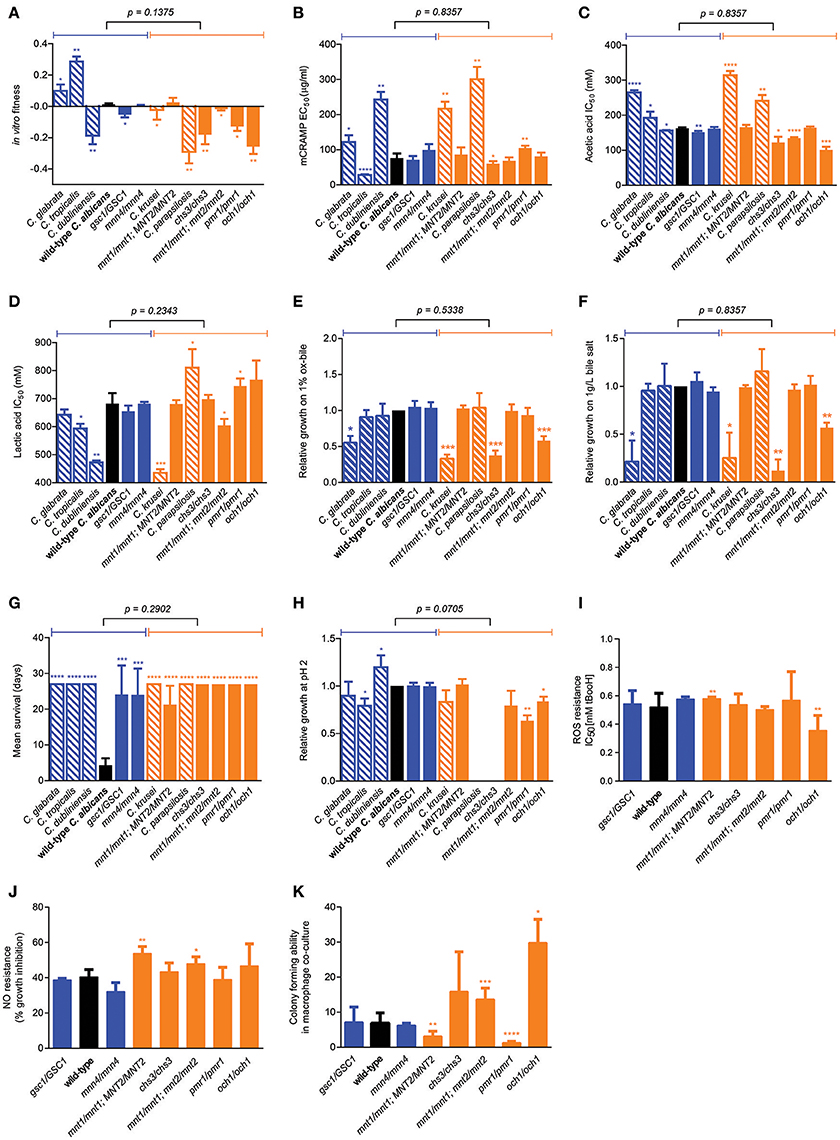
Figure 4. Factors not associated with murine GI tract fitness. (A) Wild-type C. albicans (black) and the isogenic cell wall mutants (blue and orange) were assessed for their in vitro fitness in YPD. (B–K) Using various experimental procedures, all wild-type C. albicans (black bars), isogenic C. albicans cell wall mutants (blue and orange solid bars) and Candida spp. strains (blue and orange shaded bars) were assessed for their resistance to acute exposure to (B) mCRAMP, (C) acetic acid, (D) lactic acid, (E) Ox-bile, and (F) bile-salt and for their (G) systemic virulence in wild-type C57BL/6 mice, (H) relative growth at pH 2, (I) resistance to ROS, (J) resistance to NO and (K) their survivability (colony-forming ability) after 24 h co-culture with the J774A.1 murine macrophage cell line. Strains are ranked in decreasing order of murine GI tract fitness as in Supplementary Figure 2A, and classified into two groups: fitter than or as fit as wild-type C. albicans (solid or shaded blue); less fit than wild-type C. albicans (solid or shaded orange). The p-value on top of each panel was obtained from a Mann-Whitney test between the two indicated groups. Asterisks represent significant two-tailed p-values from unpaired Welch's t-tests in comparison to wild-type C. albicans (* < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001). Each measurement was obtained from ≥3 independent biological experiments.
Role of β-Glucan/Dectin-1 Signaling Axis in Determining Fungal GI Colonization
Because GI tract fitness was not associated with any plausible microbe-intrinsic factor or phenotype that we tested so far, we reasoned that the ability of fungal cells to colonize the gut environment might be better explained by factors related to host-microbe interactions. We therefore screened our library of C. albicans cell wall mutants and Candida species for their ability to trigger signal transduction events downstream of some of the main PRRs known to be important for fungal cell recognition. To this end, we employed commercially available human cell lines expressing individual PRRs and an inducible NF-κB reporter. While TLR4 signaling was essentially undetectable in response to any tested strain (data not shown), large variation in the ability to trigger TLR2- and Dectin-2-mediated signaling was observed between the Candida strains and species; this however did not associate with differences in GI tract fitness (Figures 5A,B). Interestingly, Candida strains and species displayed a wide range of abilities to signal through the Dectin-1 receptor, and this was significantly associated with their competitive fitness in the mouse GI tract (Figure 5C). This prompted us to reevaluate our initial observation about the lack of association between GI fitness and cell wall composition, and to investigate a potential contribution not of the total amount but of the level of exposure of β-glucan (the primary PAMP for Dectin-1) on the surface of the fungal cells. Consistent with predictions, β-glucan surface exposure was significantly associated with mouse GI tract fitness across the entire range of C. albicans cell wall mutants and Candida species (Figure 5D). Overall these data indicate that the fungal cell wall may play an important role in the colonization of the mammalian GI tract, and that the level of β-glucan exposure on the cell surface correlates with a loss in competitive fitness of fungi in this host environment.
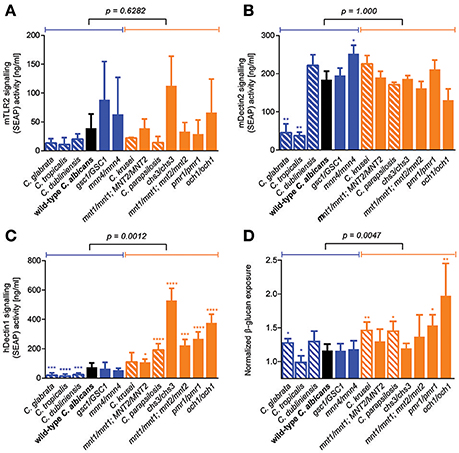
Figure 5. Role of β-glucan/Dectin-1 signaling axis in determining murine GI tract fitness. (A–D) Wild-type C. albicans (black), isogenic C. albicans cell wall mutants (blue and orange solid bars) and Candida spp. strains (blue and orange shaded bars) were assessed for their ability to activate TLR-2 receptor signaling in (A) HEK-Blue mTLR-2 cells, Dectin-2 receptor signaling in (B) HEK-Blue mDectin-2 cells, Dectin-1 receptor signaling in (C) HEK-Blue hDectin-1b cells and β-glucan exposure on their cell walls using an anti-β-glucan monoclonal antibody (D). Wild-type and mutant strains are ranked in decreasing order of murine GI tract fitness and classified into two groups: fitter than or as fit as wild-type C. albicans (solid or shaded blue); less fit than wild-type C. albicans (solid or shaded orange). The p-value on top of each panel was obtained from a Mann-Whitney test between the two indicated groups. Asterisks represent significant two-tailed p-values from unpaired Welch's t-tests in comparison to wild-type C. albicans (* < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001). Each measurement was obtained from ≥3 independent biological experiments.
Discussion
This study describes the generation of a carefully controlled set of isogenic cell wall mutants in C. albicans and provides a proof of concept of its usefulness in deciphering the role of the cell wall during in vivo host-microbe interactions. It also describes a number of optimized assays for precise quantitation of phenotypic parameters ranging from in vitro to in vivo fitness, which are expected to have broad applicability. In particular, we here show that by applying an unbiased, systematic, quantitative phenotypic characterization of a well-controlled set of Candida mutants and species, one can confidently rule in or rule out a number of hypotheses related to fungal colonization of the mammalian GI tract.
Most of the herein described mutants are disrupted in genes that had been knocked out from the C. albicans genome by other groups in the past. However, different genetic backgrounds have been used by different groups, some of which carrying auxotrophies or other mutations that might impact in vivo fitness (Bulawa et al., 1995; Hobson et al., 2004; Bates et al., 2005, 2006; Munro et al., 2005; Ben-Ami et al., 2011). Moreover, some of the laboratory strains previously used to disrupt some of these genes were later shown to carry large-scale chromosomal aberrations such as whole-chromosome aneuploidy and long-range loss-of-heterozygosity (Abbey et al., 2011). Such genomic alterations are likely to affect fungal fitness under a variety of conditions, and especially the highly stressful environment of the host (Pavelka et al., 2010; Selmecki et al., 2010). Moreover, following excision of the deletion cassette, no genetic marker was retained in the genome of our final mutants and their isogenicities were further verified by deep whole-genome sequencing. Finally, a long list of carefully quantified phenotypic information, including both in vitro and in vivo phenotypes, was systematically collected from the entire strain collection. For all these reasons, we believe that the herein described set of carefully constructed and deeply characterized C. albicans cell wall mutants will represent an important resource for the Candida research community at large, and in particular for investigations into the role of the fungal cell wall during host-microbe interactions.
The role of the fungal cell wall in host-microbe interactions has been extensively studied by several groups. C. albicans cell wall mutants have been employed to decipher the relative contribution of various cell wall components on immune responses and infection outcomes in both in vitro co-culture and systemic infection models. These studies have revealed the complexity and dynamics of the fungal cell wall and have shed light on how the host integrates information from multiple PAMPs through multiple PRRs to produce either protective or pathogenic immune responses. However, in spite of the mammalian GI tract being the main niche colonized by Candida species, little or nothing is known about the role played by the Candida cell wall during asymptomatic colonization of this niche. The most important finding in the present study is that the Candida cell wall indeed appears to play a role in determining the competitive fitness of these fungal species in the mouse GI tract.
However, GI tract fitness of the tested strains associated neither with the total amounts of individual cell wall components nor with in vitro fitness in a large number of tested environments. Two important lessons can be learned from these observations. First, predicting GI fitness from in vitro fitness in single environments is probably not possible because the GI tract is a very complex environment, which is very difficult to mimic in vitro. In fact, the GI tract is characterized by extremes of pH, gradients of nutrients and presence of a range of antimicrobial compounds like immunoglobulin A, bile salts and AMPs. The ability to survive and grow in face of any one of these individual challenges does not predict resistance to other stresses and hence does not predict overall fitness in the GI tract. The ideal colonizer most probably acts like a generalist that achieves suboptimal fitness across a wide range of individual environments rather than specializing in thriving in presence of any one given stress condition. Second, knowing the amount of chitin, mannans and glucans in the cell wall says little about how fit a Candida cell would be in the GI tract. The architecture of the cell wall, i.e., the spatial organization of various cell wall components relative to each other and to the cell surface, is far more important in determining how the host will recognize, interact with and respond to the microbe. In particular, it appears that the most critical determinant of GI tract fitness is the level of exposure of β-glucan on the cell surface, which associates with the strength of signaling through the Dectin-1 receptor on host cells. The dynamic nature of β-glucan masking and exposure was previously appreciated and shown to be linked to morphogenesis and to play an important role during systemic infection (Wheeler et al., 2008; Marakalala et al., 2013). Because of the profound negative impact of β-glucan exposure on the ability of Candida cells to colonize their main natural niche, i.e., the mammalian GI tract, we expect this phenomenon to be tightly regulated and this trait to be under strong evolutionary pressure.
Author Contributions
NP conceived, designed and supervised the study; XS, GL, AT, and GT performed microbiological, immunological, and mouse experiments; JL, KS, and FZ performed whole-genome sequencing experiments; MY, WL, MP and NP analyzed sequencing data; XS, GL, AT and NP performed statistical analyses.
Funding
This work was supported by an A*STAR Investigatorship award (JCO/1437a00117), an NMRC Bedside-and-Bench grant (NMRC/BnB/0001b/2012) and by core funding from the Singapore Immunology Network to NP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors would like to thank Robert Wheeler for the pENO1-dTom-NATR plasmid, Alvin Tan for assistance in setting up the in vivo competition assay and Giulia Rancati for useful scientific discussions. Sequencing was performed by the GIS Genome Technology and Biology Group, Singapore.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2016.00186/full#supplementary-material
References
Abbey, D., Hickman, M., Gresham, D., and Berman, J. (2011). High-resolution SNP/CGH microarrays reveal the accumulation of loss of heterozygosity in commonly used Candida albicans strains. G3 1, 523–530. doi: 10.1534/g3.111.000885
Bates, S., Hughes, H. B., Munro, C. A., Thomas, W. P., Maccallum, D. M., Bertram, G., et al. (2006). Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281, 90–98. doi: 10.1074/jbc.M510360200
Bates, S., Maccallum, D. M., Bertram, G., Munro, C. A., Hughes, H. B., Buurman, E. T., et al. (2005). Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 280, 23408–23415. doi: 10.1074/jbc.M502162200
Ben-Ami, R., Garcia-Effron, G., Lewis, R. E., Gamarra, S., Leventakos, K., Perlin, D. S., et al. (2011). Fitness and virulence costs of Candida albicans FKS1 hot spot mutations associated with echinocandin resistance. J. Infect. Dis. 204, 626–635. doi: 10.1093/infdis/jir351
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv113. doi: 10.1126/scitranslmed.3004404
Brown, G. D., Herre, J., Williams, D. L., Willment, J. A., Marshall, A. S., and Gordon, S. (2003). Dectin-1 mediates the biological effects of beta-glucans. J. Exp. Med. 197, 1119–1124. doi: 10.1084/jem.20021890
Bulawa, C. E., Miller, D. W., Henry, L. K., and Becker, J. M. (1995). Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 92, 10570–10574. doi: 10.1073/pnas.92.23.10570
Chaffin, W. L., López-Ribot, J. L., Casanova, M., Gozalbo, D., and Martínez, J. P. (1998). Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol. Mol. Biol. Rev. 62, 130–180.
Chiranand, W., McLeod, I., Zhou, H., Lynn, J. J., Vega, L. A., Myers, H., et al. (2008). CTA4 transcription factor mediates induction of nitrosative stress response in Candida albicans. Eukaryot Cell 7, 268–278. doi: 10.1128/EC.00240-07
Cottier, F., Tan, A. S., Chen, J., Lum, J., Zolezzi, F., Poidinger, M., et al. (2015a). The transcriptional stress response of Candida albicans to weak organic acids. G3 5, 497–505. doi: 10.1534/g3.114.015941
Cottier, F., Tan, A. S., Xu, X., Wang, Y., and Pavelka, N. (2015b). MIG1 regulates resistance of Candida albicans against the fungistatic effect of weak organic acids. Eukaryot Cell 14, 1054–1061. doi: 10.1128/EC.00129-15
Dijkgraaf, G. J., Brown, J. L., and Bussey, H. (1996). The KNH1 gene of Saccharomyces cerevisiae is a functional homolog of KRE9. Yeast 12, 683–692. doi: 10.1002/(SICI)1097-0061(19960615)12:7<683::AID-YEA959>3.0.CO;2-8
Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. doi: 10.1021/ac60111a017
Fan, D., Coughlin, L. A., Neubauer, M. M., Kim, J., Kim, M. S., Zhan, X., et al. (2015). Activation of HIF-1[alpha] and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med. 21, 808–814. doi: 10.1038/nm.3871
Gow, N. A., and Hube, B. (2012). Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15, 406–412. doi: 10.1016/j.mib.2012.04.005
Gow, N. A., Netea, M. G., Munro, C. A., Ferwerda, G., Bates, S., Mora-Montes, H. M., et al. (2007). Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 196, 1565–1571. doi: 10.1086/523110
Hobson, R. P., Munro, C. A., Bates, S., Maccallum, D. M., Cutler, J. E., Heinsbroek, S. E., et al. (2004). Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279, 39628–39635. doi: 10.1074/jbc.M405003200
Iliev, I. D., Funari, V. A., Taylor, K. D., Nguyen, Q., Reyes, C. N., Strom, S. P., et al. (2012). Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317. doi: 10.1126/science.1221789
Kapteyn, J. C., Ram, A. F., Groos, E. M., Kollar, R., Montijn, R. C., Van Den Ende, H., et al. (1997). Altered extent of cross-linking of beta1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall beta1,3-glucan content. J. Bacteriol. 179, 6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997
Koh, A. Y., Köhler, J. R., Coggshall, K. T., Van Rooijen, N., and Pier, G. B. (2008). Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 4:e35. doi: 10.1371/journal.ppat.0040035
Liu, G., Yong, M. Y., Yurieva, M., Srinivasan, K. G., Liu, J., Lim, J. S., et al. (2015). Gene essentiality is a quantitative property linked to cellular evolvability. Cell 163, 1388–1399. doi: 10.1016/j.cell.2015.10.069
Lowman, D. W., Greene, R. R., Bearden, D. W., Kruppa, M. D., Pottier, M., Monteiro, M. A., et al. (2014). Novel structural features in Candida albicans hyphal glucan provide a basis for differential innate immune recognition of hyphae versus yeast. J. Biol. Chem. 289, 3432–3443. doi: 10.1074/jbc.M113.529131
Maccallum, D. M. (2010). “Candida infections and modelling disease,” in Pathogenic Yeasts, eds R. Ashbee and M. E. Bignell (Berlin; Heidelberg: Springer Berlin Heidelberg), 41–67.
Marakalala, M. J., Vautier, S., Potrykus, J., Walker, L. A., Shepardson, K. M., Hopke, A., et al. (2013). Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 9:e1003315. doi: 10.1371/journal.ppat.1003315
Mio, T., Adachi-Shimizu, M., Tachibana, Y., Tabuchi, H., Inoue, S. B., Yabe, T., et al. (1997). Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis. J. Bacteriol. 179, 4096–4105. doi: 10.1128/jb.179.13.4096-4105.1997
Miranda, L. N., Van Der Heijden, I. M., Costa, S. F., Sousa, A. P., Sienra, R. A., Gobara, S., et al. (2009). Candida colonisation as a source for candidaemia. J. Hosp. Infect. 72, 9–16. doi: 10.1016/j.jhin.2009.02.009
Mora-Montes, H. M., Netea, M. G., Ferwerda, G., Lenardon, M. D., Brown, G. D., Mistry, A. R., et al. (2011). Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect. Immun. 79, 1961–1970. doi: 10.1128/IAI.01282-10
Munro, C. A., Bates, S., Buurman, E. T., Hughes, H. B., Maccallum, D. M., Bertram, G., et al. (2005). Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 280, 1051–1060. doi: 10.1074/jbc.M411413200
Netea, M. G., Brown, G. D., Kullberg, B. J., and Gow, N. A. (2008). An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6, 67–78. doi: 10.1038/nrmicro1815
Nucci, M., and Anaissie, E. (2001). Revisiting the source of candidemia: skin or gut? Clin. Infect. Dis. 33, 1959–1967. doi: 10.1086/323759
Pande, K., Chen, C., and Noble, S. M. (2013). Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat. Genet. 45, 1088–1091. doi: 10.1038/ng.2710
Pavelka, N., Rancati, G., Zhu, J., Bradford, W. D., Saraf, A., Florens, L., et al. (2010). Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325. doi: 10.1038/nature09529
Pfaller, M. A., and Diekema, D. J. (2007). Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20, 133–163. doi: 10.1128/CMR.00029-06
Prieto, D., Román, E., Correia, I., and Pla, J. (2014). The HOG pathway is critical for the colonization of the mouse gastrointestinal tract by Candida albicans. PLoS ONE 9:e87128. doi: 10.1371/journal.pone.0087128
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Quintin, J., Saeed, S., Martens, J. H., Giamarellos-Bourboulis, E. J., Ifrim, D. C., Logie, C., et al. (2012). Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232. doi: 10.1016/j.chom.2012.06.006
Rancati, G., Pavelka, N., Fleharty, B., Noll, A., Trimble, R., Walton, K., et al. (2008). Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135, 879–893. doi: 10.1016/j.cell.2008.09.039
Sasse, C., and Morschhäuser, J. (2012). Gene deletion in Candida albicans wild-type strains using the SAT1-flipping strategy. Methods Mol. Biol. 845, 3–17. doi: 10.1007/978-1-61779-539-8_1
Selmecki, A., Forche, A., and Berman, J. (2010). Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot Cell 9, 991–1008. doi: 10.1128/EC.00060-10
Strati, F., Di Paola, M., Stefanini, I., Albanese, D., Rizzetto, L., Lionetti, P., et al. (2016). Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front. Microbiol. 7:1227. doi: 10.3389/fmicb.2016.01227
Taylor, P. R., Tsoni, S. V., Willment, J. A., Dennehy, K. M., Rosas, M., Findon, H., et al. (2007). Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 8, 31–38. doi: 10.1038/ni1408
Vautier, S., Drummond, R. A., Redelinghuys, P., Murray, G. I., Maccallum, D. M., and Brown, G. D. (2012). Dectin-1 is not required for controlling Candida albicans colonization of the gastrointestinal tract. Infect. Immun. 80, 4216–4222. doi: 10.1128/IAI.00559-12
Wartenberg, A., Linde, J., Martin, R., Schreiner, M., Horn, F., Jacobsen, I. D., et al. (2014). Microevolution of Candida albicans in macrophages restores filamentation in a nonfilamentous mutant. PLoS Genet. 10:e1004824. doi: 10.1371/journal.pgen.1004824
Keywords: Candida, Candida albicans, cell wall, competitive fitness, gastrointestinal tract, beta-glucan, dectin-1
Citation: Sem X, Le GTT, Tan ASM, Tso G, Yurieva M, Liao WWP, Lum J, Srinivasan KG, Poidinger M, Zolezzi F and Pavelka N (2016) β-glucan Exposure on the Fungal Cell Wall Tightly Correlates with Competitive Fitness of Candida Species in the Mouse Gastrointestinal Tract. Front. Cell. Infect. Microbiol. 6:186. doi: 10.3389/fcimb.2016.00186
Received: 19 October 2016; Accepted: 30 November 2016;
Published: 22 December 2016.
Edited by:
Brian Wickes, University of Texas Health Science Center at San Antonio, USAReviewed by:
Karen L. Wozniak, University of Texas at San Antonio, USARobert T. Wheeler, University of Maine, USA
Copyright © 2016 Sem, Le, Tan, Tso, Yurieva, Liao, Lum, Srinivasan, Poidinger, Zolezzi and Pavelka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Norman Pavelka, bm9ybWFuX3BhdmVsa2FAaW1tdW5vbC5hLXN0YXIuZWR1LnNn
†Present Address: Marina Yurieva, The Jackson Laboratory for Genomic Medicine, Farmington, USA
Francesca Zolezzi, Galderma R&D, Sophia Antipolis, France
‡These authors have contributed equally to this work.
 XiaoHui Sem
XiaoHui Sem Giang T. T. Le
Giang T. T. Le Alrina S. M. Tan
Alrina S. M. Tan Gloria Tso
Gloria Tso Marina Yurieva
Marina Yurieva Webber W. P. Liao
Webber W. P. Liao Josephine Lum
Josephine Lum Kandhadayar G. Srinivasan
Kandhadayar G. Srinivasan Michael Poidinger
Michael Poidinger Francesca Zolezzi
Francesca Zolezzi Norman Pavelka
Norman Pavelka