Abstract
Conjugation is a key mechanism for horizontal gene transfer and plays an important role in bacterial evolution, especially with respect to antibiotic resistance. However, little is known about the role of donor and recipient cells in regulation of conjugation. Here, using an Escherichia coli (SM10λπ)-Pseudomonas aeruginosa (PAO1) conjugation model, we demonstrated that deficiency of lasI/rhlI, genes associated with generation of the quorum sensing signals N-acyl homoserine lactones (AHLs) in PAO1, or deletion of the AHLs receptor SdiA in the donor SM10λπ both facilitated conjugation. When using another AHLs-non-producing E. coli strain EC600 as recipient cells, deficiency of sdiA in donor SM10λπ hardly affect the conjugation. More importantly, in the presence of exogenous AHLs, the conjugation efficiency between SM10λπ and EC600 was dramatically decreased, while deficiency of sdiA in SM10λπ attenuated AHLs-inhibited conjugation. These data suggest the conjugation suppression function of AHLs-SdiA chemical signaling. Further bioinformatics analysis, β-galactosidase reporter system and electrophoretic mobility shift assays characterized the binding site of SdiA on the promoter region of traI gene. Furthermore, deletion of lasI/rhlI or sdiA promoted traI mRNA expression in SM10λπ and PAO1 co-culture system, which was abrogated by AHLs. Collectively, our results provide new insight into an important contribution of quorum sensing system AHLs-SdiA to the networks that regulate conjugation.
Introduction
The acquisition of antibiotic resistance by pathogenic microorganisms is a threat to public health worldwide. Horizontal gene transfer, especially conjugative transfer of plasmids that carry resistance genes, is the primary cause of bacterial antibiotic resistance and—on the larger scale—bacterial evolution (Zatyka and Thomas, 1998; Arthur et al., 2011). The self-transmissible plasmids, such as the well-studied fertility F-plasmids and IncP plasmid RP4 (also known as RK2), generally present a mobilization (MOB) region which includes the origin of transfer (oriT) and the relaxase gene. The relaxase, identified as being TraI in RP4, initiates conjugation by cleaving the oriT in a site- and strand-specific manner (Carballeira et al., 2014). Other plasmids, termed mobilizable, are incapable of initiating conjugation, but can transfer by using the conjugative apparatus of another plasmid (Zatyka and Thomas, 1998). Mobilizable plasmids are more frequently found in natural environment; therefore, replication and mobilization can be considered as important mechanisms that influence plasmid promiscuity (Fernández-López et al., 2014).
Many Gram-negative bacteria utilize N-acyl homoserine lactones (AHLs) as signal molecules to enable individual bacteria to coordinate their behavior in populations; such quorum sensing (QS) enables bacteria to not only sense members of their own species but other species as well (Smith et al., 2011). The essential constituents of QS include a signal producer, or synthase, and a cognate transcriptional regulator that responds to the accumulated signal molecules (Bassler and Losick, 2006). The opportunistic animal and plant pathogen Pseudomonas aeruginosa possesses one of the best-studied models of QS, and two different AHL systems, las and rhl, have been identified (Wagner et al., 2003). In the las QS system, the lasI gene product directs formation of the diffusible extracellular signal N-(3-oxododecanoyl)-L-HSL (3-oxo-C12-HSL), which interacts with LasR to activate a number of virulence genes including the LasA and LasB elastases, exotoxinA, and alkaline protease (Toder et al., 1991; Gambello et al., 1993; Jones et al., 1993; Passador et al., 1993). In the rhl system, the rhlI gene product catalyzes the synthesis of N-butanoyl-L-HSL (C4-HSL). This diffusible signaling molecule, together with RhlR, activates directly some virulence genes like those encoding rhamnolipids and pyocyanin, and represses those genes responsible for assembly and function of the type III secretion system (Bleves et al., 2005; Jimenez et al., 2012). Besides the fact that the las and rhl systems are hierarchically connected, both rhlR and rhlI are positively regulated by the las system (Wagner et al., 2003). The roles of QS in diverse biological processes, such as virulence, biofilm formation and metabolism in P. aeruginosa have attracted research attention (Pearson et al., 1994; Hassett et al., 1999; Whiteley et al., 1999; García-Contreras, 2016). However, as the cell-to-cell communication system, the influence of QS on inter-species conjugation remains largely unknown.
Some organisms, such as Escherichia, Klebsiella, Salmonella and Shigella lack AHL synthase and therefore do not produce AHLs; however, they possess a LuxR homolog known as SdiA that can bind AHLs produced by other microorganisms and affect gene expression(Smith and Ahmer, 2003; Yao et al., 2006; Sabag-Daigle et al., 2015). Case et al. described the phenomenon of non-AHLs-producing microorganisms binding and utilizing AHLs produced by other organisms as eavesdropping (Case et al., 2008). Although SdiA can bind to DNA and regulate transcription in the absence of AHLs, the structural studies of SdiA suggest a double mode of action for AHLs on SdiA activity, by increasing both protein stability and DNA-binding affinity (Nguyen et al., 2015; Ishihama et al., 2016). Besides, a neighbor-joining tree analysis revealed that SdiA of E. coli did not cluster with the LuxR homologs found in other enterobacterial species, but was closely related to the RhlR of P. aeruginosa (Gray and Garey, 2001).
Herein, we clarified the effect of QS on conjugation and investigated the underlying mechanisms by employing a mobilizable plasmid and E. coli-P. aeruginosa conjugation model. We found that QS signal molecules produced by P. aeruginosa inhibited interspecies conjugation by activating E. coli SdiA, resulting in decreased mRNA expression of traI in E. coli. Blockade of AHL-SdiA signaling using strains deficient in lasI, rhlI or sdiA significantly enhanced conjugative transfer. These findings provide new insight into the regulatory networks of conjugation, and offer novel potential targets for antibiotic resistance.
Materials and methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Lysogeny Broth (LB) medium or on LB plates containing 1.5% agar unless otherwise indicated. If required, antibiotics were added at the following final concentrations: ampicillin (Amp, 100 μg/mL), gentamycin (Gm, 30 μg/mL), chloramphenicol (Cm, 20 μg/mL), kanamycin (Kan, 50 μg/mL) and rifampicin (Rif, 50 μg/mL).
Table 1
| Strains/plasmids | Genotype or characteristics | Source |
|---|---|---|
| STRAINS | ||
| E. coli SM10λπ | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpir | Simon et al., 1983 |
| E. coli SM10λπΔsdiA | Mutants of E. coli SM10λπ deficient in sdiA gene | This work |
| E. coli EC600 | LacZ-, NalRr, RifR | Our lab |
| E. coli BW25113 | Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), lambda-, rph-1, Δ(rhaD-rhaB)568, hsdR514 | Our lab |
| P. aeruginosa PAO1 | Wild-type | Stover et al., 2000 |
| PAO1ΔlasI | Mutants of PAO1 deficient in lasI gene | Our lab Zeng et al., 2016 |
| PAO1ΔrhlI | Mutants of PAO1 deficient in rhlI gene | Our lab Zeng et al., 2016 |
| PLASMIDS | ||
| pKD3 | oriR6K, FRT::cat::FRT template plasmid CmR, AmpR | Datsenko and Wanner, 2000 |
| pKD46 | oriR101 repA101ts P-araB-gam-bet-exo AmpR | Datsenko and Wanner, 2000 |
| pCP20 | pSC101 temperature-sensitive repliconts, Flp(λ Rp), cI857, CmR, AmpR | Datsenko and Wanner, 2000 |
| pQF50 | Promoterless lacZ reporter plasmid, AmpR | Farinha and Kropinski, 1990 |
| pQF50-traI | pQF50 derivative, containing traI promoter region, AmpR | This work |
| pUCP24T | 370 bp oriT fragment from pCVD442 cloned into pUCP24, ori1600, GmR | Philippe et al., 2004 |
Bacterial strains and plasmids.
RifR, KmR, CmR, GmR, and AmpR stand for rifampicin, kanamycin, chloramphenicol, gentamycin and ampicillin resistance, respectively.
Growth curves
The indicated bacterial strains were cultured in LB overnight (8~10 h) at 37°C, then diluted to 0.5 MCF (McFarland standard) and 3 mL cultures were grown at 37°C with shaking at 200 rpm. The samples were collected at the indicated time points and the OD600 values were determined.
Plasmid construction
The plasmid pUCP24T was constructed by inserting the oriT fragment into pUCP24 (West et al., 1994), which contains a gene cassette (aacC1) conferring gentamycin resistance in recipient cells. As a result, pUCP24T is not able to transfer on its own, but can transfer by using the conjugative apparatus of E. coli SM10λπ. Details of construction of the plasmids used to delete sdiA gene or express SdiA are described in the Supporting Materials and Methods.
Construction of PAO1 lasI or rhlI and E. coli SM10λπ sdiA deficient mutants
The phage λ Red recombination system was employed for sdiA deletion in E. coli SM10λπ, while the sacB-based suicide vector system was adapted for knockout of lasI or rhlI in PAO1 (Zeng et al., 2016); further details are provided in the Supporting Materials and Methods.
Conjugation experiments
For the conjugation assays, the same amount (0.5 × 107 CFU/mL, counted using the Sysmex UF-1000i™ Automated Urine Particle Analyzer; Tokyo, Japan) of mid-logarithmic phase donor (E. coli SM10λπ harboring plasmid pUCP24T) and recipient cells (PAO1 or EC600) were mixed in 200 μL LB with or without the indicated HSLs in 96-well plates. After 6 h mating at 37°C, the cultures were vigorously mixed and 30 μL aliquots of each conjugation mixture were spread on LB agar containing 30 μg/mL Gm plus 100 μg/mL Amp for SM10λπ-PAO1 or 30 μg/mL Gm plus 50 μg/mL Rif for SM10λπ-EC600 transconjugants. The numbers of transconjugant colonies were counted after overnight incubation at 37°C.
Quantification of HSLs by HPLc-MS/MS
Supernatants of PAO1, PAO1ΔlasI, and PAO1ΔrhlI cultures were collected for HPLC-MS/MS detection of HSLs; full details are provided in the Supporting Materials and Methods.
β-galactosidase assays
β-Galactosidase activities were performed on cells in the mid-log phase of growth according to the modified Miller's method (Giacomini et al., 1992). All tests were performed in triplicate.
Electrophoretic mobility shift assays (EMSA)
His-SdiA fusion protein was expressed in E. coli BL21 (DE3) and purified via Ni-chelating affinity chromatography. Gel shift assays were carried out using the Lightshift Chemiluminescent EMSA kit according to the manufacturer's instructions (Thermo Scientific, Waltham, MA, USA), details are provided in the Supporting Materials and Methods.
Real-time PCR
Total RNA was extracted using total RNA isolation reagent (Promega, Madison, WI, USA). Reverse transcription (1 μg of total RNA) was performed with the PrimeScript RT reagent Kit (Takara, Dalian, Liaoning, China). The cDNA was subjected to qPCR on a ViiA™ 7 Dx system (Applied Biosystems, Foster, CA, USA) using SYBR Green qPCR Master Mixes (ThermoFisher Scientific). The expression levels of the target genes were normalized to the expression of the internal control gene (rpoD), using the 2−ΔΔCt method. The sequences of the primers are listed in Table S1.
Statistical analysis
Data are expressed as the mean ± standard error of the mean (SEM) of at least three independent experiments. The differences between groups were analyzed using the Student's t-test when two groups were compared or one-way ANOVA when more than two groups were compared. All analyses were performed using GraphPad Prism, version 5 (GraphPad Software, Inc., San Diego, CA, USA). All statistical tests were two-sided; P < 0.05 was considered statistically significant.
Results
Deficiency of lasI or rhlI in P. aeruginosa promotes SM10λπ-PAO1 conjugation
To elucidate the biological significance of the QS system in P. aeruginosa conjugation, we first constructed lasI or rhlI single gene-deficient mutants, named PAO1ΔlasI and PAO1ΔrhlI, respectively. In P. aeruginosa, lasI catalyzes the formation of 3-oxo-C12-HSL, which positively regulates the expression of RhlI. RhlI directs the synthesis of C4-HSL, which subsequently regulates pyocyanin production (O'Loughlin et al., 2013). In this study, despite the existence of rhlI in the genome of PAO1ΔlasI, both 3-oxo-C12-HSL and C4-HSL were barely detectable in the conditioned medium of this mutant strain using HPLC-MS/MS analysis. For PAO1ΔrhlI, the deficiency of rhlI in the genome led to an absence of C4-HSL in the conditioned medium of this mutant strain, whereas lasI and its product 3-oxo-C12-HSL were present at similar levels as the WT strain (Figure 1A and Figure S1). Furthermore, as a result of mutation of the QS system, both PAO1ΔlasI and PAO1ΔrhlI lost the ability to express pyocyanin, which could be rescued by exogenous addition of 3-oxo-C12-HSL or C4-HSL (Figure 1B). Taken together, these results confirmed the successful creation of PAO1 strains deficient in lasI or rhlI.
Figure 1
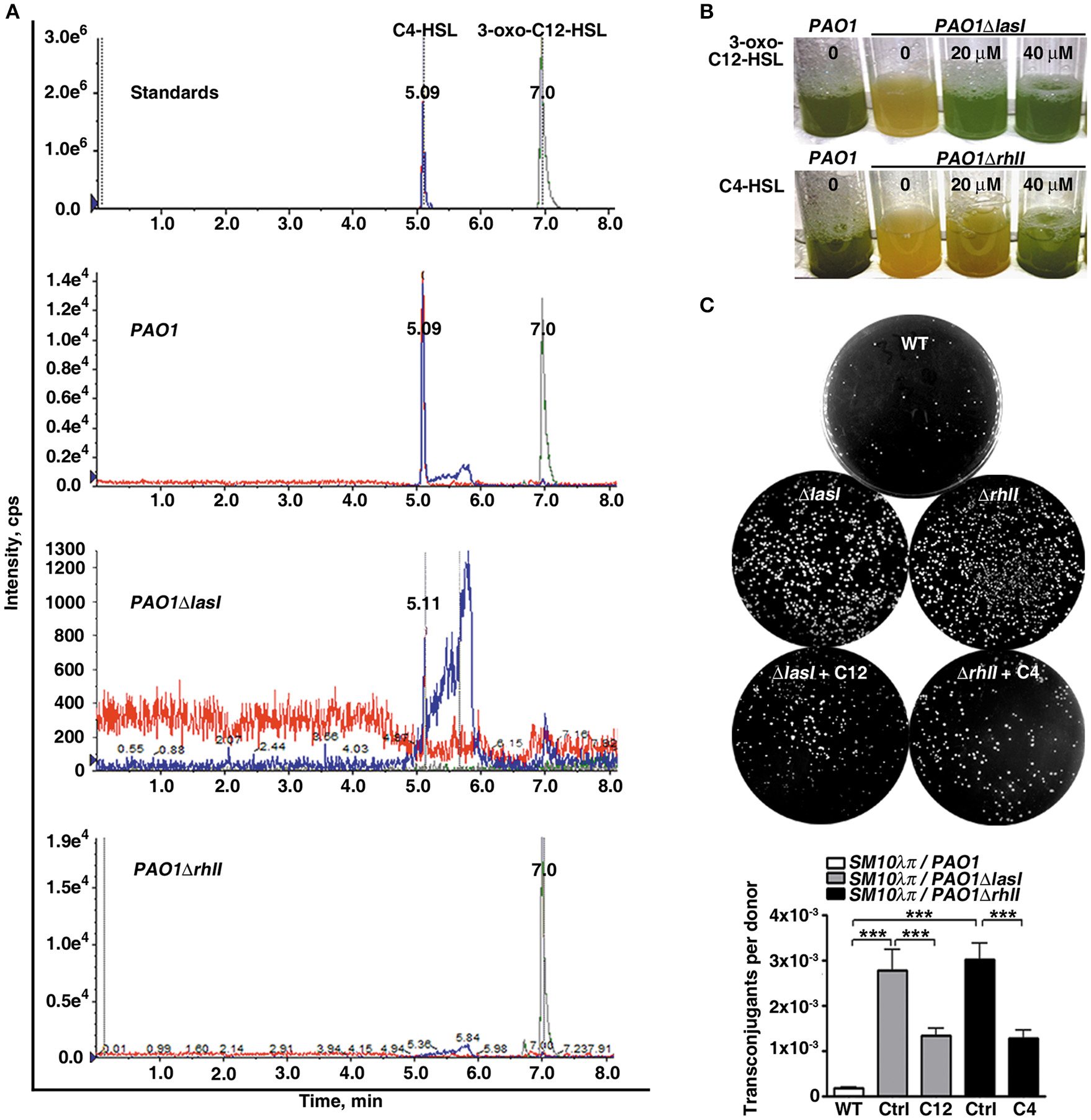
The quorum sensing system of P. aeruginosa inhibits conjugation between P. aeruginosa and E. coli. (A) Deficiency of the AHLs synthase genes rhlI or lasI in P. aeruginosa (PAO1) resulted in the absence of C4-HSL or both 3-oxo-C12-HSL and C4-HSL, respectively. The 3-oxo-C12-HSL and C4-HSL in the cell-free supernatants were extracted with ethyl acetate and redissolved in methanol, followed by HPLC-MS/MS analysis. (B) Deficiency of lasI or rhlI in P. aeruginosa abolished production of the downstream toxin of rhlI system pyocyanin. PAO1, PAO1ΔlasI and PAO1ΔrhlI were cultured in the presence or absence of 3-oxo-C12-HSL or C4-HSL as indicated for 30 h. (C) Deficiency of lasI or rhlI in P. aeruginosa significantly promoted SM10λπ-PAO1 conjugation; this effect could be abrogated by supplementation with exogenous 3-oxo-C12-HSL or C4-HSL. SM10λπ and PAO1 (107 CFU/mL each) were mated at 37°C for 6 h in the presence or absence of 40 μM of C4-HSL or 3-oxo-C12-HSL. Ctrl, control; C12, 3-oxo-C12-HSL; C4, C4-HSL. Values are mean ± SEM of at least three independent experiments; ***P < 0.001.
We subsequently examined the growth and conjugation ability of PAO1ΔlasI and PAO1ΔrhlI. Compared to the WT strain, deficiency of lasI or rhlI hardly affected the growth of PAO1 (Figure S2), but significantly promoted SM10λπ-PAO1 conjugation (Figure 1C). Furthermore, exogenous 3-oxo-C12-HSL or C4-HSL attenuated the interspecies conjugation ability of PAO1ΔlasI and PAO1ΔrhlI (Figure 1C). What's more, we counted the amount of donor SM10λπ after co-culture with PAO1, PAO1ΔlasI or PAO1ΔrhlI, and found that there is no difference among the three groups (Figure S3), indicted that the observed effect of quorum sensing on conjugation efficiency was not due to the growth suppressive effect on SM10λπ. These data suggested that the QS system may negatively regulate SM10λπ-PAO1 conjugation.
The quorum sensing system of P. aeruginosa inhibits conjugation by activating SdiA of E. coli
It is well recognized that AHLsq regulate gene transcription via binding to their receptor proteins (LuxR-like proteins). In this conjugation model, in contrast to the recipient cells PAO1, the donor E. coli SM10λπ cells lack AHL synthase and therefore do not produce AHLs; however, these cells produce a LuxR homolog known as SdiA that can bind AHLs produced by other bacterial species to regulate gene transcription. Given that the conjugative apparatus exist in donor cells, we speculated that P. aeruginosa-released AHLs may act on SdiA of E. coli. To assess whether SdiA of E. coli is involved in the ability of P. aeruginosa's AHLs to inhibit E. coli-P. aeruginosa conjugation, we constructed the sdiA deficient mutant SM10λπΔsdiA. As expected, deficiency of sdiA in SM10λπ significantly enhanced E. coli-P. aeruginosa conjugation, whereas overexpression of SdiA reversed the phenotype (Figure 2A). However, when using a AHLs-non-producing E. coli strain EC600 as the recipient cell, SM10λπΔsdiA did not increase conjugation ability compared to the WT strain (Figure 2B). More importantly, the conjugation efficiency of SM10λπ and EC600 significantly decreased in the presence of exogenous 3-oxo-C12-HSL and C4-HSL, while sdiA deletion in SM10λπ abrogated the effects of AHLs on conjugation (Figure 2B), suggesting the inhibitory effect of SdiA on E. coli-P. aeruginosa or SM10λπ-EC600 conjugation is dependent on the presence of AHLs. In addition, growth curves demonstrated that deficiency of sdiA in E. coli had no influence on cell proliferation (Figures S4, S5), confirming that the regulatory function of SdiA in conjugation in this model was not due to an altered growth rate.
Figure 2
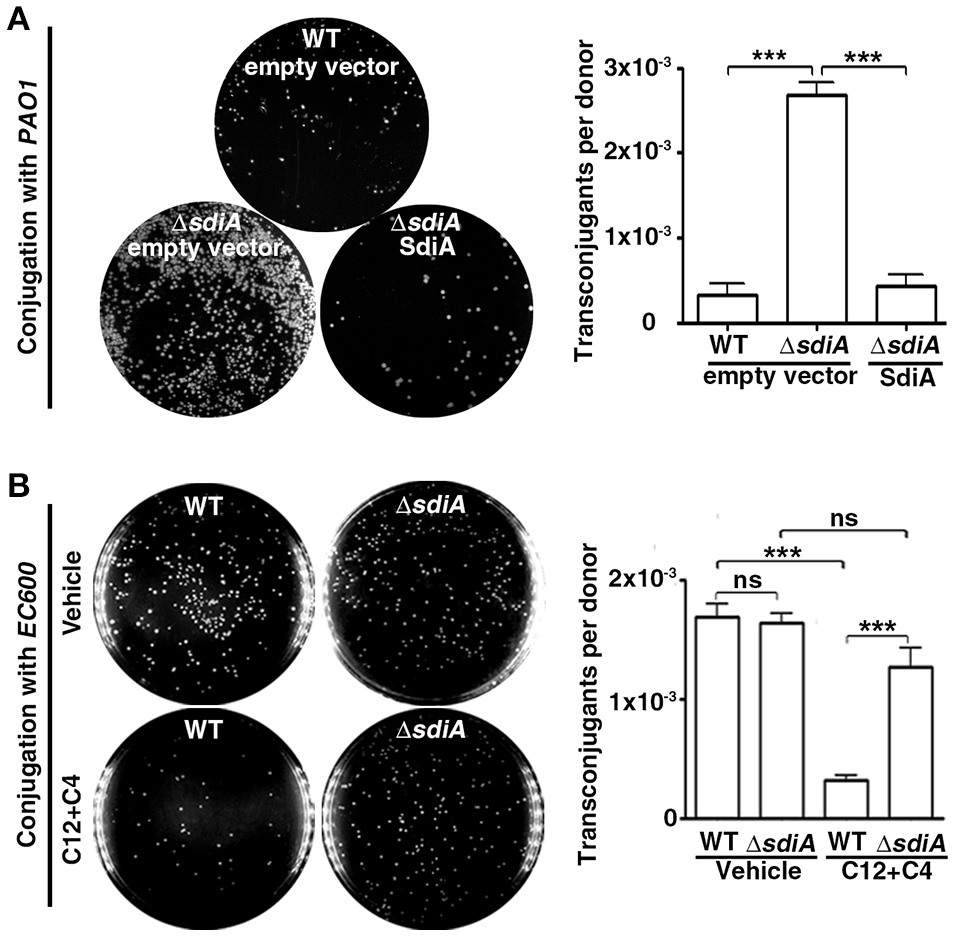
AHL inhibits SM10λπ-PAO1 conjugation via a mechanism dependent on SdiA of E. coli. (A) Deficiency of sdiA in E. coli significantly promoted SM10λπ-PAO1 conjugation. E. coli SM10λπ and P. aeruginosa PAO1 (107 CFU/mL each) were mated at 37°C for 6 h. (B) Deficiency of sdiA in SM10λπ did not significantly affect SM10λπ-EC600 conjugation, but could rescue 3-oxo-C12-HSL and C4-HSL (C4/12)-inhibited conjugation. SM10λπ was cultured in the presence of DMSO or 40 μM C4-HSL and 3-oxo-C12-HSL for 6 h, followed by conjugation with EC600. Values are mean ± SEM of at least three independent experiments; ns, not significant, ***P < 0.001.
Collectively, these data imply that AHLs produced by PAO1 may repress SM10λπ-PAO1 conjugation through binding to SdiA of E. coli.
The interaction between P. aeruginosa HSL and E. coli SdiA inhibits the expression of traI in E. coli
Mechanisms behind transcription regulation function of SdiA is being disclosed, it seems that genes with specific DNA sequences (SdiA-box) 5′-AAAAG(N8)GAAAA-3′ in the promoter region may be the potential targets of SdiA (Yamamoto et al., 2001). In view of the presence of SdiA-box in the promoter of many SdiA-regulated genes in our bioinformatics analysis (Table S2), we computationally mapped the DNA sequence in the RP4 plasmid to search for conjugation-related genes potentially regulated by SdiA. An SdiA-box sequence (5′-AAGAGcgattgagGAAAA-3′) was identified −317 bp upstream of the traI start codon (Figure S6). Subsequently, EMSA assays confirmed the interaction between SdiA and the predicted SdiA-box of the traI promoter in vitro (Figures 3A,B). We therefore further evaluated the role of SdiA in the regulation of traI transcription. DNA fragments of traI promoter carrying the predicted SdiA-box was cloned upstream of the β -galactosidase gene in the pQF50-promoter reporter. When transformed into BW25113 (another E. coli strain without endogenous β-galactosidase compared to SM10λπ), the β-galactosidase activity of pQF50-traI was greatly elevated, compared to that of the control, while addition of 3-oxo-C12-HSL and C4-HSL impaired this activity, which was severely attenuated when the sdiA was deleted (Figure 3B). Intriguing, when AHLs was absent, deletion of sdiA hardly affected β-galactosidase activity of pQF50-traI (Figure 3B), this is consistent with the phenotype shown in Figure 2. Compared with the WT strain, SM10λπΔsdiA showed higher mRNA expression of traI when cultured with PAO1 (Figure 3C). On the other hand, in the SM10λπ-PAO1 co-culture system, deficiency of lasI or rhlI in PAO1 also led to enhanced expression of traI (Figure 3D), while supplementation with exogenous 3-oxo-C12-HSL and C4-HSL significantly repressed traI expression (Figure 3E). These results suggest that repressing traI expression in the donor cells may be a critical mechanism behind the inhibitory effect of the AHLs on conjugation.
Figure 3
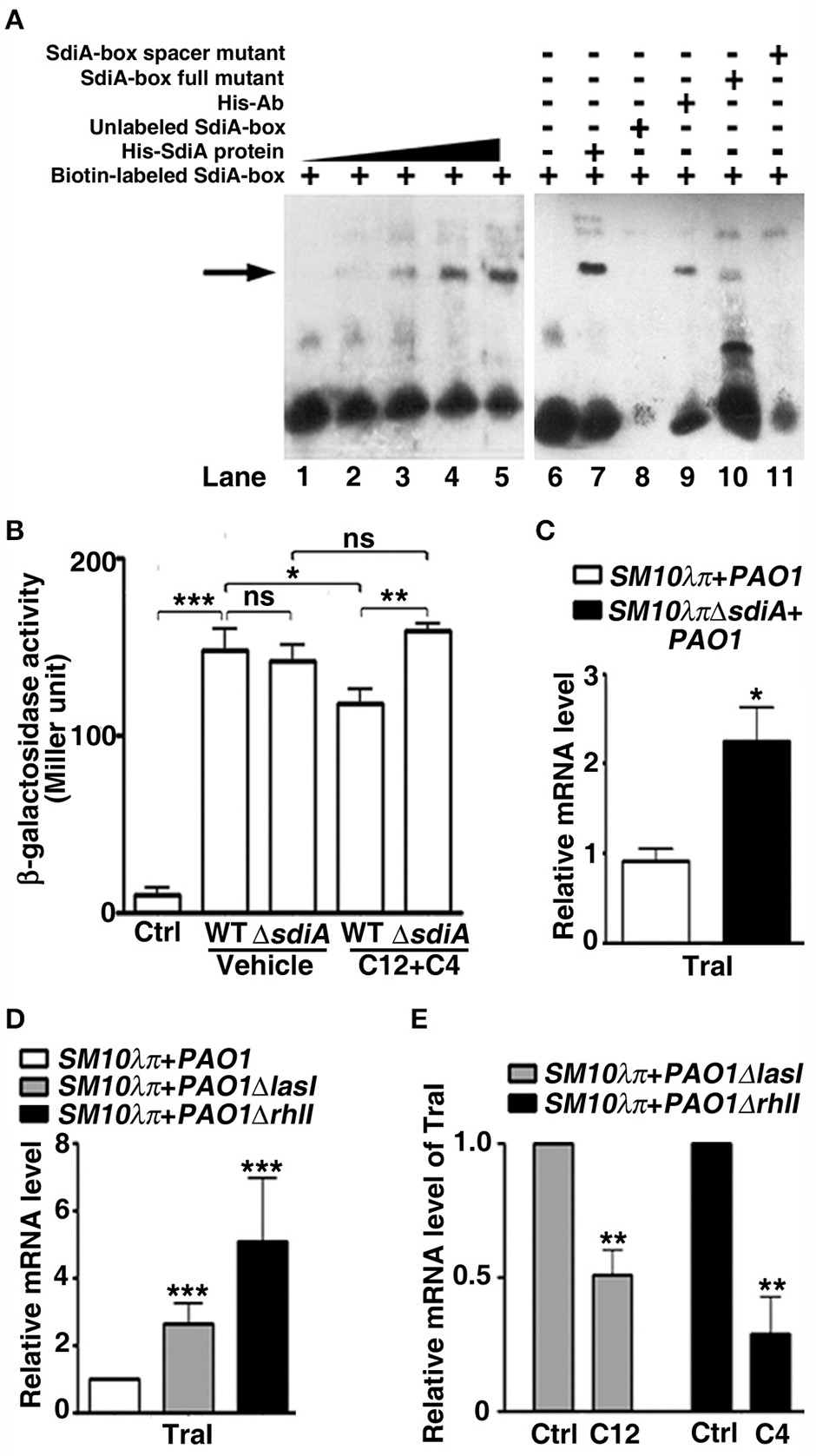
The AHL-SdiA interaction represses the expression of TraI in E. coli. (A) EMSA verified the interaction of SdiA protein with the putative SdiA-box of the traI promoter. Biotin-labeled SdiA-box (50 fmol) was added to each reaction with 0, 2, 4, 8, or 16 pmol E. coli His-SdiA fusion protein (lane 1 to lane 5, respectively). The specificity analysis (lane 6 to lane 11) was performed in the presence (−) or absence (+) of His-SdiA fusion protein (8 pmol) and biotin-labeled SdiA-box (50 fmol, lanes 7), 200-fold unlabeled SdiA-box (lanes 8), anti-His antibody (lanes 9), or 200-fold SdiA-box full or spacer mutant competitors (lane 10, 11). (B) Activity of β-galactosidase reporters containing the predicted SdiA-box in genomic regions upstream of traI under various conditions. E.coli BW25113 or E.coli BW25113ΔsdiA were cultured with SM10λπ in the presence of DMSO (Ctrl) or 40 μM C4-HSL and 3-oxo-C12-HSL for 6 h, followed by β-galactosidase activity analysis. (C) Deficiency of sdiA promoted traI expression in E. coli. (D) Deficiency of lasI/rhlI promoted traI expression in E. coli. For (C,D), 107 CFU/mL (each) of the indicated E. coli donor and recipient P. aeruginosa cells were mated at 37°C for 6 h, followed by real-time PCR analysis. The rpoD gene of E. coli was used as an internal control. (E) Exogenous 3-oxo-C12-HSL or C4-HSL inhibited traI expression in E. coli. PAO1ΔlasI or PAO1ΔrhlI were cultured with SM10λπ in the presence of DMSO (Ctrl) or 40 μM C4-HSL and 3-oxo-C12-HSL for 6 h, followed by real-time PCR analysis. The rpoD gene of E. coli was used as an internal control. Ctrl, control; C12, 3-oxo-C12-HSL; C4, C4-HSL. Values are mean ± SEM of at least three independent experiments; *P < 0.05; **P < 0.01; ***P < 0.001.
In summary, we disclosed the cooperative effect of AHLs produced by recipient P. aeruginosa cells and SdiA of donor E. coli cells in the conjugation regulation. These findings indicate that QS may inhibit conjugation and prevent the excessive dissemination of plasmid.
Discussion
Most recent publications in this field have focused on the regulatory function of QS in virulence and biofilm formation. Here, using E. coli (SM10λπ) as donor cells and AHLs-producing P. aeruginosa (PAO1) or non-AHLs producing E. coli (EC600) as recipient cells, we identified a conjugation-inhibitory effect for QS based on the following evidence. First, for SM10λπ and PAO1 co-culture system in which AHLs is normally self-sustained, deficiency of the AHLs-producing genes lasI or rhlI in PAO1 or the solo AHLs receptor SdiA in SM10λπ promoted SM10λπ-PAO1 conjugation, while supplementation with exogenous 3-oxo-C12-HSL or C4-HSL abrogated the enhanced conjugation ability of PAO1ΔlasI and PAO1ΔrhlI. On the other hand, for both non-AHLs producing SM10λπ and EC600 mixed cultures, stimulation with exogenous 3-oxo-C12-HSL and C4-HSL inhibited conjugation, while deletion of sdiA in SM10λπ attenuated this effect. Conventionally, conjugation is considered to be mainly regulated by the self-transmissible plasmids. While our results indicate that QS system of donor and recipient cells may play a role in conjugation regulation.
Conjugation enables the dissemination of virulence genes and antibiotic resistance genes, which leads to the adaption of bacteria to new circumstances (Norman et al., 2009). Therefore, the ability to inhibit conjugation may be a potentially efficacious strategy for avoiding the spread of resistance traits. Here, we demonstrate that AHL-SdiA is capable of suppressing conjugation. Most SdiA-expression bacteria, such as Escherichia, Salmonella and Shigella are enterobacteria, while many biological evidences suggest a lack of HSLs in the normal mammalian intestine (Swearingen et al., 2013), despite the presence of AHLs in bovine rumen (Hughes et al., 2010). Thus, although P. aeruginosa could be detected in stool sample in our clinical microbiology laboratory, future studies are needed to illuminate the role of AHL-SdiA signaling in pathogenic bacteria communities within the gastrointestinal tract.
To date, many SdiA regulon members have been described (Kanamaru et al., 2000; Wei et al., 2001; Dyszel et al., 2010; Sabag-Daigle et al., 2015). Here we report the identification of SdiA-regulated and AHL-responsive gene traI in the plasmid RP4. TraI is reported to function as a relaxase enzyme that creates a nick at the oriT of conjugative plasmids, which is required to initiate conjugation (Furuya and Komano, 2000). We discovered a DNA motif recognized by SdiA in the promoter region of the traI gene in the plasmid RP4, and the interaction between SdiA and the predicted SdiA-box was validated in vitro using an EMSA. However, some SdiA-regulated genes do not have this particular SdiA-box (Dyszel et al., 2010; Swearingen et al., 2013; Abed et al., 2014; Nguyen et al., 2015), there may be other conjugation-related genes repressed by AHL-SdiA. Moreover, the EMSA was performed without addition of AHLs, so it seems that high concentration of SdiA could bind to traI promoter in the absence of AHLs in vitro (Figure 3A), however, the reporter system (Figure 3B) and conjugation experiment (Figure 2B) showed that in the absence of AHLs, deletion of sdiA hardly affected the promoter activity of traI, as well as conjugation frequency in vivo. Thus, we proposed AHLs may increase both SdiA protein stability and traI promoter-binding affinity to repress traI expression.
Despite the advantages of conjugation for bacteria, the introduction of novel genes into the pre-existing, well-tuned genetic background is a source of genetic conflict, and possession of the conjugation-associated machinery also places a burden on the host arising from the energy expended to create and maintain the conjugative apparatus and its associated features (Zatyka and Thomas, 1998; Baltrus, 2013; San Millan et al., 2015). This raises the question of how host bacteria minimize the metabolic cost while obtaining the benefits provided by conjugation. In this study, we found that under normal conditions, when mobilizable plasmid containing a resistance gene was not required by PAO1 (Table S3), conjugation between SM10λπ and PAO1 was inhibited via the LasI/RhlI-HSL-SdiA pathway. These findings reveal that QS system may play a role in protecting host cells against external conjugative plasmids.
Utilizing ecological data from 2801 samples, Freilich et al. explored the ubiquitous competitive and cooperative interactions between the bacteria within natural communities (Freilich et al., 2011). Nonetheless, revealing more detail of the strategies bacteria adopt for survival in mixed cultures remains a major challenge. The E. coli-P. aeruginosa conjugation model has been widely used in studies of bacterial conjugation, and the most prevalent donor strain is E. coli SMλπ in which the RP4 plasmid is chromosomally-integrated. Thus, conjugation-associated genes, such as traI initially only exist in and are expressed by the E. coli (SM10λπ) cells, similarly rhlI and lasI are only expressed by PAO1. This makes it easy to detect the expression of these genes in E. coli (SM10λπ) and PAO1, specifically in mixed-cultures. Using this co-culture system, we found that LasI/RhlI and SdiA jointly repressed traI expression in E. coli and inhibited SM10λπ-PAO1 conjugation, indicating that the QS system may provide a mechanism of cooperative regulation between bacteria.
In conclusion, the findings of this study highlight the regulatory role for the QS system in conjugation, and expand our understanding of the bacterial communication and defense systems of P. aeruginosa.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81572058, 81672081), the Natural Science Foundation of Guangdong Province (Grant No. 2014A030313143) and the Science and Technology Planning Project of Guangdong Province (Grant No. 2016A020215236).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Statements
Author contributions
YL, JZ, XH, BH, and CC designed research; YL, JZ, BW, RC, and NZ performed research; SE and YQL contributed new reagents/analytic tools; YL, JZ, and BW analyzed data; YL, JZ, and CC wrote the paper.
Acknowledgments
We are very grateful to Prof. B.L. Wanner (Department of Biological Sciences, Purdue University, West Lafayette, USA) for generously providing the λ Red recombination system, Prof. E. Peter Greenberg (Department of Microbiology, University of Washington School of Medicine, Seattle, Washington, USA) for providing pQF50 plasmid and Prof. H. P. Schweizer (Department of Microbiology, Immunology and Pathology, College of Veterinary Medicine and Biological Sciences, Colorado State University, Fort Collins, Colorado, USA) for providing the plasmid pUCP24.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00007/full#supplementary-material
- QS
quorum sensing
- AHLs
N-acyl homoserine-lactones
- P. aeruginosa
Pseudomonas aeruginosa
- E. coli
Escherichia coli
- qPCR
quantitative real-time PCR.
Abbreviations
References
1
Abed N. Grépinet O. Canepa S. Hurtado-Escobar G. A. Guichard N. Wiedemann A. et al . (2014). Direct regulation of the pefI-srgC operon encoding the Rck invasin by the quorum-sensing regulator SdiA in Salmonella Typhimurium. Mol. Microbiol.94, 254–271. 10.1111/mmi.12738
2
Arthur D. C. Edwards R. A. Tsutakawa S. Tainer J. A. Frost L. S. Glover J. N. M. (2011). Mapping interactions between the RNA chaperone FinO and its RNA targets. Nucleic Acids Res.39, 4450–4463. 10.1093/nar/gkr025
3
Baltrus D. A. (2013). Exploring the costs of horizontal gene transfer. Trends Ecol. Evol.28, 489–495. 10.1016/j.tree.2013.04.002
4
Bassler B. L. Losick R. (2006). Bacterially speaking. Cell125, 237–246. 10.1016/j.cell.2006.04.001
5
Bleves S. Soscia C. Nogueira-Orlandi P. Lazdunski A. Filloux A. (2005). Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J. Bacteriol.187, 3898–3902. 10.1128/JB.187.11.3898-3902.2005
6
Carballeira J. D. González-Pérez B. Moncalián G. De La Cruz F. (2014). A high security double lock and key mechanism in HUH relaxases controls oriT-processing for plasmid conjugation. Nucleic Acids Res.42, 10632–10643. 10.1093/nar/gku741
7
Case R. J. Labbate M. Kjelleberg S. (2008). AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J.2, 345–349. 10.1038/ismej.2008.13
8
Datsenko K. A. Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A.97, 6640–6645. 10.1073/pnas.120163297
9
Dyszel J. L. Soares J. A. Swearingen M. C. Lindsay A. Smith J. N. Ahmer B. M. M. (2010). E. coli K-12 and EHEC genes regulated by SdiA. PLoS ONE5:e8946. 10.1371/journal.pone.0008946
10
Farinha M. A. Kropinski A. M. (1990). Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol.172, 3496–3499. 10.1128/jb.172.6.3496-3499.1990
11
Fernández-López C. Bravo A. Ruiz-Cruz S. Solano-Collado V. Garsin D. A. Lorenzo-Díaz F. et al . (2014). Mobilizable rolling-circle replicating plasmids from gram-positive bacteria: a low-cost conjugative transfer. Microbiol. Spectr.2, 1–33. 10.1128/microbiolspec.PLAS-0008-2013
12
Freilich S. Zarecki R. Eilam O. Segal E. S. Henry C. S. Kupiec M. et al . (2011). Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun.2, 589–595. 10.1038/ncomms1597
13
Furuya N. Komano T. (2000). Initiation and termination of DNA transfer during conjugation of IncI1 plasmid R64: roles of two sets of inverted repeat sequences within oriT in termination of R64 transfer. J. Bacteriol.182, 3191–3196. 10.1128/JB.182.11.3191-3196.2000
14
Gambello M. J. Kaye S. Iglewski B. H. (1993). LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun.61, 1180–1184.
15
García-Contreras R. (2016). Is quorum sensing interference a viable alternative to treat Pseudomonas aeruginosa infections?Front. Microbiol.7:1454. 10.3389/fmicb.2016.01454
16
Giacomini A. Corich V. Ollero F. J. Squartini A. Nuti M. P. (1992). Experimental conditions may affect reproducibility of the beta-galactosidase assay. FEMS Microbiol. Lett.100, 87–90. 10.1111/j.1574-6968.1992.tb05687.x
17
Gray K. M. Garey J. R. (2001). The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology147, 2379–2387. 10.1099/00221287-147-8-2379
18
Hassett D. J. Ma J. F. Elkins J. G. McDermott T. R. Ochsner U. A. West S. E. et al . (1999). Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol.34, 1082–1093. 10.1046/j.1365-2958.1999.01672.x
19
Hughes D. T. Terekhova D. A. Liou L. Hovde C. J. Sahl J. W. Patankar A. V. et al . (2010). Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U.S.A.107, 9831–9836. 10.1073/pnas.1002551107
20
Ishihama A. Shimada T. Yamazaki Y. (2016). Transcription profile of Escherichia coli: genomic SELEX search for regulatory targets of transcription factors. Nucleic Acids Res.44, 2058–2074. 10.1093/nar/gkw051
21
Jimenez P. N. Koch G. Thompson J. A. Xavier K. B. Cool R. H. Quax W. J. (2012). The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev.76, 46–65. 10.1128/MMBR.05007-11
22
Jones S. Yu B. Bainton N. J. Birdsall M. Bycroft B. W. Chhabra S. R. et al . (1993). The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J.12, 2477–2482.
23
Kanamaru K. Tatsuno I. Tobe T. Sasakawa C. (2000). SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol.38, 805–816. 10.1046/j.1365-2958.2000.02171.x
24
Nguyen Y. Nguyen N. X. Rogers J. L. Liao J. MacMillan J. B. Jiang Y. et al . (2015). Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. MBio6, 2092–2096. 10.1128/mBio.02429-14
25
Norman A. Hansen L. H. Sørensen S. J. (2009). Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B. Biol. Sci.364, 2275–2289. 10.1098/rstb.2009.0037
26
O'Loughlin C. T. Miller L. C. Siryaporn A. Drescher K. Semmelhack M. F. Bassler B. L. (2013). A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. U.S.A.110, 17981–17986. 10.1073/pnas.1316981110
27
Passador L. Cook J. M. Gambello M. J. Rust L. Iglewski B. H. (1993). Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science260, 1127–1130. 10.1126/science.8493556
28
Pearson J. P. Gray K. M. Passador L. Tucker K. D. Eberhard A. Iglewski B. H. et al . (1994). Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U.S.A.91, 197–201. 10.1073/pnas.91.1.197
29
Philippe N. Alcaraz J. P. Coursange E. Geiselmann J. Schneider D. (2004). Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid51, 246–255. 10.1016/j.plasmid.2004.02.003
30
Sabag-Daigle A. Dyszel J. L. Gonzalez J. F. Ali M. M. Ahmer B. M. M. (2015). Identification of sdiA-regulated genes in a mouse commensal strain of Enterobacter cloacae. Front. Cell. Infect. Microbiol.5:47. 10.3389/fcimb.2015.00047
31
San Millan A. Toll-Riera M. Qi Q. MacLean R. C. (2015). Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun.6, 6845–6852. 10.1038/ncomms7845
32
Simon R. Priefer U. Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol.9, 784–791. 10.1038/nbt1183-784
33
Smith J. L. Fratamico P. M. Yan X. (2011). Eavesdropping by bacteria: the role of SdiA in Escherichia coli and Salmonella enterica serovar typhimurium quorum sensing. Foodborne Pathog. Dis.8, 169–178. 10.1089/fpd.2010.0651
34
Smith J. N. Ahmer B. M. M. (2003). Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol.185, 1357–1366. 10.1128/JB.185.4.1357-1366.2003
35
Stover C. K. Pham X. Q. Erwin A. L. Mizoguchi S. D. Warrener P. Hickey M. J. et al . (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature406, 959–964. 10.1038/35023079
36
Swearingen M. C. Sabag-Daigle A. Ahmer B. M. M. (2013). Are there acyl-homoserine lactones within mammalian intestines?J. Bacteriol.195, 173–179. 10.1128/JB.01341-12
37
Toder D. S. Gambello M. J. Iglewski B. H. (1991). Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol. Microbiol.5, 2003–2010. 10.1111/j.1365-2958.1991.tb00822.x
38
Wagner V. E. Bushnell D. Passador L. Brooks A. I. Iglewski B. H. (2003). Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol.185, 2080–2095. 10.1128/JB.185.7.2080-2095.2003
39
Wei Y. Lee J. M. Smulski D. R. LaRossa R. A. (2001). Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J. Bacteriol.183, 2265–2272. 10.1128/JB.183.7.2265-2272.2001
40
West S. E. Schweizer H. P. Dall C. Sample A. K. Runyen-Janecky L. J. (1994). Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene148, 81–86. 10.1016/0378-1119(94)90237-2
41
Whiteley M. Lee K. M. Greenberg E. P. (1999). Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A.96, 13904–13909. 10.1073/pnas.96.24.13904
42
Yamamoto K. Yata K. Fujita N. Ishihama A. (2001). Novel mode of transcription regulation by SdiA, an Escherichia coli homologue of the quorum-sensing regulator. Mol. Microbiol.41, 1187–1198. 10.1046/j.1365-2958.2001.02585.x
43
Yao Y. Martinez-Yamout M. A. Dickerson T. J. Brogan A. P. Wright P. E. Dyson H. J. (2006). Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J. Mol. Biol.355, 262–273. 10.1016/j.jmb.2005.10.041
44
Zatyka M. Thomas C. M. (1998). Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol. Rev.21, 291–319. 10.1111/j.1574-6976.1998.tb00355.x
45
Zeng J. Zhang N. Huang B. Cai R. Wu B. E S. et al . (2016). Mechanism of azithromycin inhibition of HSL synthesis in Pseudomonas aeruginosa. Sci. Rep.6:24299. 10.1038/srep24299
Summary
Keywords
conjugation, N-acyl homoserine lactones, P. aeruginosa , SdiA, antibiotic resistance
Citation
Lu Y, Zeng J, Wu B, E S, Wang L, Cai R, Zhang N, Li Y, Huang X, Huang B and Chen C (2017) Quorum Sensing N-acyl Homoserine Lactones-SdiA Suppresses Escherichia coli-Pseudomonas aeruginosa Conjugation through Inhibiting traI Expression. Front. Cell. Infect. Microbiol. 7:7. doi: 10.3389/fcimb.2017.00007
Received
19 October 2016
Accepted
05 January 2017
Published
20 January 2017
Volume
7 - 2017
Edited by
Ghassan M. Matar, American University of Beirut, Lebanon
Reviewed by
Eric Déziel, Institut National de la Recherche Scientifique, Canada; Shouguang Jin, University of Florida, USA
Updates
Copyright
© 2017 Lu, Zeng, Wu, E, Wang, Cai, Zhang, Li, Huang, Huang and Chen.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Huang huangb3@mail.sysu.edu.cn
†These authors have contributed equally to this work.
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.