- Produce Safety and Microbiology Research Unit, Western Regional Research Center, Agricultural Research Service, United States Department of Agriculture, Albany, CA, United States
Mycobacterium avium subsp. paratuberculosis (MAP), the causative agent of Johne's disease in cattle, is responsible for significant economic losses to the US dairy industry. The pathogen has also been associated with chronic human diseases like Crohn's disease, type 1 diabetes and multiple sclerosis. Determining causation requires rapid characterization and source tracking the pathogen. Here, we used matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry to characterize and differentiate strains of MAP from 14 other species of Mycobacterium from bovine, human, and environmental sources. Lysates from cells disrupted by bead beating in TFA-acetonitrile solution were analyzed by MALDI-TOF. MAP strains were differentiated by mass spectral profiles that are distinct from each other and from other Mycobacterium species. Cluster analysis of spectral profiles indicates two distinct clusters, one dominated by the members of avium complex and a second group dominated by members of fortuitum and parafortuitum complexes. We believe that MALDI-TOF methods can be used to differentiate and source-track MAP strains.
Introduction
Mycobacterium avium subsp. paratuberculosis (MAP), the causative agent of Johne's disease in cattle, is responsible for an annual loss of 200–250 million dollars to the US dairy industry (Ott et al., 1999). Johne's disease is a debilitating chronic infectious enteritis of ruminants whose spread can only be controlled by culling. There is no cure. Herd level prevalence was estimated to be as high as 91% (Lombard et al., 2013) and spread of the pathogen is through the fecal-oral route and also through milk. Early pathogen detection coupled with culling and introduction of MAP-free animals into the herd is the only option for pathogen-free dairies.
MAP has been associated with different autoimmune diseases such as Crohn's disease (Sechi et al., 2005; Chiodini et al., 2012), type 1 diabetes (Sechi et al., 2008; Cossu et al., 2011; Masala et al., 2011), and multiple sclerosis (Cossu et al., 2013; Frau et al., 2013) but casual links were not established. Clinical manifestations of Crohn's disease partly resemble the clinical signs of Johne's disease in ruminants (Overduin et al., 2004). Milk contaminated with MAP has been considered to be a potential source of exposure to humans (Grant, 2005). Both live and dead cells of MAP were detected in pasteurized milk (Millar et al., 1996; Grant et al., 2002). Determining causation requires sensitive methods to detect and isolate the pathogen and methods for source tracking.
Isolation of the pathogen using traditional culture-based methods can take up to 20 weeks (Whittington, 2009). Thus, real-time quantitative PCR methods based on the detection of insertion sequence targets IS900 and ISMav2 were developed for rapid and sensitive detection of MAP (Ravva and Stanker, 2005; Sting et al., 2014). PCR methods can be coupled with magnetic-bead separations for cleanup and concentration of MAP DNA from complex fecal samples (Leite et al., 2013; Sting et al., 2014). PCR methods can rapidly detect the presence of MAP, but they cannot aid in typing the strains necessary for source tracking the pathogen.
Molecular typing methods were developed that discriminate strains of MAP. IS900 restriction fragment length polymorphism (RFLP) typing and multilocus variable-number tandem-repeat analysis (MLVA) methods were used to type MAP strains from humans and cattle (Overduin et al., 2004). In one study, the human isolates were found to be genetically indistinguishable from the cattle strains and the authors suggested that humans can be infected with strains from cattle. However, this method lacked resolution in discriminating strains as 82% of strains tested were of one MLVA type. Other studies that used tandem application of mycobacterial interspersed repetitive unit (MIRU) and multilocus short sequence repeat (MLSSR) genotyping methods were able to distinguish sheep isolates from cattle isolates (Amonsin et al., 2004) and generated 22 distinct genotypes from 38 MAP strains (Douarre et al., 2011).
Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) detection of biomolecules in cell-free extracts or culture supernatants have been used to characterize and discriminate strains of Escherichia coli O157:H7 and Campylobacter (Fagerquist et al., 2005, 2010; Mandrell et al., 2005). MALDI-TOF MS has been recently used to characterize mycobacteria (Park et al., 2016; Samli and Ilki, 2016; Zingue et al., 2016). However, much of the work was focused on differentiating pathogenic strains of Mycobacterium tuberculosis from other species of mycobacteria including some members of the M. avium complex. Although, MAP was included in a couple of studies (Pignone et al., 2006; El Khechine et al., 2011), mass spectral data was either not published or spectral data was limited to a mass range of <5,000 Da (Pignone et al., 2006). These studies also lacked uniformity in sample preparation. Samples for mass spectrometry included whole cells in matrix solution (Lin et al., 2013), cells directly applied on MALDI plates (Pignone et al., 2006), non-sonicated cell-free supernatants (Hettick et al., 2006), mechanical breaking of heat-inactivated cells followed by protein extractions with formic acid and acetonitrile (El Khechine et al., 2011) or protein extraction from culture supernatants (Marekovic et al., 2016). Cell lysates were used routinely for MALDI-TOF characterization of foodborne pathogens in our laboratory (Mandrell et al., 2005; Fagerquist et al., 2010; Penny et al., 2016).
We predicted that MALDI-TOF characterization of biomolecules in cell lysates would provide valuable data for characterization and differentiation of MAP from other mycobacteria and for typing MAP strains. Thus, MAP strains and other mycobacteria from different host species and environments were characterized by MALDI-TOF MS. Spectral data was analyzed using Bionumerics software to obtain similarities in spectral patterns and unique finger prints that differentiate Mycobacterium species and strains of MAP.
Materials and Methods
Mycobacterial Cultures
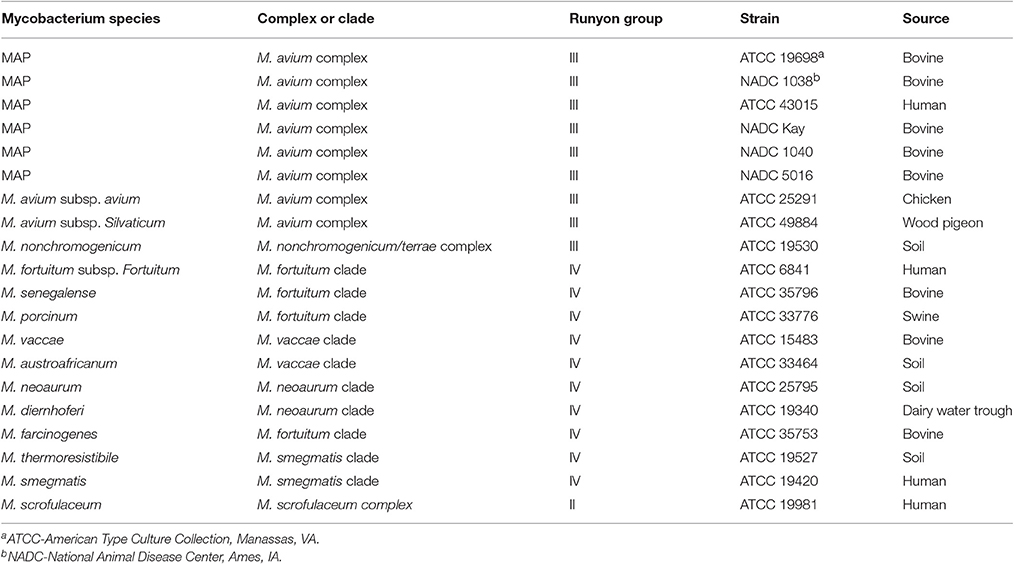
Six different strains of MAP and 14 different Mycobacterium species from different host species and environments (Table 1) were evaluated. Mycobacterial cells were grown in 50 mL of 7H9 broth supplemented with 10% (vol/vol) Middlebrook OADC growth supplement (Becton Dickinson, Franklin Lakes, NJ), 0.5% Tween 80 and 0.0002% (wt/vol) mycobactin J (Allied Monitor Inc., Fayette, MO) (Chang et al., 2002). The growth medium was supplemented with 50 μg/mL of amphotericin B and 100 μg/mL each of nalidixic acid and vancomycin to reduce contaminant growth during the 2–4 months of incubation required for MAP (Collins et al., 1990). The cultures were grown at 37°C on a gyratory shaker at 200 rpm. Mycobacteria other than MAP strains were harvested within 1–2 weeks of incubation.
Cell Lysates for MALDI-TOF MS
Protein extraction solution was prepared by combining 1:2 acetonitrile-water with 0.1% sequencing-grade trifluoroacetic acid (TFA) (Sigma-Aldrich, St. Louis, MO).
Cultured cells (0.1 mL) were centrifuged at 18,000 × g for 5 min. Pelleted cells were bead-beat (Mini-Beadbeater-16; BioSpec Products Inc., Bartlesville, OK) 3 cycles in 0.3 ml of TFA extraction solution containing ~75 mg of 0.1-mm zirconia-silica beads (BioSpec). Each cycle consisted of 1 min of bead-beating followed by 2 min of cooling on ice. After bead-beat treatment, TFA extracted cell slurry was centrifuged again at 18,000 × g for 5 min and then 0.2 mL supernatant was removed for analysis. OD280 measurements were taken using a 1:6 ratio of lysate to TFA extraction solution. Measurements ranged from 0.24 to 0.70 with an average value of 0.31, and normalized to 0.23 for mass spectral runs.
MALDI-TOF Sample Preparation and Operation
A saturated solution of trans-4-hydroxy-3-methoxy-cinammic acid (ferulic acid) (Sigma-Aldrich, St. Louis, MO) was prepared in 0.5 mL of the above TFA extraction solution. A working matrix solution was prepared by diluting 200 μL of saturated solution with 100 μL of the TFA extraction solution. The matrix solution was briefly vortexed and 0.5 μL was deposited at room temperature for each sample spot and allowed to air dry on a 7 by 7 spot homemade stainless steel MALDI target plate. For each sample of cell lysate supernatant, 0.5 μL was deposited onto a dried matrix spot and samples allowed to air dry at room temperature. All experiments were run in the positive ion mode on a Bruker Daltonics (Billerica, MA) Reflex II MALDI-TOF mass spectrometer operated in reflectron mode with delayed extraction; the delayed extraction setting was set to “medium.” Sample ionization was achieved with a nitrogen laser at 337 nm. A minimum of 200 laser shots per sample was used to generate each spectrum. The instrument was externally calibrated using the adrenocorticotropic hormone clip (peptide 18–39), angiotensin II, substance P, bovine insulin, and horse heart myoglobin as molecular weight standards (Mandrell et al., 2005). In addition, cell lysates from MAP type strain ATCC 19698 were included along with samples run on different days to determine the reproducibility of spectral profiles. Data was processed using the Bruker Flex Analysis software.
Spectral Analysis
Raw spectral data was exported as text files and analyzed using the fingerprint data module of Bionumerics v7.1 (Applied Maths, Sint-Martens-Latem, Belgium). Spectral comparisons were made using the automated workflow with standard baseline correction. Signal-to-noise ratio corrections were made for each spectrum with a value between 35 and 55 to eliminate background noise from the peaks. The noise reduction in spectral peaks did not eliminate any important banding information. Band matching analysis was used to determine peak band patterns. Spectral characterization was targeted for mass ions with mass to charge ratios of 3–11 kDa. By judicious baseline correction and noise reduction techniques, the number of spectral peaks in the molecular mass window of 3–11 KDa was limited to a maximum of 50. Relative intensity of the smallest peak was >2% of the most abundant peak. Percent distribution of peaks and most abundant peaks were calculated based on signal of peaks within the active mass range. Within the fingerprint data module, global band matching was performed to mark multiple banding patterns. Dendrograms along with similarity matrices were generated using cluster analysis with Pearson correlations. Tree stability was assessed by bootstrap analysis with 1,000 iterations. Spectral fingerprint data was overlayed to compare the mass spectra generated for different strains of MAP and species of mycobacteria.
Results
Differentiating Mycobacteria at the Species Level
MALDI-TOF analysis revealed distinct spectral profiles for each of the 15 different species of mycobacteria (Figure 1) that could be used to differentiate them from each other (Table 2). However, similarities in spectral profiles were observed between the members of M. avium complex and between all mycobacteria tested. Based on such similarities in peak profiles mycobacteria were grouped into two distinct clades (Figure 1). Clade 1 was dominated by members of M. avium complex, and clade 2 with mostly fast growing members of mycobacteria. Molecular ions 4,927, 5,331, 7,209, and 10,604 were common in spectra of most members of clade 1, whereas molecular ions 4,939, 5,531, 6,257, 6,303, 7,170, 10,298, and 10,471 were common to most in clade 2.
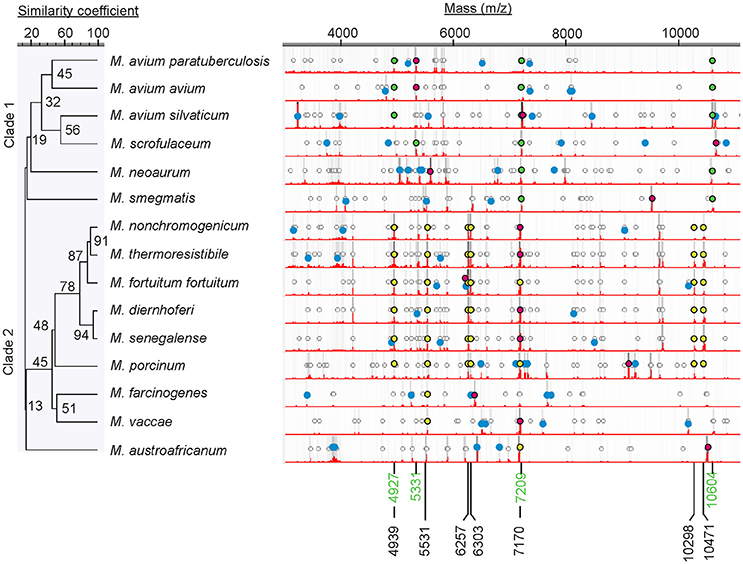
Figure 1. Dendrogram showing mass spectral similarities between different mycobacteria. Circular markings denote peaks that are >2% of the most abundant peak. Relative darkness of gray lines corresponds to relative intensity of the peaks. Spectra show most abundant peaks (red dots), peaks unique to each organism (blue dots), and finger prints common to each other within each clade (clade 1, green dots; clade 2, yellow dots). Molecular ions common to each clade are also marked (clade 1, green m/z; clade 2 black m/z). Mass spectrum for MAP reference strain ATCC 19698 is shown in this comparison.
Differentiating Strains of MAP
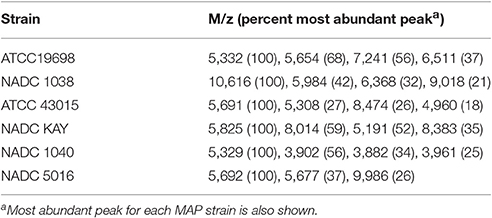
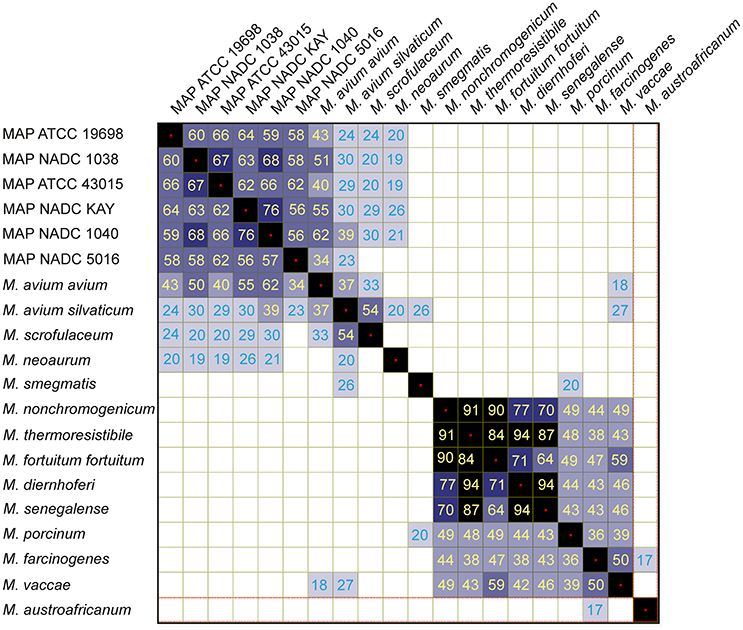
Six different strains of MAP were analyzed by MALDI-TOF and found that all spectra contained several peaks (m/z 3,861, 4,929, 5,329, 5,691, 5,785, 5,828, 7,236, 10,598, and 10,618) in common (Figure 2), but also contained peaks that were unique to each strain (Table 3). Most intense peaks were also different for different strains of MAP. In addition, the dendrogram show more than 58% similarity in peak profiles between all MAP strains, whereas expression profiles of bovine strains NADC Kay and NADC 1040 showed most similarity of 76%. Furthermore, the spectral profile of human strain ATCC 43015 was 67% similar to that of the bovine strain NADC 1038. Spectral profiles of repetitive runs for strain ATCC 19698 on different days were essentially similar.
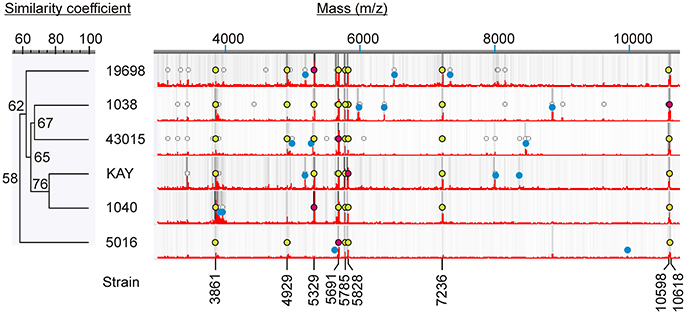
Figure 2. Mass spectral characteristics of MAP strains. Circular markings denote peaks that are >2% of the most abundant peak. Relative darkness of gray lines corresponds to relative intensity of the peaks. Mass spectra show most abundant peaks (red dots), peaks unique to each strain (blue dots) and peaks common to all (yellow dots).
Similarity of Mass Spectral Profiles of Mycobacteria
Cluster analysis with Pearson pair-wise comparisons of mass spectral profiles indicated two distinct clusters (Figure 3). Similarity in spectral profiles between MAP strains was as high as 76%, while the similarity for M. avium subsp. avium and MAP strains ranged between 34 and 62%. Members of clade 2 were highly related by the spectral profiles. For example, a 94% similarity in expression was observed between M. diernhoferi and M. thermoresistibile and between M. diernhoferi and M. senegalense. M. neoaurum, M. smegmatis, and M. austroafricanum were unrelated by cluster analysis.
Discussion
It is essential to minimize the spread of MAP because of the significant economic losses to the US dairy industry and the association of this deadly pathogen with human autoimmune diseases. Confirming the causation requires tracking the pathogen to the source. Molecular typing methods for trace back studies are available that discriminate strains of MAP (Amonsin et al., 2004; Oakey et al., 2014), but molecular typing of large collection of strains is costly and time consuming. Molecular methods require extraction and purification of DNA, PCR amplification of the target sequences and followed by sequencing the targets. Alternatively, the inexpensive and rapid MALDI-TOF MS method (Samli and Ilki, 2016) described here uses protein profiles (Park et al., 2016; Penny et al., 2016) of pure cultures to type MAP strains and further differentiate them from other mycobacteria.
Extraction of proteins while disrupting cells by bead beating in TFA extraction solution resulted in clean and resolved spectral peaks in the mass window of 3–11 KDa range. Similar extraction procedures were used to obtain quality spectral profiles for enteric pathogens E. coli O157:H7 (Fagerquist et al., 2010) and Campylobacter (Mandrell et al., 2005; Penny et al., 2016). Likewise, lysates obtained by cell disruption using glass beads were used to obtain quality profiles for clinical isolates of M. avium and M. tuberculosis complex organisms (Machen et al., 2013). However, incomplete extractions or un-extracted whole cells spotted directly on MALDI plates resulted in dissimilar spectral profiles (Hettick et al., 2006; Pignone et al., 2006; Lotz et al., 2010; Lin et al., 2013). Thus, sample preparation techniques define the quality and composition of protein expression profiles.
We were able to differentiate MAP strains from 14 other mycobacterial species from bovine, human and environmental sources. In addition, mycobacterial species were differentiated from each other based on their unique spectral profiles (Table 2). Three to eight distinct protein biomarkers were used in distinguishing mycobacteria at the species level. Cluster analysis resulted in two distinct groups (Figure 3) each with common spectral signatures (Figure 1). It is remarkable that all the slow growing members of the avium complex had expressed protein profiles that are distinctly different from fast growing members of clade 2. Furthermore, the protein profiles are so distinct that taxonomically related avium complex group subspecies paratuberculosis, avium, and silvaticum can be distinguished from each other (Table 2, Figure 1).
A greater degree of similarity in protein expression profiles of fast growing mycobacteria was found. Although DNA-DNA hybridization studies indicated that M. diernhoferi of M. parafortuitum clade and M. senegalense of M. fortuitum clade are of distinctly separate species (Baess, 1982), a 94% similarity in protein profiles was observed between both organisms. Similarly, genetically distinct strains M. nonchromogenicum and M. thermoresistibile have similar mycolic acid production patterns (Minnikin et al., 1984) and protein expression profiles (91% similar).
It is remarkable that MALDI-TOF differentiated all six strains of MAP from each other by their unique protein expression profiles (Table 3, Figure 2). Three to four distinct protein biomarkers were chosen for each strain to aid in differentiating them from each other. Currently differentiation of genotypes of MAP may have been possible only with the time consuming MLSSR genotyping coupled with MIRU-VNTR typing (Douarre et al., 2011) or by mass spectral characterization of short sequence repeats (Ahlstrom et al., 2014). Although the findings reported here need further validation using a larger collection of MAP strains, protein expression profiling by MALDI-TOF MS may aid in rapid classification of MAP genotypes. Both MALDI-TOF and DNA based typing methods could be used complementary to each other in source tracking the pathogens.
Similarity matrix data generated using protein expression profiles of mycobacteria indicate two distinct clusters (Figure 3). It is noteworthy that there is not a greater degree of similarity (<76%) in protein expression between strains of MAP and thus they could be easily distinguished from each other. It is also remarkable that the MAP human strain is 62–66% similar to the other bovine isolates. In contrast, a higher degree of similarity (94%) was noted at species level for M. diernhoferi, M. senegalense, and M. thermoresistibile. Protein profile based typing would have been difficult with only a 6% differences in spectral profiles for these organisms. Nonetheless, they were differentiated by the presence of a few unique peaks (Figure 1, Table 2).
In conclusion, we analyzed lysates of cells by MALDI-TOF MS and obtained protein expression profiles for mycobacteria. MAP strains were differentiated from each other and from various species of mycobacteria. This rapid MALDI-TOF method can be used in classifying and source-tracking MAP strains in that the spectral profiles are vastly different between isolates.
Author Contributions
SR, LH, and CS equally contributed in designing and conducting the experiments. SR has written the manuscript and LH and CS contributed in preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work was funded by the US Department of Agriculture, Agricultural Research Service CRIS project 2030-42000-050-00D. We thank Amarnath Ravva for his helpful review.
References
Ahlstrom, C., Barkema, H. W., and De Buck, J. (2014). Improved short-sequence-repeat genotyping of Mycobacterium avium subsp. paratuberculosis by using matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 80, 534–539. doi: 10.1128/AEM.03212-13
Amonsin, A., Li, L. L., Zhang, Q., Bannantine, J. P., Motiwala, A. S., Sreevatsan, S., et al. (2004). Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42, 1694–1702. doi: 10.1128/JCM.42.4.1694-1702.2004
Baess, I. (1982). Deoxyribonucleic acid relatedness among species of rapidly growing mycobacteria. Acta Pathol. Microbiol. Immunol. Scand. B 90, 371–375. doi: 10.1111/j.1699-0463.1982.tb00133.x
Chang, C. L., Park, T. S., Oh, S. H., Kim, H. H., Lee, E. Y., Son, H. C., et al. (2002). Reduction of contamination of mycobacterial growth indicator tubes with a modified antimicrobial combination. J. Clin. Microbiol. 40, 3845–3847. doi: 10.1128/JCM.40.10.3845-3847.2002
Chiodini, R. J., Chamberlin, W. M., Sarosiek, J., and McCallum, R. W. (2012). Crohn's disease and the mycobacterioses: a quarter century later. Causation or simple association? Crit. Rev. Microbiol. 38, 52–93. doi: 10.3109/1040841x.2011.638273
Collins, M. T., Kenefick, K. B., Sockett, D. C., Lambrecht, R. S., McDonald, J., and Jorgensen, J. B. (1990). Enhanced radiometric detection of Mycobacterium paratuberculosis by using filter-concentrated bovine fecal specimens. J. Clin. Microbiol. 28, 2514–2519.
Cossu, A., Rosu, V., Paccagnini, D., Cossu, D., Pacifico, A., and Sechi, L. A. (2011). MAP3738c and MptD are specific tags of Mycobacterium avium subsp. paratuberculosis infection in type I diabetes mellitus. Clin. Immunol. 141, 49–57. doi: 10.1016/j.clim.2011.05.002
Cossu, D., Masala, S., Cocco, E., Paccagnini, D., Tranquilli, S., Frau, J., et al. (2013). Association of Mycobacterium avium subsp. paratuberculosis and SLC11A1 polymorphisms in Sardinian multiple sclerosis patients. J. Infect. Dev. Ctries. 7, 203–207. doi: 10.3855/jidc.2737
Douarre, P. E., Cashman, W., Buckley, J., Coffey, A., and O'mahony, J. (2011). Molecular characterization of Mycobacterium avium subsp. paratuberculosis using multi-locus short sequence repeat (MLSSR) and mycobacterial interspersed repetitive units-variable number tandem repeat (MIRU-VNTR) typing methods. Vet. Microbiol. 149, 482–487. doi: 10.1016/j.vetmic.2010.12.001
El Khechine, A., Couderc, C., Flaudrops, C., Raoult, D., and Drancourt, M. (2011). Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS ONE 6:e24720. doi: 10.1371/journal.pone.0024720
Fagerquist, C. K., Garbus, B. R., Miller, W. G., Williams, K. E., Yee, E., Bates, A. H., et al. (2010). Rapid identification of protein biomarkers of Escherichia coli O157:H7 by matrix-assisted laser desorption ionization-time-of-flight-time-of-flight mass spectrometry and top-down proteomics. Anal. Chem. 82, 2717–2725. doi: 10.1021/ac902455d
Fagerquist, C. K., Miller, W. G., Harden, L. A., Bates, A. H., Vensel, W. H., Wang, G., et al. (2005). Genomic and proteomic identification of a DNA-binding protein used in the “fingerprinting” of campylobacter species and strains by MALDI-TOF-MS protein biomarker analysis. Anal. Chem. 77, 4897–4907. doi: 10.1021/ac040193z
Frau, J., Cossu, D., Coghe, G., Lorefice, L., Fenu, G., Melis, M., et al. (2013). Mycobacterium avium subsp. paratuberculosis and multiple sclerosis in Sardinian patients: epidemiology and clinical features. Mult. Scler. 19, 1437–1442. doi: 10.1177/1352458513477926
Grant, I. R. (2005). Zoonotic potential of Mycobacterium avium ssp. paratuberculosis: the current position. J. Appl. Microbiol. 98, 1282–1293. doi: 10.1111/j.1365-2672.2005.02598.x
Grant, I. R., Ball, H. J., and Rowe, M. T. (2002). Incidence of Mycobacterium paratuberculosis in bulk raw and commercially pasteurized cows' milk from approved dairy processing establishments in the United Kingdom. Appl. Environ. Microbiol. 68, 2428–2435. doi: 10.1128/AEM.68.5.2428-2435.2002
Hettick, J. M., Kashon, M. L., Slaven, J. E., Ma, Y., Simpson, J. P., Siegel, P. D., et al. (2006). Discrimination of intact mycobacteria at the strain level: a combined MALDI-TOF MS and biostatistical analysis. Proteomics 6, 6416–6425. doi: 10.1002/pmic.200600335
Leite, F. L., Stokes, K. D., Robbe-Austerman, S., and Stabel, J. R. (2013). Comparison of fecal DNA extraction kits for the detection of Mycobacterium avium subsp. paratuberculosis by polymerase chain reaction. J. Vet. Diagn. Invest. 25, 27–34. doi: 10.1177/1040638712466395
Lin, C. S., Su, C. C., Hsieh, S. C., Lu, C. C., Wu, T. L., Jia, J. H., et al. (2013). Rapid identification of Mycobacterium avium clinical isolates by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Microbiol. Immunol. Infect. 48, 205–212. doi: 10.1016/j.jmii.2013.08.008
Lombard, J. E., Gardner, I. A., Jafarzadeh, S. R., Fossler, C. P., Harris, B., Capsel, R. T., et al. (2013). Herd-level prevalence of Mycobacterium avium subsp. paratuberculosis infection in United States dairy herds in 2007. Prev. Vet. Med. 108, 234–238. doi: 10.1016/j.prevetmed.2012.08.006
Lotz, A., Ferroni, A., Beretti, J. L., Dauphin, B., Carbonnelle, E., Guet-Revillet, H., et al. (2010). Rapid identification of mycobacterial whole cells in solid and liquid culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48, 4481–4486. doi: 10.1128/JCM.01397-10
Machen, A., Kobayashi, M., Connelly, M. R., and Wang, Y. F. (2013). Comparison of heat inactivation and cell disruption protocols for identification of mycobacteria from solid culture media by use of vitek matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51, 4226–4229. doi: 10.1128/JCM.02612-13
Mandrell, R. E., Harden, L. A., Bates, A., Miller, W. G., Haddon, W. F., and Fagerquist, C. K. (2005). Speciation of Campylobacter coli, C. jejuni, C. helveticus, C. lari, C. sputorum, and C. upsaliensis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 71, 6292–6307. doi: 10.1128/AEM.71.10.6292-6307.2005
Marekovic, I., Bosnjak, Z., Jakopovic, M., Boras, Z., Jankovic, M., and Popovic-Grle, S. (2016). Evaluation of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in identification of nontuberculous mycobacteria. Chemotherapy 61, 167–170. doi: 10.1159/000442517
Masala, S., Paccagnini, D., Cossu, D., Brezar, V., Pacifico, A., Ahmed, N., et al. (2011). Antibodies recognizing Mycobacterium avium paratuberculosis epitopes cross-react with the beta-cell antigen ZnT8 in Sardinian type 1 diabetic patients. PLoS ONE 6:e26931. doi: 10.1371/journal.pone.0026931
Millar, D., Ford, J., Sanderson, J., Withey, S., Tizard, M., Doran, T., et al. (1996). IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cows' milk in England and Wales. Appl. Environ. Microbiol. 62, 3446–3452.
Minnikin, D. E., Minnikin, S. M., Parlett, J. H., Goodfellow, M., and Magnusson, M. (1984). Mycolic acid patterns of some species of Mycobacterium. Arch. Microbiol. 139, 225–231. doi: 10.1007/BF00402005
Oakey, J., Gavey, L., Singh, S. V., Platell, J., and Waltisbuhl, D. (2014). Variable-number tandem repeats genotyping used to aid and inform management strategies for a bovine Johne's disease incursion in tropical and subtropical Australia. J. Vet. Diagn. Invest. 26, 651–657. doi: 10.1177/1040638714547257
Ott, S. L., Wells, S. J., and Wagner, B. A. (1999). Herd-level economic losses associated with Johne's disease on US dairy operations. Prev. Vet. Med. 40, 179–192. doi: 10.1016/S0167-5877(99)00037-9
Overduin, P., Schouls, L., Roholl, P., Van Der Zanden, A., Mahmmod, N., Herrewegh, A., et al. (2004). Use of multilocus variable-number tandem-repeat analysis for typing Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 42, 5022–5028. doi: 10.1128/JCM.42.11.5022-5028.2004
Park, J. S., Choi, S. H., Hwang, S. M., Hong, Y. J., Kim, T. S., Park, K. U., et al. (2016). The impact of protein extraction protocols on the performance of currently available MALDI-TOF mass spectrometry for identification of mycobacterial clinical isolates cultured in liquid media. Clin. Chim. Acta 460, 190–195. doi: 10.1016/j.cca.2016.06.039
Penny, C., Grothendick, B., Zhang, L., Borror, C. M., Barbano, D., Cornelius, A. J., et al. (2016). A designed experiments approach to optimizing MALDI-TOF MS spectrum processing parameters enhances detection of antibiotic resistance in Campylobacter jejuni. Front. Microbiol. 7:818. doi: 10.3389/fmicb.2016.00818
Pignone, M., Greth, K. M., Cooper, J., Emerson, D., and Tang, J. (2006). Identification of mycobacteria by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. J. Clin. Microbiol. 44, 1963–1970. doi: 10.1128/JCM.01959-05
Ravva, S. V., and Stanker, L. H. (2005). Real-time quantitative PCR detection of Mycobacterium avium subsp. paratuberculosis and differentiation from other mycobacteria using SYBR Green and TaqMan assays. J. Microbiol. Methods 63, 305–317. doi: 10.1016/j.mimet.2005.04.004
Samli, A., and Ilki, A. (2016). Comparison of MALDI-TOF MS, nucleic acid hybridization and the MPT64 immunochromatographic test for the identification of M. tuberculosis and non-tuberculosis Mycobacterium species. New Microbiol. 39, 259–263.
Sechi, L. A., Gazouli, M., Ikonomopoulos, J., Lukas, J. C., Scanu, A. M., Ahmed, N., et al. (2005). Mycobacterium avium subsp. paratuberculosis, genetic susceptibility to Crohn's disease, and Sardinians: the way ahead. J. Clin. Microbiol. 43, 5275–5277. doi: 10.1128/JCM.43.10.5275-5277.2005
Sechi, L. A., Paccagnini, D., Salza, S., Pacifico, A., Ahmed, N., and Zanetti, S. (2008). Mycobacterium avium subspecies paratuberculosis bacteremia in type 1 diabetes mellitus: an infectious trigger? Clin. Infect. Dis. 46, 148–149. doi: 10.1086/524084
Sting, R., Hrubenja, M., Mandl, J., Seemann, G., Salditt, A., and Waibel, S. (2014). Detection of Mycobacterium avium subsp. paratuberculosis in faeces using different procedures of pre-treatment for real-time PCR in comparison to culture. Vet. J. 199, 138–142. doi: 10.1016/j.tvjl.2013.08.033
Whittington, R. J. (2009). Factors affecting isolation and identification of Mycobacterium avium subsp. paratuberculosis from fecal and tissue samples in a liquid culture system. J. Clin. Microbiol. 47, 614–622. doi: 10.1128/JCM.01986-08
Keywords: MALDI-TOF, Mycobacterium, Mycobacterium avium avium, Mycobacterium avium paratuberculosis, mass spectrometry
Citation: Ravva SV, Harden LA and Sarreal CZ (2017) Characterization and Differentiation of Mycobacterium avium subsp. paratuberculosis from Other Mycobacteria Using Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Front. Cell. Infect. Microbiol. 7:297. doi: 10.3389/fcimb.2017.00297
Received: 02 March 2017; Accepted: 16 June 2017;
Published: 30 June 2017.
Edited by:
Lorenza Putignani, Bambino Gesù Ospedale Pediatrico (IRCCS), ItalyReviewed by:
Paras Jain, Albert Einstein College of Medicine, United StatesBrian Weinrick, Albert Einstein College of Medicine, United States
Copyright © 2017 Ravva, Harden and Sarreal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subbarao V. Ravva, c3ViYmFyYW8ucmF2dmFAYXJzLnVzZGEuZ292
 Subbarao V. Ravva
Subbarao V. Ravva Leslie A. Harden
Leslie A. Harden Chester Z. Sarreal
Chester Z. Sarreal