- 1State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 2Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Hangzhou, China
We aimed to investigate and understand the characterization and distribution of the autB gene in Neisseria meningitidis in China. autB is flanked by two conservative genes, smpB and glcD, and it can be present in the majority of meningococcal isolates, but not in 053442 of clonal complex 4821 (CC4821) which contains a 968 bp sequence. In this study, we sequenced the intervenient region between smpB and glcD in 178 Chinese N. meningitidis strains isolated from both patients and carriers. There were 110 serogroupable strains, other 68 were non-groupable (NG). Ninety nine of the 178 strains were clustered into 13 CCs, the remaining 79 were unassigned (UA). CC4821 is one of the dominant CCs in China. Forty of the 42 CC4821 strains and 26 of the 79 UA strains were autB-null, while the remaining 12 CCs were autB-positive. According to the N-terminal sequence, most (97/112) of the autB-positive strains were clustered into AutB1 and the remaining 15 were AutB2. The autB gene and its flanking intergenic sequences was superseded by a perfectly conservative sequence of an identical 968 bp in all of the autB-null N. meningitidis strains which had no identity with the relatively conservative intergenic sequences that flanked the autB gene in autB-positive strains. There was a 10 bp DNA uptake sequence (DUS) at the beginning of the interval 968 bp sequence in the autB-null strains while there was a 9 bp Haemophilus-specific uptake sequence (hUS) at the beginning of the partial holB gene and at the end of the partial tmk gene in autB-positive strains, holB and tmk gene were flanking the autB gene in Haemophilus. In conclusion, not all pathogenic N. meningitidis strains especially CC4821 possess the autB gene in China and the corresponding spacer region of the autB-null strains was not homologous to that found in autB-positive strains. There's a hypothesis that the DUS and hUS are likely to play a key part in the mechanism of uptake or loss of the autB gene.
Introduction
As a Gram-negative diplococcus, Neisseria meningitidis is an obligate human pathogen and it usually inhabits the nasopharynx, often asymptomatically. Nonetheless, this pathogen is also a common cause of meningitis and/or sepsis and it can generate large outbreaks. Autotransporters (ATs), which are synthesized by pathogenic Gram-negative bacteria, have been implicated in forming biofilms and may also correlate with virulence (Arenas et al., 2015; Arenas and Tommassen, 2017). To date, eight types of ATs have been studied in N. meningitidis. They are proteins that are usually composed of an N-terminal signal sequence, a secreted passenger domain and a C-terminal translocator domain (Grijpstra et al., 2013). The iga gene of the immunoglobulin A1 (IgA1) protease of N. meningitidis is single-copy and polymorphic, which suggests that it was attained through interstrain horizontal gene transfer (Lomholt et al., 1992). App is an acronym for adhesion and penetration protein which is homologous to the Haemophilus adherence and penetration protein (Hap). Similarly, the NalP is a Neisseria autotransporter lipoprotein which was historically named AspA (autotransported serine protease A) (St Geme et al., 1994; Turner et al., 2002; Serruto et al., 2003; van Ulsen and Tommassen, 2006). The functions of AusI (also known as MspA) have not been elucidated and its protein expression is influenced by NalP (van Ulsen et al., 2003, 2006; Turner et al., 2006). The former 4 ATs are monomeric and have protease activity whereas NadA and NhhA (Neisseria hia homolog A) are trimeric ATs that are related to host cell adhesion (Peak et al., 2000; Capecchi et al., 2005; Scarselli et al., 2006; Arenas et al., 2013). Other than the former four monomeric ATs, AutA is not secreted into the milieu of the bacteria and it also participates in autoaggregation (Arenas et al., 2015; Arenas and Tommassen, 2017). In contrast, the passenger domain of AutB is secreted but the AutB is still attached to the cell surface due to the translocator domain (Arenas et al., 2016).
A detailed study has elucidated the expression of the autB gene in N. meningitidis, whereby the mechanism of AutB appears to affect biofilm formation and epithelial transmigration (Arenas et al., 2016). A comprehensive analysis of AutB has been conducted and it complements the study of AutB, which has firmly assessed the ATs (Arenas et al., 2016). The distribution of the autB gene is well-known in the two pathogenic Neisseria spp. N. meningitidis and N. gonorrhoeae. There is no autB gene in N. lactamica, N. flavescens, N. polysaccharea, and N. sicca while some strains of Haemophilus influenzae, H. haemolyticus, H. parainfluenzae, and H. aegyptius are autB positive (Arenas et al., 2016). The number of AAGC tetranucleotide repeats (Rn) in the autB gene has previously been published and studies have considered whether the autB gene is in frame or out of frame depends on these repeats (Arenas et al., 2016). Although the autB gene is intact in most of the meningococcal strains (105/117), it is often out of phase, suggesting conservation but a very strong negative selection for AutB expression (Arenas et al., 2016). The amino acid sequence of AutB can be divided into three parts: the N-terminal part, the linker and the C-terminal translocator domain. The variability of AutB can be demonstrated by phylogenetic analysis of the N-terminal component and the linker domain and has been designated AutB1, AutB2, and AutB3, most of the variants are associated with AutB1 from N. meningitidis (Arenas et al., 2016). The phase variation was studied in patient strains of sequence type ST-32 clonal complex (CC32) (49 isolates) and CC213 (53 isolates) collected in Netherlands and 207 genome sequences of carriage strains in public data base, only one strain 2081107 (disease isolate) of CC32 have an intact autB gene and demonstrated in Western blotting, five strains of carrier isolates were also have an undisrupted autB gene, no obvious differences in autB expression between carriage and patient strains were demonstrated (Arenas et al., 2016).
It is noteworthy that the autB gene is omnipresent in the pathogenic Neisseria spp. except for 053442 (Arenas et al., 2016). 053442 is a serogroup C strain with the unique ST-4821, which was isolated from a patient in an epidemic of cerebrospinal meningitis in China (Shao et al., 2006; Peng et al., 2007). CC4821 was first isolated in China during 2003–2004 and then rapidly spread to two-thirds of the provinces within China (Shao and Zhu, 2016). Although CC4821 is one of the dominant hyperinvasive CCs in China, it has rarely been isolated in other countries (Zhou et al., 2012). The meningococcal A+C polysaccharide-based vaccine was introduced to the Expanded Program on Immunization (EPI) in 2005 and N. meningitidis invasive serogroup B soon increased after 2006 (Zhou et al., 2012). Meanwhile, the population structure of N. meningitidis invasive serogroup B were used to be mainly CC11 and CC41/44 and it evolved into CC4821 in China, CC4821 is one of the dominant hyperinvasive CCs in China now. It has been verified that there were capsular switching from N. meningitidis CC4821 serogroup C strains to serogroup B strains (Zhu et al., 2015).
The DNA uptake sequence (DUS) is a 10-bp (5′-GCCGTCTGAA-3′) non-palindromic sequence, which mediates the transformation of species-specific DNA (Goodman and Scocca, 1988; Rotman and Seifert, 2014). Twelve base pairs sequences (5′-ATGCCGTCTGAA-3′) have also been noted in pathogenic Neisseria (termed DUS12) (Ambur et al., 2007). It was first to show the presence of DUS bordering autB gene in 2001, in which autB was characterized as lav (Davis et al., 2001). Studies have also demonstrated that the DUS is sufficient for DNA uptake and transformation in previously non-transformable plasmids, while it can also competitively inhibit transformation when a DUS is already present (Goodman and Scocca, 1988; Elkins et al., 1991; Obergfell and Seifert, 2015). Therefore, the purpose of our work is to assess the distribution and characterization of the autB gene in N. meningitidis in China, especially for CC4821.
Materials and Methods
Bacterial Strains and DNA Preparation
One hundred and seventy eight N. meningitidis strains obtained from 24 provinces in China from 1956 to 2015 were analyzed in our study. Sixty-seven of these strains were isolated from patients and the remaining 111 were isolated from healthy carriers. There were 27 strains isolated from 1956 to 2000, 31 strains from 2001 to 2005, 85 strains from 2006 to 2010, and 35 strains isolated from 2011 to 2015.
The bacteria were cultured on Blood Agar Media (Columbia) (PB0123A, Oxoid, China) at 37°C for 18–24 h in a 5% CO2 atmosphere. The genomic DNA of the strains was extracted using a Wizard Genomic DNA Purification Kit (A1125, Promega, Madison, USA) according to the protocol provided by the manufacturer.
Serotyping, Genotyping and MLST
Serotyping was performed on N. meningitidis strains by serum agglutination tests and rabbit antiserums against the 12 serogroups (BD, Sparks, USA; Remel, Kent, UK) were used (Xu et al., 2015). Genotyping was then implemented using PCR of the corresponding genes, the primers and amplification conditions have been described in former studies (Dolan-Livengood et al., 2003; Zhu et al., 2012; Xu et al., 2015). Multilocus sequence typing (MLST) was also performed on these strains according to the standard protocol (http://pubmlst.org/neisseria/) (Maiden et al., 1998; Maiden, 2006).
Identification of the autB Gene and Bioinformatics Analysis
As the genes flanking the autB gene are conserved in N. meningitidis, we gained the autB gene and its flanking sequences by PCR and sequencing of the PCR products. The dominant CCs of N. meningitidis in China are different to other countries, and the primers were redesigned according to the sequences of the smpB (NMB1526) and glcD (NMB1524) from the whole genome of N. meningitidis MC58 (Accession number NC_003112) (Tettelin et al., 2000; Shao and Zhu, 2016). The primer sequences are listed as follows: autB-F: 5′-GAAGGCTGGGAAGTCAAAG-3′ and autB-R: 5′-CGAAAACGACATCAACAGCAC-3′. To amplify the autB gene, TaKaRa LA Taq (RR02MA, Takara, Dalian, China) high fidelity polymerase was used and the annealing temperature was set to 57°C. The length of the products of MC58 ought to be 3,156 base pairs (bp) but it might fluctuate in other strains according to the number of AAGC repeats. In autB-null strains, the length of the products was considered to be ~1,539 bp according to the whole genome sequence of N. meningitidis 053442 (Accession number NC_010120). Both the PCR products with the positive and negative strains were sequenced by Beijing Tianyi Huiyuan Bioscience & Technology Inc, China. The alleles were aligned with the reference autB sequence of MC58 or the sequence of the spacer region in 053442 by MEGA (version 6.0) (http://www.megasoftware.net/). We then obtained the correct sequence of autB by excising the spare sequence (Tamura et al., 2013). The sequences of the spacer regions flanking the autB gene were obtained using the same method. The correct reading frame of autB was predicted by altering the number of AAGC repeats and removing the premature stop codons and frameshift mutations. Using the SignalP web tools (http://www.cbs.dtu.dk/services/SignalP/) to predict the cleavage site of the N-terminal signal sequence of AutB and neighbor-joining in MEGA, the phylogeny of the N-terminal domain of the passengers was analyzed (Arenas et al., 2016).
Results
Serotyping, Genotyping, and MLST
Among the 178 N. meningitidis strains in this study, 6 serogroups (A, B, C, W, E, and X) and NG were identified. Twenty-one of the strains were serogroup A, 59 were serogroup B, 23 were serogroup C, 4 were serogroup W, 1 was serogroup E, 2 were serogroup X, and the remaining 68 were non-groupable (NG) strains. The genogroups of the 110 serogroupable strains were in accordance with their serogroups. The 68 NG strains were assigned to 6 genogroups and one group of capsule null locus (cnl) using PCR: genogroup B (22 isolates), C (18 isolates), E (7 isolates), W (5 isolates), Y (5 isolates), and X (3 isolates); eight were identified as being cnl.
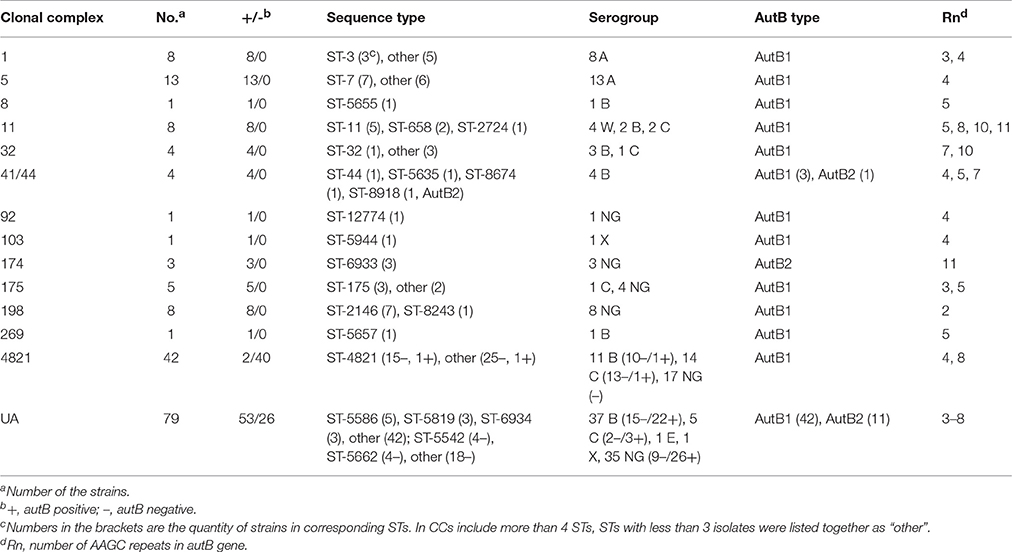
These strains could also be clustered into 120 STs; 13 CCs and unassigned (UA) groups. Fifty-eight of these STs belonged to 13 CCs and another 62 STs were UA. Eight of the strains were CC1, 13 were CC5, 42 were CC4821, 8 were CC11, 8 were CC198, 79 were UA and the remaining 20 belonged to CC8 (1), CC32 (4), CC41/44 (4), CC92 (1), CC103 (1), CC174 (3), CC175 (5), and CC269 (1).
The characterization of the strains is listed in Table 1. Most of the strains are belonged to the dominant CCs in China, such as CC4821, CC5, CC1, CC11, CC198, and UA. All of the strains in CC1 and CC5 are serogroup A while other CCs distributed into many serogroups, especially the UA strains decentralized to serogroup B, C, E, X, and NG. The STs in the UA group are also flexible as there are 62 STs of the 79 UA strains.
Distribution and Characterization of the autB Gene in China
One hundred and twelve of the 178 N. meningitidis strains were autB-positive and the other 66 were autB-null. The distribution of the autB gene is listed in Table 1 and detailed in Table S1.
The 21 strains of serogroup A were composed of eight strains of CC1 and 13 strains of CC5. All 21 strains were autB-positive.
Among the 59 strains of serogroup B, 22 strains belonged to 6 CCs, and 37 strains were UA. In the 11 strains of CC4821, only one strain of ST-4821 was autB-positive. This strain was 341215, isolated from the CSF of a patient in Anhui Province in 2012. The 11 strains of other five CCs were clustered into 11 different STs. They were all autB-positive. Among the 37 UA strains, 22 were autB-positive and distributed into 22 different STs. The remaining 15 strains of UA were autB-null strains and dispersed across 13 STs.
In serogroup C, there were 18 strains belonging to four CCs and five strains belonging to UA, all of the 23 strains were isolated after 2004. Fourteen strains belonged to CC4821. Only 440501, a ST-4831 strain, carried the autB gene. It was a clinical strain isolated from CSF in 2005. Another 13 strains without the autB gene were dispersed across 9 different STs. Three of the five UA strains were autB-positive, clustering into three STs. The remaining two were autB-null.
The four strains of serogroup W were ST-11, CC11, and autB-positive. The one strain of serogroup E was isolated from healthy carriers and clustered into ST-5586 (UA). The 2 serogroup X strains were all autB-positive.
All the 68 NG strains were isolated from healthy carriers. There were no genogroup A strains. The 22 genogroup B strains were isolated after 2005. Four of these were CC4821 and autB-null. The remaining 18 strains were UA, six of them were autB-null which belonged to six different STs. In genogroup C, all of the 11 CC4821 strains and three UA strains were autB-null, the remaining 4 UA strains were autB-positive. In the five genogroup W strains, 2 CC4821 strains were autB-null, the other 3 CC174 strains were all autB-positive. The 5 genogroup Y strains were autB-positive, 4 of them were CC175. The 7 genogroup E and 3 genogroup X strains were all UA and autB-positive. The 8 cnl strains were clustered into CC198, they were all autB-positive strains.
In conclusion, autB gene was only absent in strains of CC4821 (40 of 42) and UA (26 of 79), while an autB gene was detected in strains of the rest of CCs.
Phase Variation of the autB Gene
The 112 autB-positive strains were analyzed. Without 9, the number of AAGC repeats in the autB gene were ranged from 2 to 11. Eight of the 112 strains were in frame according to the number of the AAGC repeats, seven of them with three tetranucleotide repeats and another one with six. In the Seven, the only one strain from patient was serogroup A, CC1 and there were no frameshifts, transposase or premature stop codons in its autB gene which indicated that AutB may synthesized in this strain. Other six strains were all isolated from carriers and they were all NG, 4 of them were genogroup Y of CC175 and another 2 were genogroup B of UA. Frameshift mutation was observed in all of the six strains. The other strain with six repeats was a NG strain belonged to genogroup X, it was isolated from carrier and clustered into UA. There were nothing wrong with the autB gene when turned to codons. In the 104 strains out of frame, the 8 cnl strains were all with 2 AAGC repeats and the 20 serogroup A strains were all with 4 repeats. The number of AAGC repeats maybe not strict to different serogroups.
Structure and Variability of AutB
The gene assignment of 106 of the 112 autB-positive strains was similar to N. meningitidis MC58, which has been studied in Arenas's paper. These strains might transcribe the autB gene after phase variation. In the other six strains, this was the same as the N. gonorrhoeae NCCP11945 (Figure 1). The arrangement for all 66 autB-null strains was identical to that observed in the N. lactamica 020-06, as previously described.
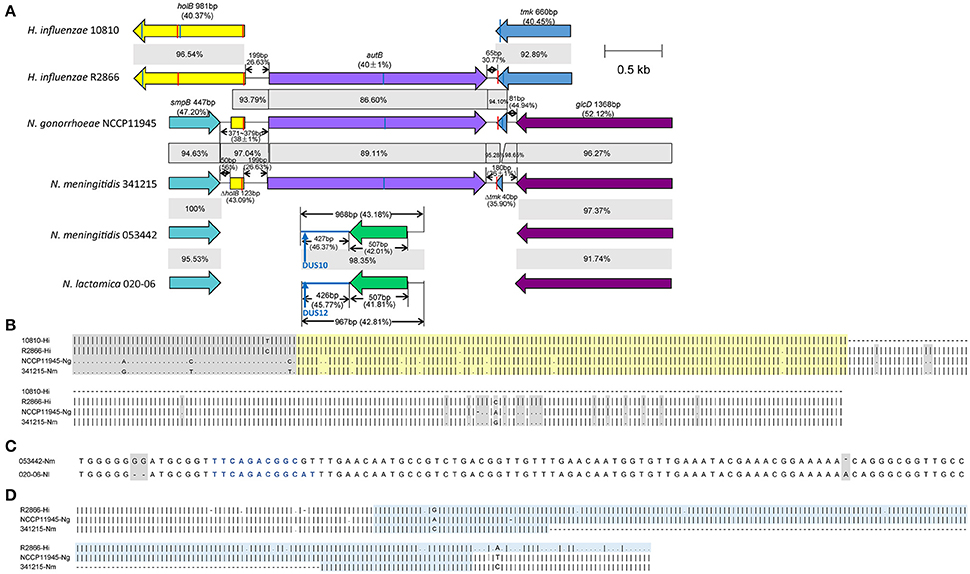
Figure 1. Comparison of the autB gene and its flanking genes. The autB gene and its flanking genes of H. influenzae 10810, H. influenzae R2866, N. gonorrhoeae NCCP11945, N. meningitidis 341215, N. meningitidis 053442, and N. lactamica 020-06 are compared. (A) The genes similar to each other are colored identically and the gray area represents the homology between the two sequences and the percentages in it represent the similarity between the two strains. The percentages in the brackets are the G+C content. The vertical bar in red represents the 9 bp hUS sequence of 5′-AAGTGCGGT-3′ and the vertical bar in blue represents the 9 bp hUS reverse complement to the former. In N. meningitidis 053442, the position of the blue arrow points from 16 to 25 of the 968 bp and is a 10 bp DUS sequence of 5′-TTCAGACGGC-3′ while in N. lactamica 020-06, it is a 12 bp DUS. (B) Alignment of the nucleotide sequence between smpB and autB, H. influenzae 10810 is autB-null, and there are ~123 bp in autB-positive strains of N. gonorrhoeae NCCP11945 and N. meningitidis 341215, which are homologous to holB in H. influenzae. This is highlighted in light yellow. The sequence in gray shadow demonstrates that nucleotides are different to each other. (C) Alignment of the nucleotide sequence between smpB and glcD in autB-null strains. There are only three nucleotide differences to each other which are in gray shadow and the nucleotides in blue are 10 bp DUS and 12 bp DUS, respectively. (D) Alignment of the nucleotide sequence between autB and glcD in autB-positive strains, shows there were 222 bp (highlighted in light blue) of the tmk gene in N. gonorrhoeae NCCP11945, which is homologous to that in H. influenzae R2866, while there were only 73 bp in N. meningitidis 341215.
The analysis of the N-terminal part of the representative AutB passengers is displayed in Table 1 and Table S1. The phylogenetic trees of the N-terminal sequence of the autB gene in the 112 autB-positive strains and 10 strains from the NCBI database included N. meningitidis MC58, H. influenzae R2866, and F3047 were analyzed. There were also three branches of the N-terminal part of the AutB passenger and all 112 autB-positive strains in this study belonged to AutB1 and AutB2. However, except for the reference sequence of H. influenzae F3047, no AutB3 were found in our study. Most (97/112) of the strains with autB gene were clustered into AutB1. There were also many secondary branches of AutB1. Only three strains of CC174 (ST-6933), 1 strain of CC41/44 (ST-8918), and 11 strains of UA were clustered into AutB2.
There were 67 strains isolated from patients; 37 of these were clustered into AutB1, three were clustered into AutB2 and 27 were autB-null strains. In the 27 autB-null strains, only three UA strains were isolated in 1985 and 1988, while the other 17 strains (CC4821 and UA) were isolated after 2004. For the remaining 111 strains isolated from carriers, 60 of these were clustered into AutB1, 12 strains in AutB2 and 39 were autB-null strains. However, the correlation of AutB variability and different isolates was not clear.
Characteristics of the Spacer Region Flanking the autB Gene
In H. influenzae R2866, the sequence of the spacer region between holB and autB was 199 bp with a G+C content of 26.63%, while between autB and tmk it was 65 bp with a G+C content of 30.77% (Figure 1). At the beginning of the holB gene, there was a 9 bp hUS within an extended 29 bp consensus sequence. At the end of the tmk gene, there was also a 9 bp hUS sequence. In H. influenzae 10810, a strain without the autB gene, the holB and tmk gene had a 4 bp overlap and there were also two hUSs on holB and tmk, respectively. In N. meningitidis MC58, which is identical to 341215 in the flanking sequences of autB gene, the sequence of the spacer region 1 between smpB and autB was 372 bp with a total G+C content of 36.29% while the spacer region two between autB and glcD was 180 bp with a total G+C content of 38.89% (Figure 1A). Both of the two spacer regions can be divided into three parts. In H. influenzae R2866, spacer region 1 consisted of 50 bp with a G+C content of 56.00%, 123 bp (43.90%) were homologous to holB gene and 199 bp (26.63%) were homologous to the sequence between holB and autB (Figure 1B). The length of spacer region 1 in the strains in China were 371, 372, or 379 bp (Figure 1A). The spacer region 2 was 180 bp and it consisted of 67 bp that was homologous to the sequence between autB and tmk with a G+C content of 32.84%; 73 bp was homologous to the tmk gene (42.47%); and 40 bp had a G+C content of 42.50% (Figure 1D). In N. meningitidis 053442 and another 66 autB-null strains in our study, the sequences between smpB and glcD were all 968 bp with a G+C content of 43.18% and were highly conserved with a 98% identity to N. lactamica 020-06 (Figure 1C). In the 968 bp sequence, there was a 10 bp DUS from 16 to 25. The 968 bp spacer region of autB-null strains was not homologous to the intergenic sequences flanking the autB gene in autB-positive strains. The autB gene and part of its flanking sequences in N. meningitidis in China are also homologous to the sequences in H. influenzae.
Discussion
Microcolony formation is one of the strategies of N. meningitidis to elude the host immune response (Sim et al., 2000). The autotransporter AutB has been demonstrated to play a part in biofilm formation (Arenas et al., 2016). The autB gene was first described as an orf2 downstream of lsi1 (rfaF), a LPS biosynthesis related gene, and was then demonstrated to be horizontally-transferred from H. influenzae to Neisseria immediately before the bifurcation of N. meningitidis and Neisseria gonorrhoeae, where it was characterized as lav (Jennings et al., 1995; Davis et al., 2001). Subsequently, Arenas et al. have extended the analysis of the distribution of the autB gene and it is worth noting that in Arenas' study, N. meningitidis strain 053442 was the only clinical isolate without the autB gene. 053442 was a CC4821 strain and this clonal lineage has only been detected in China (Zhu et al., 2015). CC4821 has been a dominant lineage in China since 2003 (Zhou et al., 2012; Shao and Zhu, 2016). On account of this, we analyzed the isolates in China to further enrich the study of the autB gene.
We have not found any explicit relationship between the autB gene and the meningococcal serogroup. In a former study, all strains of the highly pathogenic CC213 and CC32 were autB-positive and these strains were clinical isolates; all 49 isolates of CC32 were clustered into AutB1 and 52 of the 53 isolates of CC213 were AutB1, while only one CC213 isolate harbored an AutB2 variant (Arenas et al., 2016). In our study, the 67 clinical strains included 37 strains of AutB1 and 3 of AutB2, and the others were autB-null strains. The number of AAGC nucleotide repeats in our strains was 2–11 while in the former study was 3–19 (Arenas et al., 2016). With regard to the 207 carrier isolates in the former study, the number of the repeats was 2–22 and in most of the strains, the autB gene was disrupted by single-nucleotide insertions downstream of the repeats (Arenas et al., 2016). In the 111 isolates from carriers in our study, 25 isolates of CC4821 and 14 of UA were autB-null and isolated after 2005. The repeat number in the other 72 autB–positive isolates was also 2–11, where 60 harbored an AutB1 variant and the five isolates with 3 or 6 AAGC repeats were all disrupted by frame shift mutations. Meanwhile, another 12 AutB2 isolates belonged to CC174 (ST-6933) and UA. There were no AutB3 isolates in our strains (Table 1). Our study indicates that not all highly pathogenic strains are autB-positive and as expression is switched off in most N. meningitidis isolates, it can also be absent in some of the strains such as the CC4821 strains. Of 17 clinical strains of CC4821, 341215, and 440501, with the autB gene belonging to AutB1, two had repeat numbers of 8 and 4, respectively.
The sequences of the 66 autB-null strains between the smpB and glcD gene are all 968 bp long with no SNPs and these sequences are homologous to N. lactamica 020-06 (Figure 1C). The autB gene has been recognized as a pseudogene because it is not expressed in many meningococcal or gonococcal strains with the sequence (Ait-Tahar et al., 2000). Authors suggested this is probably due to the accumulation of mutations on autB, which with no need for coping with selective pressure (Ait-Tahar et al., 2000). Conversely, the high variability of the AutB passenger domain may be a reflection of the antigenic variability response to immune pressure, so that an immune response to AutB can be avoided, and where expression of the autB gene is switched off in most isolates (Arenas et al., 2016).
As has been verified in Arenas' study, the majority of N. meningitidis isolates equipped with the autB gene do not express AutB. One possible explanation is that some functions of AutB can be substituted by other ATs such as NalP, which cleaves the heparin-binding antigen NhbA or the α-peptide of IgA protease from the cell surface (Serruto et al., 2010; Roussel-Jazede et al., 2014). AutB is likely to be a backup mechanism when the concentration of these two proteins is low (Arenas et al., 2016). Nevertheless, the proportion of strains with the autB gene to autB-null strains is 24/27 (1956-2000), 20/31 (2001-2005), 49/85 (2006-2010), and 19/35 (2011-2015) in our study. In the 66 autB-null strains, only two have been isolated in 1985, one in 1988 and the others were all isolated after 2004. There are two hypothesis for this. First of this, it seems that the strains in China habitually lose the autB gene and the autB gene will not be retained as an intact gene in N. meningitidis. However, the number of strains in our study is relatively small so this hypothesis may be premature. Secondly, CC4821 strains are similar to N. lactamica, also shown in Figure 1A. In here, we have demonstrate that this is indeed a conservative feature of this CC, as we found in many isolates of this CC. Thus, in a common ancestor of this CC in China, the autB was lost by acquisition of N. lactamica sequences. Interestingly, two isolates of this CC seems to acquire autB, probably from another circulating strain as they harbor a typical and common AutB1 (see Table 1). This also supported by the similarity in the flanking regions with other CCs found in China and the predominance of this CC. Some isolates of UA, but no other CCs, do not have the autB gene but this is restricted uniquely to isolates of the same ST. Possibly, those isolates acquired sequences from the predominant clones of CC4821, which rarely have an autB gene.
It has been suggested that the autB gene in Neisseria was obtained from Haemophilus through domain shuffling and it has been proposed that the function of the 9 bp inverted repeat sequences of hUS at the beginning of the partial holB gene and the end of the partial tmk gene is recognition sites that enhance the transformation (Davis et al., 2001; Duffin and Seifert, 2010). In autB-null N. meningitidis and N. lactamica 020-06, the 10 or 12 bp DUS was located in the intergenic sequence (Figure 1C), according with the former study that most of the DUS (65%) in Neisseria is located in the intergenic sequences (Ambur et al., 2007). It has been postulated that the DUS may function to competitively inhibit transformation and this may be the mechanism of autB-null in these strains while strains without the DUS could gain the autB gene from other strains (Goodman and Scocca, 1988; Duffin and Seifert, 2010; Obergfell and Seifert, 2015).
In summary, we demonstrated the characterization and distribution of the autB gene and the variability of AutB in N. meningitidis isolates in China. Not all N. meningitidis strains were autB-positive in China. This study offers a new perspective on the autB gene and how it is possible that CC4821 and some of the UA N. meningitidis genomes could lose autB genes or acquire the alternative sequence from N. lactamica, the DUS may promote this process to some extent.
Author Contributions
ZS, BZ, and AZ designed the experiments; AZ, FS, LX, YG, and NX carried out the experiments; AZ, PZ, BZ, and ZS analyzed the experimental results; AZ wrote the manuscript; PZ, BZ, and ZS modified the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by grants from the State Key Laboratory of Infectious Disease Prevention and Control (2015SKLID502), and the National Natural Science Foundation of China (81602903).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JC declared a shared affiliation, with no collaboration, with the authors to the handling Editor.
Acknowledgments
We would like to thank the native English speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2017.00436/full#supplementary-material
References
Ait-Tahar, K., Wooldridge, K. G., Turner, D. P., Atta, M., Todd, I., and Ala'Aldeen, D. A. (2000). Auto-transporter a protein of Neisseria meningitidis: a potent CD4+ T-cell and B-cell stimulating antigen detected by expression cloning. Mol. Microbiol. 37, 1094–1105. doi: 10.1046/j.1365-2958.2000.02061.x
Ambur, O. H., Frye, S. A., and Tonjum, T. (2007). New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J. Bacteriol. 189, 2077–2085. doi: 10.1128/JB.01408-06
Arenas, J., Cano, S., Nijland, R., van Dongen, V., Rutten, L., van der Ende, A., et al. (2015). The meningococcal autotransporter AutA is implicated in autoaggregation and biofilm formation. Environ. Microbiol. 17, 1321–1337. doi: 10.1111/1462-2920.12581
Arenas, J., Nijland, R., Rodriguez, F. J., Bosma, T. N., and Tommassen, J. (2013). Involvement of three meningococcal surface-exposed proteins, the heparin-binding protein NhbA, the alpha-peptide of IgA protease and the autotransporter protease NalP, in initiation of biofilm formation. Mol. Microbiol. 87, 254–268. doi: 10.1111/mmi.12097
Arenas, J., Paganelli, F. L., Rodriguez-Castano, P., Cano-Crespo, S., van der Ende, A., van Putten, J. P., et al. (2016). Expression of the gene for autotransporter AutB of Neisseria meningitidis affects biofilm formation and epithelial transmigration. Front. Cell Infect. Microbiol. 6:162. doi: 10.3389/fcimb.2016.00162
Arenas, J., and Tommassen, J. (2017). Meningococcal biofilm formation: let's stick together. Trends Microbiol. 25, 113–124. doi: 10.1016/j.tim.2016.09.005
Capecchi, B., Adu-Bobie, J., Di Marcello, F., Ciucchi, L., Masignani, V., Taddei, A., et al. (2005). Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55, 687–698. doi: 10.1111/j.1365-2958.2004.04423.x
Davis, J., Smith, A. L., Hughes, W. R., and Golomb, M. (2001). Evolution of an autotransporter: domain shuffling and lateral transfer from pathogenic Haemophilus to Neisseria. J. Bacteriol. 183, 4626–4635. doi: 10.1128/JB.183.15.000-000.2001
Dolan-Livengood, J. M., Miller, Y. K., Martin, L. E., Urwin, R., and Stephens, D. S. (2003). Genetic basis for nongroupable Neisseria meningitidis. J. Infect. Dis. 187, 1616–1628. doi: 10.1086/374740
Duffin, P. M., and Seifert, H. S. (2010). DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J. Bacteriol. 192, 4436–4444. doi: 10.1128/JB.00442-10
Elkins, C., Thomas, C. E., Seifert, H. S., and Sparling, P. F. (1991). Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J. Bacteriol. 173, 3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991
Goodman, S. D., and Scocca, J. J. (1988). Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U.S.A. 85, 6982–6986. doi: 10.1073/pnas.85.18.6982
Grijpstra, J., Arenas, J., Rutten, L., and Tommassen, J. (2013). Autotransporter secretion: varying on a theme. Res. Microbiol. 164, 562–582. doi: 10.1016/j.resmic.2013.03.010
Jennings, M. P., Bisercic, M., Dunn, K. L., Virji, M., Martin, A., Wilks, K. E., et al. (1995). Cloning and molecular analysis of the Isi1 (rfaF) gene of Neisseria meningitidis which encodes a heptosyl-2-transferase involved in LPS biosynthesis: evaluation of surface exposed carbohydrates in LPS mediated toxicity for human endothelial cells. Microb. Pathog. 19, 391–407. doi: 10.1006/mpat.1995.0074
Lomholt, H., Poulsen, K., Caugant, D. A., and Kilian, M. (1992). Molecular polymorphism and epidemiology of Neisseria meningitidis immunoglobulin A1 proteases Proc. Natl. Acad. Sci. U.S.A. 89, 2120–2124. doi: 10.1073/pnas.89.6.2120
Maiden, M. C. (2006). Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60, 561–588. doi: 10.1146/annurev.micro.59.030804.121325
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Obergfell, K. P., and Seifert, H. S. (2015). Mobile DNA in the pathogenic Neisseria. Microbiol. Spectr. 3:MDNA3-0015-2014. doi: 10.1128/microbiolspec.MDNA3-0015-2014
Peak, I. R., Srikhanta, Y., Dieckelmann, M., Moxon, E. R., and Jennings, M. P. (2000). Identification and characterisation of a novel conserved outer membrane protein from Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 28, 329–334. doi: 10.1111/j.1574-695X.2000.tb01494.x
Peng, J., Zhang, X., Yang, E., Wang, J., Yang, J., Shao, Z., et al. (2007). Characterization of serogroup C meningococci isolated from 14 provinces of China during 1966-2005 using comparative genomic hybridization. Sci. China C Life Sci. 50, 1–6. doi: 10.1007/s11427-007-0016-2
Rotman, E., and Seifert, H. S. (2014). The genetics of Neisseria species. Annu. Rev. Genet. 48, 405–431. doi: 10.1146/annurev-genet-120213-092007
Roussel-Jazede, V., Arenas, J., Langereis, J. D., Tommassen, J., and van Ulsen, P. (2014). Variable processing of the IgA protease autotransporter at the cell surface of Neisseria meningitidis. Microbiology 160(Pt 11), 2421–2431. doi: 10.1099/mic.0.082511-0
Scarselli, M., Serruto, D., Montanari, P., Capecchi, B., Adu-Bobie, J., Veggi, D., et al. (2006). Neisseria meningitidis NhhA is a multifunctional trimeric autotransporter adhesin. Mol. Microbiol. 61, 631–644. doi: 10.1111/j.1365-2958.2006.05261.x
Serruto, D., Adu-Bobie, J., Scarselli, M., Veggi, D., Pizza, M., Rappuoli, R., et al. (2003). Neisseria meningitidis App, a new adhesin with autocatalytic serine protease activity. Mol. Microbiol. 48, 323–334. doi: 10.1046/j.1365-2958.2003.03420.x
Serruto, D., Spadafina, T., Ciucchi, L., Lewis, L. A., Ram, S., Tontini, M., et al. (2010). Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc. Natl. Acad. Sci. U.S.A. 107, 3770–3775. doi: 10.1073/pnas.0915162107
Shao, Z., Li, W., Ren, J., Liang, X., Xu, L., Diao, B., et al. (2006). Identification of a new Neisseria meningitidis serogroup C clone from Anhui province, China. Lancet 367, 419–423. doi: 10.1016/S0140-6736(06)68141-5
Shao, Z., and Zhu, B. (2016). Meningococcal disease in China. Infect. Dis. Transl. Med. 2, 48–53. doi: 10.11979/idtm.201601007
Sim, R. J., Harrison, M. M., Moxon, E. R., and Tang, C. M. (2000). Underestimation of meningococci in tonsillar tissue by nasopharyngeal swabbing. Lancet 356, 1653–1654. doi: 10.1016/S0140-6736(00)03162-7
St Geme, J. W. III., de la Morena, M. L., and Falkow, S. (1994). A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14, 217–233. doi: 10.1111/j.1365-2958.1994.tb01283.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tettelin, H., Saunders, N. J., Heidelberg, J., Jeffries, A. C., Nelson, K. E., Eisen, J. A., et al. (2000). Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287, 1809–1815. doi: 10.1126/science.287.5459.1809
Turner, D. P., Marietou, A. G., Johnston, L., Ho, K. K., Rogers, A. J., Wooldridge, K. G., et al. (2006). Characterization of MspA, an immunogenic autotransporter protein that mediates adhesion to epithelial and endothelial cells in Neisseria meningitidis. Infect. Immun. 74, 2957–2964. doi: 10.1128/IAI.74.5.2957-2964.2006
Turner, D. P., Wooldridge, K. G., and Ala'Aldeen, D. A. (2002). Autotransported serine protease A of Neisseria meningitidis: an immunogenic, surface-exposed outer membrane, and secreted protein. Infect. Immun. 70, 4447–4461. doi: 10.1128/IAI.70.8.4447-4461.2002
van Ulsen, P., Adler, B., Fassler, P., Gilbert, M., van Schilfgaarde, M., van der Ley, P., et al. (2006). A novel phase-variable autotransporter serine protease, AusI, of Neisseria meningitidis. Microbes Infect. 8, 2088–2097. doi: 10.1016/j.micinf.2006.03.007
van Ulsen, P., and Tommassen, J. (2006). Protein secretion and secreted proteins in pathogenic Neisseriaceae. FEMS Microbiol. Rev. 30, 292–319. doi: 10.1111/j.1574-6976.2006.00013.x
van Ulsen, P., van Alphen, L., ten Hove, J., Fransen, F., van der Ley, P., and Tommassen, J. (2003). A neisserial autotransporter NalP modulating the processing of other autotransporters. Mol. Microbiol. 50, 1017–1030. doi: 10.1046/j.1365-2958.2003.03773.x
Xu, Z., Du, P., Zhu, B., Xu, L., Wang, H., Gao, Y., et al. (2015). Phylogenetic study of clonal complex (CC)198 capsule null locus (cnl) genomes: a distinctive group within the species Neisseria meningitidis. Infect. Genet. Evol. 34, 372–377. doi: 10.1016/j.meegid.2015.07.013
Zhou, H., Gao, Y., Xu, L., Li, M., Li, Q., Li, Y., et al. (2012). Distribution of serogroups and sequence types in disease-associated and carrier strains of Neisseria meningitidis isolated in China between 2003 and 2008. Epidemiol. Infect. 140, 1296–1303. doi: 10.1017/S0950268811001865
Zhu, B., Xu, Z., Du, P., Xu, L., Sun, X., Gao, Y., et al. (2015). Sequence type 4821 clonal complex serogroup B Neisseria meningitidis in China, 1978-2013. Emerg. Infect. Dis. 21, 925–932. doi: 10.3201/eid2106.140687
Keywords: autB, Neisseria meningitidis, autotransporter, characteristic, distribution, DNA uptake sequence
Citation: Zhang A, Zhao P, Zhu B, Shi F, Xu L, Gao Y, Xie N and Shao Z (2017) Characterization and Distribution of the autB Gene in Neisseria meningitidis. Front. Cell. Infect. Microbiol. 7:436. doi: 10.3389/fcimb.2017.00436
Received: 27 June 2017; Accepted: 21 September 2017;
Published: 06 October 2017.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Jing-Ren Zhang, Tsinghua University, ChinaJianping Cao, Chinese Center for Disease Control and Prevention, China
Xinxiang Huang, Jiangsu University, China
Copyright © 2017 Zhang, Zhao, Zhu, Shi, Xu, Gao, Xie and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhujun Shao, c2hhb3podWp1bkBpY2RjLmNu
 Aiyu Zhang
Aiyu Zhang Pan Zhao1
Pan Zhao1 Zhujun Shao
Zhujun Shao