- 1Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL, United States
- 2Department of Biology, Texas A&M University, College Station, TX, United States
Clostridioides difficile is a Gram-positive, spore-forming, toxin-producing anaerobe, and an important nosocomial pathogen. Due to the strictly anaerobic nature of the vegetative form, spores are the main morphotype of infection and transmission of the disease. Spore formation and their subsequent germination play critical roles in C. difficile infection (CDI) progress. Under suitable conditions, C. difficile spores will germinate and outgrow to produce the pathogenic vegetative form. During CDI, C. difficile produces toxins (TcdA and TcdB) that are required to initiate the disease. Meanwhile, it also produces spores that are responsible for the persistence and recurrence of C. difficile in patients. Recent studies have shed light on the regulatory mechanisms of C. difficile sporulation and germination. This review is to summarize recent advances on the regulation of sporulation/germination in C. difficile and the corresponding therapeutic strategies that are aimed at these important processes.
Introduction
Clostridioides difficile (formerly Clostridium difficile; Lawson et al., 2016; Oren and Garrity, 2016) is a Gram-positive, spore-forming, toxin-producing, anaerobic bacterium which has established itself as a leading cause of nosocomial antibiotic-associated diarrhea in the developed countries (Sebaihia et al., 2006). It is found widely in the mammalian gastrointestinal (GI) tract and can cause toxin-mediated C. difficile infections (CDI) that range from mild diarrhea to pseudomembranous colitis and potential death (Lessa et al., 2012). C. difficile causes over 500,000 infections per year in the United States alone, resulting in an estimated 29,000 deaths and an estimated cost of $1–3 billion (Dubberke and Olsen, 2012; Lessa et al., 2015). Currently, antibiotics are the standard treatments for CDI (i.e., vancomycin, metronidazole, or fidaxomicin; Evans and Safdar, 2015). Though effective, CDI recurrence after the initial treatment can still reach up to 15–35% in treated patients (Leffler and Lamont, 2015). Though recurrence is not fully understood, one of the reasons for high recurrence rate is that C. difficile spores may still be present within the patients gut and germinate to the vegetative form after completion or discontinuation of antibiotic treatment (Cornely et al., 2012). Meanwhile, poor host immune response to C. difficile and frequent disruption of the normal gut flora may also contribute to the high recurrence rate (Johnson, 2009). Due to the inherent antibiotic resistance of C. difficile cells and high prevalence of CDI in some hospitals, the Centers for Disease Control and Prevention (CDC) has listed C. difficile as “an urgent threat” regarding the antibiotic associated threats to the United States (Centres for Disease Control and Prevention (US), 2013).
Because C. difficile is an obligate anaerobic pathogen, the vegetative cells are unable to survive outside of a host in the aerobic environment. When C. difficile cells meet certain environmental stimuli (e.g., nutrient deprivation, quorum sensing, and other unidentified stress factors), they will initiate a sporulation pathway to produce sufficient dormant spores to survive in extreme situations (Setlow, 2006; Rodriguez-Palacios and LeJeune, 2011; Deakin et al., 2012; Higgins and Dworkin, 2012). C. difficile pathogenesis relies on the formation of aerotolerant dormant spores which allows C. difficile to persist within the host and to disseminate through patient-to-patient contact/environmental contamination (Britton and Young, 2012). In the host GI tract, the dormant spores must germinate from dormancy to form the actively growing vegetative cells which produce the toxins that cause the primary symptoms of the disease. Under suitable conditions, when germinant receptors sense the presence of small molecules (germinants), spore germination will be induced (Sorg and Sonenshein, 2008).
Recent studies have focused on the regulatory mechanisms of C. difficile sporulation/germination to gain insight into these important processes. However, when compared to other well-studied organisms such as Bacillus subtilis and Clostridium perfringens, our knowledge of C. difficile spore biology still lags far behind. In this review, we will discuss recent progresses in the field of C. difficile spore biology, specifically on the sporulation and germination processes and their implications for CDI treatment.
C. difficile Sporulation
Sporulation Program
Though the signals/molecules that trigger C. difficile sporulation have not been identified, based on studies in other organisms, it is likely that environmental stimuli such as nutrient limitation, quorum sensing, and other unidentified stress factors are involved (Higgins and Dworkin, 2012). In fact, though the mechanism is not well-defined, a recent report has suggested that quorum sensing is important for C. difficile spore formation (Darkoh et al., 2016). As described in other spore-forming bacteria (e.g., B. subtilis), the main process of C. difficile sporulation contains four morphogenetic stages (Figure 1; Edwards and McBride, 2014; Gil et al., 2017): (I) an asymmetric septation generates a smaller compartment (SC) and a larger mother cell (MC); (II) the MC engulfs the SC (now the forespore) in a phagocytic-like event resulting in a forespore being wholly contained within the MC's cytoplasm; (III) the spore cortex and coat layers are assembled; (IV) the MC lyses and releases the mature spore into the surrounding environment. Though the mechanisms that initiate spore formation may differ between organisms, the overall spore architecture is conserved among endospore-forming bacteria. Located in the center of the mature spore is the core. The spore core contains the genomic DNA, mRNA, ribosomes, protein, and is very rich in pyridine-2,6-dicarboxylic acid (DPA), commonly as a calcium salt (CaDPA). The spore core is surrounded by an inner membrane, a peptidoglycan-containing germ cell wall, a specialized peptidoglycan-containing cortex, an outer membrane and layers of coat protein (Figure 1; Edwards and McBride, 2014; Gil et al., 2017). In some C. difficile strains, an exosporium layer surrounds the coat, but not all spore-forming bacteria and not all C. difficile strains have this layer (thus this layer is not shown in Figure 1).
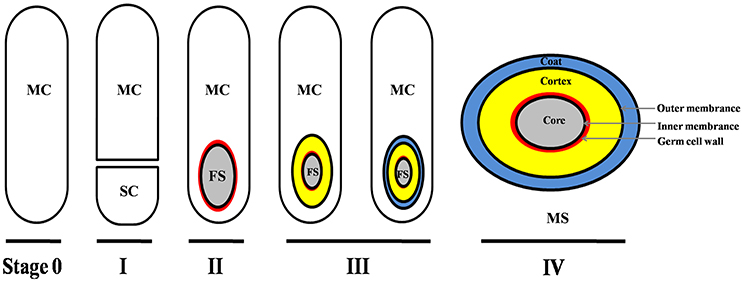
Figure 1. Main morphogenetic stages of the sporulation process and structure of C. difficile spore. This figure was drawn based on the references (Edwards and McBride, 2014; Gil et al., 2017). The main layers of spore structure are shown in this figure (IV). Some C. difficile strains have exosporium, while some have not. The exosporium structure was not shown in this figure. MC, mother cell compartment; SC, smaller compartment; FS, forespore compartment; MS, mature spore.
Regulator CodY and CcpA
Environmental stimuli (e.g., nutrient deprivation or quorum sensing) could trigger C. difficile sporulation. Previous studies in Bacillus and Clostridioides species have revealed that the CodY and CcpA nutritional sensor proteins work as negative regulators of sporulation (Figure 2; Duncan et al., 1995; Hofmeister et al., 1995; Karow et al., 1995; Londoño-Vallejo and Stragier, 1995; Antunes et al., 2012; Nawrocki et al., 2016; Serrano et al., 2016). Among the genes CodY regulates are genes involved in spore formation including spo0A, rapA, rapC, rapE, sinI/R, sigH, and kinB. Recently, Edwards et al. demonstrated that the oligopeptide permease genes app and opp, and the putative sporulation regulator genes sinI and sinR, were regulated by CodY to suppress the initiation of C. difficile sporulation (Edwards et al., 2014). Previous studies indicated that the variability of CodY-dependent regulation is an important contributor to virulence and sporulation in current epidemic isolates (Bennett et al., 2007; Majerczyk et al., 2008; van Schaik et al., 2009). But, to date, the regulatory mechanisms by which CodY affects sporulation are not fully understood because the factors that initiate sporulation in C. difficile are still being identified.
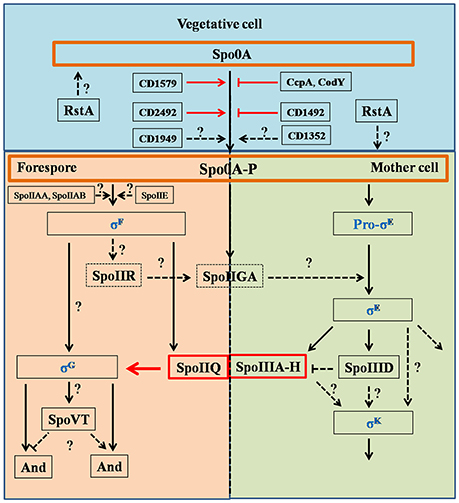
Figure 2. Regulation pathways of C. difficile sporulation. This figure was drawn based on the references (Fimlaid et al., 2013; Fimlaid and Shen, 2015). Regulation of box SpoIIQ, SpoIIIA-H, CD1579 (CD630_15790), CD1492 (CD630_14920), CD2492 (CD630_24920), CcpA, CodY, and RstA were added in this figure based on the recent advances in C. difficile sporulation. CD1492 (CD630_14920), CcpA, and CodY were the negative regulators in sporulation pathways. Function of SpoIIQ-SpoIIIAH complex was characterized by Pinho group, recently (Serrano et al., 2016). Spo0A and Spo0A-P were scheduled in orange boxes, four sigma factors were colored in blue. Dashed boxes indicate that the function of the proteins in regulation pathways has not been identified. Black arrows indicate the regulatory relationship between the factors has been confirmed, dashed arrows indicate the regulatory relationship between the factors has not been tested, red arrows, stops and boxes indicate the function of proteins and the correlation of factors has been confirmed recently. Question marks indicate that there is suggestive, but no conclusive experimental evidence.
CcpA, a LacI family DNA-binding transcriptional regulator, works as a global transcriptional regulator that responds to the availability of carbohydrates (Deutscher et al., 2006). The CcpA sequence and structure are conserved in C. difficile, and has high homology to other pathogens (identity ≥62% analyzed with NCBI website), such as Staphylococcus aureus, Clostridium perfringens, and Clostridium perfringens. CcpA represses the use of alternative carbon sources and positively regulates sugar uptake, fermentation, and amino acid metabolism (Fujita, 2009). In the past few years, CcpA has been shown to regulate several virulence-associated genes. For example, it regulates the expression of the S. aureus α-hemolysin (hla), enterotoxins A, B, and C (sea, seb, and sec) genes, the C. perfringens enterotoxin (cpe) gene, and the Bacillus anthracis atxA and protective antigen (pagA) genes (Varga et al., 2004; Seidl et al., 2006; Chiang et al., 2011). Moreover, CcpA also plays critical role in the control of colonization, antibiotic resistance, and biofilm formation (Seidl et al., 2006; Varga et al., 2008). In C. difficile, CcpA directly regulates the PaLoc genes (tcdR, tcdB, tcdA, and tcdC) to mediate glucose-dependent repression of toxin production and indirectly regulates C. difficile sporulation (Antunes et al., 2011).
Sporulation Progress
Studies have revealed the master transcriptional regulator Spo0A plays the critical role during C. difficile sporulation (Deakin et al., 2012). In all studied endospore-forming bacteria, Spo0A must be phosphorylated (Spo0A-P) by a histidine kinase to become activated. In Bacilli, these histidine kinases (Kin) are found on the plasma membrane and lead to the phosphorylation of Spo0A through Spo0F/Spo0B phosphotransfer system. C. difficile does not encode orthologs of these kinases or the phosphotransfer system. However, previous studies have demonstrated five putative orphan histidine kinases {CD1352 [CD630_13520; cprK (McBride and Sonenshein, 2011)], CD1492 (CD630_14920), CD1579 (CD630_15790), CD1949 (CD630_19490), and CD2492 (CD630_24920)} in C. difficile strain 630 genome that could potentially phosphorylate Spo0A (Figure 2; Underwood et al., 2009). A ClosTron mutation in CD2492 (CD630_24920) resulted in a decreased capacity of the resulting strain to generate spores compared to the WT parent. However, this mutant still generated spores (~4%) suggesting that other histidine kinases can phosphorylate Spo0A or lead to Spo0A phosphorylation (Underwood et al., 2009). In support of this hypothesis, CD1579 (CD630_15790) was shown to autophosphorylate and transfer a phosphate directly to Spo0A (Underwood et al., 2009). Importantly though, the authors did not complement their CD2492 ClosTron mutation, which could have polar effects on downstream genes. In contrast, Childress et al. found that a markerless deletion of CD1492 (CD630_14920) was an inhibitor of sporulation and suppresses spore formation (Childress et al., 2016). This phenotype could be complemented by expression of the wild type allele. Currently, the function of the other putative orphan histidine kinases and their ability to phosphorylate Spo0A are unclear.
Recently, RstA was found to be a novel, positive regulator of sporulation initiation in C. difficile (Figure 2; Edwards et al., 2016). RstA positively affects the initiation of C. difficile sporulation through its peptide-interacting domain (TPR), and negatively regulates toxin production and mobility by affecting the flagellar-specific sigma factor (SigD) expression. But a detailed pathway on the regulation of sporulation initiation by RstA is not fully appreciated. The authors hypothesized that RstA may be a C. difficile global transcriptional regulator, similar to the broad physiological roles that the RNPP (Rap/NprR/PlcR/PrgX) proteins play in other bacteria (Edwards et al., 2016).
Spo0A functions as a critical regulator for sporulation by regulating sporulation-specific RNA polymerase sigma factors, especially for σE, σF, σG, and σK (Fimlaid and Shen, 2015). These σ factors activate compartment-specific transcriptional regulation during B. subtilis sporulation and are also conserved in Clostridium species. σE and σK are MC-specific, and σF and σG are specific to the developing forespore. The sporulation regulatory pathway of sigma factors in C. difficile is illustrated in Figure 2: (1) σF is activated in the forespore soon after polar septation, and it controls early stages of development in this compartment. σF becomes active when the anti-sigma factor SpoIIAB (ADP form) binds to the anti-anti-sigma factor SpoIIAA in its unphosphorylated form, while SpoIIE catalyzes dephosphorylation; (2) σF activity leads to expression of SpoIIR, which interacts with the membrane-bound protease SpoIIGA (SpoIIGA is responsible for the cleavage of pro-σE through trans-septum signaling, yielding active σE); (3) after σF and σE become specifically active in the forespore and MC, respectively, the MC engulfs the forespore; (4) σE activity leads to expression of SpoIIIA-H, which works with σF-controlled SpoIIQ to form a channel in the inner and outer forespore membranes. SpoIIIAH and SpoIIQ localize to the asymmetric septum and the engulfing membranes and interact in the intermembrane space via their extracytoplasmic domains; (5) σE-controlled SpoIIID activates σK in the MC (Haraldsen and Sonenshein, 2003; Fimlaid et al., 2013; Pereira et al., 2013; Paredes-Sabja et al., 2014; Saujet et al., 2014). Though many of the factors that control spore formation are conserved in C. difficile, there are some differences in the sporulation program between C. difficile and B. subtilis. For instance (Figure 2), pro-σk is not encoded by C. difficile, but the mature σk is produced directly in C. difficile, σE activation is dispensable for σG activation, σG activation is dispensable for σK activation, and σK is responsible for transcribing the germinant receptors while σG is responsible in B. subtilis (Fimlaid et al., 2013; Pereira et al., 2013). Importantly, the FS line of gene expression occurs largely independently of the MC line of gene expression. Moreover, σG can be activated before σE and σK, indicating that the order/sequence of sigma factor activation is not as tightly controlled in C. difficile as it is in B. subtilis.
Finally, and in another departure from the model of spore formation in B. subtilis, a recent article by Ribis and colleagues used a TargeTron-based gene disruption demonstrated that the SpoVM protein is not required for spore formation/maturation (Ribis et al., 2017). SpoVM is a small protein that is expressed in the MC that recognizes the positive curvature of outer membrane of the developing forespore and embeds itself there. In B. subtilis, SpoVM recruits the SpoIVA scaffolding protein which polymerizes and surrounds the forespore. Subsequently, the coat is deposited onto the polymerized SpoIVA protein. In C. difficile, a spoVM mutation resulted in a modest defect in spore production (< 5-fold), but their resistance properties are not different from a wildtype spore. This phenotype could be complemented through chromosomal complementation of the wild type allele. However, and importantly, the mutation in spoVM lead to a mislocalization of the coat proteins to one pole of the developing forespore and the coat extended into the MC cytoplasm; SpoIVA still polymerized on the surface of the forespore.
C. difficile Spore Germination
Germination Program
In most organisms, spore germination is induced when specific germinant receptors sense the presence of small molecules (germinants; Setlow, 2003). To date, germination has been most-studied in Bacillus spp. and it contains three main steps (Paredes-Sabja et al., 2011, 2014): (I) germinant (e.g., nucleosides, sugars, amino acids, and/or ions) binding with their cognate Ger-type receptors (GerAA-AB-AC) at the inner spore membrane to trigger the release of monovalent cations (H+, Na+, and K+) and the large amount of CaDPA stored within the core, in exchange for water; (II) CaDPA release and core rehydration leads to the activation of spore cortex lytic enzymes (SCLEs) SleB and CwlJ; (III) activated SleB and CwlJ degrade the peptidoglycan cortex layer, which allows for full core rehydration and resumption of metabolism in the spore core.
Germinant Recognition/Signaling
Germination of C. difficile spores is the first step for initiating CDI. C. difficile spore germination is activated in response to certain host-derived bile salt germinants [e.g., taurocholic acid (TCA)/cholic acid derivatives] and amino acids (e.g., glycine or alanine; Sorg and Sonenshein, 2008). Chenodeoxycholic acid-derivatives (a compound structurally similar to cholic acid but lacking the 12α-hydroxyl group) are competitive inhibitors of cholic acid-mediated germination (Francis et al., 2013b). Though the Ger-type germinant receptors have been widely studied in many organisms, including C. perfringens and Clostridium botulinum/sporogenes, C. difficile does not encode orthologs of the gerA germinant. Instead, C. difficile spores use the subtilisin-like, CspC pseudoprotease as the bile acid germinant receptor (Figure 3; Paredes-Sabja et al., 2008; Francis et al., 2013a, 2015; Wang S. W. et al., 2015; Bhattacharjee et al., 2016; Francis and Sorg, 2016). C. difficile packages three subtilisin-like serine proteases proteins, CspA, CspB, and CspC, into the spore. In C. difficile, CspB, and CspA are encoded as a cspBA gene fusion, where the CspA portion of CspBA lacks an intact catalytic triad (Adams et al., 2013; Kevorkian et al., 2016). The CspBA fusion protein undergoes interdomain cleavage during spore formation, leading to the separation of CspB and CspA, which are transported into the spore by unknown mechanisms. cspC is encoded downstream of cspBA and, similar to cspA, encodes an incomplete catalytic triad. Despite the loss of apparent catalytic activity, cspC (and cspA) important for C. difficile spore germination. Interruption of the cspC coding region through ethyl methanesulfonate (EMS)-generated SNPs and TargeTron methods abrogates spore germination, and certain SNPs in the cspC sequence also affect germinant specificity (Francis et al., 2013a). Similarly, though cspA lacks an intact catalytic triad, cspA is essential for spore germination by controlling the levels of CspC into the developing spore (Francis et al., 2013a; Kevorkian et al., 2016). Only CspB contains an intact catalytic triad and, thus, is hypothesized to be important for activating the SCLE, pro-SleC, to its active, cortex-degrading form (Kevorkian et al., 2016).
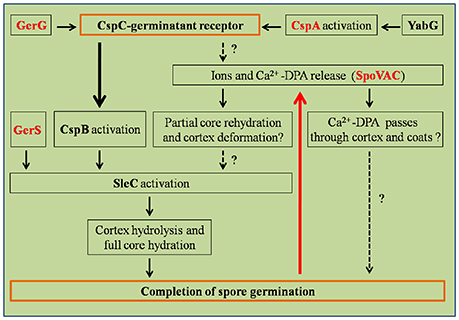
Figure 3. Regulation pathways of C. difficile spore germination. This figure was drawn based on the references (Paredes-Sabja et al., 2011; Fimlaid et al., 2013). Regulation of box GerG, GerS, CspA, and SpoVAC texted in red were drawn in this figure based on the recent advances in C. difficile spore germination. GerS, GerG, and SpoVAC proteins were characterized by Shen group, recently (Fimlaid et al., 2015; Donnelly et al., 2016, 2017). CspC-germinant receptor and completion of germination were scheduled in the orange boxes. Black arrows indicate the regulatory relationship between the factors has been confirmed, dashed arrows indicate the regulatory relationship between the factors has not been tested. Thick black/red arrow indicates central signal pathway in germination progress. Question marks indicate that there is suggestive, but no conclusive experimental evidence.
Activation of the cortex hydrolase SleC depends on the CspB protease, which cleaves the N-terminal pro sequence from the protein. Activated SleC degrades the cortex leading to CaDPA release from the spore core in response to osmotic swelling sensed at the inner spore membrane as a result of cortex degradation (Francis and Sorg, 2016). The osmotic pressure at the inner spore membrane is regulated by SpoVAC (a mechanosensing protein), which allows CaDPA release from the core (Velásquez et al., 2014; Donnelly et al., 2016; Francis and Sorg, 2016). Strikingly, inactivation of either the CspC or SleC inhibited cortex degradation and CaDPA release. These results suggest that the CspC is required for CaDPA release and that cortex degradation precedes CaDPA release, opposite to what occurs in B. subtilis (Francis and Sorg, 2016). These studies suggest that the process of C. difficile spore germination appears to occur in an outside-in manner, while in B. subtilis, the signal appears to travel from the inside-out (Francis and Sorg, 2016).
GerG and GerS Regulators of Spore Germination
Recently, GerG and GerS were identified as important players in C. difficile spore germination (Figure 3; Fimlaid et al., 2015; Donnelly et al., 2017). Donnelly et al. identified the C. difficile-specific protein GerG as an important player in the C. difficile germination process (Donnelly et al., 2017). A deletion of C. difficile gerG resulted in spores with germination defects and reduced responsiveness to bile salt germinants. This phenotype was likely due to the decrease in the incorporation of the CspC, CspB, and CspA germination proteins into spores; this phenotype could be complemented in trans. Similarly, Fimlaid et al. identified another regulator of C. difficile spore germination using TargeTron-based gene disruption (Fimlaid et al., 2015). The GerS lipoprotein functions as a critical regulator in C. difficile spore germination (Fimlaid et al., 2015). In this study, the gerS mutant has a severe germination defect and fails to degrade cortex; this phenotype could be complemented in trans. Interestingly, C. difficile gerS mutant spores still cleave pro-SleC to its active form, suggesting that either cortex is not appropriately modified for SleC-recognition or that SleC is bound to other proteins that GerS regulates (Fimlaid et al., 2015). Importantly, loss of GerS attenuated the C. difficile virulence in the hamster infection model (Fimlaid et al., 2015). Because GerG and GerS are found exclusively in C. difficile, GerG and GerS proteins could be the potential targets to develop C. difficile-specific anti-infective therapies.
Activators and Inhibitors of C. difficile Spore Germination
Bile-acid mediated germination is essential for C. difficile spore germination and CDI in mammalian GI tract. Bile acids are the end products of cholesterol metabolism in liver and are essential for lipoprotein, glucose, drug, and energy metabolism (Chiang, 2009; Howerton et al., 2011). In humans, cholic acid (CA) and chenodeoxycholic acid (CDCA) are two main primary bile acids (PBAs) that are conjugated with either taurine or glycine. Though most of the bile acids secreted into the gut are reabsorbed and recycled back to the liver to be used in other rounds of digestion, some escape hepatic recirculation and enter the large intestine where they become acted upon by the colonic microbiome. Here, the conjugated bile acids become deconjugated due to the action of bile salt hydrolases that are expressed on the cell surfaces of many different bacteria. Subsequently, a small subset of the colonic microbiome will take up and 7α-dehydroxylate the PBAs to form secondary bile acids (SBAs; Ridlon et al., 2006). About 50 different chemically distinct SBAs [e.g., deoxycholate (DCA), lithocholate (LCA), ursodeoxycholate (UDCA), isodeoxycholate (iDCA), and isolithocholate (iLCA)] can be found in human large intestine (Setchell et al., 1983). Recently, Thanissery et al. have analyzed the impact of gut microbial derived SBAs on C. difficile life cycle, specifically, the differences in inhibition efficiency of spore germination, growth, and toxin activity among of DCA, iDCA, LCA, iLCA, UDCA, ωMCA, and HDCA in clinically relevant C. difficile strains R20291 and CD196 (ribotype 027), M68 and CF5 (017), 630 (012), BI9 (001), and M120 (078) (Thanissery et al., 2017). Not surprisingly, the authors found these cholic acid- and chenodeoxycholic acid-derivatives all impacted the C. difficile life cycle; the sensitivity varied by strain and SBA.
Although bile acids are essential to activate C. difficile spore germination, they are not sufficient to activate germination on their own. Amino acid co-germinants are also required for spore germination (Sorg and Sonenshein, 2008; Howerton et al., 2011; Shrestha and Sorg, 2017; Shrestha et al., 2017). However, different amino acids function as co-germinants with different spore germination efficiencies. Glycine is the most effective co-germinant in C. difficile, while alanine is most-often used as co-germinant in B. subtilis and other organisms. In B. subtilis, L-alanine interacts with the GerAA-AB-AC germinant receptor to trigger CaDPA release from the spore core and subsequent cortex hydrolysis. However, D-alanine competitively-inhibits L-alanine-mediated spore germination in B. subtilis (Yasuda and Tochikubo, 1984). In C. difficile, L-alanine can also function as a co-germinant with TCA to stimulate spore germination (Shrestha et al., 2017). Though D-alanine is unable to inhibit L-alanine-mediated C. difficile spore germination, unlike what is observed in B. subtilis, D-alanine can work as a co-germinant to trigger C. difficile spore germination in defined medium (Shrestha and Sorg, 2017; Shrestha et al., 2017). In order for D-alanine to function as a good co-germinant, an alanine racemase (Alr2) should be present in the C. difficile spore. Alr2 interconverts L-alanine and D-alanine (Shrestha et al., 2017). Interestingly, C. difficile Alr2 can also interconvert L- and D-serine, and both of these amino acids can act as co-germinants for C. difficile spore germination (Shrestha et al., 2017). Building on this work, Shrestha et al. found that many different amino acids are co-germinants when tested at 37°C (Shrestha and Sorg, 2017). In this work, two different C. difficile strains responded to a hierarchy of amino acid co-germinants. For UK1 and M68 strains, glycine was the most effective co-germinant (EC50 = ~200 μM) and L-alanine, taurine, and L-glutamine were also good co-germinants (Shrestha and Sorg, 2017). Interestingly, amino acids that regulate important physiological processes were not co-germinants (L-isoleucine, L-leucine and L-valine).
Recently, Kochan et al. identified a critical role for Ca2+ during C. difficile spore germination (Kochan et al., 2017). In their study, they found that C. difficile spores cannot germinate in rich medium supplemented with TCA but without Ca2+, indicating that Ca2+ is indispensable for spore germination. The authors suggested that it works together with glycine to stimulate germination; however, Ca2+ may play a role in the activity of the CspB serine protease, the CspC germinant receptor, the CspA pseudoprotease, or in the activity of the cortex hydrolase. Other subtilisin-like proteases require Ca2+ for activity (Siezen and Leunissen, 1997) and some cortex-degrading enzymes also require Ca2+. Though no Ca2+ was found in the CspB crystal structure, the structures of CspC and CspA have yet to be determined. Thus, Ca2+ may not function as a co-germinant with glycine, but, rather, as an essential cofactor for C. difficile spore germination. However, and importantly, the role of Ca2+ during C. difficile spore germination was also verified in the murine model. Ex vivo assays with mouse ileal contents that were depleted with chelex resin (to remove Ca2+) did not support germination of C. difficile spores (Kochan et al., 2017). This work provided a novel potential strategy for CDI control by modulating intestinal Ca2+ concentration.
In summary, although several main components of spore sporulation/germination machinery of C. difficile have been identified and characterized, several questions remain regarding how C. difficile decides when to enter the sporulation pathway. Moreover, though the Csp pseudoproteases are important for germination, how they interact with and transmit the bile acid signal are still unknown. Further detailed work is necessary to characterize these important aspects of C. difficile physiology.
Treatments of CDI Based on Sporulation/Germination
Currently, the standard treatment of CDI is the use of vancomycin, metronidazole, or fidaxomicin, each of which has some level of recurring disease due to the continued insult to the colonic microbiome and the presence of spores within the colon/environment (Allen et al., 2013). To meet this challenge, non-antibiotic and immune-based therapies against CDI have been developed, such as anti-toxins, vaccines, fecal microbiota transplant (FMT), and anti-germination-based compounds (Gerding et al., 2008; Howerton et al., 2013; Kociolek and Gerding, 2016). Many anti-toxins and vaccines for CDI have been developed in the past two decades (Cox et al., 2013; Monteiro et al., 2013; Mathur et al., 2014; Zhao et al., 2014; Wang Y. K. et al., 2015; Yang et al., 2015; Qiu et al., 2016). Though these treatments can effectively decrease the morbidity and mortality of CDI, most of the anti-toxins and vaccines cannot suppress C. difficile colonization and kill C. difficile spores. Therefore, with these treatments, there are still risks of potential CDI relapse in the host.
Instead of merely neutralizing C. difficile toxins in host, strategies which can directly decrease C. difficile colonization, kill the vegetative cells, and suppress sporulation/germination are desirable treatments for CDI. FMT is an effective strategy to reconstruct the gut microbiota to suppress C. difficile colonization, especially for patients who have multiple bouts of recurring disease and who have failed conventional treatment methods (Borody and Khoruts, 2012; Weingarden et al., 2014; Khoruts and Sadowsky, 2016; Kim et al., 2016). Although FMT is deemed relatively safe and low-cost, the unappealing aesthetics of the procedure is often a concern of patients (Sampath et al., 2013; Varier et al., 2015). Because the C. difficile spore form is necessary for dissemination and persistence, sporulation/germination are critical steps for CDI. Thus, it is worth developing therapeutic strategies for disrupting C. difficile disease transmission and spread according to C. difficile spore biology. Basing on the progress of C. difficile spore germination, the PBA CDCA and secondary bile acids LCA, UDCA, and iLCA are potent inhibitors of C. difficile spore germination (Sorg and Sonenshein, 2010; Zhang and Klaassen, 2010; Heeg et al., 2012). Moreover, several mouse-derived bile acids, such as α-muricholic acid, β-muricholic acid, and ω-muricholic acid inhibit C. difficile spore germination and growth (Francis et al., 2013b). Excitingly, synthesized bile acid analogs, such as CAmSA, methylchenodeoxycholic acid diacetate, and compound 21b (derived from UDCA) have been identified to inhibit C. difficile spore germination (Sorg and Sonenshein, 2009; Howerton et al., 2013; Stoltz et al., 2017). Of these compounds, CAmSA showed promise in inhibiting/delaying C. difficile disease in a mouse model of CDI. These anti-germination-based strategies could work in a couple of different ways. (i) High risk patients who are to be treated with antibiotics could also take an anti-germinant to prevent the germination of spores within the host's gut. This patient continues to take the anti-germinant during and post-antibiotic treatment so that the normal, colonic, microbiome has a chance to repopulate and provide natural protection against CDI. (ii) Patients with CDI could take the recommended course of antibiotics plus the anti-germinants. This strategy would prevent recurring disease by allowing the microbiome to re-establish colonization resistance post-antibiotic treatment. Because both strategies block germination, and thus downstream events (vegetative growth, toxin production, and spore formation), anti-germination therapy would limit the presence of spores within the surrounding environment because C. difficile would not have a chance to expand in population and produce spores. In contrast germination-inducing strategies are a viable option for environmental cleanup; inducing in vivo germination has the potential for toxin-production and, thus, exacerbation of symptoms. Due to the inherent nature of the dormant spore, harsh chemicals (e.g., bleach) are required to clean environmental surfaces. But by germinating the spores in the environment, the germinated spores become susceptible to a wider range of sanitizing agents (Nerandzic and Donskey, 2010, 2013, 2017; Nerandzic et al., 2016). More studies should be investigated for further application of germination inhibitors.
Concluding and Remarks
Although much has been learned about the sporulation/germination processes of C. difficile and the different therapeutic strategies for CDI, many key questions related to regulation pathways of sporulation/germination processes remain unanswered. Thus, much work remains to be done to further understand C. difficile spore biology and develop new efficient approaches for CDI treatment: (1) It is expected that further work will allow us to fully understand the mechanisms of the initiation of sporulation by identifying the proteins that are involved in Spo0A phosphorylation; (2) Due to the relevance of spore germination with CDI progression, it is worth defining how the bile acid germinant receptor, CspC, and the unidentified glycine germinant receptor regulate CaDPA release and cortex degradation; (3) More alternative therapeutic strategies for CDI disease need to be developed based on the knowledge of C. difficile sporulation/germination.
Author Contributions
All authors listed, have made a substantial, direct, and intellectual contribution to the work; DZ wrote and revised this manuscript; JS and XS revised this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by National Institutes of Health grants (K01-DK092352, R21-AI113470, R03-DK112004, R01-AI132711) to XS, and was also supported by awards 5R01AI116895 and 1U01AI124290 to JS from the National Institute of Allergy and Infectious Diseases.
References
Adams, C. M., Eckenroth, B. E., Putnam, E. E., Doublie, S., and Shen, A. (2013). Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog. 9:e1003165. doi: 10.1371/journal.ppat.1003165
Allen, C. A., Babakhani, F., Sears, P., Nguyen, L., and Sorg, J. A. (2013). Both fidaxomicin and vancomycin inhibit outgrowth of Clostridium difficile spores. Antimicrob. Agents Ch. 57, 664–667. doi: 10.1128/AAC.01611-12
Antunes, A., Martin-Verstraete, I., and Dupuy, B. (2011). CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 79, 882–899. doi: 10.1111/j.1365-2958.2010.07495.x
Antunes, A., Camiade, E., Monot, M., Courtois, E., Barbut, F., Sernova, N. V., et al. (2012). Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 40, 10701–10718. doi: 10.1093/nar/gks864
Bennett, H. J., Pearce, D. M., Glenn, S., Taylor, C. M., Kuhn, M., Sonenshein, A. L., et al. (2007). Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63, 1453–1467. doi: 10.1111/j.1365-2958.2007.05597.x
Bhattacharjee, D., McAllister, K. N., and Sorg, J. A. (2016). Germinants and their receptors in Clostridia. J. Bacteriol. 198, 2767–2775. doi: 10.1128/JB.00405-16
Borody, T. J., and Khoruts, A. (2012). Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol. 9, 88–96. doi: 10.1038/nrgastro.2011.244
Britton, R. A., and Young, V. B. (2012). Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol. 20, 313–319. doi: 10.1016/j.tim.2012.04.001
Centres for Disease Control and Prevention (US) (2013). Antibiotic Resistance Threats in the United States, 2013. Atlanta, GA: Centres for Disease Control and Prevention, US Department of Health and Human Services.
Chiang, C., Bongiorni, C., and Perego, M. (2011). Glucose-dependent Aactivation of Bacillus anthracis toxin gene expression and virulence requires the carbon catabolite protein CcpA. J. Bacteriol. 193, 52–62. doi: 10.1128/JB.01656-09
Chiang, J. Y. (2009). Bile acids: regulation of synthesis. J. Lipid Res. 50, 1955–1966. doi: 10.1194/jlr.R900010-JLR200
Childress, K. O., Edwards, A. N., Nawrocki, K. L., Anderson, S. E., Woods, E. C., and McBride, S. M. (2016). The phosphotransfer protein CD1492 represses sporulation initiation in Clostridium difficile. Infect. Immun. 84, 3434–3444. doi: 10.1128/IAI.00735-16
Cornely, O. A., Miller, M. A., Louie, T. J., Crook, D. W., and Gorbach, S. L. (2012). Treatment of first recurrence of Clostridium difficile infection: fidaxomicin versus vancomycin. Clin. Infect. Dis. 55, S154–S161. doi: 10.1093/cid/cis462
Cox, A. D., St Michael, F., Aubry, A., Cairns, C. M., Strong, P. C., Hayes, A. C., et al. (2013). Investigating the candidacy of a lipoteichoic acid-based glycoconjugate as a vaccine to combat Clostridium difficile infection. Glycoconjugate J. 30, 843–855. doi: 10.1007/s10719-013-9489-3
Darkoh, C., Odo, C., and DuPont, H. L. (2016). Accessory gene regulator-1 locus is essential for virulence and pathogenesis of Clostridium difficile. Mbio 7:e01237. doi: 10.1128/mBio.01237-16
Deakin, L. J., Clare, S., Fagan, R. P., Dawson, L. F., Pickard, D. J., West, M. R., et al. (2012). The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 80, 2704–2711. doi: 10.1128/IAI.00147-12
Deutscher, J., Francke, C., and Postma, P. W. (2006). How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. R. 70, 939–1031. doi: 10.1128/MMBR.00024-06
Donnelly, M. L., Fimlaid, K. A., and Shen, A. (2016). Characterization of Clostridium difficile spores lacking either SpoVAC or dipicolinic acid synthetase. J. Bacteriol. 198, 1694–1707. doi: 10.1128/JB.00986-15
Donnelly, M. L., Li, W., Li, Y. Q., Hinkel, L., Setlow, P., and Shen, A. (2017). A Clostridium difficile-specific, gel-forming protein required for optimal spore germination. Mbio 8:e02085–16. doi: 10.1128/mBio.02085-16
Dubberke, E. R., and Olsen, M. A. (2012). Burden of Clostridium difficile on the healthcare system. Clin. Infect. Dis. 55, S88–SS92. doi: 10.1093/cid/cis335
Duncan, L., Alper, S., Arigoni, F., Losick, R., and Stragier, P. (1995). Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science 270, 641–644. doi: 10.1126/science.270.5236.641
Edwards, A. N., and McBride, S. M. (2014). Initiation of sporulation in Clostridium difficile: a twist on the classic model. FEMS Microbiol. Lett. 358, 110–118. doi: 10.1111/1574-6968.12499
Edwards, A. N., Nawrocki, K. L., and McBride, S. M. (2014). Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect. Immun. 82, 4276–4291. doi: 10.1128/IAI.02323-14
Edwards, A. N., Tamayo, R., and McBride, S. M. (2016). A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol. Microbiol. 100, 954–971. doi: 10.1111/mmi.13361
Evans, C. T., and Safdar, N. (2015). Current trends in the epidemiology and outcomes of Clostridium difficile infection. Clin. Infect. Dis. 60, S66–S71. doi: 10.1093/cid/civ140
Fimlaid, K. A., and Shen, A. (2015). Diverse mechanisms regulate sporulation sigma factor activity in the Firmicutes. Curr. Opin. Microbiol. 24, 88–95. doi: 10.1016/j.mib.2015.01.006
Fimlaid, K. A., Bond, J. P., Schutz, K. C., Putnam, E. E., Leung, J. M., Lawley, T. D., et al. (2013). Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet. 9:e1003660. doi: 10.1371/journal.pgen.1003660
Fimlaid, K. A., Jensen, O., Donnelly, M. L., Francis, M. B., Sorg, J. A., and Shen, A. (2015). Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog. 11:e1005239. doi: 10.1371/journal.ppat.1005239
Francis, M. B., and Sorg, J. A. (2016). Dipicolinic acid release by germinating Clostridium difficile spores occurs through a mechanosensing mechanism. Msphere 1:e00306–16. doi: 10.1128/mSphere.00306-16
Francis, M. B., Allen, C. A., Shrestha, R., and Sorg, J. A. (2013a). Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 9:e1003356. doi: 10.1371/journal.ppat.1003356
Francis, M. B., Allen, C. A., and Sorg, J. A. (2013b). Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS ONE 8:e7365310. doi: 10.1371/journal.pone.0073653
Francis, M. B., Allen, C. A., and Sorg, J. A. (2015). Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J. Bacteriol. 197, 2276–2283. doi: 10.1128/JB.02575-14
Fujita, Y. (2009). Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73, 245–259. doi: 10.1271/bbb.80479
Gerding, D. N., Muto, C. A., and Owens, R. C. (2008). Treatment of Clostridium difficile infection. Clin. Infect. Dis. 46, S32–S42. doi: 10.1086/521860
Gil, F., Lagos-Moraga, S., Calderon-Romero, P., Pizarro-Guajardo, M., and Paredes-Sabja, D. (2017). Updates on Clostridium difficile spore biology. Anaerobe 45, 3–9. doi: 10.1016/j.anaerobe.2017.02.018
Haraldsen, J. D., and Sonenshein, A. L. (2003). Efficient sporulation in Clostridium difficile requires disruption of the σK gene. Mol. Microbiol. 48, 811–821. doi: 10.1046/j.1365-2958.2003.03471.x
Heeg, D., Burns, D. A., Cartman, S. T., and Minton, N. P. (2012). Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS ONE 7:e32381. doi: 10.1371/journal.pone.0032381
Higgins, D., and Dworkin, J. (2012). Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 36, 131–148. doi: 10.1111/j.1574-6976.2011.00310.x
Hofmeister, A. E., Londonovallejo, A., Harry, E., Stragier, P., and Losick, R. (1995). Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Cell 83, 219–226. doi: 10.1016/0092-8674(95)90163-9
Howerton, A., Ramirez, N., and Abel-Santos, E. (2011). Mapping interactions between germinants and Clostridium difficile spores. J. Bacteriol. 193, 274–282. doi: 10.1128/JB.00980-10
Howerton, A., Patra, M., and Abel-Santos, E. (2013). A new strategy for the prevention of Clostridium difficile infection. J. Infect. Dis. 207, 1498–1504. doi: 10.1093/infdis/jit068
Johnson, S. (2009). Recurrent Clostridium difficile infection: a review of risk factors, treatments, and outcomes. J. Infect. 58, 403–410. doi: 10.1016/j.jinf.2009.03.010
Karow, M. L., Glaser, P., and Piggot, P. J. (1995). Identification of a gene, spoIIR that links the activation of σE to the transcriptional activity of σF during sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 92, 2012–2016. doi: 10.1073/pnas.92.6.2012
Kevorkian, Y., Shirley, D. J., and Shen, A. (2016). Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie 122, 243–254. doi: 10.1016/j.biochi.2015.07.023
Khoruts, A., and Sadowsky, M. J. (2016). Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 13, 508–516. doi: 10.1038/nrgastro.2016.98
Kim, H. B., Wang, Y., and Sun, X. (2016). A detrimental role of immunosuppressive drug, dexamethasone, during Clostridium difficile infection in association with a gastrointestinal microbial shift. J. Microbiol. Biotechnol. 26, 567–571. doi: 10.4014/jmb.1512.12017
Kochan, T. J., Somers, M. J., Kaiser, A. M., Shoshiev, M. S., Hagan, A. K., Hastie, J. L., et al. (2017). Intestinal calcium and bile salts facilitate germination of Clostridium difficile spores. PLoS Pathog. 13:e1006443. doi: 10.1371/journal.ppat.1006443
Kociolek, L. K., and Gerding, D. N. (2016). Breakthroughs in the treatment and prevention of Clostridium difficile infection. Nat. Rev. Gastroenterol. Hepatol. 13, 150–160. doi: 10.1038/nrgastro.2015.220
Lawson, P. A., Citron, D. M., Tyrrell, K. L., and Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe 40, 95–99. doi: 10.1016/j.anaerobe.2016.06.008
Leffler, D. A., and Lamont, J. T. (2015). Clostridium difficile infection. New Engl. J. Med. 373, 287–288. doi: 10.1056/NEJMra1403772
Lessa, F. C., Gould, C. V., and McDonald, L. C. (2012). Current status of Clostridium difficile infection epidemiology. Clin. Infect. Dis. 55, S65–S70. doi: 10.1093/cid/cis319
Lessa, F. C., Winston, L. G., McDonald, L. C., and Difficil, E. I. P. C. (2015). Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 2369–2370. doi: 10.1056/NEJMoa1408913
Londoño-Vallejo, J. A., and Stragier, P. (1995). Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Gene Dev. 9, 503–508. doi: 10.1101/gad.9.4.503
Majerczyk, C. D., Sadykov, M. R., Luong, T. T., Lee, C., Somerville, G. A., and Sonenshein, A. L. (2008). Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265. doi: 10.1128/JB.01545-07
Mathur, H., Rea, M. C., Cotter, P. D., Ross, R. P., and Hill, C. (2014). The potential for emerging therapeutic options for Clostridium difficile infection. Gut Microbes 5, 696–710. doi: 10.4161/19490976.2014.983768
McBride, S. M., and Sonenshein, A. L. (2011). Identification of a genetic locus responsible for antimicrobial peptide resistance in Clostridium difficile. Infect. Immun. 79, 167–176. doi: 10.1128/IAI.00731-10
Monteiro, M. A., Ma, Z. C., Bertolo, L., Jiao, Y., Arroyo, L., Hodgins, D., et al. (2013). Carbohydrate-based Clostridium difficile vaccines. Expert Rev. Vaccines 12, 421–431. doi: 10.1586/erv.13.9
Nawrocki, K. L., Edwards, A. N., Daou, N., Bouillaut, L., and McBride, S. M. (2016). CodY-dependent regulation of sporulation in Clostridium difficile. J. Bacteriol. 198, 2113–2130. doi: 10.1128/JB.00220-16
Nerandzic, M. M., and Donskey, C. J. (2010). Triggering germination represents a novel strategy to enhance killing of Clostridium difficile spores. PLoS ONE 5:e12285. doi: 10.1371/journal.pone.0012285
Nerandzic, M. M., and Donskey, C. J. (2013). Activate to eradicate: inhibition of Clostridium difficile spore outgrowth by the synergistic effects of osmotic activation and nisin. PLoS ONE 8:e54740. doi: 10.1371/journal.pone.0054740
Nerandzic, M. M., and Donskey, C. J. (2017). Sensitizing Clostridium difficile spores with germinants on skin and environmental surfaces represents a new strategy for reducing spores via ambient mechanisms. Pathog. Immun. 2, 404–421 doi: 10.20411/pai.v2i3.221
Nerandzic, M. M., Sankar, C. T., Setlow, P., and Donskey, C. J. (2016). A cumulative spore killing approach: synergistic sporicidal activity of dilute peracetic acid and ethanol at low pH against Clostridium difficile and Bacillus subtilis spores. Open Forum. Infect. Dis. 3:ofv206. doi: 10.1093/ofid/ofv206
Oren, A., and Garrity, G. M. (2016). Notification of changes in taxonomic opinion previously published outside the IJSEM. Int. J. Syst. Evol. Microbiol. 66, 2469–2470. doi: 10.1099/ijsem.0.001150
Paredes-Sabja, D., Torres, J. A., Setlow, P., and Sarker, M. R. (2008). Clostridium perfringens spore germination: characterization of germinants and their receptors. J. Bacteriol. 190, 1190–1201. doi: 10.1128/JB.01748-07
Paredes-Sabja, D., Setlow, P., and Sarker, M. R. (2011). Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19, 85–94. doi: 10.1016/j.tim.2010.10.004
Paredes-Sabja, D., Shen, A., and Sorg, J. A. (2014). Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 22, 406–416. doi: 10.1016/j.tim.2014.04.003
Pereira, F. C., Saujet, L., Tome, A. R., Serrano, M., Monot, M., Couture-Tosi, E., et al. (2013). The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet. 9:e1003782. doi: 10.1371/journal.pgen.1003782
Qiu, H. Y., Cassan, R., Johnstone, D., Han, X. B., Joyee, A. G., McQuoid, M., et al. (2016). Novel Clostridium difficile anti-toxin (TcdA and TcdB) humanized monoclonal antibodies demonstrate in vitro neutralization across a broad spectrum of clinical strains and in vivo potency in a hamster spore challenge model. PLoS ONE 11:e0157970. doi: 10.1371/journal.pone.0157970
Ribis, J. W., Ravichandran, P., Putnam, E. E., Pishdadian, K., and Shen, A. (2017). The conserved spore coat protein SpoVM is largely dispensable in Clostridium difficile spore formation. Msphere 2:e00315–17. doi: 10.1128/mSphere.00315-17
Ridlon, J. M., Kang, D. J., and Hylemon, P. B. (2006). Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259. doi: 10.1194/jlr.R500013-JLR200
Rodriguez-Palacios, A., and LeJeune, J. T. (2011). Moist-heat resistance, spore aging, and superdormancy in Clostridium difficile. Appl. Environ. Microbiol. 77, 3085–3091. doi: 10.1128/AEM.01589-10
Sampath, K., Levy, L. C., and Gardner, T. B. (2013). Fecal transplantation: beyond the aesthetic. Gastroenterology 145, 1151–1153. doi: 10.1053/j.gastro.2013.09.015
Saujet, L., Pereira, F. C., Henriques, A. O., and Martin-Verstraete, I. (2014). The regulatory network controlling spore formation in Clostridium difficile. FEMS Microbiol. Lett. 358, 1–10. doi: 10.1111/1574-6968.12540
Sebaihia, M., Wren, B. W., Mullany, P., Fairweather, N. F., Minton, N., Stabler, R., et al. (2006). The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38, 779–786. doi: 10.1038/ng1830
Seidl, K., Stucki, M., Ruegg, M., Goerke, C., Wolz, C., Harris, L., et al. (2006). Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents. Chemother. 50, 1183–1194. doi: 10.1128/AAC.50.4.1183-1194.2006
Serrano, M., Crawshaw, A. D., Dembek, M., Monteiro, J. M., Pereira, F. C., Pinho, M. G., et al. (2016). The SpoIIQ-SpoIIIAH complex of Clostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis. Mol. Microbiol. 100, 204–228. doi: 10.1111/mmi.13311
Setchell, K. D., Lawson, A. M., Tanida, N., and Sjovall, J. (1983). General-methods for the analysis of metabolic profiles of bile-acids and related-compounds in feces. J. Lipid Res. 24, 1085–1100.
Setlow, P. (2003). Spore germination. Curr. Opin. Microbiol. 6, 550–556. doi: 10.1016/j.mib.2003.10.001
Setlow, P. (2006). Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101, 514–525. doi: 10.1111/j.1365-2672.2005.02736.x
Shrestha, R., and Sorg, J. A. (2017). Hierarchical recognition of amino acid co-germinants during Clostridioides difficile spore germination. Anaerobe 49, 41–47. doi: 10.1016/j.anaerobe.2017.12.001
Shrestha, R., Lockless, S. W., and Sorg, J. A. (2017). A Clostridium difficile alanine racemase affects spore germination and accommodates serine as a substrate. J. Biol. Chem. 292, 10735–10742. doi: 10.1074/jbc.M117.791749
Siezen, R. J., and Leunissen, J. A. (1997). Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 6, 501–523. doi: 10.1002/pro.5560060301
Sorg, J. A., and Sonenshein, A. L. (2008). Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190, 2505–2512. doi: 10.1128/JB.01765-07
Sorg, J. A., and Sonenshein, A. L. (2009). Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 191, 1115–1117. doi: 10.1128/JB.01260-08
Sorg, J. A., and Sonenshein, A. L. (2010). Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 192, 4983–4990. doi: 10.1128/JB.00610-10
Stoltz, K. L., Erickson, R., Staley, C., Weingarden, A. R., Romens, E., Steer, C. J., et al. (2017). Synthesis and biological evaluation of bile acid analogues inhibitory to Clostridium difficile spore germination. J. Med. Chem. 60, 3451–3471. doi: 10.1021/acs.jmedchem.7b00295
Thanissery, R., Winston, J. A., and Theriot, C. M. (2017). Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45, 86–100. doi: 10.1016/j.anaerobe.2017.03.004
Underwood, S., Guan, S., Vijayasubhash, V., Baines, S. D., Graham, L., Lewis, R. J., et al. (2009). Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 191, 7296–7305. doi: 10.1128/JB.00882-09
van Schaik, W., Chateau, A., Dillies, M. A., Coppee, J. Y., Sonenshein, A. L., and Fouet, A. (2009). The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infect. Immun. 77, 4437–4445. doi: 10.1128/IAI.00716-09
Varga, J., Stirewalt, V. L., and Melville, S. B. (2004). The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 186, 5221–5229. doi: 10.1128/JB.186.16.5221-5229.2004
Varga, J. J., Therit, B., and Melville, S. B. (2008). Type IV Pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infect. Immun. 76, 4944–4951. doi: 10.1128/IAI.00692-08
Varier, R. U., Biltaji, E., Smith, K. J., Roberts, M. S., Kyle Jensen, M., LaFleur, J., et al. (2015). Cost-effectiveness analysis of fecal microbiota transplantation for recurrent Clostridium difficile infection. Infect. Control Hosp. Epidemiol. 36, 438–444. doi: 10.1017/ice.2014.80
Velásquez, J., Schuurman-Wolters, G., Birkner, J. P., Abee, T., and Poolman, B. (2014). Bacillus subtilis spore protein SpoVAC functions as a mechanosensitive channel. Mol. Microbiol. 92, 813–823. doi: 10.1111/mmi.12591
Wang, S., Shen, A., Setlow, P., and Li, Y. Q. (2015). Characterization of the dynamic germination of individual Clostridium difficile spores using raman spectroscopy and differential interference contrast microscopy. J. Bacteriol. 197, 2361–2373. doi: 10.1128/JB.00200-15
Wang, Y. K., Yan, Y. X., Kim, H. B., Ju, X. H., Zhao, S., Zhang, K., et al. (2015). A chimeric protein comprising the glucosyltransferase and cysteine proteinase domains of toxin B and the receptor binding domain of toxin A induces protective immunity against Clostridium difficile infection in mice and hamsters. Hum. Vacc. Immunother. 11, 2215–2222. doi: 10.1080/21645515.2015.1052352
Weingarden, A. R., Chen, C., Bobr, A., Yao, D., Lu, Y. W., Nelson, V. M., et al. (2014). Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest Liver Physiol. 306, G310–G319. doi: 10.1152/ajpgi.00282.2013
Yang, Z. Y., Ramsey, J., Hamza, T., Zhang, Y. R., Li, S., Yfantis, H. G., et al. (2015). Mechanisms of protection against Clostridium difficile infection by the monoclonal antitoxin antibodies actoxumab and bezlotoxumab. Infect. Immun. 83, 822–831. doi: 10.1128/IAI.02897-14
Yasuda, Y., and Tochikubo, K. (1984). Relation between D-glucose and L-alanine and D-alanine in the initiation of germination of Bacillus subtilis spore. Microbiol. Immunol. 28, 197–207. doi: 10.1111/j.1348-0421.1984.tb00671.x
Zhang, Y., and Klaassen, C. D. (2010). Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 51, 3230–3242. doi: 10.1194/jlr.M007641
Keywords: C. difficile, spores, germination, CDI, sporulation
Citation: Zhu D, Sorg JA and Sun X (2018) Clostridioides difficile Biology: Sporulation, Germination, and Corresponding Therapies for C. difficile Infection. Front. Cell. Infect. Microbiol. 8:29. doi: 10.3389/fcimb.2018.00029
Received: 30 October 2017; Accepted: 23 January 2018;
Published: 08 February 2018.
Edited by:
Nathan W. Schmidt, University of Louisville, United StatesReviewed by:
Paul Edward Carlson, Food and Drug Administration, United StatesPeter Mullany, University College London, United Kingdom
Copyright © 2018 Zhu, Sorg and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingmin Sun, c3VuNUBoZWFsdGgudXNmLmVkdQ==
 Duolong Zhu
Duolong Zhu Joseph A. Sorg
Joseph A. Sorg Xingmin Sun
Xingmin Sun