- 1Department of Health Sciences, Università degli Studi di Milano, Milan, Italy
- 2Institute of Biomedical Technologies, National Research Council, Segrate, Italy
- 3Department of Pediatrics, San Paolo Hospital, Università degli Studi di Milano, Milan, Italy
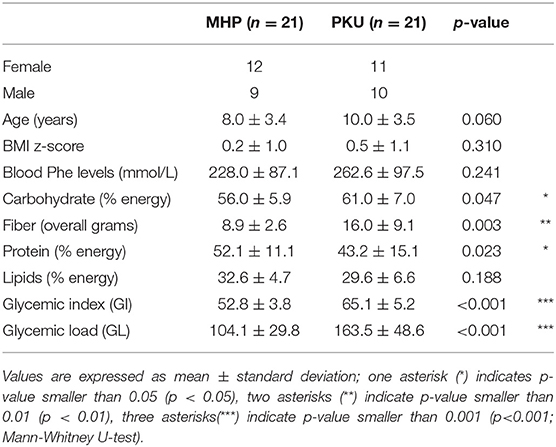
Low-phenylalanine diet, the mainstay of treatment for phenylketonuria (PKU), has been shown to increase glycemic index and glycemic load, affecting the availability of substrates for microbial fermentation. Indeed, changes in the PKU gut microbiota compared with healthy controls have been previously reported. In this study we compared the gut microbial communities of children with PKU and with mild hyperphenylalaninemia (MHP, unrestricted diet). For each group, we enrolled 21 children (4–18 years old), for a total dataset of 42 subjects. We assessed dietary intake and performed gut microbiota analysis by sequencing the V3–V4 hypervariable regions of the 16S rRNA gene. Short chain fatty acids (SCFAs) were quantified by gas chromatographic analysis. While alpha-diversity analysis showed no significant differences between PKU and MHP groups, microbial community analysis highlighted a significant separation of the gut microbiota according to both unweighted (p = 0.008) and weighted Unifrac distances (p = 0.033). Major differences were seen within the Firmicutes phylum. Indeed, PKU children were depleted in Faecalibacterium spp. and enriched in Blautia spp. and Clostridium spp (family Lachnospiraceae). We found a divergent response of members of the Firmicutes phylum with respect to daily glycemic index, higher in PKU children. Faecalibacterium prausnitzii, unclassified Ruminococcaceae and, to a lesser extent Roseburia spp. negatively correlated with glycemic index, whereas unclassified Lachnospiraceae were positively associated. Indicator species analysis suggested F. prausnitzii be related to MHP status and Ruminococcus bromii to be associated with PKU. Despite PKU children having a higher vegetable and fiber intake, resembling a vegan diet, their gut microbial profile is different from the microbiota reported in the literature for individuals consuming a high-fiber/low-protein diet. Indeed, beneficial microorganisms, such as F. prausnitzii, considered a biomarker for a healthy status and one of the main butyrate producers, are depleted in PKU gut microbiota. We suggest that both the quality and quantity of carbohydrates ingested participate in determining the observed Firmicutes shifts on the PKU population.
Introduction
Phenylketonuria (PKU; OMIM 261600) is an inherited metabolic disorder caused by a mutation in the phenylalanine hydroxylase enzyme (PAH), which converts phenylalanine (Phe) into tyrosine. As PAH activity is hampered, phenylalanine accumulates in the blood and becomes toxic to the brain (Williams et al., 2008). Allelic heterogeneity at PAH locus results in a variety of metabolic phenotypes, ranging from mild, moderate and classical PKU (blood Phe levels >360 μmol/L) to mild hyperphenylalaninemia (MHP, blood Phe levels ranging 120–360 μmol/L; Güttler and Guldberg, 1996).
Untreated PKU leads to neurodevelopmental damage and behavioral problems that are preventable by early diagnosis and dietary treatment (van Spronsen et al., 2017).
A PKU diet, started in the neonatal period and followed life-long, is characterized by low-protein natural foods (vegetables, fruits) and special low-protein products, which are low-protein variants of some foods (bread, pasta, and biscuits; Giovannini et al., 2012). Adequate protein intake is guaranteed by Phe-free amino acid mixtures (AAM) with a balanced content of amino acids and micronutrients. Despite improvements in taste, the palatability of such formulas is still less than optimal, often resulting in poor acceptance by school-aged patients. Moreover, a PKU diet has been shown to increase glycemic index and glycemic load (Moretti et al., 2017), probably due to special low-protein products frequently being enriched in sugars.
Considering the crucial role of diet in shaping the gut microbiota, i.e., the microbial community inhabiting gastrointestinal tract (Albenberg and Wu, 2014), it is not surprising that such a peculiar diet leads to microbial changes in phenylketonuric patients (Pinheiro de Oliveira et al., 2016; Verduci et al., 2018). Alterations in the gut microbiota, in turn, may influence gastrointestinal homeostasis, predispose to chronic inflammation and modulate other metabolic functions through gut-liver axis and gut-brain axis (Nieuwdorp et al., 2014).
Up to date, only a study by Pinheiro de Oliveira et al. (2016) has investigated by 16S rRNA sequencing the gut community of PKU patients. However, the small cohort (eight patients vs. ten healthy controls) and the presence of confounding factors (i.e., antibiotic treatment, age <1 year) might have mitigated the observed microbial alterations within the gut. Moreover, whether reported changes in the gut microbiota represent an effect of the disease itself or a consequence of the modified diet is still unclear.
To this end, this work aims at elucidating, by comparing the microbiota of PKU children with mild hyperphenylalaninemia, the role of the low-Phe diet as potential inducer of microbial dysbiosis.
Materials and Methods
Subject Recruitment and Sampling
A total of 42 children (21 PKU/21 MHP) were enrolled in the study at the Pediatric Department of San Paolo Hospital in Milan, Italy (Verduci et al., 2018). Inclusion criteria were: gestational age 37–42 week inclusive, Caucasian, living in Northern Italy, birth weight ≥2,500 g, single birth, diagnosis of PKU or MHP due to PAH deficiency. Exclusion criteria were: congenital malformation, endocrine disorders, chronic liver diseases, chronic or acute intestinal diseases, treatments with antibiotic, and probiotic/prebiotic (including glycomacropeptide) in the 3 months preceding the study. All PKU subjects started the diet therapy at disease diagnosis, usually within 10 days from birth.
Blood phenylalanine concentration was monthly monitored by the Guthrie test (Guthrie and Susi, 1963).
From all subjects we collected: anthropometric data (height, weight and z-score body mass index), dietary habits and stool samples, stored at −80°C until use. Three-days food diaries were filled out by a parent for each enrolled subject and processed by dieticians to calculate the average amounts of energy and nutrient intake (carbohydrates, soluble and insoluble fibers, lipids, proteins) using a commercially available software (MetaDieta®, Software version 3.1, ME.TE.DA S.r.l., San Benedetto del Tronto, Italy). For each meal, the glycemic index (GI) value and glycemic load (GL) were calculated as described by Verduci et al. (2018) using the following formulas:
GImeal = (∑ i = 1, …, n GIfoodi * grams of carbohydratesfoodi)/total grams of carbohydratesmeal
GIdaily = (∑ i = 1, …, n GImeali * grams of carbohydratesmeali)/daily total grams of carbohydrates
GLmeal = ∑ i = 1, …, n GLfoodi.
Gut Microbiota Analysis
Fecal DNA extraction was performed using the Spin stool DNA kit (Stratec Molecular, Berlin, Germany), according to manufacturer's instructions. For each sample, 25 ng of extracted DNA was used to construct the sequencing library. The V3–V4 hypervariable regions of the bacterial 16S rRNA were amplified with a two-step barcoding approach according to the Illumina 16S Metagenomic Sequencing Library Preparation (Illumina, San Diego, CA, USA). For library preparation, DNA samples were amplified with dual-index primers using a Nextera XT DNA Library Preparation Kit (Illumina). Library concentration and quantification were determined using a KAPA Library Quantification Kit (Kapa Biosystems, Woburn, MA, USA) and Agilent 2100 Bioanalyzer System (Agilent, Santa Clara, CA, USA), respectively. The libraries were pooled and sequenced with a MiSeq platform (Illumina) for 2 × 250 base paired-end reads and a total of 2.5 Gbases raw reads were obtained.
Fecal Metabolite Measurement
We performed short chain fatty acids (SCFAs) and calprotectin quantification from stool samples.
Concentrations of acetic, propionic, iso-butyric, butyric, and iso-valeric acids were assessed by gas liquid chromatography in accordance with the method proposed by Weaver et al. (1989) with slight modifications described in Borgo et al. (2017). Analyses were performed using a Varian model 3400 CX Gas-chromatograph fitted with FID detector, split/splitless injector and a SPB-1 capillary column (30 m × 0.32 mm ID, 0.25 μm film thickness; Supelco, Bellefonte, PA, USA). Results are expressed as mg/g of wet weight of feces. Quantification of the SCFAs was obtained through calibration curves of acetic, propionic, iso-butyric, butyric, and iso-valeric acid in concentrations between 0.25 and 10 mM (10 mM 2-ethylbutyric acid as internal standard). SCFA data on the same cohort have been previously described in Verduci et al. (2018).
Fecal calprotectin concentrations were measured by a commercial ELISA kit (Calprotectin ELISA Kit, Immundiagnostik, Bensheim, Germany), according to manufacturer instructions.
Absolute Quantification of Methanobrevibacter smithii
Real-time PCR was carried out using a SYBRGreen chemistry (ThermoScientific, USA) and the specific primers for Methanobrevibacter smithii (MSfw: 5′-CCGGGTATCTAATCCGGTTC-3′ and MSrev: 5′-CTCCCAGGGTAGAGGTGAAA-3′), as previously described (Borgo et al., 2017). The following thermal cycling parameters were used for amplification of DNA: 95°C for 10 min followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. A melting curve analysis was also performed to verify amplicon specificity.
The control strain M. smithii DSM-861 (DSM: Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) was used for the standard curve.
Microbiota Profiling
The 16S rRNA gene paired sequences obtained were merged using Pandaseq (release 2.5; Masella et al., 2012), then reads were filtered by trimming stretches of 3 or more low-quality bases (quality <3) and discarding the trimmed sequences whenever they were shorter than 75% of the original one. Bioinformatic analyses were conducted using the QIIME pipeline (release 1.8.0; Caporaso et al., 2010), clustering filtered reads into Operational Taxonomic Unit (OTUs) at 97% identity level and discarding singletons (i.e., OTUs having only 1 supporting read along the whole dataset) as possible chimeras. Taxonomic assignment was performed via the RDP classifier (Wang et al., 2007) against the Greengenes database (ftp://greengenes.microbio.me/greengenes_release/gg_13_8_otus).
Indicator species analysis was performed using the indicspecies package in R (De Cáceres and Legendre, 2009) on the QIIME-derived OTU table.
Alpha-diversity was computed using the Chao1, number of OTUs, Shannon diversity, and Faith's Phylogenetic Diversity whole tree (PD whole tree) metrics; statistical evaluation among alpha-diversity indices was performed by a non-parametric Monte Carlo-based test, using 9999 random permutations. Weighted and unweighted UniFrac distances and PERMANOVA (adonis function) in the R package vegan (version 2.0-10; Oksanen et al., 2013) were used to compare the microbial community structure of the PKU and MHP children.
Statistical Analysis
Statistical comparisons were performed using MATLAB software (Natick, MA, USA). Comparisons of the two groups were performed using Student's t-test for normally distributed variables and Wilcoxon test for non-normally distributed variables. For evaluating differences in relative abundances of bacterial groups, a Mann-Whitney U-test was performed. For each phylogenetic level, only the 25 most abundant taxa were considered, in order to focus on the major players of the gut microbiota. Due to multiple testing, a Benjamini-Hochberg correction was applied, considering a FDR < 0.15 as significant. For clarity, uncorrected p-values were reported in the text. Co-abundance of microbial groups, as well as correlations between taxa and nutritional values and SCFA quantities were assessed through Spearman correlation and the associated linear regression model. Unless otherwise stated, p-values < 0.05 were considered as significant.
Results
Cohort Description
Cohort characteristics are reported in Table 1. At recruitment, blood Phe levels in PKU children was slightly higher than the MHP group (p = 0.24). Compared with MHP children, PKU children showed higher dietary intakes of carbohydrates and fibers and a significant lower consume of proteins (expressed as %). Glycemic index (GI) and glycemic load (GL), evaluated for each meal, were significantly higher in PKU children compared with MHP subjects (p < 0.001). Anthropometric measurements were similar, with a BMI z-score not significantly different in the two groups (p = 0.31).
Gut Microbiota Composition in PKU and MHP Children
Five samples (2 PKU and 3 MHP subjects) were excluded from the analysis due to very low raw read quantities (with an average of 189 reads compared to an average of 159,914 reads among the other samples); the final dataset for microbiota analysis, then, consisted of 37 subjects: 19 PKU and 18 MHP.
To avoid biases related to uneven sequencing depth, samples were subsampled to 50,000 reads each. After quality filtering processes, we obtained a mean count of 49,749 ± 111 reads per sample.
Alpha-diversity analysis (data not shown) revealed no significant differences between PKU and MHP groups for any of the metrics used (number of OTUs, p = 0.306; chao1, p = 0.131; Shannon, p = 0.894; PD whole tree, p = 0.31). Beta-diversity analysis, instead, showed that the structure of the PKU fecal microbiota differed significantly from that of the MHP group according to both unweighted (p = 0.008) and weighted (p = 0.032) Unifrac distances (β-diversity, Figure 1).
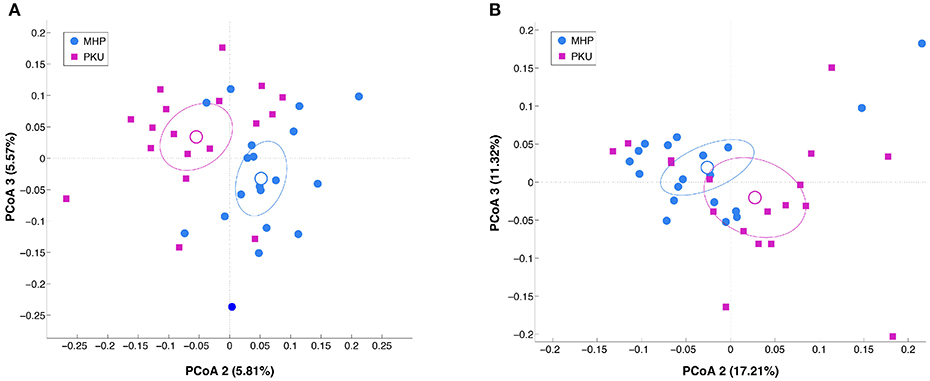
Figure 1. Beta-diversity analysis in MHP (blue) and PKU (magenta). Principal coordinates analysis (PCoA) of (A) unweighted and (B) weighted Unifrac distances. Microbial communities are statistically different (adonis test: unweighted p = 0.008, R2 = 0.040; weighted p = 0.033, R2 = 0.058). Second and third principal coordinates are shown in the plot for both distances.
The gut microbiota composition at the phylum and family levels is depicted in Figure 2 and in Table S1. The most relatively abundant phyla in PKU and MHP subjects were Firmicutes and Bacteroidetes, the latter slightly higher in MHP subjects. Among the most relatively abundant families, we found Veillonellaceae to be significantly depleted in PKU children (p = 0.002). Although not statistically significant, Ruminococcaceae were enriched in MHP and Lachnospiraceae in PKU subjects.
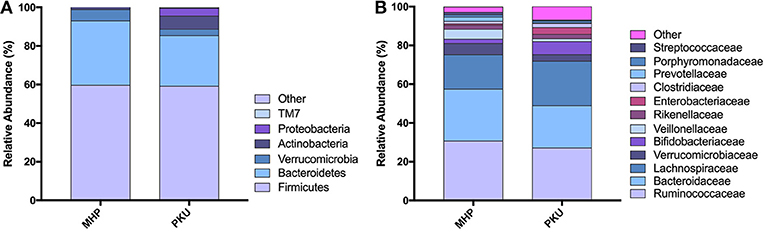
Figure 2. Gut microbiota composition in MHP and PKU children. Relative abundance at (A) phylum and (B) family level. All bacterial taxa present at <1% relative abundance were grouped into the “Other” classification.
At the genus level (Figure 3), Faecalibacterium (p = 0.001), Ruminococcaceae (other) (p = 0.03) were more relatively abundant in MHP children; although not significant, Bacteroides and Prevotella genera showed the same trend. Furthermore, we observed an increased Prevotella/Bacteroides ratio in MHP compared with PKU (0.14 ± 0.59; 0.02 ± 0.05, respectively).
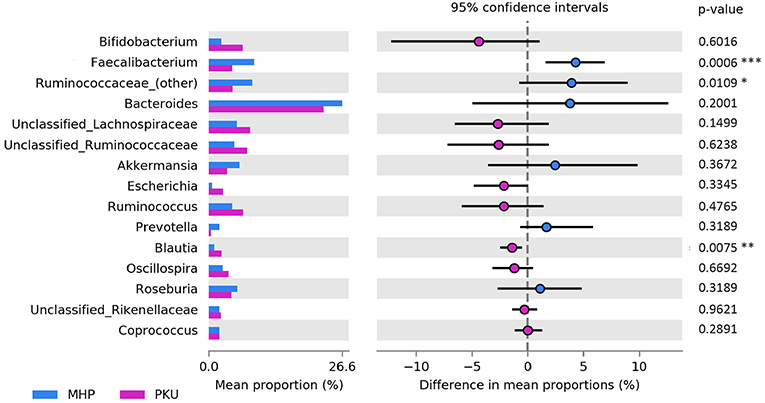
Figure 3. Bacterial abundances at genus level. Genera abundances of the 15 most abundant genera are reported as mean proportion along with significant p-values (two-sided White's non-parametric t-test) and 95% of confidence interval (bootstrap method).
In contrast, PKU children were characterized by a significant increase in the relative abundance of Blautia (p = 0.004), Clostridium (belonging to Lachnospiraceae family, p = 0.002), and Lachnospiraceae (other) (p = 0.019). Significantly altered taxa belonged to the Firmicutes phylum and are highlighted in Table 2.
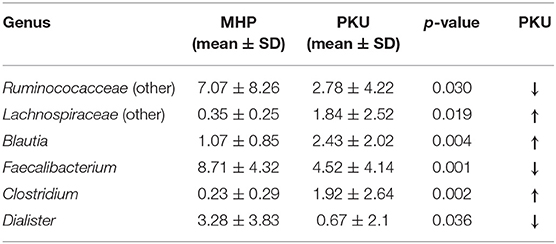
Table 2. Genera belonging to Firmicutes phylum significantly increased or depleted in PKU children (Mann-Whitney U-test, p-value < 0.05).
Because of the underestimation of Archaea by 16S rRNA gene sequencing, Real-time PCR quantification of the most abundant Archaea species in human gut microbiota, Methanobrevibacter smithii, was performed. We did not observe significant differences between PKU and MHP subjects (p = 0.40).
Indicator species analysis highlighted Akkermansia muciniphila OTU 1045 (best BLAST hit: Accession number NR_074436.1, with 97% seq. similarity over 420 bp, e-value = 0.046) and Faecalibacterium prausnitzii OTU 3793 (accession number: NR_028961.1, with 97% seq. similarity over 420 bp, e-value = 0.001) to be characteristic of MHP microbiota, whereas Ruminococcus bromii OTU 3232 (accession number: NR_025930.1, with 97% seq. similarity over 420 bp, e-value = 0.018) to be associated to PKU.
Bacterial Correlation Patterns
Because cross-feeding between species is relevant in gut microbiota dynamic, we performed correlation analysis to study interactions between different members of intestinal microbiota. Several significant bacterial connections (Table S2), both positive and negative, have been observed. In particular, MHP indicator species A. muciniphila and F. prausnitzii showed several interactions with other members of the microbial community: Akkermansia was negatively related to Unclassified Lachnospiraceae (R = −0.61) and to Blautia spp. (R = −0.40), while was positively associated to Oscillospira (R = 0.57) and to Unclassified Clostridiales (R = 0.34). On the other hand, F. prausnitzii was only found positively related to Ruminococcaceae (other) genus (R = 0.42). The PKU indicator species, Ruminococcus bromii, instead, showed no correlation to other genera itself.
Correlations Between Gut Microbiota and Nutritional Values
Statistical correlation analysis between diet and microbiota showed that Faecalibacterium spp., significantly increased in MHP group, negatively correlated with fiber intake, both soluble and insoluble fibers (R = −0.61; R = −0.37), and with GI, GL (R = −0.53 and R = −0.49, respectively). Ruminococcaceae family as well as its genus Ruminococcaceae (other), with higher relative abundance among MHP patients (Figure 4), negatively related with GI and soluble fibers (R = −0.40 and R = −0.49, respectively), while an opposite trend was observed among the Lachnospiraceae (other) genus within the same nutritional values (GI R = 0.43; soluble fibers R = 0.49). Oscillospira, slightly higher in PKU children, was positively related to energy assumption and carbohydrates (R = 0.43 and R = 0.47, respectively); Roseburia, genus enriched in MHP, was found negatively related to soluble fibers (R = −0.42) and to lipids (R = −0.42). All of these correlation coefficients were statistically significant (p < 0.05).
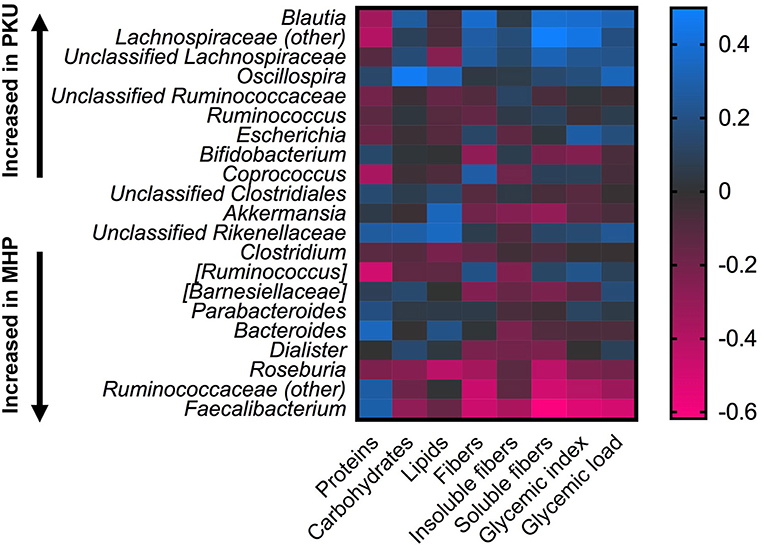
Figure 4. Correlations between microbiota and dietary information. Heatmap showing Spearman's correlations between the most abundant microbial genera and nutritional values. Color intensity represents the degree of association; positive correlations are depicted in blue and negative in magenta.
SCFAs and Gut Microbiota Correlation
As reported in our previous work (Verduci et al., 2018), a decrease in total SCFAs and in particular in butyrate production was observed in PKU children compared with MHP children (p = 0.044 and p = 0.026, respectively). Although not statistically significant, acetate (p = 0.161) was also reduced in PKU group. Propionate and the isoforms iso-butyrate and iso-valerate, instead, were similar.
Correlation analysis with gut microbiota composition revealed several significant interactions between the relative abundance of certain taxa and fecal SCFA concentrations.
Acetate was found negatively related to Coprococcus (R = −0.42) and to Blautia (R = −0.50), genera enriched in PKU. Butyrate was found inversely correlated to Lachnospiraceae (other) (R = −0.41). Propionate was found to positively correlate to Bacteroides (R = 0.49) and inversely to Unclassified Lachnospiraceae (R = −0.42) and to Blautia (R = −0.56). The branched-chain fatty acids iso-butyrate and iso-valerate were negatively related to Unclassified Lachnospiraceae (R = −0.64, R = −0.65, respectively), to Clostridium spp. (R = −0.45 and R = −0.46) and to Blautia spp (R = −0.40, only iso-butyrate) and positively correlated to Akkermansia (R = 0.47 and R = 0.44). All correlation coefficients were statistically significant (p < 0.05).
Fecal Calprotectin Concentrations in PKU and MHP Patients
To investigate whether PKU subjects are chronically inflamed in the gut, we quantified fecal calprotectin, a recognized biomarker for gastrointestinal diseases (Pang et al., 2014). No significant differences were recorded between PKU and MHP groups (24.8 ± 14.9 μg/g and 40.6 ± 28.3 μg/g, p = 0.10, respectively).
Conclusions and Discussion
The aim of this study was to investigate the impact of a low-Phe diet on microbial gut community and its possible consequences on PKU patient wellbeing. Indeed, according to European guidelines for PKU management, the recommended diet should start as early as possible, usually before the age of 10 days, to prevent neurological damage (Singh et al., 2014; van Spronsen et al., 2017). This age corresponds to a well-recognized crucial step in microbiota acquisition and maturation (Dominguez-Bello et al., 2019) on which environmental factors may have a profound impact.
To rule out a direct effect of PAH deficiency in microbiota alteration, subjects with mild hyperphenylalaninemia, under normal diet, were enrolled as control group.
Bioinformatic analyses revealed several changes in the microbial taxa inhabiting the PKU gut compared with MHP subjects, as already suggested by Pinheiro de Oliveira et al. (2016). Nevertheless, some differences were observed between their study and ours, mainly ascribable to the different enrolled control group (healthy children instead of MHP), the different age range (4 out of their 8 PKU patients were <2 years-old), the sequencing method used (Ion Torrent vs. Illumina) and the subjects' ethnicity (Brazilian vs. Italians).
In our cohort, the relative abundance of Firmicutes and Bacteroidetes was similar, with slight decrease of both phyla in the PKU group. Although not statistically significant, both Bacteroides and Prevotella, the two main genera belonging to the Bacteroidetes, were more relatively abundant in MHP children. While Bacteroides result is not surprising, probably related to the reduced protein intake (David et al., 2014) in Phe-free diet, Prevotella is usually associated with increased fiber intake (De Filippo et al., 2010), typical of the PKU diet. Similarly, the Prevotella/Bacteroides ratio was slightly higher in MHP group, in contrast to the common finding of a higher ratio in strict vegetarians/vegans (Wu et al., 2011; Franco-de-Moraes et al., 2017). Recently, a high Prevotella/Bacteroides ratio has been suggested to be associated with an improvement of glucose response, possibly preventing cardiometabolic diseases (Sandberg et al., 2018).
The most relevant shifts, however, concerned the Firmicutes phylum. Indeed, the PKU gut microbiota was enriched in Blautia spp. and Clostridium spp. and depleted in Faecalibacterium spp., as anticipated in our previous work by absolute real-time PCR quantification (Verduci et al., 2018). As discussed, it was reasonable to expect that the higher fiber intake in PKU patients would have increased Faecalibacterium spp. proliferation, but the relative abundance of this genus showed an opposite trend. However, as described by Benus et al. (2010), a fiber-supplemented diet does not necessarily increase F. prausnitzii and Ruminococcus populations compared with a normal balanced diet. Moreover, the quality of fibers (Verduci et al., 2018) as well as the supplementation of some PKU special low protein products with inulin could also impact the abundance of these genera.
It is intriguing that the indicator species analysis showed an association between MHP phenotype and F. prausnitzii, a known biomarker for health status (Lopez-Siles et al., 2017). In healthy adults, this bacterial genus commonly represents more than 5% of the total gut bacterial population (Miquel et al., 2013) and is one of the major butyrate producers. Butyrate is considered the main energy source of colonocytes and displays anti-inflammatory properties in the colonic mucosa (Flint et al., 2012). A. muciniphila also characterized the MHP gut microbiota in the indicator species analysis. A. muciniphila, a mucin-degrading bacterium, is considered a potential probiotic, that is able to maintain intestinal integrity (Zhai et al., 2018). As already reported by other authors (Arumugam et al., 2011; Cani and de Vos, 2017), A. muciniphila is positively associated with members of the family Ruminococcaceae, probably sharing nutritional requirements or cross feeding phenomena.
In contrast, PKU microbial communities were characterized by Ruminococcus bromii, a well-known starch degrader that belongs to the non-butyrate-forming Ruminococcaceae (Ze et al., 2012; Kettle et al., 2015).
Overall, in agreement with the lower total fecal SCFAs content and in particular of butyrate, the PKU gut microbiota was depleted in butyrate-producing species and enriched in genera, i.e., Blautia, that are recognized to exert a pro-inflammatory effect on gut mucosa. Indeed, Blautia spp. has been demonstrated to induce cytokines secretion, like tumor necrosis factor alpha (TNF-alpha), involved in immune acute phase response (Tuovinen et al., 2013).
Moreover, about half of PKU subjects were characterized by an increase in the relative abundance of Proteobacteria, in particular Escherichia spp. Proteobacteria is a phylum of Gram-negative microorganisms whose membrane lipopolysaccharide is a well-known inducer of innate immune responses (Hotamisligil, 2006).
As discussed for the phyla Firmicutes and Bacteroidetes, the enrichment in Proteobacteria observed in PKU children is in contrast with the reported underrepresentation of this phylum in children with rural diet, more similar to PKU vegetarian-vegan diet than MHP, compared with children under Western-diet (De Filippo et al., 2010).
Although our study did not show any alteration in calprotectin, a recognized gut inflammation marker, previous work by our group (Moretti et al., 2017) demonstrated an increase in the triglyceride glucose index (TyG index) in PKU children compared with age- and sex-matched healthy controls. TyG index is considered a marker of low-grade inflammation and of peripheral insulin resistance (Er et al., 2016). Moretti et al. (2017) showed in PKU a positive correlation of TyG index with glycemic load, reinforcing a possible link between carbohydrate quality and metabolic disorder predisposition. Indeed, PKU children showed an increased consumption of fast-absorbing carbohydrates that escape gut microbiota fermentation (Verduci et al., 2018), resulting in higher GI and GL.
It is important to note, we also found a divergent response of the Firmicutes phylum with respect to daily GI and GL. Most members of the Lachnospiraceae family, with the exception of Roseburia, were positively correlated with both indexes (with Blautia as the most relevant genus), whereas members of Ruminococcaceae family were negatively, in particular F. prausnitzii.
The quality of carbohydrates ingested by PKU children might directly affect Faecalibacterium abundance. In accordance with our data, Fava and colleagues (Fava et al., 2013) showed that a diet enriched in carbohydrates with a high glycemic index resulted in a decreased F. prausnitzii abundance in subjects at-risk of developing a metabolic syndrome. A plausible explanation is that fast-absorbing carbohydrates do not represent a suitable substrate for F. prausnitzii growth, commonly fermenting complex carbohydrates. On the other hand, short chain carbohydrates represent a good substrate for Blautia spp. (Egshatyan et al., 2016), which were more relatively abundant in the PKU group. The Blautia genus encompasses a huge number of strains with different metabolic capabilities (Eren et al., 2015), several of them considered to be acetogens. In contrast, we found an inverse correlation with fecal acetate concentration. However, this observation is in agreement with recent findings by Org et al. (2017) that investigated the relationship between the gut microbiota composition and metabolic disorder traits. The authors also suggested a positive correlation between Blautia and body mass index. Indeed, PKU patients, more than MHP subjects, are at risk for excessive weight gain (Scaglioni et al., 2004; Rocha et al., 2013; Couce et al., 2018) and insulin resistance (Moretti et al., 2017; Couce et al., 2018).
The enrichment of PKU low-protein products in simple sugars, is a consequence of their poor palatability. In the last few years, the increasing awareness of the health consequences of PKU diet led to the development of new products that could implement or complement these formulas. For example, some companies have made commercially available products based on glycomacropeptide (GMP). GMP is a protein derived from cheese whey, rich in specific essential amino acids and lacking aromatic amino acids (i.e., Phe). GMP has been recently demonstrated to have prebiotic properties with beneficial effects on the gut microbiota (Sawin et al., 2015).
In addition, new formulas enriched in prolonged-release amino acids have been recently released. Such medical foods, meant to allow a physiological-like adsorption, are strongly improved in palatability thanks to the amino acids coating layer (Giarratana et al., 2018). It still remains to be evaluated whether an improvement in current free-amino acid formulas, as well as increased attention to the management of dietary carbohydrate quality in PKU diet (with a particular focus on special low protein products), can rebalance the PKU microbial community.
Ethics Statement
The study was approved by the ethics committee (Comitato Etico Milano Area 1, Protocol number 2015/ST/135). The parents of eligible children or their legal guardian received a detailed explanation of the study and signed a consent form.
Author Contributions
EB and EV designed the study. GB, FB, and CC performed experiments and data analysis, and drafted the manuscript. CC and MS performed microbiota data analysis. VR performed subject enrollment and analyzed clinical data. EB, EV, and GM performed supervision and writing—review and editing.
Funding
This work was supported by a Università degli Studi di Milano-Linea B grant awarded to EB, by Molecular and Translational Medicine Ph.D.-Università degli Studi di Milano scholarship (to CC), and by Nutritional Sciences Ph.D.-Università degli Studi di Milano scholarship (to GB).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00101/full#supplementary-material
Raw reads are available in NCBI Short Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA447916.
References
Albenberg, L. G., and Wu, G. D. (2014). Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology 146, 1564–1572. doi: 10.1053/j.gastro.2014.01.058
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Benus, R. F., van der Werf, T. S., Welling, G. W., Judd, P. A., Taylor, M. A., Harmsen, H. J. M., et al. (2010). Association between Faecalibacterium prausnitzii and dietary fiber in colonic fermentation in healthy human subjects. Br. J. Nutr. 104, 693–700. doi: 10.1017/S0007114510001030
Borgo, F., Riva, A., Benetti, A., Casiraghi, M. C., Bertelli, S., Garbossa, S., et al. (2017). Microbiota in anorexia nervosa: the triangle between bacterial species, metabolites and psychological tests. PLoS ONE 12:e0179739. doi: 10.1371/journal.pone.0179739
Cani, P. D., and de Vos, W. M. (2017). Next-Generation beneficial microbes: the case of Akkermansia muciniphila. Front. Microbiol. 8:1765. doi: 10.3389/fmicb.2017.01765
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). Correspondence QIIME allows analysis of high throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nat. Methods 7, 335–336. doi: 10.1038/nmeth0510-335
Couce, M. L., Sánchez-Pintos, P., Vitoria, I., De Castro, M. J., Aldámiz-Echevarría, L., Correcher, P., et al. (2018). Carbohydrate status in patients with phenylketonuria. Orphanet. J. Rare Dis. 13:103. doi: 10.1186/s13023-018-0847-x
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D.B., Button, J.E., Wolfe, B.E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
De Cáceres, M., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696. doi: 10.1073/pnas.1005963107
Dominguez-Bello, M. G., Godoy-Vitorino, F., Knight, R., and Blaser, M. J. (2019). Role of the microbiome in human development. Gut gutjnl-2018-317503. doi: 10.1136/gutjnl-2018-317503
Egshatyan, L., Kashtanova, D., Popenko, A., Tkacheva, O., Tyakht, A., Alexeev, D., et al. (2016). Gut microbiota and diet in patients with different glucose tolerance. Endocr. Connect. 5, 1–9. doi: 10.1530/EC-15-0094
Er, L. K., Wu, S., Chou, H. H., Hsu, L. A., Teng, M. S., Sun, Y. C., et al. (2016). Triglyceride glucose-body mass index is a simple and clinically useful surrogate marker for insulin resistance in nondiabetic individuals. PLoS ONE 11:e0149731. doi: 10.1371/journal.pone.0149731
Eren, A. M., Sogin, M. L., Morrison, H. G., Vineis, J. H., Fisher, J. C., Newton, R. J., et al. (2015). A single genus in the gut microbiome reflects host preference and specificity. ISME J. 9, 90–100. doi: 10.1038/ismej.2014.97
Fava, F., Gitau, R., Griffin, B. A., Gibson, G. R., Tuohy, K. M., and Lovegrove, J. A. (2013). The type and quantity of dietary fat and carbohydrate alter faecal microbiome and short-chain fatty acid excretion in a metabolic syndrome ‘at-risk' population. Int J Obes. 37, 216–223. doi: 10.1038/ijo.2012.33
Flint, H. J., Scott, K. P., Duncan, S. H., Louis, P., and Forano, E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306. doi: 10.4161/gmic.19897
Franco-de-Moraes, A. C., de Almeida-Pititto, B., da Rocha Fernandes, G., Gomes, E. P., da Costa Pereira, A., and Ferreira, S. R. G. (2017). Worse inflammatory profile in omnivores than in vegetarians associates with the gut microbiota composition. Diabetol. Metab. Syndr. 9:62. doi: 10.1186/s13098-017-0261-x
Giarratana, N., Gallina, G., Panzeri, V., Frangi, A., Canobbio, A., and Reiner, G. (2018). A new Phe-free protein substitute engineered to allow a physiological absorption of free amino acids for phenylketonuria. JIEMS. 6, 1–9. doi: 10.1177/2326409818783780
Giovannini, M., Verduci, E., Salvatici, E., Paci, S., and Riva, E. (2012). Phenylketonuria: nutritional advances and challenges. Nutr. Metab. 9:7e13. doi: 10.1186/1743-7075-9-7
Guthrie, R., and Susi, A. (1963). A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 32, 318–343.
Güttler, F., and Guldberg, P. (1996). The influence of mutations on enzyme activity and phenylalanine tolerance in phenylalanine hydroxylase deficiency. Eur. J. Pediatr. 155, 6S−10S.
Hotamisligil, G. S. (2006). Inflammation and metabolic disorders. Nature 444, 860–867. doi: 10.1038/nature05485
Kettle, H., Louis, P., Holtrop, G., Duncan, S. H., and Flint, H. J. (2015). Modelling the emergent dynamics and major metabolites of the human colonic microbiota. Environ. Microbiol. 17, 1615–1630. doi: 10.1111/1462-2920.12599
Lopez-Siles, M., Duncan, S. H., Garcia-Gil, L. J., and Martinez-Medina, M. (2017). Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 11, 841–852. doi: 10.1038/ismej.2016.176
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31
Miquel, S., Martìn, R., Rossi, O., Bermù dez-Humarà, L. G, Chatel, J. M., Sokol, H., et al. (2013). Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261. doi: 10.1016/j.mib.2013.06.003
Moretti, F., Pellegrini, N., Salvatici, E., Rovelli, V., Banderali, G., Radaelli, G., et al. (2017). Dietary glycemic index, glycemic load and metabolic profile in children with phenylketonuria. Nutr. Metab. Cardiovasc. Dis. 27:176e182. doi: 10.1016/j.numecd.2016.11.002
Nieuwdorp, M., Gilijamse, P. W., Pai, N., and Kaplan, L. M. (2014). Role of the microbiome in energy regulation and metabolism. Gastroenterology 146, 1525–1533. doi: 10.1053/j.gastro.2014.02.008
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., et al. (2013). Package “Vegan”. R Package Version 2.0–8. Available online at: http://CRAN.R-project.org/package=vegan (accessed April 1, 2019).
Org, E., Blum, Y., Kasela, S., Mehrabian, M., Kuusisto, J., Kangas, A. J., et al. (2017). Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 18:70. doi: 10.1186/s13059-017-1194-2
Pang, T., Leach, S. T., Katz, T., Day, A. S., and Ooi, C. Y. (2014). Fecal biomarkers of intestinal health and disease in children. Front. Pediatr. 2:6. doi: 10.3389/fped.2014.00006
Pinheiro de Oliveira, F., Mendes, R. H., Dobbler, P. T., Mai, V., Pylro, V. S., Waugh, S. G., et al. (2016). Phenylketonuria and gut microbiota: a controlled study based on next-generation sequencing. PLoS ONE 11:e0157513. doi: 10.1371/journal.pone.0157513
Rocha, J. C., MacDonald, A., and Trefz, F. (2013). Is overweight an issue in phenylketonuria? Mol Genet Metab. 110(Suppl), S18–24. doi: 10.1016/j.ymgme.2013.08.012
Sandberg, J., Kovatcheva-Datchary, P., Björck, I., Bäckhed, F., and Nilsson, A. (2018). Abundance of gut Prevotella at baseline and metabolic response to barley prebiotics. Eur. J. Nutr. doi: 10.1007/s00394-018-1788-9. [Epub ahead of print].
Sawin, E. A., De Wolfe, T. J., Aktas, B., Stroup, B. M., Murali, S. G., Steele, J. L., et al. (2015). Glycomacropeptide is a prebiotic that reduces Desulfovibrio bacteria, increases cecal short-chain fatty acids, and is anti-inflammatory in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G590–G601. doi: 10.1152/ajpgi.00211.201
Scaglioni, S., Verduci, E., Fiori, L., Lammardo, A. M., Rossi, S., Radaelli, G., et al. (2004). Body mass index rebound and overweight at 8 years of age in hyperphenylalaninaemic children. Acta Paediatr. 93, 1596–1600.
Singh, R. H., Rohr, F., Frazier, D., Cunningham, A., Mofidi, S., Ogata, B., et al. (2014). Recommendations for the nutrition management of phenylalanine hydroxylase deficiency. Genet. Med. 16, 121–131. doi: 10.1038/gim.2013.179
Tuovinen, E., Keto, J., Nikkilä, J., Mättö, J., and Lähteenmäki, K. (2013). Cytokine response of human mononuclear cells induced by intestinal Clostridium species. Anaerobe 19, 70–76. doi: 10.1016/j.anaerobe.2012.11.002
van Spronsen, F. J., van Wegberg, A. M., Ahring, K., Bélanger-Quintana, A., Blau, N., Bosch, A. M., et al. (2017). Key European guidelines for the diagnosis and management of patients with phenylketonuria. Lancet Diabetes Endocrinol. 5, 743–756. doi: 10.1016/S2213-8587(16)30320-5
Verduci, E., Moretti, F., Bassanini, G., Banderali, G., Rovelli, V., Casiraghi, M. C., et al. (2018). Phenylketonuric diet negatively impacts on butyrate production. Nutr. Metab. Cardiovasc. Dis. 28, 385–392. doi: 10.1016/j.numecd.2018.01.004
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Weaver, G. A., Krause, J. A., Miller, T. L., and Wolin, M. J. (1989). Constancy of glucose and starch fermentations by two different human faecal microbial communities. Gut 30, 19–25.
Williams, R. A., Mamotte, C. D., and Burnett, J. R. (2008). Phenylketonuria: an inborn error of phenylalanine metabolism. Clin. Biochem. Rev. 29, 31–41.
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Ze, X., Duncan, S. H., Louis, P., and Flint, H. J. (2012). Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543. doi: 10.1038/ismej.2012.4
Keywords: phenylketonuria, mild hyperphenylalaninemia, diet, microbiota, glycemic index, Faecalibacterium prausnitzii, butyrate
Citation: Bassanini G, Ceccarani C, Borgo F, Severgnini M, Rovelli V, Morace G, Verduci E and Borghi E (2019) Phenylketonuria Diet Promotes Shifts in Firmicutes Populations. Front. Cell. Infect. Microbiol. 9:101. doi: 10.3389/fcimb.2019.00101
Received: 08 February 2019; Accepted: 25 March 2019;
Published: 16 April 2019.
Edited by:
Benoit Chassaing, Georgia State University, United StatesReviewed by:
Silvia Turroni, University of Bologna, ItalyDevin Holman, Agriculture and Agri-Food Canada, Canada
Copyright © 2019 Bassanini, Ceccarani, Borgo, Severgnini, Rovelli, Morace, Verduci and Borghi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elvira Verduci, ZWx2aXJhLnZlcmR1Y2lAdW5pbWkuaXQ=
Elisa Borghi, ZWxpc2EuYm9yZ2hpQHVuaW1pLml0
†These authors have contributed equally to this work
 Giulia Bassanini
Giulia Bassanini Camilla Ceccarani
Camilla Ceccarani Francesca Borgo
Francesca Borgo Marco Severgnini
Marco Severgnini Valentina Rovelli
Valentina Rovelli Giulia Morace
Giulia Morace Elvira Verduci
Elvira Verduci Elisa Borghi
Elisa Borghi