- 1MOE Key Laboratory of Industrial Fermentation Microbiology, College of Biotechnology, Tianjin University of Science and Technology, Tianjin, China
- 2Tianjin Engineering Research Center of Microbial Metabolism and Fermentation Process Control, Tianjin University of Science and Technology, Tianjin, China
- 3Robustnique Corporation Ltd., Tianjin, China
Streptococcus zooepidemicus is an important opportunistic pathogen of several species including humans. This organism is also well-known as the main producing strain in industrial production of hyaluronic acid (HA), which is the component of its capsule polysaccharide. How its virulence and capsule polysaccharide production is regulated remains poorly understood. Intercellular chemical signaling among bacteria provides communities of microbes the opportunity to coordinate gene expression to facilitate group behavior, such as pathogenicity, capsule polysaccharide production, etc. Yet no conserved cell-to-cell signaling system has been elucidated in S. zooepidemicus. Encoded within the genome of S. zooepidemicus is one Rgg regulator encoding gene (rgg) with low similarity to both rgg2 and rgg3 from Streptococcus pyogenes. A small ORF (named as shp) encoding a novel short hydrophobic peptide (SHP) was found in the vicinity of rgg. We found that the active form of pheromone is short and hydrophobic (LLLLKLA), corresponding to the C terminal 7 amino acids of the pre-peptide Shp, which shows divergent sequence to all peptide pheromones reported in streptococci. In response to active SHP, Rgg functions as a transcriptional activator to induce the expression of shp, forming a positive feedback circuit. Bacteria social behaviors, such as capsule polysaccharide production and biofilm formation, were significantly affected when the rgg-shp pathway was inactivated. These data provide the first demonstration that Rgg/Shp signaling pathway comprises an active quorum sensing system in S. zooepidemicus.
Introduction
Streptococcus equi subsp. zooepidemicus (commonly referred to as Streptococcus zooepidemicus) is an important opportunistic pathogen of several species, notably equines and sometimes humans (Waller, 2017). This microorganism generally inhabits the mucous membranes and skin of healthy animals, and causes severe diseases such as pneumonia, septicemia, or meningitis in some conditions. It is a rare cause of human invasive disease originating from zoonotic transmission from domesticated animals (Pelkonen et al., 2013). Why this bacterium causes persistent infection and disease under certain circumstances remains largely unknown. It is believed that the regulated expression of capsular polysaccharide in S. zooepidemicus is one of the essential factors for the pathogenicity and dissemination of this microorganism (Xu et al., 2016). The main component of capsular polysaccharide for S. zooepidemicus is hyaluronic acid (HA), which is a component that is widely used in ophthalmic surgery and as an ingredient in cosmetics. This character also makes S. zooepidemicus the first HA commercial producer on an industrial scale, and remain the major strain in the industrial production of HA till now (Liu et al., 2011). Capsular polysaccharide production is usually a social behavior of bacteria regulated by cell-cell communication. For example, capsular polysaccharide synthesis and virulence in Staphylococcus aureus are controlled by an AI-2 quorum-sensing (QS) system (Zhao et al., 2010), and extracellular polysaccharide production in Streptococcus pneumoniae is regulated by a small hydrophobic peptide (SHP) QS system (Junges et al., 2017). Despite the clinical significance and economic importance of S. zooepidemicus, the identity of a widespread, conserved intercellular signaling system remained elusive in this organism.
QS is a widespread cell-cell signaling system among bacteria, that generate population-wide responses to low molecular weight signaling molecules (also referred to as pheromones), depending on cellular density (Miller and Bassler, 2001). QS provides communities of microbes the opportunity to coordinate gene expression to facilitate group behavior, such as virulence, extracellular polysaccharide production and biofilm formation (Rutherford and Bassler, 2012). For most QS signaling in Gram-negative bacteria, a small molecule such as N-acyl homoserine lactone is used as the secreted pheromone (Papenfort and Bassler, 2016), while for QS in many species of Gram-positive bacteria, secreted short peptide signals were usually sensed by protein receptors that alter the expression of downstream target genes (Lyon and Novick, 2004). Rgg QS pathways, widely found among low G+C Gram-positive bacteria, use Rgg transcription factors as cytoplasmic pheromone receptors for SHP pheromones (Fleuchot et al., 2013). Genome-wide surveys showed that such systems are present in the majority of streptococci (Fleuchot et al., 2011). Among streptococci, Streptococcus pyogenes (Group A Streptococcus, GAS) is perhaps the best understood species in terms of the role the Rgg QS (Jimenez and Federle, 2014). The gene for the SHP pheromone (shp) and the gene for Rgg transcription factor (rgg) have a characteristic arrangement in the genome with shp being located in the vicinity of rgg in GAS. Interestingly, different Rgg regulators might exhibit diverse regulatory effects. For example, there are two Rgg protein family members (Rgg2 and Rgg3) as cytoplasmic receptors of SHP pheromones in all completely sequenced GAS genomes. In response to the SHP pheromones, Rgg2 functions as a transcription activator, while Rgg3 functions as a transcription repressor (Chang et al., 2011).
S. zooepidemicus and GAS are phylogenetically related. Both of them belong to the pyogenic group of streptococci (Kawamura et al., 1995). However, there is no report that any Rgg protein functions as a QS effector in S. zooepidemicus. Here we demonstrate for the first time that an Rgg family transcriptional regulator in S. zooepidemicus functions as QS effector protein in response to a novel peptide pheromone (SHP) encoded by a neighboring shp gene. We provide further evidence that the Rgg transcriptional regulator functions as transcription activator to induce the expression of shp, and regulates HA production and biofilm formation. Therefore, Rgg/Shp comprises the first characterized quorum-sensing pathway in S. zooepidemicus.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
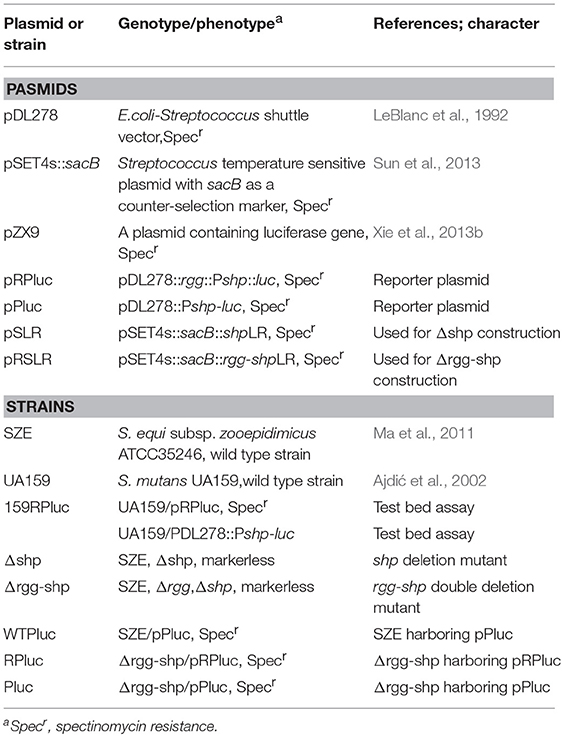
All plasmids and strains used in this study are listed in Table 1. All Streptococcus strains were grown in Todd-Hewitt medium (Difco) supplemented with 0.3% (wt/vol) yeast extract or in Chemically Defined Media (CDM) (van de Rijn and Kessler, 1980). All Streptococcus mutans strains were grown anaerobically (in a Candle Extinction Jar) at 37°C. All Streptococcus zooepidemicus strains were grown under aerobic conditions at 30°C or 37°C. When testing for HA production, cells were grown in a THYE containing 0.2% sucrose. For the selection of antibiotic-resistant colonies in S. mutans, THYE plates were supplemented with spectinomycin (Sigma) (1,000 μg ml−1). For the selection of antibiotic-resistant colonies in S. zooepidemicus THYE plates were supplemented with spectinomycin (Sigma) (200 μg ml−1).
Construction of Firefly Luciferase Reporter Plasmids
Primers used in this study are listed in Table 2. For Plasmid pRPluc (pDL278 vector containing a luciferase open reading frame driven by shp promoter and an intact rgg gene)construction, the fragment containing the rgg gene and the full intergenic region between rgg and shp was generated by PCR with the primer pair 763F/shpR from the genome of S. zooepidemicus, and luciferase open reading frame (luc) was amplified with primer pair lucF/R from plasmid pZX9 (Xie et al., 2013b). There are overlapping regions between the two amplicons, which allowed a subsequent overlapping PCR using primer pair 763F and lucR. The resulting 3-kb amplicons were digested with BamHI and EcoRI and ligated into the corresponding sites of pDL278 to obtain pRPluc. For Plasmid pPluc (pDL278 vector containing a luciferase open reading frame driven by shp promoter)construction, shp promoter was generated by PCR with the primer pair shpF/shpR, and luciferase open reading was amplified with primer pair lucF/R. There are overlapping regions between the two amplicons, which allowed a subsequent overlapping PCR using primer pair shpF and lucR. The resulting 1.6-kb amplicons were digested with BamHI and EcoRI and ligated into the corresponding sites of pDL278 to obtain pPluc.
Transformation of S. mutans and S. zooepidemicus
S. mutans was transformed using the natural transformation assay as previously described (Xie et al., 2013a). S. zooepidemicus was transformed using electroporation method as previously described (Sun et al., 2013).
Construction of shp Deletion Mutant and rgg-shp Double Deletion Mutant
The shp gene deletion mutant (Δshp) was constructed using a markerless gene-deletion system as described previously (Sun et al., 2013). Briefly, using S. zooepidemicus genomic DNA as the template, the upstream and downstream fragments of shp were amplified by PCR with primer pair shpupF/R and shpdnF2/R, respectively. There are overlapping regions between the two amplicons, which allowed a subsequent overlapping PCR using primer pair shpupF and shpdnR. The resulting amplicons were digested with EcoRI and BamHI and ligated into the corresponding sites of SET4s::sacB to obtain pSLR. pSLR was transformed into S. zooepidemicus wild type, and the resulting transformant was first grown at 30°C for 12 h and then further cultured at 37°C for another 4 h in THYE medium supplemented with 200 μg/mL spectinomycin to select for the single crossover strain. After that, the single crossover strain was cultivated in THYE medium without antibiotics addition. Thereafter, the sucrose-resistant strains were selected on THYE plate supplemented with 5% (w/v) sucrose. Finally, the sucrose-resistant and spectinomycin-sensitive clones were isolated, and shp deletion mutants were examined by PCR and further confirmed by sequencing. The same strategy was used for the construction of rgg-shp double deletion mutant (Δrgg-shp). During the construction of Δrgg-shp, the primer pair rggdnF/R and primer pair shpdnF1/R were used, and plasmid pRSLR was constructed. The primers used for the construction of gene deletion mutants are listed in Table 2.
Preparation of Synthetic Peptides
The peptides used in this experiment were purchased from GenScript (Nanjing, China) and the purity of all peptides were above 98%. Stock solutions were dissolved in DMSO or some weak acid according to the instructions at a concentration of 1 mM. Peptide sequences are the following, sSHP-C6: LLLKLA; sSHP-C7-1: LLLLKL; sSHP-C7: LLLLKLA; sSHP-C8: DLLLLKLA; sSHP-C9: HDLLLLKLA; sSHP-CN: LPYFAGCL.
S.mutans “Test-Bed” Reporter Assay
Reporter plasmids pRPluc and pPluc were transformed into S.mutans UA159 respectively, to obtain S.mutans 159RPluc and 159Pluc, respectively. The luciferase activity in response to synthetic peptides in each reporter strain was detected based on a previously described method (Shanker et al., 2016). Briefly, the reporter strain was incubated overnight in 5 ml of CDM, which were supplemented with spectinomycin (100 μg/μl). The overnight culture was diluted in multiples into fresh CDM until the OD600 reached to 0.1, then added different concentrations of the test peptide to make the final concentration of the peptide in the culture was 0, 5, 10, 25, 50, 100, and 150 nM. Thereafter, the cultures were continued to be incubated under anaerobic conditions at 37 ° C for 1 h. Finally, a 180 μl aliquot of each culture was placed in a Falcon white flat-bottom 96-well plate with the addition of 20 μl of 1 nM D-luciferin sodium salt. The optical density (OD) at 600 nm and luminescence of the culture were measured using an Infinite 200PRO multi-plate reader. Normalized luciferase activity was expressed as relative light units (RLU) (luminescence value)/OD600.
Growth Curve and Time Course Luciferase Activity of Wild-Type S. zooepidemicus Containing Plasmid pPluc
Reporter plasmids pPluc was transformed into wild-type S. zooepidemicus to obtain strain S. zooepidemicus WTPluc. The reporter strain WTPluc was grown at 37°C in CDM after dilution from the overnight culture to an OD600 of 0.01. The growth and luciferase activity of the reporter strain were monitored throughout growth by taking 180 μl aliquots every 4 h from the growing cultures. Aliquots were placed into a Falcon white flat-bottom 96-well plate and were mixed with 20 μl of 1nM D-luciferin. After that, the OD600 and Luminescence were measured using an Infinite 200PRO multi-plate reader. Normalized luciferase activity was expressed as relative light units (RLU) /OD600.
Luciferase Transcriptional Reporter Assay in S. zooepidemicus
Reporter plasmids pRPluc and pPluc were transformed into S. zooepidemicus Δrgg-shp to obtain S. zooepidemicus RPluc and Pluc, respectively. The luciferase activity in response to synthetic peptide (sSHP-C7, at a final concentration of 100 nM) for each reporter strain was measured as the method described above in S.mutans “test-bed” reporter assay.
HA Production Determination
Exopolysaccharide produced by S. zooepidemicus was identified and confirmed as HA previously (Vázquez et al., 2015). For exploring the effect of rgg-shp pathway on the exopolysaccharide production in S. zooepidemicus, S. zooepidemicus wild type and pathway deletion mutant Δrgg-shp were cultivated in THYE medium containing 0.2 % sucrose with agitation (200 rpm) at 37°C. Samples were taken at 2, 4, 6, 8, 12, 16, and 24 h, respectively. HA concentration for each sample was determined by CTAB Turbidimetric method as described previously (Chen and Wang, 2009). Briefly, the sample culture was capsule-released by adding 0.1% (w/v) SDS and centrifuged at 10,000 g for 10 min to remove cells. The supernatant was mixed with three volumes of absolute ethanol and held at 4°C for 1 h. The precipitant was collected by centrifugation at 5,000 g for 10 min and re-dissolved in one volume distilled water. Two volumes of cetyltrimethyl-ammoniun bromide (CTAB) buffer (2.5 g L−1) were added to the samples and mixed gently for a reaction time of 10 min at room temperature, followed by OD600 measurement. HA concentration was determined based on the OD600 and a standard curve, which was established using an HA stock solution prepared using standard HA (Sigma).
Biofilm Assay
Biofilm assays were performed as described before (O'Toole, 2011; Yi et al., 2014), with some modifications. Briefly, bacterial strains were grown overnight in THYE medium at 37°C, then diluted 1:10 into fresh CDM containing 1% fibrinogen, and 0.6 ml volumes were aliquoted to three separate wells of a 96-well polystyrene plate. When indicated, synthetic peptide (sSHP-C7) was added to a final concentration of 100 nM. Plates were incubated at 37°C for 24 h without shaking. Medium alone served as a negative control. Biofilm production was determined using crystal violet staining. Briefly, the biofilm sample was visualized by staining with 1 % crystal violet for 10 min after washing with PBS. After adding 200 μl of 95% ethanol, A595 was measured by an Infinite 200PRO multi-plate reader. The experiment was repeated three times.
Data Analysis
All data points shown in this study represent the average values from three independent experiments. Error Bars show standard deviations. Statistical analysis was performed by using the two-tailed Student's t-test. P < 0.05 was considered to be statistically significant. Statistical significance was concluded at *P < 0.05, **P < 0.01, ***P < 0.001.
Results
Presence of rgg-Like Gene in S. zooepidemicus
The sequences of the characterized Rgg proteins (Rgg2 and Rgg3) from S. pyogenes were used as templates for homology searching in the genome of S. zooepidemicus ATCC 35246 (GenBank accession number: NC_017582). The best matches for both searches were found in the same gene (SeseC_00763). SeseC_00763 (designated as rgg) shows 31 and 30% identities to rgg2 and rgg3, respectively. Since most Rgg regulator genes are located close to their associated pheromone-encoding genes, a closer examination of the rgg locus was performed. We identified a 78-bp Open reading frame (ORF) in the vicinity of rgg, that was not annotated in the public database. The genomic coordinates of the small ORF are at bases 632,144 to 632,221. An invert repeat motif was found in the intergenic region between rgg and the small ORF, which is predicted to be the binding site of Rgg (Figure 1A). The small ORF encodes a 25-amino-acid hydrophobic peptide and was therefore referred to shp. Shp shows no sequence similarity with SHP2 and SHP3 from GAS (Figure 1B), whereas the putative product of shp meets the criteria of short hydrophobic peptides (SHPs) established by Ibrahim et al (Ibrahim et al., 2007). Furthermore, we performed homology searches in other strains of S. equi by using Rgg as a template and found that all the 15 fully sequenced strains of S. equi available in the NCBI database, including 10 stains of S. equi ssp zooepidemicus and 5 strains of S. equi ssp equi, carry an Rgg-homologous protein with more than 98% identity. This indicates that the system is widely conserved in different strains of S. equi species.
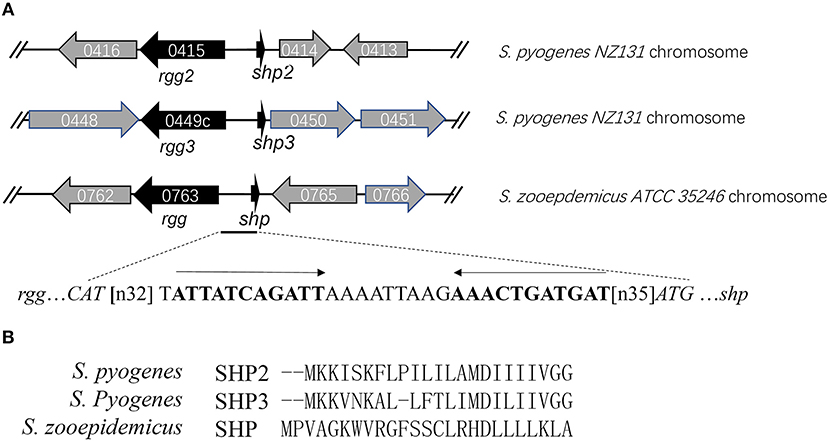
Figure 1. Rgg/SHP signaling system in the S. zooepidemicus genome. (A) The genetic organization of rgg and shp. The inverted repeat located in the intergenic region is shown in bold. (B) Alignment of pre-peptides SHP2, SHP3 (from S. pyogenes) and SHP (from S. zooepidemicus) indicating their sequence divergence.
The SHP Pheromone Acts as an Autoinducer via Rgg
The study results of Rgg quorum sensing pathways in other Gram-positive bacteria indicate that the SHP pheromones usually undergo post-translational processing steps to release the C-terminal five to eight amino acids of the peptide as the active pheromone, and that the active SHP pheromone frequently forms a positive feedback loop with the corresponding Rgg regulator, stimulating a fast response to activate (or repress) the expression of SHP encoding gene (Fleuchot et al., 2013). S. mutans has been successfully used as the heterologous expression host to study the interaction between different Rgg regulators and SHP pheromones from different species (Shanker et al., 2016). A similar assay was performed in our study to make a fast assessment on the response of Rgg to SHP peptide from S. zooepidemicus. The fact that S. mutans does not contain the shp homolog of S. zooepidemicus make it feasible to use S. mutans as the test bed.
To do the S. mutans test bed experiment, we firstly constructed two luciferase transcriptional reporter plasmids pPluc and pRPluc. In the plasmid pPluc, the full intergenic region between rgg and shp from S. zooepidemicus is fused with fire fly luciferase gene luc, which is driven by the promoter of shp (Pshp). The structure of pRPluc is similar as pPluc, except that an intact rgg is included in pRPluc. These two reporter plasmids were transferred into S.mutans UA159 to obtain strain 159Pluc and 159RPluc respectively, allowing us to test whether the Rgg could regulate the expression of Pshp in response to added synthetic SHP peptides. Based on the sizes of known SHP pheromones from other bacteria, we synthesized five peptides corresponding to truncated C-terminal derivatives of Shp. An SHP from S. thermophilus (LPYFAGCL) was also synthesized as a negative-control peptide (Fontaine et al., 2010). The peptides were tested in chemically defined medium (CDM), which contains individual amino acids instead of polypeptides. Synthetic peptides were added to low-density log-phase cultures of S.mutans 159Pluc and 159RPluc. Luciferase activity was then measured 60 min thereafter. As shown in Figure 2A, all synthetic peptides, except the control peptide (sSHP-CN) from S. thermophilus, could activate Pshp in a dose dependent manner in the reporter strain of 159RPluc, while the synthetic peptide corresponding to the C-terminal 7 amino acids of Shp (sSHP-C7) exhibited the strongest induction in reporter activity. Pshp was induced by sSHP-C7 at the concentration as low as 5 nM, and approached a maximum rate at 100 nM, suggesting that sSHP-C7 is, or closely resembles, the mature form of SHP pheromone. As shown in Figure 2B, no induction in reporter activity was observed in the reporter strain of 159Pluc, indicating the reporter induction is dependent on the presence of Rgg. Taken together, the results of the test-bed assay indicate that shp from S. zooepidemicus encodes an active pheromone which acts as an autoinducer via Rgg, and that Rgg functions a transcription activator in response to the active SHP pheromone to activate the expression of shp.
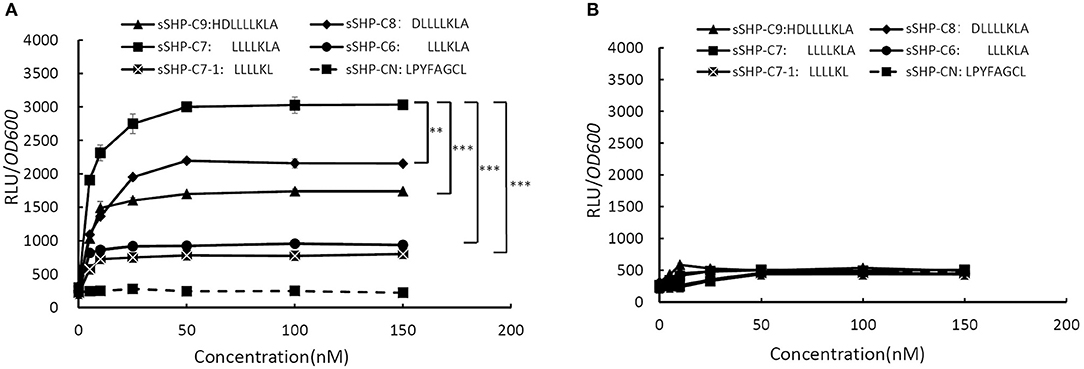
Figure 2. Effect of the sSHP pheromones on relative expression of shp promoter (Pshp) in the S. mutans test-bed assay. (A) Relative Luciferase activities (RLU/OD600) of strain 159RPluc (Streptococcus mutans UA159 containing plasmid pRPluc) in response to increasing concentrations of synthetic peptides derived from the C-terminal region of Shp (sSHP-C6, sSHP-C7-1, sSHP-C7-1, sSHP-C8, sSHP-C9) and negtive-control peptide (sSHP-CN). (B) Relative Luciferase activities (RLU/OD600) of strain 159Pluc (Streptococcus mutans UA159 containing plasmid pPluc) in response to increasing concentrations of synthetic peptides derived from the C-terminal region of Shp and negtive-control peptide (sSHP-CN). All data points shown in this figure represent the average values from three independent experiments. Error Bars show standard deviations. **P < 0.01, ***P < 0.001 (2-tailed paired t-test).
The SHP Pheromone Acts as an Autoinducer in S. zooepidemicus
The results of test bed experiment indicate that Rgg/Shp comprises an active qurom sensing system. In order to confirm it is the real case in S. zooepidemicus, we transformed the plasmid pPluc into S. zooepidemicus wild type strain. The resulting strain WTPluc were grown in CDM at 37°C, and cell density and luciferase activity were monitored throughout growth. The results presented in Figure 3A show that the expression of Pshp was low in the lag phase, and induced constantly with the increase of the OD value in the log phase. The maximum RLU/OD600 was obtained in the late-log phase and remained high when the culture entered into the stationary phase. The typical QS-regulated expression pattern of Pshp suggests that shp is regulated by a QS system in S. zooepidemicus.
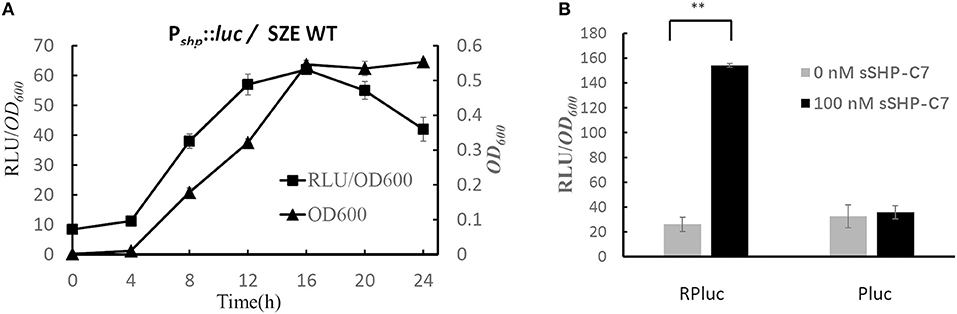
Figure 3. Rgg and Shp comprise an active autoinducing system in S. zooepidemicus. (A) The graph shows the bacterial growth (OD600) and relative luciferase activities (RLU/OD600) for strain WTPluc (wild-type S. zooepidemicus containing plasmid pPluc). The data shown are average values from three independent experiments ± standard deviations. (B) Relative Luciferase activities (RLU/OD600) of strain RPluc (S. zooepidemicusΔrgg-shp containing pRPluc) and Pluc (S. zooepidemicusΔrgg-shp containing pPluc) in response to synthetic peptide (0 nM and 100 nM). The data shown are average values from three independent experiments. Error Bars show standard deviations. **P < 0.01 (2-tailed paired t-test).
To provide direct evidence that Rgg/Shp comprise an active quorum sensing system in S. zooepidemicu, rgg/shp double deletion strain (Δrgg-shp) was constructed using the marker-less genetic manipulation system we established before (Sun et al., 2013). Luciferase reporter plasmids (pRPluc and pPluc) were transformed into Strain Δrgg-shp, respectively. The resulting strains RPluc and Pluc were cultured in CDM. Luciferase activities were measured for cultures treated with the predicted mature pheromone (sSHP-C7) and for untreated cultures. As shown in Figure 3B, gene expression of Pshp was significantly induced in cultures treated with the specific pheromone for strain Δrgg-shp containing pRPluc. While the addition of the synthetic pheromone into the strain Pluc (Δrgg-shp containing pPluc) showed no upregulation of Pshp expression compared with the untreated samples. These results indicated that Rgg/Shp is indeed a functional autoinducing system in S. zooepidemicus. Altogether, our results demonstrate that Shp is the precursor of the active pheromone, and that Rgg acts as an activator by responding to the pheromone to activate the expression of shp, forming a positive feedback regulation loop in S. zooepidemicus.
Inactivation of the Rgg/Shp System Decreases Capsule Polysaccharide Production and Biofilm Formation
With the evidence that the rgg/shp system provides S. zooepidemicus with the capability to communicate intercellularly, we next asked what social behavior does the Rgg/Shp system regulate in S. zooepidemicus. For this purpose, capsule polysaccharide (CPS) production and biofilm formation were tested and compared for wild-type and rgg/shp deletion mutants. To test if the Rgg/SHP signaling pathway is involved in the production of CPS, which is composed with HA in the case of S. zooepidemicus, HA production throughout the fermentation process was detected for wild type stain (SZE WT), shp single deletion strain (Δshp) and rgg-shp double deletion strain (Δrgg-shp). As shown in Figure 4A, both shp single deletion and rgg-shp double deletion result in lower production of HA. The final titer of Δshp and Δrgg-shp is similar, and is about 20% lower than that of wild-type, indicating Rgg/Shp signaling pathway has a positive regulation on the production of capsule polysaccharide.
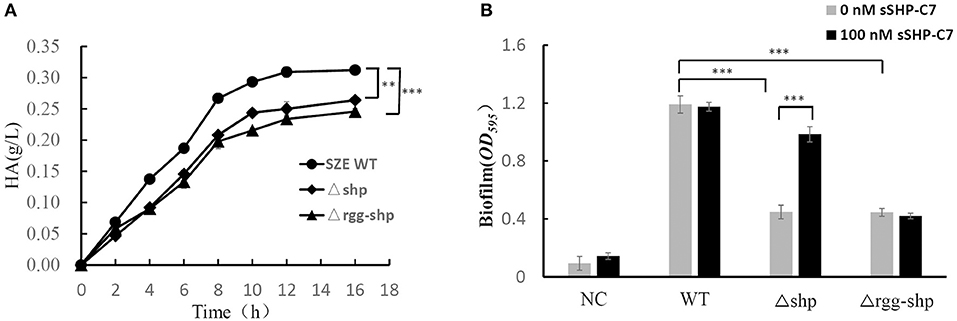
Figure 4. Rgg/Shp signaling pathway plays a key regulatory role in important social behavior processes of S. zooepidemicus. (A) HA production time course data from the wild-type S. zooepidemicus (SZE WT),deletion mutants Δrgg-shp and Δshp in flask cultures. (B) Biofilm formation of wild-type S. zooepidemicus (SZE WT), deletion mutants Δrgg-shp and Δshp when treated with 0 or 100 nM synthetic peptide, respectively. The medium without inoculation was used as the negative control (NC). All data points shown in this figure represent the average values from three independent experiments± standard deviations. **P < 0.01, ***P < 0.001 (2-tailed paired t-test).
To detect whether the Rgg/SHP signaling pathway affects the ability of biofilm formation, biofilm formation abilities of the wild-type strain, shp single deletion strain(Δshp) and rgg/shp double deletion strain(Δrgg-shp) were evaluated using a standard crystal violet staining method. As showed in Figure 4B, compared with the wild-type strain, deletion of shp results in very low production of biofilm, and rgg/shp double deletion strain exhibited similar result. We next asked if treatment with sSHP could restore the biofilm formation for those mutant strains. Biofilm formation abilities were assessed for the samples treated with 100 nM sSHP-C7. As shown in Figure 4B, exogenous addition of sSHP-C7 to wild-type did not cause an obvious change in biofilm formation; and shp deletion mutant partially restored the biofilm formation ability when sSHP-C7 was added, whereas no recovery was observed when the sSHP-C7peptide was added to rgg/shp double deletion strain. All these results indicate that Rgg/Shp signaling pathway played a positive regulatory role in the process of biofilm formation.
Taken together, our results demonstrate that the Rgg-shp system played a key regulatory role involved in important group processes, such as capsule polysaccharide production and biofilm formation.
Discussion
Intercellular signaling communication among bacteria provides communities of microbes the opportunity to coordinate gene expression to facilitate group behaviors. An established paradigm for intercellular communication among streptococci relies on peptide signaling molecules and Rgg transcriptional regulator. The signaling peptide is ribosomally-synthesized as an unmodified, linear pre-peptide that is secreted and processed to an active pheromone, then imported to the cytoplasm and recognized by Rgg regulator that alters gene expression (Fleuchot et al., 2011; Neiditch et al., 2017). However, examples of widely conserved quorum-sensing systems have largely remained unknown for S. zooepidemicus. Here we present evidence for the presence of an active QS system which is composed of Rgg regulator and a peptide signaling pheromone (encoded by shp) in S. zooepidemicus. The predicted products of shp are composited of 25 amino acids. Our data support that the mature form of signaling peptide is corresponding to the last seven amino acids of the pre-peptide Shp. So, to fulfill its putative signaling function, pre-peptide Shp must be secreted and processed through some specific pathway that releases the mature pheromone. In signal-receiving cells, mature pheromone must be imported to the cytoplasm, where it directly engages and controls the activity of Rgg transcriptional regulator. The mechanism about how Shp is processed, exported and imported remains to be studied. It should be noted that the Rgg transcriptional regulator identified in this study is conserved in all sequenced genomes of S. equi, indicating that it is a conserved signaling system in the species of S. equi. To the best of our knowledge, this is the first demonstration of Rgg-SHP mediated cell-cell signaling in the species of S. equi. It is intriguing to point out that active signal peptide (LLLLKLA) identified in S. zooepidemicus shows divergent sequence with any known SHP pheromones in other species. It is tempting to speculate that the Rgg/Shp pathway identified in this study is involved in intra-species communication, not inter-species communication.
An emerging theme in social behaviors controlled by quorum sensing is the regulation of surface polysaccharide expression. The connection between quorum sensing and extracellular polysaccharide production has been reported in a wild range of bacteria, such as S. pneumoniae (Junges et al., 2017), Staphylococcus aureus and Vibrio vulnificus (Zhao et al., 2010; Lee et al., 2013). In this study, inactivation of rgg/shp system led to a decreased exopolysaccharide (HA) production, indicating the rgg/shp system has a positive regulatory role on the capsule HA production in S. zooepidemicus. Extracellular polysaccharides have long been known to affect biofilm formation in other bacteria (Vu et al., 2009; Limoli et al., 2015). Our study did show that rgg/shp system positively affects the production of biofilm in S. zooepidemicus, however at the current stage, we cannot conclude that rgg/shp system affects the biofilm formation through the regulation of the extracellular polysaccharide. Altogether, our data support that rgg/shp system positively regulates both capsule polysaccharide production and biofilm formation in S. zooepidemicus. Although the direct target of the Rgg/Shp signaling pathaway and its mechanism in the regulation of exopolysaccharide production and biofilm formation remains unknown at current stage, we have provided the groundwork for studies aimed at revealing the role of rgg/shp system in coordinating bacterial social behavior in S. zooepidemicus.
An increased understanding of how cell-cell signaling systems work is leading to new possibilities for manipulating bacterial functions. In the clinical area, some interesting studies have been conducted recently to target QS as a tool to combat or modulate infections associated with bacterial biofilms (Aggarwal et al., 2015; Ishii et al., 2017). In biotechnology area, QS in bacteria has been applied for induction of recombinant protein expression (Tsao et al., 2010), dynamical regulation of the expression of target genes (Gupta et al., 2017), microbial biosensors designing, etc (Choudhary and Schmidt-Dannert, 2010). So far as we know, our study is the first demonstration of Rgg/SHP mediated cell-cell signaling in S. zooepidemiccus, the significant streptococci species both in the clinical area and in the industrial area. Our finding is of importance for the design of quorum sensing inhibitory strategies in controlling S. zooepidemiccus infection, and is also of importance for metabolic engineering of S. zooepdemicus in commercially producing HA products by providing an autoinducing regulation module.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files. The datasets generated for this study can be found in GenBank, accession number: NC_017582.
Author Contributions
ZX, HL, and WC designed the experiments. ZX, KM, XY, JL, and JY conducted most of the studies. CZ took part in HA production determination and data analysis. ZX, HL, and WC analyzed data. ZX, HL, and WC wrote the paper with the contributions from all other authors.
Funding
The study was funded in part by National Natural Science Foundation of China (31570050 and 21706192), a grant from Tianjin Science and Technology Committee of China (18YFZCSY01360), and a grant from Tianjin Engineering Research Center of Microbial Metabolism and Fermentation Process Control (17PTGCCX00190).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aggarwal, C., Jimenez, J. C., Lee, H., Chlipala, G. E., Ratia, K., and Federle, M. J. (2015). Identification of quorum-sensing inhibitors disrupting signaling between Rgg and short hydrophobic peptides in Streptococci. mBio 6, e00393–e00315. doi: 10.1128/mBio.00393-15
Ajdić, D., McShan, W. M., McLaughlin, R. E., Savić, G., Chang, J., Carson, M. B., et al. (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Antimicrob. Agents Chemother. 99, 14434–14439. doi: 10.1073/pnas.172501299
Chang, J. C., LaSarre, B., Jimenez, J. C., Aggarwal, C., and Federle, M. J. (2011). Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. Plos. Pathog. 7:e1002190. doi: 10.1371/journal.ppat.1002190
Chen, Y. H., and Wang, Q. (2009). Establishment of CTAB Turbidimetric method to determine hyaluronic acid content in fermentation broth. Carbohyd. Polym. 78, 178–181. doi: 10.1016/j.carbpol.2009.04.037
Choudhary, S., and Schmidt-Dannert, C. (2010). Applications of quorum sensing in biotechnology. Appl. Microbiol. Biotechnol. 86, 1267–1279. doi: 10.1007/s00253-010-2521-7
Fleuchot, B., Gitton, C., Guillot, A., Vidic, J., Nicolas, P., Besset, C., et al. (2011). Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80, 1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x
Fleuchot, B., Guillot, A., Mézange, C., Besset, C., Chambellon, E., Monnet, V., et al. (2013). Rgg-associated SHP signaling peptides mediate cross-talk in Streptococci. PLoS ONE 8:e66042. doi: 10.1371/journal.pone.0066042
Fontaine, L., Boutry, C., de Frahan, M. H., Delplace, B., Fremaux, C., Horvath, P., et al. (2010). A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192, 1444–1454. doi: 10.1128/JB.01251-09
Gupta, A., Reizman, I. M., Reisch, C. R., and Prather, K. L. (2017). Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35, 273–279. doi: 10.1038/nbt.3796
Ibrahim, M., Nicolas, P., Bessières, P., Bolotin, A., Monnet, V., and Gardan, R. (2007). A genome-wide survey of short coding sequences in streptococci. Microbiology 153, 3631–3644. doi: 10.1099/mic.0.2007/006205-0
Ishii, S., Fukui, K., Yokoshima, S., Kumagai, K., Beniyama, Y., Kodama, T., et al. (2017). High-throughput screening of small molecule inhibitors of the Streptococcus quorum-sensing signal pathway. Sci. Rep.7:4029. doi: 10.1038/s41598-017-03567-2
Jimenez, J. C., and Federle, M. J. (2014). Quorum sensing in group A streptococcus. Front. Cell. Infect. 4:127. doi: 10.3389/fcimb.2014.00127
Junges, R., Salvadori, G., Shekhar, S., Åmdal, H. A., Periselneris, J. N., Chen, T., et al. (2017). A quorum-sensing system that regulates Streptococcus pneumoniae biofilm formation and surface polysaccharide production. Msphere 2:e00324-17. doi: 10.1128/mSphere.00324-17
Kawamura, Y., Hou, X. G., Sultana, F., Miura, H., and Ezaki, T. (1995). Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J. Syst. Bacteriol. 45, 406–408. doi: 10.1099/00207713-45-2-406
LeBlanc, D. J., Lee, L. N., and Abu-Al-Jaibat, A. (1992). Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28, 130–145. doi: 10.1016/0147-619X(92)90044-B
Lee, K. J., Kim, J. A., Hwang, W., Park, S. J., and Lee, K. H. (2013). Role of capsular polysaccharide (CPS) in biofilm formation and regulation of CPS production by quorum-sensing in Vibrio vulnificus. Mol. Microbiol. 90, 841–857. doi: 10.1111/mmi.12401
Limoli, D. H., Jones, C. J., and Wozniak, D. J. (2015). Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectrum 3:MB-0011-2014. doi: 10.1128/microbiolspec.MB-0011-2014
Liu, L., Liu, Y., Li, J., Du, G., and Chen, J. (2011). Microbial production of hyaluronic acid: current state, challenges, and perspectives. Microb. Cell Fact. 10:99. doi: 10.1186/1475-2859-10-99
Lyon, G. J., and Novick, R. P. (2004). Peptide signaling in Staphylococcus aureus and other gram-positive bacteria. Peptides 25, 1389–1403. doi: 10.1016/j.peptides.2003.11.026
Ma, Z., Geng, J., Zhang, H., Yu, H., Yi, L., Lei, M., et al. (2011). Complete genome sequence of Streptococcus equi subsp. zooepidemicus strain ATCC 35246. J. Bacteriol. 193, 5583–5584. doi: 10.1128/JB.05700-11
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Neiditch, M. B., Capodagli, G. C., Prehna, G., and Federle, M. J. (2017). Genetic and structural analyses of RRNPP intercellular peptide signaling of gram-positive bacteria. Annu. Rev. Genet. 51, 311–333. doi: 10.1146/annurev-genet-120116-023507
O'Toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Visual. Exp. 47:e2437. doi: 10.3791/2437
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Pelkonen, S., Lindahl, S. B., Suomala, P., Karhukorpi, J., Vuorinen, S., Koivula, I., et al. (2013). Transmission of Streptococcus equi subspecies zooepidemicus infection from horses to humans. Emerg. Infect. Dis. 19, 1041–1048. doi: 10.3201/eid1907.121365
Rutherford, S. T., and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427
Shanker, E., Morrison, D. A., Talagas, A., Nessler, S., Federle, M. J., and Prehna, G. (2016). Pheromone recognition and selectivity by ComR proteins among streptococcus species. PLoS Pathog. 12:e1005979. doi: 10.1371/journal.ppat.1005979
Sun, X., Yang, D., Wang, Y., Geng, H., He, X., and Liu, H. (2013). Development of a markerless gene deletion system for Streptococcus zooepidemicus: functional characterization of hyaluronan synthase gene. Appl. Microbiol. Biotechnol. 97, 8629–8636. doi: 10.1007/s00253-013-5058-8
Tsao, C. Y., Hooshangi, S., Wu, H. C., Valdes, J. J., and Bentley, W. E. (2010). Autonomous induction of recombinant proteins by minimally rewiring native quorum sensing regulon of E. coli. Metab. Eng. 12, 291–297. doi: 10.1016/j.ymben.2010.01.002
van de Rijn, I., and Kessler, R. E. (1980). Growth characteristics of group A streptococci in a new chemically defined medium. Infect. Immun. 27, 444–448.
Vázquez, J. A., Pastrana, L., Piñeiro, C., Teixeira, J. A., Pérez-Martín, R. I., and Amado, I. R. (2015). Production of hyaluronic acid by Streptococcus zooepidemicus on protein substrates obtained from Scyliorhinus canicula Discards. Mar. Drugs 13, 6537–6549. doi: 10.3390/md13106537
Vu, B., Chen, M., Crawford, R. J., and Ivanova, E. P. (2009). Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554. doi: 10.3390/molecules14072535
Waller, A. S. (2017). Science-in-brief: Streptococcus zooepidemicus: a versatile opportunistic pathogen that hedges its bets in horses. Equine Vet. J. 49, 146–148. doi: 10.1111/evj.12658
Xie, Z., Qi, F., and Merritt, J. (2013a). Cloning-independent plasmid construction for genetic studies in streptococci. J. Microbiol. Methods. 94, 77–82. doi: 10.1016/j.mimet.2013.05.005
Xie, Z., Qi, F., and Merritt, J. (2013b). Development of a tunable wide-range gene induction system useful for the study of streptococcal toxin-antitoxin systems. Appl. Environ. Microbiol. 79, 6375–6384. doi: 10.1128/AEM.02320-13
Xu, B., Pei, X., Su, Y., Ma, Z., and Fan, H. (2016). Capsule of Streptococcus equi subsp. zooepidemicus hampers the adherence and invasion of epithelial and endothelial cells and is attenuated during internalization. FEMS Microbiol. Lett. 363:fnw164. doi: 10.1093/femsle/fnw164
Yi, L., Wang, Y., Ma, Z., Zhang, H., Li, Y., Zheng, J. X., et al. (2014). Biofilm formation of Streptococcus equi ssp. zooepidemicus and comparative proteomic analysis of biofilm and planktonic cells. Curr. Microbiol. 69, 227–233. doi: 10.1007/s00284-014-0574-z
Keywords: quorum sensing, capsule polysaccharide, hyaluronic acid, biofilm, Streptococcus zooepidemicus
Citation: Xie Z, Meng K, Yang X, Liu J, Yu J, Zheng C, Cao W and Liu H (2019) Identification of a Quorum Sensing System Regulating Capsule Polysaccharide Production and Biofilm Formation in Streptococcus zooepidemicus. Front. Cell. Infect. Microbiol. 9:121. doi: 10.3389/fcimb.2019.00121
Received: 15 February 2019; Accepted: 04 April 2019;
Published: 18 April 2019.
Edited by:
Suzanne R. Dawid, University of Michigan, United StatesReviewed by:
Michael S. Chaussee, University of South Dakota, United StatesKyu Hong Cho, Indiana State University, United States
Copyright © 2019 Xie, Meng, Yang, Liu, Yu, Zheng, Cao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Cao, Y2Fvd2VpMDQwMTA4QHR1c3QuZWR1LmNu
Hao Liu, bGl1aGFvQHR1c3QuZWR1LmNu
 Zhoujie Xie
Zhoujie Xie Kai Meng
Kai Meng Xiaoli Yang1
Xiaoli Yang1 Wei Cao
Wei Cao Hao Liu
Hao Liu