- 1College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
- 2Shenzhen Center for Disease Control and Prevention, Shenzhen, China
- 3College of Life Sciences, Sichuan University, Chengdu, China
- 4School of Life Sciences, Xiamen University, Xiamen, China
The dynamic nature of Vibrio parahaemolyticus epidemiology has presented a unique challenge for disease intervention strategies. Despite the continued rise of disease incidence and outbreaks of vibriosis, as well as the global emergence of pandemic clones and serovariants with enhanced virulence, there is a paucity of molecular methods for the serotyping of V. parahaemolyticus strains to improve disease surveillance and outbreak investigations. We describe the development of a multiplex ligation reaction based on probe melting curve analysis (MLMA) for the simultaneous identification of 11 clinically most common V. parahaemolyticus serotypes spanning a 10-year period. Through extensive sequence analyses using 418 genomes, specific primers and probes were designed for a total of 22 antigen gene targets for the O- and K- serogroups. Additionally, the toxR gene was incorporated into the assay for the confirmation of V. parahaemolyticus. All gene targets were detected by the assay and gave expected Tm values, without any cross reactions between the 11 clinically common serotypes or with 38 other serotypes. The limit of identification for all gene targets ranged from 0.1 to 1 ng/μL. The intra- and inter-assay standard deviations and the coefficients of variation were no more than 1°C and <1% respectively, indicating a highly reproducible assay. A multicenter double-blind clinical study was conducted using the traditional V. parahaemolyticus identification workflow and the MLMA assay workflow in parallel. From consecutive diarrheal stool specimens (n = 6118) collected over a year at 10 sentinel hospitals, a total of 153 V. parahaemolyticus isolates (2.5%) were identified by both workflows. A total agreement (kappa = 1.0) between the serotypes identified by the MLMA assay and conventional serological method was demonstrated. This is the first molecular assay to simultaneously identify multiple clinically important V. parahaemolyticus serotypes, which satisfies the acute need for a practical, rapid and robust identification of V. parahaemolyticus serotypes to facilitate the timely detection of vibriosis outbreaks and surveillance.
Introduction
Vibrio parahaemolyticus is a halophilic bacterium that thrives in estuarine, marine, and coastal waters and is recognized as the primary cause of gastroenteritis associated with the consumption of seafood and marine products worldwide (Cabrera-Garcia et al., 2004; Mclaughlin et al., 2005; Letchumanan et al., 2014). In China, V. parahaemolyticus has been the leading cause of foodborne outbreaks and infectious diarrhea among adults, particularly in coastal cities (Li Y. et al., 2014; Liu et al., 2018). According to the Foodborne Disease Outbreak Surveillance System (FDOSS) in China, a year-on-year increase in the number of outbreaks due to V. parahaemolyticus was observed between 2011 and 2016, having caused more outbreaks than other major foodborne pathogens for each given year (Liu et al., 2018). Similarly, a clear upward trend in the incidence of foodborne vibriosis was observed in the United States in recent years (Newton et al., 2012; Marder et al., 2017). Indeed, the incidence of vibriosis has been rising, and there is a growing body of evidence that this could become increasingly common due in part to the rising sea water temperatures from the effects of global warming that could favor its spread (Baker-Austin et al., 2010, 2017; Vezzulli et al., 2013). Currently, 13 O- serogroups and 71 K- serogroups are recognized based on the antigenic properties of the somatic (O) and capsular (K) antigens, respectively (Iguchi et al., 1995; Oliver and Jones, 2015). V. parahaemolyticus infections are recognized as a multiserogroup affliction caused by diverse serotypes (Bhuiyan et al., 2002), leading primarily to acute gastroenteritis, and occasionally, wound infection and septicemia (Daniels et al., 2000). In 1996, the emergence of a more virulent serotype O3:K6 was subsequently being recognized as a pandemic clone owing to its rapid dissemination and the cause of numerous gastroenteritis outbreaks globally (Nair et al., 2007). To date, the pandemic clone and its serovariants are known to represent 49 serotypes, which are widely distributed among 22 countries in Asia, Europe, America and Africa (Han et al., 2016). In China, up to 27 pandemic serovariants have been found amongst clinical samples, in which O3:K6, O4:K68, and O1:KUT predominated (Han et al., 2017). Hence, the serotyping of V. parahaemolyticus has played a pivotal role in significantly advancing our understanding of its epidemiology.
Currently, the serotyping of V. parahaemolyticus is performed routinely using the traditional serum slide agglutination method. However, this conventional serological method suffers from several major practical and technical drawbacks, including tedious and time-consuming procedures, ambiguous results arising from cross reactivity, batch-to-batch inconsistencies, and subjectivity in results interpretation. Despite these drawbacks, the development of molecular methods for the serotyping of V. parahaemolyticus has been scarce, with only a few methods described to-date. A conventional PCR-based method has been described for the detection and identification of V. parahaemolyticus O-serogroups based on O-serogroup genetic determinants (OGDs) (Chen et al., 2012), which was subsequently extended to include three additional provisional O-serogroups, but without any K- serogroups (Guo et al., 2017). Another conventional PCR was developed to detect an arbitrary marker for the identification of a single serotype O3:K6 (Yeung et al., 2003). A MALDI-ToF mass spectrometry method have also been explored for the serotyping of V. parahaemolyticus, albeit with usage limited to differentiate O4:K8 and non-O4:K8 isolates (Li et al., 2018). These methods suffer from practical limitations such as low throughput, cumbersome procedures, ambiguous results arising from gel electrophoresis, and the ability to identify limited serogroups and serotypes.
In recent years, multiplex ligation reaction based on the probe melting curve analysis (MLMA) has been shown to be a robust and high throughput method that has been successfully applied for the simultaneous detection of 10 bacterial pathogens (Jiang et al., 2017). This method combines the use of fluorescence color and melting temperature (Tm) as a virtual two-dimensional label that allows the homogenous detection of multitudes of gene targets (Liao et al., 2013). Based on the principles of MLMA, we present the first molecular assay to simultaneously identify 11 clinically most common V. parahaemolyticus serotypes. Coupled with the use of basic laboratory protocols and inexpensive equipment, the current assay offers a practical solution to facilitate the early identification of V. parahaemolyticus serotypes, which is an essential strategy for the successful prevention and control of vibriosis incidence and outbreaks.
Materials and Methods
Reference Bacterial Isolates
A total of 244 reference bacterial isolates were used for the initial development and evaluation of the MLMA assay. This comprised of reference isolates (n = 176) representing 11 clinically most common V. parahaemolyticus serotypes during a 10-year period (2007–2017) from the Shenzhen Center for Disease Control and Prevention isolates collection (Table 1); and reference isolates (n = 66) representing additional 38 serotypes for detecting cross reactions (Table S1). For completeness, two rare serogroups absent during the 10 year period (O7 and O12) were incorporated into the assay using Escherichia coli TOP10 strains (n = 2) containing serogroup-specific genes (O7-wvcN, O12-wvcP) cloned into a pUC57 vector (Sangon Biotech Co. Ltd., Shanghai, China).

Table 1. Reference isolates (n = 176) representing 11 clinically most common V. parahaemolyticus serotypes over a 10-year period (2007–2017).
Bacterial Culture and Conventional Serotyping
Stool specimens from sentinel hospitals in the multicenter double-blind study were enriched in alkaline peptone water [(pH 8.6); 3% NaCl] and incubated at 37°C for 16 h on a shaker; then streaked onto Vibrio chromogenic agar incubated at 37°C for 12 h for single colonies (Guangdong Huankai Microbial Science and Technology, Guangzhou, China).
Suspected colonies were picked and streaked onto triple sugar iron slants (Guangdong Huankai Microbial Science and Technology, Guangzhou, China) and incubated at 37°C for 16 h followed by serotyping by serum slide agglutination tests using commercial antisera (Denka Seiken, Tokyo, Japan) according to the manufacturer's protocol and the Chinese National Food Safety Standards: Food Microbiological Examination Vibrio parahaemolyticus Testing, GB 4789.7-2013. Two E. coli TOP10 strains with pUC57 vector for O7 and O12 serogroup-specific genes were cultured onto LB medium (Guangdong Huankai Microbial Science and Technology, Guangzhou, China) and incubated at 37°C on a shaker for 16 h.
DNA Extraction
DNA templates for reference and clinical isolates were prepared using the boiled lysates method. A single colony was suspended in 50 μl of distilled water, then boiled at 100°C for 8 min and centrifuged at 12,000×g for 10 min, with supernatant used as DNA template. Plasmid DNA templates were extracted from two E. coli TOP10 strains using the SanPrep Plasmid MiniPrep Kit (Sangon Biotech Co. Ltd., Shanghai, China). The concentration of all DNA templates was measured spectrophotometrically (Nanodrop ND-1000, Wilmington, Delaware, USA).
Primers and Probes Design
In the current assay, the target gene loci for of O- and K- antigens of V. parahaemolyticus were located between dgkA and gmhD genes (Chen et al., 2012) and between gmhD and rjg genes (Chen et al., 2010), respectively, as previously determined. We sequenced 418 genomes and performed sequence analyses to identify polymorphic sites specific among the serogroup antigen genes using multiple alignments (MEGA7.0). The toxR gene was incorporated into the assay as a target gene for the confirmation of V. parahaemolyticus as previously described (Jiang et al., 2017). Designed primers (Primer Premier v5.0) and probes (DNA folding software, http://unafold.rna.albany.edu/?q=mfold/DNA-Folding-Form) were evaluated using the BlastN algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Fluorogenic probes were labeled at 5′ end with reporter fluorophores carboxy fluorescein (FAM), carboxy-X-rhodamine (ROX), or indodicarbocyanine (Cy5) and with Black Hole Quencher sequestering molecule at the 3′ end. All primers and probes were synthesized by Sangon Biotech Co., Ltd. (Table 2).
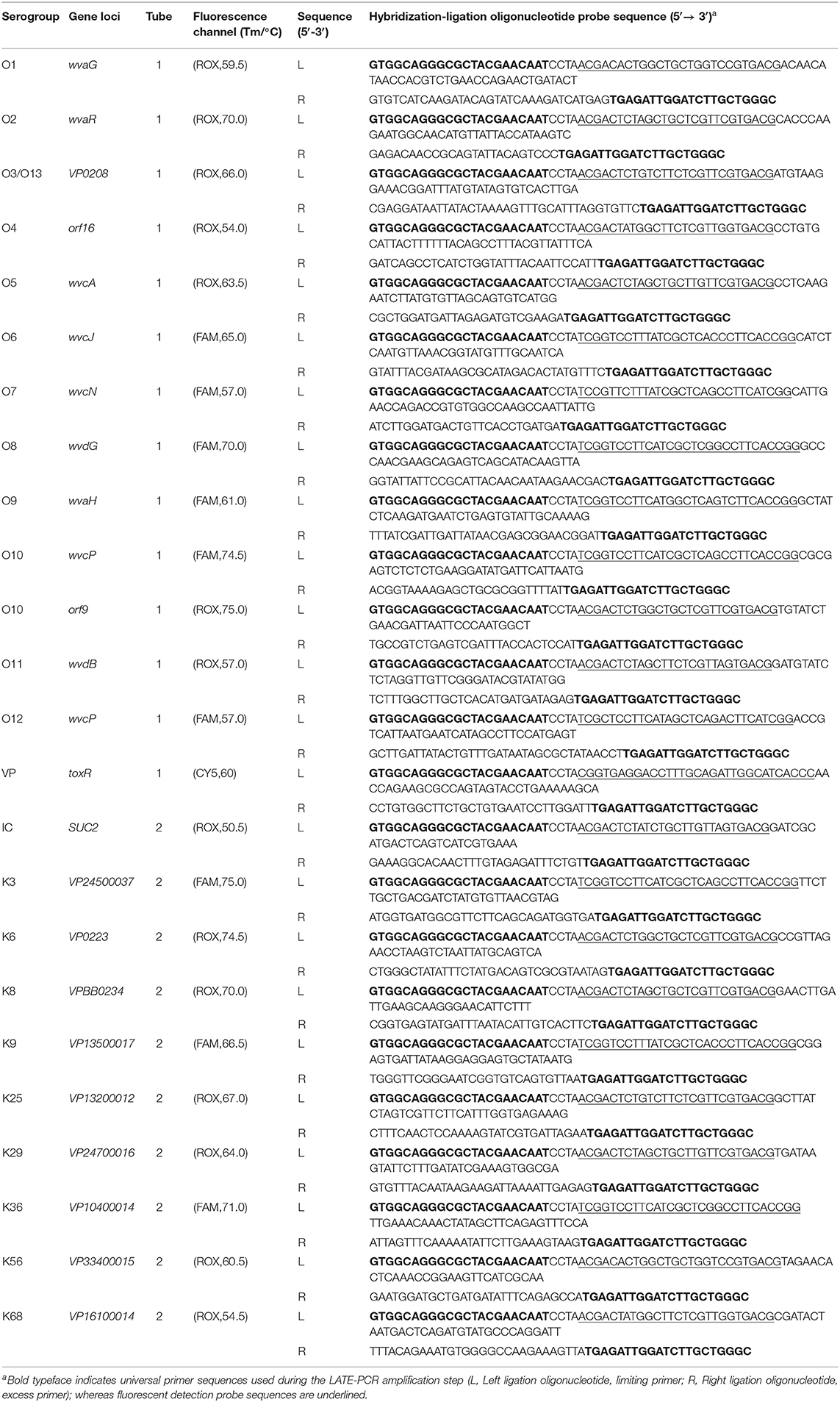
Table 2. MLMA assay for the identification of V. parahaemolyticus O- and K- serogroups in a two-tube system.
Development of the MLMA Assay
The MLMA assay involves two main steps as previously described (Jiang et al., 2017): (1) a hybridization-ligation process; and (2) a PCR amplification and melt curve analysis. During (1) hybridization-ligation, a pair of oligonucleotide probes hybridizes with the flanking sequences on either side of the target gene loci, which is ligated only upon complete hybridization. (2) Using a universal PCR primer pair via Linear After-The-Exponential (LATE) PCR, the ligated product is first amplified then hybridized with fluorescently labeled detection probes. This allows V. parahaemolyticus O- and K- serogroups to be determined based on unique melting temperatures (Tm) obtained using a melt curve analysis established by corresponding fluorescent detection probes in the respective fluorophore channels.
The hybridization and ligation reaction was performed using a T3 Thermocycler (Biometra, Germany). Each 10 μl reaction mix contained a 1.5 μl of ligation probe mix (2.5–4 fmol of each hybridization oligonucleotide probe, Table 2), 1U Taq DNA ligase and 1 μl DNA ligase buffer (New England Biolabs, Beijing, China), 1.5 μl sterilized water, and 5 μl DNA template. Reaction conditions include an initial DNA denaturation step at 95°C for 5 min containing only genomic DNA and ligation probe mix and allowed to reach 75°C before the addition of the remainder of reaction mixture (3.5 μl). This is followed by an incubation step at 60°C for 80 min and 95°C for 5 min. The PCR amplification and melting curve analysis was performed on a Bio-Rad CFX 96 real-time PCR system (Bio-Rad Inc., Hercules, CA). Each 50 μl reaction mixture consisted of 1× PCR buffer, 3 μl MgCl2 (25 mM), 4 μl of deoxynucleoside triphosphate (2.5 mM), 1U of Taq polymerase (TaKaRa Biotech Co., Dalian, China), 0.3μl limiting primer (2.5 μM), 0.3μl excess primer (50 μM), 0.20 μM fluorogenic probes (Table 2), and 5 μl of ligation product from the hybridization-ligation step. Amplification began with a denaturation step at 95°C for 3 min, followed by 38 cycles of 95°C for 5 s, 56°C for 15 s, and then 74°C for 15 s. This is followed by melt curve analysis that began with a denaturation step at 95°C for 2 min, hybridization at 40°C for 2 min, and a stepwise temperature rise from 40°C to 85°C at 1°C per step with 5 s for each step. Fluorescence from the FAM, ROX, and Cy5 channels were obtained and recorded automatically during the 38 amplification cycles, where a melt curve analysis was processed through the CFX Manager 3.0 software.
Analytical Performance of the MLMA Assay
The limit of identification was determined using a series of 10-fold dilutions in triplicates (from 10 to 0.01 ng/μl) of purified DNA templates from isolates corresponding to V. parahaemolyticus O- and K- serogroups. The reproducibility of the assay was evaluated using two sets of 10-fold serial dilutions (100–10 ng/μl) of DNA templates measured in triplicates. The standard deviations and coefficient of variation values of the intra-assay and inter-assay were calculated.
Multicenter Double-Blind Clinical Study
A multicenter double-blind clinical study was conducted throughout 2018 from 10 sentinel hospitals in Shenzhen to evaluate the MLMA assay compared to the conventional serotyping method. Suspected isolates from diarrheal stool samples were analyzed using the traditional V. parahaemolyticus identification workflow and the MLMA assay workflow in parallel and double-blinded. In the traditional identification workflow, biochemical testing, and conventional serotyping was performed; whereas in the MLMA workflow, DNA extraction, and MLMA assay followed. Procedures were performed as per protocols described above. The agreement between the MLMA assay and the conventional serological method was determined by calculating the kappa value.
Ethics
Stool samples from which suspected isolates originated for use in this study were collected as part of laboratory-based surveillance of infectious diarrheal disease in Shenzhen, Guangdong province, China in accordance to the public health mandate, therefore ethical clearance was not required. All suspected isolates derived from stool samples were de-identified and anonymous to protect patient privacy and confidentiality.
Results
Detection Scheme of V. parahaemolyticus O and K Serogroups
A two-tube system was developed with three fluorescence channels (ROX, FAM, and Cy5) in the first tube and two fluorescence channels (ROX and FAM) in the second tube. In the first tube that detects O-serogroup antigens, the ROX channel detects antigen genes for serogroups O1, O2, O3, O4, O5, O10-orf9, and O11; the FAM channel detects antigen genes for serogroups O6, O7, O8, O9, O10-wvcP, and O12; whereas the Cy5 channel detects the toxR gene. In the second tube that detects K-serogroup antigens, the ROX channel detects antigen genes for serogroups K6, K8, K25, K29, K56, K68, and the SUC2 gene as an internal control; whereas the FAM channel detects antigen genes for serogroups K3, K9, and K36. In this study, we have identified an additional gene locus, orf9, to be specific for serogroup O10 (Accession no: CNA0002415) and was included in the assay as a target in addition to wvcP for serogroup O10 identification. Table 2 details the experimental setup in each tube for the identification of all serogroups in the assay, including their gene loci, fluorophore channels, designated melting temperatures and probe sequences.
Performance Characteristics of the MLMA Assay
All targeted gene loci was detected by the MLMA assay, which yielded expected Tm values for each serogroup without any cross reaction (Figure 1). The limit of identification for all gene targets ranged from 0.1 to 1.0 ng/μl of DNA template for each serogroup. The standard deviation (SD) and coefficient of variation (CV) values for intra-assay and inter-assay were derived. The largest SD value for mean Tm was no more than 1°C and the CV values of the intra-assay and inter-assay ranged from 0 to 1% (Table 3).
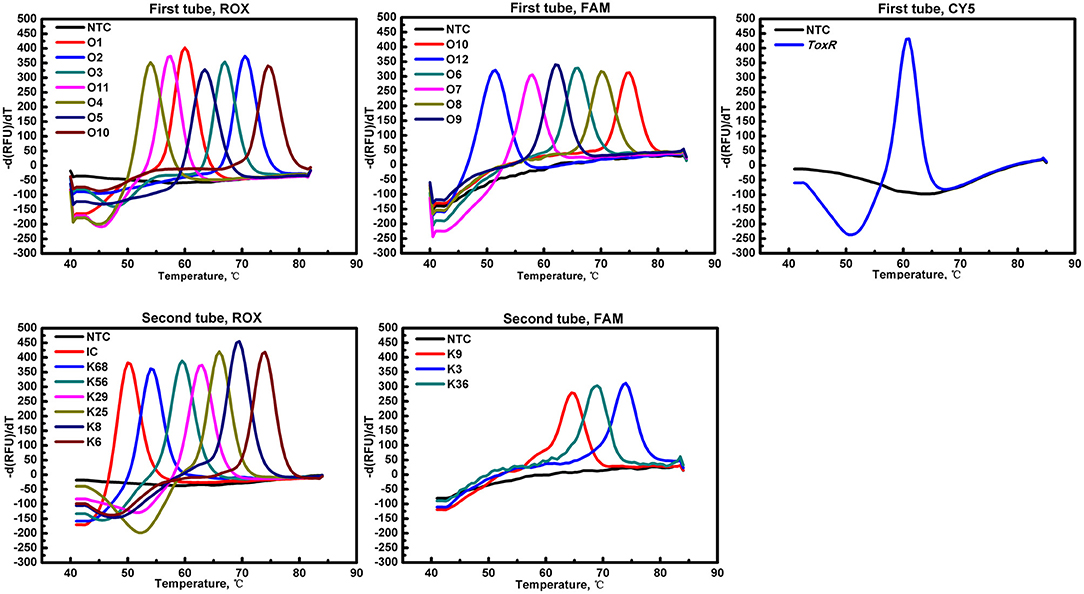
Figure 1. Probe melting curve analysis for the identification of V. parahaemolyticus O and K serogroups in the MLMA assay. Color-coded melting curves represent the different antigen genes in each of the fluorophore channels (ROX, FAM, and Cy5) in a two-tube system. IC, SUC2 gene was used as a positive internal control (IC); NTC, negative control.
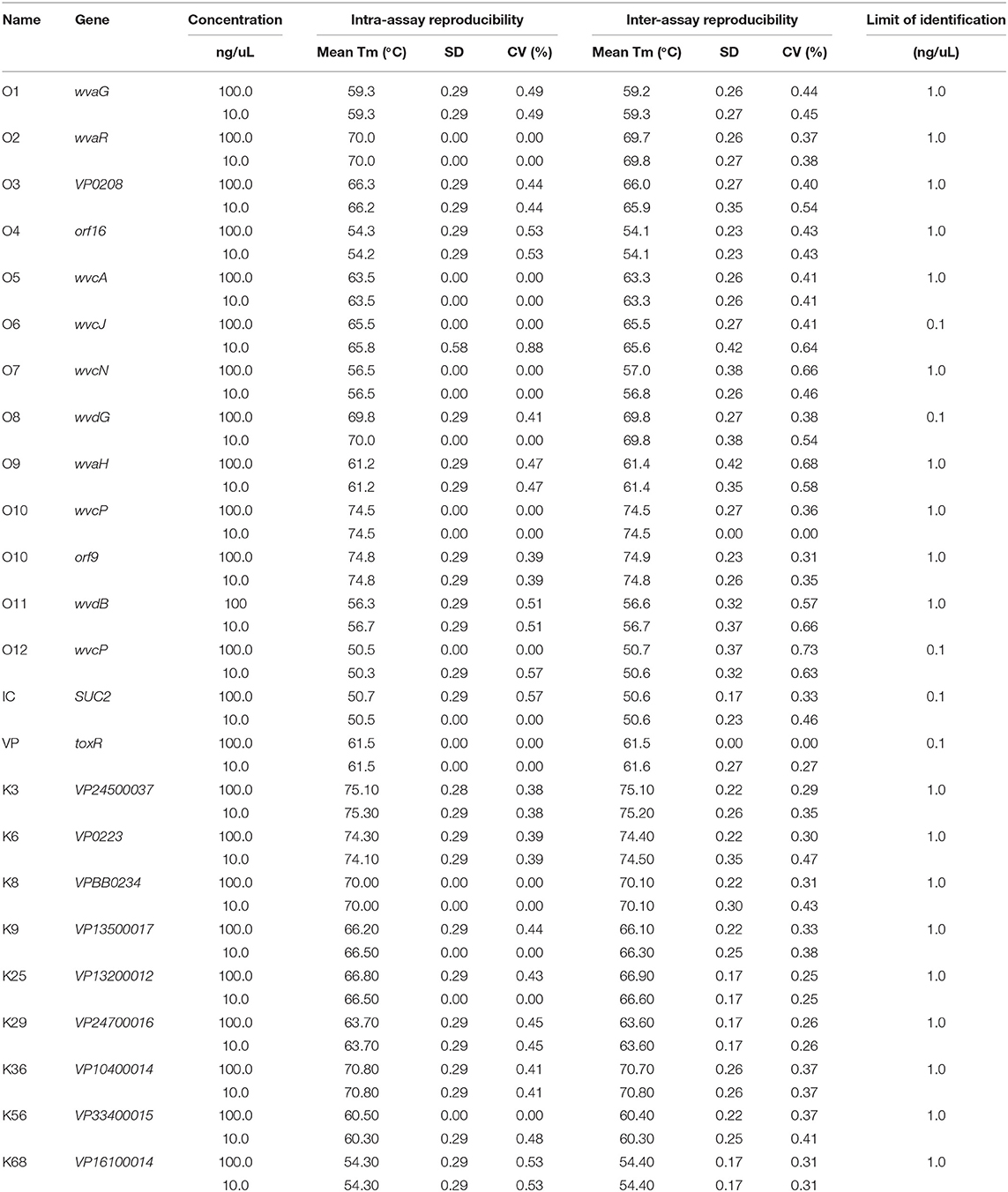
Table 3. Reproducibility of designated melting temperatures (Tm) and Limit of Identification values for each serogroup target loci in the assay.
Multicenter Double-Blind Clinical Study
From a total of 6,118 consecutive diarrheal stool specimens in 2018, 153 V. parahaemolyticus isolates (2.5%) were identified by both traditional and MLMA workflows. Among these, 149 (97.4%) isolates belonged to 9 of the 11 serotypes identifiable by the MLMA workflow, whereas 4 (2.6%) isolates belonged to 3 other serotypes. The MLMA results demonstrated kappa values of 1.0 for each of the identifiable serotypes when compared against conventional serotyping results, indicating total agreement. A full comparison of serotype identification by MLMA and conventional serotyping is available (Table 4). In the clinical setting, the MLMA assay workflow was 12–18 h faster compared to traditional workflow.
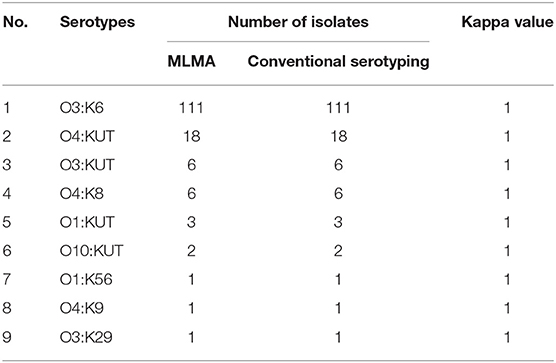
Table 4. Serotype identification by MLMA assay and conventional serotyping among clinical V. parahaemolyticus isolates (n = 149) from the multicenter double-blind study.
Discussion
The transmission dynamics of V. parahaemolyticus infections is complex. To date, the possible mechanisms behind the emergence of a virulent pandemic serotype O3:K6 clone which has disseminated rapidly worldwide since 1996 has remained elusive (Nair et al., 2007); whilst a prominent yet unexplained upward trend in disease incidence and outbreaks of vibriosis have also been observed globally in recent years (Newton et al., 2012; Marder et al., 2017; Liu et al., 2018). Since early 1970s, the tracking of disease spread has relied on conventional serological typing, which is considered the international gold standard pertaining to V. parahaemolyticus epidemiology. In light of the growing importance of this foodborne pathogen and the lack of rapid and reliable molecular serotyping methods, we have developed a novel MLMA assay for the simultaneous identification of 11 clinically common V. parahaemolyticus serotypes.
The current assay was able to detect all gene targets (Table 3) and gave expected Tm values with robust performance characteristics. The assay has shown high reproducibility where the largest SD value for mean Tm was no more than 1°C and the CVs were no more than 1% in both intra-assay and inter-assay reproducibility analyses; whereas the detection limit ranged from 0.1 to 1 ng/μl. Overall, these performance attributes are comparable to those used for the rapid identification of several other foodborne pathogens (Jiang et al., 2017). In the multicenter double-blind clinical study we conducted, the MLMA assay has shown perfect concordance with the conventional serological method for 97.4% of all clinical isolates over a year at 10 sentinel hospitals, as indicated by kappa values of 1.0 for each of the V. parahaemolyticus serotypes identified. Such superior performances of the assay can be attributed to the robust MLMA methodology, which relies on the high fidelity of DNA ligase to identify gaps in nucleotide sequences of the target loci for oligonucleotide probes to fully hybridize with the flanking nucleotides before ligation could occur. We have demonstrated that the current MLMA workflow could be completed 12–18 h faster compared to the traditional workflow that relies on biochemical tests and overnight incubation for identification. From DNA extraction, both confirmation of V. parahaemolyticus and serotyping results can be obtained in a total assay time of 3.5 h with minimal hands-on operation. Another advantage of the assay is the applicable use of a simple boiled lysates protocol for the preparation of DNA templates, as demonstrated during the clinical study on all isolates from stool specimens. This eliminates the need for laborious procedures and the high costs associated with commercial DNA extraction and purification kits. Therefore, the current assay presents a simple and pragmatic scheme for the rapid and accurate identification of V. parahaemolyticus serotypes, which is especially suited for real-time applications such as outbreak situations involving large number of samples be processed quickly in a short period of time. In developing this assay, we chose 11 V. parahaemolyticus serotypes that represented 90% of all serotypes isolated from human infections over a 10-year period (2007–2017) in Shenzhen, China; many of which are also commonly reported among infections domestically (Han et al., 2017) and worldwide (Velazquez-Roman et al., 2014; Han et al., 2016). Given its clinical relevance, the current assay could serve as an invaluable tool for clinical and public health laboratories.
For many major foodborne pathogens, such as Salmonella, a plethora of molecular serotyping methods have been developed, including the use of conventional multiplex PCRs, quantitative PCRs, bead-based suspension arrays, CRISPR-based assay and pyrosequencing assay (Munoz et al., 2010; Li R. et al., 2014; Aydin et al., 2018; Bugarel et al., 2018; Furukawa et al., 2018; Heymans et al., 2018). However, the lack of molecular serotyping described for V. parahaemolyticus has been remarkable, given the growing public health importance of this pathogen. To our knowledge, only four studies have explored the use of molecular methods for serogroup or serotype identification of V. parahaemolyticus to-date. Conventional PCR-based assays have been developed for the identification of O-serogroups (Chen et al., 2012; Guo et al., 2017) and serotype O3:K6 (Yeung et al., 2003), whereas the utility of MALDI-ToF has been explored for the identification for serotype O4:K8 (Li et al., 2018). However, the practical application of these methods has significant limitations, such as the need for cumbersome procedures associated with gel electrophoresis and the ability to only identify limited or single serogroups and serotypes simultaneously. Taken together, the lack of developments hitherto and the disadvantages of existing methods have prompted the urgent need for a pragmatic molecular serotyping method for V. parahaemolyticus.
During the development of the current assay, we chose genes encoding for the somatic (O) antigen and the capsular (K) antigen as our target genes for identification of V. parahaemolyticus O-serogroups (Chen et al., 2012) and K-serogroups (Chen et al., 2010), respectively. As such, the allelic heterogeneity indexed would closely mirror that of the conventional serological method. This has the advantage over the use of arbitrary molecular markers from other gene loci for serotype prediction or correlation. It is noteworthy that the lipopolysaccharide (LPS) in the outer membrane of V. parahaemolyticus does not possess the O-side chain (Hashii et al., 2003) that consists of oligosaccharide repeats that forms the basis of antigenic variation of the O antigen in many other Gram negative bacteria. Instead, the serological O-specificity and the determinant of V. parahaemolyticus O-serogroups is attributable to the core oligosaccharide of the LPS (Iguchi et al., 1995), referred to as the OGD (Chen et al., 2012). In reviewing the OGD for all serogroups during the study, we have discovered that in addition to the wvcP gene, the orf9 locus was also a genetic determinant for serogroup O10. Subsequently, orf9 was incorporated into our assay to complement the use of wvcP to provide a robust support for the identification of serogroup O10. Indeed, a better understanding of the OGD through genetic analyses would further improve the current O-antigenic scheme. A recent study has shown that the frequent occurrence of untypeable O-serogroups (OUTs) may have resulted from intra-species recombination events that occurred within the OGD region and evolved from the current O-serogroups, where several provisional and rare O-serogroup candidates were proposed (Guo et al., 2017). Whereas, for K- serogroups, the genetic locations for encoding K-antigen synthesis in V. parahaemolyticus (Guvener and Mccarter, 2003; Okura et al., 2003) have been a subject of debate, but was subsequently confirmed to be between gmhD and rjg using an O3:K6 isolate by construction of gene deletions (Chen et al., 2010). By using the genetic location as reference during sequence analyses for assay development, our assay was able to produce robust identification for additional K-serogroups (K8, K25, K29, K56, K68) and further corroborated these findings. Indeed, our current study presented the first molecular assay for the identification of V. parahaemolyticus K-serogroups on the basis of the K- antigen synthesis gene locus; as well as being used in conjunction of O- serogroup identification for the first time to provide the molecular identification of V. parahaemolyticus O:K serotypes.
There are a number of aspects in which the current assay could be further improved. The rare O13 serogroup was not included for identification in the current assay, owing to the 100% homology of the corresponding regions with the O3 serogroup (Makino et al., 2003), as was found in a conventional PCR method that could not differentiate O3 and O13 serogroups (Chen et al., 2012). Further work would be needed to identify heterogeneous genetic markers for the accurate identification of the O13 serogroup. Additionally, the inclusion of additional K- serogroups in response to the rapidly evolving nature of K antigens would further increase the utility of the assay. Finally, a useful aspect worth exploring would be the direct application of clinical and food samples by incorporating inhibitor resistant polymerases during the amplification of target genes, further reducing processing time from the assay workflow.
Conclusion
This is the first description of a molecular assay for the simultaneous identification of multiple clinically common V. parahaemolyticus serotypes. The current assay satisfies the urgent need for a practical, rapid, and robust identification of V. parahaemolyticus serotypes. Given the marked rise in the incidence of vibriosis in recent years, it is anticipated that the current assay could contribute significantly for the timely and effective public health intervention of vibriosis.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
ML and YJ conceived and designed the experiment. ML, YLin, YQ, LZ, YD, and ZL performed the experiments. ML, XS, YLi, MJ, YLiao, and QL contributed analysis. ML and QH wrote the paper.
Funding
This research was supported by the China National Science and Technology Major Projects Foundation (No. 2017ZX10303406), National Natural Science Foundation of China (No. 81773436), Sanming Project of Medicine in Shenzhen (No. SZSM201811071), Shenzhen health and family planning commission (SZGW2017004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to express their deepest gratitude to Dr. Patrick Kwan for critical reading of the manuscript; and thank Liqiang Li and Qiwen Li of BGI, Shenzhen, China, for their valuable technical expertise in genomic sequencing and analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00385/full#supplementary-material
Abbreviations
MLMA, Multiplex Ligation Reaction based on Probe Melting Curve Analysis; FAM, carboxy fluorescein; ROX, carboxy-X-rhodamine; Cy5, indodicarbocyanine5; LATE PCR, Linear After-The-Exponential PCR; Tm, melting temperature; IC, internal control.
References
Aydin, M., Carter-Conger, J., Gao, N., Gilmore, D. F., Ricke, S. C., and Ahn, S. (2018). Molecular identification of common Salmonella serovars using multiplex DNA sensor-based suspension array. Anal. Bioanal. Chem. 410, 2637–2646. doi: 10.1007/s00216-018-0938-5
Baker-Austin, C., Stockley, L., Rangdale, R., and Martinez-Urtaza, J. (2010). Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2, 7–18. doi: 10.1111/j.1758-2229.2009.00096.x
Baker-Austin, C., Trinanes, J., Gonzalez-Escalona, N., and Martinez-Urtaza, J. (2017). Non-cholera vibrios: the microbial barometer of climate change. Trends Microbiol. 25, 76–84. doi: 10.1016/j.tim.2016.09.008
Bhuiyan, N. A., Ansaruzzaman, M., Kamruzzaman, M., Alam, K., Chowdhury, N. R., Nishibuchi, M., et al. (2002). Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40, 284–286. doi: 10.1128/JCM.40.1.284-286.2002
Bugarel, M., Bakker, H. D., Grout, J., Vignaud, M. L., Loneragan, G. H., Fach, P., et al. (2018). CRISPR-based assay for the molecular identification of highly prevalent Salmonella serotypes. Food Microbiol. 71, 8–16. doi: 10.1016/j.fm.2017.03.016
Cabrera-Garcia, M. E., Vazquez-Salinas, C., and Quinones-Ramirez, E. I. (2004). Serologic and molecular characterization of Vibrio parahaemolyticus strains isolated from seawater and fish products of the Gulf of Mexico. Appl. Environ. Microbiol. 70, 6401–6406. doi: 10.1128/AEM.70.11.6401-6406.2004
Chen, M., Guo, D., Wong, H. C., Zhang, X., Liu, F., Chen, H., et al. (2012). Development of O-serogroup specific PCR assay for detection and identification of Vibrio parahaemolyticus. Int. J. Food Microbiol. 159, 122–129. doi: 10.1016/j.ijfoodmicro.2012.08.012
Chen, Y., Dai, J., Morris, J. G. Jr., and Johnson, J. A. (2010). Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol. 10:274. doi: 10.1186/1471-2180-10-274
Daniels, N. A., Mackinnon, L., Bishop, R., Altekruse, S., Ray, B., Hammond, R. M., et al. (2000). Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181, 1661–1666. doi: 10.1086/315459
Furukawa, M., Goji, N., Janzen, T. W., Thomas, M. C., Ogunremi, D., Blais, B., et al. (2018). Rapid detection and serovar identification of common Salmonella enterica serovars in Canada using a new pyrosequencing assay. Can. J. Microbiol. 64, 75–86. doi: 10.1139/cjm-2017-0496
Guo, X., Liu, B., Chen, M., Wang, Y., Wang, L., Chen, H., et al. (2017). Genetic and serological identification of three Vibrio parahaemolyticus strains as candidates for novel provisional O serotypes. Int. J. Food Microbiol. 245, 53–58. doi: 10.1016/j.ijfoodmicro.2017.01.010
Guvener, Z. T., and Mccarter, L. L. (2003). Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J. Bacteriol. 185, 5431–5441. doi: 10.1128/JB.185.18.5431-5441.2003
Han, C., Tang, H., Ren, C., Zhu, X., and Han, D. (2016). Sero-prevalence and genetic diversity of pandemic V. parahaemolyticus strains occurring at a global scale. Front. Microbiol. 7:567. doi: 10.3389/fmicb.2016.00567
Han, D., Yu, F., Tang, H., Ren, C., Wu, C., Zhang, P., et al. (2017). Spreading of pandemic Vibrio parahaemolyticus O3:K6 and its serovariants: a re-analysis of strains isolated from multiple studies. Front. Cell. Infect. Microbiol. 7:188. doi: 10.3389/fcimb.2017.00188
Hashii, N., Isshiki, Y., Iguchi, T., and Kondo, S. (2003). Structural analysis of the carbohydrate backbone of Vibrio parahaemolyticus O2 lipopolysaccharides. Carbohydr. Res. 338, 1063–1071. doi: 10.1016/S0008-6215(03)00078-8
Heymans, R., Vila, A., Van Heerwaarden, C. A.M., Jansen, C. C. C., Castelijn, G. A. A., Van Der Voort, M., et al. (2018). Rapid detection and differentiation of Salmonella species, Salmonella typhimurium and Salmonella enteritidis by multiplex quantitative PCR. PLoS ONE 13:e0206316. doi: 10.1371/journal.pone.0206316
Iguchi, T., Kondo, S., and Hisatsune, K. (1995). Vibrio parahaemolyticus O serotypes from O1 to O13 all produce R-type lipopolysaccharide: SDS-PAGE and compositional sugar analysis. FEMS Microbiol. Lett. 130, 287–292. doi: 10.1111/j.1574-6968.1995.tb07733.x
Jiang, Y., He, L., Wu, P., Shi, X., Jiang, M., Li, Y., et al. (2017). Simultaneous identification of ten bacterial pathogens using the multiplex ligation reaction based on the probe melting curve analysis. Sci. Rep. 7:5902. doi: 10.1038/s41598-017-06348-z
Letchumanan, V., Chan, K. G., and Lee, L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5:705. doi: 10.3389/fmicb.2014.00705
Li, P., Xin, W., Xia, S., Luo, Y., Chen, Z., Jin, D., et al. (2018). MALDI-TOF mass spectrometry-based serotyping of V. parahaemolyticus isolated from the Zhejiang province of China. BMC Microbiol. 18:185. doi: 10.1186/s12866-018-1328-z
Li, R., Wang, Y., Shen, J., and Wu, C. (2014). Development of a novel hexa-plex PCR method for identification and serotyping of Salmonella species. Foodborne Pathog. Dis. 11, 75–77. doi: 10.1089/fpd.2013.1551
Li, Y., Xie, X., Shi, X., Lin, Y., Qiu, Y., Mou, J., et al. (2014). Vibrio parahaemolyticus, Southern Coastal Region of China, 2007–2012. Emerg. Infect. Dis. 20, 685–688. doi: 10.3201/eid2004.130744
Liao, Y., Wang, X., Sha, C., Xia, Z., Huang, Q., and Li, Q. (2013). Combination of fluorescence color and melting temperature as a two-dimensional label for homogeneous multiplex PCR detection. Nucleic Acids Res. 41:e76. doi: 10.1093/nar/gkt004
Liu, J., Bai, L., Li, W., Han, H., Fu, P., Ma, X., et al. (2018). Trends of foodborne diseases in China: lessons from laboratory-based surveillance since 2011. Front. Med. 12, 48–57. doi: 10.1007/s11684-017-0608-6
Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749. doi: 10.1016/S0140-6736(03)12659-1
Marder, E. P., Cieslak, P. R., Cronquist, A. B., Dunn, J., Lathrop, S., Rabatsky-Ehr, T., et al. (2017). Incidence and trends of infections with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance - foodborne diseases active surveillance network, 10 U.S. Sites, 2013-2016. MMWR Morb. Mortal. Wkly. Rep. 66, 397–403. doi: 10.15585/mmwr.mm6615a1
Mclaughlin, J. B., Depaola, A., Bopp, C. A., Martinek, K. A., Napolilli, N. P., Allison, C. G., et al. (2005). Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N. Engl. J. Med. 353, 1463–1470. doi: 10.1056/NEJMoa051594
Munoz, N., Diaz-Osorio, M., Moreno, J., Sanchez-Jimenez, M., and Cardona-Castro, N. (2010). Development and evaluation of a multiplex real-time polymerase chain reaction procedure to clinically type prevalent Salmonella enterica serovars. J. Mol. Diagn. 12, 220–225. doi: 10.2353/jmoldx.2010.090036
Nair, G. B., Ramamurthy, T., Bhattacharya, S. K., Dutta, B., Takeda, Y., and Sack, D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48. doi: 10.1128/CMR.00025-06
Newton, A., Kendall, M., Vugia, D. J., Henao, O. L., and Mahon, B. E. (2012). Increasing rates of vibriosis in the United States, 1996–2010: review of surveillance data from 2 systems. Clin. Infect. Dis. 54 (Suppl. 5), S391–S395. doi: 10.1093/cid/cis243
Okura, M., Osawa, R., Iguchi, A., Arakawa, E., Terajima, J., and Watanabe, H. (2003). Genotypic analyses of Vibrio parahaemolyticus and development of a pandemic group-specific multiplex PCR assay. J. Clin. Microbiol. 41, 4676–4682. doi: 10.1128/JCM.41.10.4676-4682.2003
Oliver, J. D., and Jones, J. L. (2015). “Vibrio parahaemolyticus and Vibrio vulnificus,” in Molecular Medical Microbiology, 2nd Edn., eds Y. Tang, M. Sussman, D. Liu, I. Poxton, and J. Schwartzmann (London, UK: Academic Press), 1169–1186. doi: 10.1016/B978-0-12-397169-2.00066-4
Velazquez-Roman, J., Leon-Sicairos, N., De Jesus Hernandez-Diaz, L., and Canizalez-Roman, A. (2014). Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect. Microbiol. 3:110. doi: 10.3389/fcimb.2013.00110
Vezzulli, L., Colwell, R. R., and Pruzzo, C. (2013). Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb. Ecol. 65, 817–825. doi: 10.1007/s00248-012-0163-2
Keywords: Vibrio parahaemolyticus, vibriosis, molecular serotyping, serogroup, identification, multiplex ligation reaction based on probe melting curve analysis
Citation: Li M, Jiang Y, Shi X, Li Y, Jiang M, Lin Y, Qiu Y, Zuo L, Deng Y, Lin Z, Liao Y, Li Q and Hu Q (2019) Simultaneous Identification of Clinically Common Vibrio parahaemolyticus Serotypes Using Probe Melting Curve Analysis. Front. Cell. Infect. Microbiol. 9:385. doi: 10.3389/fcimb.2019.00385
Received: 01 August 2019; Accepted: 28 October 2019;
Published: 14 November 2019.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Jessica L. Jones, United States Food and Drug Administration, United StatesXin-an Jiao, Yangzhou University, China
Thandavarayan Ramamurthy, Translational Health Science and Technology Institute (THSTI), India
Copyright © 2019 Li, Jiang, Shi, Li, Jiang, Lin, Qiu, Zuo, Deng, Lin, Liao, Li and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghua Hu, aHVxaW5naHVhMDNAMTYzLmNvbQ==
 Minxu Li
Minxu Li Yixiang Jiang2
Yixiang Jiang2 Yinhua Deng
Yinhua Deng Qingge Li
Qingge Li Qinghua Hu
Qinghua Hu