- 1School of Public Health and Management, Chongqing Medical University, Chongqing, China
- 2Hangzhou Hospital for the Prevention and Treatment of Occupational Disease, Hangzhou, China
- 3Nanjing Center for Disease Control and Prevention, Nanjing, China
The iron acquisition system is an essential virulence factor for human infection and is under tight regulatory control in a variety of pathogens. Ferric-uptake regulator (Fur) is one of Fe2+-responsive transcription factor that maintains iron homeostasis, and the regulator of capsule synthesis (Rcs) is known to regulate exopolysaccharide biosynthesis. We speculate the Rcs may involve in iron-acquisition given the identified regulator box in the upstream of entC that participated in the biosynthesis of enterobactin. To study the coregulation by RcsAB and Fur of entC, we measured the β-galactosidase activity and relative mRNA expression of entC in WT and mutant strains. The RcsAB- and Fur-protected regions were identified by an electrophoretic mobility shift assay (EMSA) and a DNase I footprinting assay. A regulatory cascade was identified with which Fur repressed rcsA expression and reduced RcsAB and entC expression. Our study demonstrated that entC was coregulated by two different transcriptional regulators, namely, RcsAB and Fur, in response to iron availability in Klebsiella pneumoniae.
Introduction
Klebsiella pneumoniae is an opportunistic pathogen that causes severe infections, mainly manifesting as pneumonia, bacteremia, septicemia, and urinary and respiratory tract infections (Podschun and Ullmann, 1998). A number of virulence factors identified in K. pneumoniae are involved in pathogenicity, including capsule polysaccharide (CPS), lipopolysaccarides, type 1 and 3 fimbriae, biofilm formation-related factors, urease an the iron-acquisition system (Clegg and Murphy, 2016; Paczosa and Mecsas, 2016; Lam et al., 2018; Bengoechea and Sa Pessoa, 2019; Russo and Marr, 2019).
The regulator of capsule synthesis (Rcs) phosphorelay is a complex signal transduction pathway composed of RcsB, RcsC, and RcsD (Majdalani and Gottesman, 2005; Wall et al., 2018). RcsC, a transmembrane sensor kinase, transfers a phosphoryl group to another membrane-spanning protein, RcsD, and finally to the response regulator, RcsB (Clarke, 2010). In addition to acting alone as a transcriptional regulators (Casino et al., 2018; Filippova et al., 2018), RcsB can also combine with the accessory protein RcsA to regulate related genes (Mouslim et al., 2003; Liu et al., 2014; Fang et al., 2015; Su et al., 2018). The Rcs phosphorelay plays a major role in the regulation of CPS (Mouslim et al., 2003; Llobet et al., 2011; Pando et al., 2017; Peng et al., 2018; Walker et al., 2019), biofilm formation (Sun et al., 2012; Fang et al., 2015), flagellar biogenesis (Lehti et al., 2012; Jozwick et al., 2019). The RcsAB binding site (RcsAB box) consists of a 7-7 inverted repeat sequence, TAAGAAT-ATTCTTA (Fang et al., 2015). The promoter-proximal regions of entC contain a RcsAB box-like sequence, so we hypothesize that the expression of entC is regulated by RcsAB.
There are 10 putative iron uptake systems in K. pneumoniae strain NTUH-K2044. Among those, 4 are siderophore-dependent, namely enterobactin (entABCDEF), yersiniabactin (Yersinia HPI), aerobactin (iucABCDiutA), and salmochelin (iroNDCB) (Hsieh et al., 2008). Siderophore is considered to be an important iron acquiring strategy by K. pneumoniae, especially under iron-restricted conditions (Paczosa and Mecsas, 2016). While yersiniabactin, aerobactin, and salmochelin predominate in pyogenic live abcess-associated K. pneumoniae strains (Hsieh et al., 2008), enterobactin is ubiquitous among almost all K. pneumoniae and has the highest iron affinity as compared with other siderophores (Hsieh et al., 2008; Li et al., 2014). The entC gene encodes isochorismate synthetase, which plays a critical role in enterobactin synthesis (Liu et al., 1990; Raymond et al., 2003). Rcs phosphorelay is well-known for its function in regulating CPS, its role in iron acquisition system is unclear.
Excess iron is toxic for bacteria (Becker and Skaar, 2014). Ferric uptake regulator (Fur), is a transcriptional regulator that alters gene expression in response to iron availability, regulates iron homeostasis in many bacteria (Seo et al., 2014). In general, holo-Fur (Fur bound to Fe2+) represses gene expression, whereas apo-Fur (Fur not bound to Fe2+) de-represses gene expression (Stacy et al., 2016). In Escherichia coli, holo-Fur can directly repress regulation of entC (Brickman et al., 1990). Therefore, we postulated that holo-Fur can also function as a repressor of entC under iron-rich conditions in K. pneumoniae.
In this study, we explored how RcsAB and Fur coregulate entC under different iron conditions. Our results suggested an regulatory cascade in which Fur regulates the rcsA and entC promoters. This study provides new light on the regulons of RcsAB and the mechanisms controlling iron acquisition in iron-repletion and iron-depletion.
Materials and Methods
Bacterial Strains, Plasmids, Primers, and Media
The bacterial strains and plasmids used in this study are listed in Table 1. The primers used in this study are listed in Table 2. Bacterial strains were routinely cultured aerobically at 37°C in Luria-Bertani (LB) broth or on LB agar plates with antibiotics added as required at the following concentrations: kanamycin, 50 μg/ml; chloramphenicol, 35 μg/ml; ampicillin, 100 μg/ml. Bacterial growth was monitored by measuring the optical density of the cultures at a wavelength of 600 nm (OD600).
Construction of Gene Deletion and Complementation Strains
The mutants Kp-ΔrcsA, Kp-ΔrcsB, Kp-ΔrcsAB, and Kp-Δfur were constructed as previously described (Peng et al., 2018; Su et al., 2018). Gene deletion was done by allelic replacement. In brief, the upstream and downstream flanking regions of target gene fragments were amplified, purified, fused, and cloned into the temperature-sensitive suicide vector pKO3-Km. The resulting plasmid was transformed into NTUH-K2044 by electroporation. After the recombinant plasmid was integrated (at 30°C) and excised (at 43°C), the mutants Kp:ΔrcsA, Kp:ΔrcsB, Kp:ΔrcsAB, and Kp:Δfur were constructed and further verified by PCR and DNA sequencing.
For complementation experiments, the amplified DNA fragments were ligated to pBAD33. The recombinant plasmids were introduced into the mutant strains. The complementation strains were selected with chloramphenicol on LB agar plates and verified by PCR.
Chrome Azurol S (CAS) Assay
The CAS assay described by Schwyn and Neilands was used to check the siderophores from bacteria (Schwyn and Neilands, 1987). Briefly, bacteria were inoculated into MM9 minimal medium (which contained the following components per liter: 100 ml of 10 × MM9 minimal medium [3 g of KH2PO4, 5 g NaCl, 1 g of NH4Cl], 30 ml of deferrated casamino acids, 10 ml of 20% glucose, 1 ml of 1 M MgCl2, 1 ml of 100 mM CaCl2, 30.24 g of PIPES, 6 g of NaOH) and cultured for 16 h. OD600 was read and siderophore levels were standardized by the OD600 measurements. The supernatants were collected, diluted with MM9 medium and subjected to the CAS assay with percent siderophore units calculated as previously described (Payne, 1994).
lacZ Fusion and β-Galactosidase Assay
The putative promoter DNA region of entC was amplified by KP1_entC-lacZ-F/R and cloned into the placZ15 plasmid that harbors a promoterless lacZ reporter gene and transferred into K. pneumoniae NTUH-K2044ΔlacZ strain CCW01 and the deletion mutants. A single colony was inoculated into LB with or without 2,2-dipyridyl (Dip) and grown to logarithmic phase. Cells grown as described above were assayed for β-galactosidase activity (Luo et al., 2017), and the units of activity were calculated as described by Miller. Every sample was tested in triplicate, and the assay was repeated in at least three independent experiments.
Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
NTUH-K2044, Kp:ΔrcsA, Kp:ΔrcsB, Kp:ΔrcsAB, and Kp:Δfur were inoculated into LB liquid medium. After overnight growth, NTUH-K2044, Kp:ΔrcsA, Kp:ΔrcsB, and Kp:ΔrcsAB were diluted 1:100 in 15 ml of fresh medium with Dip (250 μM final concentration). NTUH-K2044 and Kp:Δfur were diluted 1:1,000 in 15 ml of fresh medium with FeSO4 (100 μM final concentration). When the strains grew to logarithmic phase, total RNA was extracted using the TIANGEN RNAprep Pure Cell/Bacteria Kit following the manufacture's protocol. RNA integrity was analyzed by agarose gel electrophoresis, and RNA purity and concentration were calculated by measuring the optical density of the samples at 260 and 280 nm using a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Scientific). Then, the RNA was converted to single-stranded cDNA using the PrimeScript™ RT Reagent Kit. Real-time PCR was carried out using a LightCycler® system. Relative gene expression was quantified using the Ct value-based method with rho (Gomes et al., 2018) rRNA as the internal control.
Protein Expression and Purification
RcsA and RcsB were expressed and purified as previously described (Peng et al., 2018). Briefly, The entire coding regions of rcsA and rcsB were cloned into the pMAL-C5X and pET-28a, respectively. Then, the resulting plasmids were transformed into E. coli BL21 (DE3) cells, and the recombinant proteins MBP-RcsA and His6-RcsB were overexpressed under induction by isopropyl β-D-thiogalactopyranoside (IPTG). The cells were lysed by supersonication, and the proteins were purified by column chromatography and dialyzed.
Purification of the His6-Fur fusion protein was carried out as previous study described (Gao et al., 2008). The recombinant plasmid, pET-28a-fur, was transformed into E. coli BL21 (DE3) cells. The culture was grown at 37°C in LB medium overnight and then was transferred into 500 ml LB medium. The His6-Fur was induced with 1 mM IPTG at OD600 of 0.6 and expressed for 4 h at 30°C before the cells were harvested. The pellet was suspended in 15 ml buffer A (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 10% glycerol, pH 8.0). The cells were lysed with sonication and centrifuged at 4°C. The supernatant was loaded onto a nickel column, and the column was washed with a gradient of 20–40 mM imidazole prepared in buffer A, respectively. The His6-Fur was eluted using buffer A with 250 mM imidazole. His6-Fur was further dialyzed in buffer B (10 mM NaH2PO4, 40 mM Na2HPO4, 145 mM NaCl, 30% glycerol) at 4°C. The MBP-RcsA, His6-RcsB, and His6-Fur were detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Electrophoretic Mobility Shift Assay (EMSA)
The putative promoter regions of the entC gene were amplified by PCR using primers KP1_entC-EMSA-RcsAB-F/R and KP1_entC-EMSA-Fur-F/R that listed in Table 2. Mix 25 mM fresh acetyl phosphate, His6-RcsB and binding buffer (50 mM Tris-HCl, 750 mM KCl, 0.5 mM DTT, 0.5 mM EDTA) and incubate at 37°C for 30 min to phosphorylae the His6-RcsB. The target entC promoter DNA (20 ng) was mixed with increasing amounts of MBP-RcsA or phosphorylated His6-RcsB. After incubation at 37°C for 30 min, the samples were analyzed by 4% (w/v) PAGE in 0.5 × TBE buffer (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA).
EMSAs for the entC and rcsA promoters by His6-Fur, DNA were incubated with purified His6-Fur in a 10-μl solution containing 50 mM Tris-HCl, 5 mM MgCl2, 250 mM KCl, 20% glycerol, 2.5 mM dithiothreitol, 0.25 mg/ml BSA, 500 μM MnCl2 at 37°C for 20 min. Then, the samples were examined by separation on a native 4% (w/v) polyacrylamide gels in 0.5 × TB buffer (45 mM Tris-HCl, 45 mM boric acid). A constant voltage of 150 V was applied to all gels for 2 h at 4°C. After staining with SYBR Green EMSA stain (Invitrogen), the gel was examined with a UV transilluminator.
DNase I Footprinting
The target DNA fragment of the promoter DNA region was PCR amplified using the primers M13F-47 (FAM) and M13R-48 with DNA polymerase premix from the constructed plasmid for preparation of fluorescent FAM labeled probes. DNase I footprinting assays were performed in a manner similar to the method described by Wang et al. (2012). DNase I footprinting assay for entC promoter by RcsAB, porbes were incubated with 0, 1, 3 μg of His6-RcsB for 30 min at 37°C. Then MBP-RcsA was added and this mixture incubated for 30 min at 37°C. DNase I footprinting assay for entC promoter by Fur, probes were incubated with 0, 0.5, 1 μg of His6-Fur for 30 min at 37°C. After adding DNase I (Promega), samples were extracted with phenol/chloroform, then precipitated with ethanol. Pellets were dissolved in 30 μl MiniQ water. The preparation of DNA ladder, capillary electrophoresis and data analysis were the same as described before (Wang et al., 2012), except that the GeneScan-LIZ600 size standard (Applied Biosystems) was used.
Statistical Analysis
All experiments were performed at least three times. Results were presented as means ± standard deviation (SD). GraphPad Prism 7.0 software (GraphPad Software, Inc, La Jolla, CA, USA) was used for statistical. Statistical significance was determined using one-way ANOVA for multiple comparisons and Student's t-test for comparing two groups. Asterisk indicate P values (*P < 0.05).
Results
RcsAB Affects the Iron Acquisition System
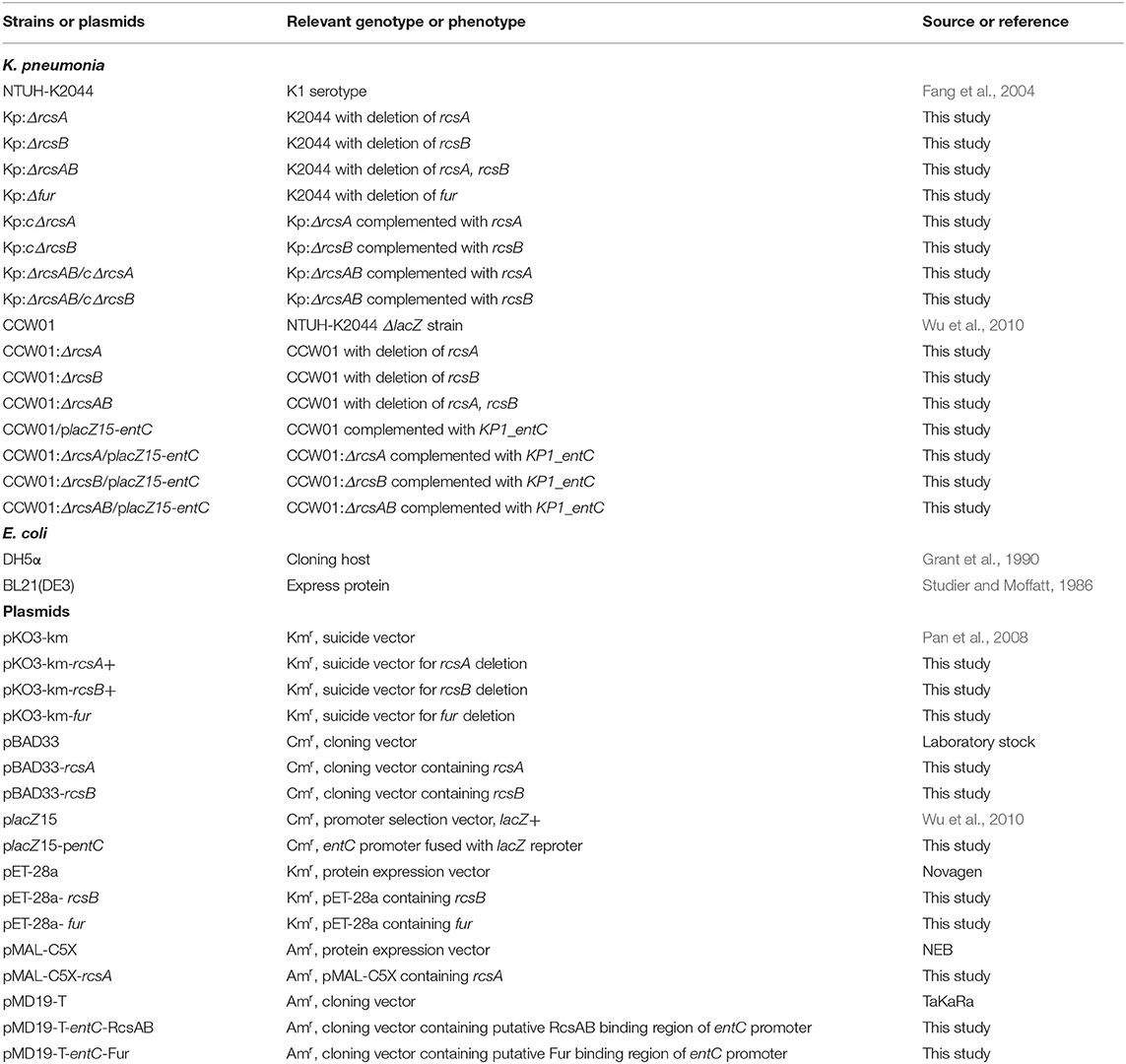
RcsAB and Fur are transcriptional regulators that can regulate various virulence factors in NTUH-K2044. And iron acquisition system is an important virulence factor. To analyze whether RcsAB and Fur affect iron acquisition in NTUH-K2044, an in vitro Chrome azurol S (CAS) assay examining the secretion of siderophores was performed. As shown in Figure 1, no statistical difference was found in WT and WT with empty plasmid. Siderophore secretion by Kp:ΔrcsA, Kp:ΔrcsB, Kp:ΔrcsAB, and Kp:Δfur were increased by 3, 8, 7, and 12-fold relative to WT, respectively. Complementation with plasmid-encoded rcsA, rcsB, and fur led to a decrease in secretion of siderophore relative to the mutant strains. Collectively, these results indicated that the iron acquisition in NTUH-K2044 was influenced by RcsAB and Fur.
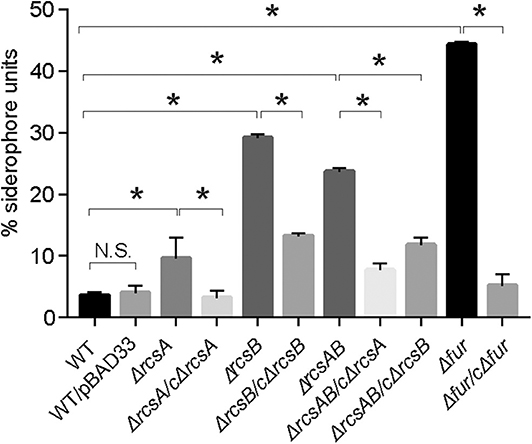
Figure 1. The K. pneumoniae iron acquisition system is modulated by RcsAB and Fur. CAS assay for K. pneumoniae NTUH-K2044 wild type, wild type with empty plasmid, mutants, and complement strains were assessed as described in Material and Methods. Siderophores secreted from strains removed iron from dye complex giving rise to a reduction in blue color of the solution. Measure the absorbance (A630) for loss of blue color. Results were the means of biological triplicates plus standard deviations. Bar graph showed percent siderophore units, calculated as [(Ar – As) / Ar] × 100, where Ar is the absorbance of MM9/CAS solution and As is the sample absorbance. And MM9 can be used as a blank. The percent siderophore units of WT was compared with mutants and the percent siderophore units of mutants were compared with its complement strains. P values were calculated by one-way ANOVA. *P < 0.05. Shown are the averages ± standard deviation (SD) from three independent experiments.
Iron Limitation Enhances the Activity of the entC Promoter
Ent, a siderophore, is enriched under iron restriction. The entC participates in the synthesis of Ent. To examine whether the restriction of environmental iron increases entC expression, we cloned the putative promoter regions upstream of entC into the lacZ reporter plasmid and transformed the constructs into CCW01. And CCW01/placZ15 was used as a control. For iron depletion, increasing amounts of the iron chelator 2,2-dipyridyl (Dip) were added to LB medium. Figure 2 shows that as the concentration of Dip increased, the activity of the entC promoter increased. The addition of 50, 100, and 250 μM Dip to the medium increased entC promoter activity by ~2, 4, and 8-fold, respectively. There were no statistical differences of β-galactosidase from CCW01/placZ15 under different concentrations of Dip (data not shown), suggesting unspecific regulation by iron is unlikely. The promoter activity of entC was activated under iron-limited conditions.
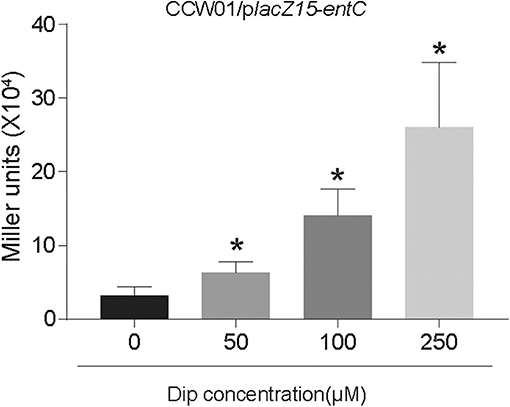
Figure 2. Iron starvation enhanced the activity of the entC promoter. The putative promoter region of entC was cloned into the placZ15 plasmid and then introduced into CCW01 to measure β-galactosidase activity (Miller units) in increasing amounts of Dip. *P < 0.05, compared with the 0 μM Dip group.
RcsAB and Fur Coregulate entC Expression
RcsAB and Fur are predicted and have been shown to impact the iron acquisition system. We sought to examine how these proteins regulated entC expression according to the level of iron in the environment. In LB medium, there are no statistical differences among CCW01/placZ15-entC, CCW01:ΔrcsA/placZ15-entC, CCW01:ΔrcsB/placZ15-entC, and CCW01:ΔrcsAB/placZ15-entC. After adding FeSO4 to LB, bacteria were in iron replete conditions and expression from entC promoter was not altered in CCW01:ΔrcsA/placZ15-entC, CCW01:ΔrcsB/placZ15-entC, although the miller units of CCW01:ΔrcsAB/placZ15-entC were higher than CCW01/placZ15-entC. However, when Dip added to LB, bacteria were in iron-restricted conditions. CCW01:ΔrcsA/placZ15-entC, CCW01:ΔrcsB/placZ15-entC, and CCW01:ΔrcsAB/placZ15-entC all led to 2–fold less β-galactosidase activity relative to CCW01/placZ15-entC (Figure 3A). These results demonstrated that RcsAB positively regulated entC transcription under iron deficient conditions.
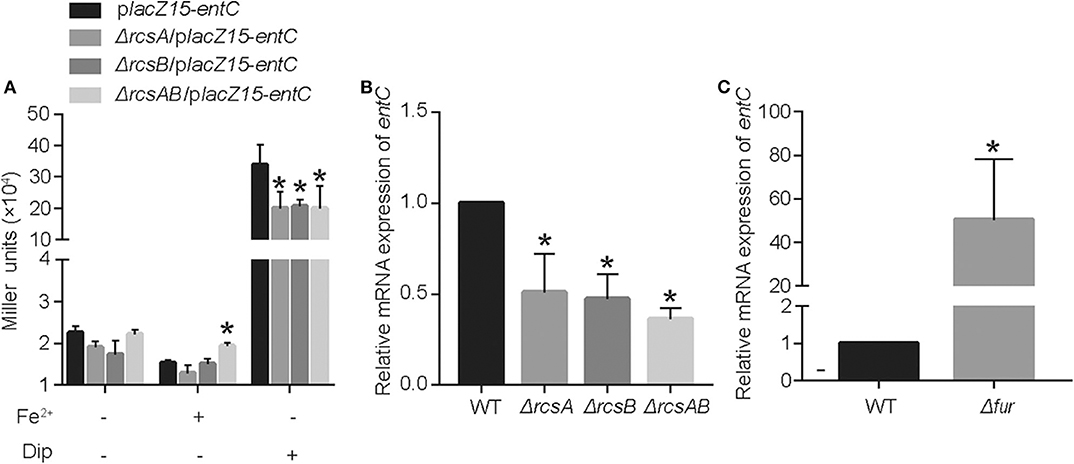
Figure 3. RcsAB and Fur coregulate entC expression under different iron levels. (A) The promoter-lacZ fusion for entC was transformed into the CCW01, CCW01:ΔrcsA, CCW01:ΔrcsB, and CCW01:ΔrcsAB that were then grown in LB, LB with 100 μM FeSO4 and LB with 250 μM Dip. (B) When 250 μM Dip added to LB. entC mRNA levels in WT, Kp:ΔrcsA, Kp:ΔrcsB, and Kp:ΔrcsAB were measured in iron-restricted conditions via RT-qPCR analysis. One-way ANOVA was performed to determine statistically significant differences between each strain and the WT. (C) When 100 μM FeSO4 added to LB, WT, and Kp:Δfur were examined for entC expression under iron-replete conditions by RT-qPCR analysis. *P < 0.05, compared with the WT. Error bars are standard deviation.
To verify further that RcsAB regulate entC expression under iron restricted conditions, we determined the mRNA levels of entC in WT, Kp:ΔrcsA, Kp:ΔrcsB, and Kp:ΔrcsAB by RT-qPCR. When LB medium was supplemented with 250 μM Dip, transcription of entC by Kp:ΔrcsA, Kp:ΔrcsB, and Kp:ΔrcsAB were decreased by 1.4, 1.8, and 2.3-fold relative to WT, respectively (Figure 3B). On the other hand, after adding FeSO4 to LB, deletion of fur resulted in a dramatic increase in expression of entC mRNA (Figure 3C). These data suggest that RcsAB and Fur coregulated entC in response to iron availability.
To investigate further whether entC served as a direct target of RcsAB and Fur, EMSAs were performed. MBP-RcsA, His6-RcsB, His6-RcsB mixed with 20 pmol MBP-RcsA and His6-Fur were subjected to EMSAs with the purified whole promoter DNA region of entC. Neither MBP-RcsA nor His6-RcsB binded to the entC upstream DNA (Figures 4A,B). However, His6-RcsB mixed with 20 pmol MBP-RcsA and His6-Fur could bind to the putative entC promoter DNA fragment (Figures 4C,D).
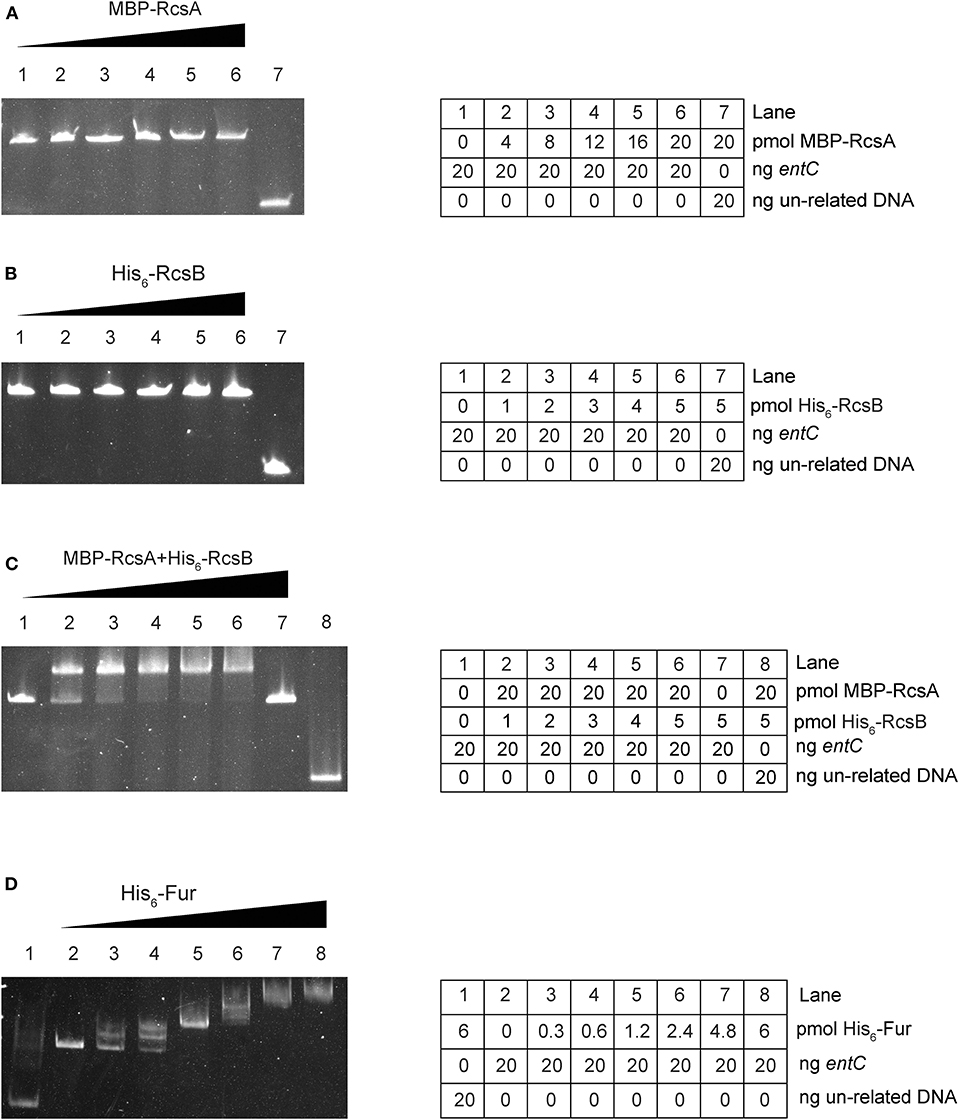
Figure 4. RcsAB and Fur bind the entC promoter. The DNA binding capacities were evaluated by EMSAs. The promoter DNA fragments of entC were incubated with increasing amounts of purified MBP-RcsA (A), His6-RcsB (B), His6-RcsB mixed with MBP-RcsA (C), and His6-Fur (D) and then subjected to polyacrylamide gel electrophoresis.
As further determined by DNase I footprinting (Figures 5A,B), both His6-RcsB mixed with MBP-RcsA and His6-Fur protected the DNA region upstream of entC, covering ~27 bases and 29 bases, respectively. Besides, the protected regions corresponded to the predicted binding sites for these proteins as indicated in Figure 6. The entC promoter was constructed with translation start site, core promoter−10 and−35 elements (Brickman et al., 1990), Shine-Dalgamo sequence, RcsAB box-like sequences, RcsAB sites, Fur box-like sequences, and Fur sites (Figure 6). Taken together, RcsAB and Fur directly coregulate entC expression.
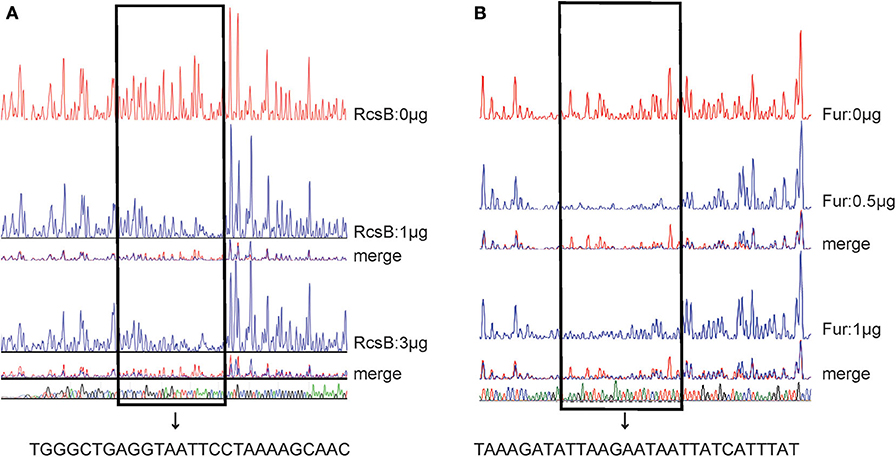
Figure 5. DNase I footprints of RcsAB and Fur at the entC promoter. The promoter DNA regions of entC were labeled with FAM and incubated with increasing amounts of purified His6-RcsB (0, 1, 3 μg) (A) and His6-Fur (B) (0, 0.5, 1 μg). The footprint regions are boxed within lines and marked.

Figure 6. Organization of entC promoter-proximal DNA regions. The DNA sequences were derived from NTUH-K2044. Shown are the translation start sites,−10 and−35 elements, Shine-Dalgamo sequence, RcsAB-binding site, RcsAB box-like sequence, Fur-binding site and Fur box-like sequence.
Fur Directly Represses rcsA Expression Under Iron Repletion
RcsA, an auxiliary activator protein, acts with RcsB as a transcription regulator. In K. pneumoniae CG43, the expression of rcsA is reportedly regulated by Fur (Lin et al., 2011). Due to the heterogeneity of K. pneumoniae genomes, we tested whether Fur repressed rcsA expression in a direct way in NTUH-K2044. A putative Fur-binding box was located at the translation start site of rcsA in NTUH-K2044. Thus, to verify whether Fur could bind to the putative promoter regions of rcsA, we performed an EMSA. As shown in Figure 7A, His6-Fur was able to bind to the regions.
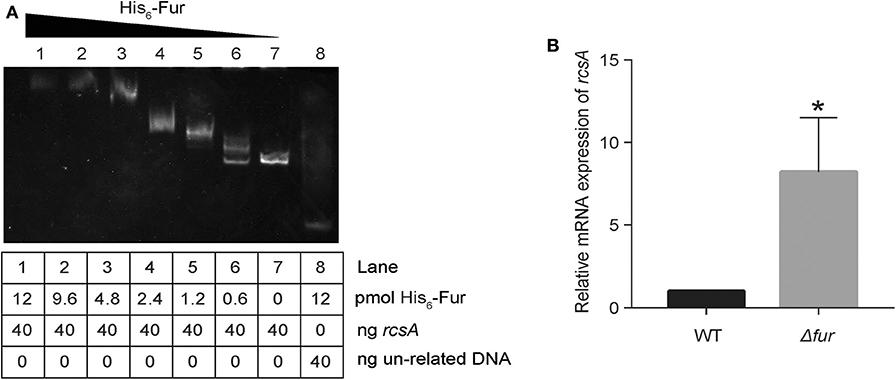
Figure 7. Fur directly represses rcsA. (A) The DNA binding capacity of Fur was evaluated by EMSA with rcsA. (B) Relative mRNA expression of rcsA in WT and Kp:Δfur was assayed by RT-qPCR in an iron-rich environment. *P < 0.05, compared with the WT.
Additionally, when iron is abundant in the environment, holo-Fur represses the regulons (Seo et al., 2014). To confirm that rcsA expression was indeed increased in Kp: Δfur under iron repletion, we measured the transcript levels of rcsA via RT-qPCR. As detailed in Figure 7B, we observed an 8-fold (p < 0.05) increase of rcsA transcript levels in Kp: Δfur. These data confirm that Fur negatively regulates rcsA expression under iron repletion and represses the biosynthesis of RcsAB in NTUH-K2044, which indirectly downregulates entC expression. Furthermore, Fur could repress entC in a direct way.
Discussion
Iron acquisition system is important for virulence of K. pneumoniae. Numerous transcriptional regulators are involved in the process of iron uptake and metabolism, such as Fur (Gao et al., 2008; Seo et al., 2014). Dorman and colleagues (Dorman et al., 2018) found Rcs phosphorelay system is a major node for transcriptional control. Intriguingly, the identification of the RcsAB box upstream of entC that plays a crucial role in the biosynthesis of Ent in K. pneumoniae suggests that RcsAB may also involve in the regulation of iron acquisition system. Our data suggested that RcsAB contribute to not only CPS and mobility but also the iron acquisition system (Figure 1).
The Rcs phosphorelay is a signal transduction system, and the phosphorylation and dephosphorylation of RcsC, RcsD, and RcsB were affected by environmental signals, such as overproduction of DjlA, YmgA, and YmgB; the presence of a solid surface; osmotic shock; acid shock; and growth at low temperature in the presence of glucose and 1 mM zinc (Sledjeski and Gottesman, 1996; Kelley and Georgopoulos, 1997; Ferrieres and Clarke, 2003; Hagiwara et al., 2003; Kannan et al., 2008; Tschowri et al., 2009). Therefore, we hypothesized that iron status may activate the Rcs system, which has never been reported in any organism.
RcsA is an unstable positive regulator required for the synthesis of CPS (Stout et al., 1991). And Fur regulate the rcsA to control the expression of CPS in K. pneumoniae CG43 (Lin et al., 2011). Our study suggested a regulatory cascade exists under iron repletion in which Fur controls rcsA expression. The reduction in rcsA levels appears to have impacts on the synthesis of RcsAB and thus affects the expression of entC. RcsB alone can regulate expression of many genes. When act as combination with other regulators, such as RcsA, RcsB can regulate the expression of wider spectrum of genes. Our result suggested that RcsB or RcsA alone is unable to regulate entC. Instead, it appears that these proteins form a complex that can bind to the promoter of entC (Figure 4). Hence, our study suggest a model in which when iron is restricted, Fur repression of rcsA is relieved. RcsA then binds with RcsB to activate entC expression (Figure 8).
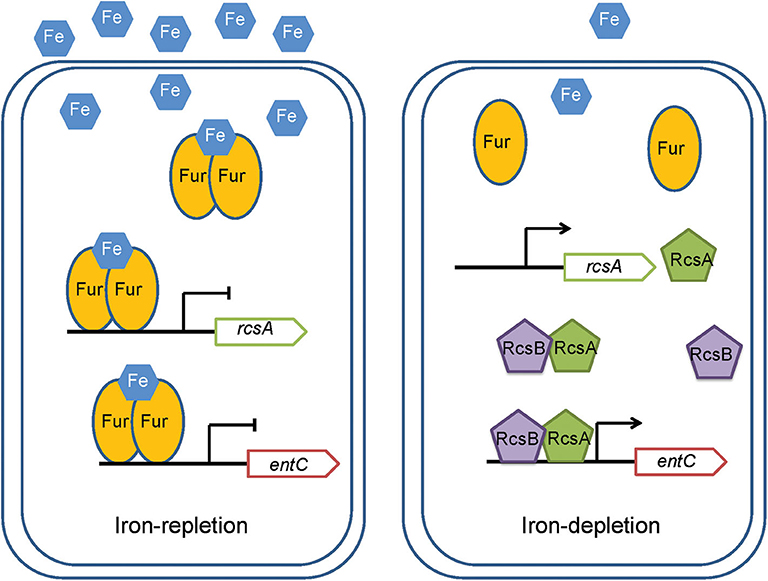
Figure 8. A model for the regulation by RcsAB and Fur of rcsA and entC in K. pneumoniae. Under iron-replete conditions, apo-Fur combines with Fe2+ to form holo-Fur, which strongly represses rcsA and entC. Under iron depletion, upon derepression by apo-Fur of rcsA and entC and activation of RcsAB, the transcription of entC increased.
Fur represses the transcription of entC when iron is sufficient, while RcsAB promotes entC expression in the absence of iron. However, under iron repletion, according to the results of the lacZ fusion assays, the β-galactosidase activity (Miller units) of entC in CCW01:ΔrcsAB was higher than that in CCW01/placZ15-pentC (Figure 3A). It is possible that the decrease in Ent expression is compensated by other transcriptional regulators due to the lack of RcsAB even in the presence of iron, as the iron acquisition system is regulated by multiple transcriptional regulators. As an example, some studies reported that cyclic AMP receptor protein (CRP) can also regulate entC in E. coli (Zhang et al., 2005; Seo et al., 2014). CRP is a global transcriptional regulator and regulates virulence-related gene expression (Xue et al., 2016). It is likely that CRP may regulate the iron acquisition system by regulating entC in K. pneumoniae.
Fur was not considered to be a regulator of rcsB as supported by Lin et al. (2011) and our preliminary result using RT-qPCR (data not shown). RcsB is the response regulator in Rcs phosphorelay. RcsB either alone or with auxiliary protein, such as RcsA, BglJ, MatA, could regulate genes expression. Importantly, RcsB serves as an essential partner. Hence, when the expression of RcsA or other auxiliary proteins have been affected by other transcriptional regulators, such as Fur, the regulons which regulated by complex proteins also can be affected. However, RcsB-dependent regulons cannot be affected.
Given that untimely expression of Ent under iron repletion conditions likely provides a detrimental energy burden, Fur represses entC transcription. Ent can be neutralized by the host-secreted molecule lipocalin-2 (Raymond et al., 2003). Hence, the expression of entC was reduced by Fur, which could protect K. pneumoniae from attacking by host immune system. Additionally, repression of rcsA expression by Fur give rise to a decrease of RcsAB, which negatively regulate fim gene cluster expression (Su et al., 2018). And fimbriae are important mediators of K. pneumoniae adhesion (Paczosa and Mecsas, 2016). Therefore, relatively high extracellular iron concentrations result in the upregulation of fimbriae, which is beneficial for adhesion and colonization of K. pneumoniae.
In summary, our study results suggested that RcsAB could modulate the iron acquisition system by directly regulating entC positively under iron starvation conditions, and Fur could directly repress entC under iron-rich conditions. Our study suggested a regulatory cascade by which Fur controls rcsA expression and the synthesis of RcsAB, and Fur also impacts entC expression. This study improves current understanding the mechanism of regulation of virulence factors by RcsAB in K. pneumoniae and the importance of the iron acquisition system in bacteria.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
JQ contributed the conception. YL and LY designed the study. XL, LD, JZ, and KS performed the experiments. PL, QH, ZZ, DP, and LS analyzed the data. LY wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 31200064].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Becker, K. W., and Skaar, E. P. (2014). Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 38, 1235–1249. doi: 10.1111/1574-6976.12087
Bengoechea, J. A., and Sa Pessoa, J. (2019). Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol. Rev. 43, 123–144. doi: 10.1093/femsre/fuy043
Brickman, T. J., Ozenberger, B. A., and McIntosh, M. A. (1990). Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212, 669–682. doi: 10.1016/0022-2836(90)90229-F
Casino, P., Miguel-Romero, L., Huesa, J., García, P., García-del Portillo, F., and Marina, A. (2018). Conformational dynamism for DNA interaction in the salmonella RcsB response regulator. Nucleic Acids Res. 46, 456–472. doi: 10.1093/nar/gkx1164
Clarke, D. J. (2010). The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. 5, 1173–1184. doi: 10.2217/fmb.10.83
Clegg, S., and Murphy, C. N. (2016). Epidemiology and virulence of Klebsiella pneumoniae. Microbiol. Spectr. 4:UTI-0005-2012. doi: 10.1128/microbiolspec.UTI-0005-2012
Dorman, M. J., Feltwell, T., Goulding, D. A., Parkhill, J., and Short, F. L. (2018). The capsule regulatory network of Klebsiella pneumoniae defined by density-TraDISort. MBio 9:e01863–18. doi: 10.1128/mBio.01863-18
Fang, C-T., Chuang, Y-P., Shun, C-T., Chang, S-C., and Wang, J-T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Fang, N., Yang, H., Fang, H., Liu, L., Zhang, Y., Wang, L., et al. (2015). RcsAB is a major repressor of yersinia biofilm development through directly acting on hmsCDE, hmsT, and hmsHFRS. Sci. Rep. 5:9566. doi: 10.1038/srep09566
Ferrieres, L., and Clarke, D. J. (2003). The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 50, 1665–1682. doi: 10.1046/j.1365-2958.2003.03815.x
Filippova, E. V., Zemaitaitis, B., Aung, T., Wolfe, A. J., and Anderson, W. F. (2018). Structural basis for DNA recognition by the two-component response regulator RcsB. MBio 9:e01993–17. doi: 10.1128/mBio.01993-17
Gao, H., Zhou, D., Li, Y., Guo, Z., Han, Y., Song, Y., et al. (2008). The iron-responsive fur regulon in Yersinia pestis. J. Bacteriol. 190, 3063–3075. doi: 10.1128/JB.01910-07
Gomes, A. E. I., Stuchi, L. P., Siqueira, N. M. G., Henrique, J. B., Vicentini, R., Ribeiro, M. L., et al. (2018). Selection and validation of reference genes for gene expression studies in Klebsiella pneumoniae using reverse transcription quantitative real-time PCR. Sci. Rep. 8:9001. doi: 10.1038/s41598-018-27420-2
Grant, S. G., Jessee, J., Bloom, F. R., and Hanahan, D. (1990). Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 4645–4649. doi: 10.1073/pnas.87.12.4645
Hagiwara, D., Sugiura, M., Oshima, T., Mori, H., Aiba, H., Yamashino, T., et al. (2003). Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185, 5735–5746. doi: 10.1128/JB.185.19.5735-5746.2003
Hsieh, P-F., Lin, T-L., Lee, C-Z., Tsai, S-F., and Wang, J-T. (2008). Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 197, 1717–1727. doi: 10.1086/588383
Jozwick, A. K. S., LaPatra, S. E., Graf, J., and Welch, T. J. (2019). Flagellar regulation mediated by the Rcs pathway is required for virulence in the fish pathogen Yersinia ruckeri. Fish Shellfish Immunol. 91, 306–314. doi: 10.1016/j.fsi.2019.05.036
Kannan, G., Wilks, J. C., Fitzgerald, D. M., Jones, B. D., Bondurant, S. S., and Slonczewski, J. L. (2008). Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 8:37. doi: 10.1186/1471-2180-8-37
Kelley, W. L., and Georgopoulos, C. (1997). Positive control of the two-component RcsC/B signal transduction network by DjlA: a member of the DnaJ family of molecular chaperones in Escherichia coli. Mol. Microbiol. 25, 913–931. doi: 10.1111/j.1365-2958.1997.mmi527.x
Lam, M. M. C., Wyres, K. L., Duchene, S., Wick, R. R., Judd, L. M., Gan, Y-H., et al. (2018). Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 9:2703. doi: 10.1038/s41467-018-05114-7
Lehti, T. A., Heikkinen, J., Korhonen, T. K., and Westerlund-Wikstrom, B. (2012). The response regulator RcsB activates expression of mat fimbriae in meningitic Escherichia coli. J. Bacteriol. 194, 3475–3485. doi: 10.1128/JB.06596-11
Li, B., Zhao, Y., Liu, C., Chen, Z., and Zhou, D. (2014). Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol. 9, 1071–1081. doi: 10.2217/fmb.14.48
Lin, C-T., Wu, C-C., Chen, Y-S., Lai, Y-C., Chi, C., Lin, J-C., et al. (2011). Fur regulation of the capsular polysaccharide biosynthesis and iron-acquisition systems in Klebsiella pneumoniae CG43. Microbiology 157, 419–429. doi: 10.1099/mic.0.044065-0
Liu, J., Quinn, N., Berchtold, G. A., and Walsh, C. T. (1990). Overexpression, purification, and characterization of isochorismate synthase (EntC), the first enzyme involved in the biosynthesis of enterobactin from chorismate. Biochemistry 29, 1417–1425. doi: 10.1021/bi00458a012
Liu, L., Fang, N., Sun, Y., Yang, H., Zhang, Y., Han, Y., et al. (2014). Transcriptional regulation of the waaAE-coaD operon by PhoP and RcsAB in Yersinia pestis biovar microtus. Protein Cell 5, 940–944. doi: 10.1007/s13238-014-0110-8
Llobet, E., Campos, M. A., Gimenez, P., Moranta, D., and Bengoechea, J. A. (2011). Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect. Immun. 79, 3718–3732. doi: 10.1128/IAI.05226-11
Luo, M., Yang, S., Li, X., Liu, P., Xue, J., Zhou, X., et al. (2017). The KP1_4563 gene is regulated by the cAMP receptor protein and controls type 3 fimbrial function in Klebsiella pneumoniae NTUH-K2044. PLoS ONE 12:e0180666. doi: 10.1371/journal.pone.0180666
Majdalani, N., and Gottesman, S. (2005). The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59, 379–405. doi: 10.1146/annurev.micro.59.050405.101230
Mouslim, C., Latifi, T., and Groisman, E. A. (2003). Signal-dependent requirement for the co-activator protein RcsA in transcription of the RcsB-regulated ugd gene. J. Biol. Chem. 278, 50588–50595. doi: 10.1074/jbc.M309433200
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Pan, Y-J., Fang, H-C., Yang, H-C., Lin, T-L., Hsieh, P-F., Tsai, F-C., et al. (2008). Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 46, 2231–2240. doi: 10.1128/JCM.01716-07
Pando, J. M., Karlinsey, J. E., Lara, J. C., Libby, S. J., and Fang, F. C. (2017). The Rcs-regulated colanic acid capsule maintains membrane potential in Salmonella enterica serovar typhimurium. MBio 8:e00808–17. doi: 10.1128/mBio.00808-17
Payne, S. M. (1994). Detection, isolation, and characterization of siderophores. Methods Enzymol. 235, 329–344. doi: 10.1016/0076-6879(94)35151-1
Peng, D., Li, X., Liu, P., Zhou, X., Luo, M., Su, K., et al. (2018). Transcriptional regulation of galF by RcsAB affects capsular polysaccharide formation in Klebsiella pneumoniae NTUH-K2044. Microbiol. Res. 216, 70–78. doi: 10.1016/j.micres.2018.08.010
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Raymond, K. N., Dertz, E. A., and Kim, S. S. (2003). Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588. doi: 10.1073/pnas.0630018100
Russo, T. A., and Marr, C. M. (2019). Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 32:e00001–19. doi: 10.1128/CMR.00001-19
Schwyn, B., and Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Seo, S. W., Kim, D., Latif, H., O'Brien, E. J., Szubin, R., and Palsson, B. O. (2014). Deciphering fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 5:4910. doi: 10.1038/ncomms5910
Sledjeski, D. D., and Gottesman, S. (1996). Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178, 1204–1206. doi: 10.1128/JB.178.4.1204-1206.1996
Stacy, A., Abraham, N., Jorth, P., and Whiteley, M. (2016). Microbial community composition impacts pathogen iron availability during polymicrobial infection. PLoS Pathog. 12:e1006084. doi: 10.1371/journal.ppat.1006084
Stout, V., Torres-Cabassa, A., Maurizi, M. R., Gutnick, D., and Gottesman, S. (1991). RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173, 1738–1747. doi: 10.1128/JB.173.5.1738-1747.1991
Studier, F. W., and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. doi: 10.1016/0022-2836(86)90385-2
Su, K., Zhou, X., Luo, M., Xu, X., Liu, P., Li, X., et al. (2018). Genome-wide identification of genes regulated by RcsA, RcsB, and RcsAB phosphorelay regulators in Klebsiella pneumoniae NTUH-K2044. Microb. Pathog. 123, 36–41. doi: 10.1016/j.micpath.2018.06.036
Sun, Y-C., Guo, X-P., Hinnebusch, B. J., and Darby, C. (2012). The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J. Bacteriol. 194, 2020–2026. doi: 10.1128/JB.06243-11
Tschowri, N., Busse, S., and Hengge, R. (2009). The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 23, 522–534. doi: 10.1101/gad.499409
Walker, K. A., Miner, T. A., Palacios, M., Trzilova, D., Frederick, D. R., Broberg, C. A., et al. (2019). A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. MBio 10:e00089–19. doi: 10.1128/mBio.00089-19
Wall, E., Majdalani, N., and Gottesman, S. (2018). The complex Rcs regulatory cascade. Annu. Rev. Microbiol. 72, 111–139. doi: 10.1146/annurev-micro-090817-062640
Wang, Y., Cen, X-F., Zhao, G-P., and Wang, J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. doi: 10.1128/JB.00989-12
Wu, C-C., Huang, Y-J., Fung, C-P., and Peng, H-L. (2010). Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology 156, 1983–1992. doi: 10.1099/mic.0.038158-0
Xue, J., Tan, B., Yang, S., Luo, M., Xia, H., Zhang, X., et al. (2016). Influence of cAMP receptor protein (CRP) on bacterial virulence and transcriptional regulation of allS by CRP in Klebsiella pneumoniae. Gene 593, 28–33. doi: 10.1016/j.gene.2016.08.006
Keywords: RcsAB, Fur, entC, iron-acquisition system, Klebsiella pneumoniae
Citation: Yuan L, Li X, Du L, Su K, Zhang J, Liu P, He Q, Zhang Z, Peng D, Shen L, Qiu J and Li Y (2020) RcsAB and Fur Coregulate the Iron-Acquisition System via entC in Klebsiella pneumoniae NTUH-K2044 in Response to Iron Availability. Front. Cell. Infect. Microbiol. 10:282. doi: 10.3389/fcimb.2020.00282
Received: 12 December 2019; Accepted: 12 May 2020;
Published: 10 June 2020.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Jin Wang, Shanghai Institute for Biological Sciences (CAS), ChinaAndrés González, University of Zaragoza, Spain
Andreas F. Haag, University of Glasgow, United Kingdom
Mark Stephen Thomas, University of Sheffield, United Kingdom
Copyright © 2020 Yuan, Li, Du, Su, Zhang, Liu, He, Zhang, Peng, Shen, Qiu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingli Li, MTE1MDU2MzY4N0BxcS5jb20=
 Lingyue Yuan
Lingyue Yuan Xuan Li1
Xuan Li1