- 1Bacteriology Laboratory, Division of Laboratory Medicine, Department of Diagnostics, Geneva University Hospitals, Geneva, Switzerland
- 2Infection Control Program, Geneva University Hospitals and Faculty of Medicine, Geneva, Switzerland
- 3Division of Laboratory Medicine, Department of Diagnostics, Geneva University Hospitals, Geneva, Switzerland
- 4Division of Laboratory Medicine, Department of Medical Specialties, Geneva University Hospitals and Faculty of Medicine, Geneva, Switzerland
- 5Genomic Research Laboratory, Division of Infectious Diseases, Department of Medical Specialties, Geneva University Hospitals and Faculty of Medicine, Geneva, Switzerland
Using computerized time-stamps, we compared the turnaround-times (TAT) for urine samples and screening ESwabs of MRSA, VRE, and ESBL carriage in the bacteriology laboratory of Geneva University Hospitals between January and December 2017 (period preceding the implementation of the WASPLabTM) with the same specimen types analyzed between January and December 2019 (period after the implementation of the automation). During both 1-year periods, a total of 98'380 specimens were analyzed (48'158 in 2017 vs. 50'222 in 2019). On the WASPLabTM, all culture plates were imaged at defined intervals each day of incubation, but the processing of the cultures (i.e., pathogen identification and antimicrobial susceptibility testing) was only performed during day shift hours (~8:00 A.M. to 4:30 P.M.). The median TAT for negative reports decreased by almost half for urine samples from 52.1 (2017) to 28.3 h (2019) (p < 0.001), and for MRSA screening specimens from 50.7 to 26.3 h (p < 0.001). The difference in median TAT for negative reports was less pronounced for screening of ESBL (50.2 vs. 43.0 h) (p < 0.001) and VRE (50.6 vs. 45.7 h) (p < 0.001). Despite a trend toward shorter result delivery for positive samples, there was no significant change in the median TAT. These results suggest that TAT for negative samples immediately benefit from automation, whereas TAT for positive samples also depend on the laboratory hours of operation and daily human resource management.
Introduction
Tremendous resources were allocated during the last two decades to develop faster assays and optimized microbiological processes, with the ultimate goal to expedite answers to the ordering clinicians, as measured by reduced turnaround-times (TAT) a major requisite for timely patient management in case of infection suspicion (Francois et al., 2007; Afshari et al., 2012). Conventionally, the workup of microbial cultures in routine clinical laboratories is tightly related to the laboratory hours of operation and to the daily human resource management, much more than driven by the growth kinetics of the microorganisms. This can be explained by the diversity of the samples that has led to a cascade of complex analytical processes, most of them performed manually. In essence, clinical samples are inoculated onto various agar media according the origin of the specimen. Culture media plates are then transferred manually into conventional incubators and removed the next morning to be inspected by technologists. This way of working, which was the norm until recently, can now be deeply revisited due to the availability of total laboratory automation (TLA) systems for culture-based bacteriology. TLA enables sample streaking and swift transfer into incubators with more stable temperature control. The availability of automated digital pictures of culture plates at defined time-points, as specified by the user, obviates the classical 24 h culture cycle. TLA had therefore an immediate and significant impact by enabling standardized and shorter durations of cultures (Dauwalder et al., 2016; Graham et al., 2016; Burckhardt et al., 2018, 2019; Cherkaoui et al., 2020a). Our laboratory implemented the WASPLabTM system in November 2018. The whole automation process was split into different steps (based on the various specimen types) allowing sequential validation of each process (e.g., defining accurate and reproducible time points for plate imaging and for closing the case), thereby providing documentation for the accreditation, a non-disruptive training of the technologists and a stepwise routine implementation (Cherkaoui et al., 2020b).
The purpose of the current study was to compare the direct impact of implementing the WASPlabTM on the TAT (i.e., the time needed to process the sample from reception in the laboratory to the delivery of the culture results to the prescribing physician) taken to complete the analysis process within the lab for urine samples, as well as for nasal and inguinal/perineal screening-ESwab for methicillin-resistant Staphylococcus aureus (MRSA), rectal screening-ESwab for extended-spectrum beta-lactamase-producers (ESBLs) and vancomycin-resistant Enterococcus (VRE).
Materials and Methods
Setting
The present study was performed in the bacteriology laboratory of Geneva University Hospitals, a Swiss tertiary care center with 1'920 beds. The bacteriology laboratory processes ~180'000 clinical samples annually including 30'000 urines, and 28'000 ESwabs for the screening of MRSA, VRE, and ESBL carriage. Current hours of operation of the bacteriology laboratory span from 07.30 A.M. to 10.00 P.M. (7/7). On the WASPLabTM, all culture plates were imaged at defined intervals each day of incubation, but the processing of the cultures [i.e., pathogen identification and antimicrobial susceptibility testing (AST)] is currently only carried out during the day shift (~8:00 A.M. to 4:30 P.M.).
Study Design
We compared the TAT for urine cultures, and for the screening of MRSA, VRE and ESBLs carriage for the specimens referred to the bacteriology laboratory between January and December 2017 and analyzed using the WASP coupled to conventional incubation and manual diagnostic (period preceding the implementation of the WASPLabTM) with the same specimens types that were analyzed using the WASPLabTM between January and December 2019. We began using automation on such specimens in November 2018. However, in order to minimize the effects of the run-in period for using the WASPLabTM, we have chosen to start our observational study on January 2019. To avoid results variability for urine cultures between 2017 and 2019, we restricted the comparison of TAT for negative specimens, the positive specimens with mixed flora, and monomicrobial cultures with the frequent uro-pathogens (Escherichia coli, and Enterococcus faecalis). All bacterial culture results and time-stamps were downloaded from our in-house laboratory information system (Unilab2, version 2.38.1). Table 1 depicts the number of samples included by specimen type, the culture procedures, and the analysis parameters defined on the WASPLabTM based on our previous studies (Cherkaoui et al., 2019a,b). During the two compared periods, the identification of the bacterial strains was performed by matrix-assisted desorption ionization time-of-flight mass spectrometry (MALDI-TOF/MS, Bruker Daltonics, Bremen, Germany) according to the manufacturer's instructions. The antimicrobial susceptibility testing was performed by disc diffusion and VITEK2 (BioMérieux) according to the EUCAST guidelines. All the negative results were validated and transmitted to the clinicians by the technologists (i.e., not auto-released by the WASPLabTM). The positive results were not reported based on specific color on chromogenic media, thus confirmatory testing was systematically performed. The presence of ESBL was confirmed by double-disc synergy tests (DDST20 and DDST30). Confirmation of MRSA and MSSA strains was performed by a previously published qPCR assay targeting femA and mecA (Francois et al., 2003). The presence of VRE was confirmed by defining the minimum inhibitory concentration (MIC) for vancomycin and teicoplanin.
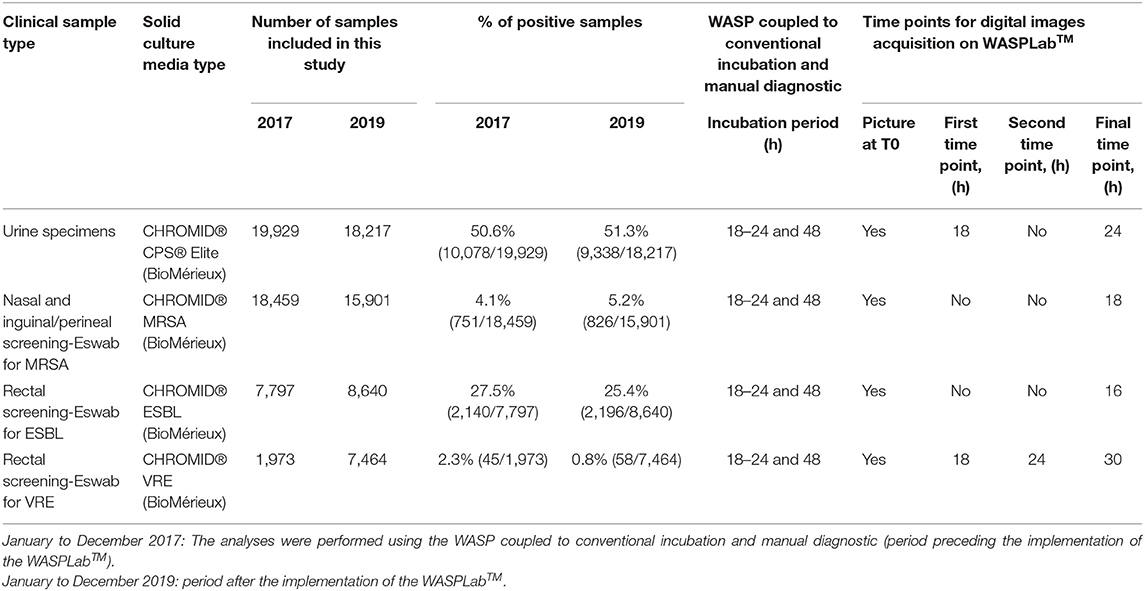
Table 1. Workup of bacterial culture, samples included in this study, and analysis parameters on the WASPLabTM based on previous studies.
Statistical Analyses
Differences in TAT were reported as the median to account for skewed distribution, and tested using Wilcoxon test. All analyses were performed using R.3.6, and RStudio software [RStudio Team (2015)].
Results
Urine Cultures
For the positive samples, only cultures positive with mixed flora, Escherichia coli, or Enterococcus faecalis were included in the analysis. In total, 19'929 urine samples were cultivated in 2017 vs. 18'217 samples in 2019. The percentage of positive samples remained stable around 51%. During the period preceding TLA, the median TAT for negative results was 52.1 h. The implementation of the WASPLabTM significantly reduced the TAT to 28.3 h (p < 0.001) in 2019 (Figure 1). Importantly, we did not see any significant decrease in the TAT for positive samples (56.2 vs. 54.0 h; p < 0.001). The culture workup (i.e., uropathogen identification and AST) was still performed on the day shifts and only during weekdays except for urine samples from infants and pregnant women (about 2–3% of the samples).
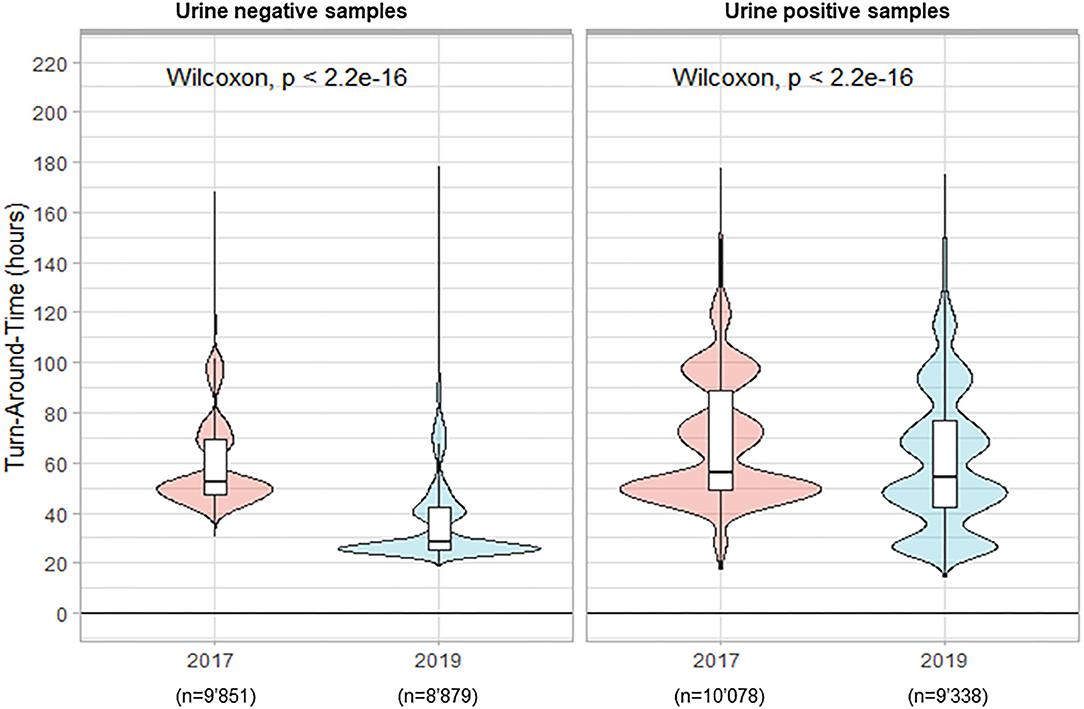
Figure 1. Turn-Around Times (from reception of samples to delivery of the culture results) for urine analysis.
Screening Specimens for MRSA Carriage
In total 18'459 nasal and inguinal/perineal samples were analyzed in 2017 to detect MRSA carriage vs. 15'901 samples in 2019. The percentage of positive samples reached 4.1% in 2017 and showed a slight increase in 2019, with 5.2% positivity. During the period preceding TLA, the median TAT for negative results was 50.7 h. The implementation of the WASPLabTM with the incubation times defined and previously validated (Cherkaoui et al., 2019a,b) enabled a significant reduction of the median TAT to 26.3 h (p < 0.001) (Figure 2). However, we did not see any decrease in the median TAT for positive samples (69.2 h in 2017 vs. 70.2 h in 2019, p = 0.053).
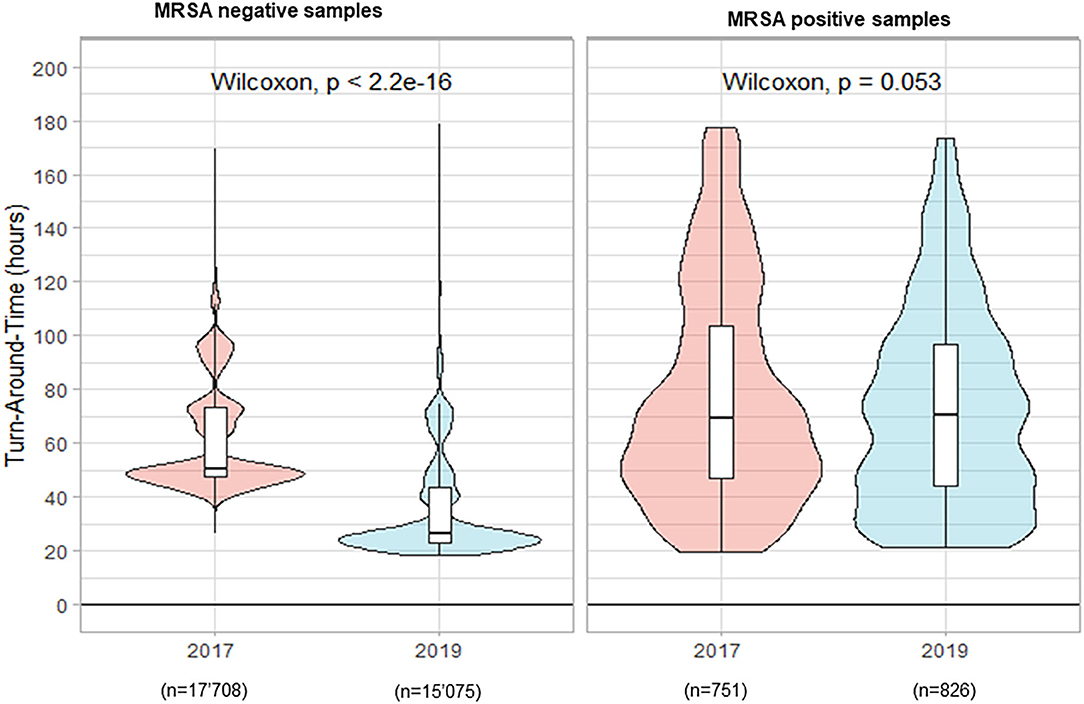
Figure 2. Turn-Around Times (from reception of samples to delivery of the culture results) for nasal and inguinal/perineal screening-ESwab for methicillin-resistant Staphylococcus aureus (MRSA) carriage by culture.
Rectal ESwabs for Detection of ESBL Carriage
In total 7'797 rectal ESwab were analyzed in 2017 for the screening of ESBL carriage vs. 8'640 samples in 2019. The percentage of positive samples was 27.5 and 25.4%, in 2017 and 2019, respectively. During the period preceding TLA, the median TAT for negative results was 50.2 h. The implementation of TLA with its previously defined timing parameters led to a statistically significantly reduced median TAT of 43.0 h (p < 0.001) in 2019 (Figure 3). Paradoxically, we witnessed an increased median TAT for positive samples (from 72.2 h in 2017 to 74.4 h in 2019; p < 0.001).
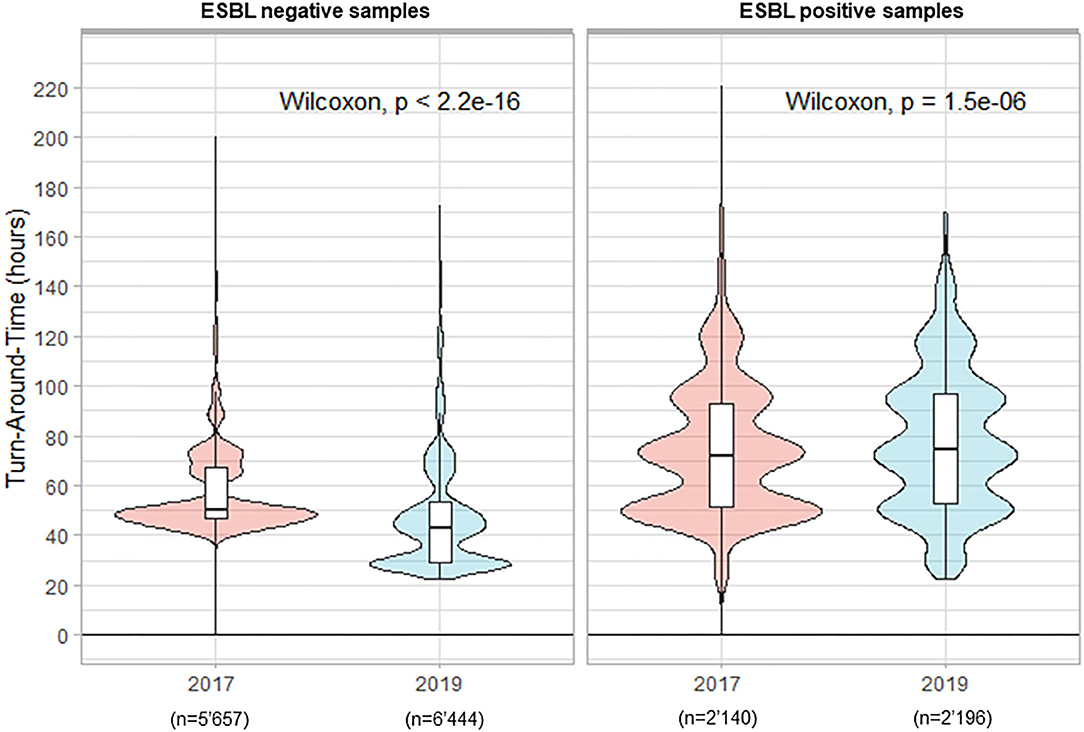
Figure 3. Turn-Around Times (from reception of samples to delivery of the culture results) for rectal screening-ESwab for extended-spectrum beta-lactamases (ESBLs) carriage by culture.
Detection of VRE From Rectal ESwab Samples
In total 1'973 rectal ESwabs were processed in 2017 for the screening of VRE carriage vs. 7'464 samples in 2019. The percentage of positive samples decreased from 2.3 to 0.8% in 2017 and 2019, respectively. The important increase in samples referred to the laboratory in 2019 was related to the implementation of a systematic screening in 2019, whereas in 2017 the search for VRE was performed only for suspected cases. This change toward a systematic screening was prompted by a nosocomial VRE outbreak in Switzerland (Moulin et al., 2018; Buetti et al., 2019). During the period preceding the automation, the median TAT for negative results was 50.6 h (Figure 4). The implementation of the WASPLabTM reduced the median TAT slightly to 45.7 (p < 0.001). Additionally, we witnessed a reduction of the median TAT for positive samples (from 102.0 to 92.2 h, in 2017 and 2019, respectively; p = 0.087), because these samples were also processed during the weekends.
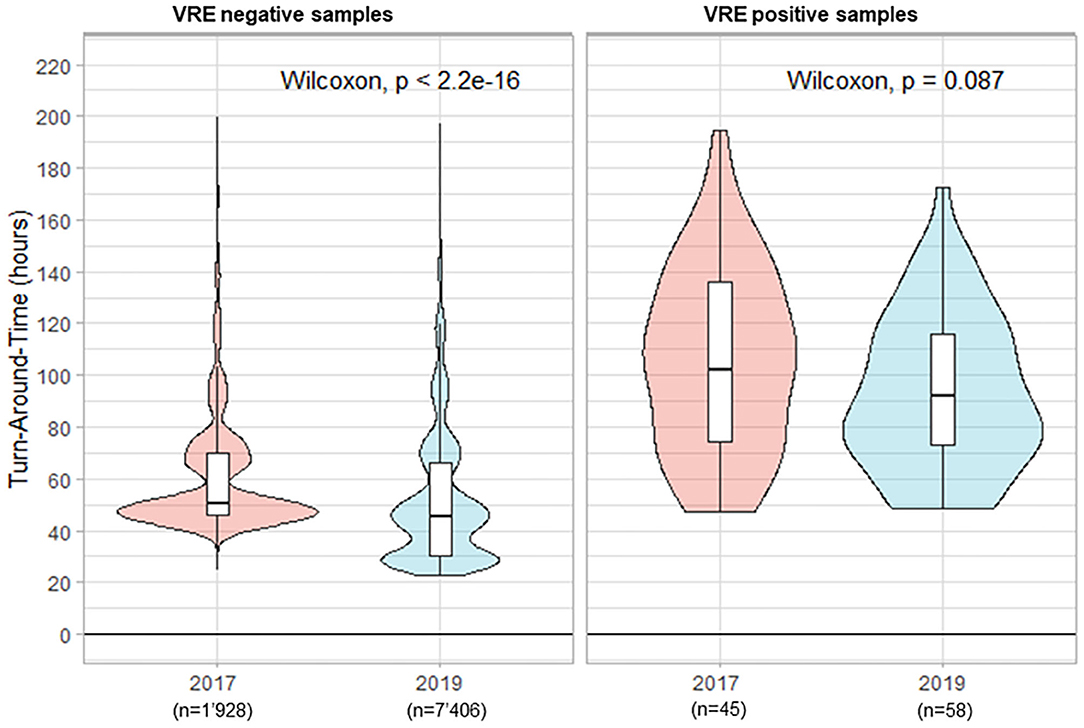
Figure 4. Turn-Around Times (from reception of samples to delivery of the culture results) for rectal screening-ESwab for vancomycin-resistant Enterococcus (VRE) carriage by culture.
Discussion
The main objective of a clinical bacteriology laboratory is to provide prompt and accurate results to clinicians and infection control specialists in order to timely target antimicrobial therapy and infection control precautions. In principle, many factors affect the speed of such culture-based results: (i) the microbial growth kinetics, (ii) the specimens' workflow in the laboratory, (iii) the laboratory hours of operation, and (iv) the daily human resource management. The advent of TLA in clinical bacteriology has brought the potential for reduced specimen processing times, more standardized culture-based testing, and decreased TAT. All of that was enabled by the TLA workflow which begins at the inoculation step to permit microorganisms to grow rapidly after inoculation because plates are automatically transferred within a couple of minutes to incubators whose temperature can be kept stable. Importantly, the three steps in the TLA workflow “screening, reading and picking” allow for more flexibility leading to a significant impact on the TAT. As negative results can be determined at the screening phase, without the need for the reading phase, this easily explains why such samples can immediately benefit from reduced TAT. The picking phase and downstream operations for positive samples will therefore rely on the lab workflow organization for improving their TAT.
In our conventional manual workflow, urine cultures and samples for MRSA, ESBL and VRE carriage screening were inoculated 6 days a week (Monday to Saturday), but the reading of culture plates was performed only on weekdays. In addition, all plates were read on two consecutive days, except if the specimen was already reported positive on day 1. This stems from the fact that the inoculation times for individual plates are not recorded, when performing routine manual operations. Additionally, if day 1 and/or day 2 happened to fall on a weekend or a public holiday, the reading of the media plates was deferred to the next working day. With the implementation of the WASPLabTM, the incubation times for MRSA, VRE, ESBL, and urines culture media plates could be significantly shortened as evidenced by our previous validation studies (Cherkaoui et al., 2019a,b). Yet, it remained necessary to assess the impact of the WASPLabTM implementation on TAT for urine cultures, and for the screening of MRSA, VRE, and ESBL by culture. Using computerized time-stamps, the duration required from sample reception to results release to the ordering provider was retrospectively assessed for each of the specimens included prior to and after implementation of the WASPLabTM. Rates for positive samples remained stable between the pre- and post-TLA study periods, except for VRE because of a Swiss outbreak in 2019 that prompted to add systematic VRE screening in patients at risk in our institution. Importantly, the laboratory hours of operation and the daily human resource management remained unchanged during the compared periods. In particular, culture media plates were read only during the day shift. The median TAT for negative reports went down for MRSA from 50.7 to 26.3 h. Similar results have been previously reported for the detection of MRSA in nasal swabs by Burckhardt et al. (2018), benefiting also from automation. The authors compared the times to report (TTR) for 16'111 nasal swabs (n = 7'620 during the classic workflow period and n = 8'491 during the TLA; BD Kiestra workflow period). The median TTR for negative reports was reduced by 50%. In contrast with this study, we have not seen any decrease in the median TAT for positive samples, which can be explained by the fact that we did not change neither the daily human resource management in 2019 (i.e., the period after the implementation of the lab automation) nor the laboratory operating hours, as would automation permit.
The culture workup (i.e., MRSA identification by qPCR and AST) was still performed on the day shift and only on weekdays. Additionally, we did not find any significant difference in TAT when we excluded samples referred to the laboratory during weekends. The same trend was observed for urine cultures. Namely, the median TAT for negative reports was reduced by 50% after the implementation of the WASPLabTM. These results are similar to findings of previous studies (Yarbrough et al., 2018; Bailey and Burnham, 2019). The difference in the median TAT for negative reports was less pronounced for VRE and ESBL (Table 2). Taken together, this decrease in TAT is directly related to the implementation of the WASPLabTM and to the possible shorter incubation times of the chromogenic agar plates. The use of artificial intelligence (AI), to identify picture plates without bacterial growth could further improve the TAT by providing an automated release of the results, pending an adequate validation of this AI tool.
Despite a downward trend, there were no significant changes in the median TAT for positive samples (i.e., organism identification and susceptibility results). The slight increase of TAT for ESBL positive samples (72.0 vs. 74.4, in 2017 and 2019, respectively), is probably explained by the introduction in 2018 of a systematic search for ESBLs and carbapenemases for non-fermenting Gram-negative bacilli. Thus, additional time was required for such additional confirmatory tests.
Shortening the TAT could positively improve the patient's outcome and the infection control measures. By providing accurate and earlier results to the physicians, one could contribute to optimize therapeutic decisions. However, the real impact of shorter TAT on medical decisions is strongly linked to the reactiveness of the medical teams when the results are delivered on the laboratory information system. Further studies focusing on the overall effectiveness of earlier reporting of culture results on the patient's outcome are therefore warranted, as well as on the potential improvements on infection control. The implementation of automated image analysis, provided cautious validation, could permit the distribution of alerts (e.g., by SMS) in quasi real-time to the infection control team, and enable to remove unnecessary temporary measures.
Conclusions
In conclusion, we established here that TLA per se contributes to significantly reduce TAT for negative samples of urine cultures and for screening ESwabs of MRSA, VRE, and ESBL carriage. However, TAT for positive samples depends largely on the laboratory organization (hours of operation and daily human resource management). That same analysis has been echoed by a recent report (Dauwalder et al., 2019). TLA may enable improved infection control, by the earlier release of negative screening samples. A more comprehensive lab organization is mandatory to deliver also shorter TAT for positive samples and for guiding better and earlier targeted treatments.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee Commission cantonale d'éthique de la recherche (CCER) https://www.hug-ge.ch/ ethique. The ethics committee waived the requirement of written informed consent for participation.
Author Contributions
AC designed the study, analyzed the data, and wrote the manuscript. GR participated in data analysis. RM performed statistical analyses. SH and NV reviewed the manuscript. JS revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported using internal funding.
Conflict of Interest
JS received restricted research grants and participated to advisory boards from BioMérieux and Debiopharm. NV received restricted research grants from Roche. SH participated in an advisory board of DNAe.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Afshari, A., Schrenzel, J., Ieven, M., and Harbarth, S. (2012). Bench-to-bedside review: Rapid molecular diagnostics for bloodstream infection–a new frontier? Crit. Care 16:222. doi: 10.1186/cc11202
Bailey, A. L., and Burnham, C. D. (2019). Reducing the time between inoculation and first-read of urine cultures using total lab automation significantly reduces turn-around-time of positive culture results with minimal loss of first-read sensitivity. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1135–1141. doi: 10.1007/s10096-019-03512-3
Buetti, N., Wassilew, N., Rion, V., Senn, L., Gardiol, C., Widmer, A., et al. (2019). Emergence of vancomycin-resistant enterococci in Switzerland: a nation-wide survey. Antimicrob. Resist. Infect. Control 8:16. doi: 10.1186/s13756-019-0466-x
Burckhardt, I., Horner, S., Burckhardt, F., and Zimmermann, S. (2018). Detection of MRSA in nasal swabs-marked reduction of time to report for negative reports by substituting classical manual workflow with total lab automation. Eur. J. Clin. Microbiol. Infect. Dis. 37, 1745–1751. doi: 10.1007/s10096-018-3308-5
Burckhardt, I., Last, K., and Zimmermann, S. (2019). Shorter incubation times for detecting multi-drug resistant bacteria in patient samples: defining early imaging time points using growth kinetics and total laboratory automation. Ann. Lab. Med. 39, 43–49. doi: 10.3343/alm.2019.39.1.43
Cherkaoui, A., Renzi, G., Azam, N., Schorderet, D., Vuilleumier, N., and Schrenzel, J. (2020a). Rapid identification by MALDI-TOF/MS and antimicrobial disk diffusion susceptibility testing for positive blood cultures after a short incubation on the WASPLab. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1063–1070. doi: 10.1007/s10096-020-03817-8
Cherkaoui, A., Renzi, G., Charretier, Y., Blanc, D. S., Vuilleumier, N., and Schrenzel, J. (2019a). Automated incubation and digital image analysis of chromogenic media using copan WASPLab enables rapid detection of vancomycin-resistant enterococcus. Front. Cell. Infect. Microbiol. 9:379. doi: 10.3389/fcimb.2019.00379
Cherkaoui, A., Renzi, G., Viollet, A., Fleischmann, M., Metral-Boffod, L., Dominguez-Amado, D., et al. (2020b). Implementation of the WASPLab and first year achievements within a university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 39, 1527–1534. doi: 10.1007/s10096-020-03872-1
Cherkaoui, A., Renzi, G., Vuilleumier, N., and Schrenzel, J. (2019b). Copan WASPLab automation significantly reduces incubation times and allows earlier culture readings. Clin. Microbiol. Infect. 25, e1435–1430.e1412. doi: 10.1016/j.cmi.2019.04.001
Dauwalder, O., Landrieve, L., Laurent, F., De Montclos, M., Vandenesch, F., and Lina, G. (2016). Does bacteriology laboratory automation reduce time to results and increase quality management? Clin. Microbiol. Infect. 22, 236–243. doi: 10.1016/j.cmi.2015.10.037
Dauwalder, O., de Martini, J. C., Sauzon, J. F., Drevard, M., Lina, G., Laurent, F., et al. (2019). “Medical value of WASPLab automation solution in a 24/7 clinical laboratory: some preliminary results,” in 29th ECCMID (Lyon).
Francois, P., Bento, M., Renzi, G., Harbarth, S., Pittet, D., and Schrenzel, J. (2007). Evaluation of three molecular assays for rapid identification of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 45, 2011–2013. doi: 10.1128/JCM.00232-07
Francois, P., Pittet, D., Bento, M., Pepey, B., Vaudaux, P., Lew, D., et al. (2003). Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 41, 254–260. doi: 10.1128/JCM.41.1.254-260.2003
Graham, M., Tilson, L., Streitberg, R., Hamblin, J., and Korman, T. M. (2016). Improved standardization and potential for shortened time to results with BD Kiestra total laboratory automation of early urine cultures: a prospective comparison with manual processing. Diagn. Microbiol. Infect. Dis. 86, 1–4. doi: 10.1016/j.diagmicrobio.2016.06.020
Moulin, E., Deggim-Messmer, V., Erard, V., Christin, L., Petignat, C., Blanc, D. S., et al. (2018). [Vancomycine-resistant enterocci (VRE) : a new reality in our hospitals]. Rev. Med. Suisse 14, 791–794.
RStudio Team (2015). RStudio: Integrated Development for R. Boston, MA: RStudio, PBC. Available online at: http://www.rstudio.com/
Keywords: turnaround-times, WASPLab, total laboratory automation, reduction of time to report, urine culture, ESBL - extended-spectrum beta-lactamase, MRSA - methicillin-resistant Staphylococcus aureus, VRE - vancomycin-resistant enterococcus
Citation: Cherkaoui A, Renzi G, Martischang R, Harbarth S, Vuilleumier N and Schrenzel J (2020) Impact of Total Laboratory Automation on Turnaround Times for Urine Cultures and Screening Specimens for MRSA, ESBL, and VRE Carriage: Retrospective Comparison With Manual Workflow. Front. Cell. Infect. Microbiol. 10:552122. doi: 10.3389/fcimb.2020.552122
Received: 15 April 2020; Accepted: 25 September 2020;
Published: 28 October 2020.
Edited by:
Max Maurin, Université Grenoble Alpes, FranceReviewed by:
Olivier Dauwalder, Hospices Civils de Lyon, FranceMatthew Leon Faron, Medical College of Wisconsin, United States
Ahmad Qasem, University of Central Florida, United States
Copyright © 2020 Cherkaoui, Renzi, Martischang, Harbarth, Vuilleumier and Schrenzel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdessalam Cherkaoui, YWJkZXNzYWxhbS5jaGVya2FvdWlAaGN1Z2UuY2g=
 Abdessalam Cherkaoui
Abdessalam Cherkaoui Gesuele Renzi1
Gesuele Renzi1 Jacques Schrenzel
Jacques Schrenzel