- 1Department of Entomology, The Pennsylvania State University, University Park, PA, United States
- 2Department of Biology and Biotechnology, University of Pavia, Pavia, Italy
- 3Huck Institutes of the Life Sciences, The Pennsylvania State University, University Park, PA, United States
- 4Center for Infectious Disease Dynamics, The Pennsylvania State University, University Park, PA, United States
The piRNA pathway is a specialized small RNA interference that in mosquitoes is mechanistically distant from analogous biology in the Drosophila model. Current genetic engineering methods, such as targeted genome manipulation, have a high potential to tease out the functional complexity of this intricate molecular pathway. However, progress in utilizing these methods in arthropod vectors has been geared mostly toward the development of new vector control strategies rather than to study cellular functions. Herein we propose that genetic engineering methods will be essential to uncover the full functionality of PIWI/piRNA biology in mosquitoes and that extending the applications of genetic engineering on other aspects of mosquito biology will grant access to a much larger pool of knowledge in disease vectors that is just out of reach. We discuss motivations for and impediments to expanding the utility of genetic engineering to study the underlying biology and disease transmission and describe specific areas where efforts can be placed to achieve the full potential for genetic engineering in basic biology in mosquito vectors. Such efforts will generate a refreshed intellectual source of novel approaches to disease control and strong support for the effective use of approaches currently in development.
Introduction
The piRNA pathway (PIWI-interacting RNA pathway) is a fascinating biological system that allows an organism to identify parasitic nucleic acids in its cells and create a heritable, genetic memory of these exogenous sequences (Figures 1A–D, Siomi et al., 2011). Arrays of degraded or partial sequences from transposons and viruses are clustered into discrete genomic loci termed piRNA clusters. Very short ribonucleic acids (>24 nt in length), now called piRNAs are encoded from piRNA clusters (Aravin et al., 2006; Vagin et al., 2006; Brennecke et al., 2007). Mature piRNAs, in concert with PIWI (P-element induced wimpy testis) proteins target homologous RNA sequences for destruction (Lin and Spradling, 1997). The activity against a specific target can be amplified by ping-pong amplification, where-by additional piRNAs are generated by interaction with the target RNA and PIWI paralogs (Brennecke et al., 2007; Czech and Hannon, 2016). Complexity of function within an organism is added by protein binding partners that interact with the PIWI/piRNA complex and direct or influence its localization and activity (Brower-Toland et al., 2007; Czech and Hannon, 2011; Arkov, 2018). Many specific features of this system are unknown. For example, piRNA clusters harbor sequences that represent a history of exposure to these invasive agents that is different among species and perhaps across populations of the same species, but we have little understanding of how foreign sequences are integrated into piRNA clusters (Blair et al., 2020).
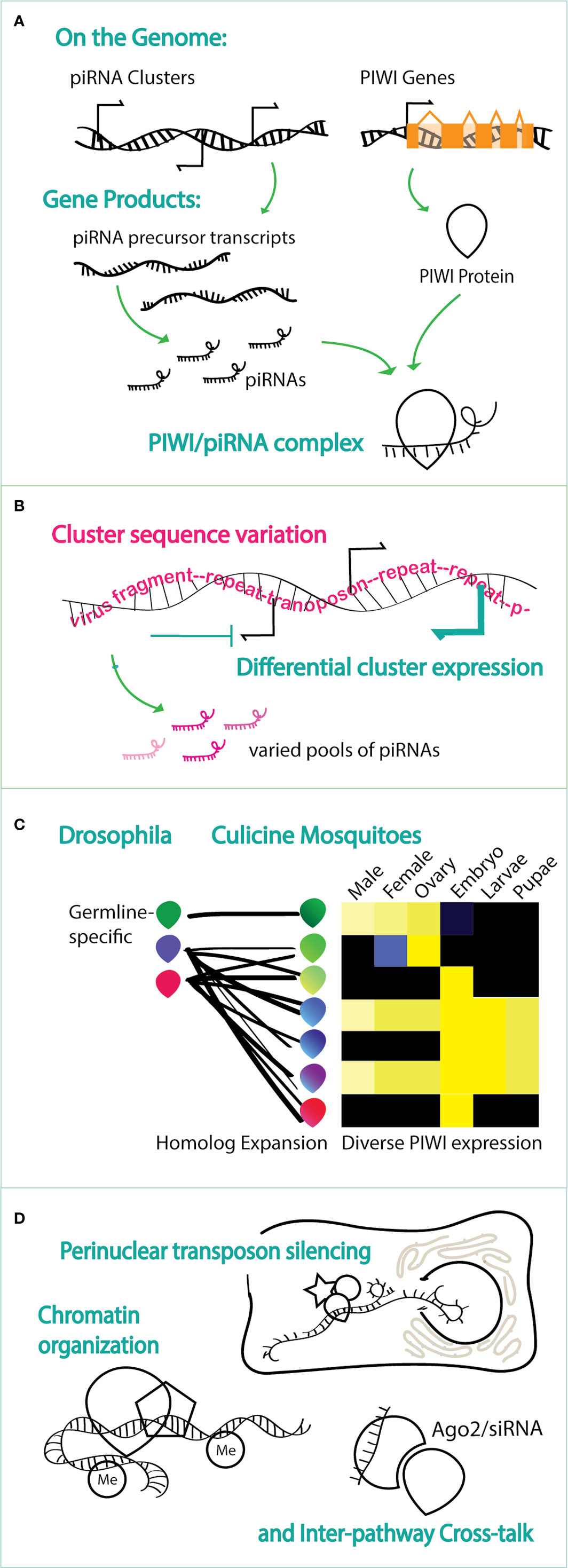
Figure 1 Schematics of endogenous piRNA pathway activity, highlighting the molecular phenomena that contribute to functional complexity. (A) piRNAs, from piRNA clusters, and PIWI proteins, encoded by PIWI genes, are the basic functional unit whereby a target is recognized by piRNA sequence. (B) piRNAs present in a given context vary and are dictated by the sequences present in piRNA clusters (as depicted in pink) and by whether and how much a cluster is expressed in that context (depicted in blue). (C) Increased functional complexity in Culicine mosquitoes is added by the expansion of PIWI proteins in mosquitoes with respect to Drosophila and by the varied expression across tissues and stages in Culicine mosquitoes compared to primarily germ-line specific expression of PIWIs in Drosophila. Heat map schema is based on data from Akbari et al. (2013). (D) Protein binding partners add a level functional complexity, including identified roles of the PIWI/piRNAs complex in with perinuclear proteins for transcript silencing, interaction with protein complexes that add marks to the chromatin, association with the siRNA pathway and, in mosquitoes, with viruses.
The specialization of piRNA pathways among species has captivated the minds of scientists who study diverse organisms with questions about their activity in somatic tissues and roles such as in stem cell maintenance, genome architecture and integrity, behavior, evolution and cancer (Gibson et al., 2015; Lewis et al., 2018; Waldron et al., 2018; Liu et al., 2019; Ozata et al., 2019). Although mosquitoes share hallmark piRNA pathway features with Drosophila, such as phased biogenesis and ping-pong signatures resulting in 1U and 10A trends on piRNAs (Gainetdinov et al., 2018), specialized features of piRNA pathway function is evident in mosquitoes (Gamez et al., 2020). A prominent difference is the antiviral activity of the piRNA pathway observed in mosquitoes, but not Drosophila (Petit et al., 2016). piRNAs that may regulate virus replications are generated both from the virus genome (Hess et al., 2011; Morazzani et al., 2012; Vodovar et al., 2012; Léger et al., 2013; Miesen et al., 2015; Miesen et al., 2016; Göertz et al., 2019) and from Endogenous Virus Elements (EVEs) in the mosquito genome, fragments of viruses often integrated into piRNA clusters (Lequime and Lambrechts, 2017; Palatini et al., 2017; Suzuki et al., 2017; Whitfield et al., 2017; ter Horst et al., 2018; Aguiar et al., 2020; Blair et al., 2020; Suzuki et al., 2020). Further specialization in mosquitoes is evidenced in both Aedes and Anopheles species: 1) PIWI genes and other piRNA pathway genes have expanded and diverged in Aedes mosquitoes (Bernhardt et al., 2012; Miesen et al., 2015; Marconcini et al., 2019; Ma et al., 2020). 2) Cluster content, number and genomic distribution is diverged in both genera [(George et al., 2015; Crava et al., 2020; Palatini et al., 2020). 3) A strong activity of PIWIs and piRNAs in the soma and 4] multiple distinct roles in gene expression are evident in both genera (Belda et al., 2019; Halbach et al., 2020). As such, the piRNA pathway represents an enigmatic biological system in mosquitoes, warranting a more aggressive study of the system in mosquito species.
The piRNA Pathway Exemplifies Basic Biology in Mosquitoes That is Important, But Enigmatic
Knowledge of the basic interactions between pathogens and hosts serves as basis for the development of strategies to interrupt those interactions. From what is already understood about the piRNA pathway, several possible applications to disease control can be imagined. The piRNA pathway genetically encodes targeted destruction of RNA. This brings forward several questions: is it possible that we could engineer the pathway to recognize and repress viral RNA, preventing transmission to humans? Knowing that this biology targets foreign and integrated mobile DNA elements, is it possible that genetically modified mosquitoes could also recognize transgenes, such as those being developed and deployed for public health efforts to mitigate mosquito-borne diseases? If the answer to either of these questions is yes, a myriad of additional mechanistic questions will follow. To answer these, we need a much better handle on the PIWI/piRNA activity in individual cells, across tissues and even up to the level of population effects in diverse environmental contexts.
Despite the intellectual draw of untangling the complexity of the piRNA biology in mosquitoes and its relevance to host-virus interactions, little is known about this pathway. The current knowledge has mostly been generated in model organisms by rigorous and elegant combinations of immunohistochemistry and next-generation sequencing methods. The availability of many antibodies has enabled the characterization of protein-protein and protein-small RNA interactions and the characterization of cellular localization of PIWI proteins. Well-annotated model genomes along with increasingly robust and affordable small RNA sequencing have been applied to identify sequences and patterns of piRNAs and piRNA clusters as well as to compare piRNA populations among experimental conditions. This body of knowledge provides a basis for studies in non-model organisms. Unfortunately, studies in non-model arthropods suffer from a paucity of specific antibodies and, while sequencing can be used in various experimental contexts, the absence of good reference genomes for several arthropods and Aedes species, except for Ae. aegypti, makes high-precision bioinformatic analyses difficult (Richards, 2018).
Excellent work in cell lines has enabled us to make headway in light of these impediments (Schnettler et al., 2013; Varjak et al., 2017; Joosten et al., 2019; Ma et al., 2020), but the expansion of mosquito PIWIs from germ-line restricted functions (Figure 1C) leaves us with many questions about multi-tissue functions such as might be relevant to virus regulation that are better answered in live mosquitoes (Akbari et al., 2013; Wang et al., 2018). It is therefore valuable to develop our ability to perturb the function of this pathway in vivo, but direct manipulation of endogenous piRNA sequences for direct hypothesis testing is not yet straight-forward in any organism. Although transgenesis and targeted genome editing using Cas9 in mosquitoes were accomplished by 1998 and 2015 respectively, opening up the possibility use genome modifications to explore the piRNA biology, the first report to do so came out this year (Coates et al., 1998; Jasinskiene et al., 1998; Basu et al., 2015; Dong et al., 2015; Kistler et al., 2015; Suzuki et al., 2020).
Similar to many processes that are involved in pathogen transmission in mosquitoes, such as host-seeking, blood-meal acquisition and digestion, and pathogen traversal of the gut, the piRNA pathway is sufficiently specialized that the closest model organism, Drosophila melanogaster, offers little for extrapolation to mosquitoes (Nouzova et al., 2019). This exacerbates the investigational lag induced by technological disadvantages and provides impetus to aggressively pursue new discoveries in disease vector biology. Because arthropod host stages of human disease pathogens are the most tractable targets for pathogen control, studies into specialized mosquito functions have led to novel approaches to disease control (Shaw and Catteruccia, 2019). Thus, the piRNA-pathway biology is an apt example of a pool of knowledge that, while potentially beneficial to efforts to control mosquito-borne disease, is just out of reach. We submit that a more aggressive application of genetic engineering will lengthen that reach and that existing genetic tools are not being applied to their full potential.
The Particular Necessity of Genetic Engineering in Mosquitoes for Pirna Research
We can imagine many ways in which molecular genetics, especially genetic engineering and sequencing, can allow us to answer the outstanding questions in piRNA biology (Figure 2). For example, direct targeting of mosquito genomes by programmable endonucleases will allow us to not only encode piRNA sequences within clusters, but also to test elements of the clusters that may be involved in expression, such as promoters, enhancers and repressors (Figure 2A) (Olovnikov et al., 2014). Target mutations in PIWI genes sequences can allow us to produce specific protein modifications to inhibit or alter function (Figure 2B). The scarcity of specific antibodies also can be addressed using genetic engineering; targeted transgenesis, insertion of new DNA into a gene, can be used to add fluorescence or epitope tags, such as FLAG or Myc, onto the endogenous PIWI protein. These tags can be visualized or immunoprecipitated using well characterized antibodies to characterize proteins localization or determine proteins and piRNA populations bound to a specific PIWI under different experimental conditions (Figure 2B) (Stadler et al., 2013; Tassetto et al., 2019). Similarly, transgenesis can be used to encode functional domains onto PIWI genes, such an RNA tether domain that can be used to detect PIWI cleavage activity on synthetic reporter transcript (Keryer-Bibens et al., 2008). Genetic manipulation can allow us to overcome the challenges of genes suppression and ablation in the germline where the piRNA pathway is greatly active, but which is mostly inaccessible to RNAi-based injection methods of knockdown and where much of the gene function is necessary for reproduction (Huvenne and Smagghe, 2010; Balakrishna Pillai et al., 2017).
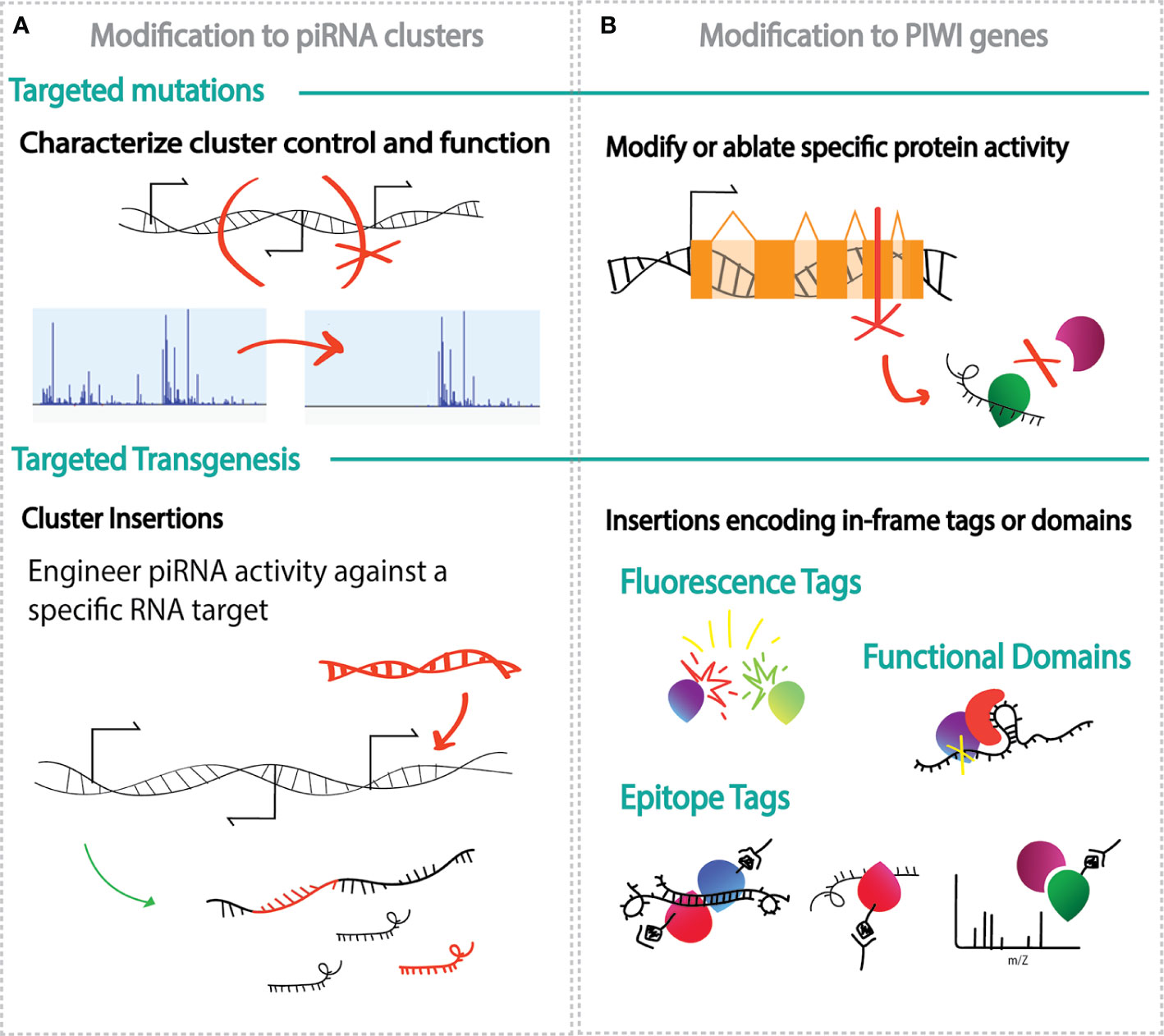
Figure 2 Schematic examples of genetic engineering applications to explore ethe biology of the piRNA pathway. (A) Modifications of the piRNA clusters through target mutations (top) and insertion of new DNA sequence (transgenesis, bottom) to identify regulatory elements within the cluster or generate piRNAs against chosen targets; (B) modification to the PIWI genes through targeted mutations (top) and/or transgenesis (bottom) and paired with methods such as fluorescence microscopy, immunoprecipitation, mass spectrometry and sequencing, to uncover in specialization of each PIWI proteins and protein partners.
The General Impetus to Expand Genetic Engineering for Molecular Biology in Disease Vectors
In general, molecular genetic applications in mosquitoes is largely being moved forward by laboratories with defined, direct objectives for developing genetic technologies for mosquito control, but there is a real advantage in applying resources to vector biology with the explicit goal of understanding the basic biology of the vector. Technology-driven research should remain a major focus, but laboratories that are developing applications in disease control have largely driven discovery. Resources should also be put toward exploring basic biology for the sake of understanding unique cellular mechanisms of disease vectors, with the implications for disease control in mind. This will lay a strong foundation for inspiration and development of new technologies. A timely example from bacteriology is that investigations into CRISPR/Cas biology led to one of the most widespread revolutions in genetic technologies (Ishino et al., 2018). Taking advantage of the ingenuity of curious researchers to answer difficult and interesting questions, a serious commitment to basic biology will produce technological innovations.
Similarly, the more complete our fundamental understanding of mosquitoes and pathogenesis at the molecular level, the more likely our new efforts to manipulate mosquitoes and pathogens at that level for disease control will succeed. From our example of the piRNA pathway, if organisms can learn to recognize DNA in their genome as foreign (and putatively deleterious), it is possible that mosquitoes identify certain parts of transgenes as foreign and repress them (Adelman et al., 2004; Franz et al., 2009). If we understand what contributes to such recognition, we can engineer transgenic mosquitoes to avoid it. When we face an immense and long-standing problem like vector-borne disease, an earnest pursuit of lasting solutions requires efforts and resources to complete our knowledge of the basic biology of disease vectors.
We highlight the need for a concerted effort to translate genetic tools currently available in model organisms to mosquito and other disease vectors. The potential shift in momentum that we are hoping will occur will require coordination and persistence. An encouraging example of this is the creation of the field of Disease Vector Biology which was inspired by a coincidence of the resurgence of mosquito-borne diseases, a recognition that we were critically under-equipped to address this situation and a conviction that an application of modern genetic tools would lead to innovations that would bolster the resources available (Beaty et al., 2009). A collaborative and well-organized effort to translate techniques that were available for Drosophila genetics into mosquitoes paid off: in the early 2000s a renewed effort was made to eradicated malaria and leishmania and incredible gains have been made in decreasing the burden of disease spread caused by arthropod vectors in the decades that followed (Alonso and Noor, 2017; Benelli and Beier, 2017). Remembering the gains made in the first malaria eradication agenda and the resurgence that followed, we can reflect that the gains that aren’t made can reverse progress.
In light of the advancement of molecular genetic methods in model organisms, the current lag in the translation of molecular genetic methods in mosquitoes highlights major knowledge gaps in our understanding of vector biology. We propose the following areas of focus to support the robust application of molecular genetics to fill these gaps:
1. Pursue and Support Difficult Projects for New Discovery in the Biology of Mosquitoes. These projects are often difficult and long, but the investment is worthwhile as novel methods will be required for specialized experimentation in arthropod-borne diseases. An analogous moment in the history of the field of vector biology is the accomplishment of Drosophila genome transformation using the P-element (Bachmann and Knust, 2008) and subsequent efforts to translate this technology into mosquitoes. What was not widely known or demonstrated at the time was that transposable elements generally behave differently in distinct organisms and in this case the P-element is not effective for mosquito genome transformation. When this became apparent, assays to find and test the activity of alternate transposable elements led to the genome transformation in mosquitoes, 16 years after genome transformation in D. melanogaster (Spradling and Rubin, 1982; Coates et al., 1998; Jasinskiene et al., 1998). The lesson here is that the technologies we need will not necessarily be directly translatable from one system to another and in extension that time, energy and funding should be allocated to make necessary discoveries.
2. Improve and Publish Information on Insect Rearing and Handling Generally, and in Relationship to Genetic Manipulation and Screening Methods. The expansion of genetic methods to laboratories that are not specialized in mosquito genetics will be supported by development and publication of methods with low-cost and straight-forward implementation, such as those available for Anopheles from MR4 (Methods in Anopheles Research Manual). Several groups develop methods that work for their laboratory to efficiently manage mosquitoes with limited equipment or insect-rearing space, for example the oviplate and glytube (Costa-da-Silva et al., 2013; Ioshino et al., 2018) but many of these may not be published. This can be supported by an expansion of current information hubs to support small methods publications, foster discussion and questions that can support this effort and by introducing more descriptive methods sections in the primary literature.
3. Expand Non-Embryo Injection Strategies for RNAi and Genetic Modification to Include More Species and Transgenesis. The translation of Cas9-mediated gene editing using adult injections have been demonstrated to efficiently support targeted mutagenesis in several species (Chaverra-Rodriguez et al., 2018; Chaverra‐Rodriguez et al., 2020; Heu et al., 2020; Macias et al., 2020). This is supported by robust and affordable PCR and sequencing techniques (Ran et al., 2013; Bell et al., 2014; Carballar-Lejarazú et al., 2020). Expansion of this will include both translation to more vector species and engineering the techniques for DNA and RNA delivery for heritable transgenesis and RNA interference methods.
4. Improve the Efficiency of Targeted Insertions. In vector species in which embryo injections methods are available, rigorous testing of methods currently used in other systems to improve the efficiency of homology-directed repair will improve the manipulation of genes of interest through the introduction of a visible marker of mutation by transgenesis. A goal in mosquito species that could serve as vector model organisms, such as Ae. aegypti and Anopheles stephensi, would be to mirror the situation for D. melanogaster in which a transgenic line exists for most genes of interest with a variety of endogenous tags.
5. Improve Genome Annotations and Expand Vector Sequencing. As traditional vector control methods such as insecticides have become less effective, alternative methods involving genetic manipulation of mosquitoes are being investigated. Reliable and accurate genome assemblies of vector species and genome resequencing data of individuals from different populations and sub-species are not only essential to develop newer genetic editing approaches but also a collective tool to advance our understanding of vector species biology, gene expression, immunity and global variability.
6. Expand Training Among Vector Biologists in Current and Emerging Genetic Methods and Use Them. Many methods for genome investigation and manipulation are currently available for mosquitoes, but not widely used (Adelman et al., 2008; O’Brochta et al., 2012; Adelman et al., 2016; Häcker et al., 2017; Adolfi and Lycett, 2018; Reid et al., 2018). Efforts to remedy this are, for example, the course on the Biology of Disease Vectors, a technical short course on Insect Genetic Techniques and peer-to-peer training opportunities available through the IGTRCN. An important part of expanding this training may be for labs to recognize that many methods already exist for use by groups that have not traditionally used genetic techniques. For laboratories in countries directly impacted by vector-borne disease, genetic technologies can be combined with accessibility to relevant samples and colonies. These are more likely to conduct field tests with the knowledge gained and so should be a focus for training and support in genetic engineering methods. In parallel, labs globally that are already using existing techniques and that leverage genetic methods for their biological investigations, such as those studying mosquito chemo-sensation, are taking an important part in building momentum (DeGennaro et al., 2013; McMeniman et al., 2014; Matthews et al., 2019). These goals are supported by infrastructures and networks such as INFRAVEC-2 that provides coordination of resources and projects in vector-borne disease. Developing genetic engineering methods and coordinating the expansion of these methods will open the door to a phase of research that can freshly energize our efforts against vector-borne disease with new knowledge and new technologies.
With these areas of focus in mind, we can move the state of mosquito transgenic technologies forward enough to support a more robust effort to approach questions, as in those surrounding the piRNA biology in mosquitoes, that will grant us access to a molecular mechanisms currently out of reach and likely to produce insight and innovation for the control of vector-borne diseases. Doing so will provide basic knowledge and molecular methods that will feed inspiration and innovation for new approaches to vector and pathogen control and will bolster current control applications.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding author.
Author Contributions
VM, UP, MB, and JR wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by a Huck Innovative and Transformative Seed Fund award to VM, by NIH grants R01AI150251, R01AI128201, and R01AI116636 to JR, by NSF grant 1645331 to JR, by USDA Hatch funds (Accession #1010032; Project #PEN04608) to JR; by a Human Frontier Science Program Research grant (RGP0007/2017) to MB, by the Italian Ministry of Education, University and Research FARE-MIUR project R1623HZAH5 to MB, by a European Research Council Consolidator Grant (ERC-CoG) under the European Union’s Horizon 2020 Programme (Grant Number ERC-CoG 682394) to MB; and by the Italian Ministry of Education, University and Research (MIUR): Dipartimenti di Eccellenza Program (2018–2022) to the Dept. of Biology and Biotechnology “L. Spallanzani,” University of Pavia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Anthony A. James for fruitful discussion of the content of this manuscript.
References
Adelman Z. N., Jasinskiene N., Vally K. J. M., Peek C., Travanty E., Olson K. E., et al. (2004). Formation and loss of large, unstable tandem arrays of the piggyBac transposable element in the yellow fever mosquito, Aedes aegypti. Transgenic Res. 13, 411–425. doi: 10.1007/s11248-004-6067-2
Adelman Z. N., Anderson M. A. E., Morazzani E. M., Myles K. M. (2008). A transgenic sensor strain for monitoring the RNAi pathway in the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 38, 705–713. doi: 10.1016/j.ibmb.2008.04.002
Adelman Z. N., Basu S., Myles K. M. (2016). “Gene insertions and deletion in mosquitoes,” in Genetic Control of Malaria and Dengue. Ed. Adelman Z. N. (Cambridge, MA: Academic Press), 139–168.
Adolfi A., Lycett G. J. (2018). Opening the toolkit for genetic analysis and control of Anopheles mosquito vectors. Curr. Opin. Insect Sci. 30, 8–18. doi: 10.1016/j.cois.2018.07.014
Aguiar E. R. G. R., de Almeida J. P. P., Queiroz L. R., Oliveira L. S., Olmo R. P., da Silva De Faria I. J., et al. (2020). A single unidirectional piRNA cluster similar to the flamenco locus is the major source of EVE-derived transcription and small RNAs in Aedes aegypti mosquitoes. RNA 26, 581–594. doi: 10.1261/rna.073965.119
Akbari O. S., Antoshechkin I., Amrhein H., Williams B., Diloreto R., Sandler J., et al. (2013). The Developmental Transcriptome of the Mosquito Aedes aegypti, an Invasive Species and Major Arbovirus Vector. G3 (Bethesda) 3, 1493–1509. doi: 10.1534/g3.113.006742
Alonso P., Noor A. M. (2017). The global fight against malaria is at crossroads. Lancet (London England) 390, 2532–2534. doi: 10.1016/S0140-6736(17)33080-5
Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. doi: 10.1038/nature04916
Arkov A. L. (2018). RNA Selection by PIWI Proteins. Trends Biochem. Sci. 43, 153–156. doi: 10.1016/j.tibs.2017.12.007
Bachmann A., Knust E. (2008). The use of P-element transposons to generate transgenic flies. Methods Mol. Biol. 420, 61–77. doi: 10.1007/978-1-59745-583-1_4
Balakrishna Pillai A., Nagarajan U., Mitra A., Krishnan U., Rajendran S., Hoti S. L., et al. (2017). RNA interference in mosquito: understanding immune responses, double-stranded RNA delivery systems and potential applications in vector control. Insect Mol. Biol. 26, 127–139. doi: 10.1111/imb.12282
Basu S., Aryan A., Overcash J. M., Samuel G. H., Anderson M. A. E., Dahlem T. J., et al. (2015). Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti. Proc. Natl. Acad. Sci. 112, 4038–4043. doi: 10.1073/pnas.1502370112
Beaty B. J., Prager D. J., James A. A., Jacobs-Lorena M., Miller L. H., Law J. H., et al. (2009). From Tucson to genomics and transgenics: The vector biology network and the emergence of modern vector biology. PloS Negl. Trop. Dis. 3, e343. doi: 10.1371/journal.pntd.0000343
Belda E., Nanfack-Minkeu F., Eiglmeier K., Carissimo G., Holm I., Diallo M., et al. (2019). De novo profiling of RNA viruses in Anopheles malaria vector mosquitoes from forest ecological zones in Senegal and Cambodia. BMC Genomics 20, 664. doi: 10.1186/s12864-019-6034-1
Bell C. C., Magor G. W., Gillinder K. R., Perkins A. C. (2014). A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC Genomics 15, 1002. doi: 10.1186/1471-2164-15-1002
Benelli G., Beier J. C. (2017). Current vector control challenges in the fight against malaria. Acta Trop. 174, 91–96. doi: 10.1016/j.actatropica.2017.06.028
Bernhardt S. A., Simmons M. P., Olson K. E., Beaty B. J., Blair C. D., Black W. C. (2012). Rapid Intraspecific Evolution of miRNA and siRNA Genes in the Mosquito Aedes aegypti. PloS One 7, e44198. doi: 10.1371/journal.pone.0044198
Blair C. D., Olson K. E., Bonizzoni M. (2020). The Widespread Occurrence and Potential Biological Roles of Endogenous Viral Elements in Insect Genomes. Curr. Issues Mol. Biol. 34, 13–30. doi: 10.21775/cimb.034.013
Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., Sachidanandam R., et al. (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. doi: 10.1016/j.cell.2007.01.043
Brower-Toland B., Findley S. D., Jiang L., Liu L., Yin H., Dus M., et al. (2007). Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21, 2300–2311. doi: 10.1101/gad.1564307
Carballar-Lejarazú R., Kelsey A., Pham T. B., Bennett E. P., James A. A. (2020). Digital droplet PCR and IDAA for the detection of CRISPR indel edits in the malaria species Anopheles stephensi. Biotechniques 68, 172–179. doi: 10.2144/BTN-2019-0103
Chaverra-Rodriguez D., Macias V. M., Hughes G. L., Pujhari S., Suzuki Y., Peterson D. R., et al. (2018). Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 9, 3008. doi: 10.1038/s41467-018-05425-9
Chaverra-Rodriguez D., Dalla Benetta E., Heu C. C., Rasgon J. L., Ferree P. M., Akbari O. S. (2020). Germline mutagenesis of Nasonia vitripennis through ovarian delivery of CRISPR-Cas9 ribonucleoprotein. Insect Mol. Biol. 29, 569–577. doi: 10.1111/imb.12663
Coates C. J., Jasinskiene N., Miyashiro L., James A. A. (1998). Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 95, 3748–3751. doi: 10.1073/pnas.95.7.3748
Costa-da-Silva A. L., Navarrete F. R., Salvador F. S., Karina-Costa M., Ioshino R. S., Azevedo D. S., et al. (2013). Glytube: A Conical Tube and Parafilm M-Based Method as a Simplified Device to Artificially Blood-Feed the Dengue Vector Mosquito, Aedes aegypti. PloS One 8, e53816. doi: 10.1371/journal.pone.0053816
Crava C., Varghese F. S., Pischedda E., Halbach R., Palatini U., Marconcini M., et al. (2020). Immunity to infections in arboviral vectors by integrated viral sequences: an evolutionary perspective. bioRxiv. doi: 10.1101/2020.04.02.022509
Czech B., Hannon G. J. (2011). Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 12, 19–31. doi: 10.1038/nrg2916
Czech B., Hannon G. J. (2016). One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem. Sci. 41, 324–337. doi: 10.1016/j.tibs.2015.12.008
DeGennaro M., McBride C. S., Seeholzer L., Nakagawa T., Dennis E. J., Goldman C., et al. (2013). orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 498, 487–491. doi: 10.1038/nature12206
Dong S., Lin J., Held N. L., Clem R. J., Passarelli A. L., Franz A. W. E. (2015). Heritable CRISPR/Cas9-Mediated Genome Editing in the Yellow Fever Mosquito, Aedes aegypti. PloS One 10, e0122353. doi: 10.1371/journal.pone.0122353
Franz A. W. E., Sanchez-Vargas I., Piper J., Smith M. R., Khoo C. C. H., James A. A., et al. (2009). Stability and loss of a virus resistance phenotype over time in transgenic mosquitoes harbouring an antiviral effector gene. Insect Mol. Biol. 18, 661–672. doi: 10.1111/j.1365-2583.2009.00908.x
Gainetdinov I., Colpan C., Arif A., Cecchini K., Zamore P. D. (2018). A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals Unified piRNA biogenesis model. Mol. Cell 71, 775–790.e5. doi: 10.1016/j.molcel.2018.08.007
Gamez S., Srivastav S., Akbari O. S., Lau N. C. (2020). Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways. Cells 9, 2180. doi: 10.3390/cells9102180
George P., Jensen S., Pogorelcnik R., Lee J., Xing Y., Brasset E., et al. (2015). Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster. Epigenet. Chromatin 8, 50. doi: 10.1186/s13072-015-0041-5
Gibson J. D., Arechavaleta-Velasco M. E., Tsuruda J. M., Hunt G. J. (2015). Biased Allele Expression and Aggression in Hybrid Honeybees may be Influenced by Inappropriate Nuclear-Cytoplasmic Signaling. Front. Genet. 6, 343. doi: 10.3389/fgene.2015.00343
Göertz G. P., Miesen P., Overheul G. J., van Rij R. P., van Oers M. M., Pijlman G. P. (2019). Mosquito Small RNA Responses to West Nile and Insect-Specific Virus Infections in Aedes and Culex Mosquito Cells. Viruses 11, 271. doi: 10.3390/v11030271
Häcker I., Harrell Ii R. A., Eichner G., Pilitt K. L., O’brochta D. A., Handler A. M., et al. (2017). Cre/lox-Recombinase-Mediated Cassette Exchange for Reversible Site-Specific Genomic Targeting of the Disease Vector, Aedes aegypti OPEN. Sci Rep 7, 43883. doi: 10.1038/srep43883
Halbach R., Miesen P., Joosten J., Taşköprü E., Rondeel I., Pennings B., et al. (2020). A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 580, 274–277. doi: 10.1038/s41586-020-2159-2
Hess A. M., Prasad A. N., Ptitsyn A., Ebel G. D., Olson K. E., Barbacioru C., et al. (2011). Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 11, 45. doi: 10.1186/1471-2180-11-45
Heu C. C., Mccullough F. M., Luan J., Rasgon J. L. (2020). CRISPR-Cas9-Based Genome Editing in the Silverleaf Whitefly (Bemisia tabaci). Cris J. 3, 89–96. doi: 10.1089/crispr.2019.0067
Huvenne H., Smagghe G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 56, 227–235. doi: 10.1016/j.jinsphys.2009.10.004
Ioshino R. S., Carvalho D. O., Marques I. C. S., Fernandes E. S., Capurro M. L., Costa-Da-Silva A. L. (2018). Oviplate: A convenient and space-saving method to perform individual oviposition assays in Aedes aegypti. Insects 9, 103. doi: 10.3390/insects9030103
Ishino Y., Krupovic M., Forterre P. (2018). History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J. Bacteriol. 200, e00580-17. doi: 10.1128/JB.00580-17
Jasinskiene N., Coates C. J., Benedict M. Q., Cornel A. J., Rafferty C. S., James A. A., et al. (1998). Stable transformation of the yellow fever mosquito, Aedes aegypti, with the Hermes element from the housefly. Proc. Natl. Acad. Sci. U. S. A. 95, 3743–3747. doi: 10.1073/pnas.95.7.3743
Joosten J., Miesen P., Tas¸köprü E., Tas¸k T., Tas¸köpr T., Tas¸köprü T., et al. (2019). The Tudor protein Veneno assembles the ping-pong amplification complex that produces viral piRNAs in Aedes mosquitoes. Nucleic Acids Res. 47, 2546–2559. doi: 10.1093/nar/gky1266
Keryer-Bibens C., Barreau C., Osborne H. B. (2008). Tethering of proteins to RNAs by bacteriophage proteins. Biol. Cell 100, 125–138. doi: 10.1042/BC20070067
Kistler K. E., Vosshall L. B., Matthews B. J. (2015). Genome Engineering with CRISPR-Cas9 in the Mosquito Aedes aegypti. Cell Rep. 11, 51–60. doi: 10.1016/j.celrep.2015.03.009
Léger P., Lara E., Jagla B., Sismeiro O., Mansuroglu Z., Coppée J. Y., et al. (2013). Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J. Virol. 87, 1631–1648. doi: 10.1128/JVI.02795-12
Lequime S., Lambrechts L. (2017). Discovery of flavivirus-derived endogenous viral elements in Anopheles mosquito genomes supports the existence of Anopheles-associated insect-specific flaviviruses. Virus Evol. 3, 1–8. doi: 10.1093/ve/vew035
Lewis S. H., Quarles K. A., Yang Y., Tanguy M., Frézal L., Smith S. A., et al. (2018). Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2, 174–181. doi: 10.1038/s41559-017-0403-4
Lin H., Spradling A. C. (1997). A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovar. Development 124, 2463–2476.
Liu Y., Dou M., Song X., Dong Y., Liu S., Liu H., et al. (2019). The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 18, 123. doi: 10.1186/s12943-019-1052-9
Ma Q., Srivastav S. P., Gamez S., Feitosa-Suntheimer F., Patterson E. II, Johnson R., et al. (2020). An integrated mosquito small RNA genomics resource reveals dynamic evolution and host responses to viruses and transposons. bioRxiv. doi: 10.1101/2020.04.25.061598 2020.04.25.061598.
Macias V. M., Mckeand S., Chaverra-Rodriguez D., Hughes G. L., Fazekas A., Pujhari S., et al. (2020). Cas9-Mediated Gene-Editing in the Malaria Mosquito Anopheles stephensi by ReMOT Control. G3 10, 1353–1360. doi: 10.1534/g3.120.401133
Marconcini M., Hernandez L., Iovino G., Houé V., Valerio F., Palatini U., et al. (2019). Polymorphism analyses and protein modelling inform on functional specialization of Piwi clade genes in the arboviral vector Aedes albopictus. PloS Negl. Trop. Dis. 13, e0007919. doi: 10.1371/journal.pntd.0007919
Matthews B. J., Younger M. A., Vosshall L. B. (2019). The ion channel ppk301 controls freshwater egg-laying in the mosquito Aedes aegypti. Elife 8, e43963. doi: 10.7554/eLife.43963
McMeniman C., Corfas R., Matthews B., Ritchie S. (2014). Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell 156, 1060–1071. doi: 10.1016/j.cell.2013.12.044
Methods in Anopheles Research Manual. Available at: https://www.beiresources.org/AnophelesProgram/TrainingMethods.asp (Accessed September 28, 2020).
Miesen P., Girardi E., van Rij R. P. (2015). Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 43, 6545–6556. doi: 10.1093/nar/gkv590
Miesen P., Joosten J., van Rij R. P. (2016). PIWIs Go Viral: Arbovirus-Derived piRNAs in Vector Mosquitoes. PloS Pathog. 12, e1006017. doi: 10.1371/journal.ppat.1006017
Morazzani E. M., Wiley M. R., Murreddu M. G., Adelman Z. N., Myles K. M. (2012). Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PloS Pathog. 8, e1002470. doi: 10.1371/journal.ppat.1002470
Nouzova M., Clifton M. E., Noriega F. G. (2019). Mosquito adaptations to hematophagia impact pathogen transmission. Curr. Opin. Insect Sci. 34, 21–26. doi: 10.1016/j.cois.2019.02.002
Olovnikov I., Le Thomas A., Aravin A. A. (2014). “A Framework for piRNA Cluster Manipulation,” in Methods in Molecular Biology 1093. Ed. Siomi M, (Totowa, NJ: Humana Press). doi: 10.1007/978-1-62703-694-8_5
O’Brochta D. A., Pilitt K. L., Harrell R. A., Aluvihare C., Alford R. T. (2012). Gal4-based enhancer-trapping in the malaria mosquito anopheles stephensi. G3 Genes Genomes Genet. 2, 1305–1315. doi: 10.1534/g3.112.003582
Ozata D. M., Gainetdinov I., Zoch A., O’Carroll D., Zamore P. D. (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108. doi: 10.1038/s41576-018-0073-3
Palatini U., Miesen P., Carballar-Lejarazu R., Ometto L., Rizzo E., Tu Z., et al. (2017). Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics 18, 512. doi: 10.1186/s12864-017-3903-3
Palatini U., Masri R. A., Cosme L. V., Koren S., Thibaud-Nissen F., Biedler J. K., et al. (2020). Improved reference genome of the arboviral vector Aedes albopictus. Genome Biol. 21, 215. doi: 10.1186/s13059-020-02141-w
Petit M., Mongelli V., Frangeul L., Blanc H., Jiggins F., Saleh M.-C. (2016). piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc. Natl. Acad. Sci. 113, E4218–E4227. doi: 10.1073/pnas.1607952113
Ran F. A., Hsu P. D., Lin C.-Y., Gootenberg J. S., Konermann S., Trevino A. E., et al. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389. doi: 10.1016/j.cell.2013.08.021
Reid W., Pilitt K., Alford R., Cervantes-Medina A., Yu H., Aluvihare C., et al. (2018). An anopheles stephensi promoter-trap: Augmenting genome annotation and functional genomics. G3 Genes Genomes Genet. 8, 3119–3130. doi: 10.1534/g3.118.200347
Richards S. (2019). Arthropod Genome Sequencing and Assembly Strategies. Methods Mol. Biol. 1858, 1–14. doi: 10.1007/978-1-4939-8775-7_1
Schnettler E., Donald C. L., Human S., Watson M., Siu R. W. C., McFarlane M., et al. (2013). Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol. 94, 1680–1689. doi: 10.1099/vir.0.053850-0
Shaw W. R., Catteruccia F. (2019). Vector biology meets disease control: using basic research to fight vector-borne diseases. Nat. Microbiol. 4, 20–34. doi: 10.1038/s41564-018-0214-7
Siomi M. C., Sato K., Pezic D., Aravin A. A. (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12, 246–258. doi: 10.1038/nrm3089
Spradling A. C., Rubin G. M. (1982). Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218, 341–347. doi: 10.1126/science.6289435
Stadler C., Rexhepaj E., Singan V. R., Murphy R. F., Pepperkok R., Uhlén M., et al. (2013). Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat. Methods 10, 315–323. doi: 10.1038/nmeth.2377
Suzuki Y., Frangeul L., Dickson L. B., Blanc H., Verdier Y., Vinh J., et al. (2017). Uncovering the Repertoire of Endogenous Flaviviral Elements in Aedes Mosquito Genomes. J. Virol. 91, e00571–e00517. doi: 10.1128/JVI.00571-17
Suzuki Y., Baidaliuk A., Miesen P., Van Rij R. P., Lambrechts L., Saleh M.-C. (2020). Non-retroviral Endogenous Viral Element Limits Cognate Virus Replication in Aedes aegypti Ovaries CFAV-EVE Efficient viral replication CFAV. Curr. Biol. 30, 3495–3506.e6. doi: 10.1016/j.cub.2020.06.057
Tassetto M., Kunitomi M., Whitfield Z. J., Dolan P. T., Sánchez-Vargas I., Garcia-Knight M., et al. (2019). Control of RNA viruses in mosquito cells through the acquisition of vDNA and endogenous viral elements. Elife 8, e41244. doi: 10.7554/eLife.41244
ter Horst A. M., Nigg J. C., Dekker F. M., Falk B. W. (2018). Endogenous Viral Elements Are Widespread in Arthropod Genomes and Commonly Give Rise to PIWI-Interacting RNAs. J. Virol. 93, e02124-18. doi: 10.1128/jvi.02124-18
Vagin V. V., Sigova A., Li C., Gvozdev V., Zamore P. D. (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Sci. (80-) 313, 320–325. doi: 10.1126/science.1129333
Varjak M., Donald C. L., Mottram T. J., Sreenu V. B., Merits A., Maringer K., et al. (2017). Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. PloS Negl. Trop. Dis. 10, e0006010. doi: 10.1371/journal.pntd.0006010
Vodovar N., Bronkhorst A. W., van Cleef K. W. R., Miesen P., Blanc H., van Rij R. P., et al. (2012). Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PloS One 7, e30861. doi: 10.1371/journal.pone.0030861
Waldron F. M., Stone G. N., Obbard D. J. (2018). Metagenomic sequencing suggests a diversity of RNA interference-like responses to viruses across multicellular eukaryotes. PloS Genet. 14, e1007533. doi: 10.1371/journal.pgen.1007533
Wang Y., Jin B., Liu P., Li J., Chen X., Gu J. (2018). piRNA Profiling of Dengue Virus Type 2-Infected Asian Tiger Mosquito and Midgut Tissues. Viruses 10, 213. doi: 10.3390/v10040213
Keywords: vector-borne disease, Piwi-interacting RNAs, mosquito basic biology, host-pathogen interactions, genetic engineering technologies
Citation: Macias VM, Palatini U, Bonizzoni M and Rasgon JL (2021) Leaning Into the Bite: The piRNA Pathway as an Exemplar for the Genetic Engineering Need in Mosquitoes. Front. Cell. Infect. Microbiol. 10:614342. doi: 10.3389/fcimb.2020.614342
Received: 05 October 2020; Accepted: 30 November 2020;
Published: 14 January 2021.
Edited by:
Saravanan Thangamani, Upstate Medical University, United StatesReviewed by:
Peiwen Liu, Southern Medical University, ChinaIgor V. Sharakhov, Virginia Tech, United States
Copyright © 2021 Macias, Palatini, Bonizzoni and Rasgon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vanessa M. Macias, dnptMTBAcHN1LmVkdQ==
 Vanessa M. Macias
Vanessa M. Macias Umberto Palatini
Umberto Palatini Mariangela Bonizzoni
Mariangela Bonizzoni Jason L. Rasgon
Jason L. Rasgon