- 1Heibei Key Laboratory of Chinese Medicine Research on Cardiocerebrovascular Disease, Hebei University of Chinese Medicine, Shijiazhuang, China
- 2Food Microbiology and Animal Quarantine Laboratory, Technology Center of Shijiazhuang Customs, Shijiazhuang, China
- 3School of Public Health, Key Laboratory of Environment and Human Health, Hebei Medical University, Shijiazhuang, China
Salmonella spp. is among the main foodborne pathogens which cause serious foodborne diseases. An isothermal real-time recombinase polymerase amplification (RPA) and lateral flow strip detection (LFS RPA) were used to detect Salmonella spp. targeting the conserved sequence of invasion protein A (invA). The Real-time RPA was performed in a portable florescence scanner at 39°C for 20 min. The LFS RPA was performed in an incubator block at 39°C for 15 min, under the same condition that the amplifications could be inspected by the naked eyes on the LFS within 5 min. The detection limit of Salmonella spp. DNA using real-time RPA was 1.1 × 101 fg, which was the same with real-time PCR but 10 times higher than that of LFS RPA assay. Moreover, the practicality of discovering Salmonella spp. was validated with artificially contaminated lamb, chicken, and broccoli samples. The analyzing time dropped from 60 min to proximately 5–12 min on the basis of the real-time and LFS RPA assays compared with the real-time PCR assay. Real-time and LFS RPA assays’ results were equally reliable. There was no cross-reactivity with other pathogens in both assays. In addition, the assays had good stability. All of these helped to show that the developed RPA assays were simple, rapid, sensitive, credible, and could be a potential point-of-need (PON) test required mere resources.
Introduction
Salmonella spp. is a Gram-negative bacterium belonging to the family of Enterobacteriaceae. Salmonella spp. is a major cause of the foodborne pathogen around the world (Nassib et al., 2003). It is widespread in nature and proliferates under ambient temperature with low nutritional demands (Li et al., 2013). Salmonella spp. infections attract much attention in public health especially in food safety. Salmonella spp. causes food poisoning, typhoid fever, gastrointestinal inflammation, and septicemia for both humans and animals (McGuinness et al., 2009; Rukambile et al., 2019). Currently, there are over 80 million cases of foodborne salmonellosis in the world (Li et al., 2019). Additionally, reports revealed that outbreaks caused by Salmonella spp. were largely associated with animal derived products such as poultry, egg, and chicken, and contamination is common in retail raw meats (Nassib et al., 2003; Foley and Lynne, 2008; Foley et al., 2008). An accurate and fast diagnosis is needed in order to prevent Salmonella spp. infections.
Salmonella spp. is currently detected in foods primarily through traditional laboratory methods. These traditional laboratory methods are inconvenient, time-consuming and takes over 3 days to obtain the result following multiple analytical steps (Li et al., 2019; Hong et al., 2020). Moreover, the complexity of the samples had a great effect on the bacterial morphology colony. The cross-reactivity among bacteria in Enterobacteriaceae restricted the specificity and sensitivity of the test (Nassib et al., 2003; Li et al., 2019). Upgrading molecular diagnostics provides powerful means for detecting Salmonella spp. in light of sensitivity and specificity. Currently, many nucleic acid amplification-based assays have gained popularity such as polymerase chain reaction (PCR), real-time PCR, multiplex PCR, reverse transcriptase PCR (RT-PCR), and loop-mediated isothermal amplification (LAMP) (Hirose et al., 2002; Yeh et al., 2002; Techathuvanan et al., 2010; Domesle et al., 2018). Real-time PCR is extensively applied for the quantitative detection of Salmonella spp. However, PCR requires sophisticated thermal cyclers with trained personnel which makes its use difficult in underequipped laboratories and low-resource field settings (Techathuvanan et al., 2011). PCR assays’ application is limited within the walls of the laboratories (Techathuvanan et al., 2011). In addition to the PCR assays, state-of-the-art isothermal amplification technologies such as LAMP, have been used for early and rapid detection of Salmonella spp. Simpler and more convenient techniques are required imperatively for the point-of-need (PON) diagnosis of Salmonella spp. in field conditions.
Recombinase polymerase amplification (RPA) acts as one of isothermal gene amplification techniques. RPA has the merit of amplification at a relatively low temperature (37–42°C) within 10–20 min (Piepenburg et al., 2006; Geng et al., 2018). The use of RPA-based methods has been proved to be a success in detecting pathogenic bacteria and viruses in clinical and food samples (Euler et al., 2012; Abd El Wahed et al., 2015; Wang et al., 2020). RPA-based methods have been designed to be a miniaturized diagnostic device that includes all the components for the RPA assay (Asiello and Baeumner, 2011). RPA assay was a rapid, stable, and promising assay for the on-site detection. The objective of the study was to develop the real-time and LFS RPA assays using the exo probe and nfo probe combined with lateral flow strip respectively as a way of rapidly detecting Salmonella spp. in food samples.
Materials and Methods
Bacterial Strains and DNA Extraction
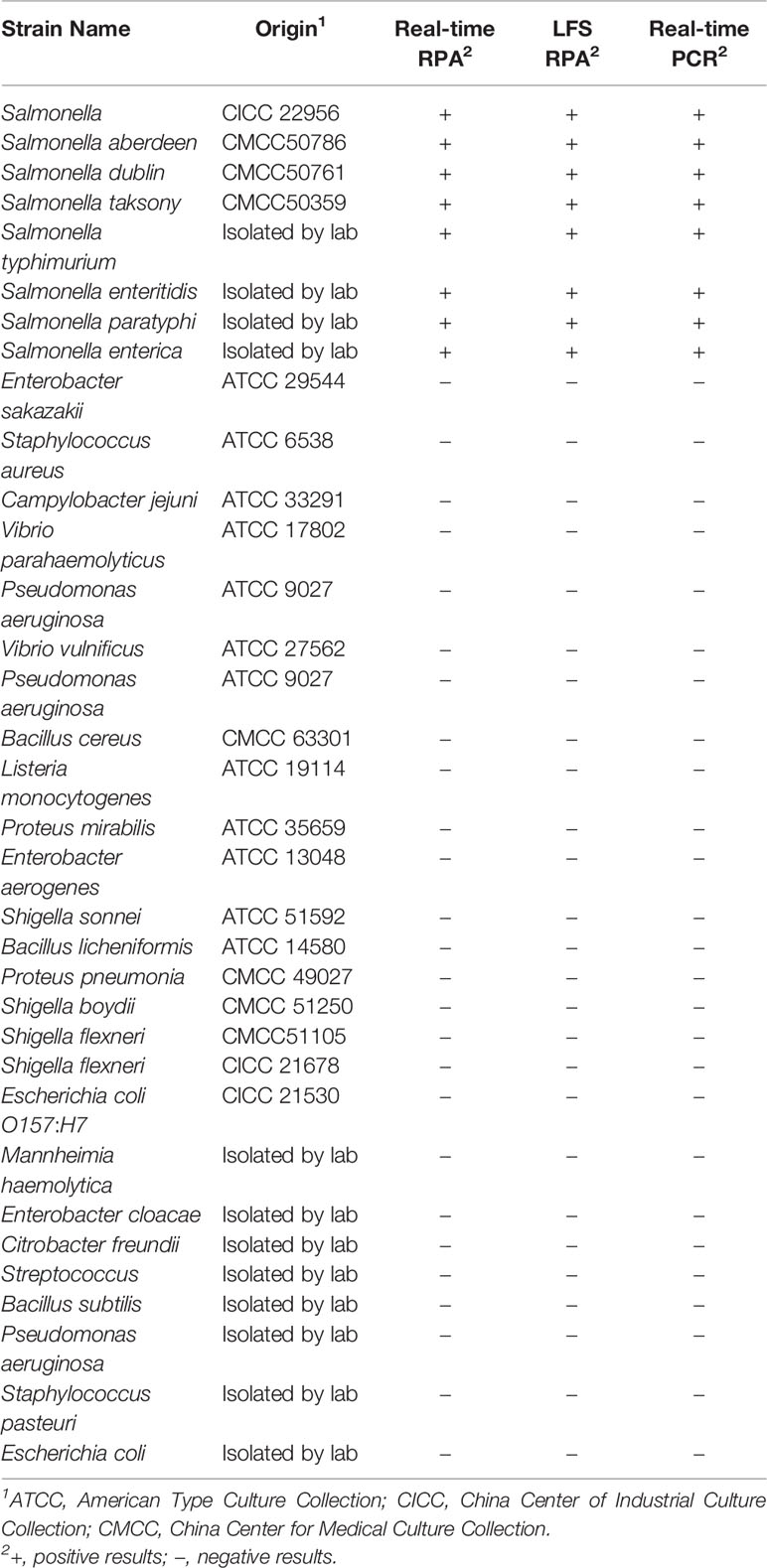
A total of 34 common pathogenic bacteria strains were used to validate the techniques employed in this study (Table 1). These pathogenic bacteria were purchased from the American Type Culture Collection (ATCC), China Center of Industrial Culture Collection (CICC), China Center for Medical Culture Collection (CMCC) or isolated in the lab. All strains were reserved in the lab. Stock cultures were stored at −80°C in 0.8 ml of Nutrient broth (Beijing Land Bridge Technology Co., Ltd., Beijing, China) and 0.2 ml of 80% glycerol. The DNA templates were extracted using the TIANamp Bacteria DNA Kit (Tiangen, Beijing, China). These DNA samples were stored at −20°C before the assays.
RPA Primers and Probe
Nucleotide sequence data for Salmonella spp. strains from GenBank were aligned to identify conserved regions. Based on the reference sequences of different Salmonella spp. genotypes (accession numbers: AY594273, AY594271, DQ644632, DQ644633, EU348367, EU348368, JF951188, and JN982040), three pairs of primers targeting the conserved region of invA were designed (Rahn et al., 1992; Gonzalez-Escalona et al., 2009). RPA, Real-time RPA primers, and probes were then selected through testing the combination that yielded the highest sensitivity (Table 2). Primers and exo probes were synthesized by Sangon (Sangon, Shanghai, China).
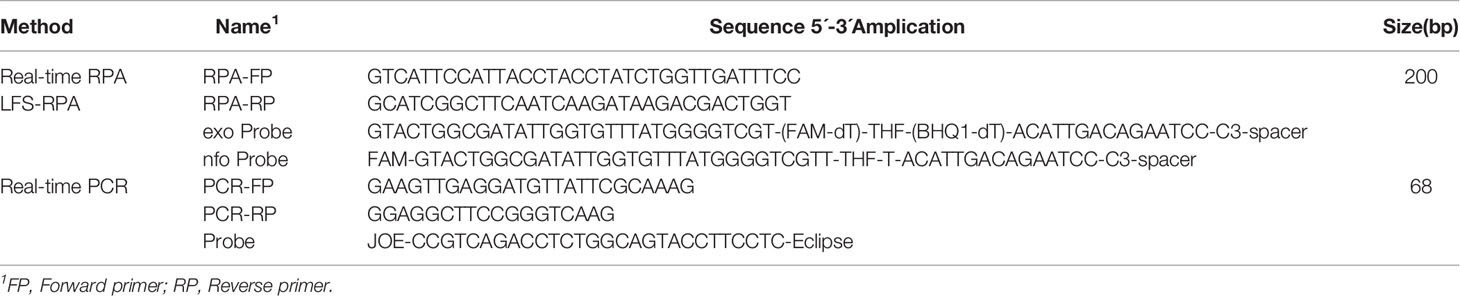
Table 2 Primer and probe sequences for Salmonella spp. Real-time PCR, RPA, real-time RPA and LFS RPPA assays.
Real-Time RPA Assay
Real-time RPA was accomplished in the tube with 50 μl reaction volume, including 40.9 μl of Buffer A (rehydration buffer), 2.0 μl of each RPA primers (Sa-exo-F and Sa-exo-R, 10 μmol/L), 0.6 μl of exo probe (Sa-exo-P,10 μmol/L), and 2.5 μl of Buffer B (magnesium acetate, 280 mmol/L). Furthermore, 1 μl of genomic DNA was used for the specificity and sensitivity analysis, or 2 μl of sample DNA was used for the clinical sample diagnosis. In the process, the Genie III scanner device (OptiGene Limited, West Sussex, UK) and TwistAmpTM exo kit (TwistDX, Cambridge, UK) were applied.
LFS RPA Assay
Moreover, the LFS RPA assay was performed according to the given instructions. The commercial TwistAmp™ nfo kit (TwistDX, Cambridge, UK) was used in the LFS RPA. The reactions were performed in a 50 μl volume with 29.5 μl of rehydration buffer with 2.5 μl of magnesium acetate (280 mM) included. Other components contained 420 nM RPA primer, 120 nM exo probe, and 1 μl of bacterial genomic DNA or 5 μl of sample DNA. The assay was performed in an incubator block at 39°C for 15 min and the lateral flow strips (Ustra Biotec GmbH, Germany) were employed to discover the RPA amplicons dual-labeled with FAM and biotin. Testing samples were considered positive when both the test line and the control line were visible. The testing sample was considered negative when the control line was visible. However, the sample was considered invalid when the control line was invisible.
Real-Time PCR
Real-time PCR was performed on the ABI 7500 instrument in which premix Ex Taq TM (Takara Co., Ltd., Dalian, China) was employed (Geng et al., 2019). The reaction was performed as follows: 95°C for 30 s, followed by 35 cycles of 95°C for 10 s and then 60°C for 34 s. The sequences of the primers and probes used for real-time PCR were listed in Table 2. The reporter and fluorescence quencher were marked with 6-FAM (6-CarboxyFluorescein) and BHQ1 (Black Hole Quencher 1) respectively.
Analytical Specificity and Analytical Sensitivity Analysis
For the food security, the real-time RPA and LFS RPA assays were evaluated to amplify the nucleic acid of some important pathogens. Five independent reactions were performed.
The genomic DNA of Salmonella spp. varying from 1.1 × 108 to 1.1 × 100 fg was diluted in nuclease-free water for the analytical sensitivity analysis of the RPA. One microliter of each DNA dilution was amplified by both RPA assays to determine the limit of detection (LOD). The culture of Salmonella spp. was diluted in sterile water (ranging from 1.4 × 107 to 1.4 × 100 CFU) and counted by plate counting in 37°C overnight. The sensitivity of the real-time RPA and LFS RPA method were assessed with Salmonella spp. in pure culture. The analytical sensitivity analysis was repeated for five times.
Validation With Artificially Contaminated Samples
The pure colony of Salmonella spp. was picked into a tube containing 1 ml sterile saline. The solution was vortexed for 30 s and the turbidity was measured to 1.00 using a turbidimeter. Sterile saline was used for 10-fold gradient dilution until it attained 10−8 dilution. With 10−5, 10−6, and 10−7 diluents of 200 μl on the chromogenic medium of Salmonella spp., the initial concentration of the pure culture bacteria was calculated using three parallels.
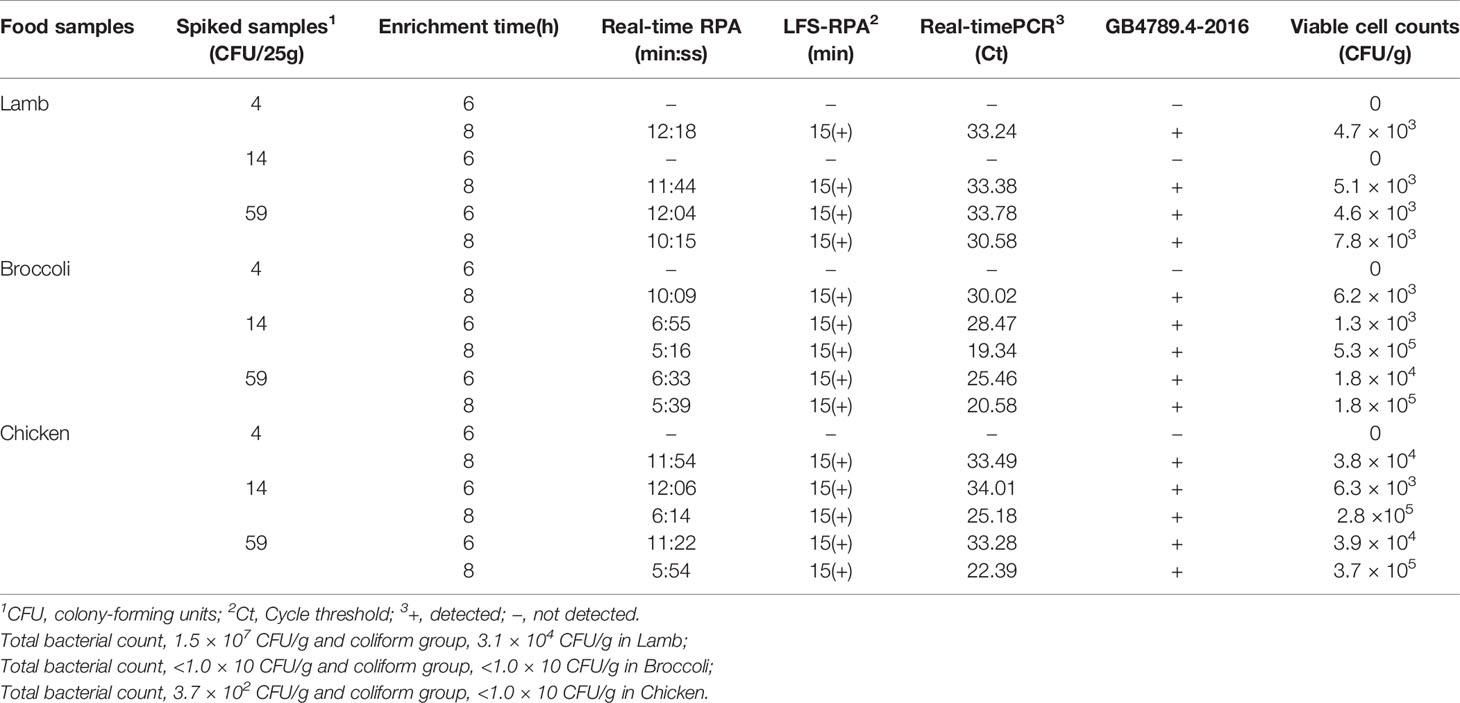
Commercially available chicken/lamb/broccoli were purchased from a local supermarket free of Salmonella spp. to assess the potential use and suitability of the developed RPA assays. Testing of the samples was done according to the Chinese national standard (GB 4789.4-2016). A total of 4, 14, and 59 CFU/25g of Salmonella spp. with chicken, lamb, and broccoli respectively, were added into a sterile stomaching bag containing 225 ml Nutrient broth. These samples were mixed well to get homogenous samples and incubated for 6 or 8 h at 37°C to increase the bacterial concentrations to attain detectable levels. The Bacterial genomic DNA extraction, the real-time RPA, and LFS RPA reactions were performed, and each experiment was repeated for no less than three times to attain results.
Results
Analytical Specificity and Sensitivity of the Real-Time RPA and LFS-RPA Assay
The invA gene coding of the invasion protein of Salmonella spp. is the most used specific gene for the discovery of many different Salmonella spp. serotypes. The RPA primers and probes were designed according to the invA gene of Salmonella spp. in this study. Both RPA assays provide excellent results at 39°C within 20 min. This was faster than any other common nucleic acid amplification method. The analytical sensitivity of RPA methods was evaluated by employing Salmonella spp. genomic DNA and bacterial pure culture as templates from 1.1 × 108 to 1.1 × 100 fg and from 1.4 × 107 to 1.4 × 100 CFU. The data on the analytical sensitivity of RPA methods was presented in Figures 1A, 2A. The detection limit (LOD) of real-time RPA was 1.1 × 101 fg similar to that of real-time PCR (the data was not shown) (Figure 1A). The limit of detection of the LFS RPA method was 1.1 × 102 fg for genomic DNA (Figure 2A), 10-times lower compared with the real-time PCR. The LOD of the real-time RPA and LFS RPA was 1.4 × 102 CFU for bacteria in pure culture (shown in Figures 1B, 2B).
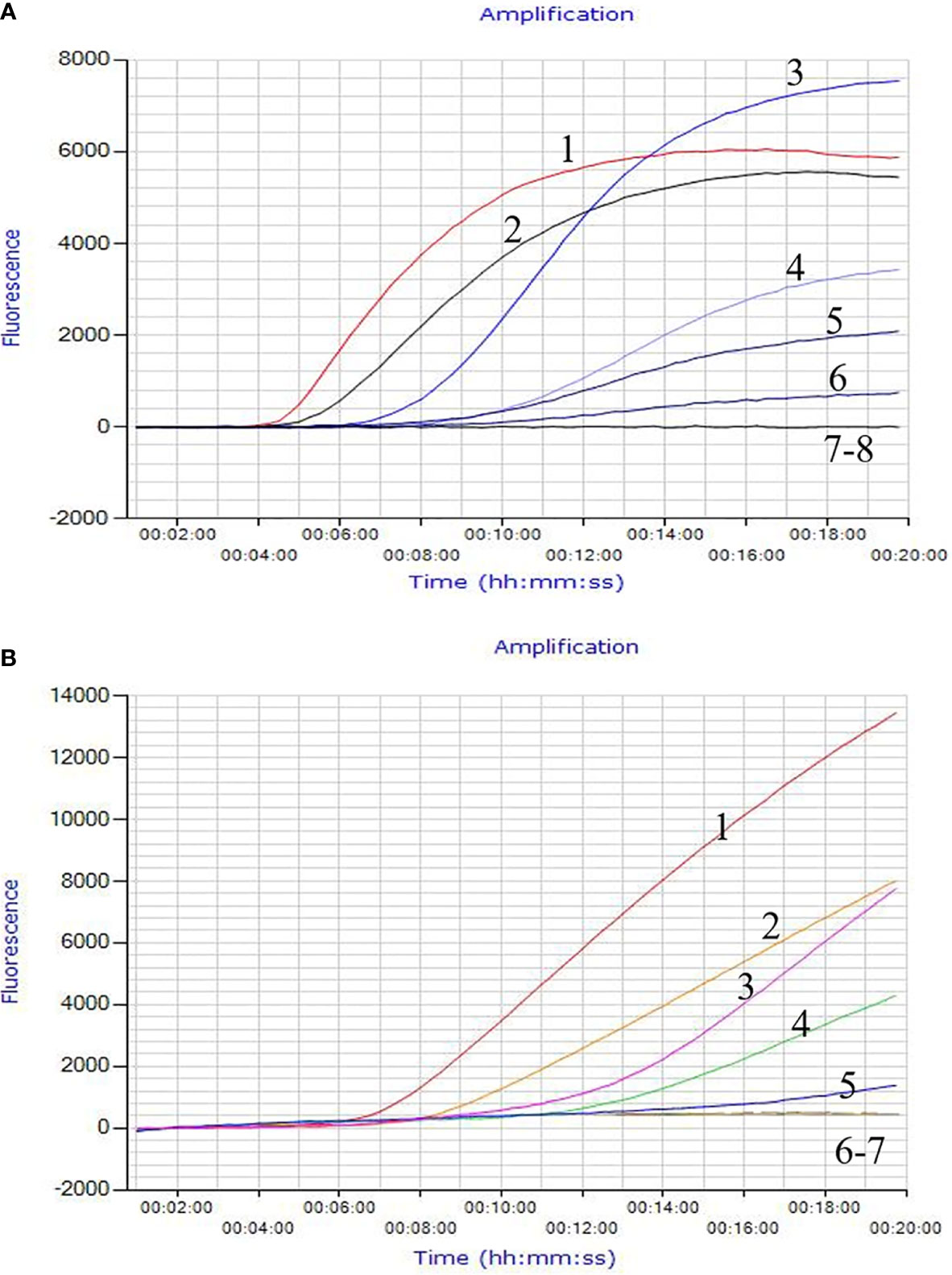
Figure 1 Analytical Sensitivity of the real-time RPA assay. The LOD of the real-time RPA was 1.1 × 101 fg/μl of Salmonella spp. standard DNA and 1.4 × 102 CFU/ml for bacteria in pure culture. Fluorescence development over time using a dilution range of 1.1 × 106–1.1 × 100 fg of Salmonella spp. genomic DNA. For (A): Curve 1, 1.1 × 106 fg; Curve 2, 1.1 × 105 fg; Curve 3, 1.1 × 104 fg; Curve 4, 1.0 × 103 fg; Curve 5, 1.1 × 102 fg; Curve 6, 1.1 × 101 fg; Curve 7, 1.1 × 100 fg; Curve 8, ddH2O. For (B): Curve 1, 1.4 × 106 CFU; Curve 2, 1.4 × 105 CFU; Curve 3, 1.4 × 104 CFU; Curve 4, 1.4 × 103CFU; Curve 5, 1.4 × 102 CFU; Curve 6, 1.4 × 101CFU; Curve 7, 1.4 × 100 CFU.
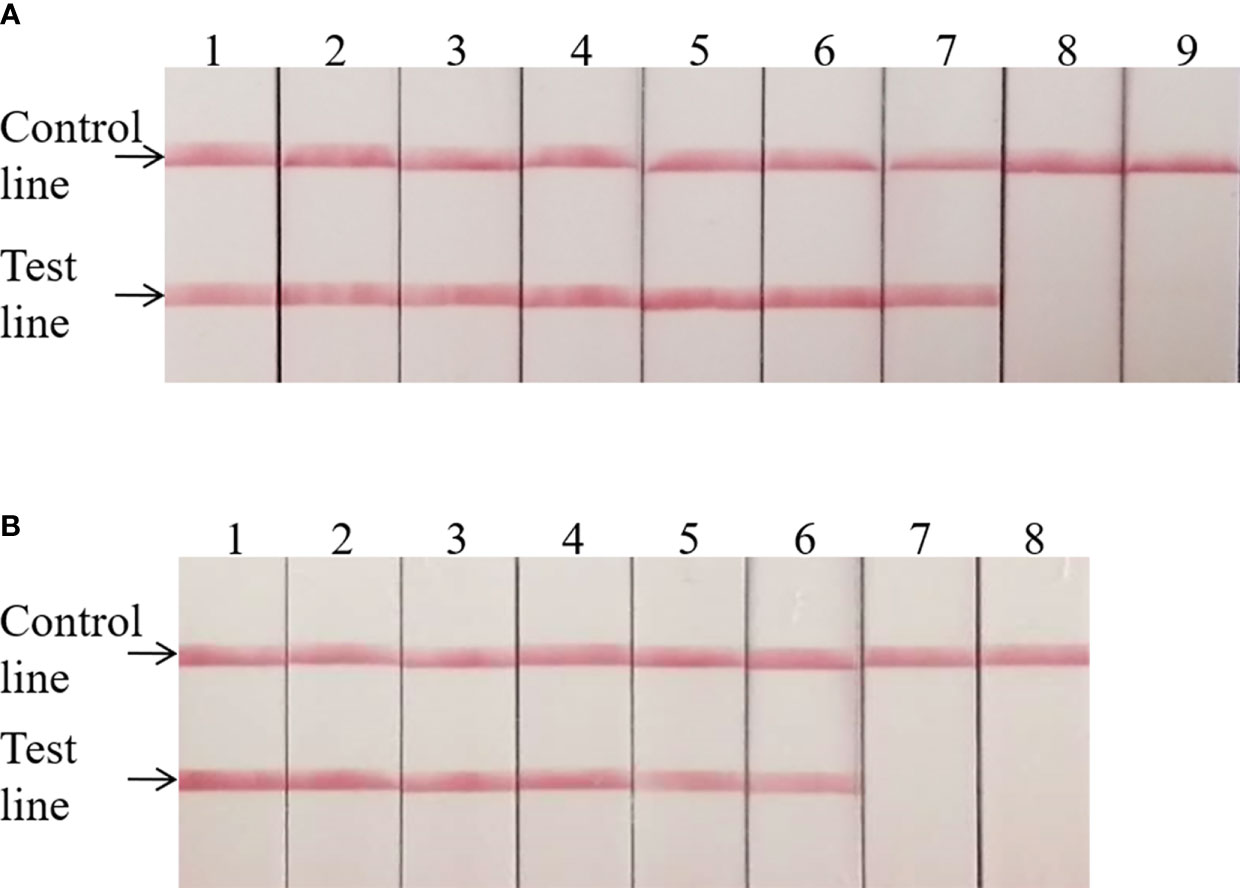
Figure 2 Analytical sensitivity of the LFS RPA assay. The LOD of the LFS RPA method was 1.1 × 102 fg for genomic DNA and 1.4 × 102 CFU/ml for bacteria in pure culture. For (A): Sample 1, 1.1 × 108 fg; Sample 2, 1.1 × 107fg; Sample 3, 1.1 × 106 fg; Sample 4, 1.1 × 105 fg; Sample 5, 1.1 × 104 fg; Sample 6, 1.1 × 103 fg; Sample 7, 1.1 × 102 fg; Sample 8, 1.1 × 101 fg; Sample 9, 1.1 × 100 fg. For (B): Sample 1, 1.4 × 107 CFU; Sample 2, 1.4 × 106 CFU; Sample 3, 1.4 × 105 CFU; Sample 4, 1.4 × 104 CFU; Sample 5, 1.4 × 103 CFU; Sample 6, 1.4 × 102 CFU; Sample 7, 1.4 × 101 CFU; Sample 8, 1.4 × 100 CFU.
Regarding specificity, only amplification signal was observed at the control line with Salmonella spp. and no cross-detection of other pathogens were shown in both real-time RPA and LFS-RPA assays (Table 1). Five independent reactions were repeated and similar results were obtained. This manifests the high specificity of the assays.
Evaluation With Artificially Contaminated Samples After Enrichment
The diagnostic performance of the developed RPA assays was compared with other detection approaches and was shown in Table 3. This was done while detecting Salmonella spp. in artificially contaminated food samples. The National Standard GB 4789.4-2016 method was used as a reference to guarantee that the food samples were successfully contaminated. However, after 6-h enrichment of food samples contaminated with 4 CFU/25 g of Salmonella spp., no Salmonella spp. was discovered through any of the detection methods. This signaled no false-positive results from samples containing low levels of Salmonella spp. All contaminated food samples were detected and showed increasing values of CFU/25 g of spiked samples and enrichment time. However, lamb contaminated with 14 CFU/25 g of Salmonella spp. was different from the other contaminated food samples and was enriched for 6 h. Furthermore, a diagnostic agreement of 100% with real-time PCR and the traditional method was indicated in the developed real-time RPA and LFS RPA assays. Moreover, the speed of the RPA assays outstripped. The time required to attain positive results with real-time RPA and LFS RPA was 5–12 min and 15 min respectively. Real-time PCR with CT values at 19.34–34.01 required approximately 20–35 min. Therefore, the results demonstrated that, with the equal sensitivity, the real-time and LFS RPA assays was faster than the real-time PCR.
Discussion
The disease induced by foodborne pathogens remains a major public health issue worldwide despite ongoing measurements to ensuring food safety. Salmonella spp. frequently leads to infections and worldwide outbreaks accounting for huge economic costs and life loss every year (Foley and Lynne, 2008). Rapid and reliable diagnostic techniques play an important part to efficiently detect Salmonella spp. from contaminated specimens and make appropriate measures for preventing and controlling the risk of Salmonella spp. infection as early as possible.
The real-time RPA and LFS RPA assays are good choices for detecting Salmonella spp. as demonstrated in this report. These assays are specific, sensitive, and simple to perform. In the specificity analysis, both the real-time RPA and LFS RPA only amplified the genomic DNA of Salmonella spp. used in the study. This indicated high specificity of these assays. However, other more Salmonella strains are needed to further examine the cross-reactivity of these RPA assays. The real-time RPA had equal sensitivity (limit of detection) as real-time PCR in this study. This was 10 times higher than the LFS assay. However, it is possible for a varied reaction mechanism and enzyme kinetics between the different methods. The reaction time of RPA assays was much shorter than real-time PCR. The diagnostic performances of the developed real-time RPA and LFS RPA assays has been further assessed. These assays were proved to be a success in the detection of the artificially contaminated food samples, and performed better than the real-time PCR in light of the detecting speed. However, a pre-enrichment step was necessary when the level of pathogen contamination was low. A similar ideal result was obtained using the direct water boiling method to extract the bacterial DNA as the template of RPA reaction. Direct boiling method was used to extract the Salmonella spp. genomic DNA as the template of the RPA reaction. The LFS strip were combined to facilitate the detection of Salmonella spp. at quarantine stations, ports, or the site of outbreak by the PRA assay based on nfo-probe.
RPA was first introduced in 2006 and represented an innovative DNA isothermal detecting technology beyond the reach of PCR or traditional culture-based methods (Piepenburg et al., 2006; James and Macdonald, 2015). RPAs have successfully been practiced in the discovery of pathogenic bacteria (Hong et al., 2020), fungus (Ahmed et al., 2014), and viruses (Boyle et al., 2013). The reagents in RPA are available in lyophilized form for long-term storage and are conveniently transported even without cold-chain (Wang et al., 2020). Moreover, under the prerequisite that the testing results were visible, the real-time RPA assay was accomplished on the user-friendly PON (point of need) detection platform (Genie III) with battery power. The developed LFS RPA assay only needed a simple incubator block. Therefore, these two pieces of equipment were portable, lightweight, and less expensive than the equipment for LAMP/PCR. As other isothermal DNA amplification methods, Loop-mediated isothermal amplification (LAMP) and the cross-priming amplification assay (CPA) have been adopted for rapidly and sensitively detecting Salmonella spp. Both RPA assays have the merits of amplification at a relatively lower temperature and within shorter time than that of LAMP and CPA assays. Both RPA reactions could be done at 37–42°C within 10–20 min. However, the optimum time and temperature were approximately 60 min and above 60°C respectively which were required for LAMP and CPA (Domesle et al., 2018; Wang et al., 2018). Moreover, several reports have shown that RPA is tolerant to mismatches, background DNA, and most of PCR inhibitors (Daher et al., 2016; Li et al., 2019; Liu et al., 2019). All of these outstanding characteristics make the assays readily suitable for the field, PON (point-of-need), or diagnosis of infectious diseases with poor environmental resources.
In conclusion, the current study proved that, the developed RPA assays with high specificity and sensitivity was convenient, rapid, and reliable for Salmonella spp. detection. In addition, the simple devices and easy operation protocol helped to improve the efficiency of detection. Among the isothermal amplification techniques, real-time RPA and LFS RPA assays play an outstanding role in preventing and controlling of Salmonella spp. especially in the settings with limited resources.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
YG, JCW, and WG designed and conducted the experiment. LZ, XS, JFW, XX, MD, Y-nG, YW, and PC performed the experiments and analyzed the data. YG drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the program of Traditional Chinese Medicine Scientific Research foundation in Hebei Administration of Traditional Chinese Medicine (2020142, Hebei, China), the Project of Excellent Young Teacher Fundamental Research (YQ2019003) and Doctoral Foundation (BSZ2019009) of Hebei University of Chinese Medicine.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abd El Wahed A., Weidmann M., Hufert F. T. (2015). Diagnostics-in-a-Suitcase: Development of a portable and rapid assay for the detection of the emerging avian influenza A (H7N9) virus. J. Clin. Virol. 69, 16–21. doi: 10.1016/j.jcv.2015.05.004
Ahmed A., van der Linden H., Hartskeerl R. A. (2014). Development of a recombinase polymerase amplification assay for the detection of pathogenic Leptospira. Int. J. Environ. Res. Public Health 11, 4953–4964. doi: 10.3390/ijerph110504953
Asiello P. J., Baeumner A. J. (2011). Miniaturized isothermal nucleic acid amplification, a review. Lab. Chip. 11, 1420–1430. doi: 10.1039/c0lc00666a
Boyle D. S., Lehman D. A., Lillis L., Peterson D., Singhal M., Armes N., et al. (2013). Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. mBio. 4, e00135-13. doi: 10.1128/mBio.00135-13
Daher R. K., Stewart G., Boissinot M., Bergeron M. G. (2016). Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 62, 947–958. doi: 10.1373/clinchem.2015.245829
Domesle K. J., Yang Q., Hammack T. S., Ge B. (2018). Validation of a Salmonella loop-mediated isothermal amplification assay in animal food. Int. J. Food Microbiol. 264, 63–76. doi: 10.1016/j.ijfoodmicro.2017.10.020
Euler M., Wang Y., Otto P., Tomaso H., Escudero R., Anda P., et al. (2012). Recombinase polymerase amplification assay for rapid detection of Francisella tularensis. J. Clin. Microbiol. 50, 2234–2238. doi: 10.1128/JCM.06504-11
Foley S. L., Lynne A. M. (2008). Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86, E173–E187. doi: 10.2527/jas.2007-0447
Foley S. L., Lynne A. M., Nayak R. (2008). Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 86, E149–E162. doi: 10.2527/jas.2007-0464
Geng Y., Liu S., Wang J., Nan H., Liu L., Sun X., et al. (2018). Rapid Detection of Staphylococcus aureus in Food Using a Recombinase Polymerase Amplification-Based Assay. Food Anal. Methods 11, 2847–2856. doi: 10.1016/j.mimet.2018.12.017
Geng Y., Liu G., Liu L., Deng Q., Zhao L., Sun X. X., et al. (2019). Real-time recombinase polymerase amplification assay for the rapid and sensitive detection of Campylobacter jejuni in food samples. J. Microbiol. Methods 157, 31–36. doi: 10.1007/s12161-018-1267-1
Gonzalez-Escalona N., Hammack T. S., Russell M., Jacobson A. P., De Jesus A. J., Brown E. W., et al. (2009). Detection of live Salmonella sp. cells in produce by a TaqMan-based quantitative reverse transcriptase real-time PCR targeting invA mRNA. Appl. Environ. Microbiol. 75, 3714–3720. doi: 10.1128/AEM.02686-08
Hirose K., Itoh K., Nakajima H., Kurazono T., Yamaguchi M., Moriya K., et al. (2002). Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars Typhi and Paratyphi A. J. Clin. Microbiol. 40, 633–636. doi: 10.1128/jcm.40.02.633-636.2002
Hong H., Sun C., Wei S., Sun X., Mutukumira A., Wu X. (2020). Development of a real-time recombinase polymerase amplification assay for rapid detection of Salmonella in powdered infant formula. Int. Dairy J. 102. doi: 10.1016/j.idairyj.2019.104579
James A., Macdonald J. (2015). Recombinase polymerase amplification: Emergence as a critical molecular technology for rapid, low-resource diagnostics. Expert Rev. Mol. Diagn. 15, 1475–1489. doi: 10.1586/14737159.2015.1090877
Li R., Lai J., Wang Y., Liu S., Li Y., Liu K., et al. (2013). Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int. J. Food Microbiol. 163, 14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020
Li J., Ma B., Fang J., Zhi A., Chen E., Xu Y., et al. (2019). Recombinase Polymerase Amplification (RPA) Combined with Lateral Flow Immunoassay for Rapid Detection of Salmonella in Food. Foods. 9, 27. doi: 10.3390/foods9010027
Liu L., Li R., Zhang R., Wang J., An Q., Han Q., et al. (2019). Rapid and sensitive detection of Mycoplasma hyopneumoniae by recombinase polymerase amplification assay. J. Microbiol. Methods 159, 56–61. doi: 10.1016/j.mimet.2019.02.015
McGuinness S., McCabe E., O’Regan E., Dolan A., Duffy G., Burgess C., et al. (2009). Development and validation of a rapid real-time PCR based method for the specific detection of Salmonella on fresh meat. Meat Sci. 83, 555–562. doi: 10.1016/j.meatsci.2009.07.004
Nassib T. A., El-Din M. Z., El-Sharoud W. M. (2003). Assessment of the presence of Salmonella spp. in Egyptian dairy products using various detection media. Lett. Appl. Microbiol. 37, 405–409. doi: 10.1046/j.1472-765X.2003.01420.x
Piepenburg O., Williams C. H., Stemple D. L., Armes N. A. (2006). DNA detection using recombination proteins. PLoS Biol. 4, e204. doi: 10.1371/journal.pbio.0040204
Rahn K., De Grandis S. A., Clarke R. C., McEwen S. A., Galan J. E., Ginocchio C., et al. (1992). Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell Probes. 6, 271–279. doi: 10.1016/0890-8508(92)90002-f
Rukambile E., Sintchenko V., Muscatello G., Kock R., Alders R. (2019). Infection, colonization and shedding of Campylobacter and Salmonella in animals and their contribution to human disease: A review. Zoonoses Public Health 66, 562–578. doi: 10.1111/zph.12611
Techathuvanan C., Draughon F. A., D’Souza D. H. (2010). Real-time reverse transcriptase PCR for the rapid and sensitive detection of Salmonella typhimurium from pork. J. Food Prot. 73, 507–514. doi: 10.4315/0362-028x-73.3.507
Techathuvanan C., Draughon F. A., D’Souza D. H. (2011). Comparison of reverse transcriptase PCR, reverse transcriptase loop-mediated isothermal amplification, and culture-based assays for Salmonella detection from pork processing environments. J. Food Prot. 74, 294–301. doi: 10.4315/0362-028X.JFP-10-306
Wang Y. X., Zhang A. Y., Yang Y. Q., Lei C. W., Cheng G. Y., Zou W. C., et al. (2018). Sensitive and rapid detection of Salmonella enterica serovar Indiana by cross-priming amplification. J. Microbiol. Methods 153, 24–30. doi: 10.1016/j.mimet.2018.08.003
Wang J., Li R., Sun X., Liu L., Hao X., Wang J., et al. (2020). Development and validation of the isothermal recombinase polymerase amplification assays for rapid detection of Mycoplasma ovipneumoniae in sheep. BMC Vet. Res. 16, 172. doi: 10.1186/s12917-020-02387-3
Keywords: Salmonella, invA gene, real-time RPA, lateral flow strip (LFS), isothermal amplification
Citation: Zhao L, Wang J, Sun XX, Wang J, Chen Z, Xu X, Dong M, Guo Y-n, Wang Y, Chen P, Gao W and Geng Y (2021) Development and Evaluation of the Rapid and Sensitive RPA Assays for Specific Detection of Salmonella spp. in Food Samples. Front. Cell. Infect. Microbiol. 11:631921. doi: 10.3389/fcimb.2021.631921
Received: 21 November 2020; Accepted: 19 January 2021;
Published: 25 February 2021.
Edited by:
Jianmin Zhang, South China Agricultural University, ChinaReviewed by:
Jun Tang, China Agricultural University, ChinaQianru Yang, United States Food and Drug Administration, United States
Baowei Yang, Northwest A and F University, China
Copyright © 2021 Zhao, Wang, Sun, Wang, Chen, Xu, Dong, Guo, Wang, Chen, Gao and Geng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijuan Gao, NTAyNjc4NTYwQHFxLmNvbQ==; Yunyun Geng, Z2VuZ3l1bnl1bjIwMTVAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Liwei Zhao1†
Liwei Zhao1† Weijuan Gao
Weijuan Gao Yunyun Geng
Yunyun Geng