- 1Department of Medical Parasitology and Mycology, School of Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
- 2Department of Medical Parasitology and Mycology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
- 3Infectious Diseases and Tropical Medicine Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
- 4Department of Otolaryngology, Isfahan University of Medical Sciences, Isfahan, Iran
- 5Department of Environmental Health Engineering, School of Health, Isfahan University of Medical Sciences, Isfahan, Iran
- 6Core Facilities Laboratory (CFL), Mycology Reference Laboratory, Isfahan University of Medical Sciences, Isfahan, Iran
Background: Considering the importance of differential diagnosis of infectious otitis externa (OE), a stepwise PCR-based assay using universal and genus- or species-specific primers for the detection/identification of the most prevalent bacterial and fungal OE was developed and evaluated on the ear aspiration specimens of clinically suspected patients.
Methods and Materials: A total of 120 ear aspiration specimens with otomycosis suspicion were subjected to manual DNA extraction using phenol–chloroform extraction after tissue digestion with a lysis buffer. The multiplex PCR was initially performed using pan-fungal and bacterial homemade primers. Pseudomonas and Staphylococcus specific primers were simultaneously used in one reaction mixture to identify the bacterial genera. Furthermore, for the identification of fungal agents, Candida species-specific multiplex primers targeting the most clinically important Candida species causing OE (i.e., C. albicans, C. parapsilosis, and C. auris), as well as Aspergillus related multiplex PCR identifying the most prevalent Aspergillus species were used in two separate reaction mixtures. All the results of multiplex PCR were interpreted based on the amplicon size.
Results: The overall multiplex PCR-based detection rate of bacterial (n = 88; 73.3%) and fungal (n = 97; 81%) OE was documented to be 100% along with and complete consistency with the results of direct examination and Giemsa staining. Double amplicon bands of bacterial and fungal pathogens were evidenced in 76 specimens (63.3%). Moreover, the positivity rate of pan-fungal PCR was higher than that of the culture result. Out of 88 pan-bacterial positive PCR specimens, 66 and 47 ones were positive for Staphylococcus and Pseudomonas, respectively. In addition, 30 samples exhibited mixed infection of both, and five specimens remained negative. Out of 97 pan-fungal positive PCR specimens, 67 and 51 ones contained Candida and Aspergillus species, respectively. It should be noted that dual amplicon bands of Candida and Aspergillus-related multiplex PCR were yielded in 30 specimens.
Conclusion: The stepwise multiplex PCR assay proved to be more sensitive, more rapid, as well as less cumbersome in detection and identification of fungal and bacterial OE, compared to culture.
Introduction
Otitis externa (OE) describes a group of acute or chronic inflammatory disorders of the external auditory canal and auricle following disruption of the protective squamous epithelial layer of the ear canal (Thorne and Wetmore, 2009). The inflammation may have arisen from an allergic (reactive) or infective (bacteria, fungi, and viruses) origin (Salyer, 2007; Adegbiji et al., 2017). Considering an annual incidence rate of 1.2% (Rowlands et al., 2001), OE is a common infectious condition for which patients attend Ear, Nose, and Throat clinics to undergo medication therapy. Different types of risk factors can predispose individuals to develop OE, including water trapping in the ear canal due to swimming, humidity, or sweating; cerumen, and epithelial disintegration as a result of increase in pH; cerumen removal; hearing aids and instrumentation or itching; canal obstruction because of foreign body or sebaceous cyst; anatomic abnormalities; dermatologic conditions, such as eczema, psoriasis, or seborrhea; prolonged antibiotic usage; and diabetes status (Schaefer and Baugh, 2012). Pseudomonas aeruginosa and Staphylococcus aureus are frequently listed as the most predominant agents causing an acute form of OE (Schaefer and Baugh, 2012). On the other hand, fungal agents, most commonly Aspergillus and Candida species, are associated with prolonged antibiotic usage and chronic OE (Sarwestani et al., 2018; Aboutalebian et al., 2019; Punia et al., 2019). Complaints of acute onset of unilateral ear pain, itching, and a sense of aural fullness are the typical manifestations of acute otitis externa caused by bacteria. However, in fungal ear infections (otomycosis), pruritus with or without associated otorrhea are frequently presented, the symptoms of which are more chronic than bacterial infections; moreover, pain is usually not a prominent sign (Thorne and Wetmore, 2009). Given the different management for diverse microbial agents causing ear infections, precise detection/identification of causative agents is a matter of great importance for accurate prognosis prediction and optimizing effective therapy (Schaefer and Baugh, 2012). Although the diagnosis of ear infection is routinely established through a clinical examination and history review (Thorne and Wetmore, 2009), the correct diagnosis is hampered in case of uncooperative patients (i.e., children), unspecific clinical diagnosis, need for targeted therapy, refractory OE, co-infection, and inexperienced clinicians (Chonmaitree and Henrickson, 2000; Blomgren and Pitkäranta, 2005). Therefore, laboratory-based methods for reliable diagnosis are required in these circumstances. Direct examination and culture are still the cornerstones of bacterial and fungal diagnosis. Nevertheless, direct or histopathological examinations take a longer time to establish the diagnosis and face difficulties in accurate identification of the infecting agent and may need personal expertise. In the former study, it has been evidenced that the culture positivity rate of the agents causing fungal infections is as low as 50%, especially when Malassezia species or Mucorales are highly suspected (Roden et al., 2005). In many cases, fungi fail to grow due to pretreatment, insufficient volume of specimens, or lack of specific culture medium (Zhao et al., 2011; Zaman et al., 2017). The development of rapid molecular-based techniques, mainly polymerase chain reaction (PCR) assays, has provided and improved the possibility for the detection of small numbers of microbial genomes in fresh specimens, particularly for tissue with negative culture or slow-growing pathogens (Jalali et al., 2008; Gruber et al., 2015). Therefore, to overcome the low isolation rate of culture and difficulties in the identification of causative agents by clinical evaluation and direct examination, PCR-based methods have been recommended in different studies (Bialek et al., 2005; Rickerts et al., 2007; Hammond et al., 2011). Although sequencing of common barcoding regions has been proved to be the most reliable pathogen identification tool in developed countries that is used routinely in laboratories, its provision is not easy to afford in developing and low resources countries. Moreover, serially performing a separate PCR assay as routine use for the detection of individual pathogens is labor-intensive, time-consuming, and expensive, particularly if a various panel of potential causative agents should be tested. Although several infectious diseases have been differentially diagnosed by multiplex PCR, differential diagnosis of agents causing OE has not yet been performed via multiplex PCR directly from clinical specimens. Therefore, this study aimed to evaluate the usefulness of novel homemade multiplex PCR capable of simultaneously detecting and identifying the most commonly isolated species of bacteria and fungi directly in the ear aspiration specimens obtained from patients suspected of OE. Moreover, it was attempted to compare the results with those of the direct microscopic examination and tissue culture techniques.
Materials and Methods
Patient and Isolates
A total of 120 patients clinically suspected of OE attending the otorhinolaryngology clinic of Al-Zahra Teaching Hospital, Isfahan, Iran, whose ear aspirations or ear swab specimens documented infective fungal and/or bacterial elements on direct microscopic examination by 10% Potassium Hydroxide and Giemsa mounts, were included in this study. For the identification of the yeasts isolated on CHROM agar Candida medium, the internal transcribed spacer (ITS) PCR-RFLP using the restriction enzymes MspI was performed as described in our previous report (Aboutalebian et al., 2019). Aspergillus/Penicillium species which grew on the medium Sabouraud’s dextrose agar supplemented with 50 mg L–1 chloramphenicol, were identified by partial β-tubulin gene sequencing. Unidentified yeasts or molds (other than Aspergillus/Penicillium) were subjected to the ITS sequencing (Aboutalebian et al., 2019). Furthermore, for the identification of bacterial agents, neither culture-based methods nor molecular procedures were applied; therefore, they were blindly investigated in the present study. Written informed consent and detailed clinical data of the patient have been presented in our former report (Aboutalebian et al., 2019). The study protocol was approved by the Research Ethics Committee of Iran National Institute for Medical Research Development (IR.NIMAD.REC.1398.063).
Multiplex PCR Designing
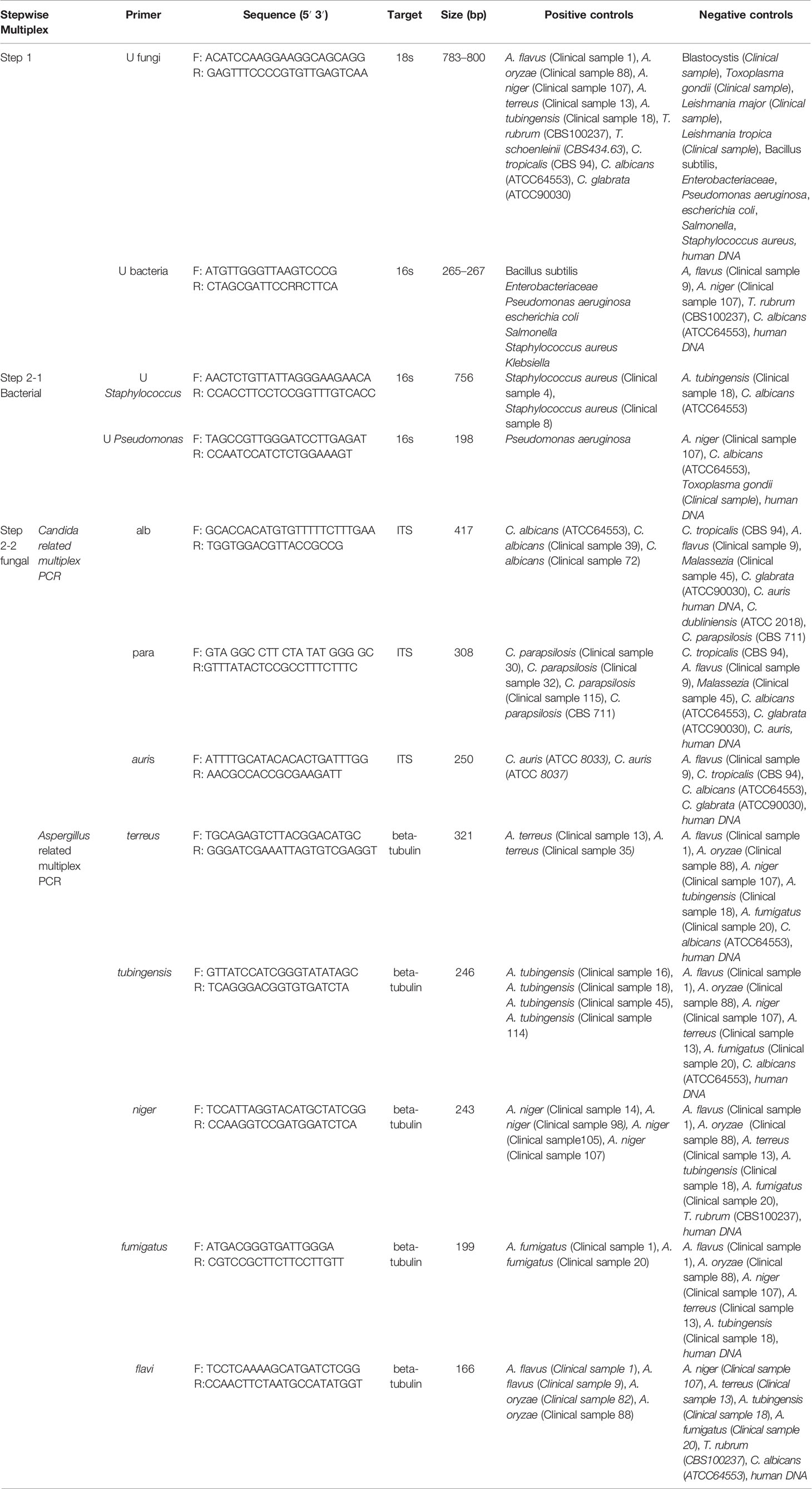
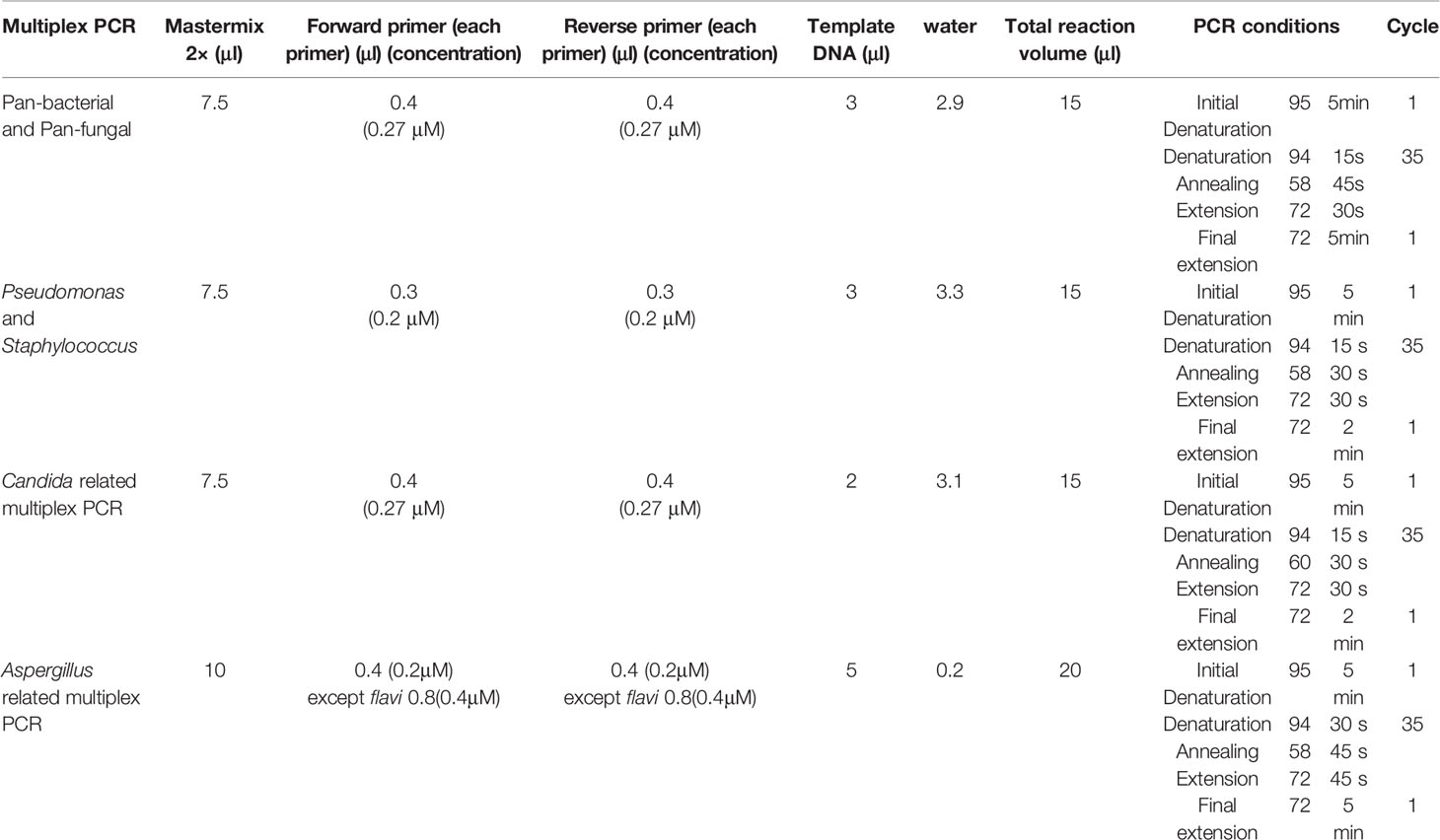
In order to diagnose a panel of most common bacteria and fungi causing OE, a step-by-step multiplex PCR assay was designed for simultaneous detection and differentiation of the etiological agents. The DNA sequences of the candidate genetic target for designing the primers were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/nucleotide/). In silico analyses and the criteria for the selection of primers were reasoned as follows: 1) no cross-reactivity with the other species, other loci, or other primers and similar annealing features of the compatible primers, 2) compatibility of amplicon sizes and sequence complexity of one target species with the rest of target species in the same multiplex PCR, 3) compatibility of melting temperature of primers within the same multiplex PCR, and 4) primer annealing avoidance with nonspecific sequences and other species by locating the gaps and mismatches in the 3′ end of primers. Potential interfering sequences and species were assigned as a negative control for each primer pair. The 18SrDNA, internal transcribed spacer 1 (ITS1) or ITS2 regions, and β-tubulin gene were the subject loci for universal fungal detection, Candida species identification, and Aspergillus species identification, respectively. Additionally, the 16SrDNA region was used for designing the primers for the pan bacterial screening of bacterial agents causing OE and also for specific discrimination between Pseudomonas and Staphylococcus. Online free Primer blast and Geneious software (https://www.geneious.com) were used for primer designing. Table 1 tabulates all primers, target loci, and estimated amplicon size of the primers. The specific primers for Staphylococcus spp. was choose according to Karimi et al. (2020).
DNA Extraction
The genomic DNA was manually extracted from each ear aspiration specimen as follows: About 5 mm3 of the clinical specimens were kept at −20°C and transferred to a 1.5-mL Eppendorf tube containing 300 mg of glass beads (0.5 mm in diameter, 400 μl of lysis buffer (100 mM Tris, pH 8; 10 mM EDTA; 100 mM NaCl; 1% sodium dodecyl sulfate; 1% Triton X-100) and 300 μl phenol–chloroform, and severely homogenized for 3 × 60 s at 6,000 rpm by a homogenizer (Bertin Instrument, Precelleys 24). After centrifugation at 5000 for 5 min, the supernatant was transferred to a new tube and 300 μl of phenol/chloroform was added, and centrifuged for 5 min at 5,000 rpm, and the supernatant was transferred to a new tube; moreover, the identical volume of the chloroform was added and centrifuged for 5 min at 5,000 rpm. In the next stage, 2.5 volumes of absolute ethanol and a 0.1-volume of 3 M sodium acetate (pH 5.2) were added to the supernatant in a new tube and incubated at −20°C for 1 h, followed by centrifugation for 10 min at 12,000 rpm. The supernatant was then discarded, and the precipitate was washed with cold ethanol (70%), dried in the air, dissolved in 30 μl of distilled water, and stored at −20°C as the purified DNA until PCR performance.
PCR Workflow
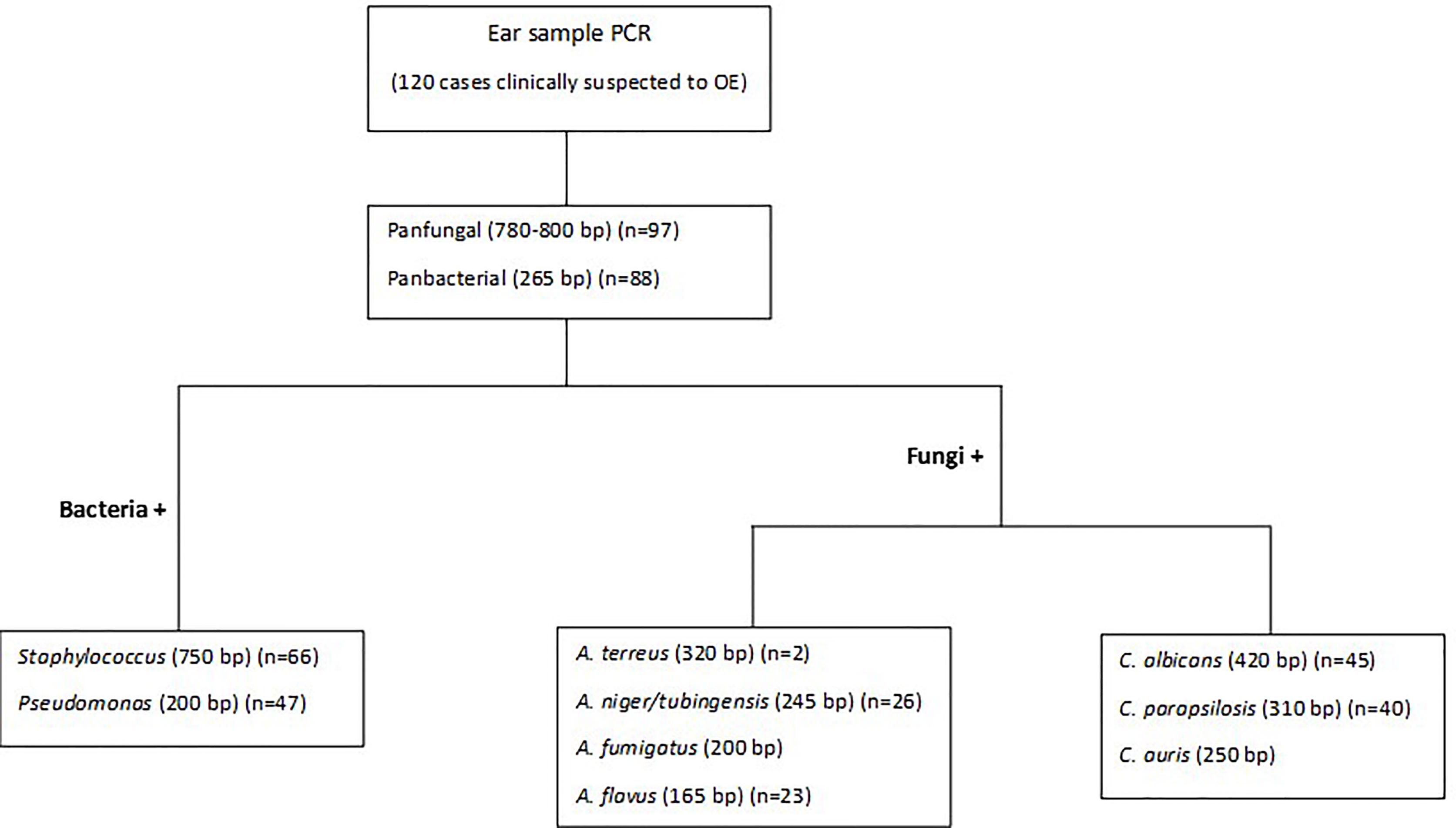
Detection/Identification of bacterial and fungal agents causing OE was performed in two or three multiplex PCRs (Figure 1). The first PCR reaction detects and simultaneously differentiates the fungal from bacterial agents using two pan-fungal and pan-bacterial primers (duplex PCR). According to the positive result of each universal primer and amplicon size (pan-fungal: 783–800 base pair, pan-bacterial: 265–267 bp), the second PCR was selectively run (Table 1, Figure 1). Accordingly, if the first PCR was positive by pan-bacterial primer, then the second duplex PCR was applied to differentiate the most prevailing causal agents of bacterial OE, including Pseudomonas and Staphylococcus based on amplicon size (Table 1). On the other hand, if the first PCR was positive by the universal fungi primer (amplicon size of 783–800 bp), fungal species-specific multiplex PCRs were run, such as Candida multiplex PCR targeting the most clinically important Candida species causing OE (i.e., C. albicans, C. parapsilosis, and C. auris), and Aspergillus multiplex PCR identifying the most prevalent Aspergillus species (i.e., A. niger, A. tubingensis, A. flavus, A. fumigatus, and A. terreus) (Tables 1 and 2).
Specificity Testing
For optimizing, validating, and evaluating the specificity of the multiplex PCR, some CBS and ATCC reference strains as the target species, as well as some human DNAs, strains, or DNAs of clinically important fungal, bacterial, and protozoan species as non-target, were tested and served as the panel of positive and negative (Table 1). The DNA extracted from healthy ears was also tested into the assay system to evaluate the possible false-positive result. The results achieved by each multiplex PCR were compared with those of species identity of each sample, direct microscopy, and culture. Eventually, the reproducibility of each obtained result was practically tested to assess the precision of each multiplex PCR. In order to monitor the presence of inhibitor or potential cross-contamination, an appropriate DNA and sterile distilled water serving as positive and negative control were employed for each PCR run.
Results
Specificity of the Primers
In silico analyses of the designed primers showed no primer mismatches with non-target organisms. Moreover, no cross-reactivity was recorded when a wide range of clinically important or environmental non-target fungal, bacterial, protozoan species, and human DNA were tested. In addition, the assay yielded the appropriate results using target DNA and sterile distilled water serving as a positive and negative control, respectively.
PCR Assay Resolution for Species Identification
Subjecting CBS and ATCC reference strains and previously identified clinical isolates to the stepwise multiplex PCR assay led to consistent identification of each species. Additionally, as expected, cryptic species belonging to Aspergillus section Nigri, Aspergillus section Flavi, and Aspergillus section Terrei were identified as A. niger/A. tubingensis, A. flavus/A. oryzae, and A. terreus, respectively. However, neither cryptic species of C. parapsilosis complex nor rare species belonging to Aspergillus section Fumigati were tested to evaluate the designed primers identifying C. parapsilosis species complex and A. fumigatus-related species.
PCR Versus Direct Examination for Detection/Identification of Bacterial and Fungal OE
A total of 120 frozen specimens from patients suspected of microbial OE which were previously shown to contain pure fungal and bacterial elements in 21 and 12 specimens, respectively, along with 76 specimens which were demonstrated to be simultaneously infected with bacterial (n = 88) and fungal (n = 97) elements using direct examination or Giemsa staining were subjected to DNA extraction and PCR in a blinded way. Out of 120 specimens included in this study, the overall multiplex PCR based detection rate of bacterial (n = 88; 73.3%) and fungal (n = 97; 81%) OE documented is 100%, of which 21 and 12 specimens yielded single amplicon band of 783–800 and 265–267 bp corresponding to fungal and bacterial targets, respectively (Figure 2A). Accordingly, the co-infection of bacterial and fungal pathogens (two amplicon bands were simultaneously yielded) by the first step of multiplex PCR was demonstrated in 76 specimens (63.3%) (Figure 2A). Therefore, the detection rate (sensitivity) of pan-fungal/pan-bacterial multiplex PCR showed complete consistency with the previously documented results of the direct examination. Considering the results of the first step multiplex PCR, an amplicon size of 265–267 bp implying bacterial agents was revealed in 88 specimens. Therefore, the specimens whose etiological agents were diagnosed to be bacterial OE were subjected to a second stepwise bacterial multiplex PCR identifying Staphylococcus and Pseudomonas genera. Out of 88 specimens with positive pan-bacterial PCR, the identification of agents was successfully achieved for 83 specimens. Accordingly, analysis of the amplicon size showed that Staphylococcus and Pseudomonas were the causative agents in 36 and 17 specimens, respectively. In addition, for 30 specimens, two amplification bands were simultaneously observed implying the co-infection of Staphylococcus and Pseudomonas (Figure 2B). The identification of the rest of the specimens (~6%) could not be provided. Therefore, Staphylococcus was evidenced as the most predominantly identified genus causing bacterial OE accounting for 75% of the cases with bacterial OE.
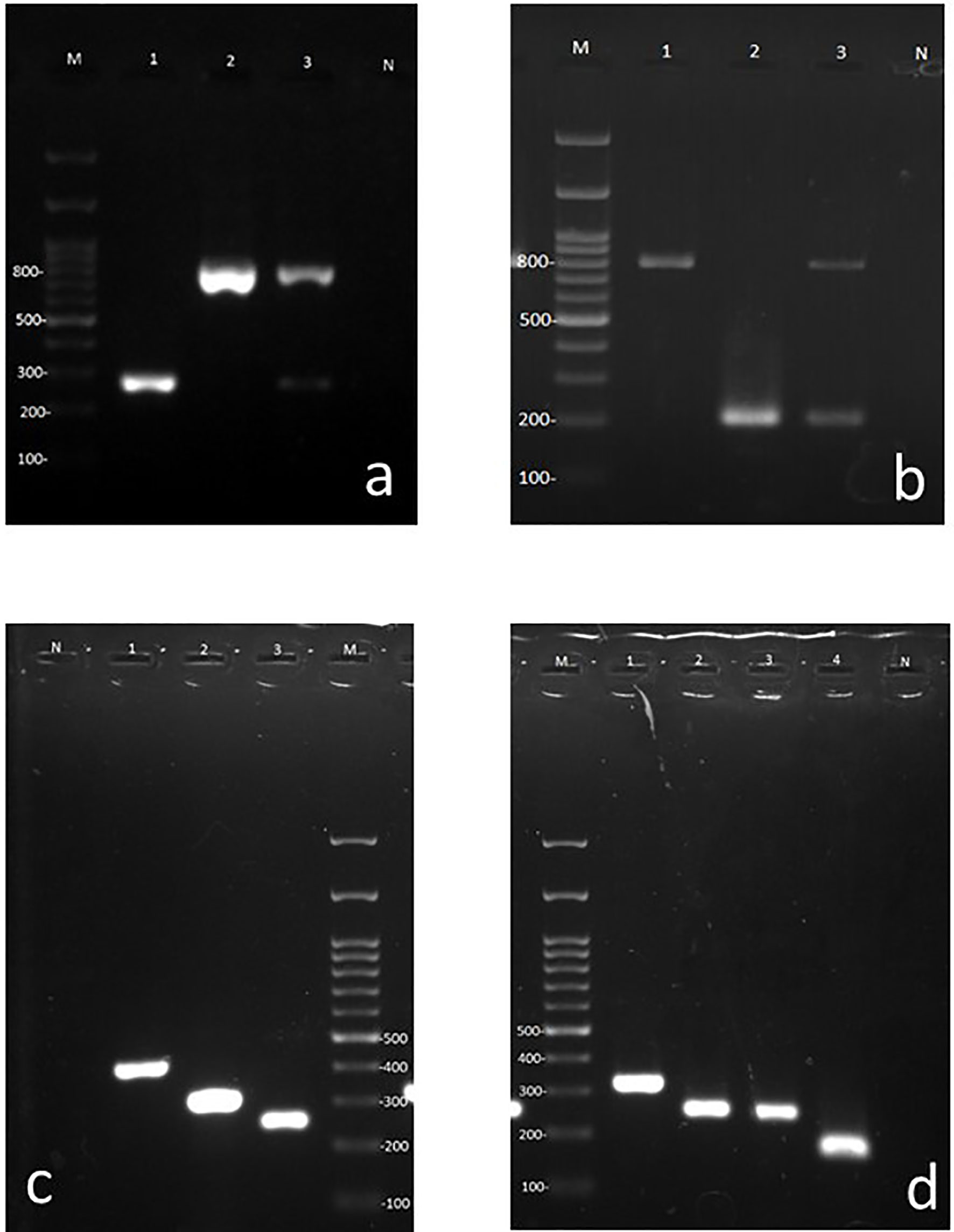
Figure 2 Agarose gels containing representative amplicons. (A) First step multiplex PCR using universal bacterial and fungal primers. Lane M 100 bp ladder, Lane 1: Bacterial (265–267 bp band), Lane 2: Fungal (783–800 bp band), Lane 3: Fungal and bacterial co-infection (dual bands of 265–267 and 783–800 bps), Lane N: negative control for PCR. (B) Pseudomonas and Staphylococcus specific primer pairs (bacterial duplex PCR), Lane M: 100 bp ladder, Lane 1: Staphylococcus, Lanes 2: Pseudomonas, 3: mixed of Pseudomonas and Staphylococcus (dual bands of 198 and 756 bp, respectively), Lane N: negative control for PCR. (C) Candida species specific multiplex primers. Lane N: negative control for PCR, Lane1: C. albicans, Lane 2: C. parapsilosis, Lane 3: C. auris, Lane M: 100 bp ladder. (D) Aspergillus species specific multiplex primer. Lane M: 100 bp ladder, Lane 1: A. terreus, Lane 2: A. tubingensis, Lane 3: A. niger, Lane 4: A. flavi, Lane N: negative control for PCR.
PCR Versus Culture for Detection/Identification of Bacterial and Fungal OE
Regarding the previously documented results of direct examination, which showed fungal elements in 97 specimens, the positivity rate of the first step of the PCR detecting fungal agents causing OE showed to be higher than that in the results previously obtained by culture (positive culture in 81 specimens). All 81 (83.5%) samples with culture positive for fungi were pan-fungal PCR positive and showed complete agreement. However, specimens that demonstrated positive bacterial elements in direct examinations in our previous study had not been investigated by culture media; therefore, the related results of culture were not available for comparison with the positivity rate of pan-bacterial PCR. It should be noted that 16 specimens with negative fungal culture were also successfully amplified in pan fungal PCR. Direct examinations of 12 specimens showed the clusters of single and broad base budding yeast cells, implying Malassezia species; moreover, four cases revealed septate hyphal elements that became Aspergillus positive in PCR.
Basic Molecular Versus Multiplex PCR for Fungal Identification
The identification of fungal agents which were investigated through the second stepwise multiplex PCR was documented to be 92.4% (47 Aspergillus and 26 Candida species). Moreover, it was consistent with the identification results obtained previously by PCR-RFLP, ITS, or β-tubulin gene sequencing of 79 isolates (53 Aspergillus and 26 Candida) recovered from the culture. Nonetheless, the Aspergillus-related multiplex PCR of six out of 53 specimens with culture positive for Aspergillus species did not yield an amplicon band. On the other hand, our designed Aspergillus-related multiplex PCR was able to identify four culture-negative specimens with direct examination indicating fungal hyphae. Considering the amplicon size of the first multiplex PCR covering the detection of fungal and bacterial OE, 97 specimens showed a 783–800 bp size band corresponding to fungal pathogens. Subsequently, the specimens whose etiological agents were diagnosed as fungi were subjected to Candida- and Aspergillus-related multiplex PCR, separately. In total, 67 and 51 specimens contained Candida and Aspergillus species, respectively (Figures 2C, D), among which 30 specimens showed dual amplicon bands and were simultaneously positive with both Candida and Aspergillus related multiplex PCR. However, according to the previously documented results of ear aspiration cultures, 27 and 53 isolates of Candida and Aspergillus species were recovered and molecularly identified, respectively (Aboutalebian et al., 2019). According to the results of the second fungal multiplex PCR, C. albicans, C. parapsilosis, and A. Section Nigri were recorded as the most predominantly identified species in descending frequency among fungal agents infecting the ear canal. However, this frequency should be interpreted with caution since Candida species can be a member of normal flora or colonizer of the external ear canal and the population of Candida may be increased during antibacterial therapy.
Discussion
Delayed or blind treatment of infectious EO due to misdiagnosis or delayed detection may cause myringitis, auricular cellulitis, and otitis media (Adegbiji et al., 2017). Therefore, timely and definitive diagnosis through fast, reliable, and sensitive methods enables the tailoring of appropriate treatment, thereby abolishing avoidable complications (Musso and Crews, 2016; Adegbiji et al., 2017). Given the great number of mixed fungal and bacterial ear infections, the current study investigated the diagnostic value of pan-fungal/pan-bacterial multiplex PCR in order to differentially detect fungal and bacterial OE in the ear specimens aspirated from patients previously proven to carry fungal and bacterial ear infections. One by one pathogen targeting is time-consuming, and contamination-susceptible PCR set-ups make the approaches infeasible for routine laboratory (Matar et al., 1998). One of the great advantages of the multiplex PCR is that this technique provides the simultaneous detection of several species and mixed infections with smaller amounts of reagents and samples (Hendolin et al., 2000). Due to the substantial importance of accurate identification of the causal agent for targeted therapy (Blomgren and Pitkäranta, 2005; Ayub et al., 2015; Adegbiji et al., 2017), Staphylococcus and Pseudomonas specific primers, as well as Candida and Aspergillus species-specific primers, the most frequently encountered agents implicated in OE infection (Aboutalebian et al., 2019) were assessed in a stepwise multiplex PCR manner in this study. No similar studies have investigated so far to molecularly differentiate the fungal and bacterial agents affecting the ear canal. To the best of our knowledge, the current study is the first report aimed to discriminate bacterial and fungal etiology of ear infections through a stepwise multiplex PCR manner. The distinction of the etiological agents affecting the ear canal is a crucial issue to ensure the accurate choice of drug warranting the successful therapy considering the different management and variations of the in vitro antimicrobial susceptibility patterns among different fungal and bacterial genera (even different species belonging to the same genus), emergence of infections caused by C. auris, fewer susceptible species to routinely used antifungal agents, and differential diagnosis (Adegbiji et al., 2017; Abastabar et al., 2019; Aboutalebian et al., 2020). The efficiency of the multiplex PCR assay for simultaneous detection of fungi and bacteria was addressed by comparing its results with those of direct examination and culture. Since a great number of our samples showed bacterial and fungal co-infection via universal primers, combined antifungal and antibacterial approach therapy may be necessary in these patients. The overall multiplex PCR-based detection rate of bacterial (n = 88; 73.3%) and fungal (n = 97; 81%) OE was documented to be 100%. Moreover, the detection rate (sensitivity) of pan-fungal/pan-bacterial multiplex PCR showed complete consistency with the previously documented results of direct examination and Giemsa staining (Aboutalebian et al., 2019). Furthermore, the positivity rate of the first step of the PCR detecting fungal agents showed to be higher than that in the results previously obtained by culture (100 vs. 83.5%). Culture-based therapy is essential for the successful resolution of agents causing the infection in most cases particularly those who have an inadequate response to treatment (Gruber et al., 2015; Musso and Crews, 2016). The PCR assay was demonstrated to be more sensitive, more rapid, and less cumbersome than culture (74 vs. 21%) (Matar et al., 1998). Nevertheless, in case of slow-growing or fastidious bacteria or fungi, pretreatment antimicrobial, low bacterial or fungal count, unavailability of fungi and bacteria due to biofilm formation, or/and polymicrobial colonization of the ear canal, the results of culture can be misleading and less sensitive (Post, 2001; Jalali et al., 2008; Hasibi et al., 2017). In our previous study, bacterial media culture was not used to recover bacterial agents; however, 83.5% of 97 specimens with positive direct examination yielded fungal growth in fungal media culture (Aboutalebian et al., 2019). Culture is necessary to recover the infecting strain, identify the species level, and assess its antifungal susceptibility pattern (Rios-Yuil, 2016; Aboutalebian et al., 2019). However, obligate lipid-dependent Malassezia species cannot easily be recovered by the routinely used fungal culture (Konaté et al., 2017). In our previous study (Aboutalebian et al., 2019), 12 specimens had clusters of single and budding yeast-like cells mixed with short and narrow hyphal fragment in direct examination suggestive of Malassezia species; moreover, their culture remained negative but pan-fungal PCR provided positive results for them. Therefore, the necessity for culture-based molecular identification will be eliminated with a direct PCR on the clinical specimen (Rios-Yuil, 2016). In our study, about 16.5% discordance was noted between the pan-fungal PCR and the fungal culture data, which confirms greater sensitivity of PCR compared to culture-based methods. These results are supported by the finding of other studies, as it was demonstrated that 77.3% of the ear aspiration specimens were PCR positive for one or more organisms in tests with viral and bacterial primers (Post et al., 1998). In another study, a two-step PCR-based assay using pan-bacterial and Haemophilus, as well as Streptococcus and Moraxella catarrhalis specific primers was performed to detect the bacterial etiologies of otitis media with effusion in Lebanese children. According to the results, 53% discordance was revealed between PCR results and those of culture (Matar et al., 1998). Similarly, although only 20–30% of ear effusions yielded positive results through culture, up to 75% of the specimens had positive PCR for pathogenic bacteria (Hendolin et al., 2000; Gok et al., 2001). In another study, the PCR performed for the detection of fungal DNA in the middle ear effusion of patients suffering from otitis media showed to be more sensitive (29 vs. 8%) than fungal culture (Jalali et al., 2008). Our results imply that the multiplex PCR is more efficient than culture in detecting the DNA of bacterial and fungal agents in EA samples. However, since both are viable and fossil of the organism’s genetic material can be amplified by PCR, residual DNA from a previous episode of OE infection is probably detected as well (Post et al., 1996). The experimental model has shown that the bacterial DNA in the ear aspiration samples disintegrates within two days following bacterial death (Matar et al., 1998). Noteworthy, in all positive pan-bacterial PCR (n = 88), bacterial elements were evidenced with Giemsa staining. Therefore, it is probable that the bacterial DNA detected by the universal primers in our EA samples might have originated from viable bacteria implicated in disease and not residual DNA persisting from a former episode of OE infection. In the same line, our results have shown the presence of fungal and bacterial DNA in 81 and 73.3% of the samples (n = 120), respectively. The rate of fungal OE was higher than that of bacterial OE, and it is also above the rate reported earlier (Lee et al., 2008). The high rate of fungal OE can be attributed to the fact that the cases suspected to carry otomycosis were recruited in our previous study (Aboutalebian et al., 2019). However, some of the specimens showed to be infected with either bacterial agent as mono-caused OE infection or mixed fungal and bacterial infection. Considering the results of species identification through our genus and species-specific primers, our finding showed that about 95% of bacterial OE is caused by Staphylococcus and Pseudomonas. The identification of Staphylococcus and Pseudomonas genera was only investigated in this study since previous studies have reported that 92% of patients with bacterial EO were implicated by Staphylococcus and Pseudomonas genera (Clark et al., 1997). The isolation rate of non-Staphylococcus/Pseudomonas genera is documented in only 5–20% of bacterial OE (Clark et al., 1997; Musso and Crews, 2016). The high rate of PCR positive by Staphylococcus specific primer (75%) could be attributed to the bacterial flora residing in the ear canal (Musso and Crews, 2016); however, since in all experiments, control panels, including ear samples, obtained from healthy individuals (served as negative control) were tested into the assay system, the likelihood of false positivity is disregarded. Moreover, our subjects were clinically relevant cases of ear infection (Aboutalebian et al., 2019). Regarding the prognosis of Aspergillus and Pseudomonas OE that are more associated with the spread and involve the temporal bone, skull base, and multiple cranial nerves, especially in susceptible hosts, and cause a disorder termed as necrotizing (malignant) OE (Musso and Crews, 2016), the importance of identification of the species causing the disease is doubled. Aspergillus and Candida species particularly those belonging to the complexes A. Section Nigri, A. section flavi, and C. parapsilosis, are the most prevailing causes of otomycosis (Musso and Crews, 2016; Aboutalebian et al., 2019). Additional fungi less commonly reported to be implicated in otomycosis include Cladosporium, Penicillium, Alternaria, Cryptococcus, Talaromyces (Aboutalebian et al., 2020), and C. auris (Abastabar et al., 2019). According to the results of the second fungal multiplex PCR, C. albicans, C. parapsilosis, and A. Section Nigri were recorded as the most predominantly identified species in descending frequency. However, culture-based molecular identification showed that species under A. Section Nigri and then C. parapsilosis were the most frequent causal agents (Aboutalebian et al., 2019). Considering the lack of significant difference in the antifungal susceptibility patterns of different species categorized in different Aspergillus sections (Szigeti et al., 2012), the identification of the related species up to section level was regarded in this study. The high rate of positive Candida-related multiplex PCR and specimens simultaneously showed dual amplicon bands of Candida and Aspergillus species possibly due to Candida overgrowth and colonization in unhealthy and humidity accumulated ear (Musso and Crews, 2016; Aboutalebian et al., 2019) or dual infection. In cases of co-infection, multiplex PCR assays targeting different genera or species-specific regions have been demonstrated to be more sensitive than culture for accurate discrimination and identification of the fungi (Zaman et al., 2017). Given the increasing reports of otomycosis due to C. auris (Abastabar et al., 2019; Jung et al., 2020), as well as the emerging multidrug-resistant yeast that was erroneously identified as C. parapsilosis, C. haemulonii, C. famata, and Rhodotorula glutinis through conventional and biochemical assays, specific primers were designed capturing C. auris in the heart of Candida related multiplex PCR. This is one of the strengths of the present study. Interestingly, two ear samples were positive for C. auris in the species-specific multiplex PCR of Candida; however, C. auris colonies were not isolated in this study, and they require more investigations before reporting. The sensitivity of identification of Aspergillus species directly from fresh specimens was lower than culture-based molecular identification, indicating the possibility that in clinical specimens, DNA is damaged during tissue processing and DNA extraction. Molecular diagnosis and species identification by universal and species-specific primers had a lower turnaround time (~10 h), compared to culture, sequencing, and histopathology. This finding is important for early diagnosis followed by targeted therapy (Blomgren and Pitkäranta, 2005; Thorne and Wetmore, 2009; Ayub et al., 2015; Musso and Crews, 2016).
Conclusion
This study revealed simultaneous detection and partial identification of fungal, bacterial, and mixed OE infections from clinical ear specimens with smaller amounts of reagents and samples using pan-fungal/pan-bacterial multiplex PCR. The sole dependence on conventional and phenotypical assays could result in misdiagnosis and erroneous identification and oblivion of emerging yeast species, such as C. auris. The necessity for culture based-molecular identification will be eliminated through stepwise molecular detection/identification directly from clinical samples; accordingly, misidentification, turnaround time, and expenses will be reduced in this regard. The stepwise multiplex PCR assay is recommended as a better detector of agents implicated in fungal and bacterial OE.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
This study was reviewed and approved by the national institute for medical research development (NIMAD) by the grant number: 982931, and the ethics code: IR.NIMAD.REC.1398.063. The patients provided their written informed consent to participate in the study.
Author Contributions
SA and HM have designed the study. SA did the experiments. AO contributed in patient selection, clinical diagnosis, and treatment of the subjects. KA, HF, and HM participated in draft preparation and revision. All authors assisted in paper edition and revision. MN, JC, and HM have provided the isolates, participated in carrying out the experiments, and assisted in paper revision. SA participated in performing multiplex PCR. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abastabar M., Haghani I., Ahangarkani F., Rezai M. S., Taghizadeh Armaki M., Roodgari S., et al. (2019). Candida auris otomycosis in Iran and review of recent literature. Mycoses 62, 101–105. doi: 10.1111/myc.12886
Aboutalebian S., Mahmoudi S., Mirhendi H., Okhovat A., Abtahi H., Chabavizadeh J. (2019). Molecular epidemiology of otomycosis in Isfahan revealed a large diversity in causative agents. J. Med. Microbiol. 68, 918–923. doi: 10.1099/jmm.0.000985
Aboutalebian S., Mahmoudi S., Okhovat A., Khodavaisy S., Mirhendi H. (2020). Otomycosis Due to the Rare Fungi Talaromyces purpurogenus, Naganishia albida and Filobasidium magnum. Mycopathologia 185, 569–575. doi: 10.1007/s11046-020-00439-8
Adegbiji W. A., Aremu S. K., Olatoke F., Olajuyin A. O., Ogundipe K. O. (2017). Epidemiology of Otitis externa in developing country. Int. J. Recent Sci. Res. 8, 18023–18027. doi: 10.12691/ajmsm-5-3-4
Ayub M., Islam A., Moiz A., Fahad M. (2015). Acute otitis media: identification of causative pathogens with antimicrobial comparative efficacy. J. Appl. Pharm. 7, 2. doi: 10.4172/1920-4159.1000205
Bialek R., Konrad F., Kern J., Aepinus C., Cecenas L., Gonzalez G., et al. (2005). PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J. Clin. Pathol. 58, 1180–1184. doi: 10.1136/jcp.2004.024703
Blomgren K., Pitkäranta A. (2005). Current challenges in diagnosis of acute otitis media. Int. J. Pediatr. Otorhinolaryngol. 69, 295–299. doi: 10.1016/j.ijporl.2004.09.012
Chonmaitree T., Henrickson K. J. (2000). Detection of respiratory viruses in the middle ear fluids of children with acute otitis media by multiplex reverse transcription: polymerase chain reaction assay. Pediatr. Infect. Dis. J. 19, 258–260. doi: 10.1097/00006454-200003000-00020
Clark W. B., Brook I., Bianki D., Thompson D. H. (1997). Microbiology of otitis externa. Otolaryngol. Head Neck Surg. 116, 23–25. doi: 10.1016/S0194-5998(97)70346-2
Gok U., Bulut Y., Keles E., Yalcin S., Doymaz M. Z. (2001). Bacteriological and PCR analysis of clinical material aspirated from otitis media with effusions. Int. J. Pediatr. Otorhinolaryngol. 60, 49–54. doi: 10.1016/S0165-5876(01)00510-9
Gruber M., Roitman A., Doweck I., Uri N., Shaked-Mishan P., Kolop-Feldman A., et al. (2015). Clinical utility of a polymerase chain reaction assay in culture-negative necrotizing otitis externa. Otol. Neurotol. 36, 733–736. doi: 10.1097/MAO.0000000000000563
Hammond S. P., Bialek R., Milner D. A., Petschnigg E. M., Baden L. R., Marty F. M. (2011). Molecular methods to improve diagnosis and identification of mucormycosis. J. Clin. Microbiol. 49, 2151–2153. doi: 10.1128/JCM.00256-11
Hasibi M., Ashtiani M. K., Motassadi Zarandi M., Yazdani N., Borghei P., Kuhi A., et al. (2017). A treatment protocol for management of bacterial and fungal malignant external otitis: a large cohort in Tehran, Iran. Ann. Otol. Rhinol. Laryngol. 126, 561–567. doi: 10.1177/0003489417710473
Hendolin P. H., Paulin L., Ylikoski J. (2000). Clinically applicable multiplex PCR for four middle ear pathogens. J. Clin. Microbiol. 38, 125–132. doi: 10.1128/JCM.38.1.125-132.2000
Jalali M., Rezaie S., Kousha A., Saadat F., Banan R. (2008). Detection of fungal DNA in the middle ear effusion of patients suffering from otitis media with effusion. Iranian J. Public Health 37, 109–113.
Jung J., Kim M. J., Kim J. Y., Lee J.-Y., Kwak S. H., Hong M. J., et al. (2020). Candida auris colonization or infection of the ear: A single-center study in South Korea from 2016 to 2018. Med. Mycol. 58, 124–127. doi: 10.1093/mmy/myz020
Karimi H., Nikaeen M., Gholipour S., Hatamzadeh M., Hassanzadeh A., Hajizadeh Y. (2020). PM 2.5-associated bacteria in ambient air: Is PM 2.5 exposure associated with the acquisition of community-acquired staphylococcal infections? J. Environ. Health Sci. Eng. 18 (2), 1007–1013. doi: 10.1007/s40201-020-00522-8
Konaté A., Barro-Kiki P. C. M., Kassi K. F., Angora K. E., Vanga-Bosson H., Djohan V., et al. (2017). Pityriasis versicolor profiles among outpatients of the Dermatology Department of the Teaching Hospital of Yopougon Abidjan, Cote dIvoire. Afr. J. Microbiol. Res. 11, 1573–1577. doi: 10.5897/AJMR2017.8659
Lee J. E., Chon K. M., Lee J. W., Lee J. K., Chang C. (2008). Detection of Fungus and Bacteria in Otitis Media with Effusion of Children Using Polymerase Chain Reaction and Its Correlation of Clinical Factors. Korean J. Otorhinolaryngol. Head Neck Surg. 51, 960–966. doi: 10.3342/kjorl-hns.2008.51.11.960
Matar G. M., Sidani N., Fayad M., Hadi U. (1998). Two-step PCR-based assay for identification of bacterial etiology of otitis media with effusion in infected Lebanese children. J. Clin. Microbiol. 36, 1185–1188. doi: 10.1128/JCM.36.5.1185-1188.1998
Musso M. F., Crews J. D. (2016). “Infections of the external ear,” in Infectious Diseases in Pediatric Otolaryngology (Springer), 15–28. doi: 10.1007/978-3-319-21744-4_2
Post J. C., Aul J. J., White G. J., Wadowsky R. M., Zavoral T., Tabari R., et al. (1996). PCR-based detection of bacterial DNA after antimicrobial treatment is indicative of persistent, viable bacteria in the chinchilla model of otitis media. Am. J. Otolaryngol. 17, 106–111. doi: 10.1016/S0196-0709(96)90005-8
Post J. C., White G. J., Liederman E. M., Aul J. J., Buchman C. A., Sirko D. A., et al. (1998). Analysis of adult otitis media: polymerase chain reaction versus culture for bacteria and viruses. Ann. Otol. Rhinol. Laryngol. 107, 10–16. doi: 10.1177/000348949810700103
Post J. C. (2001). Candidate’s Thesis: direct evidence of bacterial biofilms in otitis media. Laryngoscope 111, 2083–2094. doi: 10.1097/00005537-200112000-00001
Punia R. S., Singhal S. K., Kundu R., Das A., Chander J. (2019). Fungal Suppurative Otitis Media (Histopathology) Among Patients in North India. Head Neck Pathol. 13, 149–153. doi: 10.1007/s12105-018-0918-2
Rickerts V., Mousset S., Lambrecht E., Tintelnot K., Schwerdtfeger R., Presterl E., et al. (2007). Comparison of histopathological analysis, culture, and polymerase chain reaction assays to detect invasive mold infections from biopsy specimens. Clin. Infect. Dis. 44, 1078–1083. doi: 10.1086/512812
Rios-Yuil J. M. (2016). Pityriasis Versicolor: Clinical Spectrum and Diagnosis. Curr. Fungal Infect. Rep. 10, 121–125. doi: 10.1007/s12281-016-0261-6
Roden M. M., Zaoutis T. E., Buchanan W. L., Knudsen T. A., Sarkisova T. A., Schaufele R. L., et al. (2005). Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin. Infect. Dis. 41, 634–653. doi: 10.1086/432579
Rowlands S., Devalia H., Smith C., Hubbard R., Dean A. (2001). Otitis externa in UK general practice: a survey using the UK General Practice Research Database. Br. J. Gen. Pract. 51, 533–538.
Salyer S. W. (2007). Essential emergency medicine: for the healthcare practitioner (Elsevier Health Sciences).
Sarwestani Z. K., Hashemi S., Rezaie S., Shoar M. G., Mahmoudi S., Elahi M., et al. (2018). Species identification and in vitro antifungal susceptibility testing of Aspergillus section Nigri strains isolated from otomycosis patients. J. Mycol. Med. 28 (2), 279–284. doi: 10.1016/j.mycmed.2018.02.003
Schaefer P., Baugh R. F. (2012). Acute otitis externa: an update. Am. Family Physician 86, 1055–1061.
Szigeti G., Kocsubé S., Dóczi I., Bereczki L., Vágvölgyi C., Varga J. (2012). Molecular identification and antifungal susceptibilities of black Aspergillus isolates from otomycosis cases in Hungary. Mycopathologia 174, 143–147. doi: 10.1007/s11046-012-9529-8
Thorne M. C., Wetmore R. F. (2009). “External Otitis,” in Pediatric Otolaryngology for the Clinician (Springer), 51–54. doi: 10.1007/978-1-60327-127-1_6
Zaman K., Rudramurthy S. M., Das A., Panda N., Honnavar P., Kaur H., et al. (2017). Molecular diagnosis of rhino-orbito-cerebral mucormycosis from fresh tissue samples. J. Med. Microbiol. 66, 1124–1129. doi: 10.1099/jmm.0.000560
Keywords: Otitis externa, detection and identification, multiplex PCR, fungi, bacteria
Citation: Aboutalebian S, Ahmadikia K, Fakhim H, Chabavizadeh J, Okhovat A, Nikaeen M and Mirhendi H (2021) Direct Detection and Identification of the Most Common Bacteria and Fungi Causing Otitis Externa by a Stepwise Multiplex PCR. Front. Cell. Infect. Microbiol. 11:644060. doi: 10.3389/fcimb.2021.644060
Received: 19 December 2020; Accepted: 01 March 2021;
Published: 25 March 2021.
Edited by:
Rodolfo García-Contreras, National Autonomous University of Mexico, MexicoReviewed by:
Mahdi Abastabar, Mazandaran University of Medical Sciences, IranShahram Mahmoudi, Iran University of Medical Sciences, Iran
Manouchehr Khazandi, University of South Australia, Australia
Copyright © 2021 Aboutalebian, Ahmadikia, Fakhim, Chabavizadeh, Okhovat, Nikaeen and Mirhendi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hossein Mirhendi, cy5oLm1pcmhlbmRpQGdtYWlsLmNvbQ==
 Shima Aboutalebian
Shima Aboutalebian Kazem Ahmadikia
Kazem Ahmadikia Hamed Fakhim
Hamed Fakhim Javaher Chabavizadeh
Javaher Chabavizadeh Ahmadreza Okhovat4
Ahmadreza Okhovat4 Hossein Mirhendi
Hossein Mirhendi