- 1Guangdong Province Key Laboratory of Microbial Signals and Disease Control, Integrative Microbiology Research Centre, South China Agricultural University, Guangzhou, China
- 2Women and Children’s Health Institute, Guangdong Women and Children Hospital, Guangzhou, China
- 3Department of Microbiology, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong, Hong Kong SAR, China
Pseudomonas aeruginosa can cause various types of infections and is one of the most ubiquitous antibiotic-resistant pathogens found in healthcare settings. It is capable of adapting to adverse conditions by transforming its motile lifestyle to a sessile biofilm lifestyle, which induces a steady state of chronic infection. However, mechanisms triggering the lifestyle transition of P. aeruginosa strains with clinical significance are not very clear. In this study, we reported a recently isolated uropathogenic hyper-biofilm producer PA_HN002 and characterized its genome to explore genetic factors that may promote its transition into the biofilm lifestyle. We first showed that high intracellular c-di-GMP content in PA_HN002 gave rise to its attenuated motilities and extraordinary strong biofilm. Reducing the intracellular c-di-GMP content by overexpressing phosphodiesterases (PDEs) such as BifA or W909_14950 converted the biofilm and motility phenotypes. Whole genome sequencing and comprehensive analysis of all the c-di-GMP metabolizing enzymes led to the identification of multiple mutations within PDEs. Gene expression assays further indicated that the shifted expression profile of c-di-GMP metabolizing enzymes in PA_HN002 might mainly contribute to its elevated production of intracellular c-di-GMP and enhanced biofilm formation. Moreover, mobile genetic elements which might interfere the endogenous regulatory network of c-di-GMP metabolism in PA_HN002 were analyzed. This study showed a reprogrammed expression profile of c-di-GMP metabolizing enzymes which may promote the pathoadaption of clinical P. aeruginosa into biofilm producers.
Introduction
Pseudomonas aeruginosa is a ubiquitous opportunistic pathogen that can cause a variety of infections such as the bloodstream infection, urinary tract infection, burn wound infection and surgical site infection in human bodies (Litwin et al., 2021). Prevention and treatment of its infections are frequently hindered owing to its remarkable capacity of resisting antibiotics, and recurrence of its infections is the leading cause of morbidity and mortality in immunocompromised patients (Moradali et al., 2017; Pang et al., 2019). P. aeruginosa has a large genome which generally consists of ~6,000 genes with a high proportion of them encoding regulatory proteins, metabolic enzymes, outer membrane proteins involved in adhesion, motility, transportation, and signal sensing, enabling a surprising ecological versatility and adaptability to the environmental changes (Stover et al., 2000; Klockgether et al., 2011; Cao et al., 2017; Rossi et al., 2021). In addition, P. aeruginosa possesses a large number of genes encoding virulence factors such as phenazines, siderophores, elastase, proteases, exotoxins and cytotoxins, flagella and pili, and all these contribute significantly to its pathogenesis (Ahator and Zhang, 2019; Alonso et al., 2020).
P. aeruginosa is a motile microorganism and its motility is mainly driven by the polar monotrichous flagellum or the type IV pili (Toutain Christine et al., 2005; Burrows, 2012). Directed motility known as chemotaxis is beneficial for bacteria to migrate towards nutrients or evade from adverse environments such as the presence of toxins and predators. Motility represents an important virulence factor in P. aeruginosa to cause acute infection because it allows the pathogen to rapidly attach to and colonize surfaces of host tissues (Josenhans and Suerbaum, 2002; Kühn et al., 2021). There are three well-characterized types of motilities in P. aeruginosa: twitching, swarming and swimming. Twitching is a type IV pili-dependent form of multicellular motility by which P. aeruginosa cells move over solid surfaces (Kearns Daniel et al., 2001). Swarming occurs on semisolid surfaces (0.5-0.7% agar) which is also a multicellular phenomenon but depends on the functional flagellum as well as two biosurfactants rhamnolipids (RL) and 3-hydroxyalkanoic acids (HAA) (Ha et al., 2014a). In contrast, swimming is a unicellular movement behavior in liquid or low-viscosity conditions (up to 0.3% agar) which only requires a functional flagellum with its motor-stator complex (Ha et al., 2014b). Motilities in P. aeruginosa are tightly regulated. For example, the flagellum-based swimming and swarming motilities are largely dependent on the intracellular level of the global second messenger cyclic di-GMP (c-di-GMP) which further interconnects a broad spectrum of regulatory pathways such as the FleS/FleR two-component system, Gac/Rsm signaling cascade, Chp chemotaxis system, Wsp chemosensory pathway and the HptB pathway (Wolfe Alan and Visick Karen, 2008; Porter et al., 2011; Xin et al., 2019; Zhou et al., 2021).
As a member of the highly virulent and antibiotic-resistant bacterial group ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp.) (Mulani et al., 2019), P. aeruginosa exhibits resistance to a broad spectrum of antibiotics including β-lactams, quinolones and aminoglycosides with its intrinsic, acquired and adaptive resistance machineries (Hancock and Speert, 2000). For instance, P. aeruginosa intrinsically develops restricted outer membrane permeability, synthesizes a number of antibiotic-inactivating enzymes such as β-lactamases, and produces abundant efflux systems to minimize or obliterate the intruded antibiotics (Breidenstein et al., 2011). What’s worse, P. aeruginosa frequently elevates the refractoriness to antibiotic treatments by forming biofilms, leading to a higher rate of therapeutic failure and a state of recurrent or chronic infections (Mulcahy et al., 2014). A biofilm is described as an aggregation of microorganisms that are encased within a self-produced matrix of extracellular polymeric substances (EPSs) and adhere to each other on biotic or abiotic surfaces (Chang, 2018). The matrix of EPSs in P. aeruginosa biofilms is basically composed of polysaccharides, proteins, extracellular DNA (eDNA) and rhamnolipids (Mann and Wozniak, 2012). Formation of biofilm represents one of the best characterized mechanisms of adaptive resistance in P. aeruginosa, which protects the microbial cells inside to resist antibiotics with up to 1,000-fold greater compared to the planktonic cells and is regarded as one of the typical features during chronic infections (Mah et al., 2003; Taylor et al., 2014). Biofilm formation in P. aeruginosa are known to be regulated by a variety of cellular factors such as c-di-GMP, cAMP, quorum sensing (QS) systems and two-component systems (Sakuragi and Kolter, 2007; Dong et al., 2008; Almblad et al., 2019). In recent years, increased intracellular c-di-GMP level has been reported to promote biofilm formation of P. aeruginosa in a range of clinically important models and delay healing of wound infections caused by P. aeruginosa (Wei et al., 2019; Zhang et al., 2020; Li et al., 2021).
So far, extensive efforts have been made to understand the mechanisms underlying the regulation of motility and biofilm formation in P. aeruginosa, and c-di-GMP is found to play a critical role in the transition between the motile and sessile lifestyles, leading to the acute-to-chronic virulence switch. For instance, a high c-di-GMP level promotes sessile biofilm lifestyle while a low level of c-di-GMP is associated with the active motility (Li et al., 2017). However, genetic basis and environmental cues that trigger the shift of intracellular c-di-GMP levels and consequently induce the transition of lifestyles are still largely unknown especially in the isolates with clinical significance. In this study, we isolated a uropathogenic P. aeruginosa strain PA_HN002 which was found as a hyper-biofilm producer and its biofilm formation was confirmed to be dependent on the high intracellular c-di-GMP concentration. We analyzed its genome and performed a comprehensive analysis of c-di-GMP metabolizing enzymes between PA_HN002 and the reference strain PAO1, which realized that the altered expression profile of c-di-GMP metabolizing enzymes in PA_HN002 mainly induced its biofilm lifestyle in this strain. These findings highlighted the complex regulation of c-di-GMP metabolism and its importance to promote P. aeruginosa to adapt and persist to the clinically relevant environments.
Materials and methods
Strains and growth conditions
P. aeruginosa PA_HN002 was isolated from a mid-stream urine sample of a one-year-old pediatric patient in Guangdong Women and Children Hospital (Guangzhou, China) with the approvement from the Medical Ethical Committee of the hospital under number of 202201011. P. aeruginosa strains were routinely grown at 37°C in Luria-Bertani (LB) broth (Tryptone 10g/L, Yeast extract 5g/L, NaCl 10g/L). Gentamycin was supplemented in the medium when necessary: 25 μg/mL for E. coli DH5α and 50 μg/mL for P. aeruginosa.
Genome sequencing and bioinformatic analysis
Genomic DNA of PA_HN002 was sequenced on an Illumina Hiseq platform (Sangon, China). Genome sequences were assembled, and gap filling was performed by SPAdes, GapFiller and PrInSeS-G (Massouras et al., 2010; Nadalin et al., 2012; Prjibelski et al., 2020). Gene annotation was conducted using Prokka v1.14.5 (Seemann, 2014). Protein sequences of PA_HN002 were searched against the Clusters of Orthologous Groups (COG) database with an e-value<1e-5 using blastp and results were parsed using in-house python script to summarize proteins present exclusively or shared between different P. aeruginosa strains. Venn diagram was generated by Venny v2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/index.html). The MLST and serotype of PA_HN002 was predicted based on the PubMLST database and the Pseudomonas aeruginosa serotyper (PAst) tool, respectively (Thrane Sandra et al., 2016; Jolley et al., 2018). The phylogenetic tree was built based on the alignment of 400 marker genes in PA_HN002 and 6,800 P. aeruginosa genomes retrieved from GenBank with fasttree using PhyloPhlAn version 3.0 and visualized using iTOL (Asnicar et al., 2020; Letunic and Bork, 2021). IME was predicted with ICEberg 2.0 (Liu et al., 2019). Insertion sequences in the PA_HN002 genome were predicted by ISFinder (Siguier et al., 2006). Prophages were predicted using the online software PHASTER (Arndt et al., 2016). Genomic islands (GIs) were predicted using IslandViewer 4 (Bertelli et al., 2017). Domain architectures of the c-di-GMP metabolizing enzymes were determined based on the SMART algorithm (Letunic et al., 2021).
Construction of plasmids for gene expression
Genes for complementation or overexpression were amplified by PCR with the primers in Table S1 and ligated into the downstream of the lac promoter of pBBR1-MCS5 between the HindIII and BamHI sites. The constructed plasmids were verified by PCR and DNA sequencing. Constructed plasmids were introduced into PA_HN002 by electroporation. Successful plasmid delivery into PA_HN002 was confirmed by PCR.
Motility assay
Different agar plates were prepared for swimming and swarming motilities. For swimming motility, semisolid agar plates were prepared with 5 g/L Becto-peptone, 3 g/L Yeast extract and 2 g/L Becto-agar. For swarming motility, plates were prepared by 8 g/L Nutrient broth, 5 g/L Glucose and 6 g/L Becto-agar. 15 ml agar medium was poured into a 90-mm petri dish and then solidified at room temperature. Overnight bacterial culture was 1:50 diluted and grown to OD600 of 1.0. 2 μl cell culture was spotted into the center of the pre-dried swimming and swarming plates. The plates were incubated at 37°C for 48 hours, and images were recorded. The assays were performed in triplicates.
Biofilm assay
Biofilm assay was performed as previously described (Cao et al., 2019). Overnight bacterial culture was 1:50 diluted and grown to OD600 of 1.0. After 1:50 dilution, After 1:50 dilution, 1 ml cell culture was transferred to the 24-well plate and incubated statically at 37°C for 24 h. Planktonic cells were discarded and the biofilm cells was washed carefully with MilliQ water. The plate was air dried and then stained with 2 ml 0.1% crystal violet for 15 min. After the crystal violet was removed, biofilm was washed with MilliQ water , air dried, and dissolved in 2 ml of a mixture of glacial acetic acid and 95% ethanol (1:4). Absorbance at 595 nm was measured after 10-fold dilution in a microplate reader (BioTek).
c-di-GMP measurement by liquid chromatography-mass spectrometry
c-di-GMP was measured following a previous study with slight modifications (Chen et al., 2022). Overnight bacterial culture was 1:50 diluted and grown to OD600 of 1.0. 1 ml cell culture was harvested. Cell lysis was performed by adding perchloric acid (70%, v/v) with a final concentration of 0.6 M for 30 min at 4°C. Samples were centrifuged at 12,000 rpm for 10 min and the supernatants were collected. After the pH was neutralized by adding 1/5 volume of 2.5 M KHCO3, samples were centrifuged at 4,000 rpm for 10 min at 4°C and the supernatants were collected for LC-MS analysis. LC-MS was undertaken using a Q Exactive Focus Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) with a 1.8 μm, 100×2.1 mm high-strength silica (HSS) T3 column (Waters). 5 μl sample was injected and the m/z 691>248 transition was used for quantification.
Reverse transcription-quantitative PCR
RT-qPCR was performed as previously described (Xu et al., 2019). Primers used for this assay were listed in Table S1. After the overnight culture was 1:50 diluted and grown to OD600 of 1.0, 1 ml cell culture was collected. Total RNA was extracted using the Eastep® Super Total RNA Extraction Kit (Promega) following the manufacturer’s instructions. Reverse transcription was conducted to obtain cDNA using the TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen). qPCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme). Relative gene expression was calculated based on the 2-ΔΔCt method with the recA gene as the internal reference control (Schmittgen and Livak, 2008).
Galleria mellonella Killing assay
Time-killing of G. mellonella was performed to evaluate the acute virulence of P. aeruginosa following a previous study with slight modifications (Si et al., 2017). Overnight bacterial culture was 1:50 diluted and grown to OD600 of 1.0. Cells were harvested and resuspended with fresh LB medium to a final concentration of 105 cells per ml. Fifteen G. mellonella larvae were injected with 10 μl cell suspension at the secondary right proleg using a syringe and then incubated at 30°C. The survival rate of G. mellonella was recorded at different time points within 2 days.
Results
A red-pigmented clinical P. aeruginosa isolate PA_HN002 showed chronic infection phenotypes
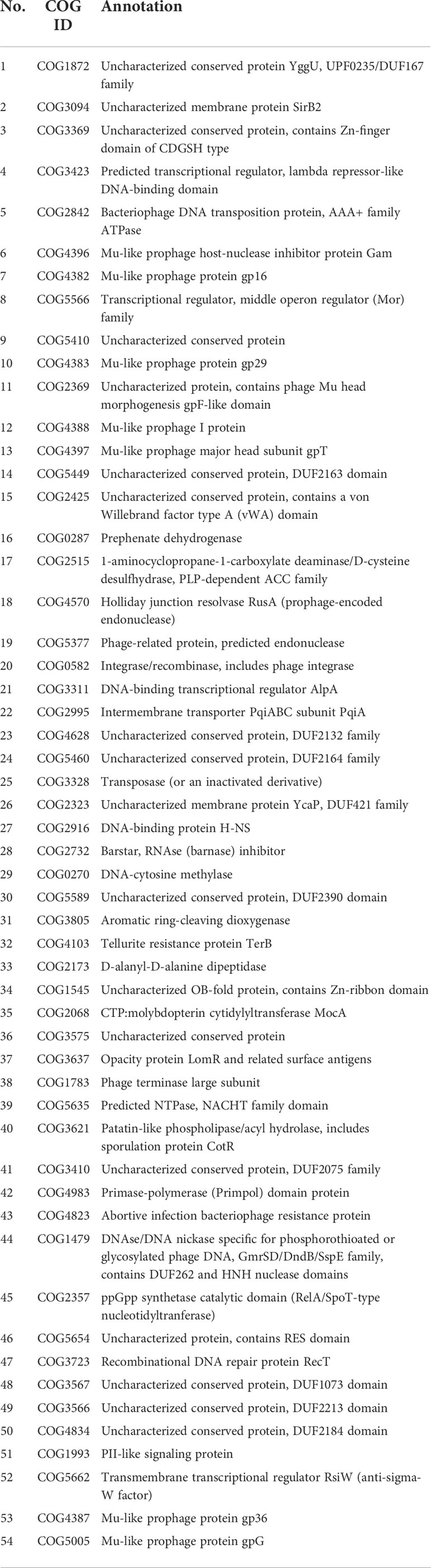
A P. aeruginosa strain PA_HN002 was recently isolated from a mid-stream urine sample of a one-year-old pediatric patient in Guangdong Women and Children Hospital (Guangzhou, China). The strain was identified as P. aeruginosa by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) and 16S rDNA sequencing. Unlike common blue/green-colored P. aeruginosa cultures due to the production of pyocyanin, PA_HN002 was a rarely seen P. aeruginosa isolate producing a red/brown-colored pigment (Figure 1A) which was supposed to be pyorubin (red) or pyomelanin (brown) as described previously (Rodríguez-Rojas et al., 2009; Orlandi et al., 2015). Compared to the reference strain PAO1, PA_HN002 displayed significantly attenuated in vivo virulence when examined using the acute infection model of Galleria mellonella (Figure 1B). Although twitching motility was comparable between PA_HN002 and PAO1 (Figure S1), flagellum-based swimming and swarming motilities of PA_HN002 were substantially diminished (Figure 1C). In contrast, extraordinary greater capacity of biofilm formation was observed in PA_HN002 with approximately 7-fold higher of biofilm biomass than that in PAO1 (Figure 1D). These traits indicated that PA_HN002 was associated with chronic urinary tract infection (UTI) and could serve as an important model to investigate molecular mechanisms underlying chronic adaptation of clinical P. aeruginosa strains.
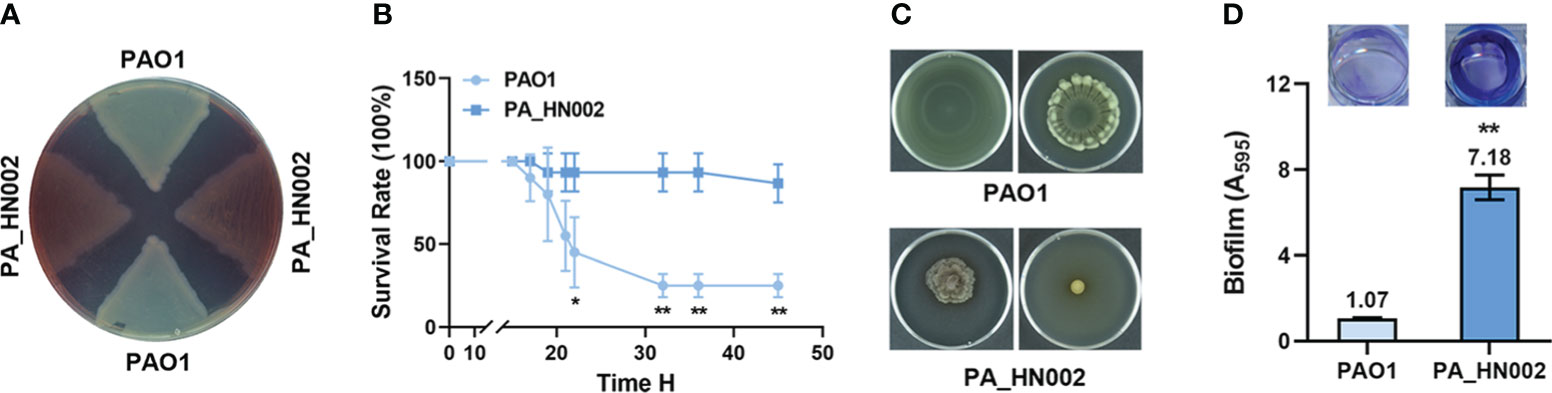
Figure 1 Phenotypic comparison between P. aeruginosa PAO1 and PA_HN002. (A) PA_HN002 produced red-colored pigments. (B) Pathogenicity of PAO1 and PA_HN002 was measured in a G. mellonella acute infection model. (C) Swimming (left) and swarming (right) motilities were measured in PAO1 and PA_HN002. Representative pictures of motilities for each strain were shown. (D) Biofilm formation of PAO1 and PA_HN002. *P<0.05, **P<0.01 based on Student’s t-test.
The high intracellular c-di-GMP level contributed to the transition from motile to sessile lifestyles in PA_HN002
Motile and sessile lifestyles are the dominant characteristics of acute and chronic infections in P. aeruginosa, respectively, which are regulated by a variety of cellular factors and dynamics of the intracellular second messenger c-di-GMP pool was known to play a critical role in the transition of these two lifestyles (Rasamiravaka et al., 2015; Valentini and Filloux, 2016; Ciofu and Tolker-Nielsen, 2019). As speculated, LC-MS measurement showed that the intracellular c-di-GMP content in PA_HN002 was about 3-fold higher than PAO1 (Figure 2A). To validate whether the attenuated motilities and strong biofilm phenotypes of PA_HN002 were ascribed to its high intracellular c-di-GMP content, we introduced two plasmids expressing two well-characterized c-di-GMP hydrolysis proteins BifA and W909_14950, respectively, into PA_HN002. BifA is a hybrid protein containing both a c-di-GMP biosynthesis and hydrolysis domains from PAO1 but only exhibits the degradation activity and the other enzyme W909_14950 from Dickeya zeae EC1 only contains a c-di-GMP hydrolysis domain (Kuchma Sherry et al., 2007; Chen et al., 2020b). Activities of both enzymes to reduce intracellular c-di-GMP content were confirmed in two recent studies (Zhou et al., 2021; Zhou et al., 2022). As shown in Figure 2A, introduction of either bifA or W909_14950 resulted in 4~5-fold decrease of the intracellular c-di-GMP content in PA_HN002. Recovered swimming and swarming motilities and inhibited biofilm formation were observed along with the decrease of intracellular c-di-GMP content (Figures 2B, C). These results demonstrated that high intracellular c-di-GMP content is the key in PA_HN002 to induce the transition from its motile lifestyle to the aggregated biofilm lifestyle.
Figure 2 PA_HN002 displayed a c-di-GMP-dependent transition of motile-sessile lifestyles. (A) LC-MS quantification of the intracellular c-di-GMP content in PAO1, PA_HN002 and PA_HN002 expressing plasmid-carried PDE genes bifA or W909_14950 (14950). Relative content was shown in the PA_HN002 strains compared to PAO1. (B) Swimming (left) and swarming (right) motilities of PA_HN002 and PA_HN002 with expression of the PDE genes bifA and W909_14950 (14950). (C) Biofilm formation of PA_HN002 and PA_HN002 with expression of the PDE genes bifA and W909_14950 (14950). **P<0.01, ***P<0.001 based on Student’s t-test in (A) and one-way ANOVA compared to the EV (empty vector) group in (C).
Whole genome sequencing and general genomic characteristics of PA_HN002
To understand the molecular basis of high c-di-GMP production in PA_HN002, genomic DNA of PA_HN002 was extracted and sequenced. Generally, a 6,380,366-bp genome with 66.33% GC content was assembled (Figure 3A), which shared a similar GC content but was ~0.12-Mb larger than the PAO1 genome (PAO1: 66.6% GC content and 6,26-Mb genome size). The PA_HN002 genome was predicted to contain 5,880 protein coding genes with 4,947 of them were annotated in COG (Clusters of Orthologous Groups of proteins). We compared COG annotations of PA_HN002 with those of PAO1 and another two widely used P. aeruginosa reference strains PA7, a non-respiratory human isolate from Argentina (Roy et al., 2010), and PA14, a highly virulent clinical isolate (He et al., 2004). A total of 26 COGs were exclusively present in PA_HN002 (Figure 3B). PubMLST analysis showed that PA_HN002 did not match any existing multiple locus sequence type (MLST), but with one variant allele from ST804 and ST907 which contained isolates from sputum and urinary tract infection, respectively, in Australia according to the allelic profiles. ST804 was noted as a predominant clonal complex in 1990s with an obvious phenotype of multidrug resistance (De Oliveira Santos et al., 2019), while the other one was lacking of documentation. PAst analysis showed that PA_HN002 and PAO1 belonged to the same serotype O5 which is prevalent in burn wound infections (Nasrin et al., 2022). To understand the phylogenetic relationship of PA_HN002 with other P. aeruginosa strains, we conducted a comprehensive phylogenetic analysis of PA_HN002 with 6,800 assembled genomes from GenBank (data not shown) and 172 genomes (Table S2) which included the most related 168 genomes to PA_HN002 and three reference genomes of PAO1, PA7 and PA14 were presented in Figure 4. It showed that PA_HN002 was closely related to six clinically isolated strains from Canada (AUS430: ST907), USA (Patient_3_Baseline: same ST as PA_HN002; VNMU089: ST218; MRSN5498: ST3014) and China (WCHPA075015: ST2968; WCHPA075017: ST2968).

Figure 3 General features of the PA_HN002 genome. (A) General features of the PA_HN002 genome including genome size, GC content, protein coding genes, RNA genes, MLST and serotype. (B) Venn diagram showing the number of shared and exclusive genes among PA_HN002, PAO1, PA7 and PA14. The numbers indicated the unique genes, shared genes among two, three or four strains based on the COG gene annotations.
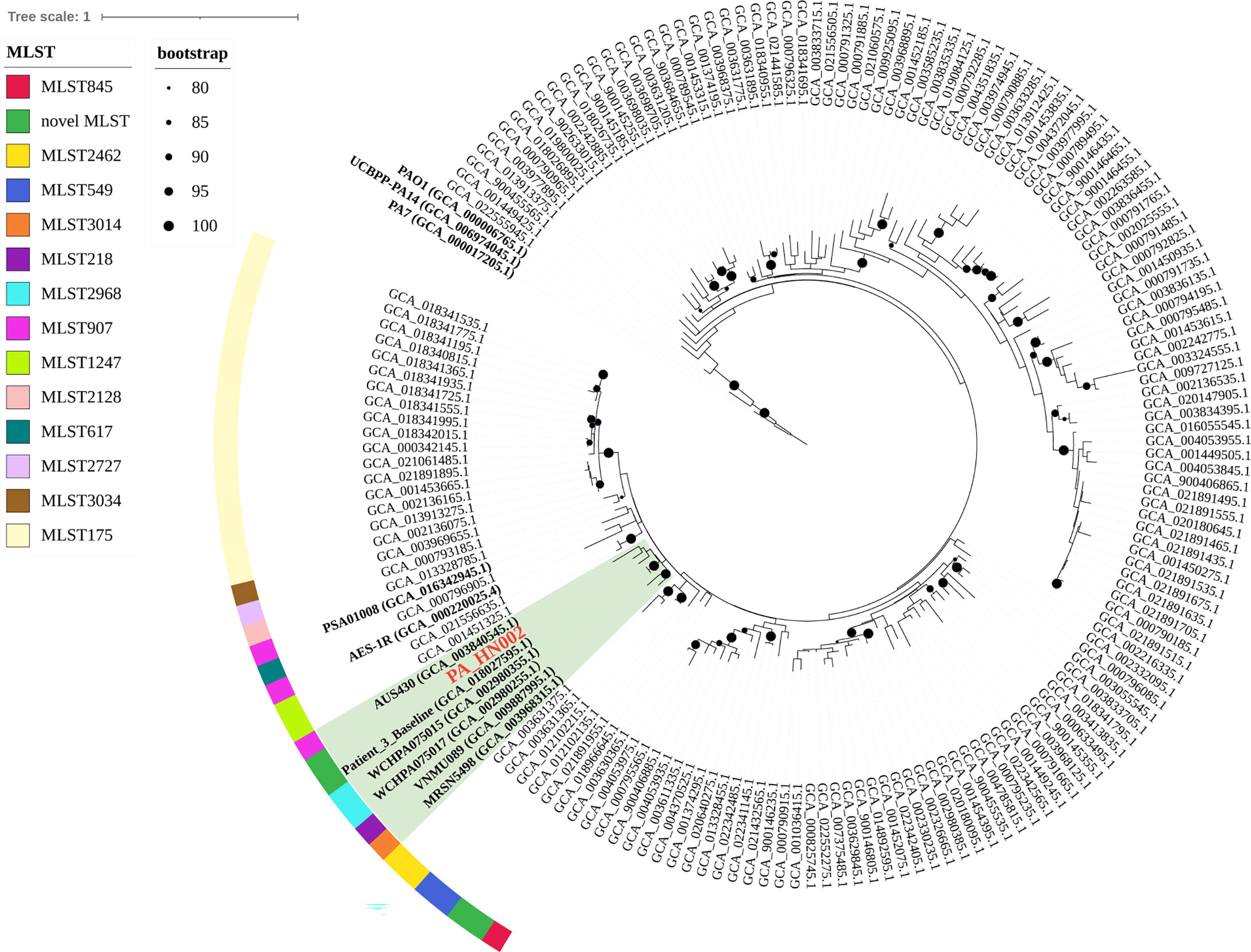
Figure 4 Phylogenetic position of PA_HN002. The phylogenetic tree was constructed based on 6,800 P. aeruginosa genomes from GenBank using PhyloPhlAn version 3.0 and the most related 168 genomes to PA_HN002 were presented. Three widely used reference genomes of PAO1, PA7 and PA14 were included and shown. Genomes located within the same clade as PA_HN002 were shaded in green. MLST types were shown in different colors for the genomes that were close to PA_HN002. Black dots on the nodes were bootstrap values and those greater than 80 were shown.
Mutations identified in the c-di-GMP metabolizing enzymes in PA_HN002 did not lead to its enhanced biofilm formation
Intracellular c-di-GMP level is modulated by the rate of two reactions. One is its synthesis by condensation of two guanosine triphosphate (GTP) molecules and the other is hydrolysis of it into 5′-phosphoguanylyl-(3′-5′)-guanosine (pGpG) or guanosine monophosphate (GMP). Synthesis of c-di-GMP is achieved by diguanylate cyclases (DGCs) which contain a characteristic conserved GGDEF domain that forms the active site of the enzymes while hydrolysis requires the PDEs which contain an EAL or an HD-GYP domain in the active site (Ryjenkov Dmitri et al., 2005; Schmidt Andrew et al., 2005; Tchigvintsev et al., 2010; Stelitano et al., 2013). In total 41 genes encoding the c-di-GMP metabolizing enzymes in PA_HN002 were predicted from its sequenced genome based on Pfam blast using the above-mentioned three domains and the reported c-di-GMP metabolizing enzymes in PAO1 (Table S3). A systematic comparative analysis of these genes between PA_HN002 and PAO1 was conducted, which revealed 10 amino acid substitutions in the GGDEF or EAL domains within six GGDEF and EAL hybrid enzymes (Figure 5A). Specifically, three mutations were found in the GGDEF domains (A448G in PA0861 (RbdA), I112V and A166S in PA3258) and seven mutations were found in the EAL domains (M476L and T509A in PA4959 (FimX), E514D in PA0285 (PipA), V456I and E535D in PA3258, E630Q in PA3311 (NbdA), L1085M in PA0575 (RmcA)). Interestingly, all these enzymes were reported to exhibit PDE activities and some of them have been demonstrated to be associated with the biofilm formation in P. aeruginosa (Kazmierczak et al., 2006; Monds et al., 2007; An et al., 2010; Li et al., 2013; Katharios-Lanwermeyer et al., 2021; Cai et al., 2022). For example, RbdA and NbdA have been reported as being important for biofilm dispersion and RmcA mutant displayed a great ability to initiate a robust biofilm (Li et al., 2013; Katharios-Lanwermeyer et al., 2021). PA3258 is a putative orthologue of RapA in P. fluorescens which was shown to be involved in c-di-GMP degradation and inhibition of biofilm formation in P. aeruginosa (Monds et al., 2007; Haddad et al., 2009). FimX is also a c-di-GMP binding protein promoting type IV pili assembly and twitching motility, which is involved in the process of biofilm development (O’toole and Kolter, 1998; Jain et al., 2017). PA0285 was recently renamed as PipA (Phage inducing phosphodiesterase A) owing to its function to positively regulate phage production in PAO1 in addition to inhibit biofilm formation (Cai et al., 2022).
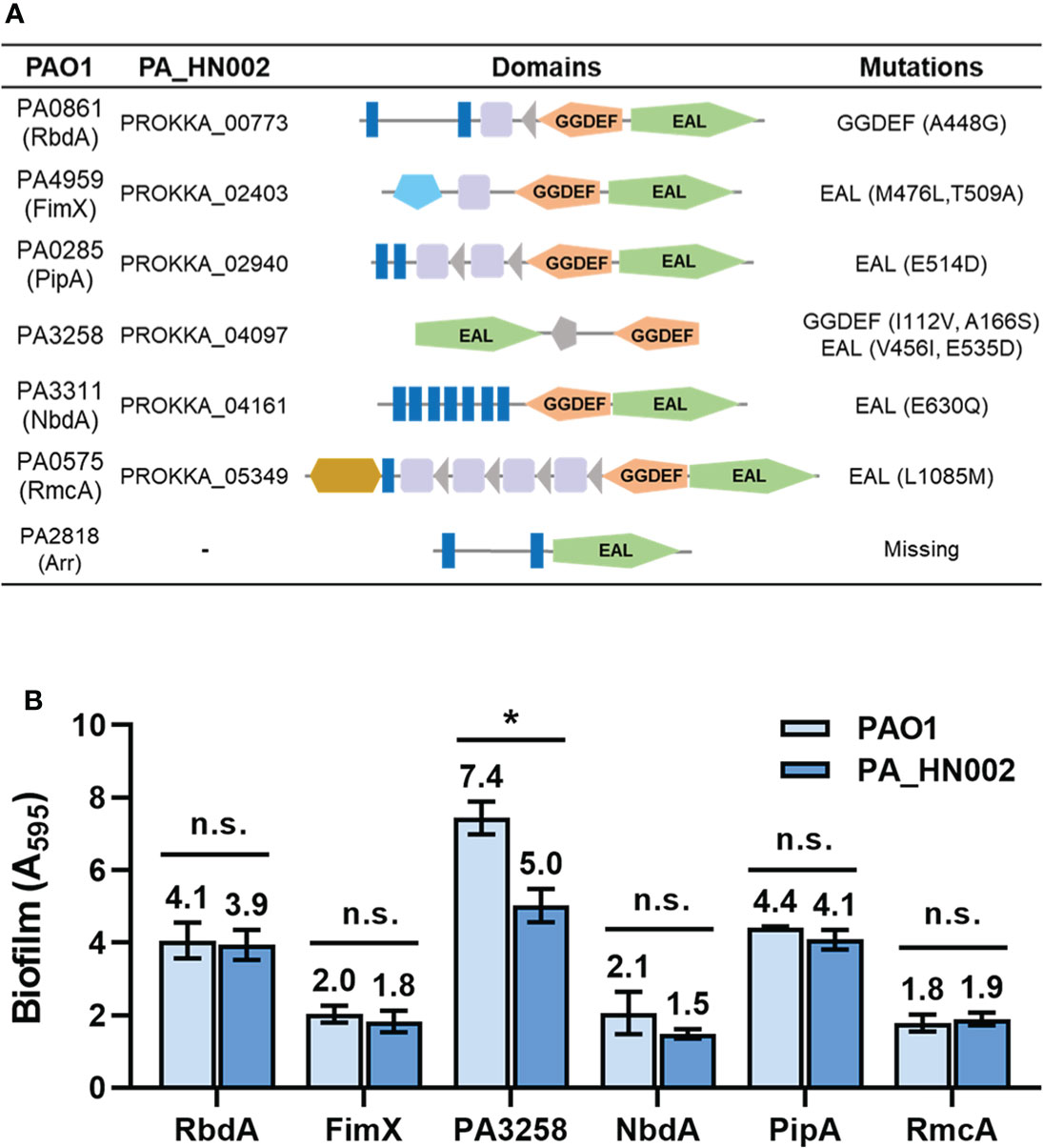
Figure 5 Mutations in the c-di-GMP metabolizing enzymes from PA_HN002 relative to those from PAO1. (A) A summary of mutations within c-di-GMP metabolizing enzymes PA0861 (RbdA), PA4959 (FimX), PA0285 (PipA), PA3258, PA3311 (NbdA), PA0575 (RmcA) and PA2818 (Arr) in PA_HN002 relative to the sequences in PAO1. (B) Biofilm formation of the PA_HN002 strain with overexpression of the PA0861 (RbdA), PA4959 (FimX), PA0285 (PipA), PA3258, PA3311 (NbdA), PA0575 (RmcA) encoding genes from PAO1 and PA_HN002, respectively. n.s., not significant, *P<0.05 based on Student’s t-test.
These mutations especially those in the EAL domains were speculated to have high possibilities of reducing the PDE activities and consequently enhancing biofilm formation. To verify whether these identified mutations contributed to the enhanced biofilm formation in PA_HN002, we cloned the six pairs of genes from PAO1 and PA_HN002, respectively, and overexpressed them in the PA_HN002 strain. If these enzymes were involved in reducing or inhibiting biofilm formation and the mutations were critical for their activity, biofilm formation in PA_HN002 should be greater when it overexpresses its endogenous enzyme compared to the homolog from the PAO1 strain. As shown in Figure 5B, overexpression of RbdA, FimX, NbdA, PipA and RmcA from both strains did not display significant differences on the biofilm biomass of PA_HN002, which suggested that mutations in these enzymes were not associated with the hyper-biofilm production in PA_HN002. Interestingly, overexpression of PA3258 from PA_HN002 showed significantly lower biofilm biomass than the PA3258 enzyme from PAO1, indicating the mutations presented in PA3258 of PA_HN002 indeed led to the inhibition of biofilm formation rather than the promotion of biofilm formation as we expected. Since mutations of I112V and A166S were located in the GGDEF domain, it was possible that PA3258 exhibited DGC activity and these two positions I112 and A166 were important for the enzymatic activity.
In addition to the substitutions of amino acids, it was noted that the entire PDE gene arr (PA2818) was absent in the PA_HN002 genome (Figure 5A). This gene together with its upstream sequence (Region-1: 289 bp of the PA2819 gene and 67 bp of the intergenic region between arr and PA2819) were replaced by a large 6,879-bp sequence (Region-2) containing 10 genes in PA_HN002 (Figure 6A). arr was reported only in a very limited number (5 of 20) of P. aeruginosa strains and supposed not to be a component of the P. aeruginosa core genome (Kulesekara et al., 2006). We also searched Arr orthologs based on its amino acids sequence within 4,955 genomes from different geographic regions and isolation sources in the Pseudomonas Genome Database using DIAMOND BLASTP (Buchfink et al., 2015; Winsor et al., 2016). Consistently, only 1155 (23.3%) hits of Arr were detected in these genomes (Figure 6B), indicating the rare distribution of Arr in the clinically and environmentally relevant strains. We next sought to figure out whether missing of Arr in PA_HN002 led to its high intracellular c-di-GMP content and strong biofilm formation. We cloned the whole Region-1 (Figure 6A) from the PAO1 genome but found that introduction of this sequence into PA_HN002 to complement Arr expression did not affect the intracellular c-di-GMP content as well as the biofilm biomass (Figures 6C, D). Further overexpression of Arr with a constitutive promoter Plac did not alter the biofilm biomass of PA_HN002 as well (Figure 6D). These results indicated that higher intracellular c-di-GMP content and biofilm formation found in PA_HN002 than PAO1 was not owing to the absence of Arr and the occurrence of identified mutations in the c-di-GMP metabolizing enzymes.
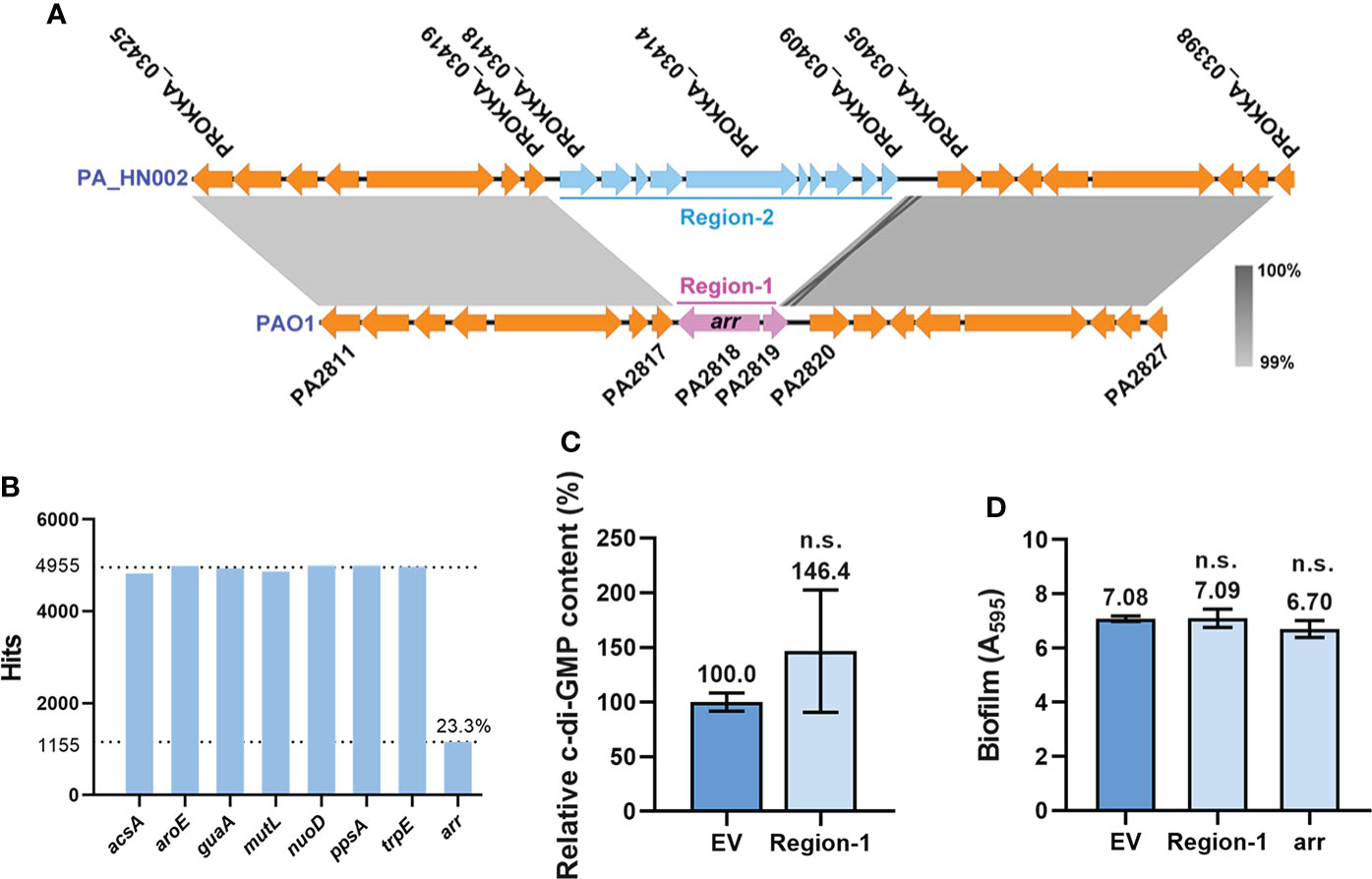
Figure 6 Loss of Arr did not affect intracellular c-di-GMP content and biofilm formation. (A) Replacement of the arr gene-containing region (Region-1) by a fragment containing 10 coding sequences (Region-2) was detected in the PA_HN002 genome. The light, gray-shaded regions showed more than 99% sequence identity. (B) Abundance of the arr gene in 4,955 P. aeruginosa genomes. A set of house-keeping genes acsA, aroE, guaA, mutL, nuoD, ppsA and trpE used for MLST analysis were selected as controls to show their distributions in almost 100% genomes. (C) Relative intracellular c-di-GMP content measured by LC-MS in the PA_HN002 strain complemented with the fragment of Region-1 from PAO1. (D) Biofilm formation of the PA_HN002 strain and PA_HN002 with the complementation of the Region-1 and overexpression of the arr gene. n.s., not significant based on Student’s t-test. EV, empty vector.
Differentially expressed c-di-GMP metabolizing enzymes in PA_HN002 compared to PAO1
Besides the mutations present in the c-di-GMP hydrolysis genes, an increased level of c-di-GMP biosynthesis and a reduced level of c-di-GMP hydrolysis owing to the changed expression levels of DGC and PDE genes represent additional important factors causing accumulation of intracellular c-di-GMP. To evaluate the expression level of enzymes involved in c-di-GMP metabolism, relative expression of all the c-di-GMP metabolizing genes between PAO1 and PA_HN002 was measured by RT-qPCR. Genes in PA_HN002 with more than 2-fold transcriptional changes relative to their homologs in PAO1, i.e. > 2-fold upregulation of DGC genes and downregulation of PDE genes, were selected for further discussion. For the GGDEF domain-containing DGCs, only the PA0169 (SiaD) was expressed 4.4-fold higher in PA_HN002 than that in PAO1 (Figure 7A). Overexpression of SiaD to increase intracellular c-di-GMP content and enhance biofilm formation was demonstrated recently (Zhou et al., 2021). For the EAL or HD-GYP domain-containing PDEs, four enzymes PA2567, PA2752, PA4108 and PA4781 in PA_HN002 besides Arr were expressed at lower levels which were 30%, 10%, 20% and 10% of that in PAO1 (Figure 7B). PA2567, PA4108 and PA4781 were confirmed enzymes with PDE activity while PA2752 did not exhibit PDE activity but was implicated with biofilm formation (Ryan et al., 2009; Kimura et al., 2017). For enzymes containing both GGDEF and EAL domains, PA0861 (RbdA) and PA5017 (DipA) in PA_HN002 were expressed at more than 2-fold lower than that in PAO1 (Figure 7C). Both enzymes were demonstrated to exhibit PDE activity (An et al., 2010; Roy Ankita et al., 2012). These results showed a dramatically different expression profile of c-di-GMP metabolizing enzymes between PA_HN002 and PAO1, which might be the major cause of the elevated c-di-GMP content and the strong biofilm formation in PA_HN002.
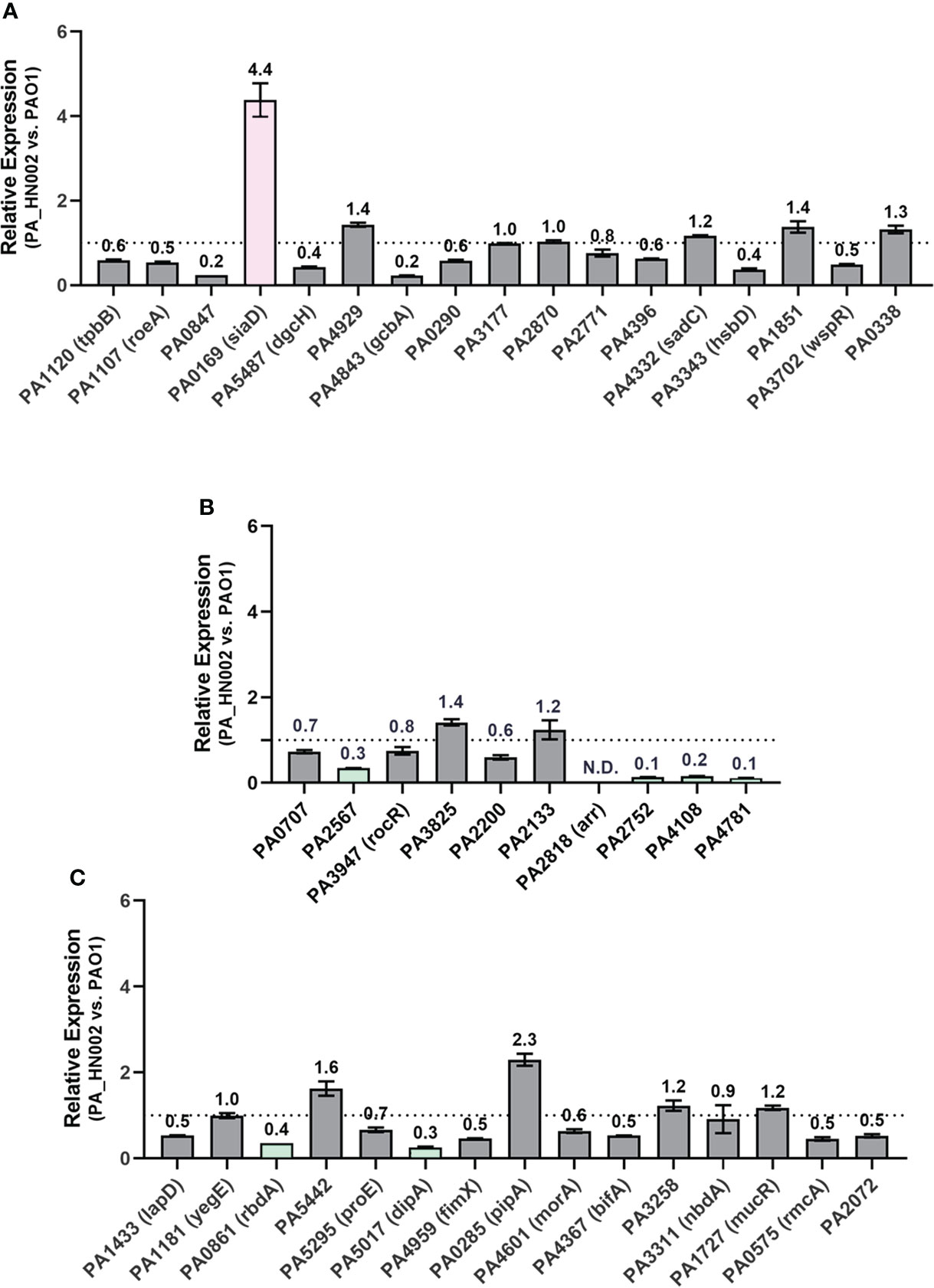
Figure 7 Relative expression of the genes encoding c-di-GMP metabolizing enzymes in PA_HN002 and PAO1. (A) Enzymes only contain the GGDEF domain. (B) Enzymes contain either EAL or HY-GYP domain. (C) Enzymes contain both the GGDEF and EAL domains. The DGC-encoding gene siaD in PA_HN002 was highlighted in pink while the genes encoding PDE enzymes with more than 2-fold changes such as PA2567, PA4108, PA4781, PA0861 (rbdA), PA5017 (dipA) and PA0285 were highlighted in green. N.D., Not detected.
Mobile genetic elements and unique COGs in PA_HN002
We next sought to investigate the factors altering the expression pattern of c-di-GMP metabolizing enzymes. P. aeruginosa genome is highly plastic and complex. It frequently incorporates exogenous sequences by horizontal gene transfers into its genome. These MGEs including genomic islands (GIs), plasmids, integrative and conjugative elements (ICEs), integrative and mobilizable elements (IMEs), insertion sequences (ISs), prophages etc. comprise the accessory genome of P. aeruginosa (Shen et al., 2006; Silby et al., 2011; Ozer et al., 2014; Subedi et al., 2018). These elements were considered as a major group of contributors to P. aeruginosa genome evolution by introducing new catabolic pathways, encoding novel regulators, or disrupting endogenous gene functions and regulatory circuits, conferring the pathogen specific phenotypes to adapt to diverse environmental conditions through shifting the gene expression profile.
To facilitate the understanding of specific regulatory mechanisms of c-di-GMP metabolism in PA_HN002, we analyzed MGEs presented in its genome. Firstly, twenty GIs were identified in the PA_HN002 genome and some genes in the GIs were assigned to functional groups related to gene regulation and signal sensing (Table S4). There was an IME element found in the PA_HN002 genome which contained 8 coding genes therein. In addition to an integrase and a relaxase, others were hypothetic proteins. Sequence alignment suggested a DNA-binding protein, a RES domain-containing protein, a sce7725 family protein and a sce7726 family protein therein (Figure S2). In total five complete ISs, namely ISPa22, ISPa6, ISPa1, ISPa57 and ISPa32, were found in the genome of PA_HN002 (Table S5). Notably, multiple incomplete copies of IS222 were found at the end of several contigs (data not shown). PHASTER analysis revealed four potential prophages with 47.8, 36.2, 32.3 and 7.7-kb in length (Table S6). Next, genes with unique COG annotations in the PA_HN002 genome compared to PAO1 were summarized, showing that 54 COGs were identified exclusively in PA_HN002 and they were basically from the MGEs (Table 1). Together, presence of the abundant MGEs and unique COGs might account for the changed expression profile of c-di-GMP metabolizing enzymes and strong biofilm formation in PA_HN002.
Discussion
P. aeruginosa is a highly motile opportunistic pathogen. Its motility is a key determinant for its colonization within the host, which promotes the establishment of numerous acute infections at different human tissues. Loss of motility and inducing the pathogen into a sessile-biofilm lifestyle reflect an adaptative process under human immune pressures or anti-infective interventions (Balloy et al., 2007). The structured biofilm is extremely resistant to antimicrobial-based treatments and eradication of the biofilm pathogen is almost impossible. Unfortunately, genetic determinants that drive the formation of biofilms in clinically significant strains remain largely unknown, which impedes the precise control and prevention of chronic infections in hospitals. PA_HN002 isolated in this study is a representative adapted strain of chronic urinary tract infection, which was phylogenetically close to the strains isolated from different countries such as China, Canada and USA. This strain provided us an important model to investigate how P. aeruginosa shifted its lifestyle to persist in the host. Some previous studies have reported that loss of the flagellar structural proteins or the stator complexes in the flagellar motor is an adaptive strategy conferring P. aeruginosa resistance to host phagocytosis by neutrophils and macrophages (Amiel et al., 2010; Lovewell et al., 2011). Here, we further demonstrated in PA_HN002 that the increased level of the second messenger c-di-GMP played a critical role in the transition from its motile lifestyle to the sessile biofilm lifestyle. Instead of the complete abolishment of swimming and swarming owing to the loss of functions of flagellar biosynthetic genes, the motile lifestyle in PA_HN002 could be reversed when its intracellular concentration of c-di-GMP was decreased.
High-throughput sequencing technique has facilitated the in-depth characterization of bacterial genetic determinants that contribute to physiological traits or biological processes with interests in the past decades. With the high-throughput genome sequencing technique, we identified a list of mutations in the c-di-GMP metabolizing enzymes in PA_HN002. The potential effects of these mutations on biofilm formation were examined and, however, we found that none of them was associated with the enhanced biofilm formation in PA_HN002. Intracellular c-di-GMP dynamics has been extensively investigated in the past decade and numerous environmental cues or cellular regulatory pathways that modulate intracellular c-di-GMP content have been revealed. For example, Proteins encoded by siaABCD regulated c-di-GMP production and biofilm formation in response to the toxic surfactant sodium dodecylsulfate (SDS) in P. aeruginosa (Colley et al., 2016; Chen et al., 2020a). We also found recently that the two-component regulatory system FleS/FleR controlled biofilm formation and type VI secretion system by modulating the intracellular c-di-GMP content (Zhou et al., 2021; Zhou et al., 2022). These achievements greatly advanced our understanding in the adaptive mechanisms of P. aeruginosa and highlighted the importance of the signaling molecule c-di-GMP during this adaptative process by controlling the transition of lifestyles between motility and sessility in P. aeruginosa.
Quantification of the c-di-GMP metabolizing gene expression in PA_HN002 showed lots of them were differentially expressed compared to their homologs in PAO1. Most of these genes with changed expression level were previously confirmed to play roles in regulating biofilm formation. Noticeably, SiaD was substantially overexpressed in PA_HN002. A comprehensive analysis of overexpressing DGC-encoding genes in PAO1 revealed that SiaD and another enzyme SadC were two dominant DGCs to produce remarkably more c-di-GMP than other DGCs (Bhasme et al., 2020). These results together indicated a plausible connection between the overexpression of SiaD and the high intracellular c-di-GMP content in PA_HN002. In addition to Arr, putative PDEs such as PA2567, PA2752, PA4108 and PA4781 were expressed at lower levels than their homologs in PAO1. PA2567 is an azithromycin responsive EAL domain-containing PDE which was repressed in the presence of azithromycin and deletion of PA2567 has been reported to reduce the quorum sensing activity and virulence of the P. aeruginosa strain (Kimura et al., 2017). PA4108 and PA4781 are two PDEs that contain a HD-GYP domain rather than the EAL domain. The role of these two PDEs in c-di-GMP hydrolysis was confirmed because mutations of PA4108 and PA4781 led to an increase in the level of c-di-GMP in P. aeruginosa (Ryan et al., 2009). Strains with PA4108 and PA4781 mutations showed different biofilm architectures but, interestingly, mutation of PA4108 led to a reduced biofilm biomass (Ryan et al., 2009). PA2572 encodes a non-canonical variant YN-GYP which was reported to display undetectable activity in c-di-GMP degradation but was associated with biofilm formation in P. aeruginosa (Ryan et al., 2009). Two more enzymes PA0861 (RbdA) and PA5017 (DipA) containing both the GGDEF and EAL domains but only exhibiting PDE activity were found differentially expressed with more than 2-fold lower compared to their homologs in PAO1 (An et al., 2010; Roy Ankita et al., 2012). Techniques to precisely control the expression of these c-di-GMP metabolizing enzymes and efficiently screen their regulators are highly desired for the in-depth understanding of the special regulation of c-di-GMP metabolism in PA_HN002 and other clinical strains.
P. aeruginosa possesses a complicated gene regulatory network to exquisitely control its virulence as well as its survival in response to the diverse environmental conditions. This is heavily dependent on its large genome and the abundant transcriptional factors encoded from it. It was estimated that there are about 600 transcriptional factors encoded from a P. aeruginosa genome with many of them are associated with the dynamics of intracellular c-di-GMP content (Stover et al., 2000). Moreover, a P. aeruginosa genome is generally composed of a large percentage of accessory genome which includes a large number of MGEs acquired by horizontal gene transfer from different sources (Ozer et al., 2014). These elements and genes therein are known to impact the native expression of host genes as well as the host virulence and antibiotic resistance. In PA_HN002, a set of MGEs and genes with unique COG annotations were identified in its genome. In the future, targeted molecular dissections to unveil the mechanisms on the regulation of c-di-GMP metabolizing enzymes and evolutionary analysis on the transmission of MGEs may provide more clinically relevant insights into the pathoadaptation of P. aeruginosa causing chronic infections and help to identify new targets for precise anti-pseudomonal treatments.
Arr is a membrane-associated PDE which was initially found to be essential for aminoglycoside-dependent induction of biofilm formation and reported to promote biofilm formation during the DNA replication stress (Hoffman et al., 2005; Gotoh et al., 2008). These were contradictory to the general concept that PDEs function to reduce c-di-GMP content and therefore prevent biofilm formation (Ha et al., 2015). We initially speculated that loss of Arr in PA_HN002 was a contributor to its high intracellular c-di-GMP content and the hyper-biofilm. However, expression of arr in the PA_HN002 strain did not influence its c-di-GMP content. This result suggested that Arr might not be involved in c-di-GMP metabolism despite it contains an EAL domain. Moreover, inconsistent with the previous reports that Arr promoted biofilm formation, we did not observe increased biofilm formation in PA_HN002 with the expression of Arr. This discrepancy indicated that Arr could be a stress-responsive regulator since our examinations were performed under non-stressed conditions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, JAMXFG000000000.
Author contributions
ZX designed the study; SL, SC, HC, TL, MH, LLiao performed experiments; LLi isolated the clinical strain; SL, SC drafted the manuscript; LLi, L-HZ, ZX revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Guangzhou Basic and Applied Basic Research Foundation (No. 202201010613), National Natural Science Foundation of China (No. 32100020) and Guangdong Basic and Applied Basic Research Foundation (No. 2022A1515010194).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.956445/full#supplementary-material
References
Ahator, S. D., Zhang, L. (2019). Small is mighty-chemical communication systems in Pseudomonas aeruginosa. Annu. Rev. Microbiol.73, 559–578. doi: 10.1146/annurev-micro-020518-120044
Almblad, H., Rybtke, M., Hendiani, S., Andersen, J. B., Givskov, M., Tolker-Nielsen, T. (2019). High levels of cAMP inhibit Pseudomonas aeruginosa biofilm formation through reduction of the c-di-GMP content. Microbiology165, 324–333. doi: 10.1099/mic.0.000772
Alonso, B., Fernández-Barat, L., Di Domenico, E. G., Marín, M., Cercenado, E., Merino, I., et al. (2020). Characterization of the virulence of Pseudomonas aeruginosa strains causing ventilator-associated pneumonia. BMC Infect. Dis.20, 909. doi: 10.1186/s12879-020-05534-1
Amiel, E., Lovewell Rustin, R., O'toole George, A., Hogan Deborah, A., Berwin, B. (2010). Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression. Infect. Immun.78, 2937–2945. doi: 10.1128/IAI.00144-10
An, S., Wu, J. E., Zhang, L.-H. (2010). Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol.76, 8160–8173. doi: 10.1128/AEM.01233-10
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res.44, W16–W21. doi: 10.1093/nar/gkw387
Asnicar, F., Thomas, A. M., Beghini, F., Mengoni, C., Manara, S., Manghi, P., et al. (2020). Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun.11, 2500. doi: 10.1038/s41467-020-16366-7
Balloy, V., Verma, A., Kuravi, S., Si-Tahar, M., Chignard, M., Ramphal, R. (2007). The role of flagellin versus motility in acute lung disease caused by Pseudomonas aeruginosa. J. Infect. Dis.196, 289–296. doi: 10.1086/518610
Bertelli, C., Laird, M. R., Williams, K. P., Simon fraser university research computing group, Lau, B. Y., Hoad, G., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res.45, W30–W35. doi: 10.1093/nar/gkx343
Bhasme, P., Wei, Q., Xu, A., Naqvi, S. T. A., Wang, D., Ma, L. Z. (2020). Evaluation and characterization of the predicted diguanylate cyclase-encoding genes in Pseudomonas aeruginosa. MicrobiologyOpen9, e975. doi: 10.1002/mbo3.975
Breidenstein, E. B. M., de la Fuente-Núñez, C., Hancock, R. E. W. (2011). Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol.19, 419–426. doi: 10.1016/j.tim.2011.04.005
Buchfink, B., Xie, C., Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods12, 59–60. doi: 10.1038/nmeth.3176
Burrows, L. L. (2012). Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu. Rev. Microbiol.66, 493–520. doi: 10.1146/annurev-micro-092611-150055
Cai, Y. M., Yu, K. W., Liu, J. H., Cai, Z., Zhou, Z.-H., Liu, Y., et al. (2022). The c-di-GMP phosphodiesterase PipA (PA0285) regulates autoaggregation and Pf4 bacteriophage production in Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol.88, e00039–e00022. doi: 10.1128/aem.00039-22
Cao, H., Lai, Y., Bougouffa, S., Xu, Z., Yan, A. (2017). Comparative genome and transcriptome analysis reveals distinctive surface characteristics and unique physiological potentials of Pseudomonas aeruginosa ATCC 27853. BMC Genomics18, 459. doi: 10.1186/s12864-017-3842-z
Cao, H., Xia, T., Li, Y., Xu, Z., Bougouffa, S., Lo Yat, K., et al. (2019). Uncoupled quorum sensing modulates the interplay of virulence and resistance in a multidrug-resistant clinical Pseudomonas aeruginosa isolate belonging to the MLST550 clonal complex. Antimicrob. Agents Chemother.63, e01944–e01918. doi: 10.1128/AAC.01944-18
Chang, C. Y. (2018). Surface sensing for biofilm formation in Pseudomonas aeruginosa. Front. Microbiol.8, 2671. doi: 10.3389/fmicb.2017.02671
Chen, G., Gan, J., Yang, C., Zuo, Y., Peng, J., Li, M., et al. (2020a). The SiaA/B/C/D signaling network regulates biofilm formation in Pseudomonas aeruginosa. EMBO J.39, e103412. doi: 10.15252/embj.2020105997
Chen, Y., Lv, M., Liang, Z., Liu, Z., Zhou, J., Zhang, L. H. (2022). Cyclic di-GMP modulates sessile-motile phenotypes and virulence in Dickeya oryzae via two PilZ domain receptors. Mol. Plant Pathol.23, 870–884. doi: 10.1111/mpp.13200
Chen, Y., Zhou, J., Lv, M., Liang, Z., Parsek Matthew, R., Zhang, L. H. (2020b). Systematic analysis of c-di-GMP signaling mechanisms and biological functions in Dickeya zeae EC1. mBio11, e02993–e02920. doi: 10.1128/mBio.02993-20
Ciofu, O., Tolker-Nielsen, T. (2019). Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents–how P. aeruginosa can escape antibiotics. Front. Microbiol.10, 913. doi: 10.3389/fmicb.2019.00913
Colley, B., Dederer, V., Carnell, M., Kjelleberg, S., Rice, S. A., Klebensberger, J. (2016). SiaA/D interconnects c-di-GMP and RsmA signaling to coordinate cellular aggregation of Pseudomonas aeruginosa in response to environmental conditions. Front. Microbiol.7, 179. doi: 10.3389/fmicb.2016.00179
De Oliveira Santos, I. C., Pereira De Andrade, N. F., Da Conceição Neto, O. C., Da Costa, B. S., De Andrade Marques, E., Rocha-De-Souza, C. M., et al. (2019). Epidemiology and antibiotic resistance trends in clinical isolates of Pseudomonas aeruginosa from Rio de janeiro - Brazil: Importance of mutational mechanisms over the years, (1995–2015). Infect. Genet. Evol.73, 411–415. doi: 10.1016/j.meegid.2019.05.015
Dong, Y., Zhang, X.-F., An, S.-W., Xu, J.-L., Zhang, L.-H. (2008). A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun. Integr. Biol.1, 88–96. doi: 10.4161/cib.1.1.6717
Gotoh, H., Zhang, Y., Dallo, S. F., Hong, S., Kasaraneni, N., Weitao, T. (2008). Pseudomonas aeruginosa, under DNA replication inhibition, tends to form biofilms via arr. Res. Microbiol.159, 294–302. doi: 10.1016/j.resmic.2008.02.002
Haddad, A., Jensen, V., Becker, T., Häussler, S. (2009). The pho regulon influences biofilm formation and type three secretion in Pseudomonas aeruginosa. Environ. Microbiol. Rep.1, 488–494. doi: 10.1111/j.1758-2229.2009.00049.x
Ha, D.-G., Kuchma, S. L., O’toole, G. A. (2014a). “"Plate-based assay for swarming motility in pseudomonas aeruginosa,",” in Pseudomonas methods and protocols. Eds. Filloux, A., Ramos, J.-L. (New York, NY: Springer New York), 67–72.
Ha, D.-G., Kuchma, S. L., O’toole, G. A. (2014b). “"Plate-based assay for swimming motility in pseudomonas aeruginosa,",” in Pseudomonas methods and protocols. Eds. Filloux, A., Ramos, J.-L. (New York, NY: Springer New York), 59–65.
Hancock, R. E. W., Speert, D. P. (2000). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updates3, 247–255. doi: 10.1054/drup.2000.0152
Ha, D.-G., O'toole George, A., Ghannoum, M., Parsek, M., Whiteley, M., Mukherjee, P. (2015). C-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr.3, MB–0003-2014. doi: 10.1128/9781555817466.ch15
He, J., Baldini Regina, L., Déziel, E., Saucier, M., Zhang, Q., Liberati Nicole, T., et al. (2004). The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci.101, 2530–2535. doi: 10.1073/pnas.0304622101
Hoffman, L. R., D'argenio, D. A., Maccoss, M. J., Zhang, Z., Jones, R. A., Miller, S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature436, 1171–1175. doi: 10.1038/nature03912
Jain, R., Sliusarenko, O., Kazmierczak, B. I. (2017). Interaction of the cyclic-di-GMP binding protein FimX and the type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog.13, e1006594. doi: 10.1371/journal.ppat.1006594
Jolley, K., Bray, J., Maiden, M. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res.3, 124. doi: 10.12688/wellcomeopenres.14826.1
Josenhans, C., Suerbaum, S. (2002). The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol.291, 605–614. doi: 10.1078/1438-4221-00173
Katharios-Lanwermeyer, S., Whitfield, G. B., Howell, P. L., O’toole, G. A. (2021). Pseudomonas aeruginosa uses c-di-GMP phosphodiesterases RmcA and MorA to regulate biofilm maintenance. mBio12, e03384–e03320. doi: 10.1128/mBio.03384-20
Kazmierczak, B. I., Lebron, M. B., Murray, T. S. (2006). Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol.60, 1026–1043. doi: 10.1111/j.1365-2958.2006.05156.x
Kearns Daniel, B., Robinson, J., Shimkets Lawrence, J. (2001). Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J. Bacteriol.183, 763–767. doi: 10.1128/JB.183.2.763-767.2001
Kimura, S., Mori, N., Kai, T., Ishii, Y., Yamaguchi, K., Tateda, K. (2017). Azithromycin modulates 3', 5'-cyclic diguanylic acid signaling in Pseudomonas aeruginosa. J. Infect. Chemother.23, 550–555. doi: 10.1016/j.jiac.2017.05.002
Klockgether, J., Cramer, N., Wiehlmann, L., Davenport, C., Tümmler, B. (2011). Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol.2, 150. doi: 10.3389/fmicb.2011.00150
Kuchma Sherry, L., Brothers Kimberly, M., Merritt Judith, H., Liberati Nicole, T., Ausubel Frederick, M., O'toole George, A. (2007). BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol.189, 8165–8178. doi: 10.1128/JB.00586-07
Kühn, M. J., Talà, L., Inclan, Y. F., Patino, R., Pierrat, X., Vos, I., et al. (2021). Mechanotaxis directs Pseudomonas aeruginosa twitching motility. Proc. Natl. Acad. Sci.118, e2101759118. doi: 10.1073/pnas.2101759118
Kulesekara, H., Lee, V., Brencic, A., Liberati, N., Urbach, J., Miyata, S., et al. (2006). Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci.103, 2839–2844. doi: 10.1073/pnas.0511090103
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293–W296. doi: 10.1093/nar/gkab301
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res.49, D458–D460. doi: 10.1093/nar/gkaa937
Li, Y., Heine, S., Entian, M., Sauer, K., Frankenberg-Dinkel, N. (2013). NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by an MHYT domain-coupled phosphodiesterase. J. Bacteriol.195, 3531–3542. doi: 10.1128/JB.01156-12
Litwin, A., Rojek, S., Gozdzik, W., Duszynska, W. (2021). Pseudomonas aeruginosa device associated – healthcare associated infections and its multidrug resistance at intensive care unit of university hospital: polish, 8.5-year, prospective, single-centre study. BMC Infect. Dis.21, 180. doi: 10.1186/s12879-021-05883-5
Liu, M., Li, X., Xie, Y., Bi, D., Sun, J., Li, J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res.47, D660–D665. doi: 10.1093/nar/gky1123
Li, K., Yang, G., Debru, A. B., Li, P., Zong, L., Li, P., et al. (2017). SuhB regulates the motile-sessile switch in Pseudomonas aeruginosa through the Gac/Rsm pathway and c-di-GMP signaling. Front. Microbiol.8, 1045. doi: 10.3389/fmicb.2017.01045
Li, D., Zhang, L., Liang, J., Deng, W., Wei, Q., Wang, K. (2021). Biofilm formation by Pseudomonas aeruginosa in a novel septic arthritis model. Front. Cell. Infect. Microbiol.11, 724113. doi: 10.3389/fcimb.2021.724113
Lovewell, R. R., Collins, R. M., Acker, J. L., O'toole, G. A., Wargo, M. J., Berwin, B. (2011). Step-wise loss of bacterial flagellar torsion confers progressive phagocytic evasion. PLoS Pathog.7, e1002253. doi: 10.1371/journal.ppat.1002253
Mah, T. F., Pitts, B., Pellock, B., Walker, G. C., Stewart, P. S., O'toole, G. A. (2003). A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature426, 306–310. doi: 10.1038/nature02122
Mann, E. E., Wozniak, D. J. (2012). Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev.36, 893–916. doi: 10.1111/j.1574-6976.2011.00322.x
Massouras, A., Hens, K., Gubelmann, C., Uplekar, S., Decouttere, F., Rougemont, J., et al. (2010). Primer-initiated sequence synthesis to detect and assemble structural variants. Nat. Methods7, 485–486. doi: 10.1038/nmeth.f.308
Monds, R. D., Newell, P. D., Gross, R. H., O'toole, G. A. (2007). Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol.63, 656–679. doi: 10.1111/j.1365-2958.2006.05539.x
Moradali, M. F., Ghods, S., Rehm, B. H. A. (2017). Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol.7, 39. doi: 10.3389/fcimb.2017.00039
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., Pardesi, K. R. (2019). Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol.10, 539. doi: 10.3389/fmicb.2019.00539
Mulcahy, L. R., Isabella, V. M., Lewis, K. (2014). Pseudomonas aeruginosa biofilms in disease. Microbial. Ecol.68, 1–12. doi: 10.1007/s00248-013-0297-x
Nadalin, F., Vezzi, F., Policriti, A. (2012). GapFiller: a de novo assembly approach to fill the gap within paired reads. BMC Bioinf.13, S8. doi: 10.1186/1471-2105-13-S14-S8
Nasrin, S., Hegerle, N., Sen, S., Nkeze, J., Sen, S., Permala-Booth, J., et al. (2022). Distribution of serotypes and antibiotic resistance of invasive Pseudomonas aeruginosa in a multi-country collection. BMC Microbiol.22, 13. doi: 10.1186/s12866-021-02427-4
O'toole, G. A., Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol.30, 295–304. doi: 10.1046/j.1365-2958.1998.01062.x
Orlandi, V. T., Bolognese, F., Chiodaroli, L., Tolker-Nielsen, T., Barbieri, P. (2015). Pigments influence the tolerance of Pseudomonas aeruginosa PAO1 to photodynamically induced oxidative stress. Microbiology161, 2298–2309. doi: 10.1099/mic.0.000193
Ozer, E. A., Allen, J. P., Hauser, A. R. (2014). Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools spine and AGEnt. BMC Genomics15, 737. doi: 10.1186/1471-2164-15-737
Pang, Z., Raudonis, R., Glick, B. R., Lin, T. J., Cheng, Z. (2019). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv.37, 177–192. doi: 10.1016/j.biotechadv.2018.11.013
Porter, S. L., Wadhams, G. H., Armitage, J. P. (2011). Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol.9, 153–165. doi: 10.1038/nrmicro2505
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., Korobeynikov, A. (2020). Using SPAdes De novo assembler. Curr. Protoc. Bioinf.70, e102. doi: 10.1002/cpbi.102
Rasamiravaka, T., Labtani, Q., Duez, P., El Jaziri, M. (2015). The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. BioMed. Res. Int.2015, 759348. doi: 10.1155/2015/759348
Rodríguez-Rojas, A., Mena, A., Martín, S., Borrell, N., Oliver, A., Blázquez, J. (2009). Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology155, 1050–1057. doi: 10.1099/mic.0.024745-0
Rossi, E., La Rosa, R., Bartell, J. A., Marvig, R. L., Haagensen, J., Sommer, L. M., et al. (2021). Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol.19, 331–342. doi: 10.1038/s41579-020-00477-5
Roy Ankita, B., Petrova Olga, E., Sauer, K. (2012). The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol.194, 2904–2915. doi: 10.1128/JB.05346-11
Roy, P. H., Tetu, S. G., Larouche, A., Elbourne, L., Tremblay, S., Ren, Q., et al. (2010). Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One5, e8842. doi: 10.1371/journal.pone.0008842
Ryan, R. P., Lucey, J., O'donovan, K., Mccarthy, Y., Yang, L., Tolker-Nielsen, T., et al. (2009). HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ. Microbiol.11, 1126–1136. doi: 10.1111/j.1462-2920.2008.01842.x
Ryjenkov Dmitri, A., Tarutina, M., Moskvin Oleg, V., Gomelsky, M. (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J. Bacteriol.187, 1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005
Sakuragi, Y., Kolter, R. (2007). Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J. Bacteriol.189, 5383–5386. doi: 10.1128/JB.00137-07
Schmidt Andrew, J., Ryjenkov Dmitri, A., Gomelsky, M. (2005). The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: Enzymatically active and inactive EAL domains. J. Bacteriol.187, 4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc.3, 1101–1108. doi: 10.1038/nprot.2008.73
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, K., Sayeed, S., Antalis, P., Gladitz, J., Ahmed, A., Dice, B., et al. (2006). Extensive genomic plasticity in Pseudomonas aeruginosa revealed by identification and distribution studies of novel genes among clinical isolates. Infect. Immun.74, 5272–5283. doi: 10.1128/IAI.00546-06
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res.34, D32–D36. doi: 10.1093/nar/gkj014
Silby, M. W., Winstanley, C., Godfrey, S., Levy, S. B., Jackson, R. W. (2011). Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev.35, 652–680. doi: 10.1111/j.1574-6976.2011.00269.x
Si, M., Zhao, C., Burkinshaw, B., Zhang, B., Wei, D., Wang, Y., et al. (2017). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci.114, E2233–E2242. doi: 10.1073/pnas.1614902114
Stelitano, V., Giardina, G., Paiardini, A., Castiglione, N., Cutruzzolà, F., Rinaldo, S. (2013). C-di-GMP hydrolysis by Pseudomonas aeruginosa HD-GYP phosphodiesterases: Analysis of the reaction mechanism and novel roles for pGpG. PLoS One8, e74920. doi: 10.1371/journal.pone.0074920
Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature406, 959–964. doi: 10.1038/35023079
Subedi, D., Vijay, A. K., Kohli, G. S., Rice, S. A., Willcox, M. (2018). Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites. Sci. Rep.8, 15668. doi: 10.1038/s41598-018-34020-7
Taylor, P. K., Yeung, A. T. Y., Hancock, R. E. W. (2014). Antibiotic resistance in Pseudomonas aeruginosa biofilms: Towards the development of novel anti-biofilm therapies. J. Biotechnol.191, 121–130. doi: 10.1016/j.jbiotec.2014.09.003
Tchigvintsev, A., Xu, X., Singer, A., Chang, C., Brown, G., Proudfoot, M., et al. (2010). Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol.402, 524–538. doi: 10.1016/j.jmb.2010.07.050
Thrane Sandra, W., Taylor Véronique, L., Lund, O., Lam Joseph, S., Jelsbak, L., Carroll, K. C. (2016). Application of whole-genome sequencing data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J. Clin. Microbiol.54, 1782–1788. doi: 10.1128/JCM.00349-16
Toutain Christine, M., Zegans Michael, E., O'toole George, A. (2005). Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol.187, 771–777. doi: 10.1128/JB.187.2.771-777.2005
Valentini, M., Filloux, A. (2016). Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem.291, 12547–12555. doi: 10.1074/jbc.R115.711507
Wei, Q., Zhang, Z., Luo, J., Kong, J., Ding, Y., Chen, Y., et al. (2019). Insulin treatment enhances Pseudomonas aeruginosa biofilm formation by increasing intracellular cyclic di-GMP levels, leading to chronic wound infection and delayed wound healing. Am. J. Trans. Res.11, 3261–3279.
Winsor, G. L., Griffiths, E. J., Lo, R., Dhillon, B. K., Shay, J. A., Brinkman, F. S. L. (2016). Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res.44, D646–D653. doi: 10.1093/nar/gkv1227
Wolfe Alan, J., Visick Karen, L. (2008). Get the message out: Cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol.190, 463–475. doi: 10.1128/JB.01418-07
Xin, L., Zeng, Y., Sheng, S., Chea, R. A., Liu, Q., Li, H. Y., et al. (2019). Regulation of flagellar motor switching by c-di-GMP phosphodiesterases in Pseudomonas aeruginosa. J. Biol. Chem.294, 13789–13799. doi: 10.1074/jbc.RA119.009009
Xu, Z., Wang, P., Wang, H., Yu, Z. H., Au-Yeung, H. Y., Hirayama, T., et al. (2019). Zinc excess increases cellular demand for iron and decreases tolerance to copper in Escherichia coli. J. Biol. Chem.294, 16978–16991. doi: 10.1074/jbc.RA119.010023
Zhang, L., Li, J., Liang, J., Zhang, Z., Wei, Q., Wang, K. (2020). The effect of cyclic-di-GMP on biofilm formation by Pseudomonas aeruginosa in a novel empyema model. Ann. Trans. Med.8, 1146. doi: 10.21037/atm-20-6022
Zhou, T., Huang, J., Liu, Z., Lin, Q., Xu, Z., Zhang, L. H. (2022). The two-component system FleS/FleR represses H1-T6SS via cyclic di-GMP signaling in Pseudomonas aeruginosa. Appl. Environ. Microbiol.88, e01655–e01621. doi: 10.1128/AEM.01655-21
Keywords: Pseudomonas aeruginosa, motility, biofilm, c-di-GMP, DGC, PDE
Citation: Lin S, Chen S, Li L, Cao H, Li T, Hu M, Liao L, Zhang L-H and Xu Z (2022) Genome characterization of a uropathogenic Pseudomonas aeruginosa isolate PA_HN002 with cyclic di-GMP-dependent hyper-biofilm production. Front. Cell. Infect. Microbiol. 12:956445. doi: 10.3389/fcimb.2022.956445
Received: 30 May 2022; Accepted: 12 July 2022;
Published: 02 August 2022.
Edited by:
Philippe Sessou, Université d’Abomey-Calavi, BeninReviewed by:
Herman Sintim, Purdue University, United StatesWang Ke, Guangxi Medical University, China
Copyright © 2022 Lin, Chen, Li, Cao, Li, Hu, Liao, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Li, bGlsaS0xNDA2QDE2My5jb20=; Zeling Xu, emVsaW5neHVAc2NhdS5lZHUuY24=
†These authors have contributed equally to this work
 Siying Lin
Siying Lin Shuzhen Chen1†
Shuzhen Chen1† Huiluo Cao
Huiluo Cao Ting Li
Ting Li Ming Hu
Ming Hu Lisheng Liao
Lisheng Liao Lian-Hui Zhang
Lian-Hui Zhang Zeling Xu
Zeling Xu