- 1Department of Laboratory Medicine, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Department of Clinical Microbiology, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 3Department of Scientific Affairs, Hugobiotech Co., Ltd., Beijing, China
- 4Department of Clinical Laboratory, The Second Affiliated Hospital of Xiamen Medical College, Xiamen, China
- 5Department of Microbiology, The University of Hongkong, Hong Kong, Hong Kong SAR, China
- 6Department of Infection Control, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 7Department of Emergency, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
Introduction: Polymyxin-resistant Enterobacterales poses a significant threat to public health globally, but its prevalence and genomic diversity within a sole hospital is less well known. In this study, the prevalence of polymyxin-resistant Enterobacterales in a Chinese teaching hospital was investigated with deciphering of their genetic determinants of drug resistance.
Methods: Polymyxin-resistant Enterobacterales isolates identified by matrix-assisted laser desorption were collected in Ruijin Hospital from May to December in 2021. Both the VITEK 2 Compact and broth dilution methods were used to determine polymyxin B (PMB) susceptibility. Polymyxin-resistant isolates were further characterized by molecular typing using PCR, multi-locus sequence typing, and sequencing of the whole genome.
Results: Of the 1,216 isolates collected, 32 (2.6%) across 12 wards were polymyxin-resistant (minimum inhibitory concentration (MIC) range, PMB 4–256 mg/ml, and colistin 4 ≥ 16 mg/ ml). A total of 28 (87.5%) of the polymyxin-resistant isolates had reduced susceptibility to imipenem and meropenem (MIC ≥ 16 mg/ml). Of the 32 patients, 15 patients received PMB treatment and 20 survived before discharge. The phylogenetic tree of these isolates showed they belonged to different clones and had multiple origins. The polymyxin-resistant Klebsiella pneumoniae isolates belonged to ST-11 (85.72%), ST-15 (10.71%), and ST-65 (3.57%), and the polymyxin-resistant Escherichia coli belonged to four different sequence types, namely, ST-69 (25.00%), ST-38 (25.00%), ST-648 (25.00%), and ST-1193 (25.00%). In addition, six mgrB specific mutations (snp_ALT c.323T>C and amino acid change p.Val8Ala) were identified in 15.6% (5/32) of the isolates. mcr-1, a plasmid-mediated polymyxin-resistant gene, was found in three isolates, and non-synonymous mutations including T157P, A246T, G53V, and I44L were also observed.
Discussion: In our study, a low prevalence of polymyxin-resistant Enterobacterales was observed, but these isolates were also identified as multidrug resistant. Therefore, efficient infection control measures should be implemented to prevent the further spread of resistance to last-line polymyxin therapy.
Introduction
The emergence of carbapenem-resistant Enterobacterales represents a major public health threat worldwide (Nordmann et al., 2012). Currently, periodic outbreaks or endemics of Enterobacterales that are not susceptible to carbapenems have now been documented in hospital settings and the wider community (Mizrahi et al., 2020), and also Enterobacterales have been designated by WHO as high-priority pathogens (Kopotsa et al., 2019), which limits the effectiveness of treatment of Enterobacterales infections. In 2021, it was estimated that 9.4%–12.5% of Enterobacterales isolates were carbapenem-resistant in China (http://www.chinets.com/). It is more serious that Enterobacterales has been included among a group of multiple drug resistance (MDR) pathogens that “ESKAPE” the actions of commonly used antibiotics (Rice, 2008).
Colistin and polymyxin B (PMB), first- generation polymyxins, were first introduced into clinical practice in the late 1950s but subsequently abandoned due to concerns about nephrotoxicity (Nang et al., 2021). Given their ever-increasing resistance to all other antibiotics, such as carbapenems and aminoglycosides, polymyxins were reintroduced into treatment regimens in the early 2000s for problematic Gram-negative “superbugs” (Velkov et al., 2019). They remain an important last-line treatment as they have excellent activity against many of these problematic pathogens (Jeannot et al., 2017). Worryingly, resistance to polymyxins of numerous bacteria, including Enterobacterales, has been reported in both humans and animals at an alarming rate (Gales et al., 2011; Lekunberri et al., 2017; Sherry and Howden, 2018). For example, in 2021, the China Antimicrobial Surveillance Network (http://www.chinets.com/) estimated that only 4.6% of isolates in China were resistant to colistin and PMB. In many countries, polymyxins are the only accessible or affordable therapeutic option for carbapenem-resistant organisms (Satlin et al., 2020). The emergence of bacteria resistant to polymyxins, notably Enterobacterales such as Klebsiella pneumoniae (K. pneumoniae) and Escherichia coli (E. coli), has been attributed to their widespread usage to treat infections (Stefaniuk and Tyski, 2019). Bacterial isolates resistant to polymyxin are of great concern worldwide because they can cause life-threatening infections, particularly those that incorporate antimicrobial resistance genes (ARGs), such as extended-spectrum β-lactamases (ESBLs) and metallo-β-lactamases (MBLs), harboring mainly blaKPC, blαNDM, and blαoxA-48 like genes which have become a major challenge to public health because of limited antibiotic choice and high case-fatality rates (Capone et al., 2013; Gao et al., 2019).
The acquisition of polymyxin resistance has been attributed to mutations of phoP/phoQ and pmrA/pmrB, which are two-component regulatory systems (TCS) (Capone et al., 2013; Jayol et al., 2015; Stefaniuk and Tyski, 2019). Constitutive upregulation of the pmrHFIJKLM-ugd operon is known to be triggered by highly specific mutations, which lead to the covalent attachment of 4-amino-4-deoxy-L-arabinose to the lipid A component of lipopolysaccharide that is located in the outer membrane of bacteria (Moffatt et al., 2010; Jones-Dias et al., 2016; Abdul Momin et al., 2017). The mcr (1–10) are polymyxin-resistant genes that have been identified in Enterobacterales isolates, and horizontal transfer of these genes through plasmid can alter bacterial resistance to polymyxin (Antoniadou et al., 2007; Jayol et al., 2015; Baraniak et al., 2016; Bardet et al., 2017; Borowiak et al., 2017; AbuOun et al., 2018; Carroll et al., 2019; Xu et al., 2022; Zhou et al., 2022). The aims of the present study were to determine the prevalence, molecular characteristics, and antibiotic susceptibility of polymyxin-resistant Enterobacterales isolated from Chinese patients in a tertiary teaching hospital.
Methods and materials
Bacterial isolates and antibiotic susceptibility testing
The study was based on retrospective samples collected from the Department of Clinical Microbiology of Ruijin Hospital, a 3,624-bed tertiary care teaching hospital located in east China, with approximately 130,000 hospital admissions per year. Ruijin Hospital has five intensive care unit (ICU) wards, and each ward has two sections. All Enterobacterales isolates were collected from May to December during 2021. The isolates were analyzed for species identification using the MALDI-TOF MS system (bioMérieux, Missouri, France) and then susceptibility tests using the AST-N335 VITEK 2 Compact system (bio Mérieux, France). The antimicrobial agents investigated were amikacin (aminoglycosides), aztreonam (monobactam), cefepime, cefoperazone/sulbactam, ceftazidime, ciprofloxacin, and levofloxacin (quinolones), colistin, doxycycline, and minocycline (tetracyclines), meropenem and imipenem (carbapenems), piperacillin/tazobactam, tigecycline (glycylcycline), ticarcillin/clavulanic acid, tobramycin, and trimethoprim/sulfamethoxazole. Minimum inhibitory concentrations (MICs) of PMB and colistin were determined using broth microdilution with E. coli ATCC 25922 used as the quality control strain. All results (except for polymyxins) were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. For patients with multi polymyxin-resistant isolates, only the first one was used for genome sequencing in the present study. A total of 28 K. pneumoniae and four E. coli isolates were identified to be polymyxin-resistant, and we performed whole- genome sequencing on all of them. The public genomes used in this study are summarized in Table S1.
Genome sequencing, assembly, and annotation
Genomic DNA of each polymyxin-resistant isolate was extracted using the method of cetyltrimethylammonium bromide (Clarke, 2009). The quantity and quality of DNA were determined using a Qubit Fluorometer (Invitrogen, USA), and the integrity was checked using the NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The sequencing libraries were constructed using a TruSeq DNA Sample Preparation Kit (Illumina, San Diego, USA) and a Template Prep Kit (Pacific Biosciences, Menlo Park, California, USA). Genome sequencing was carried out by Shanghai Personal Biotechnology (Shanghai, China) using an Illumina NovaSeq platform with a PE150 model. De novo genome assembly was conducted using A5-Miseq (v20160825) and SPAdes (v3.12.0), followed by base correction using Pilon (v1.23). The gene models were predicted by Glimmer 3.02 (http://ccb.jhu.edu/software/glimmer/index.shtml). Function annotation was completed by searching databases including NR (Non-Redundant Protein Database), KEGG (Kyoto Encyclopedia of Gene and Genomes), and COG (Cluster of Orthologous Groups of proteins) using BLAST. Subsequently, the VFDB (Virulence Factors of Pathogenic Bacteria) and CARD (the Comprehensive Antibiotic Resistance Databases) were interrogated to retrieve pathogenicity genes and antibiotic-resistant genes, respectively.
Multilocus sequence typing
The PubMLST database (https://pubmlst.org/organisms/) for multilocus sequence typing (MLST) was utilized. Seven housekeeping genes, namely, gapA, infB, mdh, pgi, phoE, rpoB, and tonB, were used for K. pneumoniae typing, whereas adk, fumC, gyrB, icd, mdh, purA, and recA were adopted for E. coli typing.
Variant calling and phylogenetic analyses
Taking the genome of K. pneumoniae (GCA_008728695.1) and E. coli (GCA_003018455.1) as references, Snippy (https://github.com/tseemann/snippy) was employed to identify SNPs across 378 complete genomes of K. pneumoniae and 404 complete genomes of E. coli from the GenBank database (Supplementary Table S1). Core SNPs were concatenated and aligned using a snippy-multi script. The SNPs were further annotated by using SnpEff (https://pcingola.github.io/SnpEff/). Subsequently, a core-SNP- based maximum likelihood tree was constructed employing IQ-TREE with a GTR+I+G model and bootstrap values of 1,000 (Minh et al., 2020), and the tree was visualized using iTOL (https://itol.embl.de/) (Letunic and Bork, 2021).
Results
Patient demographics and characteristics of the polymyxin-resistant Enterobacterales isolates
The hospitalization information, polymyxin treatment history, and amount of separated polymyxin-resistant Enterobacterales isolates for each patient are given in Figure 1. Only 28.6% (8/28) of the patients had isolates with polymyxin-resistant K. pneumoniae once, and the other patients had isolates multiple times. In particular, two patients were isolated with polymyxin-resistant K. pneumoniae isolates from one location 10 and 16 times during their hospitalization, demonstrating a persistent- infection procedure. Two patients had isolates with polymyxin-resistant E. coli only once; an other two patients had polymyxin-resistant E. coli isolates three times from one location during their hospitalization.
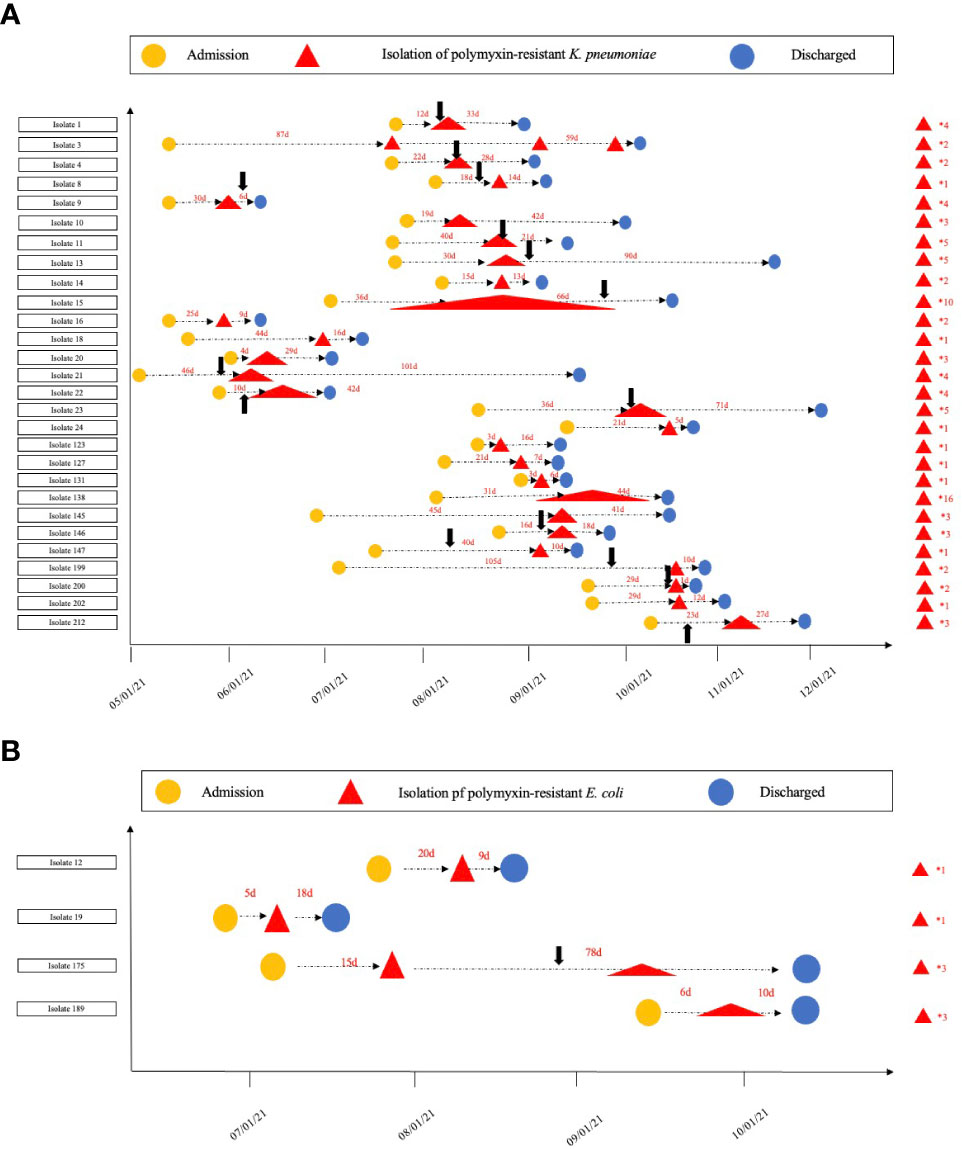
Figure 1 Timeline for the isolation of polymyxin-resistant (K) pneumoniae and (E) coli. (A). The timeline for polymyxin-resistant (K) pneumoniae isolation in each patient; (B). the timeline for the polymyxin-resistant isolation (E) coli in each patient. The numbers on the right of the red triangles show the number of polymyxin-resistant isolates from each patient. The black arrows show when polymyxin treatment commenced.
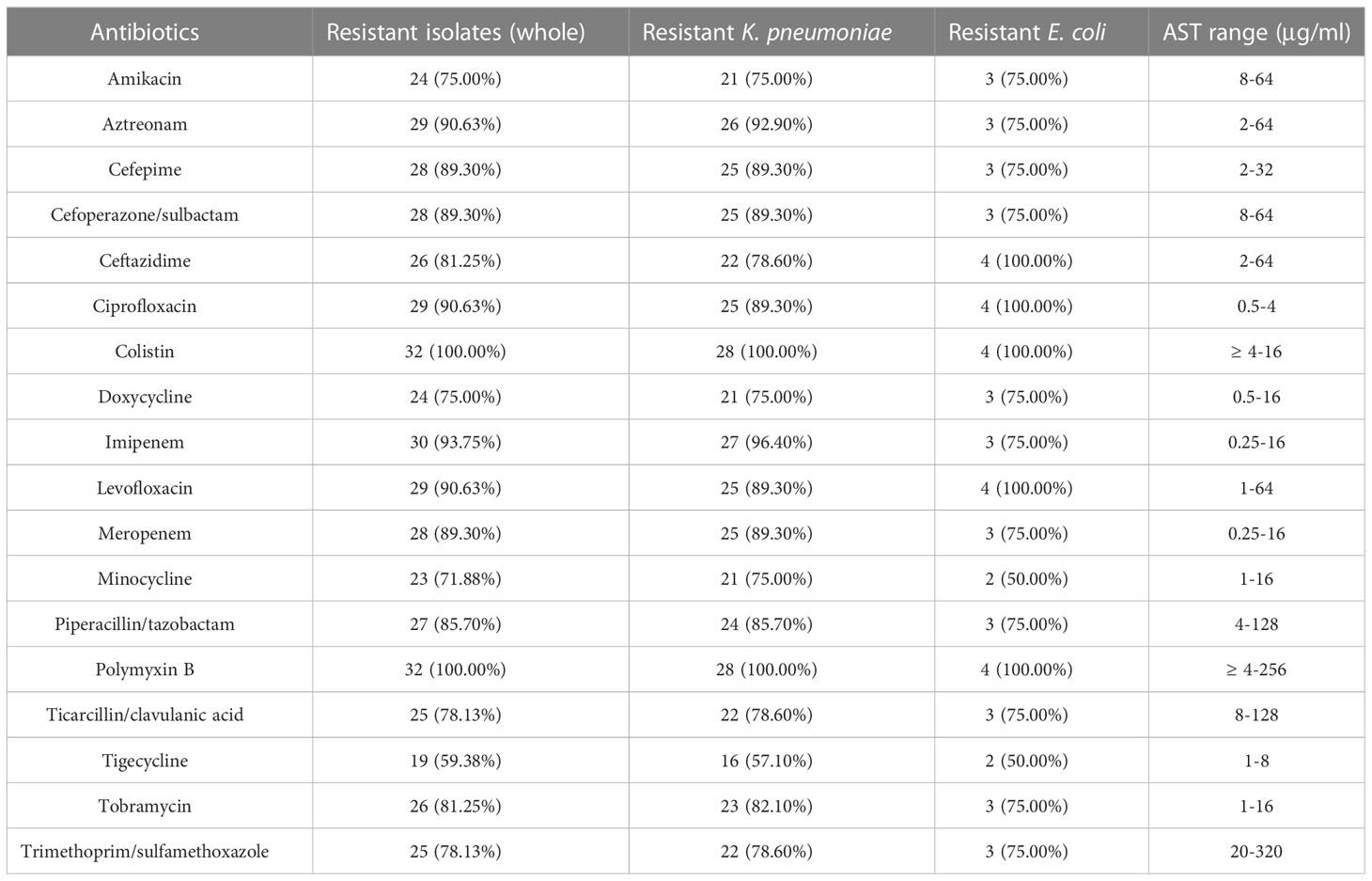
A total of 458 K. pneumoniae and 758 E. coli isolates were collected across the study period. Finally (28/458, 6.11%), polymyxin-resistant K. pneumoniae and (4/758, 0.53%) polymyxin-resistant E. coli strains were isolated from separate patients in 12 different wards (Table 1). Isolates with colistin MICs (29/32, 90.7%) > 4 μg/ml or PMB MICs that ranged from 4 to 256 μg/ml were considered to be resistant (Table 2). Most of the polymyxin-resistant isolates were collected from sputum (9/32, 28.1%), followed by throat swabs (4/32, 12.5%), wounds (4/32, 12.5%), blood (3/32, 9.4%), midstream urine (3/32, 9.4%), extravasate fluid (3/32, 9.4%), and bile (2/32, 6.2%). The ages of the patients ranged from 33 to 96 years (range 60.1 ± 14.3), and the majority were adults with multiple complicated comorbidities. The ward with the most frequent occurrence was the emergency intensive care unit (EICU) (9/32, 28.1%), followed by the burns ward (8/32, 25.0%), critical care unit (CCU) (4/32, 12.5%), respiratory intensive care unit (RICU) (4/32, 12.5%), gastrointestinal surgery ward (1/32, 0.1%), hematology ward (1/32, 3.1%), and urology surgery ward (1/32, 3.1%). The average length of a hospital stay was 43 days (CI: 32.25 to 64.75). Five patients (5/32,15.6%) were given tropical PMB treatment (7–22 days) and 10 patients (10/32, 31.2%) intravenous (i.v.) PMB (25–150 mg) sulfate (2–30 days). Fifteen patients (15/32, 46.9%) did not receive any polymyxin- related therapy during their hospitalization. Three patients (3/32, 9.3%) were given i.v. polymyxin E (15–30 mg/day) aerosol/sulfate therapy (4–18 days), and 2 patients (2/32, 6.2%) received PMB (50–150 mg/day) sulfate aerosol therapy (7–12 days). Most (14/15, 93.3%) polymyxin-resistance isolates originated from patients that had received polymyxin- related therapy were isolated after the polymyxin treatment (2–50 days) and only one isolate (1/15, 6.67%) of polymyxin-resistant K. pneumoniae was isolated before polymyxin therapy. Death due to any cause occurred in 10/32 patients, giving a mortality rate of 31.25%. Most of the polymyxin-resistant K. pneumoniae isolates were ST-11 (23/28, 82.14%), and the others were ST-65 (2, 7.14%) and ST-15 (3/28, 10.71%). Polymyxin-resistant E. coli isolates belonged to four different MLSTs, namely, ST-69, ST-38, ST-648, and ST-1193.
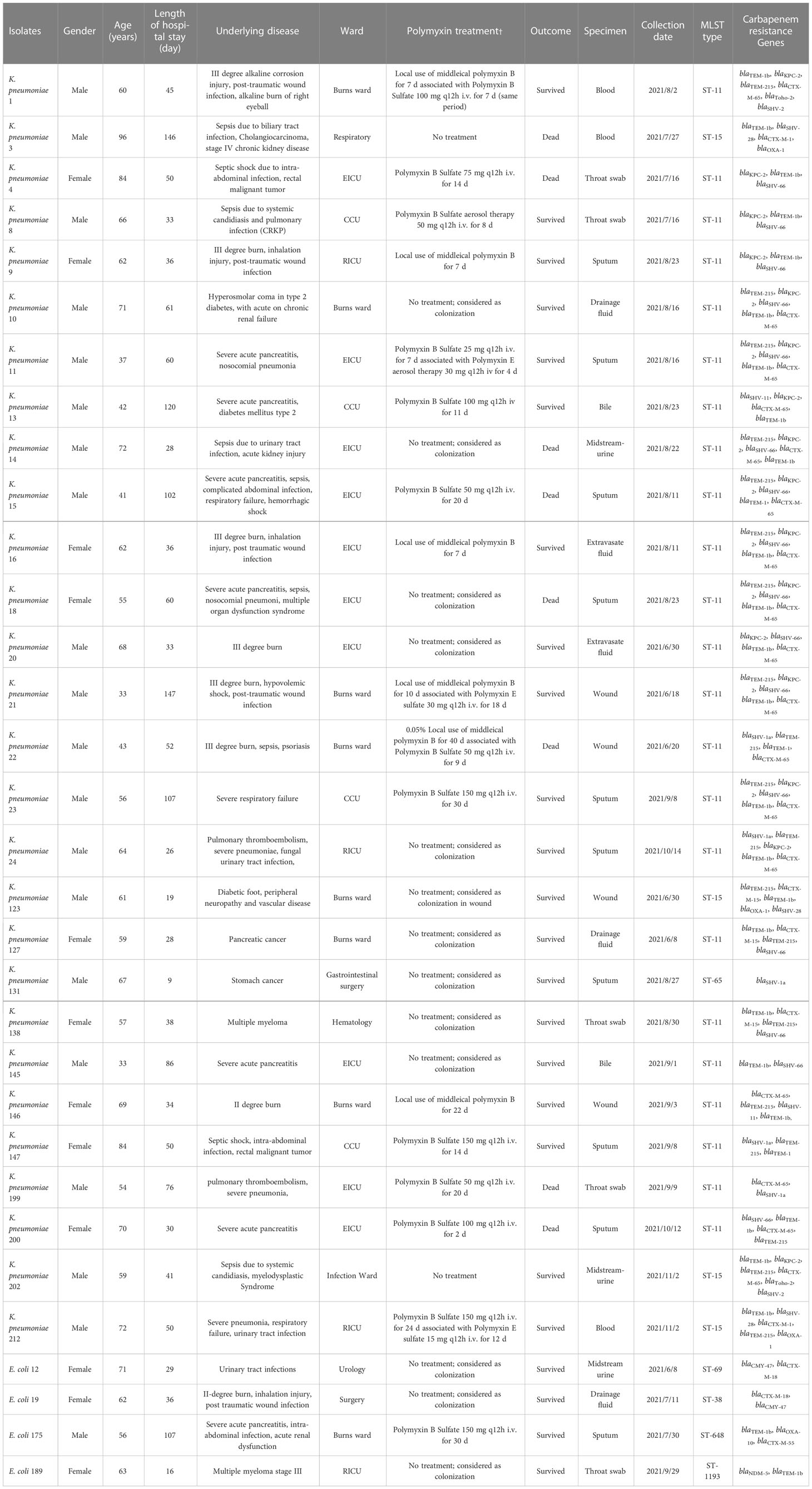
Table 1 Patient demographics and main characteristics of polymyxin-resistant K. pneumoniae and E. coli.
Genetic determinants of antibiotic resistance in polymyxin-resistant isolates
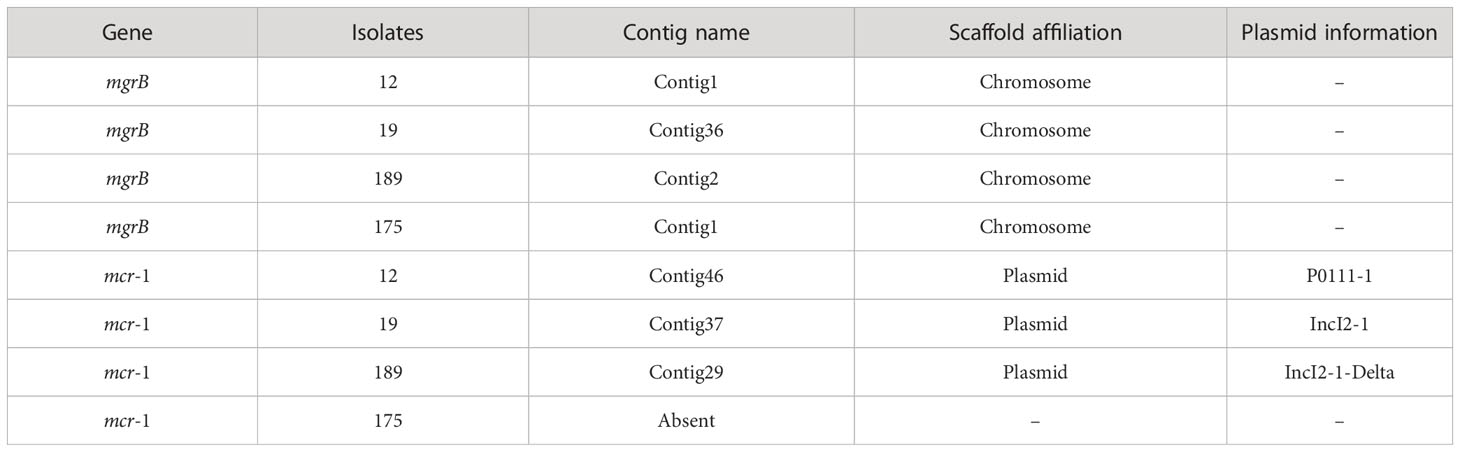
All the polymyxin-resistant isolates (32/32,100%) possessed carbapenemase genes (Table 1). No mcr genes were detected in polymyxin-resistant K. pneumoniae isolates, showing that mcr genes were not the cause of polymyxin resistance in these strains. In contrast, three of four polymyxin-resistant E. coli isolates carried mcr-1 polymyxin-resistant genes (isolates 12, 175, and 189).
Specific mutations in mgrB were identified in 15.6% (5/32) of the isolates, including one K. pneumonia (isolate 123) and four E. coli (isolates 12, 19, 175, and 189). Taking a polymyxin-susceptible genome RJBS176 (Xu et al., 2021) as the reference, 18 mutations of pmrA/pmrB and phoP/phoQ were identified in polymyxin-resistant K. pneumoniae isolates, among which nine were synonymous. T157P (Bir et al., 2022) and A246T in pmrB and G53V in pmrA (Nirwan et al., 2021) were considered as variations that potentially contributed to polymyxin resistance in K. pneumoniae isolates (Tables 3, S2, S3).
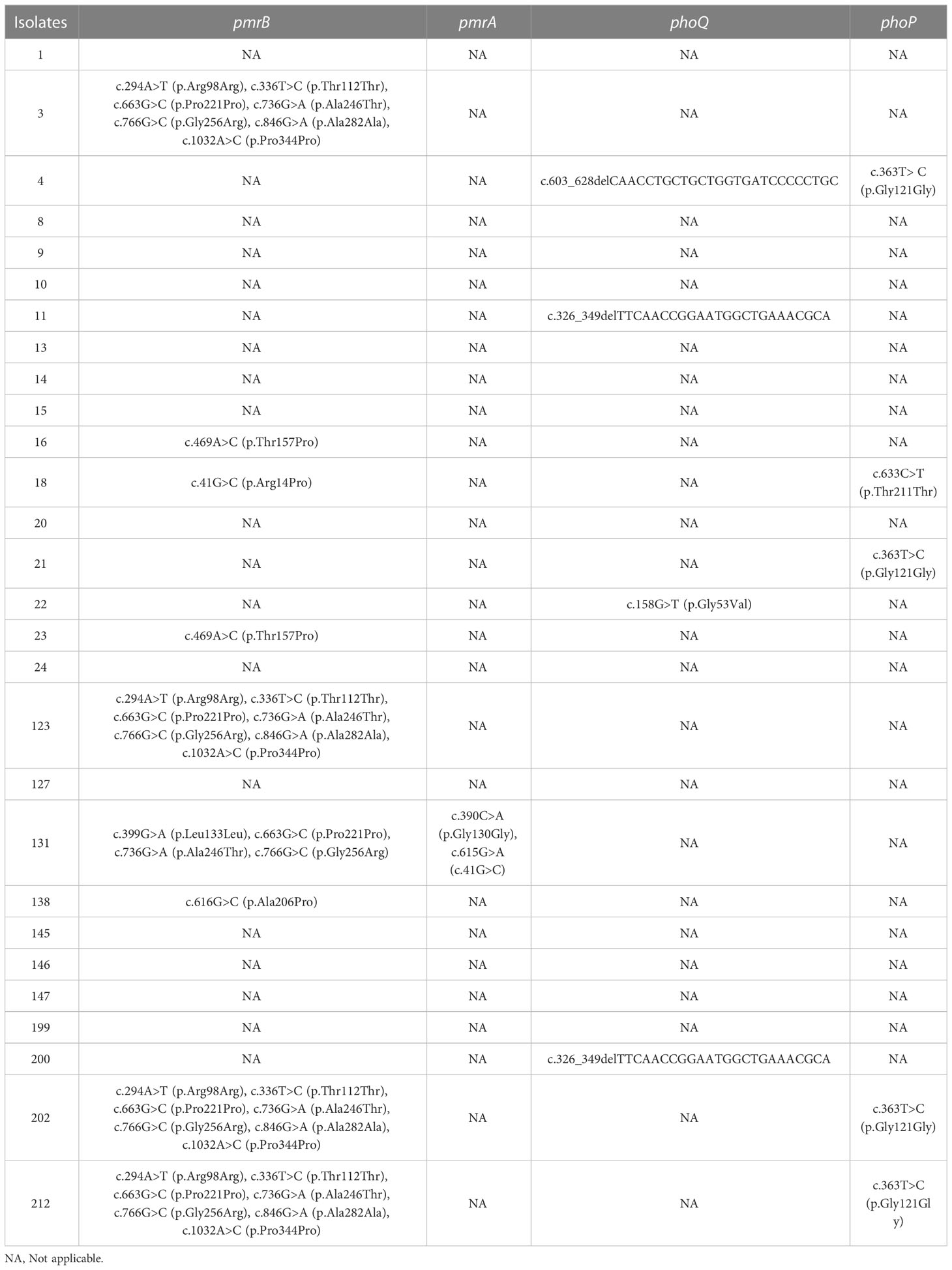
Table 3 Genetic variations in pmrA, pmrB, phoQ, and phoP of the polymyxin-resistant K. pneumoniae isolates.
When the polymyxin-resistant E. coli isolates’ genomes were compared with a polymyxin-susceptible reference genome (E. coli EC1390) (Thuy et al., 2022), we identified 137 mutations in mgrB, pmrA/pmrB, and phoP/phoQ including 63 synonymous ones. Among the genetic variations, I44L (Sánchez et al., 2021; Huang et al., 2021a) in phoP has been reported as a variation that potentially contributes to polymyxin-resistant in E. coli isolates. In addition, a non-synonymous mutation c.323T>C that resulted in p.Val8Ala was identified in mgrB of isolate 12 (Tables 4, S4, S5).
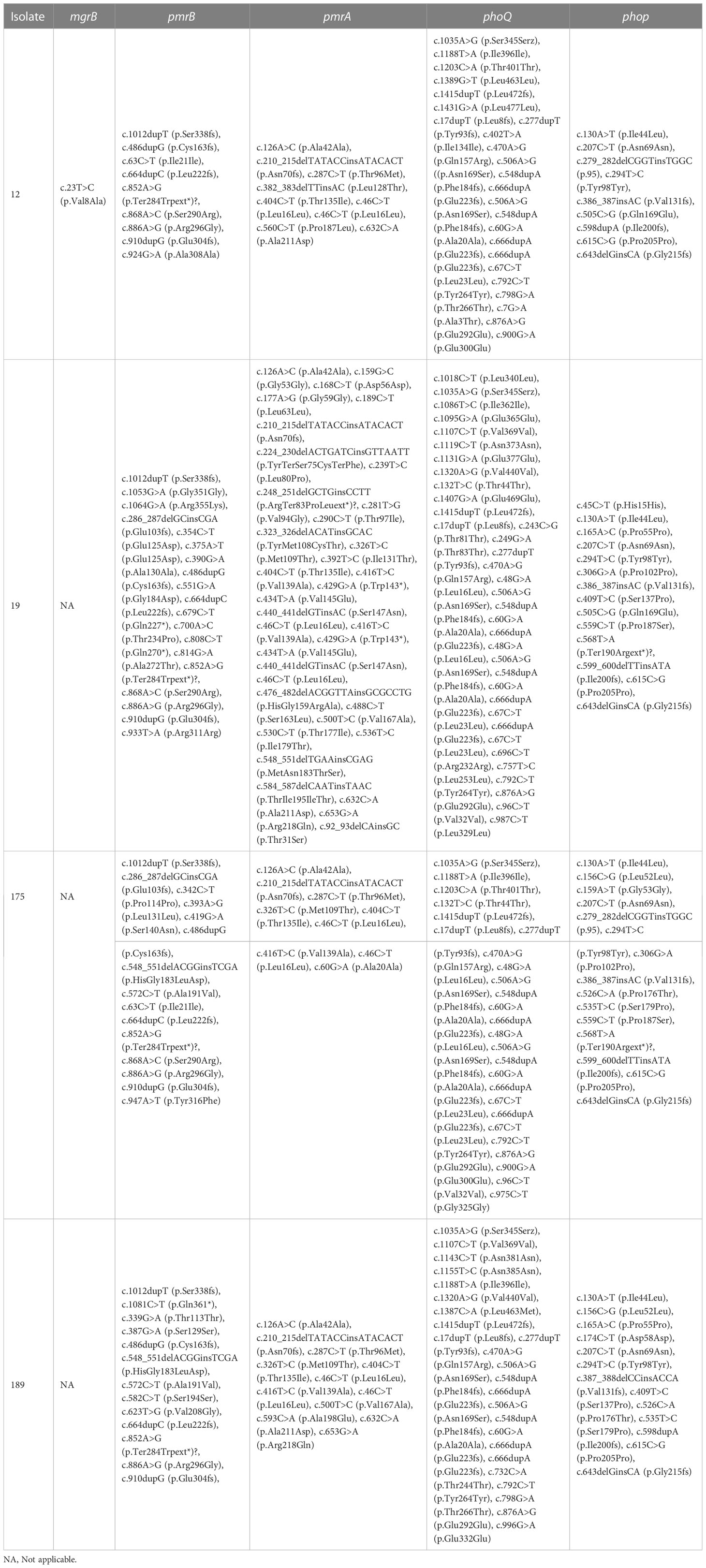
Table 4 Genetic variations in pmrA, pmrB, phoQ, phoP, and mgrB of the polymyxin-resistant E. coli isolates.
Antimicrobial susceptibility and MDR of polymyxin-resistant Enterobacterales
The antibiotic susceptibility test result of the Enterobacterales isolates is shown in Table 2. According to the CLSI (Satlin et al., 2020), most of the polymyxin-resistant K. pneumoniae also showed reduced susceptibility to carbapenems, aminoglycosides, and fluoroquinolones, with 96.4% (27/28) isolates resistant to imipenem and (25/28, 89.3%) to meropenem and 75.0% (21/28) isolates to amikacin and 89.3% (25/28) to ciprofloxacin and levofloxacin. A much lower frequency (16/28, 57.1%) of tigecycline and minocycline resistance was observed. Additionally, polymyxin-resistant E. coli also showed decreased susceptibility to carbapenems, aminoglycosides, and fluoroquinolones. 75.0% (3/4) of isolates were resistant to imipenem and meropenem, 75.0% (3/4) to amikacin, and all of the isolates to ciprofloxacin and levofloxacin. It is also noteworthy that 50% (2/4) of the isolates were sensitive to tigecycline and minocycline. Only one polymyxin-resistant K. pneumoniae (isolate 199) had PMB MICs at 256 μg/ml.
Phylogenetic analysis
Phylogenomic trees of the 28 polymyxin-resistant K. pneumoniae and four polymyxin-resistant E. coli isolates are shown in Figure S1, S2. The phylogenetic tree of E. coli isolates revealed that four polymyxin-resistant isolates did not belong to the same clone but had multiple origins, demonstrating their parallel evolutions. The same phenomenon was observed for K. pneumoniae isolates. The difference was that most of the K. pneumoniae isolates belonged to ST-11 with a small genetic distance. To further explore the polymyxin-resistant mechanism(s) of K. pneumoniae, we compared their phylogenetic relationships with polymyxin-susceptible isolates from 46 isolates and aligned antibiotic resistance genes with their phylogenetic tree (Figure 2). The tree shows a close relationship of isolates within the same ST.
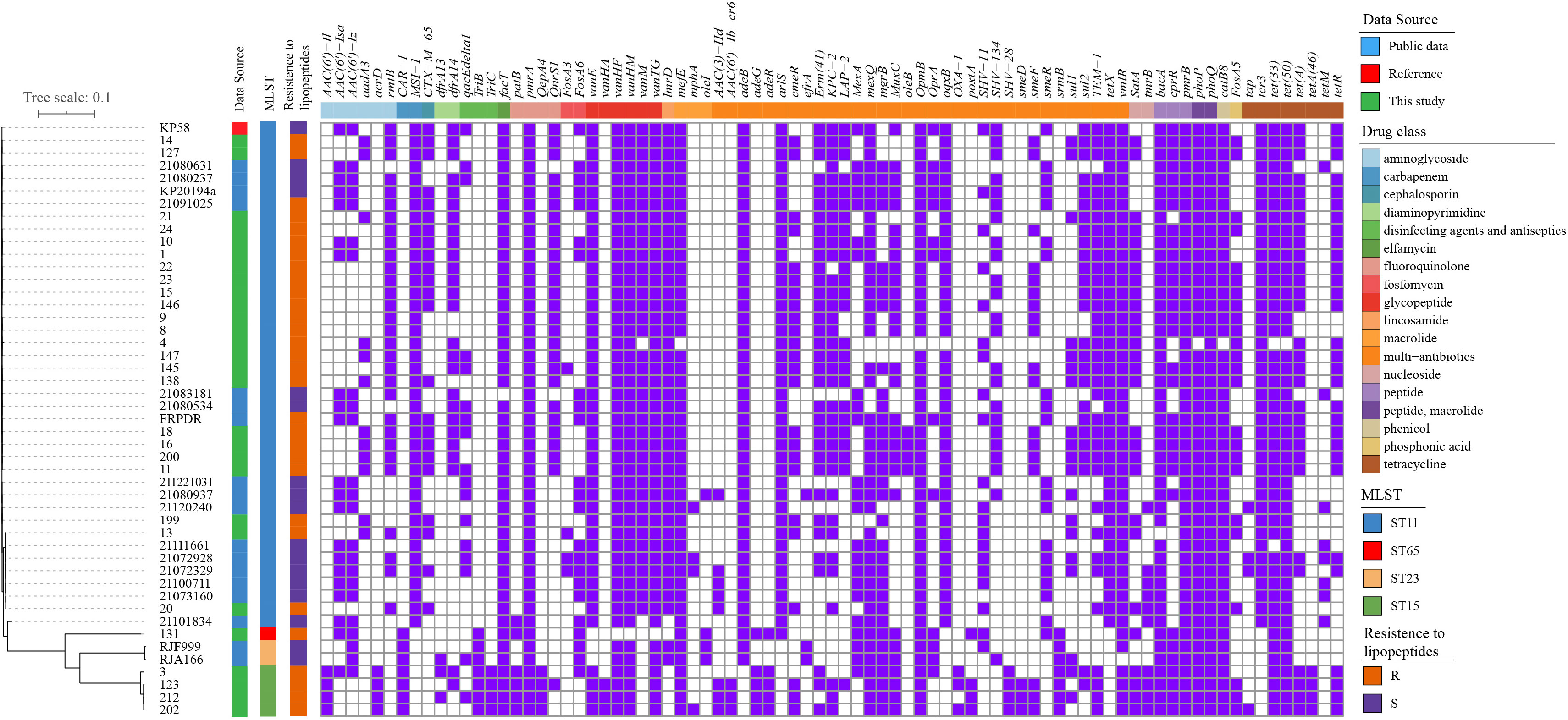
Figure 2 Phylogenetic tree and distribution of resistance genes of K. pneumoniae isolates (A) core-genome-based ML phylogenetic tree showing the relationship of 30 polymyxin-resistant and 17 polymyxin-sensitive K. pneumoniae isolates constructed taking KP58 as the reference. Apart from 28 strains sequenced in this study, the other 19 genomes were obtained from public data. The antibiotic- resistant genes were mapped on the right, with carried genes painted in purple and classified according to the ResFinder database.
A high- frequency appearance of pmrA/pmrB, phoP/phoQ, cprR, and bacA was observed in all isolates. Interestingly, fosA5, satA, and cmeR were observed in polymyxin-resistant isolates with high frequencies but were absent in all polymyxin-sensitive isolates. In reverse, aac(6’)-Iz, aac(6’)-Isa, fosA6, mexA, and smeR had a high frequency in polymyxin-resistant isolates but a low appearance in polymyxin-sensitive isolates. These genes may also be related to the polymyxin-resistant mechanism(s), but further molecular experiments will need to be performed for unequivocal verification.
82.14% (23/28) of the polymyxin-resistant K. pneumoniae isolates were detected with mgrB genes (Table 5). All the polymyxin-resistant E. coli isolates were detected with the mgrB gene, and 75% (3/4) were identified with mcr-1, which are located in the plasmids. Isolate 12 harbors a P0111_1 plasmid, isolate 19 harbors a IncI2_1 plasmid, and isolate 189 harbors a IncI2_1_Delta plasmid. There have been studies showing that mcr-1 harboring IncI plasmids were prevalent in various Enterobacterales in China, including animal- originated E. coli isolates (Tang et al., 2022; Wang et al., 2022), Escherichia fergusonii, and Salmonella (Tang et al., 2020; Tang et al., 2023), which were also observed in our study.
Discussion
The world is facing the great challenge of bacterial resistance to carbapenems. This is because tigecycline and ceftazidime/avibactam have only been registered in a small number of countries and that polymyxins are often the only effective antibiotic against multidrug-resistant organisms. However, due to the widespread use of PMB and colistin against MDR organisms, such as Acinetobacter baumannii, Pseudomonas aeruginosa (P. aeruginosa), K. pneumoniae, and E. coli, the current low levels of polymyxin resistance are changing. Our hospital started using PMB, colistin, and corresponding susceptibility tests in late 2020 to evaluate the effectiveness of therapy that was used to treat carbapenem-resistant Gram-negative bacterial infections. There have been previous reports of infections caused by polymyxin-resistant K. pneumoniae (Jayol et al., 2015) and E. coli which carried mcr genes (He et al., 2017), and other studies have pooled data from multiple institutions to determine regional susceptibility to polymyxins (Wi et al., 2017; Baek et al., 2020). In the present study, we reported an investigation of the prevalence of polymyxin-resistant Enterobacterales during 2021 in our hospital, analyzed the isolates’ genomics and phenotypes, and compared the clinical characteristics of the patients from which they were isolated.
In our study in a single hospital, the prevalence of polymyxin resistance was 2.6% (32/1216), which is a little higher than reported (1.9%) by the China Antimicrobial Surveillance Network in 2021. Most polymyxin-resistant isolates have low MICs to PMB and colistin, which is not surprising given their structural similarity, differing only by a single amino acid (Gales et al., 2011). Of concern is that all the isolates contained OXA and β-lactamases, which confer resistance to imipenem and meropenem. Using the definitions for MDR, extensively drug-resistant (XDR), and pan drug-resistant (PDR) (Magiorakos et al., 2012), half of the polymyxin-resistant isolates were PDR and resistant to all antibiotics tested, whereas the others were XDR or MDR. ST-11, one of the first identified pandemic K. pneumoniae clones, has been described in Asia, Australia, USA, and Europe (Chen et al., 2014; Tian et al., 2019; Tumbarello et al., 2021; Huang et al., 2021b). Several studies have suggested that a specific ST-11 subclone with different β-lactamases might have increased its epidemic potential (Giske et al., 2012; Sonnevend et al., 2013; Tan et al., 2019). ST-11 K. pneumoniae was also the most dominant strain prevalent in China and exhibited reduced susceptibility to most available antibiotics (Liu et al., 2022; Ouyang et al., 2022), which is consistent with our findings. The K. pneumoniae ST-15 and ST-65 were reported to be prevalent mainly in Asia and also carry multiple resistance genes (Lin et al., 2014; Zhao et al., 2021). In our study, most of the prevalence isolates (ST-11) got similar plasmid types, which carry carbapenemase resistance genes. Polymyxin-resistant E. coli isolates in the present study were composed of multiple MLST types. ST-156 or ST-167 clones have been reported at a higher frequency in China (Xia et al., 2017), whereas ST-69 was found reported in Korea (Kim et al., 2020), ST-38 in the United Kingdom (Greig et al., 2018), ST-648 in Kerman, Iran (Kalantar-Neyestanaki et al., 2020), and ST-1193 in Canada (Izydorczyk et al., 2020). All carried multiple ARGs findings consistent with the results of the present study.
Changes in the two-component systems pmrA/pmrB (E. coli, K. pneumoniae, Salmonella spp.), phoP/phoQ (K. pneumoniae, Salmonella spp.), parR/parS (P. aeruginosa), colR/colS (P. aeruginosa), and cprR/cprS (Campylobacter jejuni) are associated with polymyxin resistance in Gram-negative bacteria (Baron et al., 2016). In the present study, all the polymyxin-resistant isolates had mutations in pmrA and/or pmrB, a major TCS in Gram-negative bacteria, which are responsible for polymyxin resistance. Our study indicates that the mgrB alteration may be a common mechanism associated with polymyxin resistance for the clinical treatment of K. pneumoniae infection (Cannatelli et al., 2013). However, only one of the polymyxin-resistant K. pneumoniae isolates contains mgrB resistance genes, and an intriguing aspect is that all four isolates of polymyxin-resistant E. coli carried mgrB genes and β-lactamases. Thus far, other studies have reported 10 variants of the mcr genes, mcr (1–10) in various Enterobacterales (Antoniadou et al., 2007; Jayol et al., 2015; Baraniak et al., 2016; Bardet et al., 2017; Borowiak et al., 2017; AbuOun et al., 2018; Carroll et al., 2019; Xu et al., 2022; Zhou et al., 2022). Here, we have identified mcr-1 genes in three polymyxin-resistant E. coli isolates. The co-appearance of mcr-1 and other resistant genes, especially β-lactamases, is worrying, because of limited therapeutic options.
Several studies have shown an association between the use of polymyxin and the emergence of polymyxin resistance in K. pneumoniae in hospitals, although the results remain controversial in different countries (Antoniadou et al., 2007; Lee et al., 2009; Zarkotou et al., 2010; Xu et al., 2022; Zhou et al., 2022) In our study, 17 patients received polymyxin therapy during their hospitalizations, 15.6% received topical treatment that involved the use of polymyxin therapy for 7–22 days, 31.2% received PMB IV (25–150 mg) treatment for 2–30 days according to the International Consensus Guidelines for the Optimal Use of polymyxins (Tsuji et al., 2019), and 9 patients were given polymyxin therapy for more than 15 days. Polymyxin-resistant isolates were obtained from 16 of these patients after polymyxin treatment for 2 to 50 days, and only one patient was isolated with the polymyxin-resistant K. pneumoniae strain before polymyxin therapy was administered. We also found a divergent prevalence in patient isolates with polymyxin- resistant strains after multiple-time or once- only analysis. In patients’ isolates with multi polymyxin- resistant strains, 15 out of 22 (68.18%) patients were given polymyxin treatment. In reverse, in patients who had polymyxin- resistant strains isolated only once, only 2 of 10 patients (20%) were given polymyxin treatment. From these findings, we were able to demonstrate an association between the use of polymyxin and the emergence of polymyxin-resistant Enterobacterales. There is a great risk that long-time dosing might lead to polymyxin resistance among various Enterobacterales isolates. Monotherapy with polymyxins should be avoided, with better ways to use them in combination with drugs with synergistic effects or an i.v. spray. According to the host factors of all the collected patients, we concluded that the patients were mainly adults, with multiple complicated comorbid conditions to highly infection-prone environments, and also underwent longtime hospitalization. Fifteen patients who were not given polymyxin therapy also had isolates containing polymyxin-resistant Enterobacterales. We may attribute this to their long-term exposure to the environment during hospitalization. Since this was a retrospective study, we did not have the corresponding data about the nosocomial environments. Further studies are therefore needed to demonstrate the prevalence of polymyxin-resistant Enterobacterales in hospitals.
Conclusions
A low prevalence of polymyxin-resistant Enterobacterales was found soon after polymyxin therapy was introduced into a Chinese tertiary teaching hospital. The polymyxin-resistant Enterobacterales pose a real threat to public health. It is very important for clinical laboratories to detect polymyxin- resistant genes and to characterize the epidemiological trends of high-risk polymyxin-resistant Enterobacterales in order to optimize the use of the last-line class of antibiotics. Furthermore, effective infection control measures are urgently needed to prevent further transmission of polymyxin resistance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Shanghai Jiao Tong University School of Medicine (protocol code RJ2022055 and 1 March 2022 approval).
Author contributions
Project planning, writing of the original draft, CX, XL; genomics analysis, software, and investigation, LZH, XL, HX; project administration; review and editing, LJH, ZY. All authors have read and agreed to the version of the manuscript submitted for publication.
Funding
This work was supported by the Shanghai Hospital Development Center—Exploration and Application of Scientific, Three-Dimensional and Accurate Management Mode of Antibiotics (3D-AMS) Program (SHDC 22020203).
Conflict of interest
XL and HX were employed by Hugobiotech Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1118122/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree showing the genomic relationship of the polymyxin-resistant K. pneumoniae isolates and public isolates. The phylogenetic tree was constructed based on core genome SNPs and the isolates sequenced in this study was marked with red color in the circle. Altogether, 378 high quality genomes were collected from GenBank database and 28 K. pneumoniae polymyxin-resistant genomes with GCA_008728695 K. pneumoniae genome as a reference, resulting in 3 distinct clades. The MLST typing results and country information of the isolates were shown in the outer circles from outside to inside.
Supplementary Figure 2 | Phylogenetic tree showing the genomic relationship of the polymyxin-resistant E. coli isolates and public isolates. The phylogenetic tree was constructed based on core genome SNPs and the isolates sequenced in this study are marked with black triangles. Altogether, 400 high quality genomes were collected from GenBank database and 4 polymyxin-resistant E. coli and genomes with E. coli GCA_003018455.1_ASM301845v1 genome as a reference. The MLST typing results and country information of the isolates are shown on the right.
References
Abdul Momin, M. H. F., Bean, D. C., Hendriksen, R. S., Haenni, M., Phee, L. M., Wareham, D. W. (2017). CHROMagar COL-APSE: a selective bacterial culture medium for the isolation and differentiation of colistin-resistant gram-negative pathogens. J. Med. Microbiol. 66 (11), 1554–1561. doi: 10.1099/jmm.0.000602
AbuOun, M., Stubberfield, E. J., Duggett, N. A., Kirchner, M., Dormer, L., Nunez-Garcia, J., et al. (2018). mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in great Britain from 2014 to 2015. J. Antimicrob. Chemother. 73 (10), 2904. doi: 10.1093/jac/dky272
Antoniadou, A., Kontopidou, F., Poulakou, G., Koratzanis, E., Galani, I., Papadomichelakis, E., et al. (2007). Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: first report of a multiclonal cluster. J. Antimicrob. Chemother. 59 (4), 786–790. doi: 10.1093/jac/dkl562
Baek, M. S., Chung, E. S., Jung, D. S., Ko, K. S. (2020). Effect of colistin-based antibiotic combinations on the eradication of persister cells in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 75 (4), 917–924. doi: 10.1093/jac/dkz552
Baraniak, A., Izdebski, R., Fiett, J., Gawryszewska, I., Bojarska, K., Herda, M., et al. (2016). NDM-producing Enterobacteriaceae in polan -14: inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J. Antimicrob. Chemother. 71 (1), 85–91. doi: 10.1093/jac/dkv282
Bardet, L., Le Page, S., Leangapichart, T., Rolain, J. M. (2017). LBJMR medium: a new polyvalent culture medium for isolating and selecting vancomycin and colistin-resistant bacteria. BMC Microbiol. 17 (1), 220. doi: 10.1186/s12866-017-1128-x
Baron, S., Hadjadj, L., Rolain, J. M., Olaitan, A. O. (2016). Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents 48 (6), 583–591. doi: 10.1016/j.ijantimicag.2016.06.023
Bir, R., Gautam, H., Arif, N., Chakravarti, P., Verma, J., Banerjee, S., et al. (2022). Analysis of colistin resistance in carbapenem-resistant Enterobacterales and XDR Klebsiella pneumoniae. Ther. Adv. Infect. Dis. 9, 20499361221080650. doi: 10.1177/20499361221080650
Borowiak, M., Fischer, J., Hammerl, J. A., Hendriksen, R. S., Szabo, I., Malorny, B. (2017). Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar paratyphi b. J. Antimicrob. Chemother. 72 (12), 3317–3324. doi: 10.1093/jac/dkx327
Cannatelli, A., D'Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57 (11), 5521–5526. doi: 10.1128/AAC.01480-13
Capone, A., Giannella, M., Fortini, D., Giordano, A., Meledandri, M., Ballardini, M., et al. (2013). High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin. Microbiol. Infect. 19 (1), E23–E30. doi: 10.1111/1469-0691.12070
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., Wiedmann, M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio 10 (3), e00853-19. doi: 10.1128/mBio.00853-19
Chen, L., Mathema, B., Chavda, K. D., DeLeo, F. R., Bonomo, R. A., Kreiswirth, B. N. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22 (12), 686–696. doi: 10.1016/j.tim.2014.09.003
Clarke, J. D. (2009). Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harb. Protoc. 2009 (3), pdb.prot5177. doi: 10.1101/pdb.prot5177
Gales, A. C., Jones, R. N., Sader, H. S. (2011). Contemporary activity of colistin and polymyxin b against a worldwide collection of gram-negative pathogens: results from the SENTRY antimicrobial surveillance progra -09). J. Antimicrob. Chemother. 66 (9), 2070–2074. doi: 10.1093/jac/dkr239
Gao, B., Li, X., Yang, F., Chen, W., Zhao, Y., Bai, G., et al. (2019). Molecular epidemiology and risk factors of ventilator-associated pneumonia infection caused by carbapenem-resistant Enterobacteriaceae. Front. Pharmacol. 10. doi: 10.3389/fphar.2019.00262
Giske, C. G., Fröding, I., Hasan, C. M., Turlej-Rogacka, A., Toleman, M., Livermore, D., et al. (2012). Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the united kingdom. Antimicrob. Agents Chemother. 56 (5), 2735–2738. doi: 10.1128/aac.06142-11
Greig, D. R., Dallman, T. J., Hopkins, K. L., Jenkins, C. (2018). MinION nanopore sequencing identifies the position and structure of bacterial antibiotic resistance determinants in a multidrug-resistant strain of enteroaggregative Escherichia coli. Microb. Genom 4 (10), e000213. doi: 10.1099/mgen.0.000213
He, Q. W., Xu, X. H., Lan, F. J., Zhao, Z. C., Wu, Z. Y., Cao, Y. P., et al. (2017). Molecular characteristic of mcr-1 producing escherichia coli in a Chinese university hospital. Ann. Clin. Microbiol. Antimicrob. 16 (1), 32. doi: 10.1186/s12941-017-0207-z
Huang, C., Shi, Q., Zhang, S., Wu, H., Xiao, Y. (2021a). Acquisition of the mcr-1 gene lowers the target mutation to impede the evolution of a high-level colistin-resistant mutant in Escherichia coli. Infect. Drug Resist. 14, 3041–3051. doi: 10.2147/idr.S324303
Huang, Y., Sokolowski, K., Rana, A., Singh, N., Wang, J., Chen, K., et al. (2021b). Generating genotype-specific aminoglycoside combinations with Ceftazidime/Avibactam for KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 65 (9), e0069221. doi: 10.1128/aac.00692-21
Izydorczyk, C., Waddell, B., Edwards, B. D., Greysson-Wong, J., Surette, M. G., Somayaji, R., et al. (2020). Epidemiology of E. coli in cystic fibrosis airways demonstrates the capacity for persistent infection but not patient-patient transmission. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.00475
Jayol, A., Nordmann, P., Brink, A., Poirel, L. (2015). Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 59 (5), 2780–2784. doi: 10.1128/AAC.05055-14
Jeannot, K., Bolard, A., Plésiat, P. (2017). Resistance to polymyxins in gram-negative organisms. Int. J. Antimicrob. Agents 49 (5), 526–535. doi: 10.1016/j.ijantimicag.2016.11.029
Jones-Dias, D., Manageiro, V., Ferreira, E., Barreiro, P., Vieira, L., Moura, I. B., et al. (2016). Architecture of class 1, 2, and 3 integrons from gram negative bacteria recovered among fruits and vegetables. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01400
Kalantar-Neyestanaki, D., Mansouri, S., Kandehkar Ghahraman, M. R., Tabatabaeifar, F., Hashemizadeh, Z. (2020). Dissemination of different sequence types lineages harboring blaCTX-M-15 among uropathogenic Escherichia coli in kerman, Iran. Iran J. Basic Med. Sci. 23 (12), 1551–1557. doi: 10.22038/ijbms.2020.47520.10940
Kim, S., Kim, H., Kim, Y., Kim, M., Kwak, H., Ryu, S. (2020). Antimicrobial resistance of Escherichia coli from retail poultry meats in Korea. J. Food Prot 83 (10), 1673–1678. doi: 10.4315/jfp-20-150
Kopotsa, K., Osei Sekyere, J., Mbelle, N. M. (2019). Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann. N Y Acad. Sci. 1457 (1), 61–91. doi: 10.1111/nyas.14223
Lee, J., Patel, G., Huprikar, S., Calfee, D. P., Jenkins, S. G. (2009). Decreased susceptibility to polymyxin b during treatment for carbapenem-resistant Klebsiella pneumoniae infection. J. Clin. Microbiol. 47 (5), 1611–1612. doi: 10.1128/jcm.02466-08
Lekunberri, I., Balcázar, J. L., Borrego, C. M. (2017). Detection and quantification of the plasmid-mediated mcr-1 gene conferring colistin resistance in wastewater. Int. J. Antimicrob. Agents 50 (6), 734–736. doi: 10.1016/j.ijantimicag.2017.08.018
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi: 10.1093/nar/gkab301
Lin, J. C., Koh, T. H., Lee, N., Fung, C. P., Chang, F. Y., Tsai, Y. K., et al. (2014). Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 6, 21. doi: 10.1186/1757-4749-6-21
Liu, C., Yang, P., Zheng, J., Yi, J., Lu, M., Shen, N. (2022). Convergence of two serotypes within the epidemic ST11 KPC-producing Klebsiella pneumoniae creates the "Perfect storm" in a teaching hospital. BMC Genomics 23 (1), 693. doi: 10.1186/s12864-022-08924-8
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 (3), 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–1534. doi: 10.1093/molbev/msaa015
Mizrahi, A., Delerue, T., Morel, H., Le Monnier, A., Carbonnelle, E., Pilmis, B., et al. (2020). Infections caused by naturally AmpC-producing Enterobacteriaceae: Can we use third-generation cephalosporins? a narrative review. Int. J. Antimicrob. Agents 55 (2), 105834. doi: 10.1016/j.ijantimicag.2019.10.015
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54 (12), 4971–4977. doi: 10.1128/AAC.00834-10
Nang, S. C., Azad, M. A. K., Velkov, T., Zhou, Q. T., Li, J. (2021). Rescuing the last-line polymyxins: Achievements and challenges. Pharmacol. Rev. 73 (2), 679–728. doi: 10.1124/pharmrev.120.000020
Nirwan, P. K., Chatterjee, N., Panwar, R., Dudeja, M., Jaggi, N. (2021). Mutations in two component system (PhoPQ and PmrAB) in colistin resistant Klebsiella pneumoniae from north Indian tertiary care hospital. J. Antibiot (Tokyo) 74 (7), 450–457. doi: 10.1038/s41429-021-00417-2
Nordmann, P., Dortet, L., Poirel, L. (2012). Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18 (5), 263–272. doi: 10.1016/j.molmed.2012.03.003
Ouyang, P., Jiang, B., Peng, N., Wang, J., Cai, L., Wu, Y., et al. (2022). Characteristics of ST11 KPC-2-producing carbapenem-resistant hypervirulent Klebsiella pneumoniae causing nosocomial infection in a Chinese hospital. J. Clin. Lab. Anal. 36 (6), e24476. doi: 10.1002/jcla.24476
Rice, L. B. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197 (8), 1079–1081. doi: 10.1086/533452
Sánchez, F., Fuenzalida, V., Ramos, R., Escobar, B., Neira, V., Borie, C., et al. (2021). Genomic features and antimicrobial resistance patterns of shiga toxin-producing Escherichia coli strains isolated from food in Chile. Zoonoses Public Health 68 (3), 226–238. doi: 10.1111/zph.12818
Satlin, M. J., Lewis, J. S., Weinstein, M. P., Patel, J., Humphries, R. M., Kahlmeter, G., et al. (2020). Clinical and laboratory standards institute and European committee on antimicrobial susceptibility testing position statements on polymyxin b and colistin clinical breakpoints. Clin. Infect. Dis. 71 (9), e523–e529. doi: 10.1093/cid/ciaa121
Sherry, N., Howden, B. (2018). Emerging gram negative resistance to last-line antimicrobial agents fosfomycin, colistin and ceftazidime-avibactam - epidemiology, laboratory detection and treatment implications. Expert Rev. Anti Infect. Ther. 16 (4), 289–306. doi: 10.1080/14787210.2018.1453807
Sonnevend, A., Al Baloushi, A., Ghazawi, A., Hashmey, R., Girgis, S., Hamadeh, M. B., et al. (2013). Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the united Arab Emirates. J. Med. Microbiol. 62 (Pt 7), 1044–1050. doi: 10.1099/jmm.0.059014-0
Stefaniuk, E. M., Tyski, S. (2019). Colistin resistance in Enterobacterales strains - a current view. Pol. J. Microbiol. 68 (4), 417–427. doi: 10.33073/pjm-2019-055
Tan, D., Zhang, Y., Cheng, M., Le, S., Gu, J., Bao, J., et al. (2019). Characterization of Klebsiella pneumoniae ST11 isolates and their interactions with lytic phages. Viruses 11 (11), 1080. doi: 10.3390/v11111080
Tang, B., Chang, J., Luo, Y., Jiang, H., Liu, C., Xiao, X., et al. (2022). Prevalence and characteristics of the mcr-1 gene in retail meat samples in zhejiang province, China. J. Microbiol. 60 (6), 610–619. doi: 10.1007/s12275-022-1597-y
Tang, B., Chang, J., Zhang, L., Liu, L., Xia, X., Hassan, B. H., et al. (2020). Carriage of distinct mcr-1-Harboring plasmids by unusual serotypes of Salmonella. Adv. Biosyst. 4 (3), e1900219. doi: 10.1002/adbi.201900219
Tang, B., Guan, C., Lin, H., Liu, C., Yang, H., Zhao, G., et al. (2023). Emergence of co-existence of mcr-1 and blaNDM-5 in escherichia fergusonii. Int. J. Antimicrob. Agents 61 (3), 106742. doi: 10.1016/j.ijantimicag.2023.106742
Thuy, T. T. D., Lu, H. F., Kuo, P. Y., Lin, W. H., Lin, T. P., Lee, Y. T., et al. (2022). Whole-genome-sequence-based characterization of an NDM-5-producing uropathogenic Escherichia coli EC1390. BMC Microbiol. 22 (1), 150. doi: 10.1186/s12866-022-02562-6
Tian, X., Huang, C., Ye, X., Jiang, H., Zhang, R., Hu, X., et al. (2019). Molecular epidemiology of and risk factors for extensively drug-resistant Klebsiella pneumoniae infections in southwestern China: A retrospective study. Front. Pharmacol. 10. doi: 10.3389/fphar.2019.01307
Tsuji, B. T., Pogue, J. M., Zavascki, A. P., Paul, M., Daikos, G. L., Forrest, A., et al. (2019). International consensus guidelines for the optimal use of the polymyxins: Endorsed by the American college of clinical pharmacy (ACCP), European society of clinical microbiology and infectious diseases (ESCMID), infectious diseases society of America (IDSA), international society for anti-infective pharmacology (ISAP), society of critical care medicine (SCCM), and society of infectious diseases pharmacists (SIDP). Pharmacotherapy 39 (1), 10–39. doi: 10.1002/phar.2209
Tumbarello, M., Raffaelli, F., Giannella, M., Mantengoli, E., Mularoni, A., Venditti, M., et al. (2021). Ceftazidime-avibactam use for klebsiella pneumoniae carbapenemase-producing K. pneumoniae infections: A retrospective observational multicenter study. Clin. Infect. Dis. 73 (9), 1664–1676. doi: 10.1093/cid/ciab176
Velkov, T., Thompson, P. E., Azad, M. A. K., Roberts, K. D., Bergen, P. J. (2019). History, chemistry and antibacterial spectrum. Adv. Exp. Med. Biol. 1145, 15–36. doi: 10.1007/978-3-030-16373-0_3
Wang, J., Tang, B., Lin, R., Zheng, X., Ma, J., Xiong, X., et al. (2022). Emergence of mcr-1- and blaNDM-5-harbouring IncHI2 plasmids in Escherichia coli strains isolated from meat in zhejiang, China. J. Glob Antimicrob. Resist. 30, 103–106. doi: 10.1016/j.jgar.2022.06.002
Wi, Y. M., Choi, J. Y., Lee, J. Y., Kang, C. I., Chung, D. R., Peck, K. R., et al. (2017). Emergence of colistin resistance in Pseudomonas aeruginosa ST235 clone in south Korea. Int. J. Antimicrob. Agents 49 (6), 767–769. doi: 10.1016/j.ijantimicag.2017.01.023
Xia, L., Liu, Y., Xia, S., Kudinha, T., Xiao, S. N., Zhong, N. S., et al. (2017). Prevalence of ST1193 clone and IncI1/ST16 plasmid in E-coli isolates carrying blaCTX-M-55 gene from urinary tract infections patients in China. Sci. Rep. 7, 44866. doi: 10.1038/srep44866
Xu, L., Wan, F., Fu, H., Tang, B., Ruan, Z., Xiao, Y., et al. (2022). Emergence of colistin resistance gene mcr-10 in Enterobacterales isolates recovered from fecal samples of chickens, slaughterhouse workers, and a nearby resident. Microbiol. Spectr. 10 (2), e0041822. doi: 10.1128/spectrum.00418-22
Xu, Y., Zhang, J., Wang, M., Liu, M., Liu, G., Qu, H., et al. (2021). Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 13 (1), 119. doi: 10.1186/s13073-021-00936-5
Zarkotou, O., Pournaras, S., Voulgari, E., Chrysos, G., Prekates, A., Voutsinas, D., et al. (2010). Risk factors and outcomes associated with acquisition of colistin-resistant KPC-producing Klebsiella pneumoniae: a matched case-control study. J. Clin. Microbiol. 48 (6), 2271–2274. doi: 10.1128/jcm.02301-09
Zhao, J., Zhang, Y., Fan, Y., Han, J., Xiong, Z., Liu, X., et al. (2021). Characterization of an NDM-5-producing hypervirulent Klebsiella pneumoniae sequence type 65 clone from a lung transplant recipient. Emerg. Microbes Infect. 10 (1), 396–399. doi: 10.1080/22221751.2021.1889932
Zhou, H., Wang, S., Wu, Y., Dong, N., Ju, X., Cai, C., et al. (2022). Carriage of the mcr-9 and mcr-10 genes in clinical strains of the Enterobacter cloacae complex in China: a prevalence and molecular epidemiology study. Int. J. Antimicrob. Agents 60 (4), 106645. doi: 10.1016/j.ijantimicag.2022.106645
Keywords: Enterobacterales, polymyxin-resistance, prevalence, phenotypes, comparative genomics
Citation: Xiao C, Li X, Huang L, Cao H, Han L, Ni Y, Xia H and Yang Z (2023) Prevalence and molecular characteristics of polymyxin-resistant Enterobacterales in a Chinese tertiary teaching hospital. Front. Cell. Infect. Microbiol. 13:1118122. doi: 10.3389/fcimb.2023.1118122
Received: 07 December 2022; Accepted: 29 March 2023;
Published: 18 April 2023.
Edited by:
Biao Tang, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Jian-cang Zhou, Zhejiang University, ChinaAna Daniela Ferreira, Wellcome Sanger Institute (WT), United Kingdom
Jade LL Teng, The University of Hong Kong, Hong Kong SAR, China
Copyright © 2023 Xiao, Li, Huang, Cao, Han, Ni, Xia and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhitao Yang, eWFuZ3poaXRhb0Bob3RtYWlsLmZy
†These authors have contributed equally to this work
 Chenlu Xiao
Chenlu Xiao Xuming Li
Xuming Li Lianjiang Huang4†
Lianjiang Huang4† Huiluo Cao
Huiluo Cao Han Xia
Han Xia Zhitao Yang
Zhitao Yang