- 1Zhejiang University-University of Edinburgh (ZJU-UoE) Institute, Zhejiang University, Haining, Zhejiang, China
- 2Department of Infectious Diseases, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3Key Laboratory of Microbial Technology and Bioinformatics of Zhejiang Province, Hangzhou, China
- 4Regional Medical Center for National Institute of Respiratory Diseases, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, China
Acinetobacter baumannii is widely distributed in nature and in hospital settings and is a common pathogen causing various infectious diseases. Currently, the drug resistance rate of A. baumannii has been persistently high, showing a worryingly high resistance rate to various antibiotics commonly used in clinical practice, which greatly limits antibiotic treatment options. Tigecycline and polymyxins show rapid and effective bactericidal activity against CRAB, and they are both widely considered to be the last clinical line of defense against multidrug resistant A. baumannii. This review focuses with interest on the mechanisms of tigecycline resistance in A. baumannii. With the explosive increase in the incidence of tigecycline-resistant A. baumannii, controlling and treating such resistance events has been considered a global challenge. Accordingly, there is a need to systematically investigate the mechanisms of tigecycline resistance in A. baumannii. Currently, the resistance mechanism of A. baumannii to tigecycline is complex and not completely clear. This article reviews the proposed resistance mechanisms of A. baumannii to tigecycline, with a view to providing references for the rational clinical application of tigecycline and the development of new candidate antibiotics.
1 Background
Acinetobacter baumannii is a nonfermenting gram-negative bacterium that is one of the most important members of the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) group of microorganisms. It is an internationally notorious hospital pathogen, posing a serious threat to public health (Antimicrobial Resistance, 2022). It has been reported that A. baumannii is closely related to the occurrence and development of ventilator-associated pneumonia, wound and urinary tract infections, bloodstream infections, endocarditis and meningitis, with very high morbidity and mortality, especially in intensive care unit (ICU) patients (Moubareck and Halat, 2020; Ibrahim et al., 2021; Tokur et al., 2022).
Due to multiple factors, such as irrational use of antibiotics, cross-infection among inpatients and the transmission of resistance genetic elements, an increasing number of strains of A. baumannii have evolved into multidrug resistant (MDR), extensive drug resistant (XDR) and even pandrug resistant (PDR) strains (Ibrahim, 2019), which limits the clinical treatment options of antibiotics. At present, carbapenems are commonly used for the treatment of A. baumannii; however, in recent years, carbapenem-resistant A. baumannii (CRAB) has been widely reported (Zhu et al., 2022).
Tigecycline is a third-generation tetracycline derivative, and the main difference between tetracycline and tigecycline is that ring D of tigecycline is linked to 7-dimethylamido and 9-t-butylglycylamido moieties (Figure 1). Due to the stacking interaction of 9-t-butylglycylamido moieties with C1054 of 16S rRNA, tigecycline has an increased affinity for ribosomes, thereby overcoming TetM-mediated tetracycline resistance and exhibiting superior antibacterial activity (Jenner et al., 2013). In addition, the unique chemical structure of tigecycline increases the lipid solubility of the drug and also prevents it from being pumped out of the cell by most membrane-bound efflux proteins by creating a steric hindrance. More importantly, tigecycline also overcomes other antibiotic resistance mechanisms such as drug target modification, enzymatic degradation, and DNA gyrase mutations, making it a promising antibacterial drug for a wide range of applications (Šeputienė et al., 2010).
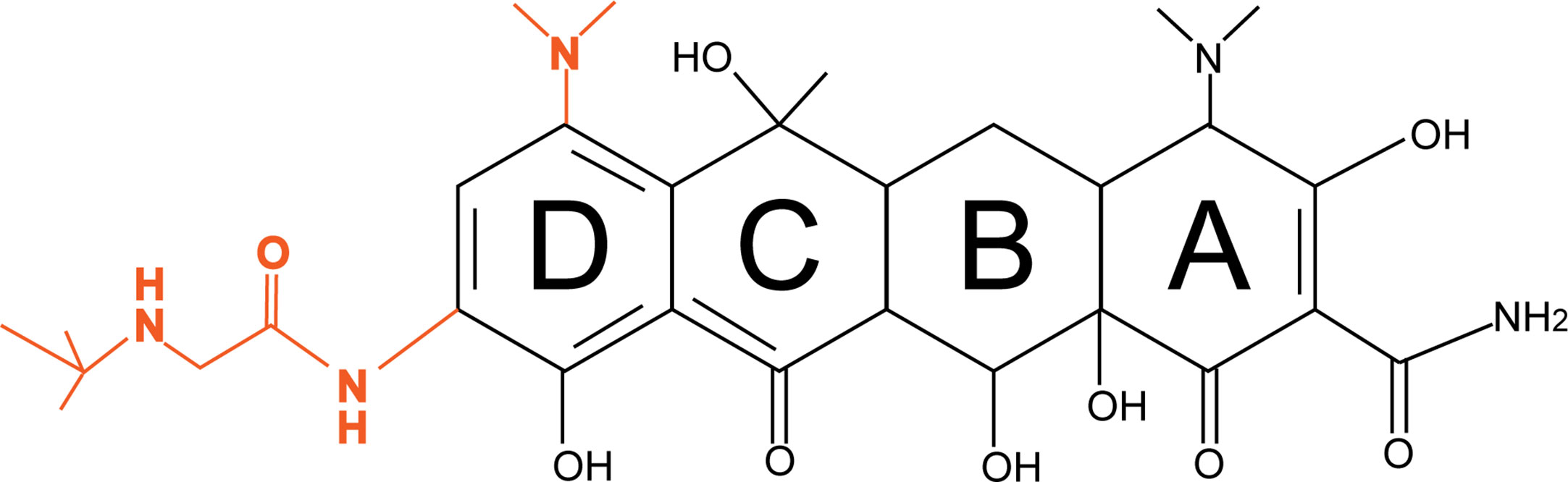
Figure 1 Diagrams of the chemical structures of tetracycline and tigecycline. The chemical structure of tetracycline is shown in black; the only difference between tigecycline and tetracycline is the attachment of 7-dimethylamino and 9-t-butylglycylamido moieties to the D ring, shown in orange.
Gamal et al. collected non-reproducible A. baumannii from Vietnam and Germany and performed susceptibility testing of 18 drugs, showing ultra-high resistance rates to fosfomycin (96%), chloramphenicol (95%) and cefotaxime (81%); moderate resistance rates to imipenem (24%) and meropenem (24%), which are carbapenem antibiotics; and low resistance rates to tigecycline (12%) and colistin (3%) (Wareth et al., 2020). Meanwhile, Harald et al. reported data from 2014-2016 for tigecycline and eight commonly used antimicrobial drugs in Africa, Asia, Europe, North America and South America. The results showed that carbapenem resistance rates in all regions were higher in A. baumannii isolates, at 65.8%. Since FDA and CLSI breakpoints do not apply to tigecycline, this article does not show data on tigecycline resistance rates, but MIC90 values of 1-2 mg/L in all regions could indicate the efficient antibacterial ability of tigecycline (Seifert et al., 2018). The first reported appearance of tigecycline-resistant A. baumannii was in 2007. A recent prevalence survey of A. baumannii showed that tigecycline resistance was less than 5.5% in Korea, India and China (Chen et al., 2023); nevertheless, clinical cases of A. baumannii infection resistant to tigecycline are increasing (Sun et al., 2013), and such a dilemma is worrisome and needs to be taken seriously (Navon-Venezia et al., 2007).
The generation and development of tigecycline resistance mechanisms have attracted widespread attention. Analysis of tigecycline resistance and the resistance mechanism of A. baumannii can provide theoretical guidance for the clinical formulation of treatment strategies and control of A. baumannii outbreaks.
2 Resistance mechanisms
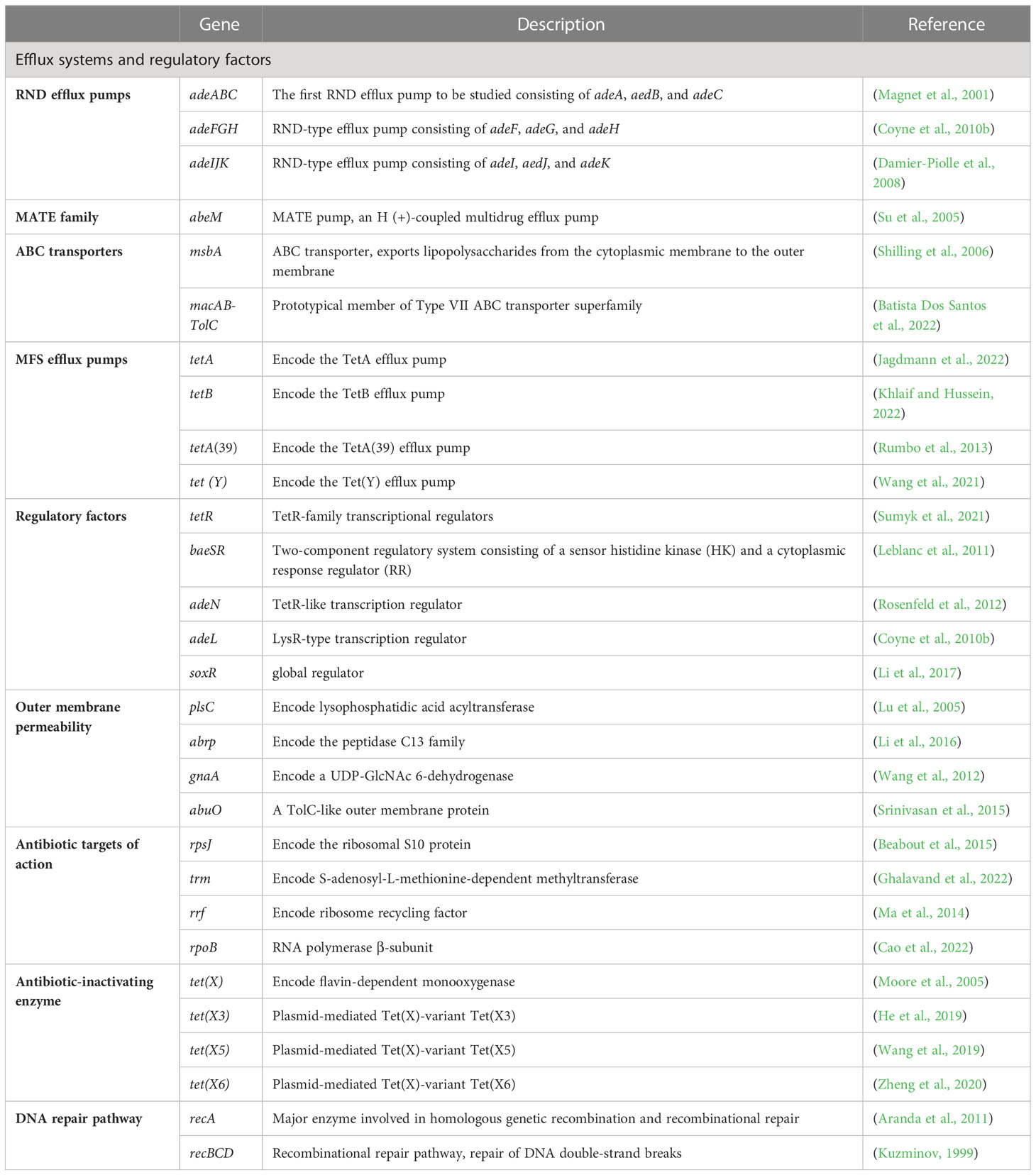
The main mechanisms of tigecycline resistance in A. baumannii can be divided into five categories: overexpression of efflux pumps, altered outer membrane permeability, altered tigecycline targets of action, production of tigecycline-inactivating enzymes, and repair pathways mediating tigecycline resistance after DNA damage (Table 1) (Figure 2).
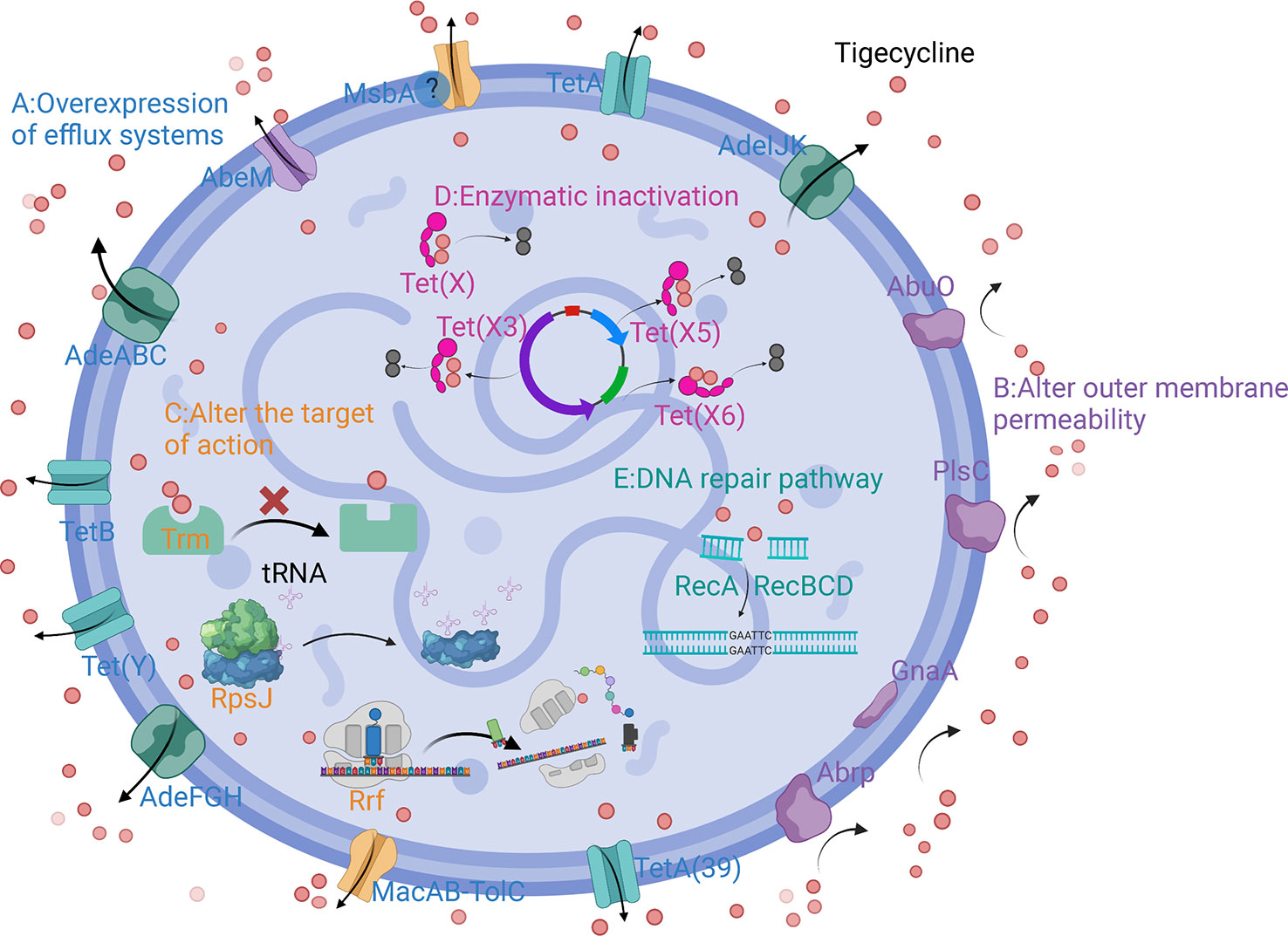
Figure 2 Schematic diagram of the mechanism of tigecycline resistance in A. baumannii. (A) Overexpression of efflux systems; (B) Altered outer membrane permeability; (C) Altered target of action; (D) Enzymatic inactivation; (E) DNA repair pathway.
2.1 Efflux pumps systems
Overexpression of efflux pumps is a key mechanism responsible for drug resistance in A. baumannii. Efflux pump excretes antimicrobial drugs from cells, which leads to the decrease of drug concentration and drug resistance (Meyer et al., 2022). There are five superfamilies associated with drug resistance in A. baumannii: the resistance-nodulation-cell division (RND) family, multidrug and toxic compound extrusion (MATE) family, the ATP-binding cassette (ABC) transporters, the major facilitator superfamily (MFS) and the small multidrug resistance (SMR) family, of which the first four have been reported to mediate tigecycline resistance (Rafiei et al., 2022).
2.1.1 RND efflux pumps and regulatory factors
The RND efflux pump is the most prevalent efflux pump for MDR A. baumannii and contains three major members: AdeABC, AdeFGH and AdeIJK (Figure 3). They are all triplets consisting of three structural proteins encoded by structural genes. AdeABC is the first RND system studied in A. baumannii and consists of adeA (major fusion protein), adeB (multidrug transporter) and adeC (outer membrane protein) (Figure 4) (Coyne et al., 2011). Many previous studies have shown that overexpression of AdeABC contributes to reduced tigecycline susceptibility (Hornsey et al., 2010; Yoon et al., 2013). The expression levels of adeA, adeB and adeC were diverse in different studies. However, most researchers prefer to consider adeB as the most important gene in adeABC, as it plays a major resistance role in A. baumannii (Yoon et al., 2016). Yin et al. screened over 1000 clinical strains of A. baumannii and found that overexpression of the AdeABC efflux pump was the major mechanism for increased tigecycline resistance; furthermore, the expression level of adeB showed a linear relationship with the minimum inhibitory concentration (MIC) of tigecycline (Yuhan et al., 2016). Privita et al. demonstrated the essential role of adeC in the AdeABC efflux pump by inhibiting adeC and achieving complete blockage of the efflux process when designing a potent inhibitor of the AdeABC efflux pump against A. baumannii (Verma and Tiwari, 2018). However, in another study, the recombinant A. baumannii strain of inactivated adeC showed no difference in the efflux function of AdeABC compared to wild-type strains, which seems to indicate that adeC is not required to exert resistance (Marchand et al., 2004). Therefore, the role of adeC in AdeABC needs to be further explored.
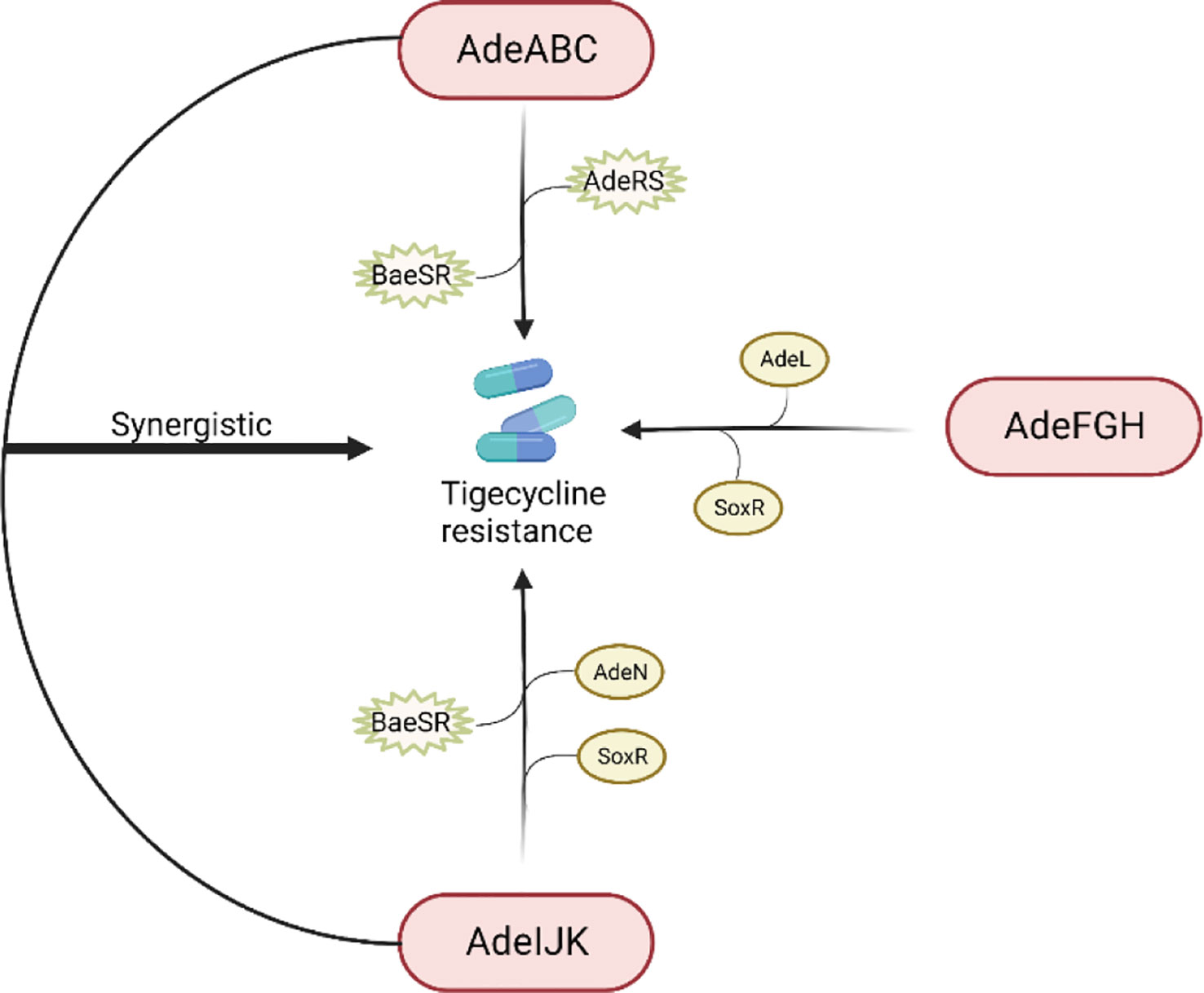
Figure 3 Regulation of tigecycline resistance in Acinetobacter baumannii by AdeABC, AdeFGH and AdeIJK efflux pumps. The AdeABC efflux pump is regulated by the two-component system AdeRS and BaeSR; the AdeIJK efflux pump is controlled by the two-component system BaeSR, the global regulator SoxR and the TetR-like transcription regulator AdeN; the expression of AdeFGH is regulated by the LysR-type transcription regulator AdeL and the global regulator SoxR; and AdeABC and AdeIJK are synergistically involved in the production of tigecycline resistance.
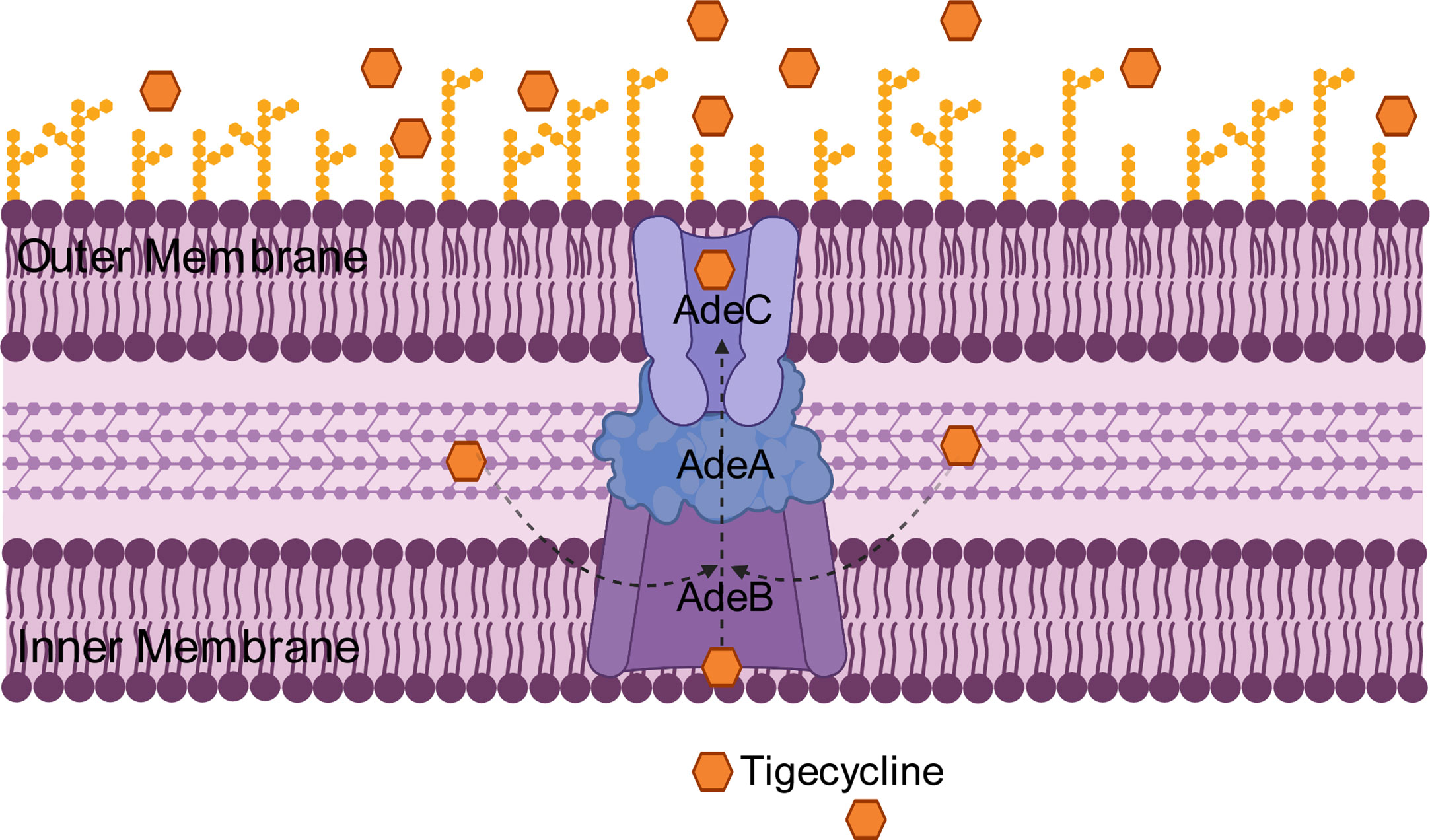
Figure 4 Schematic diagram of the tigecycline resistance mechanism of the AdeABC efflux pump in A. baumannii: adeA encodes major fusion protein, adeB encodes multidrug transporter and adeC encodes outer membrane protein, which together form the AdeABC triplet structure. AdeB, assisted by adeA, effluxes tigecycline from the inner membrane or cytoplasm out of the membrane through adeC to exert drug resistance.
Previous studies have shown that AdeABC expression is regulated by the downstream two-component system AdeRS, which consists of a sensor kinase adeS and a response regulator adeR. Mutations in AdeRS, including A94 V, S8A, H189T, I252S, and T156 M in AdeS (Yoon et al., 2013; Lucassen et al., 2021b; Zhang et al., 2022) and P56S, L192R, E219A, and D26 N in AdeR (Yoon et al., 2013; Lucassen et al., 2021b), increased the expression level of AdeABC, leading to decreased susceptibility to tigecycline. Jeongwoo et al. (Jo and Ko, 2021) identified the insertion of ISAba1 in adeS of the tigecycline-resistant subpopulation, and they observed that adeB gene expression was upregulated and a soluble truncated adeS protein was produced, which contributed to the overexpression of the AdeABC efflux pump and ultimately caused the tigecycline-resistant outcome, and a similar study also obtained tigecycline-resistant bacteria with ISAba1 (Zhang et al., 2022). Another two-component system, BaeSR, has been reported to affect tigecycline susceptibility in A. baumannii by positively regulating the adeA and adeB genes, but the relationship between BaeSR and AdeRS remains unclear (Lin et al., 2014).
Overexpression of AdeFGH was proven to be another mechanism of reduced tigecycline sensitivity in A. baumannii and is modulated by AdeL, an encoded LysR-type transcriptional regulator encoded upstream of AdeFGH (Coyne et al., 2010b). However, Deng et al. found no significant difference in the expression levels of the adeG gene between tigecycline-resistant and tigecycline-susceptible bacteria (Deng et al., 2014). Therefore, additional tigecycline resistance mechanisms must exist.
Unlike AdeABC and AdeFGH, AdeIJK is present in all A. baumannii isolates and leads to intrinsic resistance to tigecycline. The investigators constructed AdeIJK overexpressing bacteria, which resulted in significant inhibition of bacterial growth, and they concluded that AdeIJK pump overexpression inhibits the growth of A. baumannii and causes toxic effects (Damier-Piolle et al., 2008). However, Coyne et al. applied RT-PCR and microarray experiments and found that the expression level of AdeIJK in strains with low levels of tigecycline resistance was always lower than that of AdeABC, suggesting that AdeIJK can only be overexpressed in a restricted compartment and then become toxic to the host, which indicates that AdeIJK contributes less to antibiotic resistance. However, the mechanism of AdeIJK toxicity to the host has not been revealed yet and needs to be further elucidated (Coyne et al., 2010a). It has also been shown that AdeN, a transcriptional regulator belonging to the TetR family, is upstream of AdeIJK and can inhibit the expression of AdeIJK (Rosenfeld et al., 2012). In addition, the BaeSR system may also be involved in the regulation of AdeIJK. Transcriptomic data from an LPS-deficient A. baumannii strain showed increased expression levels of the baeS and baeR genes, as well as increased expression levels of the AdeIJK genes (Henry, 2012).
AdeABC, AdeFGH and AdeIJK are implicated in tigecycline resistance in A. baumannii. Yoon et al. evaluated the relative contribution of these three efflux pumps in the clinical setting and concluded that AdeABC plays a major role in resistance, while there was no necessary association between the tigecycline MIC and AdeABC expression levels (Yoon et al., 2013). Laurence et al. found that mutants with dual inactivation of the AdeABC and AdeIJK efflux pump genes were much more susceptible to tigecycline than wild-type strains and mutants with inactivation of either single efflux pump gene, suggesting that AdeABC and AdeIJK can be synergistically involved in tigecycline resistance (Damier-Piolle et al., 2008). In 2017, a study from Beijing first proposed that the global regulator SoxR is a negative regulator of efflux pump gene expression. By constructing soxR overexpression mutants, they interestingly found that soxR overexpression resulted in statistically significant reduced expression levels of adeJ, adeG, and adeS and increased (2-fold) susceptibility to tigecycline, implying that SoxR is involved in tigecycline resistance regulation in A. baumannii (Li et al., 2017).
2.1.2 (MATE) family
MATE transporters can drive various toxic and harmful substances out of the membrane through Na+ or H+ as a coupling ion. It is an important defense mechanism for bacteria to resist external environmental pressure (Zhao et al., 2021). Here, AbeM is an H+-coupled multidrug efflux pump that belongs to the MATE transporter family (Su et al., 2005). Recently, Mina et al. reported that abeM gene expression was increased 16- to 256-fold in tigecycline-resistant A. baumannii compared to controls, and an increase in the abeM gene was observed in 70% of CCCP-positive tests, (CCCP is an abbreviation for carbonyl cyanide m-chlorophenyl hydrazone, a known pump inhibitor, the purpose of the CCCP-positive test is to screen for potential efflux pumps that are not inhibited by CCCP and play a role in drug resistance), indicating that abeM is involved in tigecycline resistance (Owrang et al., 2018). In contrast, Deng et al. found that tigecycline-resistant A. baumannii showed only 1-fold higher abeM gene expression levels than sensitive bacteria, and the difference was not significant (Deng et al., 2014). What these inconsistencies can indicate is that the resistance of A. baumannii to tigecycline is not unique, and there are additional mechanisms.
2.1.3 ABC transporters
The ATP-binding cassette (ABC) transporter differs from other family transporters in that it uses the energy generated by ATP binding and hydrolysis to perform efflux functions (Okada and Murakami, 2022). MsbA was the first ATP-binding cassette transporter to be crystallized and analyzed (Shilling et al., 2006). Chen et al. transferred the wild-type msbA gene into a tigecycline-resistant mutant (msbA A84V), but they found no change in MIC, and the mutant strain had a smaller colony size than the parental strain, so they concluded that msbA did not contribute to tigecycline resistance but could reduce fitness (Chen et al., 2014). In another study, in contrast, msbA had a highly significant p value in the tigecycline-resistant group; they considered msbA mutations to be gain-of-function mutations and speculated that a possible resistance mechanism is that these mutations expand pump specificity, facilitating tigecycline efflux (Hammerstrom et al., 2015). Overall, the function of msbA needs to be re-evaluated.
MacAB-TolC is another important efflux pump system involved in tigecycline resistance. Lin et al. reported a statistically significant increase in the expression level of the macB gene in tigecycline-resistant strains compared to susceptible strains (Lin et al., 2017). In addition, the BaeSR system regulates macAB-TolC expression (Henry, 2012).
2.1.4 MFS efflux pumps
The MFS efflux pump is a proton-dependent antimicrobial drug efflux system that includes passive transporters and secondary active transport systems (Stephen et al., 2023). Most reports on TetA(39) have been on resistance to tetracycline and doxycycline. The study by Rumbo et al. proposed that tigecycline-resistant A. baumannii may be due to a novel efflux pump system, TetA(39) (Rumbo et al., 2013). TetA is an important member of the MFS family, and its expression is controlled by TetR of the TetR-family transcriptional regulators (TFR) (Sumyk et al., 2021). Foong et al. reported that all strains possessing pBAV1K_TetA, including adeAB and adeIJ knockout strains, showed increased MICs to tigecycline compared to controls (without the tetA gene). Furthermore, when exposed to tigecycline, the expression levels of tetA in A. baumannii AYE showed remarkable elevation, while adeB and adeJ genes were expressed at only considerable levels. Their findings demonstrate that TetA is a predominant determinant in tigecycline resistance and synergizes with AdeABC and AdeIJK (Foong et al., 2020). Recently, however, another study reported that all tigecycline-nonsusceptible A. baumannii isolates in their collection did not have the tetA gene but instead found the tetB gene (Khlaif and Hussein, 2022). Like TetA, TetB is also a non-chromosomally encoded MFS efflux pump (Verma et al., 2021). Overall, there is a correlation between TetB and TetA and tigecycline resistance, and more research is urgently needed to reveal the underlying mechanisms.
Tet(Y) originally encodes tetracycline resistance and is a member of the tetracycline-specific efflux pump (Fang et al., 2020). Wang et al. first observed that the plasmid-mediated tet(Y) gene is a determinant of tigecycline resistance in A. baumannii. They introduced the plasmid containing the Tet(Y) gene into the host bacterium, resulting in a 4-fold increase in tigecycline MIC. Moreover, they found that 2016GDAB1 (The tet(Y) and tetA(39) genes coexist on the p2016GDAB1 plasmid of A. baumannii, both of which are followed by a tetR gene) showed a 128-fold increase in MIC, suggesting that tet(Y) and tetA(39) together synergistically confer high levels of tigecycline resistance (Wang et al., 2021).
2.2 Outer membrane permeability
To prevent the entry of antimicrobial agents into cells, reducing the permeability of the outer membrane is another common mechanism of resistance.
PlsC, encoding 1-acyl-sn-glycero-3-phosphate acyltransferase, catalyzes the acylation of lysophosphatidic acid to form phosphatidic acid, thereby playing a role in maintaining epidermal permeability barrier function (Lu et al., 2005). Li et al. identified a strain of A. baumannii, 19606-M24, with high levels of resistance to tigecycline (MIC = 24 mg/L), and whole genome comparison revealed a mutation in plsC. Complementation studies revealed that strains introduced with the wild-type plsC gene restored susceptibility to tigecycline, indicating that the frameshift mutation of plsC gene is associated with tigecycline resistance. In subsequent flow cytometry, the highest membrane potential was observed for 19606-M24, while a decrease in membrane potential was also observed in complementary strains. These results convincingly demonstrate that plsC mediates resistance to tigecycline by affecting cell membrane permeability (Li et al., 2015).
Soon thereafter, Li et al. identified a new gene, abrp, encoding the peptidase C13 family, with a frameshift mutation. Compared to the abrp knockout strain, the wild-type strain showed reduced susceptibility to a variety of antibiotics, including tigecycline. Furthermore, they found that the abrp deletion strain increased cell membrane permeability accompanied by impaired bacterial growth. Thus, it can be concluded that abrp increases resistance to tigecycline by altering cell membrane permeability (Li et al., 2016).
Another gene, gnaA, has also been reported to be associated with tigecycline resistance (Hammerstrom et al., 2015; Lucassen et al., 2021a). GnaA encodes an enzyme involved in catalyzing the initial step of UDP-D-MANPNAC3NACA synthesis and was first identified in Escherichia coli (Wang et al., 2012). In 2019, Xu et al. identified tigecycline-resistant mutant strains with an insertion of ISAba16 in the gnaA gene of unknown function. The membrane potential of gnaA-deficient bacteria was significantly higher than that of the wild-type strain, and complementation with the wild-type gnaA gene resulted in a reduced membrane potential, indicating that the barrier function of the cell membrane was restored. Moreover, the researchers revealed that the gnaA gene affects lipooligosaccharide (LOS) and capsular polysaccharide (CPS) synthesis and that disruption of gnaA also affects the virulence, morphology, and susceptibility of A. baumannii to other classes of antibiotics. Unfortunately, no changes in tigecycline MIC were seen in gnaA complementary experiments, this could still suggest that the gnaA gene may be involved in tigecycline resistance by altering cell membrane permeability (Xu et al., 2019). In conclusion, the resistance of gnaA to tigecycline needs to be supported by more studies.
Outer membrane protein (OMP) is a specific channel protein present in the lipid bilayer structure of gram-negative bacteria, located on the surface of the cell membrane or embedded in it, which can serve as a channel for antibiotics to enter the cell (Tokuda, 2009). Vijaya et al. uncovered a putative OMP, abuO, associated with antimicrobial and oxidative stress resistance in A. baumannii. Compared with the wild-type strain, the abuO knockout strain showed increased susceptibility to different types of antibiotics, including tigecycline, with MIC values changing from 2 mg/L to 0.75 mg/L. Meanwhile, the MIC of the complementary abuO isolate again returned to the same level as the wild-type strain. This may indicate a role for abuO in tigecycline resistance (Srinivasan et al., 2015). In addition, RT−PCR results showed that the abuO-deficient strain not only increased the expression levels of efflux pump-related genes but also changed the expression of other membrane proteins, which implies that abuO plays an integral role in A. baumannii and that its relationship with tigecycline resistance mechanisms should still be investigated in depth.
2.3 Antibiotic targets of action
Changing the target of antibiotic action is also one of the mechanisms by which bacteria develop resistance. Bacteria prevent antibiotic binding by altering the target structure, thereby creating resistance.
The S10 protein, which is closest to the tigecycline binding pocket, consists of 53-60 amino acid residues encoded by the rpsJ gene and helps to participate in maintaining the normal structure of the tigecycline binding site. Researchers found that rpsJ mutations emerged in a variety of pathogens under tigecycline exposure and conferred resistance to tigecycline. They believe that mutations in rpsJ caused a structural change in the S10 loop, which in turn changes the conformation of the 16S RNA, ultimately leading to reduced binding affinity for tigecycline and ribosomes. Another model is that reduced tigecycline susceptibility promotes entry of tRNA and increases binding to ribosomes (Beabout et al., 2015). Additionally, since rpsJ mutations have a low fitness cost and occur widely in a variety of pathogens, all tested strains had a mutation at amino acid position 57 of S10, while gram-positive bacteria have a higher variation at amino acid position 60 (Figure 3). These results suggest that rspJ has promise as an important marker for detecting whether pathogens are resistant to tigecycline. Another result likewise reported that rspJ plays an important role in tigecycline resistance (Hammerstrom et al., 2015).
The gene trm (tigecycline-related-methyltransferase) encodes S-adenosyl-L-methionine-dependent methyltransferase. Chen et al. obtained the resistant strain 19606-T8 (MIC = 8 mg/L) with a deletion mutation in the trm gene and then transformed the wild-type trm gene into 19606-T8. Interestingly, it restored the sensitivity to tigecycline, MIC dropping from 8 mg/L to 1 mg/L, implying that trm plays an important role in reducing A. baumannii susceptibility to tigecycline (Chen et al., 2014). However, how methyltransferase mediates the reduction in sensitivity to tigecycline has not been investigated. Another study also reported diverse amino acid alterations in trm of tigecycline-resistant isolates, such as S94A, V121A, C285F, K291R, G310C, H312P, T321I, T323S, and M378K (Ghalavand et al., 2022). The mechanism by which trm mediates resistance to tigecycline is unclear. As a methyltransferase, trm may act as ArmA, RmtA and NpmA to methylate the ribosomal target, hinder the binding of tigecycline to its target, and mediate tigecycline resistance (Fritsche et al., 2008; Chen et al., 2014).
The rrf gene encodes the production of ribosome recycling factor (RRF). RRF is an essential component of protein synthesis and is involved in the release of polypeptides from the ribosome for a new round of translation (Ma et al., 2014). In an earlier study, Hammerstrom et al. identified mutations in the rrf gene in tigecycline-resistant A. baumannii. They proposed that mutations in rrf (M1 V, N3N) might reduce translation of mRNA and in return affect the binding of tigecycline to the ribosome, but they did not proceed to verify this hypothesis (Hammerstrom et al., 2015).
Subsequently, Hua et al. revealed the mechanism of rrf involvement in tigecycline resistance. They constructed recombinant mutants XH1457 (MDR-ZJ06 rrfH33P) and XH1458 (MDR-ZJ06 rpoBG136D), which showed only a slight increase in tigecycline tolerance compared to the parental strain, but Raman spectrometry provided a strong complement to the tigecycline resistance conferred by these two mutant genes. Furthermore, it was confirmed that mutations in rrf decreased the expression of RRF proteins, affecting the ribosome recycling process and ultimately exhibiting a tigecycline-resistant phenotype. In addition, the transcriptomic data also revealed that rpoBG136D could regulate the expression level of trm (Hua et al., 2021).
2.4 Antibiotic-inactivating enzyme
Resistance to antibiotics by degrading and modifying antibiotics to make them inactive is an important resistance mechanism for bacteria.
Several studies have recently reported the widespread dissemination of Tet(X) and its variants through plasmid mediation in A. baumannii, giving rise to resistance to a variety of tetracyclines, which has drawn the attention of researchers. Tet(X) encodes flavin-dependent monooxygenase that modifies first- and second-generation tetracycline. Tigecycline is one of the substrates of Tet (X), which requires activation in the presence of FAD, NADPH, O2, and Mg2+, Tet (X) modifies tigecycline to 11a-hydroxytigecycline, which alters the physical properties of tigecycline by weakening its binding to magnesium and decreasing its affinity to ribosomes, this inactivates tigecycline, thus allowing bacteria to develop resistance (Moore et al., 2005).
To date, eight genes associated with high levels of resistance to tigecycline have been reported in the Tet(X) family, namely Tet(X), Tet(X1), Tet(X2), Tet(X3), Tet(X4), Tet(X5), Tet(X6), Tet(X7), where Tet(X1) is a truncated variant with no enzymatic activity. These Tet(X) and its variants have been detected in various pathogens such as Klebsiella pneumoniae, Pseudomonas aeruginosa and Escherichia coli. Among them, Tet(X), Tet(X3), Tet(X5), and Tet(X6) have been reported to confer tigecycline resistance in A. baumannii (Cui et al., 2021). In addition, the Tet(X) was originally identified in the obligate anaerobe Bacteroides fragilis (Yang et al., 2004). As reported in the literature, compared to Tet(X) (M37699), Tet(X3) (MK134375), Tet(X5) (CP040912), Tet(X6) (CP044517) showed 86%, 90%, 84% amino acid identities, respectively (Cui et al., 2021). He et al. demonstrated that Tet(X3) catalyzes the oxygenation of tigecycline at C11a to form 11a-hydroxyltigecycline, resulting in tigecycline inactivation. Whereas the MIC of tigecycline was substantially increased (64-fold) in strains possessing the Tet(X3)-carrying plasmid, mouse infection models likewise revealed that Tet(X3) impaired the therapeutic efficacy of tigecycline in vivo and ultimately led to drug resistance (He et al., 2019). Wang et al. identified another Tet(X) variant, Tet(X5), which is structurally and functionally similar to other variants and hydroxylates tigecycline. Tet(X5) shows increased resistance to a wide range of tetracyclines (Wang et al., 2019). Not coincidentally, scientists have also found that Tet(X5) and Tet(X6) in A. baumannii are resistant to tigecycline through hydroxylation and that ISCR2-mediated transposition and recombination exacerbate the spread of Tet(X5) (Chen et al., 2021). Notably, ISCR2 is an insertion sequence type element capable of transposing neighboring DNA sequences, including antibiotic resistance genes, through a process of rolling-circle transposition (Chen et al., 2021). ISCR2 shares 65% amino acid identity with ISCR1, which is a mobile element involved in the spread of resistance genes (Xu et al., 2017).
Overall, the clonal transmission of Tet(X) and its variants should not be underestimated and deserves attention.
2.5 DNA repair pathway
It has been demonstrated that DNA damage under antibiotic induction can mediate the killing of A. baumannii (Sampson et al., 2012), and recA and recBCD are important pathways for DNA repair. RecA is the major enzyme involved in gene homologous recombination and recombination repair (Aranda et al., 2011), and recBCD is involved in DNA double-stranded repair (Kuzminov, 1999). Taofeek et al. found increased susceptibility of recA- and recBCD-inactivated A. baumannii to various antibiotics, including tigecycline, suggesting that the recA- and recBCD-mediated repair pathway may protect A. baumannii from antibiotic killing, leading to drug resistance (Ajiboye et al., 2018).
2.6 Other potential mechanisms
In addition to the abovementioned mechanisms related to tigecycline resistance in A. baumannii, there are some potential resistance genes that need to be further investigated. Nogbou et al. found that A. baumannii with mutations in both the parC and gyrA genes had higher levels of resistance to tigecycline than strains without mutations (Nogbou et al., 2021). Hammerstrom et al. used a bioinformatics approach to infer that rnpA, rpoD, rppH, wzc, and pcaF are candidate tigecycline resistance genes (Hammerstrom et al., 2015). Whether these genes can indeed mediate tigecycline resistance and by what mechanism they cause resistance needs to be investigated more thoroughly in the future.
3 Conclusions
Tigecycline-resistant A. baumannii is a global public health terrorist. To adapt to the pressures of the external environment, bacteria have evolved multiple resistance strategies. Many studies have shown that overexpression of the RND efflux pump is the main cause of tigecycline resistance in A. baumannii and that the expression level of the efflux pump-related gene adeB is positively correlated with the MIC value of tigecycline (Yuhan et al., 2016), but some researchers have reported that there is no necessary link between the two (Yoon et al., 2013). The expression of the efflux pump is also regulated by various regulatory factors, and the efflux pumps can interact with each other. Therefore, the mechanism of the active efflux pump is diverse and complex, and a comprehensive study of the mechanism of action of the efflux pump is needed.
The lipid bilayer is an important barrier for A. baumannnii, and loss of channel proteins or genetic mutations that reduce outer membrane permeability can lead to difficulties in tigecycline entry into the cell and an increase in the minimum inhibitory concentration of bacteria. Therefore, focusing on weakening lipopolysaccharides bilayer synthesis to make bacteria sensitive to antibiotics is the conventional direction of thinking. In addition, a deeper understanding of the function and properties of channel proteins, as well as the identification of regulatory factors that may affect outer membrane permeability and channel protein closure, will facilitate the targeted development of novel drugs and improve current clinical drug therapies.
As described above, rpsJ and rrf can be mutated to reduce the ability to bind ribosomal proteins (Beabout et al., 2015; Hammerstrom et al., 2015; Hua et al., 2021), while trm may alter the site of action of the drug through enzymatic modifications, all of which ultimately lead to resistance to tigecycline (Ghalavand et al., 2022). Importantly, the high-frequency site of tigecycline resistance due to rpsJ mutations is amino acid position 57, which occurs not only in A. baumannii but also in a wide range of pathogens (Beabout et al., 2015). This provides a unique way to identify tigecycline resistance by screening for rpsJ mutations in the future.
Tet(X) can inactivate tigecycline and develop resistance. Worryingly, novel Tet (X) variants continue to be discovered and they can spread rapidly through horizontal gene transfer in a wide range of pathogens and appear to be on the verge of a global epidemic (Fang et al., 2020). Recently, Tet(X) and its variants have been found to confer high levels of tigecycline resistance and cause clonal transmission in different species of bacteria through plasmid-mediated and widespread association with other resistance genes and transposons such as blaOXA-72, blaOXA-58, blaNDM-1, and ISCR2, posing a great safety risk to the health care system (Zheng et al., 2020; Chen et al., 2021; Hsieh et al., 2021). This calls for effective measures to intervene in the occurrence of such epidemic events. For example, we need to investigate in depth the origin, structure and evolutionary mechanisms of Tet(X) variants, as well as design inhibitors and disruptors against this enzyme to overcome the high levels of tigecycline resistance mediated by Tet(X) and variants.
RecA and recBCD are important factors involved in the repair of DNA after damage. RecA was shown to protect A. baumannii from UV light, heat shock, desiccation and several classes of antibiotics including tigecycline in the study by Aranda et al. In addition, in vitro and in vivo virulence assays showed that the virulence of the A. baumannii recA mutant was significantly reduced. Another study similarly confirmed that recA and recBCD deletions contribute to tigecycline-mediated lethality against A. baumannii. Accordingly, future attention could be focused on the inhibition of recA, recBCD for the purpose of treating A. baumannii infections.
In summary, A. baumannii can produce resistance to tigecycline through RND efflux pumps (AdeABC, AdeFGH, AdeIJK) (Magnet et al., 2001; Damier-Piolle et al., 2008; Coyne et al., 2010b), MATE family (AbeM) (Su et al., 2005), ABC transporters (MsbA, MacAB-TolC) (Shilling et al., 2006; Batista Dos Santos et al., 2022), and MFS efflux pumps (TetA, TetB, TetA(39), Tet (Y)) (Rumbo et al., 2013; Wang et al., 2021; Jagdmann et al., 2022; Khlaif and Hussein, 2022). In addition, A. baumannii also reduces membrane permeability (plsc, abrp, gnaA, abuO) (Lu et al., 2005; Wang et al., 2012; Srinivasan et al., 2015; Li et al., 2016), alters antibiotic targets (rpsJ, trm, rrf, rpoB) (Ma et al., 2014; Beabout et al., 2015; Hua et al., 2021; Ghalavand et al., 2022), produces tigecycline inactivating enzymes (Tet(X) and its variants Tet (X3), Tet(X5), Tet(X6)) (Moore et al., 2005; He et al., 2019; Chen et al., 2021) and DNA repair pathways (recA and recBCD) (Kuzminov, 1999; Aranda et al., 2011) mediating tigecycline resistance. In addition, these resistance mechanisms can be regulated by various regulatory factors.
As described in the review, the resistance mechanisms of A. baumannii to tigecycline are diverse and complex, and the emergence of resistance is also very complex. Different A. baumannii cause tigecycline resistance by different mechanisms, and several resistance mechanisms can even coexist in the same bacterium. These resistance mechanisms can also be regulated by various regulatory factors. By screening for resistance-associated genetic mutations to identify tigecycline resistance and applying targeted inhibitors against A. baumannii infection according to these described resistance mechanisms, we hope to provide clinicians with scientific treatment schemes.
Author contributions
CS, XH, and YY conceived the review, CS wrote the manuscript. XH critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China. (Grant 32270183 and U22A20338).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ajiboye, T. O., Skiebe, E., Wilharm, G. (2018). Contributions of RecA and RecBCD DNA repair pathways to the oxidative stress response and sensitivity of acinetobacter baumannii to antibiotics. Int. J. Antimicrob. Agents. 52 (5), 629–636. doi: 10.1016/j.ijantimicag.2018.07.022
Antimicrobial Resistance, C. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 399 (10325), 629–655. doi: 10.1016/S0140-6736(21)02724-0
Aranda, J., Bardina, C., Beceiro, A., Rumbo, S., Cabral, M. P., Barbe, J., et al. (2011). Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J. Bacteriol. 193 (15), 3740–3747. doi: 10.1128/JB.00389-11
Batista Dos Santos, W., Souabni, H., Picard, M. (2022). Corseting a tripartite ABC transporter to make it fit for transport. Biochimie 205, 117–123. doi: 10.1016/j.biochi.2022.11.012
Beabout, K., Hammerstrom, T. G., Perez, A. M., Magalhaes, B. F., Prater, A. G., Clements, T. P., et al. (2015). The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 59 (9), 5561–5566. doi: 10.1128/AAC.00547-15
Cao, G., Qiu, Y., Long, K., Ma, Y., Luo, H., Yang, M., et al. (2022). Rapid and ultrasensitive approach for the simultaneous detection of multilocus mutations to distinguish rifampicin-resistant mycobacterium tuberculosis. Anal. Chem. 94 (50), 17653–17661. doi: 10.1021/acs.analchem.2c04399
Chen, C., Cui, C. Y., Wu, X. T., Fang, L. X., He, Q., He, B., et al. (2021). Spread of tet(X5) and tet(X6) genes in multidrug-resistant acinetobacter baumannii strains of animal origin. Vet. Microbiol. 253, 108954. doi: 10.1016/j.vetmic.2020.108954
Chen, Q., Li, X., Zhou, H., Jiang, Y., Chen, Y., Hua, X., et al. (2014). Decreased susceptibility to tigecycline in acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J. Antimicrob. Chemother. 69 (1), 72–76. doi: 10.1093/jac/dkt319
Chen, C. H., Wu, P. H., Lu, M. C., Ho, M. W., Hsueh, P. R. (2023). Geographic patterns of carbapenem-resistant, multi-drug-resistant and difficult-to-treat acinetobacter baumannii in the Asia-pacific region: results from the antimicrobial testing leadership and surveillance (ATLAS) program, 2020. Int. J. Antimicrob. Agents. 61 (2), 106707. doi: 10.1016/j.ijantimicag.2022.106707
Coyne, S., Courvalin, P., Perichon, B. (2011). Efflux-mediated antibiotic resistance in acinetobacter spp. Antimicrob. Agents Chemother. 55 (3), 947–953. doi: 10.1128/AAC.01388-10
Coyne, S., Guigon, G., Courvalin, P., Perichon, B. (2010a). Screening and quantification of the expression of antibiotic resistance genes in acinetobacter baumannii with a microarray. Antimicrob. Agents Chemother. 54 (1), 333–340. doi: 10.1128/AAC.01037-09
Coyne, S., Rosenfeld, N., Lambert, T., Courvalin, P., Perichon, B. (2010b). Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in acinetobacter baumannii. Antimicrob. Agents Chemother. 54 (10), 4389–4393. doi: 10.1128/AAC.00155-10
Cui, C. Y., He, Q., Jia, Q. L., Li, C., Chen, C., Wu, X. T., et al. (2021). Evolutionary trajectory of the Tet(X) family: critical residue changes towards high-level tigecycline resistance. mSystems 6 (3), e00050–21. doi: 10.1128/mSystems.00050-21
Damier-Piolle, L., Magnet, S., Bremont, S., Lambert, T., Courvalin, P. (2008). AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in acinetobacter baumannii. Antimicrob. Agents Chemother. 52 (2), 557–562. doi: 10.1128/AAC.00732-07
Deng, M., Zhu, M. H., Li, J. J., Bi, S., Sheng, Z. K., Hu, F. S., et al. (2014). Molecular epidemiology and mechanisms of tigecycline resistance in clinical isolates of acinetobacter baumannii from a Chinese university hospital. Antimicrob. Agents Chemother. 58 (1), 297–303. doi: 10.1128/AAC.01727-13
Fang, L. X., Chen, C., Cui, C. Y., Li, X. P., Zhang, Y., Liao, X. P., et al. (2020). Emerging high-level tigecycline resistance: novel tetracycline destructases spread via the mobile Tet(X). Bioessays. 42 (8), e2000014. doi: 10.1002/bies.202000014
Foong, W. E., Wilhelm, J., Tam, H. K., Pos, K. M. (2020). Tigecycline efflux in acinetobacter baumannii is mediated by TetA in synergy with RND-type efflux transporters. J. Antimicrob. Chemother. 75 (5), 1135–1139. doi: 10.1093/jac/dkaa015
Fritsche, T. R., Castanheira, M., Miller, G. H., Jones, R. N., Armstrong, E. S. (2008). Detection of methyltransferases conferring high-level resistance to aminoglycosides in enterobacteriaceae from Europe, north America, and Latin America. Antimicrob. Agents Chemother. 52 (5), 1843–1845. doi: 10.1128/AAC.01477-07
Ghalavand, Z., Eslami, G., Hashemi, A., Sadredinamin, M., Yousefi, N., Dehbanipour, R. (2022). Characterization of sequence types and mechanisms of resistance to tigecycline among acinetobacter baumannii isolated from children. Curr. Microbiol. 79 (9), 285. doi: 10.1007/s00284-022-02976-5
Hammerstrom, T. G., Beabout, K., Clements, T. P., Saxer, G., Shamoo, Y. (2015). Acinetobacter baumannii repeatedly evolves a hypermutator phenotype in response to tigecycline that effectively surveys evolutionary trajectories to resistance. PloS One 10 (10), e0140489. doi: 10.1371/journal.pone.0140489
He, T., Wang, R., Liu, D., Walsh, T. R., Zhang, R., Lv, Y., et al. (2019). Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 4 (9), 1450–1456. doi: 10.1038/s41564-019-0445-2
Henry, R. (2012). Colistin-resistant, lipopolysaccharide-deficient acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrobial Agents chemotherapy. 56 (1), 59–69. doi: 10.1128/AAC.05191-11
Hornsey, M., Ellington, M. J., Doumith, M., Thomas, C. P., Gordon, N. C., Wareham, D. W., et al. (2010). AdeABC-mediated efflux and tigecycline MICs for epidemic clones of acinetobacter baumannii. J. Antimicrob. Chemother. 65 (8), 1589–1593. doi: 10.1093/jac/dkq218
Hsieh, Y. C., Wu, J. W., Chen, Y. Y., Quyen, T. L. T., Liao, W. C., Li, S. W., et al. (2021). An outbreak of tet(X6)-carrying tigecycline-resistant acinetobacter baumannii isolates with a new capsular type at a hospital in Taiwan. Antibiotics (Basel) 10 (10):1239. doi: 10.3390/antibiotics10101239
Hua, X., He, J., Wang, J., Zhang, L., Zhang, L., Xu, Q., et al. (2021). Novel tigecycline resistance mechanisms in acinetobacter baumannii mediated by mutations in adeS, rpoB and rrf. Emerg. Microbes Infect. 10 (1), 1404–1417. doi: 10.1080/22221751.2021.1948804
Ibrahim, M. E. (2019). Prevalence of acinetobacter baumannii in Saudi Arabia: risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance. Ann. Clin. Microb. Anti 18(1):1. doi: 10.1186/s12941-018-0301-x
Ibrahim, S., Al-Saryi, N., Al-Kadmy, I. M. S., Aziz, S. N. (2021). Multidrug-resistant acinetobacter baumannii as an emerging concern in hospitals. Mol. Biol. Rep. 48 (10), 6987–6998. doi: 10.1007/s11033-021-06690-6
Jagdmann, J., Andersson, D. I., Nicoloff, H. (2022). Low levels of tetracyclines select for a mutation that prevents the evolution of high-level resistance to tigecycline. PloS Biol. 20 (9), e3001808. doi: 10.1371/journal.pbio.3001808
Jenner, L., Starosta, A. L., Terry, D. S., Mikolajka, A., Filonava, L., Yusupov, M., et al. (2013). Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc. Natl. Acad. Sci. U S A. 110 (10), 3812–3816. doi: 10.1073/pnas.1216691110
Jo, J., Ko, K. S. (2021). Tigecycline heteroresistance and resistance mechanism in clinical isolates of acinetobacter baumannii. Microbiol. Spectr. 9 (2), e0101021. doi: 10.1128/Spectrum.01010-21
Khlaif, M. M., Hussein, N. H. (2022). Sequencing analysis of tigecycline resistance among tigecycline non-susceptible in three species of G-ve bacteria isolated from clinical specimens in baghdad. Mol. Biol. Rep. 49 (12), 11811–11820. doi: 10.1007/s11033-022-07997-8
Kuzminov, A. (1999). Recombinational repair of DNA damage in escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63 (4), 751–813. doi: 10.1128/MMBR.63.4.751-813.1999
Leblanc, S. K., Oates, C. W., Raivio, T. L. (2011). Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of escherichia coli. J. Bacteriol. 193 (13), 3367–3375. doi: 10.1128/JB.01534-10
Li, X., Liu, L., Ji, J., Chen, Q., Hua, X., Jiang, Y., et al. (2015). Tigecycline resistance in acinetobacter baumannii mediated by frameshift mutation in plsC, encoding 1-acyl-sn-glycerol-3-phosphate acyltransferase. Eur. J. Clin. Microbiol. Infect. Dis. 34 (3), 625–631. doi: 10.1007/s10096-014-2272-y
Li, X., Quan, J., Yang, Y., Ji, J., Liu, L., Fu, Y., et al. (2016). Abrp, a new gene, confers reduced susceptibility to tetracycline, glycylcine, chloramphenicol and fosfomycin classes in acinetobacter baumannii. Eur. J. Clin. Microbiol. Infect. Dis. 35 (8), 1371–1375. doi: 10.1007/s10096-016-2674-0
Li, H., Wang, Q., Wang, R., Zhang, Y., Wang, X., Wang, H. (2017). Global regulator SoxR is a negative regulator of efflux pump gene expression and affects antibiotic resistance and fitness in acinetobacter baumannii. Med. (Baltimore). 96 (24), e7188. doi: 10.1097/MD.0000000000007188
Lin, M. F., Lin, Y. Y., Tu, C. C., Lan, C. Y. (2017). Distribution of different efflux pump genes in clinical isolates of multidrug-resistant acinetobacter baumannii and their correlation with antimicrobial resistance. J. Microbiol. Immunol. Infect. 50 (2), 224–231. doi: 10.1016/j.jmii.2015.04.004
Lin, M. F., Lin, Y. Y., Yeh, H. W., Lan, C. Y. (2014). Role of the BaeSR two-component system in the regulation of acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol. 14, 119. doi: 10.1186/1471-2180-14-119
Lu, B., Jiang, Y. J., Man, M. Q., Brown, B., Elias, P. M., Feingold, K. R. (2005). Expression and regulation of 1-acyl-sn-glycerol- 3-phosphate acyltransferases in the epidermis. J. Lipid Res. 46 (11), 2448–2457. doi: 10.1194/jlr.M500258-JLR200
Lucassen, K., Muller, C., Wille, J., Xanthopoulou, K., Hackel, M., Seifert, H., et al. (2021a). Prevalence of RND efflux pump regulator variants associated with tigecycline resistance in carbapenem-resistant acinetobacter baumannii from a worldwide survey. J. Antimicrob. Chemother. 76 (7), 1724–1730. doi: 10.1093/jac/dkab079
Lucassen, K., Xanthopoulou, K., Wille, J., Wille, T., Wen, Y., Hua, X., et al. (2021b). Characterization of amino acid substitutions in the two-component regulatory system AdeRS identified in multidrug-resistant acinetobacter baumannii. mSphere. 6 (6), e0070921. doi: 10.1128/msphere.00709-21
Ma, Z., Tao, L., Bechthold, A., Shentu, X., Bian, Y., Yu, X. (2014). Overexpression of ribosome recycling factor is responsible for improvement of nucleotide antibiotic-toyocamycin in streptomyces diastatochromogenes 1628. Appl. Microbiol. Biotechnol. 98 (11), 5051–5058. doi: 10.1007/s00253-014-5573-2
Magnet, S., Courvalin, P., Lambert, T. (2001). Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45 (12), 3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001
Marchand, I., Damier-Piolle, L., Courvalin, P., Lambert, T. (2004). Expression of the RND-type efflux pump AdeABC in acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48 (9), 3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004
Meyer, C., Lucabetaen, K., Gerson, S., Xanthopoulou, K., Wille, T., Seifert, H., et al. (2022). Contribution of RND-type efflux pumps in reduced susceptibility to biocides in acinetobacter baumannii. Antibiotics (Basel) 11(11), 1635. doi: 10.3390/antibiotics11111635
Moore, I. F., Hughes, D. W., Wright, G. D. (2005). Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry. 44 (35), 11829–11835. doi: 10.1021/bi0506066
Moubareck, C. A., Halat, D. H. (2020). Insights into acinetobacter baumannii: a review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics-Basel 9 (3), 119. doi: 10.3390/antibiotics9030119
Navon-Venezia, S., Leavitt, A., Carmeli, Y. (2007). High tigecycline resistance in multidrug-resistant acinetobacter baumannii. J. Antimicrob. Chemother. 59 (4), 772–774. doi: 10.1093/jac/dkm018
Nogbou, N. D., Nkawane, G. M., Ntshane, K., Wairuri, C. K., Phofa, D. T., Mokgokong, K. K., et al. (2021). Efflux pump activity and mutations driving multidrug resistance in acinetobacter baumannii at a tertiary hospital in Pretoria, south Africa. Int. J. Microbiol. 2021, 9923816. doi: 10.1155/2021/9923816
Okada, U., Murakami, S. (2022). Structural and functional characteristics of the tripartite ABC transporter. Microbiol. (Reading) 168 (11). doi: 10.1099/mic.0.001257
Owrang, M., Karimi, A., Azimi, L., Nezhad, R. M., Fallah, F. (2018). Relative gene expression of RND-type efflux pumps in tigecycline resistant acinetobacter baumannii isolated from training hospitals in Tehran, Iran. Int. J. Pediatr-Massha. 6 (12), 8669–8674. doi: 10.22038/ijp.2018.33108.2922
Rafiei, E., Shahini Shams Abadi, M., Zamanzad, B., Gholipour, A. (2022). The frequency of efflux pump genes expression in acinetobacter baumannii isolates from pulmonary secretions. AMB Express. 12 (1), 103. doi: 10.1186/s13568-022-01444-4
Rosenfeld, N., Bouchier, C., Courvalin, P., Perichon, B. (2012). Expression of the resistance-nodulation-cell division pump AdeIJK in acinetobacter baumannii is regulated by AdeN, a TetR-type regulator. Antimicrob. Agents Chemother. 56 (5), 2504–2510. doi: 10.1128/AAC.06422-11
Rumbo, C., Gato, E., Lopez, M., Ruiz de Alegria, C., Fernandez-Cuenca, F., Martinez-Martinez, L., et al. (2013). Contribution of efflux pumps, porins, and beta-lactamases to multidrug resistance in clinical isolates of acinetobacter baumannii. Antimicrob. Agents Chemother. 57 (11), 5247–5257. doi: 10.1128/AAC.00730-13
Sampson, T. R., Liu, X., Schroeder, M. R., Kraft, C. S., Burd, E. M., Weiss, D. S. (2012). Rapid killing of acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob. Agents Chemother. 56 (11), 5642–5649. doi: 10.1128/AAC.00756-12
Seifert, H., Blondeau, J., Dowzicky, M. J. (2018). In vitro Activity of tigecycline and comparators (2014-2016) among key WHO ‘priority pathogens’ and longitudinal assessment (2004-2016) of antimicrobial resistance: a report from the T.E.S.T. study. Int. J. Antimicrob. Agents. 52 (4), 474–484. doi: 10.1016/j.ijantimicag.2018.07.003
Šeputienė, V., Povilonis, J., Armalytė, J., Sužiedėlis, K., Pavilonis A, E. S. (2010). Tigecycline – how powerful is it in the fight against antibiotic-resistant bacteria? Medicina (Kaunas) 46 (4), 240–8. doi: 10.3390/medicina46040033
Shilling, R. A., Venter, H., Velamakanni, S., Bapna, A., Woebking, B., Shahi, S., et al. (2006). New light on multidrug binding by an ATP-binding-cassette transporter. Trends Pharmacol. Sci. 27 (4), 195–203. doi: 10.1016/j.tips.2006.02.008
Srinivasan, V. B., Vaidyanathan, V., Rajamohan, G. (2015). AbuO, a TolC-like outer membrane protein of acinetobacter baumannii, is involved in antimicrobial and oxidative stress resistance. Antimicrob. Agents Chemother. 59 (2), 1236–1245. doi: 10.1128/AAC.03626-14
Stephen, J., Salam, F., Lekshmi, M., Kumar, S. H., Varela, M. F. (2023). The major facilitator superfamily and antimicrobial resistance efflux pumps of the ESKAPEE pathogen staphylococcus aureus. Antibiotics (Basel) 12 (2), 343. doi: 10.3390/antibiotics12020343
Su, X. Z., Chen, J., Mizushima, T., Kuroda, T., Tsuchiya, T. (2005). AbeM, an h+-coupled acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 49 (10), 4362–4364. doi: 10.1128/AAC.49.10.4362-4364.2005
Sumyk, M., Himpich, S., Foong, W. E., Herrmann, A., Pos, K. M., Tam, H. K. (2021). Binding of tetracyclines to acinetobacter baumannii TetR involves two arginines as specificity determinants. Front. Microbiol. 12, 711158. doi: 10.3389/fmicb.2021.711158
Sun, Y., Cai, Y., Liu, X., Bai, N., Liang, B., Wang, R. (2013). The emergence of clinical resistance to tigecycline. Int. J. Antimicrob. Agents. 41 (2), 110–116. doi: 10.1016/j.ijantimicag.2012.09.005
Tokuda, H. (2009). Biogenesis of outer membranes in gram-negative bacteria. Biosci. Biotechnol. Biochem. 73 (3), 465–473. doi: 10.1271/bbb.80778
Tokur, M. E., Korkmaz, P., Alkan, S., Yildiz, E., Arik, O., Renders, D. P., et al. (2022). Mortality predictors on the day of healthcare-associated acinetobacter baumanni bacteremia in intensive care unit. J. Infect. Dev. Ctries. 16 (9), 1473–1481. doi: 10.3855/jidc.16902
Verma, P., Tiwari, V. (2018). Targeting outer membrane protein component AdeC for the discovery of efflux pump inhibitor against AdeABC efflux pump of multidrug resistant acinetobacter baumannii. Cell Biochem. Biophys. 76 (3), 391–400. doi: 10.1007/s12013-018-0846-5
Verma, P., Tiwari, M., Tiwari, V. (2021). Efflux pumps in multidrug-resistant acinetobacter baumannii: current status and challenges in the discovery of efflux pumps inhibitors. Microb. Pathog. 152, 104766. doi: 10.1016/j.micpath.2021.104766
Wang, Z., Li, H., Zhang, J., Wang, X., Zhang, Y., Wang, H. (2021). Identification of a novel plasmid-mediated tigecycline resistance-related gene, tet(Y), in acinetobacter baumannii. J. Antimicrob. Chemother. 77 (1), 58–68. doi: 10.1093/jac/dkab375
Wang, L., Liu, D., Lv, Y., Cui, L., Li, Y., Li, T., et al. (2019). Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical acinetobacter baumannii isolate. Antimicrob. Agents Chemother. 64 (1), e01326–19. doi: 10.1128/AAC.01326-19
Wang, Q., Perepelov, A. V., Beutin, L., Senchenkova, S. N., Xu, Y., Shashkov, A. S., et al. (2012). Structural and genetic characterization of the escherichia coli O180 O antigen and identification of a UDP-GlcNAc 6-dehydrogenase. Glycobiology. 22 (10), 1321–1331. doi: 10.1093/glycob/cws098
Wareth, G., Linde, J., Hammer, P., Nguyen, N. H., Nguyen, T. N. M., Splettstoesser, W. D., et al. (2020). Phenotypic and WGS-derived antimicrobial resistance profiles of clinical and non-clinical acinetobacter baumannii isolates from Germany and Vietnam. Int. J. Antimicrob. Agents. 56 (4), 106127. doi: 10.1016/j.ijantimicag.2020.106127
Xu, Q., Chen, T., Yan, B., Zhang, L., Pi, B., Yang, Y., et al. (2019). Dual role of gnaA in antibiotic resistance and virulence in acinetobacter baumannii. Antimicrob. Agents Chemother. 63 (10), e00694–19. doi: 10.1128/AAC.00694-19
Xu, Y., Wang, C., Zhang, G., Tian, J., Liu, Y., Shen, X., et al. (2017). ISCR2 is associated with the dissemination of multiple resistance genes among vibrio spp. and pseudoalteromonas spp. isolated from farmed fish. Arch. Microbiol. 199 (6), 891–896. doi: 10.1007/s00203-017-1365-2
Yang, W., Moore, I. F., Koteva, K. P., Bareich, D. C., Hughes, D. W., Wright, G. D. (2004). TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 279 (50), 52346–52352. doi: 10.1074/jbc.M409573200
Yoon, E. J., Balloy, V., Fiette, L., Chignard, M., Courvalin, P., Grillot-Courvalin, C. (2016). Contribution of the ade resistance-Nodulation-Cell division-type efflux pumps to fitness and pathogenesis of acinetobacter baumannii. mBio 7 (3), e00697–16. doi: 10.1128/mBio.00697-16
Yoon, E. J., Courvalin, P., Grillot-Courvalin, C. (2013). RND-type efflux pumps in multidrug-resistant clinical isolates of acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 57 (7), 2989–2995. doi: 10.1128/AAC.02556-12
Yuhan, Y., Ziyun, Y., Yongbo, Z., Fuqiang, L., Qinghua, Z. (2016). Over expression of AdeABC and AcrAB-TolC efflux systems confers tigecycline resistance in clinical isolates of acinetobacter baumannii and klebsiella pneumoniae. Rev. Soc. Bras. Med. Trop. 49 (2), 165–171. doi: 10.1590/0037-8682-0411-2015
Zhang, J., Xie, J., Li, H., Wang, Z., Yin, Y., Wang, S., et al. (2022). Genomic and phenotypic evolution of tigecycline-resistant acinetobacter baumannii in critically ill patients. Microbiol. Spectr. 10 (1), e0159321. doi: 10.1128/spectrum.01593-21
Zhao, J., Xie, H., Mehdipour, A. R., Safarian, S., Ermler, U., Munke, C., et al. (2021). The structure of the aquifex aeolicus MATE family multidrug resistance transporter and sequence comparisons suggest the existence of a new subfamily. Proc. Natl. Acad. Sci. U.S.A. 118 (46), e2107335118. doi: 10.1073/pnas.2107335118
Zheng, X. R., Zhu, J. H., Zhang, J., Cai, P., Sun, Y. H., Chang, M. X., et al. (2020). A novel plasmid-borne tet(X6) variant co-existing with blaNDM-1 and blaOXA-58 in a chicken acinetobacter baumannii isolate. J. Antimicrob. Chemother. 75 (11), 3397–3399. doi: 10.1093/jac/dkaa342
Keywords: Acinetobacter baumannii, resistance mechanism, resistance, tigecycline, antibiotic
Citation: Sun C, Yu Y and Hua X (2023) Resistance mechanisms of tigecycline in Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 13:1141490. doi: 10.3389/fcimb.2023.1141490
Received: 10 January 2023; Accepted: 14 April 2023;
Published: 09 May 2023.
Edited by:
Yi-Wei Tang, Cepheid, United StatesReviewed by:
Xu Jia, Chengdu Medical College, ChinaJuan Carlos Vázquez-Ucha, Institute of Biomedical Research of A Coruña (INIBIC), Spain
Copyright © 2023 Sun, Yu and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoting Hua, eGlhb3RpbmdodWFAemp1LmVkdS5jbg==
 Chunli Sun
Chunli Sun Yunsong Yu
Yunsong Yu Xiaoting Hua
Xiaoting Hua