- 1Respiratory Department, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
- 2Henan Key Laboratory of Precision Diagnosis of Respiratory Infectious Diseases, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
- 3Zhengzhou Key Laboratory of Precision Diagnosis of Respiratory Infectious Diseases, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
Lung cancer has the highest mortality rate among all cancers worldwide. The 5-year overall survival rate for non-small cell lung cancer (NSCLC) is estimated at around 26%, whereas for small cell lung cancer (SCLC), the survival rate is only approximately 7%. This disease places a significant financial and psychological burden on individuals worldwide. The symbiotic microbiota in the human body has been significantly associated with the occurrence, progression, and prognosis of various diseases, such as asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis. Studies have demonstrated that respiratory symbiotic microorganisms and their metabolites play a crucial role in modulating immune function and contributing to the pathophysiology of lung cancer through their interactions with the host. In this review, we provide a comprehensive overview of the microbial characteristics associated with lung cancer, with a focus on the respiratory tract microbiota from different locations, including saliva, sputum, bronchoalveolar lavage fluid (BALF), bronchial brush samples, and tissue. We describe the respiratory tract microbiota’s biodiversity characteristics by anatomical region, elucidating distinct pathological features, staging, metastasis, host chromosomal mutations, immune therapies, and the differentiated symbiotic microbiota under the influence of environmental factors. Our exploration investigates the intrinsic mechanisms linking the microbiota and its host. Furthermore, we have also provided a comprehensive review of the immune mechanisms by which microbiota are implicated in the development of lung cancer. Dysbiosis of the respiratory microbiota can promote or inhibit tumor progression through various mechanisms, including DNA damage and genomic instability, activation and regulation of the innate and adaptive immune systems, and stimulation of epithelial cells leading to the upregulation of carcinogenesis-related pathways.
1 Introduction
Globally, lung cancer remains the leading cause of cancer-related deaths, representing a substantial proportion of total cancer fatalities (18.4%) (Siegel et al., 2022). This disease inflicts significant suffering upon patients and imposes a tremendous burden on society. Although the implementation of health education interventions such as smoking cessation programs and early CT screening, along with advancements in immunotherapy, has led to a reduction in mortality rates among lung cancer patients (Howlader et al., 2020; Giaquinto et al., 2022; Xia et al., 2022), the overall five-year survival rate remains low (Society, 2022). The five-year survival rate for NSCLC is 26%, while for SCLC, it is 7%. Furthermore, the specific mechanisms underlying various subtypes of lung cancer remain unclear, and there is still a limited number of therapeutic targets for clinical translation. These factors impose significant burdens on society’s economy and the psychological well-being of patients. Therefore, further exploration of the mechanisms underlying lung cancer and the identification of potential therapeutic targets are of utmost importance.
Traditional beliefs have held that the lower respiratory tract is sterile (Dickson et al., 2016; Huffnagle et al., 2017), as it exhibits lower microbial abundance than other body sites. However, limitations in sampling techniques and the culturing methodology have restricted the identification of bacteria through culture-based methods, resulting in only approximately 1% of bacteria being detectable (Torsvik and Øvreås, 2002; Martiny, 2019). Consequently, many microorganisms remain undetectable (Moffatt and Cookson, 2017). With the continuous development of molecular biology techniques, such as DNA sequencing (Singh et al., 2022), independent of culture-based methods, it has been discovered that the lower respiratory tract of healthy individuals harbors a microbial community. The lung microbiota represents a dynamic assemblage of bacteria, fungi, viruses, and other microorganisms that colonize the lungs through inhalation and different routes (Huxley et al., 1978; Gleeson et al., 1997). Previous research has reported their involvement in the occurrence and progression of various lung diseases, such as asthma, cystic fibrosis, and COPD (Hilty et al., 2010; Silveira et al., 2021; Yagi et al., 2021; Amati et al., 2022; Madapoosi et al., 2022).
The significance of lung microbiota in the occurrence and progression of lung cancer is increasingly recognized by researchers. Previous studies have predominantly focused on investigating the mechanisms underlying gastrointestinal cancers, including gastric cancer and colorectal cancer, concerning the gut microbiota. For instance, Helicobacter pylori (HP) has been identified as a risk factor for gastric cancer (Cheung et al., 2018a; Cheung et al., 2018b; Conteduca et al., 2013), while Fusobacterium nucleatum(Fn) has been implicated in the development of colorectal cancer (Rubinstein et al., 2013; Guo et al., 2020; Liang et al., 2020). Recently, there has been an increasing focus on the lung cancer microbiome. Studies have indicated that the microbial communities in lung cancer patients are dysregulated (Liu et al., 2020; Meng et al., 2023), and specific bacteria or bacterial groups are associated with immune dysregulation in lung cancer (Tsay et al., 2021; Khan et al., 2022). These microorganisms directly or indirectly contribute to the occurrence and development of lung cancer. Extensive research on the lung cancer microbiome highlights its significant role in lung cancer and its potential as a biomarker for early diagnosis and a prognostic indicator for evaluating treatment outcomes.
Currently, comprehensive reviews on the microbial composition in different regions of the lower respiratory tract with lung cancer are lacking. Furthermore, the precise role of specific bacteria in the immune mechanisms of lung cancer remains insufficiently understood. This review provides a comprehensive review of the microbial characteristics in lung cancer patients, focusing on various sample types, including saliva, sputum, BALF, brush samples, and tissue (Figure 1). The review further categorizes the microbial features within each sample type based on different factors such as pathological types, metastasis, PD-L1 treatment, smoking, and coal combustion. It explores the potential immune mechanisms by which the microbial community participates in the early diagnosis and treatment of lung cancer, while also revealing the relationship between respiratory microbiota dysbiosis and regulatory factors such as the environment. By offering a more encompassing overview of lung microbiota, this review is a valuable resource for researchers, facilitating further studies and comparisons. Ultimately, it provides a foundation for a better understanding and improvement of the diagnosis and treatment of lung cancer patients.
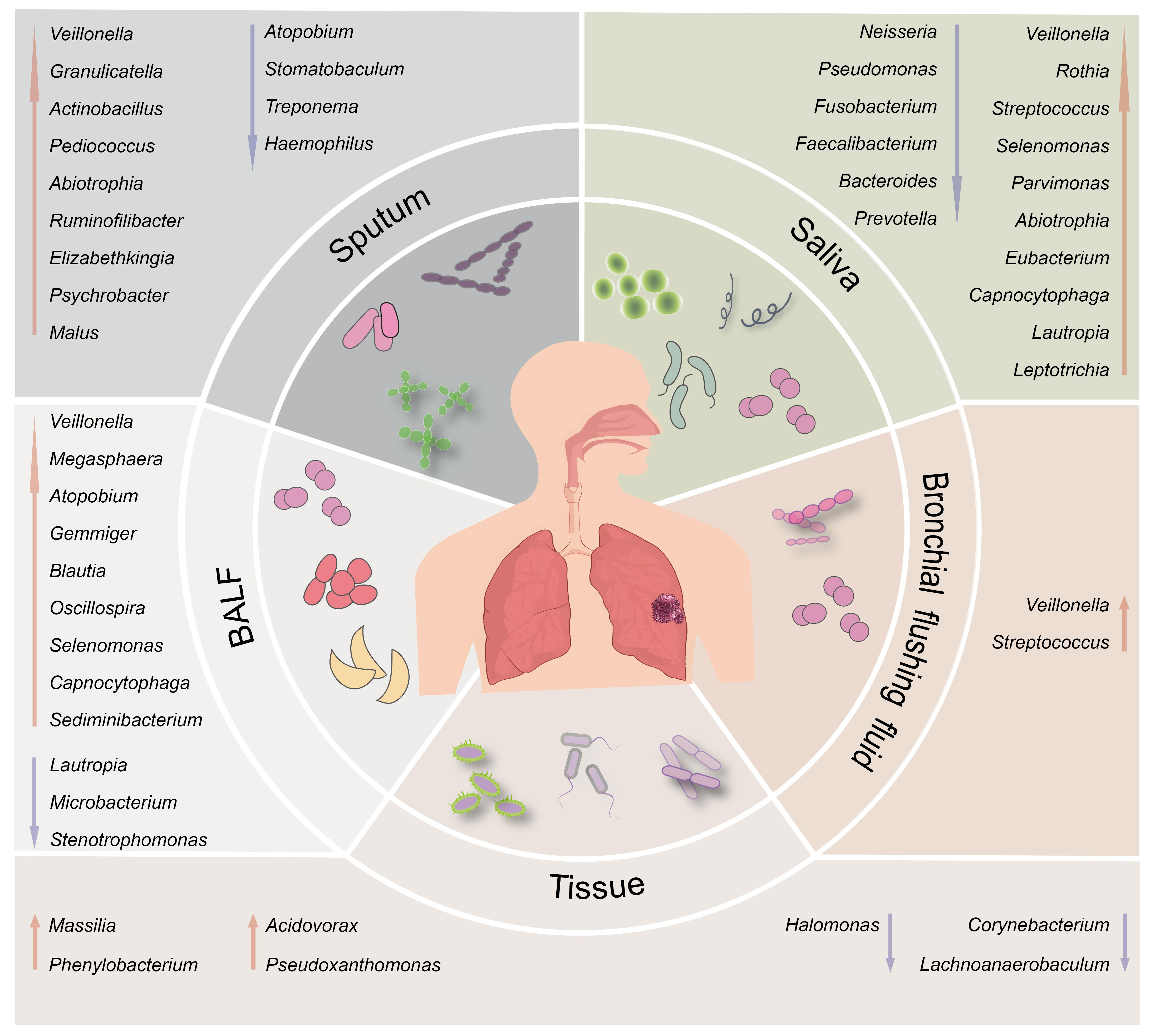
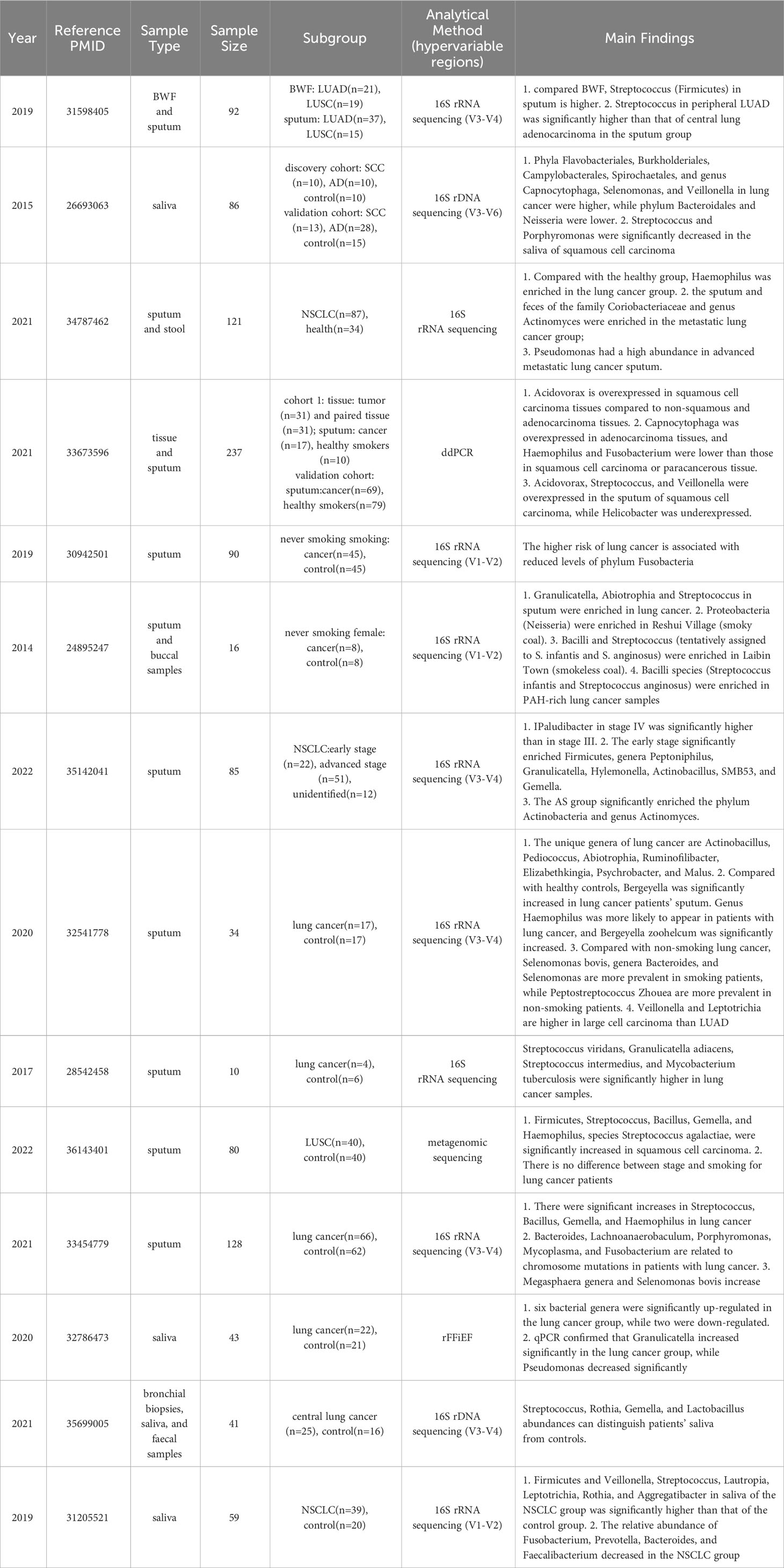
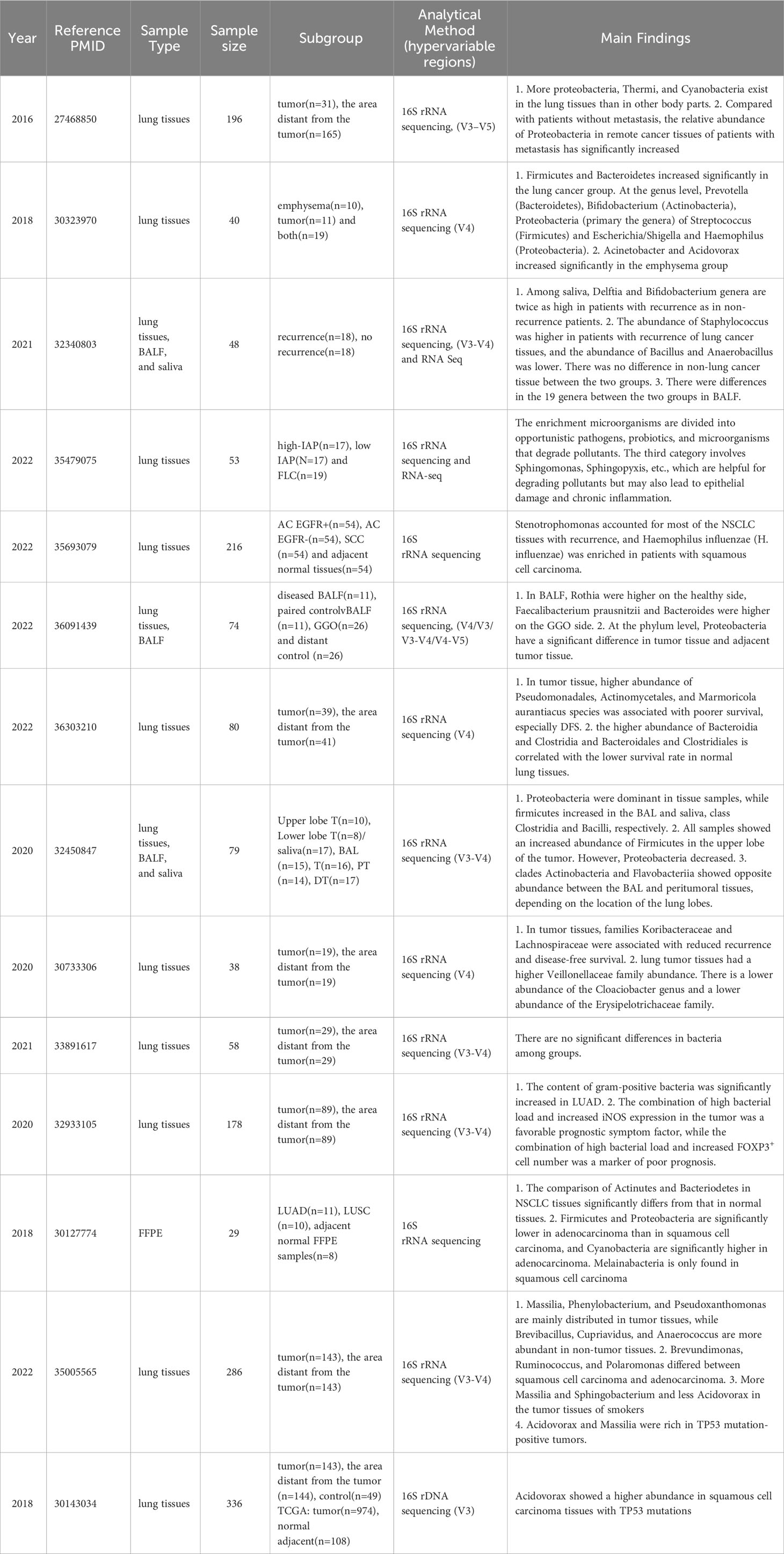
Figure 1 Schematic illustration of the differential respiratory microbiota across different anatomical sites between individuals with lung cancer and healthy subjects. The upward red arrows indicate representative microbiota that exhibit significantly higher abundance in the lung cancer group than the healthy control group. In contrast, the downward blue arrows indicate representative microbiota with decreased abundance.
2 The microbiota community in the respiratory tract of lung cancer
2.1 Microbiome features of the respiratory tract in lung cancer patients
2.1.1 Differential microbes between lung cancer and healthy controls
2.1.1.1 Upper respiratory tract
Multiple studies on lung cancer currently concentrate on genes with Somatic and Germline variants (Li et al., 2018; Shen et al., 2019). However, genetic factors can only account for 3-15% of hereditary issues (Czene et al., 2002; Dai et al., 2017), while non-genetic environmental factors also play a crucial role, such as respiratory tract symbiotic microbiota (Lee et al., 2016). Although BALF and tissue microbes have considerable research potential, these procedures are invasive. On the other hand, sputum and saliva provide non-invasive means to access respiratory microbiota, making sample collection easier. Therefore, investigating microbiota’s predictive and prognostic value in sputum and saliva is of great significance in understanding the association between microbiota and lung cancer.
Human buccal microorganisms comprise more than 600 bacteria, which play distinct functions under different etiologies (Chen et al., 2010; Dewhirst et al., 2010). In various studies, significant differences have been observed between the bacterial compositions of saliva and sputum samples from individuals with lung cancer compared to healthy controls. Xinmin Yan et al. elucidated the intricate association between the salivary microbial community and lung cancer, presenting a comprehensive account for the first time. They found that the phyla Flavobacteriales, Burkholderiales, Campylobacterales, and Spirochaetales, the family Veillonellaceae, and the genera Capnocytophaga, Selenomonas, and Veillonella are prevalent in lung cancer, although Neisseria is less dominant (Yan et al., 2015).A recent study reported similar findings when comparing saliva samples from lung cancer patients and healthy individuals. The authors observed moderate differences at the phylum level between the two groups. At the genus level, Prevotella_7 (Bacteroidetes), Neisseria (Proteobacteria), Streptococcus (Firmicutes), Veillonella (Firmicutes), and Haemophilus (Proteobacteria) were identified as the most prevalent genera in the oral samples (Sun et al., 2023).
Using recycling free-flow isoelectric focusing (RFFiEF) technology, Xiaoteng Jiang and colleagues are concentrating on analyzing salivary bacteria. Neisseria, Pseudomonas, Serratia, Lactobacillus, and Acinetobacter were more prevalent in healthy subjects, while Rothia, Granulicatella, Parvimonas, Abiotrophia, and Eubacterium significantly increased in the lung cancer group. After the qPCR test, Granulicatella considerably rose, whereas Pseudomonas significantly declined (Jiang et al., 2020).
According to research by Weiquan Zhang et al., the presence of the Firmicutes genera Veillonella, Streptococcus, Lautropia, Leptotrichia, Rothia, and Aggregatibacter was considerably more fabulous in the saliva of NSCLC patients than in the control group. Additionally, the relative abundance of Fusobacterium, Prevotella, Bacteroides, and Faecalibacterium decreased overall in the NSCLC group (Zhang et al., 2019).
Simon J. S. Cameron et al. analyzed sputum samples from ten participants, including four with lung cancer and six without. The lung cancer samples exhibited significantly higher levels of Streptococcus viridans, Granulicatella adiacens, Streptococcus intermedius, and Mycobacterium tuberculosis (Cameron et al., 2017).
Elizaveta Baranova et al. analyzed sputum samples from 80 individuals, consisting of 40 cases with squamous cell carcinoma (LUSC) and 40 healthy controls. Their investigation revealed a significant increase in the phylum Firmicutes, as well as the genera Streptococcus, Bacillus, Gemella, and Haemophilus, including the species Streptococcus agalactiae, among those with squamous cell carcinoma. Intriguingly, no variations in the microbiome were observed to pathological stage or smoking status among lung cancer patients (Baranova et al., 2022).
According to Druzhinin et al., sputum from lung cancer patients exhibited unique taxa, including Actinobacillus, Pediococcus, Abiotrophia, Ruminofilibacter, Elizabethkingia, Psychrobacter, and Malus. Among these, Bergeyella showed a significant increase in lung cancer sputum compared to that of healthy controls. Conversely, the genera Atopobium, Stomatobaculum, and Treponema demonstrated a considerable reduction. The genus Haemophilus was more prevalent in lung cancer, and the species Bergeyella zoohelcum exhibited a considerable increase. Additionally, Atopobium rimae, Treponema amulovorum, and Prevotella (P. histicola and P. sp. oral clone DO014) experienced a considerable decline (Druzhinin et al., 2020).
2.1.1.2 Lower respiratory tract
Lower respiratory tract samples are less susceptible to contamination and are closely associated with lung tissue than saliva and sputum samples. Therefore, studying the correlation between lower respiratory tract bacteria and lung cancer can significantly advance our understanding of this disease’s underlying mechanisms. This section focuses on the microbiological distinctions observed in BALF between lung cancer patients and healthy individuals.
In a study by Wang et al., a comparison was made between BALF and salivary bacteria from lung cancer patients (n = 52) and healthy individuals (n = 15), revealing significant differences. The lung cancer group exhibited considerable variations in the levels of Firmicutes and Fusobacteria in their BALF, as well as significantly different levels of Actinobacteria in their saliva, in comparison to the healthy individuals (Wang et al., 2019).
Identifying potential differences between lung cancer and benign samples using microorganisms can aid in distinguishing between suspicious and confirmed cases. In a study conducted by Sang Hoon Lee and colleagues, they examined 28 BALF samples, including 20 from lung cancer patients and eight from individuals with benign illnesses. The researchers found a significant elevation of Veillonella, Megasphaera, Atopobium, and Selenomonas in the lung cancer group (Lee et al., 2016).
The sample size was expanded to include 54 individuals, comprising 32 lung cancer patients and 22 individuals with benign lung disease. The results of this study revealed significant increases in six genera (c: TM7-3, Capnocytophaga, Sediminibacterium, Gemmiger, Blautia, and Oscillospira), while four genera (Microbacterium, Stenotrophomonas, Lautropia, and f: Pseudonadaceae) exhibited significant decreases. These ten genera and three tumor markers (CEA, NSE, and CYFRA21-1) were used to construct a random forest model. The model demonstrated promising predictive capabilities, with an area under the curve (AUC) value of 84.52% for lung cancer, indicating its potential for clinical application (Cheng et al., 2020).
Wen Zeng et al. conducted a study involving BALF samples from 46 NSCLC patients and 29 individuals with benign illnesses. The researchers found that Prevotella, Alloprevotella, and Veillonella were significantly elevated in the lung cancer group. Additionally, positive associations were observed between Prevotella and Megasphaera, Alloprevotella, and Actinomyces. Subcutaneous transplantation and endotracheal instillation experiments were conducted to further investigate the impact of Veillonella parvula. The results showed that subcutaneous transplantation significantly promoted lung cancer, while endotracheal instillation did not lead to tumor formation. These findings suggest that the influence of microorganisms on lung cancer may vary in terms of chronicity, duration, or adequacy. These numerous genera could serve as diagnostic indicators and therapeutic targets (Zeng et al., 2022). For brushing samples, lung cancer patients have a higher abundance of Veillonella and Streptococcus compared to healthy individuals (Tsay et al., 2018). However, it is essential to note that different investigations have reported varying bacterial compositions, which could be attributed to the limited sample sizes. Hence, these findings should be validated through large-scale studies.
2.1.1.3 The biodiversity
The diversity of sputum microorganisms was found to be lower in lung cancer patients compared to healthy controls (Hosgood et al., 2014; Hosgood et al., 2019; Lu et al., 2021). However, V.G. Druzhinin et al. reported no significant change in the diversity of sputum microorganisms between lung cancer patients and healthy individuals (Druzhinin et al., 2020; Druzhinin et al., 2021). These studies highlight notable disparities in β diversity. Furthermore, Salvador Bello et al. found no difference in the α diversity of salivary microbes between lung cancer patients and healthy individuals. Still they observed a substantial difference in the β diversity of salivary bacteria (Bello et al., 2021). However, Weiquan Zhang et al. have presented arguments highlighting significant variations in both α diversity and β diversity between the two groups when considering saliva (Zhang et al., 2019).
There are conflicting findings regarding the microbial diversity in BALF observed in different studies. Sílvia Gomes, Wen Zeng et al. found that the BALF microbial α-diversity in lung cancer patients was higher than in non-lung cancer patients (Gomes et al., 2019; Zeng et al., 2022). However, Sang Hoon Lee et al. reached the opposite conclusion (Lee et al., 2016). Several other studies have also reported no significant differences in microbial α- and β-diversity between lung cancer and benign disease groups in BALF (Tsay et al., 2018; Wang et al., 2019; Cheng et al., 2020; Zhuo et al., 2020). However, Sang Hoon Lee et al. suggested a significant difference in β-diversity between cancer and benign control groups (Cheng et al., 2020; Zeng et al., 2022).
2.1.2 Microbiome under different pathological types
2.1.2.1 Upper respiratory tract
It is essential to distinguish different pathologic types after comparing overall lung cancer with healthy controls. A comparative analysis of salivary microbes in LUAD and LUSC revealed distinct patterns. Specifically, in the saliva of squamous cell carcinoma, Streptococcus and Porphyromonas were significantly lower, while Prevotella showed no significant changes between the two groups (Yan et al., 2015). On the other hand, Streptococcus, Capnocytophaga, and Actinomyces were enriched in the saliva of lung cancer patients. The LSCC group demonstrated a higher enrichment of Capnocytophaga and Actinomyces than the healthy group. Additionally, the SCLC group exhibited a higher concentration of Streptococcus, while Rothia was significantly higher in the adenocarcinoma group (Wang et al., 2019).
In a study conducted by Qixin Leng et al., 25 bacteria in adenocarcinoma and squamous cell carcinoma sputum and tissues were compared using ddPCR. The results revealed significant increases in Capnocytophaga in adenocarcinoma samples and Acidovorax in squamous cell carcinoma samples, with both sputum and tissue demonstrating similar trends. However, adenocarcinoma tissues exhibited exclusive elevations of Haemophilus and Fusobacterium, contradicting previous research indicating Haemophilus accumulation in lung cancer sputum (Lu et al., 2021). Squamous cell carcinoma sputum, on the other hand, contained only Streptococcus and Veillonella. Also, Helicobacter levels were reduced solely in adenocarcinoma sputum (Leng et al., 2021).
Danhui Huang et al. investigated the microbiological variations between sputum and bronchial flushing fluid (BWF) samples and reported the microorganisms of squamous cell cancer and adenocarcinoma, respectively. The levels of Streptococcus (Firmicutes) in sputum were higher in lung cancer patients than in BWF (benign lung disease with fibrosis). Moreover, peripheral lung cancer exhibited higher levels of Streptococcus (Firmicutes) in sputum than central lung adenocarcinoma (Huang et al., 2019), which contradicts earlier research (Yan et al., 2015). Additionally, large-cell carcinoma demonstrated higher levels of Veillonella and Leptotrichia than adenocarcinoma (Druzhinin et al., 2020). Regarding sputum α microbial diversity, no significant differences were found between lung adenocarcinoma and lung squamous cell carcinoma (Huang et al., 2019).
2.1.2.2 Lower respiratory tract
A recent study showed that the populations of Haemophilus parainfluenza, Neisseria subflava, Porphyromonas endodontalis, and Fusobacterium nucleatum were significantly higher in the adenocarcinoma group when compared to squamous cell carcinoma (Jang et al., 2023). In the study conducted by Ke Wang et al., it was found that Veillonella and Capnocellhaga exhibited significantly higher levels in the BALF of patients with lung squamous cell carcinoma (LUSC). Conversely, Lactobacillus showed a marked increase in the BALF of individuals with small-cell lung carcinoma (SCLC) (Wang et al., 2019).
In the study conducted by Minglei Zhuo et al., significant increases in the genera Spiroplasma and Weissella were observed in the BALF of the lung cancer group when compared to the healthy side of the lung affected by cancer (Zhuo et al., 2020). Furthermore, Slvia Gomes et al. examined the BALF samples from individuals with adenocarcinoma, squamous cell carcinoma, and healthy controls. They also used RNA sequencing data from the TCGA database to validate their findings. Sphingomonas, Brevundimonas, Acinetobacter, and Methylobacterium were exclusively found in lung adenocarcinoma (LUAD), while Enterobacter, Morganella, Kluyvera, and Capnocytophaga were predominantly present in lung squamous cell carcinoma (LUSC). The prevalence of a proteobacteria-dominated microbiome in the BALF of squamous cell carcinoma is associated with a progressive decline in survival rates (Gomes et al., 2019).
Danhui Huang et al. conducted a study to identify the microbiological colonies in squamous cell carcinoma and adenocarcinoma and the microbial differences between sputum and bronchial washing fluid (BWF) samples in lung cancer patients. The results showed that BWF samples exhibited a significantly higher abundance of Proteobacteria and a lower abundance of Firmicutes than sputum. Comparing the bronchial washing fluids (BWFs) across various pathological types, Firmicutes and the genera Veillonella, Megasphaera, Actinomyces, and Arthrobacter were found to be more prevalent in NSCLC with no distant metastasis (AD-M0) than in squamous cell carcinoma with no distant metastasis (SC-M0). Conversely, Capnocytophaga and Rothia showed a significant decrease in AD-M1 compared to SC-M1, and no association was observed between these distinct bacteria and smoking status. AD-M1 demonstrated a lower abundance of Firmicutes and Streptococcus in BWF than SCC-M1. Furthermore, SCC-M1 exhibited a higher presence of Veillonella and Rothia than SCC-M0 (Huang et al., 2019).
There is also controversy regarding the diversity comparison among different pathological types. Danhui Huang et al. suggested that microbial diversity is not associated with pathological types (Huang et al., 2019), while Sílvia Gomes et al. found that the α-diversity of microbiota was higher in squamous cell carcinoma compared to adenocarcinoma (Gomes et al., 2019). Interestingly, Erin A Marshall et al. found, through a 10-year follow-up, that there was no significant difference in α-diversity between individuals with a previous history of lung cancer, those who would develop lung cancer in the future, and non-lung cancer patients (Marshall et al., 2022).
2.1.3 The microbiota community associated with lung cancer metastasis
Metastasis of lung cancer is a substantial contributor to death and recurrence in NSCLC (Siegel et al., 2022). Several studies have revealed that microbes play a role in tumor metastasis. In a survey conducted by Hui Lu et al., a comparison of microbes in feces and sputum was performed among 121 participants, including 87 with NSCLC and 34 in good health. The findings revealed that gut microbes were exclusively associated with lung cancer that did not metastasize, whereas sputum microbes showed a connection to lung cancer regardless of metastasis. Higher concentrations of Haemophilus (specifically, Haemophilus parainfluenzae and Haemophilus influenzae) were observed in lung cancer samples compared to healthy controls. Additionally, sputum and feces from individuals with metastatic lung cancer exhibited enrichment of Coriobacteriaceae and Actinomyces. The sputum of advanced metastatic lung cancer showed a higher abundance of Pseudomonas, while Campylobacter was more abundant in both the regular group and advanced metastatic lung cancer. An analysis of a random forest model determined that the sputum model provided better predictive accuracy than intestinal microbes. Hence, sputum microorganisms were more closely associated with the development of lung cancer than intestinal microbes (Lu et al., 2021).
Dan Hui Huang et al. examined 85 sputum samples obtained from newly diagnosed patients with NSCLC. Their analysis revealed that in early-stage lung cancer (stage I and stage II), the phylum Firmicutes and the genera Peptoniphilus, Granulicatella, Hylemonella, Actinobacillus, SMB53, and Gemella were more abundant. On the other hand, in advanced-stage lung cancer (stage III and stage IV), the phyla Actinobacteria and the genus Actinomyces exhibited more significant enrichment. Compared to the non-metastatic group, the metastatic group revealed significant enrichment of the genera Peptostreptococcus, Peptococcus, Parabacteroides, and Escherichia. Moreover, in the lymph node metastasis group, there was a considerable increase in the genera Parvimonas and Pseudomonas. The EGFR mutant group exhibited significant enrichment of Bacteroidetes and Tenericutes, along with the genera Sharpea, Prevotella, Porphyromonas, Parvimonas, Desulfovibrio, Mycoplasma, Actinobacillus, Dialister, and Eikenella. Compared to stage III lung cancer, only Paludibacter showed a significant increase. The specific airway genus and metabolic function of sputum microbiota were found to undergo changes associated with tumor stage, intrathoracic metastasis, lymph node metastasis, and EGFR mutation (Huang et al., 2022). Porphyromonas endodontalis was significantly more abundant in the sputum of patients with stages I-II than those with stages III-IV. Additionally, patients with metastases exhibited higher levels of Capnocytophaga and lower levels of Atopobium rimae than those without metastasis (Druzhinin et al., 2020).
Significant differences in microbial diversity were observed in sputum samples when comparing lung cancer’s early and advanced stages.However, microbial diversity was found to be unrelated to stages III and IV, intrathoracic and lymph node metastases, and EGFR mutation (Huang et al., 2022).
2.2 The relationship between microbiota and the host genome
Lung microbes and genes collaborate to facilitate the onset and progression of lung cancer. In their study, V. G. Druzhinin et al. investigated sputum samples obtained from lung cancer patients and controls. They focused on exploring the connection between microbes and the occurrence of chromosomal mutations. Interestingly, their findings revealed a significant decrease in the genus Atopobium, while Alloprevotella exhibited a significant increase among patients with a high frequency of chromosomal aberrations (CA) (Druzhinin et al., 2020). Moreover, V. G. Druzhinin et al. expanded the sample size to include 66 lung cancer patients and 62 healthy subjects. Their investigation revealed significant increases in Streptococcus, Bacillus, Gemella, and Haemophilus among lung cancer patients. Additionally, they found that Bacteroides, Lachnoanaerobaculum, Porphyromonas, Mycoplasma, and Fusobacterium were associated with the frequency of chromosomal aberrations. Interestingly, the genera Megasphaera and Selenomonas bovis negatively correlated with micronuclei (MN) (Druzhinin et al., 2021).
2.3 The relationship between microbiota and environmental factors
2.3.1 Upper respiratory tract
Lung microorganisms can be influenced by environmental factors such as coal combustion and smoking, which have implications for the development and progression of lung cancer. In his study, H. Dean Hosgood, III examined the impact of microbiota in sputum and buccal samples on lung cancer among never-smoking women. Additionally, the study explored the effect of coal on these associations. The findings revealed that the diversity of bacterial communities in buccal samples was comparable between the case and control groups. However, significant differences were observed in sputum samples. Specifically, Granulicatella, Abiotrophia, and Streptococcus were abundant in lung cancer patients’ sputum. Lung microorganisms can be influenced by environmental factors such as coal combustion and smoking, which have implications for the development and progression of lung cancer. In his study, H. Dean Hosgood, III examined the impact of microbiota in sputum and buccal samples on lung cancer among never-smoking women.
Additionally, the study explored the effect of coal on these associations. The findings revealed that the diversity of bacterial communities in buccal samples was comparable between the case and control groups. However, significant differences were observed in sputum samples. Specifically, Granulicatella, Abiotrophia, and Streptococcus were abundant in lung cancer patients’ sputum. In smoking-related lung cancer patients, the species Selenomonas bovis, genus Bacteroides, and genus Selenomonas were found to be more abundant compared to never-smoking lung cancer patients.
Conversely, the genus Peptostreptococcus exhibited higher abundance in never-smoking lung cancer patients. Among healthy smokers, the species Bulleidia moorei, genus Granulicatella, and genus Bulleidea were prevalent, whereas the genus Neisseria was abundant in healthy nonsmokers (Druzhinin et al., 2020). For more detailed information, please refer to Table 1.
2.3.2 Lower respiratory tract
Lung cancer and smoking are widely acknowledged to be closely related. However, only a small percentage of smokers, approximately 10-15%, will develop lung cancer. This presents a clinical challenge in predicting which smokers are at risk (Bruder et al., 2018; Herbst et al., 2018). Moreover, smoking has been shown to impair the function of the epithelial barrier, increasing the likelihood of lung resident bacteria infiltrating the lungs and contributing to the onset and progression of disease (Hou et al., 2019; Wong et al., 2020). Therefore, a comprehensive exploration of the interplay between smoking and lung bacteria has the potential to enhance the early detection of lung cancer.
Erin A Marshall et al. conducted a study where 400 bronchial brush samples were subjected to 16S microbiota sequencing. The samples were divided into an exploration group and a validation group. Over 10 years, the participants were closely monitored, and differentiated microbial communities were analyzed using linear differentiation analysis and the establishment of a linear model. The results revealed that patients with higher scores had a greater risk of developing lung cancer, and the onset occurred earlier, indicating the potential of microbiota in predicting the occurrence of lung cancer (Marshall et al., 2022).
Jun-Chieh J Tsay et al. compared oral and bronchial brush samples from individuals with lung cancer (n=39), benign lung nodules (n=36), and healthy individuals (n=10). They found that the lung cancer group exhibited an abundance of Streptococcus and Veillonella, the benign lung nodule group showed enrichment of Streptophyta, Moraxellaceae, and Stenotrophomonas. At the same time, the samples from healthy individuals were enriched with Acholeplasma and Acidocella (Tsay et al., 2018). Please refer to Table 2 for more details.
3 The association between intratumoral microbiota and lung cancer
3.1 The intratumoral microbiota characteristics in lung cancer
Studies have described on the association between bacteria in alveolar lavage fluid, lung brush samples, sputum, and saliva and lung cancer (Hilty et al., 2010; Charlson et al., 2011; Erb-Downward et al., 2011; Pragman et al., 2012; Borewicz et al., 2013; Segal et al., 2013; Willner et al., 2013; Dickson et al., 2014; Bassis et al., 2015; Dickson et al., 2015), but due to the invasiveness of lung tissue, difficult to get samples. The low microbial abundance, there are few microorganism studies in lung cancer tissues.
The location of the tumor correlates with its aggressiveness, and the 5-year survival rate after pulmonary lobectomy in the lower lobe is poorer than that of the tumor in the upper lobe (Hayakawa et al., 1996; Inoue et al., 2004; Ou et al., 2007). Rea Bingula et al. analyzed 18 cases of NSCLC saliva, BALF (directly obtained from the resected lung lobe), cancerous tissue, paracancerous tissue, and distant cancer tissue and discovered that the microbiomes in saliva, BALF, and tissue were distinct. The tissues were dominated by Proteobacteria, while BALF and saliva were dominated by Firmicutes. All samples showed an increase in the abundance of Firmicutes in lower lobe tumors and a decrease in Proteobacteria. In addition, depending on the location of the tumor, Actinobacteria and Flavobacteriia have opposing abundances in BAL and extratumor tissues. Although tumor microbiota appears to be least impacted by location, paracancerous tissues exhibit the most susceptibility, with a considerable increase in resemblance to BAL microbiota in the upper lobe (Bingula et al., 2020). See Table 3 for details.
The diversity of microorganisms reflects the complexity of the entire microbiome, and existing research has studied the diversity of microorganisms in lung tissue from various subgroups. The diversity of tumor tissue is less than that of paired distant cancer tissues (Yu et al., 2016; Peters et al., 2019; Chen et al., 2022; Kim et al., 2022), Advanced lung cancer has less diversity than early lung cancer, which has less diversity than normal tissue (Kim et al., 2022), benign lung tissue is less diverse than cancer tissue (Chen et al., 2022), and the diversity of lung tissue in healthy individuals is greater than that in cancer patients, regardless of tumor tissue or tumor-adjacent tissue (Greathouse et al., 2018). The diversity of lung tissue in healthy individuals is greater than that in cancer patients (Bingula et al., 2020; Kovaleva et al., 2020; Dong et al., 2022; Peters et al., 2022), and there is no significant difference between adenocarcinoma and squamous cell carcinoma tissues (Apopa et al., 2018; Kovaleva et al., 2020). Nathan Dumont-Leblond and colleagues hypothesized that the α diversity of malignant tissues is more significant than that of neighboring tissues (Dumont-Leblond et al., 2021). There were no significant differences between GGO and paracancerous tissue in α and β diversity (Wu et al., 2022). Various studies have found that smoking has inconsistent effects on the biodiversity of tumor tissues. The majority of studies indicate that smoking reduces the α diversity of lung tissue (Liu et al., 2018; Patnaik et al., 2021; Chen et al., 2022); BALF microorganisms are intermediate between saliva and lung cancer tissue, and the α and β diversity of saliva and BALF microorganisms are more similar in patients with high amylase of BALF, possibly due to microinhalation (Patnaik et al., 2021).
3.2 The correlation between alterations in intratumoral microbial communities and tumor progression
3.2.1 Tissue microorganisms of GGO
Low-dose computed tomography (LDCT) is widely employed as the principal tool for lung cancer screening programs globally, and an increasing number of ground-glass opacities (GGOs) are being discovered (National Lung Screening Trial Research et al., 2011). Persistent GGO with a certain morphology is regarded as malignant and most likely an indolent, slowly progressing lung cancer (Chang et al., 2013). Consequently, it is essential to investigate the microecological characteristics of lung tissue with lung ground-glass nodules. Zhigang Wu et al. compared the differences between GGO and contralateral lower respiratory tract microorganisms and lung tissue microorganisms. The genus Rothia was abundant in the contralateral BALF, while the species Faecalibacterium prausnitzii and Bacteroides uniforms were abundant on the GGO side. GGO tissue and adjacent lung tissue have significant differences in microbiota composition at the level of class, order, family, genus, and species; interestingly, the majority of these are enriched in normal lung tissue, with an AUC of 91.05 percent (95 percent confidence interval: 81.93 to 100 percent) produced by 10 different genera, which has proven effective in detecting lung cancer (Wu et al., 2022).
3.2.2 Microbial communities of the area of distant cancer tissues
Guoqin Yu et al. were the first to research the lung microbial characteristics of distant cancer tissues (n = 165) in lung cancer patients. They discovered that the microbial community of lung tissues was distinct from microorganisms in other body areas and comprised a separate bacterial colony. At the phylum level, the dominant microorganisms in distant cancer tissues were Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, whereas at the genus level, the dominant Bacilli were Bacillus and Bacteroides. In distant cancer tissue, the relative abundance of Thermus is greater in adenocarcinoma than in squamous cell carcinoma, whereas Ralstonia is diminished (Yu et al., 2016).
3.2.3 Differentiated microbial communities of lung cancer tissue versus adjacent tissues
Compared to distant metastatic tissues, the microbial differences between lung cancer tissue and adjacent tissue provide a more accurate reflection of lung cancer.Hui Dong et al. discovered Massilia, Phenylobacterium, and Pseudoxanthomonas were enriched in lung cancer tissue; Brevibacillus, Cupriavidus, and Anaerococcus were more prevalent in neighboring tissues; and Brevundimonas, Ruminococcus, and Polaromonas were significantly different between squamous cell and adenocarcinoma carcinomas. The tumor tissues of smokers include more Massilia and Sphingobacterium and fewer Acidovorax, whereas TP53 mutation-positive tumors are rich in Acidovorax and Massilia (Dong et al., 2022). Acidovorax was also found to be more prevalent in lung cancer tissue from TP53-mutated squamous cell carcinoma patients, according to K Leigh Greathouse et al. (Greathouse et al., 2018). The recent study aimed to characterize the taxonomic profiles of the microbiota in oral saliva, cancerous, and paracancerous tissues of Chinese patients diagnosed with lung adenocarcinoma. The Shannon index of cancerous tissues (CT) was significantly higher compared to that of paracancerous tissues (PT) and saliva. The observed increased relative abundance of Promicromonosporacea and Chloroflexi, coupled with the decreased relative abundance of Enterococcaceae and Enterococcus in lung tissues, may potentially be associated with the risk of developing lung adenocarcinoma (Zhou et al., 2023).
Multiple factors, including hereditary and environmental exposures, contribute to the development of lung cancer. COPD patients are three to ten times more likely to develop lung cancer than healthy smokers (Etzel et al., 2008). Despite the fact that there are overlapping susceptibility genes (Young et al., 2008), there are also individual differences. Given that smoking is the most prevalent risk factor for lung cancer (Chaturvedi et al., 2010; Yu et al., 2014; Zhou et al., 2017), Yanhong Liu et al. collected 40 lung tissues (10 pulmonary bullae, 11 lung tumors, and 19 both lung cancer and bullae) to assess if environmental factors such as smoking influence changes in respiratory bacteria. Proteobacteria (genera Acinetobacter and Acidovorax) are more frequent in the emphysema group (Liu et al., 2018). However, the tissue sampled in this study is distant from tumor tissue, and there is a space between the cancer tissue and the sampled tissue.
3.2.4 Lung cancer tissue microbes and recurrence
Lung tissue bacteria are capable of predicting the recurrence of lung cancer. The five-year survival rate for stage IA lung cancer is 83%, whereas the five-year survival rate for stage IIB lung cancer is 53% (Kay et al., 2017), and early detection of lung cancer recurrence can lessen its mortality rate. Current research on lung cancer recurrence focuses on the microenvironment of the tumor, including mutational burden (Owada-Ozaki et al., 2018), immune cell infiltration (O’Callaghan et al., 2015) and local gene expression (Planck et al., 2013; Kratz et al., 2021; Wang et al., 2021). Researchers are gradually becoming aware of the function of lung microorganisms in the occurrence and progression of lung cancer; thus, it is vital to investigate the association between early lung cancer recurrence and lung bacteria. Santosh K. Patnaik et al. compared the lung microorganisms of saliva, alveolar lavage fluid, and tissue in patients with stage I NSCLC and performed RNA sequencing on lung tissue. They discovered that Delftia and Bifidobacterium, two salivary bacteria, rose considerably in recurrent patients. Staphylococcus was more common in the lung cancer tissues of relapsed patients, but Bacillus and Anaerobacillus were more common in non-recurrent patients. There were no significant differences between the two groups in non-cancerous tissues, but there were differences in 19 genera in the BALF sample. Phingomonas, Psychromonas, and Serratia genera increased in the lung cancer recurrence group, whereas Cloacibacterium, Geobacillus, and Brevibacterium genera decreased. Lower respiratory bacteria are capable of identifying NSCLC patients with surgical recurrence, as indicated by the area under the curve value of 0.77 (Patnaik et al., 2021).
Brandilyn A. Peters et al. investigated the relationship between microbiome and recurrence in tumor and distant cancer tissue specimens (39 lung and 41 distant cancer tissues) from 46 individuals with NSCLC at stage II. There was no difference in α diversity and β diversity between cancerous and noncancerous tissues discovered. However, both α-diversity and β-diversity of the microbiota in lung cancer tissues were found to be associated with disease-free survival (DFS). In contrast, the α-diversity and β-diversity of the microbiota in adjacent tissues showed no association with DFS, recurrence-free survival (RFS), or overall survival (OS). A high abundance of species Pseudomonadales and Actinomycetales and Marmoricola aurantiacus in lung cancer tissues was negatively associated with disease-free survival (DFS). In contrast, high abundance of Bacteroides and Clostridia and orders Bacteroidales and Clostridiales in distant cancer tissues were negatively associated with survival. Survival was favorably linked with ASV of Alphaproteobacteria and Betaproteobacteria, Burkholderiales, Neisseriales, and Mycobacterium vaccae. Marmoricola aurantiacus is abundant in lung cancer tissue from individuals who have a poor prognosis (Peters et al., 2022).
Ock-Hwa Kim et al. evaluated the relationship between microbes and prognosis (recurrence) of lung cancer tissues of various pathological types and discovered that in the tissues of patients with relapsed NSCLC, the genus Stenotrophomonas accounted for the vast majority, whereas H. influenzae was enriched in patients with squamous cell carcinoma (Kim et al., 2022). Gram-positive bacteria are significantly elevated in lung adenocarcinoma, and a high bacterial load in tumors combined with increased iNOS expression is a favorable prognostic symptom factor. In contrast, a high bacterial load in tumors combined with increased FOXP3+ cell numbers is an indicator of a poor prognosis (Kovaleva et al., 2020).
Brandilyn A. Peters et al. demonstrated for the first time the association between normal lung tissue and lung cancer prognosis, discovering that the diversity and overall microbial composition of normal tissue were associated with decreased recurrence-free survival and disease-free survival. The more prosperous family Koribacteraceae was associated with improved recurrence-free and disease-free survival in normal tissues. Still, the Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae families were inversely associated with recurrence-free survival. The diversity and composition of tumor tissue are not associated with relapse-free survival. The richness and diversity of tumor tissue are lower than that of paired normal tissue (Peters et al., 2019).
3.3 The association between intratumoral microbiota and factors like IAP, smoking, and family inheritance
Y. Chen et al. compared the microbiome of lung cancer patients affected by familial lung cancer (FLC) and indoor air pollution (IAP). Additionally, they describe potential links between host gene expression patterns and their microbiome. The research revealed that smoking and IAP significantly reduced the biodiversity of specific OTUs, particularly in normal lung tissue. Enhanced microorganisms include Sphingomonas and Sphingopyxis, which can decompose pollutants but can also induce epithelial injury and promote chronic inflammation. RNA sequencing data emphasize the IL17, Ras, MAPK, and Notch pathways in the FLC and IAP groups, which are associated with carcinogenesis and reduced immune function (Chen et al., 2022).
4 The interaction between lung microbiota and immune therapy
Only approximately 20% of patients with NSCLC derive therapeutic benefits from the anti-PD-1 antibody nivolumab. Hence, the identification of more accurate predictors becomes imperative. In their study, Chufeng Zhang et al. examined and compared the differences and correlations in gut microbes and salivary microorganisms following PD-1/PD-L1 immunotherapy. Interestingly, they observed no significant difference in respiratory microbial alpha diversity between the effective (R) and ineffective (NR) groups. However, there were notable variations in beta diversity and the abundance of Streptococcus (a respiratory flora) was found to be associated with progression-free survival (PFS) (Zhang et al., 2021). In terms of sputum α diversity, no significant difference was observed between immunotherapy-responsive and non-responsive patients, although a significant difference was noted in terms of β diversity (Zhang et al., 2021).
Gaining insight into the relationship between lower respiratory tract bacteria and therapeutic efficacy can enhance our understanding of potential markers that accurately predict the effectiveness of clinical immunotherapy. Previous studies have established a connection between alterations in lung microorganisms and immune evasion mediated by PD-L1-dependent Treg cells (Gollwitzer et al., 2014). Hye Jin Jin Jang et al. investigated the correlation between microorganisms in lung BALF and PD-L1 expression, as well as the association between microbes and the efficacy of immunotherapy. Their findings revealed a higher prevalence of Veillonella dispar (Firmicutes) in the BALF of patients exhibiting high PD-L1 expression, while Neisseria (Proteobacteria) was more commonly found in the BALF of patients with low PD-L1 expression. The immune response group had a higher abundance of Bacteroidetes and a lower abundance of Proteobacteria (Masuhiro et al., 2022). Conversely, the non-response group exhibited higher levels of the phyla Proteobacteria and Bacteroidetes, as well as the genera Haemophilus and species Haemophilus influenzae and Neisseria perflava. Additionally, the non-response group showed a lower presence of Firmicutes.
Hye Jin Jang et al. studied the lower respiratory tract microorganisms in 84 patients, including 59 patients with low PD-L1 expression and 25 patients with high PD-L1 expression. Their findings revealed a higher prevalence of Veillonella, specifically the species V. dispar, in the immune response group (Jang et al., 2021). Consequently, further research should prioritize investigating the mechanisms associated with the genus Veillonella in the immune response.
The responder group to immune checkpoint inhibitors (ICIs) exhibited higher α-diversity compared to the non-responder group, while there was no difference in β-diversity between the two groups (Masuhiro et al., 2022). Hye Jin Jang et al. also reported that there were no significant differences in α- and β-diversity between the two groups with high and low PD-L1 expression levels (Jang et al., 2021).
5 Potential mechanisms through which commensal microorganisms could impact the host’s immune system in relation to lung cancer development
There is growing evidence that symbiotic microbial communities of the host are linked to the onset, progression, and therapeutic efficacy of cancer. The current literature focuses on the gut microbiome, but how the pulmonary symbiotic microbiota and the distal gut microbiota are engaged in the malignant transformation of lung cancer cells and how they simultaneously manage the balance between pro-tumor inflammation and anti-tumor immunity are not well understood. We will comprehensively overview the potential immune mechanisms involved in commensal microorganisms across four important topics (Figure 2).
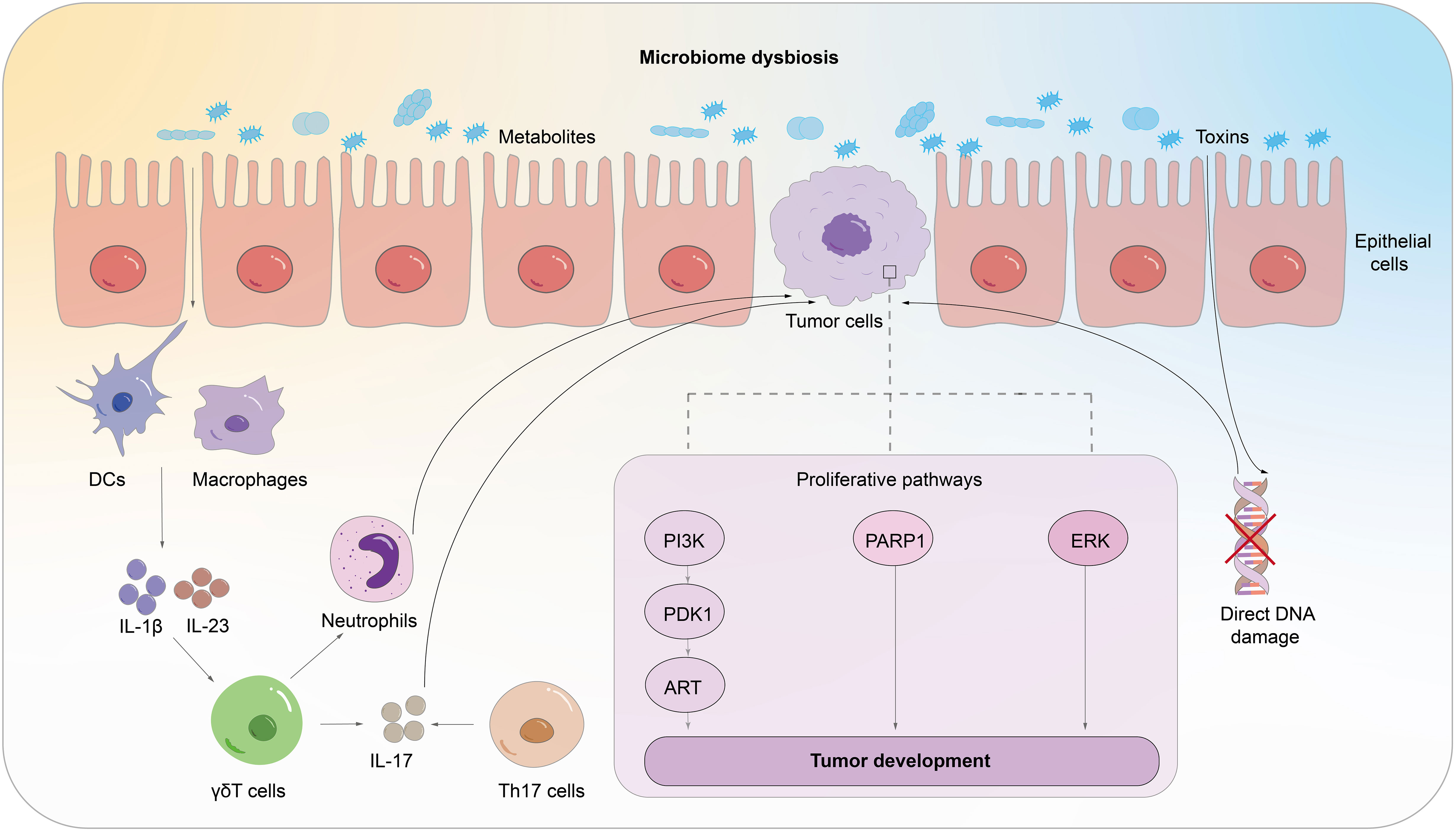
Figure 2 The potential mechanism through which commensal microorganisms could impact the host’s immune system in lung cancer pathogenesis. The microbiota generates cytotoxicity which induces host cell DNA damage, abnormal activation of proliferation and transformation pathways, and aberrant immunological pathways.
5.1 The microbes inducing DNA damage and genomic instability
Genotoxins and metabolites produced by bacteria possess the capability to directly inflict damage upon the host’s DNA and instigate genomic instability via the generation of reactive oxygen or nitrogen species and the activation of innate immune receptors. When the cumulative effects of such damage exceed the host’s self-regulation capacity, it can lead to the carcinogenic impacts (Espinoza and Minami, 2018; Goto, 2020; Goto, 2022). Studies have shown that reactive oxygen species produced by Porphyromonas, hydrogen sulfide produced by Clostridium cholephilum, and superoxide dismutase produced by symbiotic bacteria can cause genomic instability and increase susceptibility to lung cancer (Attene-Ramos et al., 2006; Carbonero et al., 2012). Additionally, microbial dysbiosis can lead to increased reactive oxygen species associated with DNA damage (Mao et al., 2018).
The enrichment of bacteria such as Massilia and Acidovorax in the pulmonary system is consistent with the trends observed in DNA recombination and repair pathways (Dong et al., 2022). Folate produced by Lactobacillus and Bifidobacteria promotes the production of 6-methyltetrahydrofolate, which influences DNA methylation (Kothapalli et al., 2005; Hassan and Zempleni, 2006; Rossi et al., 2011; Zempleni et al., 2012). These studies demonstrate that microorganisms can directly or indirectly contribute to DNA damage or genomic instability.
5.2 Activation and regulation of the innate immune system in response to microbial influence
Microbial dysbiosis can lead to the activation of the host’s innate immune system and sustained chronic inflammation through the interaction of microbial-associated molecular patterns (MAMPs) with pattern recognition receptors (PRRs) (Evavold and Kagan, 2019; Fitzgerald and Kagan, 2020; Li and Wu, 2021). Antigen-presenting cells, such as alveolar macrophages, dendritic cells, and epithelial cells, can recognize microbial stimulation through PRRs, triggering the expression of immune-related genes and initiating innate and adaptive immune responses (Sellge and Kufer, 2015). Specifically, the activation of Toll-like receptors (TLRs) by microbial products induces the activation of alveolar macrophages and neutrophils (Chang et al., 2014; Segal et al., 2016; Jin et al., 2019). Studies have also demonstrated the upregulation of TLR-4 and TLR-9 expression in lung cancer tissue (Zhang et al., 2009; Goto, 2022). Stimulation of TLR4 using heat-inactivated Escherichia coli has been shown to activate the p38 MAPK and ERK1/2 signaling pathways, leading to adhesive, migratory, and metastatic behavior of non-small cell lung cancer (NSCLC) cells in vivo (Chow et al., 2015).
A reduction in microbial diversity hampers the stimulation of antigen-presenting cells, thereby impeding immune responses against tumor antigens (Bingula et al., 2017). Moreover, bacterial overgrowth triggers excessive mucosal immune responses, leading to pro-inflammatory reactions. These reactions, often mediated by the Th17 mechanism, can result in uncontrolled cell proliferation. Excessive activation of the innate immune system results in an expansion of regulatory T cells (Tregs) and M2 macrophages. These immune cells release various molecules, including prostaglandin E2 (PGE2), transforming growth factor-beta (TGF-B), and interleukin-10 (IL-10), which are known for their immunosuppressive properties. Additionally, there is an upregulation of programmed death-ligand 1 (PD-L1), contributing to immune tolerance and tumor evasion (Soroosh et al., 2013; Hussell and Bell, 2014).
5.3 Activation and functional regulation of the adaptive immune system in response to microbes
C.Jin et al. noted that pulmonary symbiotic microorganisms can stimulate the expression of IL-1β and IL-23, thereby inducing the proliferation of Vγ6+ Vδ1+γδ T cells that produce IL-17 and other activated molecules, promoting inflammation and tumor cell proliferation (Jin et al., 2019). Additionally, excessive bacterial overgrowth leads to overactivation of the adaptive immune system and proliferation of Th17 cells, contributing to the occurrence and progression of lung cancer (Chang et al., 2014).
Research has established that certain bacteria present in non-small cell lung cancer (NSCLC) are positively correlated with the presence of Th17 and Th1 cells. This correlation suggests a potential link between the microbiome and immune response in NSCLC (Ma et al., 2017). Pasteurella is significantly positively correlated with cytotoxic CD8+ tumor-infiltrating lymphocytes (TILs) and negatively associated with M2 macrophages. However, Coriobacteriaceae is significantly positively correlated with M2 macrophages and negatively correlated with CD8+ TILs (Zheng et al., 2020). Dysbiosis can trigger the expression of PD-L1 on CD11b+CD103- dendritic cells in the lungs and modulate the activation of regulatory T cells (Gollwitzer et al., 2014).
Moreover, short-chain fatty acids (SCFAs) as microbial metabolites (Segal et al., 2017) induce the upregulation of the forkhead box P3 (FoxP3) in CD4+ lymphocytes, leading to the development of regulatory T cells (Treg) and subsequent immune tolerance (Trompette et al., 2014). However, elevated SCFAs levels in the lungs may inhibit the production of interferon-gamma (IFN-γ) by CD4+ and CD8+ T cells, resulting in T cell exhaustion and suppression of cytotoxicity against malignant cells (Segal et al., 2017).
5.4 The interaction between microbes and epithelial cells
The stimulation of epithelial cells by microorganisms can lead to abnormal activation of proliferation and transformation pathways, resulting in carcinogenic effects. The presence of specific microbial species in non-small cell lung cancer (NSCLC) has been found to be associated with the dysregulation of oncogenic transcriptional systems. Research findings indicate that Prevotella, Streptococcus, Veillonella, and tiny Vibrio can stimulate airway epithelial cells, activating the ERK and PI3K signaling pathways. Excitingly, Cyanobacteria-produced microcystins promote cell proliferation by inducing PARP1 overexpression (Apopa et al., 2018).
Stimulation of epithelial cells by nontypeable Haemophilus influenzae (NTHi) leads to the expression of IL17C, which in turn promotes neutrophil infiltration and chronic inflammation and facilitates tumor progression (Jungnickel et al., 2017). Furthermore, interleukin-6 (IL-6) has been shown to promote lung cancer growth by inducing inflammatory responses (Ochoa et al., 2011). Furthermore, CD36 is a crucial link between lung microbiota and cancer. It regulates microcystin processing in the alveoli, increasing PARP1 expression and promoting non-small cell lung cancer development (Apopa et al., 2018).
6 Conclusion
In this review, we evaluated the link between respiratory tract flora and lung cancer, focusing on the characteristics of microbial flora in different locations of lung cancer, including saliva, sputum, BALF, bronchial brushing samples, and tissues, and analyzing the relationship between microbial flora in other parts and genes, environmental factors, pathological types, stages, treatment, and prognosis. The studies on the potential role of microorganisms in the pathogenesis and progression of lung cancer were reported in this article.
So far, the following conclusions can be drawn: Firstly, independent of locations, the microbial composition of lung cancer patients differed considerably from that of healthy individuals, indicating that the microbial flora has the potential to act as a biomarker for the diagnosis and prognosis of lung cancer. Secondly, microorganisms found in the lower respiratory tract, such as BALF, BWF, and tissues, are more suggestive of lung cancer. Finally, in lung cancer patients, pulmonary symbiotic bacteria directly or indirectly promote or inhibit tumor progression via the pro-inflammatory and anti-inflammatory balance, immune response dysfunction, metabolic pathway, DNA damage, and genomic instability. In conclusion, pulmonary microbiomes may be biomarkers for predicting lung cancer’s onset, progression, and prognosis. The causal association between respiratory microbes and lung cancer must be established through more mechanistic research.
The result contradicts between samples reflects the dynamic change of microbiota in carcinogenesis, suggesting that specific bacteria may play different roles at different time points and different host locations (Ramirez-Labrada et al., 2020), which may also be due to the small sample size, regional, dietary, and other factors to resulting in the identical examination of several bacteria. To determine the microbiological characteristics of lung cancer, large-scale microbial investigations on lung cancer with control of intervention factors will be required in the future.
Nowadays, lung cancer is the most lethal tumor, placing a substantial burden on the lives and economies of people all over the world. Therefore, research into the complete pathophysiology of lung cancer, the quest for new therapeutic techniques, and the development of specialized therapeutic medications remain essential for the prevention and treatment of this condition. Due to the intense research interest in the mechanism of action of intestinal microbes on the incidence of colorectal cancer and the effect of pharmacological therapy, the association between pulmonary microbial flora and lung cancer has been increasingly recognized, but it remains a descriptive study. Future studies on the mechanism, prevention, and therapy of microbial involvement are urgently required. Numerous confirmatory cellular and animal experiments must be conducted, which will likely open up new avenues for researching lung cancer disease mechanisms.
Author contributions
JC: Writing – original draft, Writing – review & editing. LZ: Funding acquisition, Writing – review & editing. HW: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded financially by the Health Commission of Henan Province (No. LHGJ20210394) and the Science and Technology Department of Henan Province (No. 222102310699).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Amati, F., Stainer, A., Mantero, M., Gramegna, A., Simonetta, E., Suigo, G., et al. (2022). Lung microbiome in idiopathic pulmonary fibrosis and other interstitial lung diseases. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23020977
Apopa, P. L., Alley, L., Penney, R. B., Arnaoutakis, K., Steliga, M. A., Jeffus, S., et al. (2018). PARP1 is up-regulated in non-small cell lung cancer tissues in the presence of the cyanobacterial toxin microcystin. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01757
Attene-Ramos, M. S., Wagner, E. D., Plewa, M. J., Gaskins, H. R. (2006). Evidence that hydrogen sulfide is a genotoxic agent. Mol. Cancer Res. 4, 9–14. doi: 10.1158/1541-7786.MCR-05-0126
Baranova, E., Druzhinin, V., Matskova, L., Demenkov, P., Volobaev, V., Minina, V., et al. (2022). Sputum microbiome composition in patients with squamous cell lung carcinoma. Life (Basel) 12. doi: 10.3390/life12091365
Bassis, C. M., Erb-Downward, J. R., Dickson, R. P., Freeman, C. M., Schmidt, T. M., Young, V. B., et al. (2015). Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 6, e00037. doi: 10.1128/mBio.00037-15
Bello, S., Vengoechea, J. J., Ponce-Alonso, M., Figueredo, A. L., Mincholé, E., Rezusta, A., et al. (2021). Core microbiota in central lung cancer with streptococcal enrichment as a possible diagnostic marker. Archivos bronconeumologia 57, 681–689. doi: 10.1016/j.arbr.2020.05.017
Bingula, R., Filaire, E., Molnar, I., Delmas, E., Berthon, J. Y., Vasson, M. P., et al. (2020). Characterisation of microbiota in saliva, bronchoalveolar lavage fluid, non-malignant, peritumoural and tumour tissue in non-small cell lung cancer patients: a cross-sectional clinical trial. Respir. Res. 21, 129. doi: 10.1186/s12931-020-01392-2
Bingula, R., Filaire, M., Radosevic-Robin, N., Bey, M., Berthon, J. Y., Bernalier-Donadille, A., et al. (2017). Desired turbulence? Gut-lung axis, immunity, and lung cancer. J. Oncol. 2017, 5035371. doi: 10.1155/2017/5035371
Borewicz, K., Pragman, A. A., Kim, H. B., Hertz, M., Wendt, C., Isaacson, R. E. (2013). Longitudinal analysis of the lung microbiome in lung transplantation. FEMS Microbiol. Lett. 339, 57–65. doi: 10.1111/1574-6968.12053
Bruder, C., Bulliard, J. L., Germann, S., Konzelmann, I., Bochud, M., Leyvraz, M., et al. (2018). Estimating lifetime and 10-year risk of lung cancer. Prev. Med. Rep. 11, 125–130. doi: 10.1016/j.pmedr.2018.06.010
Cameron, S. J. S., Lewis, K. E., Huws, S. A., Hegarty, M. J., Lewis, P. D., Pachebat, J. A., et al. (2017). A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PloS One 12, e0177062. doi: 10.1371/journal.pone.0177062
Carbonero, F., Benefiel, A. C., Alizadeh-Ghamsari, A. H., Gaskins, H. R. (2012). Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 3. doi: 10.3389/fphys.2012.00448
Chang, B., Hwang, J. H., Choi, Y. H., Chung, M. P., Kim, H., Kwon, O. J., et al. (2013). Natural history of pure ground-glass opacity lung nodules detected by low-dose CT scan. Chest 143, 172–178. doi: 10.1378/chest.11-2501
Chang, S. H., Mirabolfathinejad, S. G., Katta, H., Cumpian, A. M., Gong, L., Caetano, M. S., et al. (2014). T helper 17 cells play a critical pathogenic role in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 111, 5664–5669. doi: 10.1073/pnas.1319051111
Charlson, E. S., Bittinger, K., Haas, A. R., Fitzgerald, A. S., Frank, I., Yadav, A., et al. (2011). Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 184, 957–963. doi: 10.1164/rccm.201104-0655OC
Chaturvedi, A. K., Caporaso, N. E., Katki, H. A., Wong, H. L., Chatterjee, N., Pine, S. R., et al. (2010). C-reactive protein and risk of lung cancer. J. Clin. Oncol. 28, 2719–2726. doi: 10.1200/JCO.2009.27.0454
Chen, T., Yu, W. H., Izard, J., Baranova, O. V., Lakshmanan, A., Dewhirst, F. E. (2010). The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010, baq013. doi: 10.1093/database/baq013
Chen, Y., Huang, Y., Ding, X., Yang, Z., He, L., Ning, M., et al. (2022). A multi-omics study of familial lung cancer: microbiome and host gene expression patterns. Front. Immunol. 13. doi: 10.3389/fimmu.2022.827953
Cheng, C., Wang, Z., Wang, J., Ding, C., Sun, C., Liu, P., et al. (2020). Characterization of the lung microbiome and exploration of potential bacterial biomarkers for lung cancer. Transl. Lung Cancer Res. 9, 693–704. doi: 10.21037/tlcr-19-590
Cheung, K. S., Chan, E. W., Wong, A. Y. S., Chen, L., Seto, W. K., Wong, I. C. K., et al. (2018a). Aspirin and risk of gastric cancer after helicobacter pylori eradication: a territory-wide study. J. Natl. Cancer Inst 110, 743–749. doi: 10.1093/jnci/djx267
Cheung, K. S., Chan, E. W., Wong, A. Y. S., Chen, L., Wong, I. C. K., Leung, W. K. (2018b). Long-term proton pump inhibitors and risk of gastric cancer development after treatment for Helicobacter pylori: a population-based study. Gut 67, 28–35. doi: 10.1136/gutjnl-2017-314605
Chow, S. C., Gowing, S. D., Cools-Lartigue, J. J., Chen, C. B., Berube, J., Yoon, H. W., et al. (2015). Gram negative bacteria increase non-small cell lung cancer metastasis via Toll-like receptor 4 activation and mitogen-activated protein kinase phosphorylation. Int. J. Cancer 136, 1341–1350. doi: 10.1002/ijc.29111
Conteduca, V., Sansonno, D., Lauletta, G., Russi, S., Ingravallo, G., Dammacco, F. (2013). H. pylori infection and gastric cancer: state of the art (review). Int. J. Oncol. 42, 5–18. doi: 10.3892/ijo.2012.1701
Czene, K., Lichtenstein, P., Hemminki, K. (2002). Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int. J. Cancer 99, 260–266. doi: 10.1002/ijc.10332
Dai, J., Shen, W., Wen, W., Chang, J., Wang, T., Chen, H., et al. (2017). Estimation of heritability for nine common cancers using data from genome-wide association studies in Chinese population. Int. J. Cancer 140, 329–336. doi: 10.1002/ijc.30447
Dewhirst, F. E., Chen, T., Izard, J., Paster, B. J., Tanner, A. C., Yu, W. H., et al. (2010). The human oral microbiome. J. Bacteriol 192, 5002–5017. doi: 10.1128/JB.00542-10
Dickson, R. P., Erb-Downward, J. R., Freeman, C. M., Mccloskey, L., Beck, J. M., Huffnagle, G. B., et al. (2015). Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann. Am. Thorac. Soc. 12, 821–830. doi: 10.1513/AnnalsATS.201501-029OC
Dickson, R. P., Erb-Downward, J. R., Martinez, F. J., Huffnagle, G. B. (2016). The microbiome and the respiratory tract. Annu. Rev. Physiol. 78, 481–504. doi: 10.1146/annurev-physiol-021115-105238
Dickson, R. P., Martinez, F. J., Huffnagle, G. B. (2014). The role of the microbiome in exacerbations of chronic lung diseases. Lancet 384, 691–702. doi: 10.1016/S0140-6736(14)61136-3
Dong, H., Tan, Q., Xu, Y., Zhu, Y., Yao, Y., Wang, Y., et al. (2022). Convergent alteration of lung tissue microbiota and tumor cells in lung cancer. iScience 25, 103638. doi: 10.1016/j.isci.2021.103638
Druzhinin, V. G., Matskova, L. V., Demenkov, P. S., Baranova, E. D., Volobaev, V. P., Minina, V. I., et al. (2020). Taxonomic diversity of sputum microbiome in lung cancer patients and its relationship with chromosomal aberrations in blood lymphocytes. Sci. Rep. 10, 9681. doi: 10.1038/s41598-020-66654-x
Druzhinin, V. G., Matskova, L. V., Demenkov, P. S., Baranova, E. D., Volobaev, V. P., Minina, V. I., et al. (2021). Genetic damage in lymphocytes of lung cancer patients is correlated to the composition of the respiratory tract microbiome. Mutagenesis 36, 143–153. doi: 10.1093/mutage/geab004
Dumont-Leblond, N., Veillette, M., Racine, C., Joubert, P., Duchaine, C. (2021). Non-small cell lung cancer microbiota characterization: Prevalence of enteric and potentially pathogenic bacteria in cancer tissues. PloS One 16, e0249832. doi: 10.1371/journal.pone.0249832
Erb-Downward, J. R., Thompson, D. L., Han, M. K., Freeman, C. M., Mccloskey, L., Schmidt, L. A., et al. (2011). Analysis of the lung microbiome in the "healthy" smoker and in COPD. PloS One 6, e16384. doi: 10.1371/journal.pone.0016384
Espinoza, J. L., Minami, M. (2018). Sensing bacterial-induced DNA damaging effects via natural killer group 2 member D immune receptor: from dysbiosis to autoimmunity and carcinogenesis. Front. Immunol. 9. doi: 10.3389/fimmu.2018.00052
Etzel, C. J., Kachroo, S., Liu, M., D'amelio, A., Dong, Q., Cote, M. L., et al. (2008). Development and validation of a lung cancer risk prediction model for African-Americans. Cancer Prev. Res. (Phila) 1, 255–265. doi: 10.1158/1940-6207.CAPR-08-0082
Evavold, C. L., Kagan, J. C. (2019). Inflammasomes: threat-assessment organelles of the innate immune system. Immunity 51, 609–624. doi: 10.1016/j.immuni.2019.08.005
Fitzgerald, K. A., Kagan, J. C. (2020). Toll-like receptors and the control of immunity. Cell 180, 1044–1066. doi: 10.1016/j.cell.2020.02.041
Giaquinto, A. N., Miller, K. D., Tossas, K. Y., Winn, R. A., Jemal, A., Siegel, R. L. (2022). Cancer statistics for African American/black people 2022. CA Cancer J. Clin. 72, 202–229. doi: 10.3322/caac.21718
Gleeson, K., Maxwell, S. L., Eggli, D. F. (1997). Quantitative aspiration during sleep in normal subjects. Chest 111, 1266–1272. doi: 10.1378/chest.111.5.1266
Gollwitzer, E. S., Saglani, S., Trompette, A., Yadava, K., Sherburn, R., Mccoy, K. D., et al. (2014). Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 20, 642–647. doi: 10.1038/nm.3568
Gomes, S., Cavadas, B., Ferreira, J. C., Marques, P. I., Monteiro, C., Sucena, M., et al. (2019). Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci. Rep. 9, 12838. doi: 10.1038/s41598-019-49195-w
Goto, T. (2020). Airway microbiota as a modulator of lung cancer. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21093044
Goto, T. (2022). Microbiota and lung cancer. Semin. Cancer Biol. 86, 1–10. doi: 10.1016/j.semcancer.2022.07.006
Greathouse, K. L., White, J. R., Vargas, A. J., Bliskovsky, V. V., Beck, J. A., Von Muhlinen, N., et al. (2018). Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19, 123. doi: 10.1186/s13059-018-1501-6
Guo, S., Chen, J., Chen, F., Zeng, Q., Liu, W. L., Zhang, G. (2020). Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut. 70, 1507–1519. doi: 10.1136/gutjnl-2020-321187
Hassan, Y. I., Zempleni, J. (2006). Epigenetic regulation of chromatin structure and gene function by biotin. J. Nutr. 136, 1763–1765. doi: 10.1093/jn/136.7.1763
Hayakawa, K., Mitsuhashi, N., Saito, Y., Furuta, M., Nakayama, Y., Katano, S., et al. (1996). Impact of tumor extent and location on treatment outcome in patients with stage III non-small cell lung cancer treated with radiation therapy. Jpn J. Clin. Oncol. 26, 221–228. doi: 10.1093/oxfordjournals.jjco.a023218
Herbst, R. S., Morgensztern, D., Boshoff, C. (2018). The biology and management of non-small cell lung cancer. Nature 553, 446–454. doi: 10.1038/nature25183
Hilty, M., Burke, C., Pedro, H., Cardenas, P., Bush, A., Bossley, C., et al. (2010). Disordered microbial communities in asthmatic airways. PloS One 5, e8578. doi: 10.1371/journal.pone.0008578
Hosgood, H. D., 3rd, Mongodin, E. F., Wan, Y., Hua, X., Rothman, N., Hu, W., et al. (2019). The respiratory tract microbiome and its relationship to lung cancer and environmental exposures found in rural China. Environ. Mol. Mutagen 60, 617–623. doi: 10.1002/em.22291
Hosgood, H. D., 3rd, Sapkota, A. R., Rothman, N., Rohan, T., Hu, W., Xu, J., et al. (2014). The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutagen 55, 643–651. doi: 10.1002/em.21878
Hou, W., Hu, S., Li, C., Ma, H., Wang, Q., Meng, G., et al. (2019). Cigarette smoke induced lung barrier dysfunction, EMT, and tissue remodeling: a possible link between COPD and lung cancer. BioMed. Res. Int. 2019, 2025636. doi: 10.1155/2019/2025636
Howlader, N., Forjaz, G., Mooradian, M. J., Meza, R., Kong, C. Y., Cronin, K. A., et al. (2020). The effect of advances in lung-cancer treatment on population mortality. New Engl. J. Med. 383, 640–649. doi: 10.1056/NEJMoa1916623
Huang, D. H., He, J., Su, X. F., Wen, Y. N., Zhang, S. J., Liu, L. Y., et al. (2022). The airway microbiota of non-small cell lung cancer patients and its relationship to tumor stage and EGFR gene mutation. Thorac. Cancer 13, 858–869. doi: 10.1111/1759-7714.14340
Huang, D., Su, X., Yuan, M., Zhang, S., He, J., Deng, Q., et al. (2019). The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am. J. Cancer Res. 9, 2047–2063.
Huffnagle, G. B., Dickson, R. P., Lukacs, N. W. (2017). The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 10, 299–306. doi: 10.1038/mi.2016.108
Hussell, T., Bell, T. J. (2014). Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 14, 81–93. doi: 10.1038/nri3600
Huxley, E. J., Viroslav, J., Gray, W. R., Pierce, A. K. (1978). Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am. J. Med. 64, 564–568. doi: 10.1016/0002-9343(78)90574-0
Inoue, M., Sawabata, N., Takeda, S., Ohta, M., Ohno, Y., Maeda, H. (2004). Results of surgical intervention for p-stage IIIA (N2) non-small cell lung cancer: acceptable prognosis predicted by complete resection in patients with single N2 disease with primary tumor in the upper lobe. J. Thorac. Cardiovasc. Surg. 127, 1100–1106. doi: 10.1016/j.jtcvs.2003.09.012
Jang, H. J., Choi, J. Y., Kim, K., Yong, S. H., Kim, Y. W., Kim, S. Y., et al. (2021). Relationship of the lung microbiome with PD-L1 expression and immunotherapy response in lung cancer. Respir. Res. 22, 322. doi: 10.1186/s12931-021-01919-1
Jang, H. J., Lee, E., Cho, Y.-J., Lee, S. H. (2023). Subtype-based microbial analysis in non-small cell lung cancer. Tuberc Respir. Dis. 86, 294–303. doi: 10.4046/trd.2022.0149
Jiang, X., Liu, S., Zhang, Y., Ji, Y., Sohail, A., Cao, C., et al. (2020). Free-flow isoelectric focusing for comprehensive separation and analysis of human salivary microbiome for lung cancer. Anal. Chem. 92, 12017–12025. doi: 10.1021/acs.analchem.0c02627
Jin, C., Lagoudas, G. K., Zhao, C., Bullman, S., Bhutkar, A., Hu, B., et al. (2019). Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 176, 998–1013.e1016. doi: 10.1016/j.cell.2018.12.040
Jungnickel, C., Schmidt, L. H., Bittigkoffer, L., Wolf, L., Wolf, A., Ritzmann, F., et al. (2017). IL-17C mediates the recruitment of tumor-associated neutrophils and lung tumor growth. Oncogene 36, 4182–4190. doi: 10.1038/onc.2017.28
Kay, F. U., Kandathil, A., Batra, K., Saboo, S. S., Abbara, S., Rajiah, P. (2017). Revisions to the Tumor, Node, Metastasis staging of lung cancer (8(th) edition): Rationale, radiologic findings and clinical implications. World J. Radiol. 9, 269–279. doi: 10.4329/wjr.v9.i6.269
Khan, F. H., Bhat, B. A., Sheikh, B. A., Tariq, L., Padmanabhan, R., Verma, J. P., et al. (2022). Microbiome dysbiosis and epigenetic modulations in lung cancer: From pathogenesis to therapy. Semin. Cancer Biol. 86, 732–742. doi: 10.1016/j.semcancer.2021.07.005
Kim, O. H., Choi, B. Y., Kim, D. K., Kim, N. H., Rho, J. K., Sul, W. J., et al. (2022). The microbiome of lung cancer tissue and its association with pathological and clinical parameters. Am. J. Cancer Res. 12, 2350–2362.
Kothapalli, N., Camporeale, G., Kueh, A., Chew, Y. C., Oommen, A. M., Griffin, J. B., et al. (2005). Biological functions of biotinylated histones. J. Nutr. Biochem. 16, 446–448. doi: 10.1016/j.jnutbio.2005.03.025
Kovaleva, O., Podlesnaya, P., Rashidova, M., Samoilova, D., Petrenko, A., Zborovskaya, I., et al. (2020). Lung microbiome differentially impacts survival of patients with non-small cell lung cancer depending on tumor stroma phenotype. Biomedicines 8. doi: 10.3390/biomedicines8090349
Kratz, J. R., Li, J. Z., Tsui, J., Lee, J. C., Ding, V. W., Rao, A. A., et al. (2021). Genetic and immunologic features of recurrent stage I lung adenocarcinoma. Sci. Rep. 11, 23690. doi: 10.1038/s41598-021-02946-0
Lee, S. H., Sung, J. Y., Yong, D., Chun, J., Kim, S. Y., Song, J. H., et al. (2016). Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer 102, 89–95. doi: 10.1016/j.lungcan.2016.10.016
Leng, Q., Holden, V. K., Deepak, J., Todd, N. W., Jiang, F. (2021). Microbiota biomarkers for lung cancer. Diagnostics 11. doi: 10.3390/diagnostics11030407
Li, D., Wu, M. (2021). Pattern recognition receptors in health and diseases. Signal Transduct Target Ther. 6, 291. doi: 10.1038/s41392-021-00687-0
Li, Y., Xiao, X., Han, Y., Gorlova, O., Qian, D., Leighl, N., et al. (2018). Genome-wide interaction study of smoking behavior and non-small cell lung cancer risk in Caucasian population. Carcinogenesis 39, 336–346. doi: 10.1093/carcin/bgx113
Liang, J. Q., Li, T., Nakatsu, G., Chen, Y. X., Yau, T. O., Chu, E., et al. (2020). A novel faecal Lachnoclostridium marker for the non- invasive diagnosis of colorectal adenoma and cancer. Gut 69, 1248–1257. doi: 10.1136/gutjnl-2019-318532
Liu, N. N., Ma, Q., Ge, Y., Yi, C. X., Wei, L. Q., Tan, J. C., et al. (2020). Microbiome dysbiosis in lung cancer: from composition to therapy. NPJ Precis Oncol. 4, 33. doi: 10.1038/s41698-020-00138-z
Liu, Y., O'brien, J. L., Ajami, N. J., Scheurer, M. E., Amirian, E. S., Armstrong, G., et al. (2018). Lung tissue microbial profile in lung cancer is distinct from emphysema. Am. J. Cancer Res. 8, 1775–1787.
Lu, H., Gao, N. L., Tong, F., Wang, J., Li, H., Zhang, R., et al. (2021). Alterations of the human lung and gut microbiomes in non-small cell lung carcinomas and distant metastasis. Microbiol. Spectr. 9, e0080221. doi: 10.1128/Spectrum.00802-21
Ma, Q. Y., Huang, D. Y., Zhang, H. J., Wang, S., Chen, X. F. (2017). Upregulation of bacterial-specific Th1 and Th17 responses that are enriched in CXCR5(+)CD4(+) T cells in non-small cell lung cancer. Int. Immunopharmacol 52, 305–309. doi: 10.1016/j.intimp.2017.09.024
Madapoosi, S. S., Cruickshank-Quinn, C., Opron, K., Erb-Downward, J. R., Begley, L. A., Li, G., et al. (2022). Lung microbiota and metabolites collectively associate with clinical outcomes in milder stage chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 206, 427–439. doi: 10.1164/rccm.202110-2241OC
Mao, Q., Jiang, F., Yin, R., Wang, J., Xia, W., Dong, G., et al. (2018). Interplay between the lung microbiome and lung cancer. Cancer Lett. 415, 40–48. doi: 10.1016/j.canlet.2017.11.036
Marshall, E. A., Filho, F. S. L., Sin, D. D., Lam, S., Leung, J. M., Lam, W. L. (2022). Distinct bronchial microbiome precedes clinical diagnosis of lung cancer. Mol. Cancer 21, 68. doi: 10.1186/s12943-022-01544-6
Martiny, A. C. (2019). High proportions of bacteria are culturable across major biomes. ISME J. 13, 2125–2128. doi: 10.1038/s41396-019-0410-3
Masuhiro, K., Tamiya, M., Fujimoto, K., Koyama, S., Naito, Y., Osa, A., et al. (2022). Bronchoalveolar lavage fluid reveals factors contributing to the efficacy of PD-1 blockade in lung cancer. JCI Insight 7. doi: 10.1172/jci.insight.157915
Meng, Y., Mao, Y., Tang, Z., Qiu, X., Bajinka, O., Tan, Y., et al. (2023). Crosstalk between the lung microbiome and lung cancer. Microbial Pathogenesis 178, 106062. doi: 10.1016/j.micpath.2023.106062
Moffatt, M. F., Cookson, W. O. (2017). The lung microbiome in health and disease. Clin. Med. (London England) 17, 525–529. doi: 10.7861/clinmedicine.17-6-525
National Lung Screening Trial Research, Aberle, T., D. R., Adams, A. M., Berg, C. D., Black, W. C., Clapp, J. D., et al. (2011). Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl. J. Med. 365, 395–409. doi: 10.1056/NEJMoa1102873
O’Callaghan, D. S., Rexhepaj, E., Gately, K., Coate, L., Delaney, D., O'donnell, D. M., et al. (2015). Tumour islet Foxp3+ T-cell infiltration predicts poor outcome in nonsmall cell lung cancer. Eur. Respir. J. 46, 1762–1772. doi: 10.1183/13993003.00176-2014
Ochoa, C. E., Mirabolfathinejad, S. G., Ruiz, V. A., Evans, S. E., Gagea, M., Evans, C. M., et al. (2011). Interleukin 6, but not T helper 2 cytokines, promotes lung carcinogenesis. Cancer Prev. Res. (Phila) 4, 51–64. doi: 10.1158/1940-6207.CAPR-10-0180
Ou, S. H., Zell, J. A., Ziogas, A., Anton-Culver, H. (2007). Prognostic factors for survival of stage I nonsmall cell lung cancer patients : a population-based analysis of 19,702 stage I patients in the California Cancer Registry from 1989 to 2003. Cancer 110, 1532–1541. doi: 10.1002/cncr.22938
Owada-Ozaki, Y., Muto, S., Takagi, H., Inoue, T., Watanabe, Y., Fukuhara, M., et al. (2018). Prognostic impact of tumor mutation burden in patients with completely resected non-small cell lung cancer: brief report. J. Thorac. Oncol. 13, 1217–1221. doi: 10.1016/j.jtho.2018.04.003
Patnaik, S. K., Cortes, E. G., Kannisto, E. D., Punnanitinont, A., Dhillon, S. S., Liu, S., et al. (2021). Lower airway bacterial microbiome may influence recurrence after resection of early-stage non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 161419–429.e416. doi: 10.1016/j.jtcvs.2020.01.104
Peters, B. A., Hayes, R. B., Goparaju, C., Reid, C., Pass, H. I., Ahn, J. (2019). The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol. Biomarkers Prev. 28, 731–740. doi: 10.1158/1055-9965.EPI-18-0966
Peters, B. A., Pass, H. I., Burk, R. D., Xue, X., Goparaju, C., Sollecito, C. C., et al. (2022). The lung microbiome, peripheral gene expression, and recurrence-free survival after resection of stage II non-small cell lung cancer. Genome Med. 14, 121. doi: 10.1186/s13073-022-01126-7
Planck, M., Isaksson, S., Veerla, S., Staaf, J. (2013). Identification of transcriptional subgroups in EGFR-mutated and EGFR/KRAS wild-type lung adenocarcinoma reveals gene signatures associated with patient outcome. Clin. Cancer Res. 19, 5116–5126. doi: 10.1158/1078-0432.CCR-13-0928
Pragman, A. A., Kim, H. B., Reilly, C. S., Wendt, C., Isaacson, R. E. (2012). The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PloS One 7, e47305. doi: 10.1371/journal.pone.0047305
Ramirez-Labrada, A. G., Isla, D., Artal, A., Arias, M., Rezusta, A., Pardo, J., et al. (2020). The influence of lung microbiota on lung carcinogenesis, immunity, and immunotherapy. Trends Cancer 6, 86–97. doi: 10.1016/j.trecan.2019.12.007
Rossi, M., Amaretti, A., Raimondi, S. (2011). Folate production by probiotic bacteria. Nutrients 3, 118–134. doi: 10.3390/nu3010118
Rubinstein, M. R., Wang, X., Liu, W., Hao, Y., Cai, G., Han, Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206. doi: 10.1016/j.chom.2013.07.012
Segal, L. N., Alekseyenko, A. V., Clemente, J. C., Kulkarni, R., Wu, B., Gao, Z., et al. (2013). Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome 1, 19. doi: 10.1186/2049-2618-1-19
Segal, L. N., Clemente, J. C., Li, Y., Ruan, C., Cao, J., Danckers, M., et al. (2017). Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-drug-treated HIV patients. Cell Host Microbe 21530–537.e534. doi: 10.1016/j.chom.2017.03.003
Segal, L. N., Clemente, J. C., Tsay, J. C., Koralov, S. B., Keller, B. C., Wu, B. G., et al. (2016). Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 1, 16031. doi: 10.1038/nmicrobiol.2016.31
Sellge, G., Kufer, T. A. (2015). PRR-signaling pathways: Learning from microbial tactics. Semin. Immunol. 27, 75–84. doi: 10.1016/j.smim.2015.03.009
Shen, H., Zhu, M., Wang, C. (2019). Precision oncology of lung cancer: genetic and genomic differences in Chinese population. NPJ Precis Oncol. 3, 14. doi: 10.1038/s41698-019-0086-1
Siegel, R. L., Miller, K. D., Fuchs, H. E., Jemal, A. (2022). Cancer statistics 2022. CA: A Cancer J. Clin. 72, 7–33. doi: 10.3322/caac.21708
Silveira, C. B., Cobián-Güemes, A. G., Uranga, C., Baker, J. L., Edlund, A., Rohwer, F., et al. (2021). Multi-omics study of keystone species in a cystic fibrosis microbiome. Int. J. Mol. Sci. 22, 12050. doi: 10.3390/ijms222112050
Singh, S., Natalini, J. G., Segal, L. N. (2022). Lung microbial-host interface through the lens of multi-omics. Mucosal Immunol. 15, 837–845. doi: 10.1038/s41385-022-00541-8
Society, A. C. (2022) Lung cancer survival rates. Available at: https://www.cancer.org/cancer/lung-cancer/detection-diagnosis-staging/survival-rates.html (Accessed October 27 2022).
Soroosh, P., Doherty, T. A., Duan, W., Mehta, A. K., Choi, H., Adams, Y. F., et al. (2013). Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med. 210, 775–788. doi: 10.1084/jem.20121849
Sun, Y., Liu, Y., Li, J., Tan, Y., An, T., Zhuo, M., et al. (2023). Characterization of lung and oral microbiomes in lung cancer patients using culturomics and 16S rRNA gene sequencing. Microbiol. Spectr. 11, e00314–e00323. doi: 10.1128/spectrum.00314-23
Torsvik, V., Øvreås, L. (2002). Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5, 240–245. doi: 10.1016/s1369-5274(02)00324-7
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Tsay, J. J., Wu, B. G., Badri, M. H., Clemente, J. C., Shen, N., Meyn, P., et al. (2018). Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am. J. Respir. Crit. Care Med. 198, 1188–1198. doi: 10.1164/rccm.201710-2118OC
Tsay, J. J., Wu, B. G., Sulaiman, I., Gershner, K., Schluger, R., Li, Y., et al. (2021). Lower airway dysbiosis affects lung cancer progression. Cancer Discovery 11, 293–307. doi: 10.1158/2159-8290.CD-20-0263
Wang, K., Huang, Y., Zhang, Z., Liao, J., Ding, Y., Fang, X., et al. (2019). A preliminary study of microbiota diversity in saliva and bronchoalveolar lavage fluid from patients with primary bronchogenic carcinoma. Med. Sci. Monit 25, 2819–2834. doi: 10.12659/MSM.915332
Wang, Q., Zhou, D., Wu, F., Liang, Q., He, Q., Peng, M., et al. (2021). Immune microenvironment signatures as biomarkers to predict early recurrence of stage Ia-b lung cancer. Front. Oncol. 11. doi: 10.3389/fonc.2021.680287
Willner, D. L., Hugenholtz, P., Yerkovich, S. T., Tan, M. E., Daly, J. N., Lachner, N., et al. (2013). Reestablishment of recipient-associated microbiota in the lung allograft is linked to reduced risk of bronchiolitis obliterans syndrome. Am. J. Respir. Crit. Care Med. 187, 640–647. doi: 10.1164/rccm.201209-1680OC
Wong, L. M., Shende, N., Li, W. T., Castaneda, G., Apostol, L., Chang, E. Y., et al. (2020). Comparative analysis of age- and gender-associated microbiome in lung adenocarcinoma and lung squamous cell carcinoma. Cancers (Basel) 12. doi: 10.3390/cancers12061447
Wu, Z., Tang, J., Zhuang, R., Meng, D., Zhang, L., Gu, C., et al. (2022). The microbiome of lower respiratory tract and tumor tissue in lung cancer manifested as radiological ground-glass opacity. Front. Bioeng Biotechnol. 10. doi: 10.3389/fbioe.2022.892613
Xia, C., Dong, X., Li, H., Cao, M., Sun, D., He, S., et al. (2022). Cancer statistics in China and United States 2022: profiles, trends, and determinants. Chin. Med. J. (Engl), 135, 584–590. doi: 10.1097/CM9.0000000000002108
Yagi, K., Huffnagle, G. B., Lukacs, N. W., Asai, N. (2021). The lung microbiome during health and disease. Int. J. Mol. Sci. 22. doi: 10.3390/ijms221910872
Yan, X., Yang, M., Liu, J., Gao, R., Hu, J., Li, J., et al. (2015). Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 5, 3111–3122.
Young, R. P., Hopkins, R. J., Hay, B. A., Epton, M. J., Black, P. N., Gamble, G. D. (2008). Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur. Respir. J. 32, 1158–1164. doi: 10.1183/09031936.00093908
Yu, G., Gail, M. H., Consonni, D., Carugno, M., Humphrys, M., Pesatori, A. C., et al. (2016). Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 17, 163. doi: 10.1186/s13059-016-1021-1
Yu, X., Rao, J., Lin, J., Zhang, Z., Cao, L., Zhang, X. (2014). Tag SNPs in complement receptor-1 contribute to the susceptibility to non-small cell lung cancer. Mol. Cancer 13, 56. doi: 10.1186/1476-4598-13-56
Zempleni, J., Teixeira, D. C., Kuroishi, T., Cordonier, E. L., Baier, S. (2012). Biotin requirements for DNA damage prevention. Mutat. Res. 733, 58–60. doi: 10.1016/j.mrfmmm.2011.08.001
Zeng, W., Zhao, C., Yu, M., Chen, H., Pan, Y., Wang, Y., et al. (2022). Alterations of lung microbiota in patients with non-small cell lung cancer. Bioengineered 13, 6665–6677. doi: 10.1080/21655979.2022.2045843
Zhang, Y. B., He, F. L., Fang, M., Hua, T. F., Hu, B. D., Zhang, Z. H., et al. (2009). Increased expression of Toll-like receptors 4 and 9 in human lung cancer. Mol. Biol. Rep. 36, 1475–1481. doi: 10.1007/s11033-008-9338-9
Zhang, W., Luo, J., Dong, X., Zhao, S., Hao, Y., Peng, C., et al. (2019). Salivary microbial dysbiosis is associated with systemic inflammatory markers and predicted oral metabolites in non-small cell lung cancer patients. J. Cancer 10, 1651–1662. doi: 10.7150/jca.28077
Zhang, C., Wang, J., Sun, Z., Cao, Y., Mu, Z., Ji, X. (2021). Commensal microbiota contributes to predicting the response to immune checkpoint inhibitors in non-small-cell lung cancer patients. Cancer Sci. 112, 3005–3017. doi: 10.1111/cas.14979
Zheng, L., Xu, J., Sai, B., Zhu, Y., Wang, L., Yin, N., et al. (2020). Microbiome related cytotoxically active CD8+ TIL are inversely associated with lung cancer development. Front. Oncol. 10. doi: 10.3389/fonc.2020.531131
Zhou, J., Qu, Z., Sun, F., Han, L., Li, L., Yan, S., et al. (2017). Myeloid STAT3 promotes lung tumorigenesis by transforming tumor immunosurveillance into tumor-promoting inflammation. Cancer Immunol. Res. 5, 257–268. doi: 10.1158/2326-6066.CIR-16-0073
Zhou, Y., Zeng, H., Liu, K., Pan, H., Wang, B., Zhu, M., et al. (2023). Microbiota profiles in the saliva, cancerous tissues and its companion paracancerous tissues among Chinese patients with lung cancer. BMC Microbiol. 23, 237. doi: 10.1186/s12866-023-02882-1
Keywords: lung cancer, respiratory tract, commensal microbiome dysbiosis, immune mechanisms, chromosome aberration
Citation: Cheng J, Zhou L and Wang H (2024) Symbiotic microbial communities in various locations of the lung cancer respiratory tract along with potential host immunological processes affected. Front. Cell. Infect. Microbiol. 14:1296295. doi: 10.3389/fcimb.2024.1296295
Received: 18 September 2023; Accepted: 18 January 2024;
Published: 02 February 2024.
Edited by:
Tao Lin, Baylor College of Medicine, United StatesReviewed by:
Kaiser Jamil, Bhagwan Mahavir Medical Research Centre, IndiaSoraya Mezouar, Aix-Marseille University, France
Copyright © 2024 Cheng, Zhou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaqi Wang, d2hxMjAwNEAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
 Jiuling Cheng1†
Jiuling Cheng1† Lujia Zhou
Lujia Zhou Huaqi Wang
Huaqi Wang