- 1TIFAC CORE in Cyber Security, Amrita School of Engineering, Amrita Vishwa Vidyapeetham, Coimbatore, India
- 2Division of Computer Science and Engineering, Karunya Institute of Technology and Sciences, Coimbatore, India
- 3Department of ISE, CMR Institute of Technology, Bengaluru, India
- 4Computer Science and Engineering, Sri Krishna College of Engineering and Technology, Coimbatore, India
- 5Department of Information, Communication and Electronics Engineering, Catholic University of Korea, Korea, Republic of Korea
The computer-aided diagnosis helps medical professionals detect and classify lung diseases from chest X-rays by leveraging medical image processing and central server-based machine learning models. These technologies provide real-time assistance to analyze the input and help efficiently detect the abnormalities at the earliest. However, traditional learning models are not suitable for live scenarios that require privacy, data diversity, and decentralized processing. The Federated learning-based model facilitates the protection of medical data privacy while processing a large volume of medical images, aiming to improve the overall efficiency of the model. This paper proposes a Federated Learning based Ensemble Model (FLEM) framework for an efficient diagnosis of lung diseases. The FLEM utilizes explainable AI techniques, including SHAP, Grad-CAM, and Differential Privacy, to provide transparency and interpretability of predictions while maintaining the privacy and security of medical data. We applied InceptionV3, Conv2D, VGG16, and ResNet-50 models on the COVID-19, TB, and pneumonia datasets and analysed the performance of the models in FLEM and Central Server-based Learning Model (CSLM). The performance analysis shows that the FLEM model outperformed the traditional CSLM model in terms of accuracy, training time, and bandwidth consumption. CSLM witnesses a quicker convergence time than FLEM. Although the CSLM model converged after a considerable number of epochs, it resulted in a 5, 8, 9, and 10% accuracy reduction compared to the FLEM-based training of InceptionV3, Conv2D, VGG16, and ResNet50 that achieved accuracies of 91.8, 88, 92.5, and 95.5%, respectively.
1 Introduction
Respiratory disorders, also known as lung problems, refer to various illnesses that affect the airways and other components of the lungs. Some well-known examples of these disorders include Covid-19, pneumonia, and tuberculosis (Ouyang et al., 2020). Recent research indicates significant mortality rates associated with these conditions, with approximately 334 million deaths attributed to Covid-19, one billion to tuberculosis, and 1.6 million to lung cancer (Asswin et al., 2023; Ganeshkumar et al., 2023; Phogat et al., 2023). Pneumonia has also caused millions of deaths and placed a considerable burden on healthcare systems worldwide. Lung diseases consistently rank among the leading causes of death globally. Early identification is crucial for increasing long-term survival rates and improving the likelihood of recovery. Traditionally, lung disorders have been diagnosed using skin tests, blood tests, and chest X-rays (Al-qaness et al., 2024). These approaches are useful, but they often lack precision, rapidity, and the ability to detect subtle changes in lung conditions. To address these challenges, deep learning has emerged as a transformative approach in this field. By leveraging advanced algorithms and large datasets, deep learning enhances diagnostic accuracy and discovers complex patterns in imaging data that traditional methods may disregard. Deep learning algorithms have emerged as a comprehensive approach for diagnosing lung infections caused by Covid-19, tuberculosis, and pneumonia, which are widely recognized as the leading causes of severe illness and mortality associated with the respiratory system on a global scale. Previous research has predominantly focused on the identification and detection of these specific lung diseases (Ganeshkumar et al., 2023). COVID-19, commonly known as corona virus illness, is a respiratory virus (Phogat et al., 2023; Al-qaness et al., 2024). Timely and accurate analysis of medical images is required for early detection of abnormalities, which facilitates recovery from life-threatening conditions.
Recent technological advances have changed the e-healthcare environment as AI-powered healthcare (Durga et al., 2024). AI systems require centralized data collection and compute-intensive processing, leading to poor quality of experience due to network delay and data privacy issues (Ashwini et al., 2024; Sabry et al., 2024). Federated Learning (FL), an emerging distributed collaborative AI paradigm, coordinates the training of models within the proximity of data sources. It promises healthcare facilities without sharing private data, which is mandatory in today’s intelligent healthcare sector (Ouyang et al., 2020). In the innovative healthcare sector, medical image analysis uses deep learning algorithms to process large amounts of health data and to detect chronic diseases early (Arun Prakash et al., 2023; Durga et al., 2024). This paper focuses on using an FL-based framework for identifying lung diseases such as COVID-19, TB, and pneumonia. The motivation of this research is that FL differs from traditional machine learning models in terms of participating clients and dataset parameters. The FL-based efficient framework is a collaborative learning environment aiming for consistent solutions. In an FL-based hospital environment, hospital local servers are the participating devices that can have heterogeneous hardware.
The key highlights of the paper are, (1) to classify the lung diseases such as COVID-19, TB, and pneumonia we build FLEM, a federated learning based efficient framework for medical image processing. (2) Deep learning models such as InceptionV3, Convolution2D, VGG16, and ResNet50 have been analyzed on the proposed framework to find which one accurately detects lung disease from the given input. (3) The pre-trained version of these models with learned features was deployed at Hospital server. The detailed workflow at the local device of each hospital and central server was given. (4) Experimental comparison of the performances of FLEM and CSLM in terms of Accuracy, Training time, convergence time and Bandwidth consumption was presented. (5) The proposed FLEM-XAI framework employs explainable AI to provide transparency and interpretability of predictions while maintaining the privacy and security of medical data. The rest of the paper is organized as follows: Section 2 details the related work. Section 3 discusses the proposed FL-based efficient architecture, algorithm, and flow of work. Section 4 presents the results and performance evaluation. Section 5 concludes the paper with possible directions towards future research.
2 Related work
Automated accurate chest X-ray lesion detection is challenging. These images often exhibit lesions with blurred boundaries, varying sizes, irregular shapes, and uneven density, making it challenging to identify them accurately (Li et al., 2020; Deshmukh et al., 2021). Additionally, traditional convolutional neural networks (CNNs) used for this task are comprised of convolution units that have limitations when it comes to sampling irregular shapes. Consequently, CNNs struggle to extract the intricate details and refined features necessary for detecting and characterizing chest X-ray lesions effectively (Hu et al., 2020; Li et al., 2020). Xi et al. developed the dual-sampling attention network to automatically recognize COVID-19 from pneumonia (Ouyang et al., 2020). They use a 3D CNN and a novel online attention module to diagnose lung infections. In detecting COVID-19 images, this system has an AUC of 0.944, accuracy of 87.5%, sensitivity of 86.9%, specificity of 90.1%, and F1-score of 82.0%. A supervised deep learning architecture was suggested for COVID-19 detection. A huge dataset from many hospitals worldwide was used to evaluate this technology. The framework correctly identified 95.12% of COVID-19 X-rays. COVID-19 cases were distinguished from normal and severe acute respiratory syndrome cases with 97.91% sensitivity, 91.87% specificity, and 93.36% precision (Hu et al., 2020).
Deep learning-based recommender systems utilizing deep CNN architectures (Sethi et al., 2020) have proven to be effective in identifying important biomarkers associated with COVID-19, among other diseases. These systems achieve an impressive overall classification accuracy of 87.66% across seven different classes. Moreover, when specifically detecting COVID-19, this method achieves exceptional accuracy rates of 99.18%, sensitivity of 97.36%, and specificity of 99.42%. Transfer learning and picture augmentation were used to train and validate many pre-trained deep CNNs in (Chowdhury et al., 2020). These networks learned to distinguish between normal and COVID-19 pneumonia and between viral and COVID-19 pneumonia with and without image enhancement. The first scenario has 99.7% classification accuracy, precision, sensitivity, and specificity, while the second had 97.9, 97.95, 97.9, and 98.8%. The results reported in (Apostolopoulos et al., 2020) indicate that training of CNN from scratch can discover biomarkers for a variety of diseases, each of which includes COVID-19. The average accuracy of categorizing the presentations into the seven classes was 87.66%. This method achieved an accuracy of 99.18%, with a sensitivity of 97.36% and a specificity averaging 42%. Overall, this approach proved to be efficient in identifying the virus.
Based on deep CNN, the DeTraC which stands for Decompose, Transfer, and Compose is a transfer learning method (Abbas et al., 2021). This model transfers knowledge from broad item identification to the specific domain tasks effectively. Analyzing the obtained experimental results, it was proved that DeTraC is capable of detecting COVID-19 based on the test dataset of global hospital images. DeTraC effectively segmented COVID-19 X-rays with 93.1% accuracy. These photos reserved these cases different from normal and severe acute respiratory syndrome cases with 100% sensitivity. The COVID-19 infection can be detected by deep learning in two stages. The first process involves Segment-Level categorizing by employing deep learning architectures and multi-tasking with Slice-Level categorizing as its primary task. This allowed the trained models to classify CT scan parts. Another one is the two-stage method of identifying and classifying COVID-19 cases with satisfactory outcomes indicated (Bougourzi et al., 2021). The models developed earlier for COVID-19 and CAP infections were used together with XGBoost classifier to analyze the CT scan and classify it to be normal, COVID-19 or pneumonia infected (Serte and Demirel, 2021; Aggarwal et al., 2022; Mahbub et al., 2022; Jin et al., 2023).
Prior to performing more detailed classifications using DenseNet and EfficientNet in the Two-Stage CNN (Chaudhary et al., 2021), the detection networks are aligned based on the precise regions of interest to be focused on. de Moura et al. (2022) proposed an automated classification technique to detect COVID-19 at an early stage using chest X-Ray images. The study conducted by Nasari et al. utilizes DenseNet169 for feature extraction, whereas XG Boost is employed to provide precise results (Nasiri and Hasani, 2022). Federated Learning (FL) makes centrally controlled trials less time-consuming, increases the possibility to trace the data, and makes the evaluations of changes in algorithms easier. The first untrained model is uploaded to many servers or nodes in the client–server architecture of FL. These nodes partially learn with the data that they own and send back the result to a central federated server. If there are intentions to train the models conjointly without the raw information the above process continues to the realization of the planned goal. A federated learning system (Feki et al., 2021) employs deep learning for the early identification of COVID 19 on chest X rays. This approach makes it possible for the medical institutions to communicate among themselves at the same time protecting patient’s privacy. Similar to a centralized method, this learning method has a comparably good result attributable to not sharing or accumulating large amounts of private information. In distributed learning, the different medical facilities can build models, and carry out precise testing on COVID-19 in a way that does not violate the patient right to privacy. The FL based architecture was proved as a resilient and a comprehensive model (Dayan et al., 2021). In particular, the new global FL model–EXAM involved the data of CXR and EMR of many institutes. In the case of using FL it was possible not to store the data in one place which in its turn contributed to the protection of privacy of the members including the member universities. The distributed data structure on the other hand makes the cooperative model training possible while at the same time protecting the data.
3 Federated learning based ensemble model (FLEM) with explainable AI framework
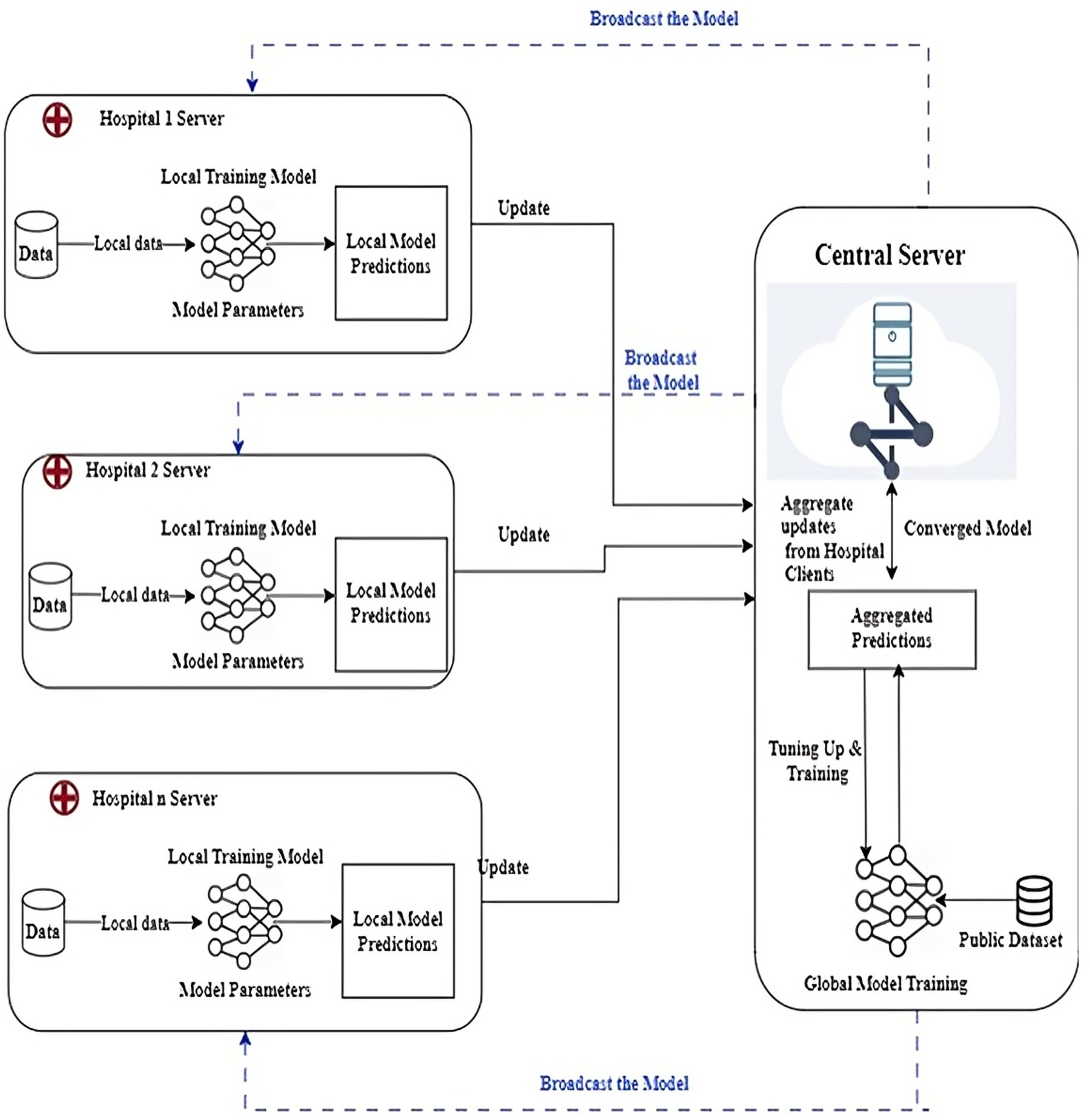
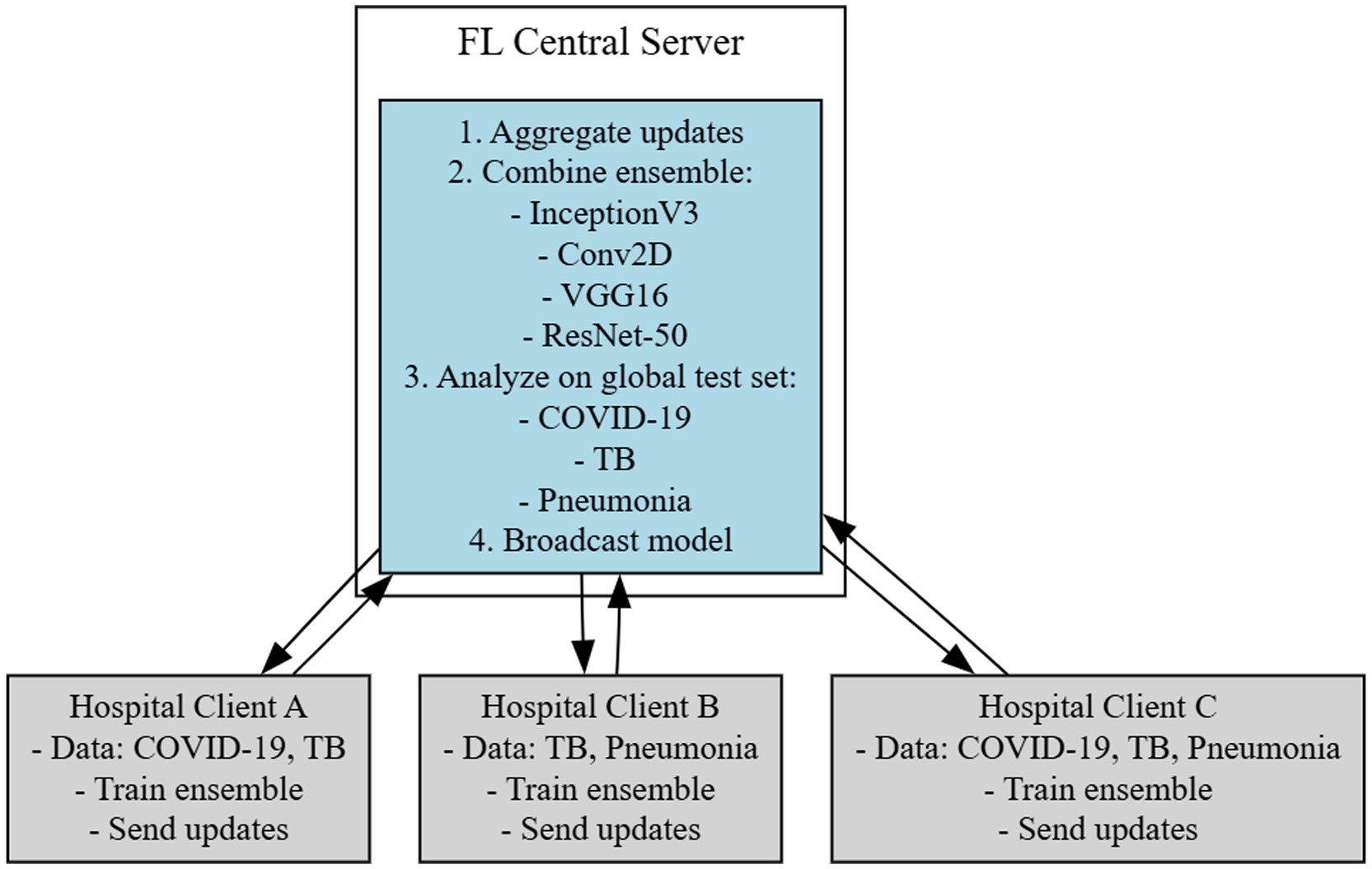
This section describes the FL-based efficient framework used for detecting lung diseases from chest X-ray images. Figure 1 shows the FL based architecture employed for diagnosing lung diseases. The proposed FLEM follows a Centralized Federated Learning Architecture which includes Client nodes, server nodes, and a communication channel. Client nodes own and protect data. Each node trains a model using its local dataset. Server node that coordinates the federated learning process by aggregating the updates received from the client devices and updating a global model without accessing the local datasets. A Communication Channel employs a secure communication protocol to transmit updates between clients and the central server. In Federated Learning, the communication protocol consists of multiple rounds of client–server communication. First, the central server initializes a global model and sends it to all participating client nodes. Next, each client node receives the global model parameters and trains the model using its local dataset for a specified number of epochs. After completing local training, the clients compute updates, such as gradients and send these updates back to the central server. Notably, only model parameters and not raw data are transmitted, preserving data privacy. The proposed framework consists of a hospital server and central server processing.
3.1 FLEM XAI framework setup
The proposed Federated learning framework as shown in Figure 1 was employed where lung disease data is distributed across multiple healthcare institutions (hospitals). Each hospital trains its model locally. The FLEM framework consists of InceptionV3, Conv2D, VGG16, and ResNet-50 models. Each participating hospital server could potentially use the model architectures based on the data available. A central server aggregates the updates from all clients, refines the global model, and then redistributes it. The training process involves the following
• Local Training: Each hospital server trains its local model on its own dataset and updates its model based on the local loss function targeting lung disease diagnosis (e.g., classification of chest X-ray images as normal or indicative of a disease).
• Aggregation: We use Federated Averaging (FedAvg) to combine the model weights from different clients into a robust global model while minimizing data exposure.
3.2 Processing at the central server
Processing at the central server includes 3 steps as follows. (1) Collecting data: A list of hospitals and other data sources are used to gather chest X-ray images with and without lung illness which is well-labeled and pre-processed data. (2) Model training and central server: A deep learning model is developed by employing CNNs and tagged data. (3) Federated learning configuration: Partner hospitals have an edge server configured to receive a model broadcast from the central server. The trained model is split and deployed on the edge server. The server begins to transfer subsets of the model to the pre-designated edge server to commence training.
3.3 Processing at the hospital edge server
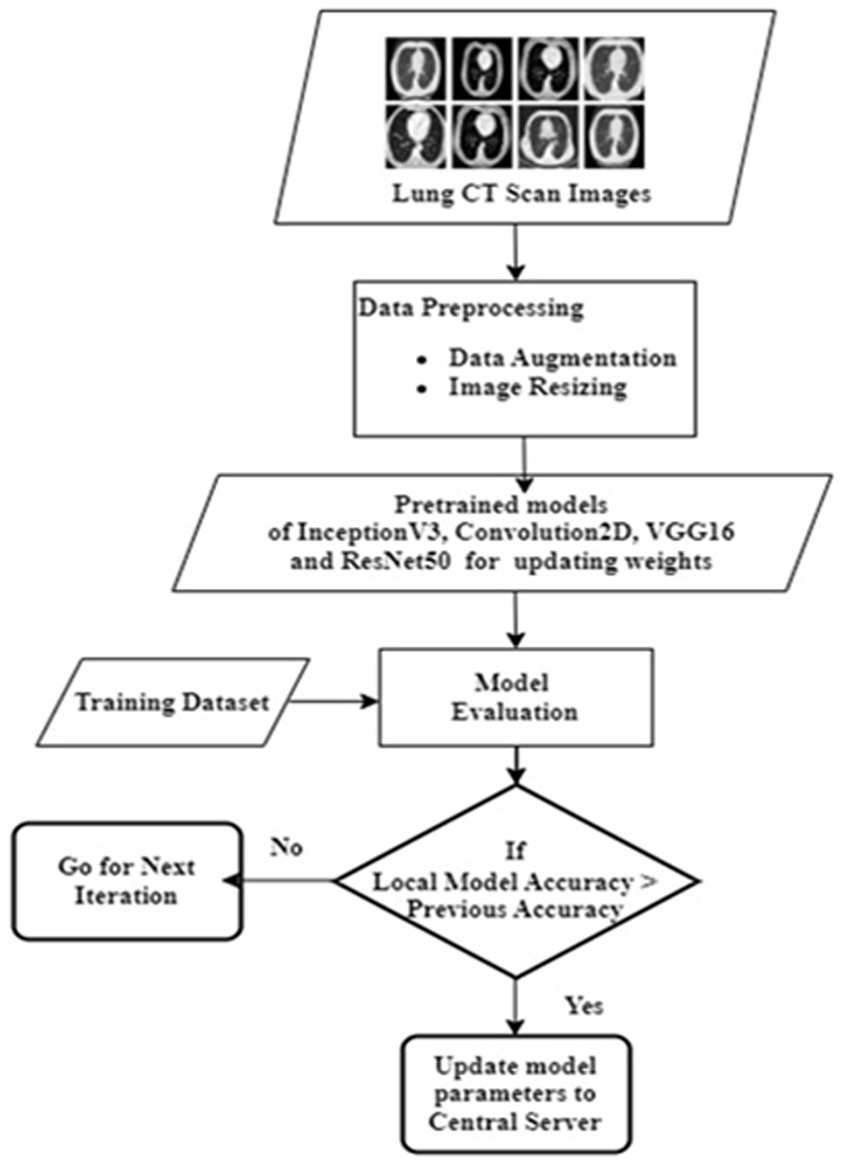
Every participating edge server receives the latest model through the model broadcast. The hospital server initiates the development of its particular section of the model with local information. It trains the model using local data. The model that is updated centrally is tested for precision using a dataset different from the one used in constructing it. The training process is continued until the model is appropriately accurate. After the model training, the hospital edge server uploads the encrypted gradient to a central server. Subsequently, the server applies a merge procedure to integrate the individual model components into the total model. The central server aggregates the gradient update model parameters of each participating edge server. The central server sends the final updated model to each participant. The final model at the edge devices diagnoses lung diseases such as COVID-19, TB, and pneumonia from chest X-rays. The flowchart of the client side processing is shown in Figure 2. It shows the step by step process along with output as loading path, pre-processing of images, augmentation of images label encoder, resNet50, algorithm selected, train resnet50 algorithm by giving input to the trained model the classifier the lung disease.
4 Materials and methods
4.1 Deep learning models employed
Deep learning trains computational models to classify lung disease images. Figure 3 shows the steps involved in lung disease classification. Advanced deep learning systems may outperform humans in accuracy. Models must be trained with massive categorised data and neural network structure. Deep learning methods VGG16, Resnet 50, InceptionV3, and Convolution2d were used to recognize and analyze data. Federated transfer learning is employed to develop collaborative training of models across multiple participative hospital servers. Local model training happens by adapting the pre-trained model to their local dataset. As shown in Figure 1, each hospital server updates the model parameters to the central server, continuing to set up the base model for consistent results. Each model underwent training for a fixed duration of 100 epochs. The architecture encompasses multiple layers and components, which are carefully designed and optimized for the task at hand.
4.1.1 VGG16
The VGG16 CNN architecture is among the most efficient for vision models. This convolution and max pool layer layout were carried over into the design. Finally, it features two FC (fully connected layers) and asoftmax for output. VGG16 denotes that it has 16 weighted layers. This network is quite extensive.
4.1.2 ResNet-50
ResNet50 has skip-connected residual blocks that speed up gradient flow. The network can be exceedingly deep without experiencing concerns like fading gradients. Individual components in unused blocks receive the gradient. Our 50-layer convolutional neural network, RESNET-50, utilised ImageNet architecture with pre-trained layers. This architecture can classify photos into 1,000 separate items.
4.1.3 Convolution2D
2D convolution is the process of convolving two signals that exist in two different orthogonal planes. We can generalize this concept to accommodate multidimensional signals, leading to multidimensional convolution. Digital convolution entails point-by-point multiplication and adding the immediate values of two inputs, of which one is inverted and sampled together with the other at successive intervals. To expand the 1D convolution method to 2D convolution, one of the input signals must be reversed once more. Digital image processing convolves a discrete digital image with a smaller 2D matrix, known as the kernel. This convolution procedure preprocesses image data and extracts low-level features.
4.1.4 InceptionV3
Inception v3 was demonstrated to achieve up to 78.1 overall accuracies on the Image dataset. The model has symmetric and asymmetric construction components such convolutions, average pooling, max pooling, concatenations, dropouts, and entirely linked layers. The model regularly batch normalises activation inputs. The softmax function is utilised to calculate the loss.
4.2 FLEM XAI model and dataset
FL ensures that third parties do not get a chance to mine patients’ sensitive information. As more edge devices arrive, training is faster, which also implies fast model updating. The model and analysis of the proposed approach are also equally simple, where only a chest X-ray image is needed to identify lung disease. Hospitals can use automated software applications to revolutionize healthcare for efficient diagnosis and healthcare assistance. The proposed FLEM framework could be used in such applications that also provide a user interface to get recommendations for doctors, specialists and nearby hospitals.
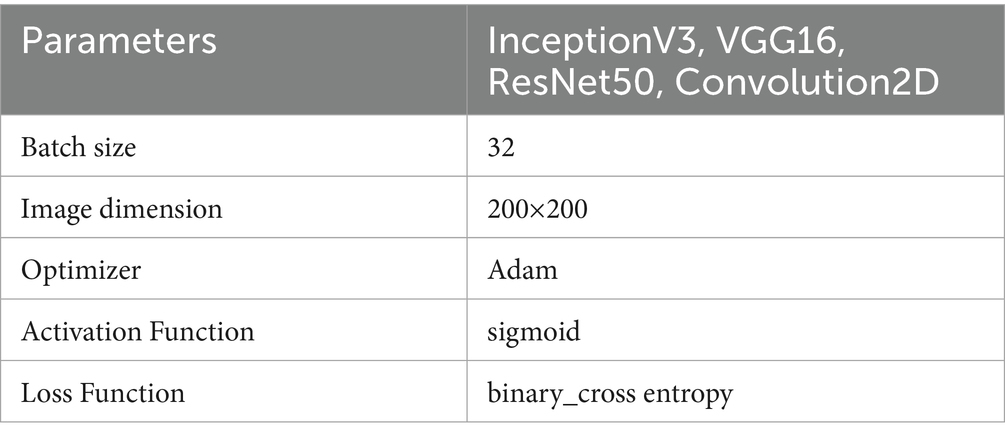
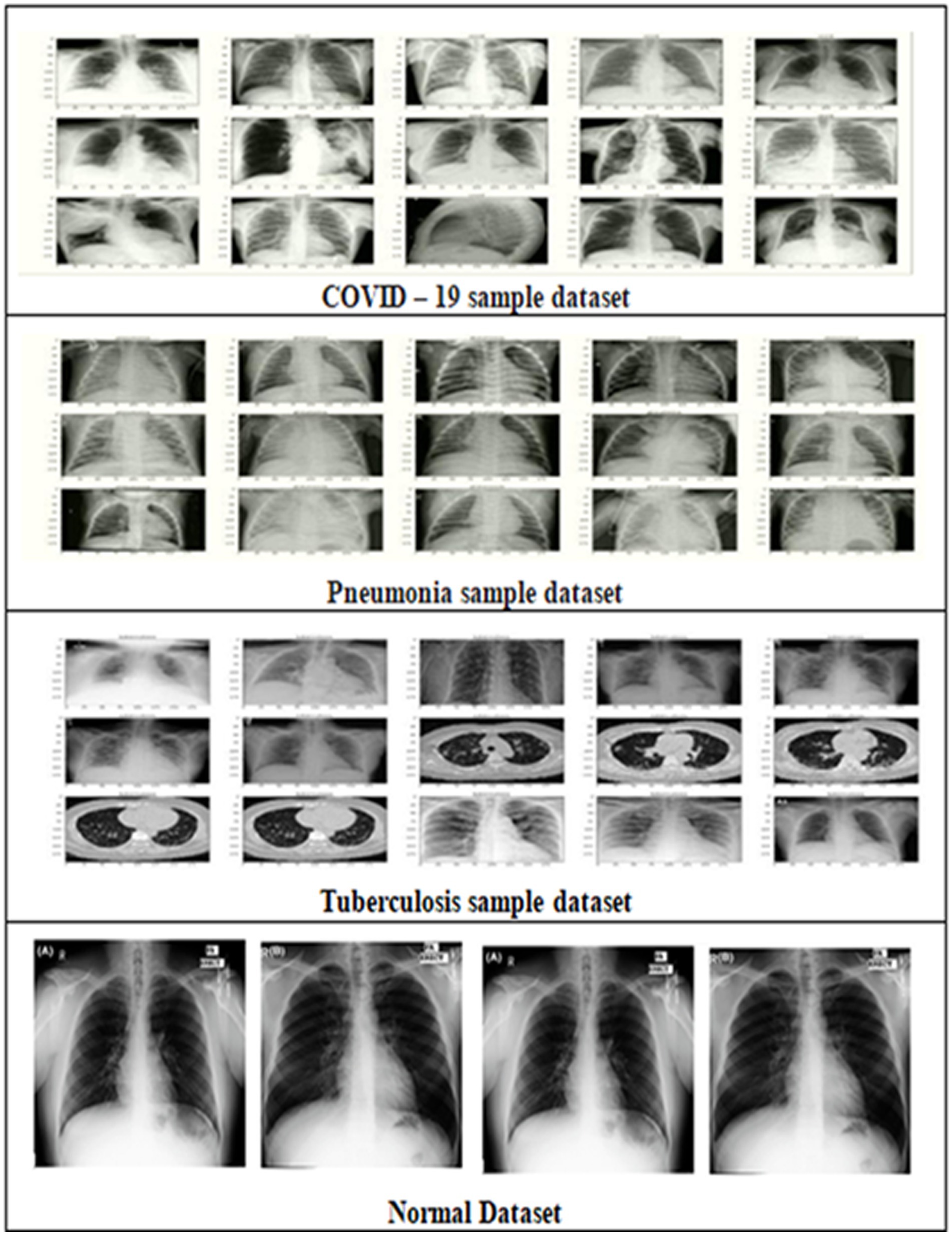
Kaggle was used to obtain 11,000 chest X-RAY images for COVID-19, PNEUMONIA, and TUBERCULOSIS samples (Kaggle, 2021). We then trained a model on these datasets. The images represent various demographics including age and gender, and variations in disease presentation. Images were collected under diverse equipment and patient positioning. The pixel values have been adjusted to fall within a standard range 0–1) for the neural networks to learn effectively. The images were resized to 200×200 pixels and the details are shown in Table 1. Data Augmentation techniques such as rotation and zooming have been applied to improve dataset size and prevent overfitting. The processed dataset was divided into training, validation, and test sets.
The CNN architectures we used for training data included InceptionV3, ResNet50, Convolution2D, and VGG16. The selection of InceptionV3, Conv2D, VGG16, and ResNet-50 for the proposed framework is based on the unique advantages of each model, providing a diversified approach that can capture a wide range of features. InceptionV3 can capture multi-scale features due to its inception modules, which contain filters of different sizes. This architecture is effective in handling varying sizes and styles of images, thus improving performance on diverse datasets. Conv2D model can serve as a baseline for performance benchmarking. VGG16 has 3×3 convolutions and permits deep feature extraction despite its depth. ResNet50 model is robust in scenarios with large amounts of data and complex features, making it a strong candidate for ensuring that intricate relationships within data are captured effectively. We used weighted averaging ensemble and assigned weights to each model based on their individual performance such as accuracy and F1 score.
Deep learning algorithms have been used to diagnose lung illness. This section describes the creation of a chest X-ray detection model for COVID-19 and other lung diseases, such as pneumonia and tuberculosis. Hospital professionals can feed the scanned images to the web user interface, which provides lung disease classification results and possible suggestions on specialty hospitals. Figure 4 shows the block diagram of FLEM server side processing. FL central server holds a global CNN ensemble model comprising architectures like VGG, ResNet50, InceptionV3, Conv2D. This global model is split by parameter segments) and selectively dispatched to partner hospitals’ edge servers. Each hospital clients receives its subset of the model and trains using local hospital data. After local training, each Hospital server sends its updated weights back to the central server securely. The central server aggregates updates using Federated Averaging (FedAvg). The proposed FLEM-XAI framework employs explainable AI to provide transparency and interpretability of predictions while maintaining the privacy and security of medical data. An XAI added FLEM model comprises of the following:
(1) Model Training & Federated Learning Setup: Each participating hospital trains its own local model. Updates are aggregated at a central server using Federated Averaging (FedAvg). The FLEM framework consists of InceptionV3, Conv2D, VGG16, and ResNet-50 models.
(2) Incorporating Explainability Methods: The following XAI techniques have been used to interpret the predictions made by the FLEM model: SHAP (Shapley Additive Explanations) is employed to analyze the importance of different features, Grad-CAM (Gradient-weighted Class Activation Mapping) is used to visualize regions in X-ray images that influence model decisions and, Federated XAI (FedXAI) offers privacy-preserving explanations.
(3) Ensuring Security and Privacy in FLEM-XAI: The proposed FLEM-XAI uses Differential Privacy (DP) method where noise injection is added to gradients to ensure privacy and the noise scale is configured as noise_scale = 0.01. Secure Aggregation (SA) techniques can be applied to mask sensitive data. We use TenSEAL (CKKS encryption) to encrypt model updates.
5 Simulation and performance analysis
This section describes the simulation environment, results, and performance analysis of FLEM and the tested setup and result analysis of XAI-FELM with RESNET50. The proposed FLEM framework for lung disease classification was tested using four distinct deep-learning models: InceptionV3, Convolution2D, VGG16, and ResNet50. The proposed work focuses on analyzing and comparing the performance of the four employed models and identifying the best model for the target environment.
5.1 Simulation setup
The simulations were carried out in Python language using the Keras and TensorFlow libraries. We executed the framework on Windows 10 with 16GB RAM, Intel i5 processor, and NVIDIA GeForce GTX GPU 1080 Ti with 11GB memory. The opted GTX GPU has the support of CUDA 8. This study incorporated pre-processing of the data, dividing it into training and validation sets and a subsequent testing set. We used CNN frameworks such as InceptionV3, ResNet50, VGG16, and Convolution2D to construct the diagnostic models. We chose to focus on the TensorFlow system in conjunction with the Keras library (Durga et al., 2025). We simulated the environment, which involves four machines: one acting as a central server and three as hospital local servers, each with different configurations, as given below.
• Hospital local servers 1,2 and 3:
• GPU: GTX 1080 Ti
• RAM: 11GB
• Python: 3.9
• CUDA: 8
• Server:
• GPU: GTX 1080 Ti
• RAM: 11GB
• Python: 3.9
• CUDA: 10.1
5.2 Dataset used
Kaggle was used to obtain the chest X-ray images for the COVID-19, TB, and pneumonia samples datasets. The collection includes 11,000 chest X-rays comprising COVID-19, pneumonia, tuberculosis, and normal non-diseased images. The dataset includes Anterior-posterior chest X-rays of diseased people in PNG format. The sample dataset is shown in Figure 5. We trained 65% of our dataset and tested 35%. Unlike centralized deep learning model training, the FL need not collect all the data on a single server. The FLEM framework enables model training using diversified datasets. Thus, it promises data privacy and minimizes the need for a huge amount of data transfer. In the current work, the central server model training has been completed with 3000 no. of chest x-ray images. Four models considered are InceptionV3, Convolution2D, VGG16 and ResNet50 for the classes of covid19, Pneumonia and tuberculosis diseases (Jasmine Pemeena Jasmine Pemeena Priyadarsini et al., 2023; Akinwamide et al., 2024) The dataset is divided into three portions, and the three hospital servers will use each portions for local model training. The dataset division ensures that a variable number of images with varied sizes are allocated to each HLS, ensuring a diligent dataset for the investigation. The FLEM initial training session starts by assigning HLS1 15c00 images of size 107.81 MB, HLS2 1000 images of size 100.2 MB, and HLS3 2200 images of size 175 MB, totaling 4700 images. The subsequent model training uses 1000 images of size 89 MB on HLS1, 1650 images of size 120 MB on HLS 2, and 2000 images of size 150 MB on HLS3, totaling 4650 images. As shown in the Figure 4, in client-side processing, the model parameters were updated with the central server to refine the central model. In the third level of training, HLS1, 2, and 3 were assigned 2000, 3500, and 4000 no. of images with a total size of 165MB, 190MB, and 220MB, respectively.
The first stage in the workflow is data pre-processing; the second is training before image augmentation. We conduct validation to check the accuracy of the models after pre-processing a portion of the datasets for 100 epochs. Next, we perform K-fold cross-validation using different test data to evaluate the models' ability to categorize lung infections. Users receive the model outputs to guide their interpretation of X-ray images for infection and to enhance the accuracy of this assessment. This process is essential for a variety of reasons, primarily because it enables healthcare decision-making and contributes to the timely diagnosis of diseases. The parameters used for training the model is shown in Table 1.
5.3 Performance metrics
The performance metrics such as Precision (P), Recall (R), F1-score (F1Score) and Accuracy (A) are used to compare the performances of the employed InceptionV3, Convolution2D, VGG16 and ResNet50 models. The details are described as follows,
Precision is computed as the ratio of actual positive cases correctly classified to the total number of positive cases classified. Equation 1 shows the precision.
Where denotes True Positive cases that are positive that have been typed as positive and is the cases that are actually negative but have been typed as positive. Sensitivity, which is also referred to as recall or true positive rate, is obtained by comparing the number of actually positive patients that were correctly diagnosed on the part of a total number of actually positive patients. Recall is described by Equation 2.
In Equation 2, False Negative represents the false negative instances, which means they are identified to be negative while they are positive. The F1-score obtained from the average of the precision and recall, which gives an equal measure of the performance of the model. Equation 3 shows the F1-score calculation.
Accuracy is the percentage of correct predictions out of the total predictions made. Equation 4 demonstrates the accuracy. Accuracy quantifies and evaluates the overall model performance across all classes.
All these metrics give a measure of the ability of the model in the identification of COVID-19, pneumonia, and tuberculosis from the chest X-ray images. Higher values are more desirable in distinguishing between the affected and non-affected instances, which is critical for decision-making in clinical practice.
5.3.1 Performance metrics of the FLEM framework
FLEM framework differs from Central Server-based Learning Model (CSLM) frameworks in terms of performance measures such as training time, convergence time and bandwidth consumption. Each of these performance metrics is described as follows.
CSLM training time ( is the sum of the time required to load the dataset in the server and the time expended for training the model at central server() and is described in Equation 5.
FLEM server training time is defined as the sum of time required for parallel training of local models on every hospital server (, time required to aggregate the models and the communication time consumed between hospital local servers and the central server ).
CSLM convergence time is the time taken by the deployed model to obtain a certain level of accuracy in the central server. Equation 7 shows the formula of this convergence time.
Where, e is the total no. of epochs needed for convergence and is the time taken for the nth epoch.
FLEM convergence time ) is defined as the time taken by the model to converge based on FL-based training. Equation 8 shows the formula of the FLEM convergence time.
In Equation 8, r is the number of communication rounds needed for convergence, is the training time at hospital local server for the nth round, is the aggregation time for the nth round and is the communication time for the nth round.
The total bandwidth consumption of the central server based deep learning framework refers to the two way communication bandwidth of the central server and the client.
Equation 9 shows the formula of computing the BW consumption of the CSLM framework. In Equation 9, D is the number of data samples and is the size of each data sample.
The total bandwidth consumption of FLEM is defined by using equation 10.
In Equation 10, r is the number of communication rounds and is the total size of the model parameters in Bytes.
5.4 Results and discussion
This study used four distinct models: InceptionV3, Convolution2D, VGG16, and ResNet50. Three data sets were used to train the FLEM models to ensure the functioning of FL. The experiments were conducted with three rounds of training, and the results obtained by the employed models were analyzed to find a suitable model for lung disease classification. Table 2 presents a comprehensive summary of the results attained by the employed models on the proposed FLEM framework. The results were obtained after updating the third round of training.
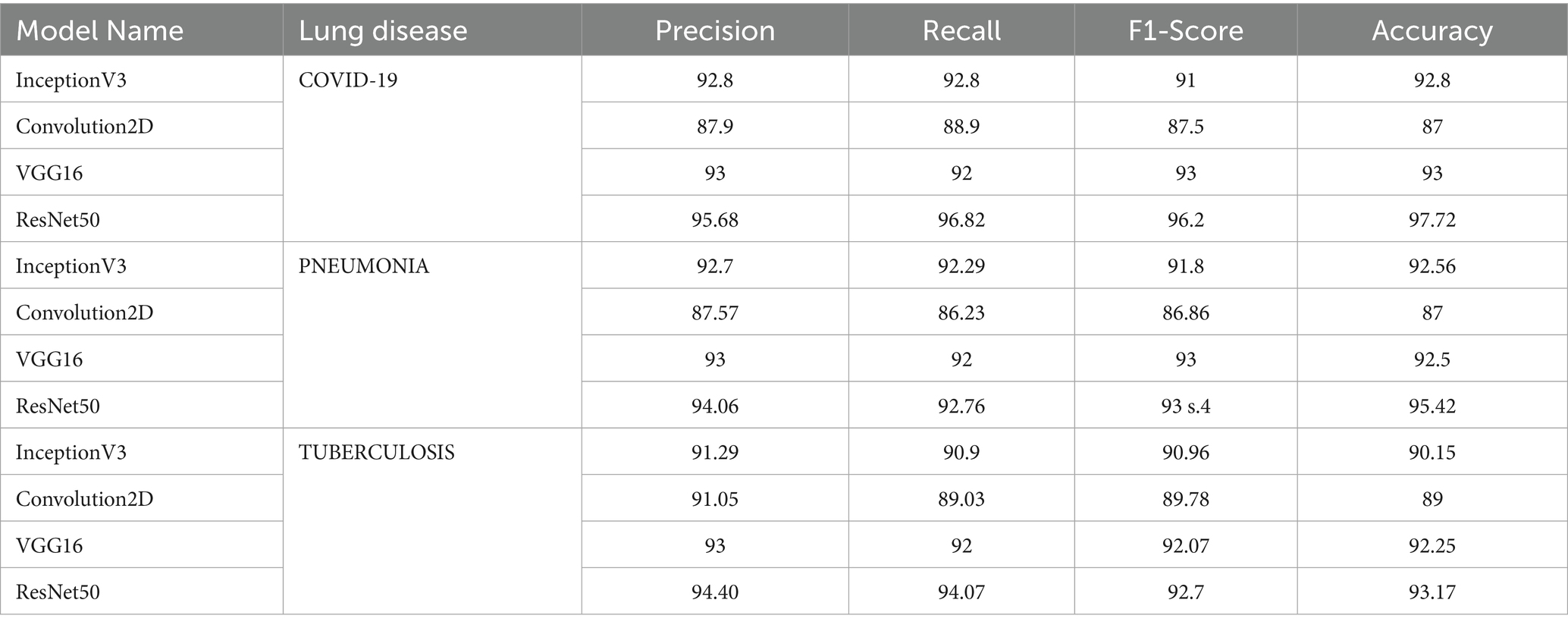
Table 2. Performance evaluation of InceptionV3, Convolution2D, VGG16 and ResNet50 models on COVID-19, PNEUMONIA and TUBERCULOSIS classification.
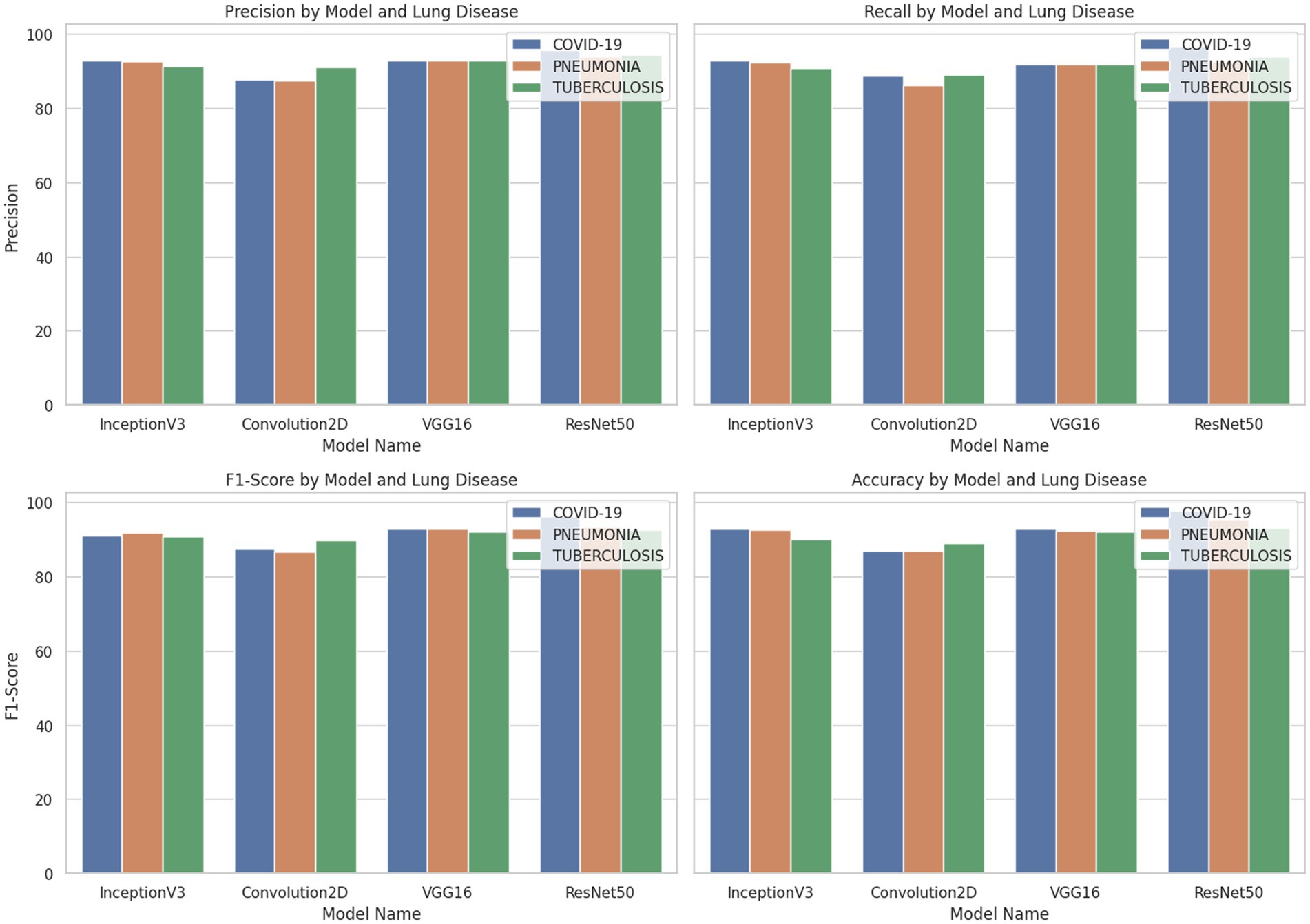
Figure 6 shows the graph comparing the performance of the employed model in terms of precision, recall, F1 score and accuracy. ResNet50 showed outstanding performance in metrics for COVID-19 classification, with an accuracy of 97.72%. VGG16 also performed well, while Convolution2D had the lowest scores. Inception V3 achieved better results with 6.67% more accuracy than Conv2D due to the use of an inception module that helps capture different levels of features by using multiple filter sizes. Overall, RESNET 50 achieved noticeable results, with a 5.3 and 6% increase in accuracy compared to inception V3 and VGG16. The reason is ResNet50’s effectiveness in learning complex features. Conv2D represents a simpler architecture with fewer parameters and layers than the others, which limits its capacity to learn complex patterns, resulting in lower performance.
InceptionV3 model implementation revealed 5.85, 7.03, 5.69 and 6.39% increases in precision, recall and F1-Score and accuracy, respectively, for Pneumonia classification. ResNet50 again showed superior performance for Pneumonia classification with an accuracy of 95.42%, which is 3.09% higher than InceptionV3 and 3.6% higher than VGG16. ResNet50 achieved a high accuracy of 93.17% for Tuberculosis classification when compared with the other three models. InceptionV3 and VGG16 were also effective, producing an accuracy of 90.15 and 92.25%, while Convolution2D had the lowest performance with 89% accuracy.
Figures 7A–D shows the confusion matrix, ROC curve, Epochs vs. accuracy and Epochs vs. loss graph of the ResNet50 model, respectively. The Epochs versus testing data accuracy of Inceptron V3, Convolution2D, VGG-16 and ResNet50 models are shown in Figure 8. Figure 8A shows the testing data accuracy of CNN models such as InceptionV3, Convolution2D, VGG16, and ResNet50 for a COVID-19 dataset with varying numbers of epochs. The accuracy of InceptionV3 increases as the number of epochs increases. The results revealed that the highest accuracy, 92.8%, was achieved at 100 epochs, and it decreased to 78% at 20 epochs. Both Convolution2D and VGG16 models also show a drop in accuracy with less no. of epochs. With Convolution2D and VGG16 models, the proposed method achieved the highest accuracy of 87 and 93% at 100 epochs, respectively, whereas, at 20 epochs, Conv2D and VGG16 models produced an accuracy of 69.1 and 82%, respectively. From the results shown in Figure 8A, the ResNet50 model produced a high accuracy of 97.72% compared to the other three models at 100 epochs, even though it started up with 84.69% accuracy with 20 epochs.
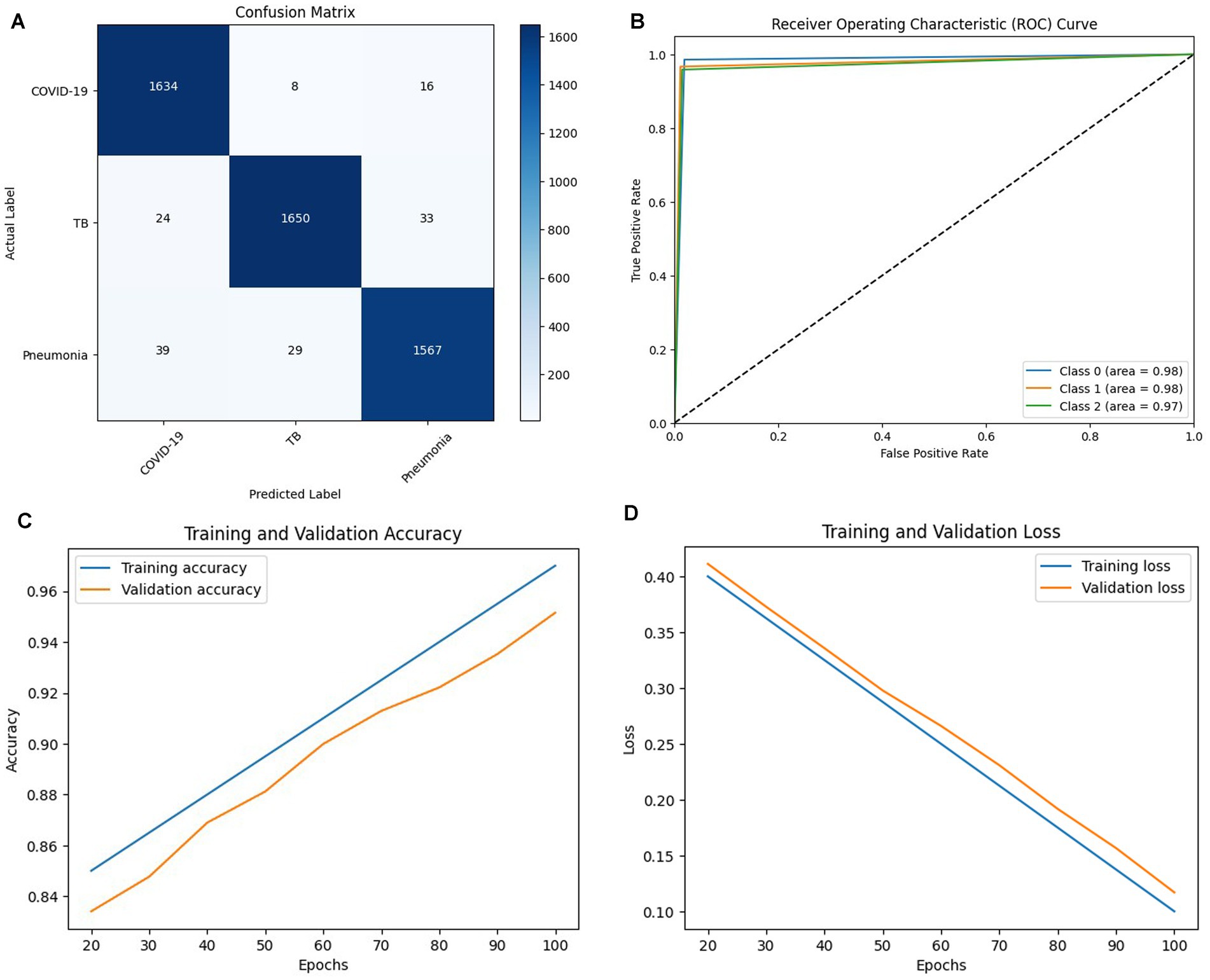
Figure 7. (A) Confusion matrix, (B) ROC curve, (C) Epochs vs. Accuracy graph (D) Epochs vs. Loss graph of RESNET 50 Model.
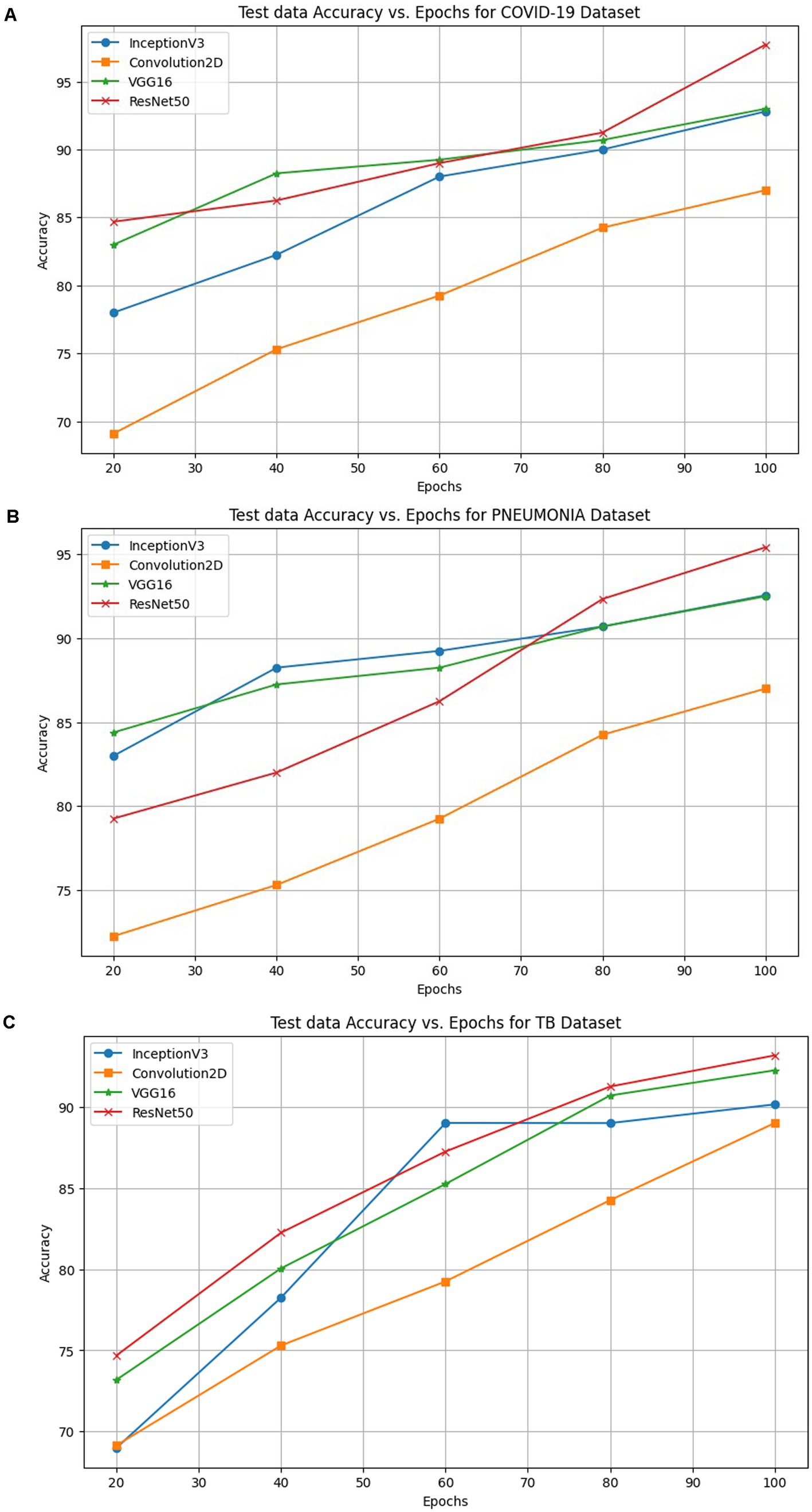
Figure 8. Accuracy of INCEPTIONV3, Conv2D, VGG16 and ResNet50 models on the proposed FLEM framework for (A) COVID-19, (B) Pneumonia and (C) TB classification.
The performance comparison in terms of accuracy computed with varying epochs is shown in Figure 8B for the pneumonia dataset. ResNet50 achieved a higher accuracy on the proposed FLEM framework than the other three models tested on the proposed framework. The model’s test accuracy increases as the no. of epochs increases. InceptionV3, Conv2D, and VGG16 showed an accuracy of 92.56, 87, and 92% at 100 epochs, and the accuracy significantly decreased to 83, 72.25, and 84% at 20 epochs, respectively.
Figure 8C shows the accuracy percentage comparison of the employed models on the proposed FLEM framework for the Tuberculosis dataset for the different epochs from 20 to 100. The inceptionV3 model showed a significant rise in accuracy from 69 to 90.15% for epochs 20 and 100. The accuracy increases with fewer epochs. Results revealed the progressive accuracy increase for increasing epochs, and the Conv2D model produced 89% accuracy at 100 epochs. VGG16 showed gradual performance improvement from an accuracy value of 73 to 92.25%. As the graphs show, the ResNet50 model outperformed the other three models with the highest accuracy of 94%.
Table 3 shows the comparative measures in terms of training time, convergence time, and bandwidth consumption between the proposed FLEM framework and the CSLM framework. The description of the performance measures is given in Section. The performance records of the FLEM and CSLM framework vary depending upon several factors, such as the total size of the dataset, the number of HLS, the computation power and storage capacity of the machines and the network speed.
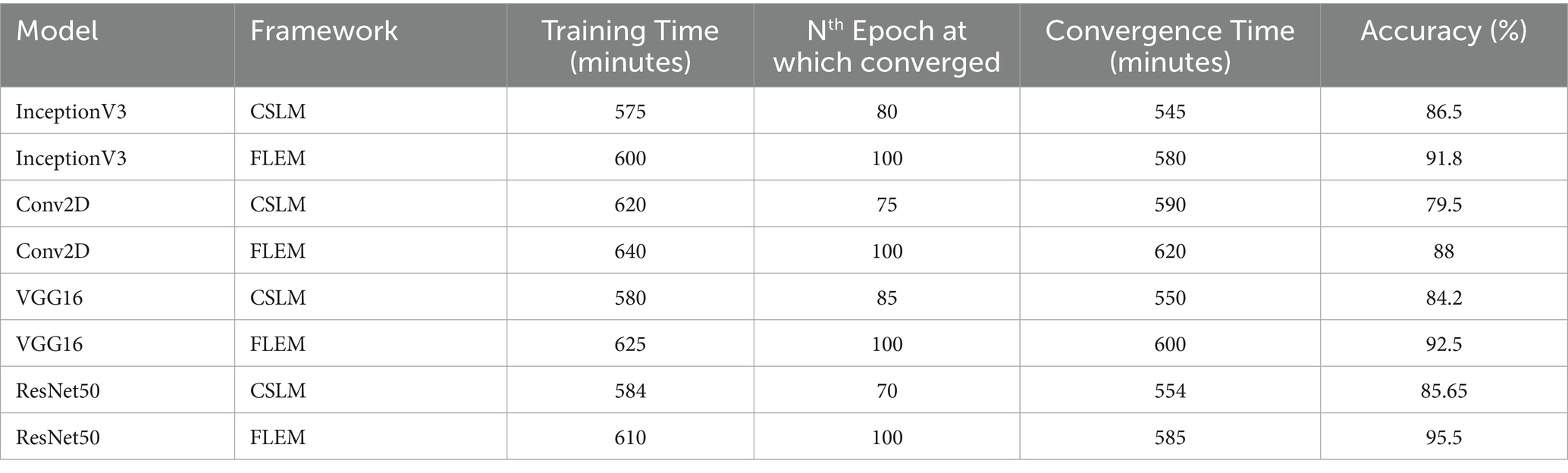
Table 3. comparative measures in terms of training time, convergence time, and bandwidth consumption between the proposed FLEM framework and the CSLM framework.
The total training time is measured by recording the total experiment time and it is measured using Equation 5 and 6. Each HLS connected with the configured Central server with an uplink speed of 16 Mbps and downlink speed of 26.5Mbps. The investigations were carried out with 100 epochs. InceptionV3, Conv2D, VGG16 and ResNet50 models were deployed and tested with variety of dataset on both CSLM and FLEM framework. With FLEM framework, parallel processing at HLS can save the training time with an assumption of stable network conditions. The training time is considerably high due to the initial loading time of the dataset and huge processing at central server.
In the FLEM framework, the overall convergence took longer than the convergence time at CSLM due to the distributed nature and potential variability in local training performance. The convergence time for CSLM and FLEM is measured using Equation 7 and 8 respectively. It often requires multiple communication rounds to reach convergence. A shorter convergence time resulted in a CSLM environment as training is centralized and consistent on an average of 22.5% higher convergence time in the FLEM environment than in the CSLM environment. Though the CSLM system converges more quickly with a certain level of accuracy than FLEM, the obtained accuracy is significantly less in the CSLM environment. The table results showed that though the CSLM system converged with a considerable number of epochs, it resulted in a 5, 8, 9, and 10% accuracy reduction when compared to the FLEM-based training of InceptionV3, Conv2D, VGG16, and ResNet50, respectively. Thus, the FLEM framework ensured better accuracy than the central server-based learning model.
The bandwidth consumption of the CSLM framework was computed using Equation 9. We consider 5,000 chest X-ray images, each approximately 95 KB. By substituting the value BW = 2 × 5,000 × 95,000 = 950,000,000 bytes = 950 MB per sec. The bandwidth consumption at the FLEM environment was computed using Equation 10. We assume the average dataset size at each HLS is 180 MB, and the number of rounds is 3. The approximate size of the model update for the three employed models includes 3*9 MB = 27 MB. The total bandwidth consumption of the FLEM model is 2*3*27 = 162 MB per sec. FLEM framework consumed less bandwidth by sharing only the model update parameters rather than sharing the massive volume of raw data. Federated learning effectively minimizes the bandwidth and storage needs for data transmission. However, frequent communication rounds can increase bandwidth usage. CSLM resulted in higher bandwidth consumption due to the transfer of large volumes of raw data to the central server.
5.5 XAI-FLEM test bed setup and results
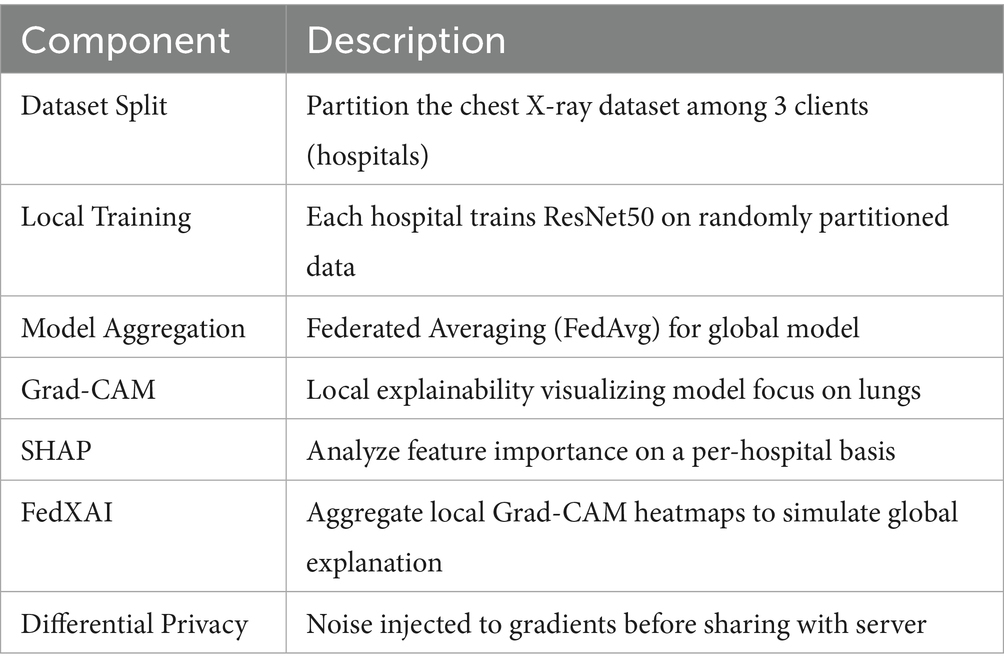
From the observations on accuracy and related performance measures such as Bandwidth consumption, training time, and convergence time explained in Section 5.4, the FL framework with the ResNet50 model outperformed the other learning models. Hence we created a test bed that simulates the XAI-FLEM using ResNet50 for lung disease diagnosis, along with Explainability using Grad-CAM, SHAP, FedXAI, and Differential Privacy. This simulation involves: 3 virtual hospitals (participating client nodes), Shared ResNet50 model structure, Simulated local data partitions, Model training on local data, Explainability per node (Grad-CAM, SHAP), Federated Aggregation (FedAvg), FedXAI (aggregated Grad-CAM) and Differential Privacy (Gaussian noise to gradients) (Alkhanbouli et al., 2025; Mohale and Obagbuwa, 2025). Table 4 shows the description of the Test bed components.
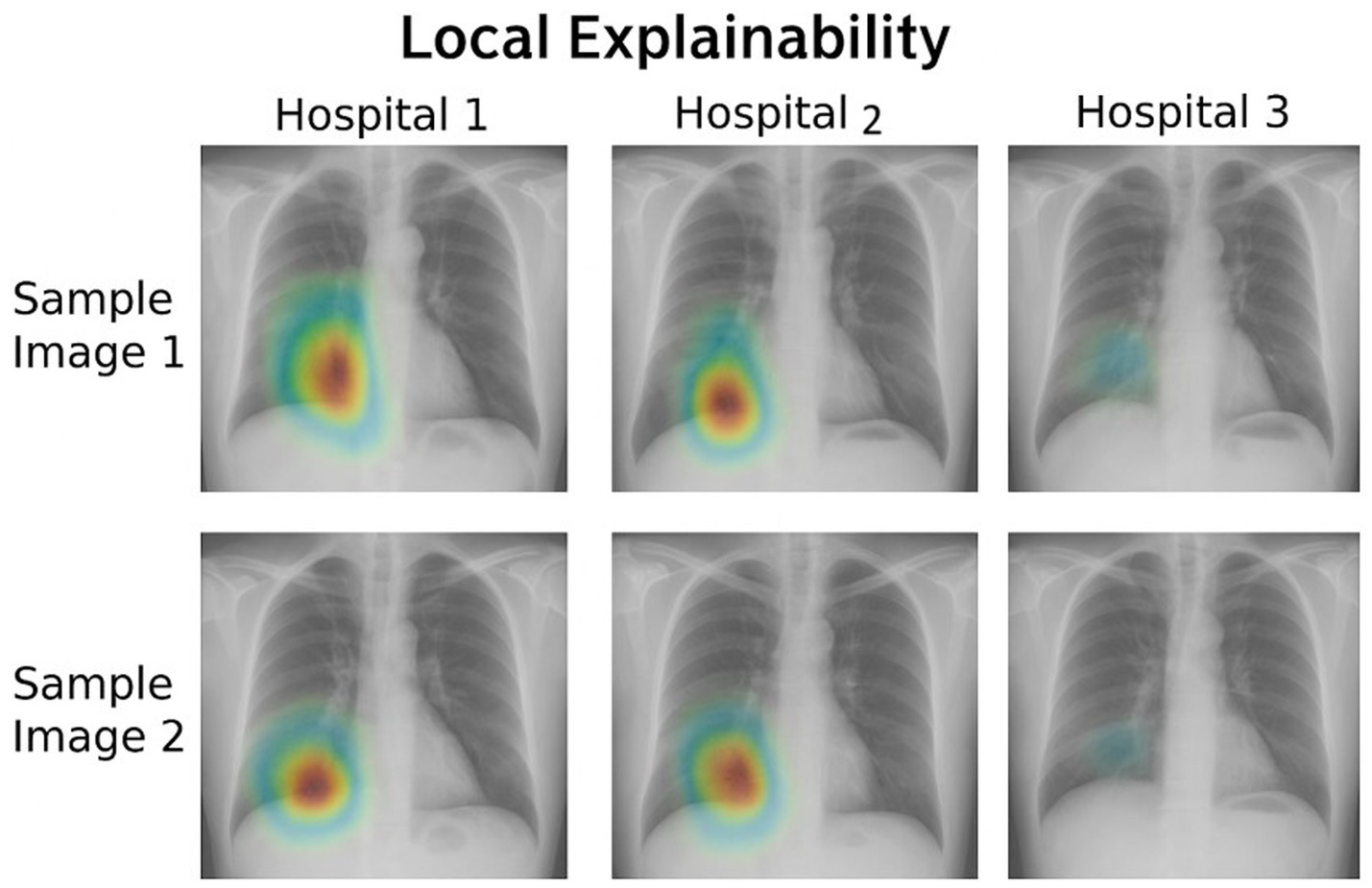
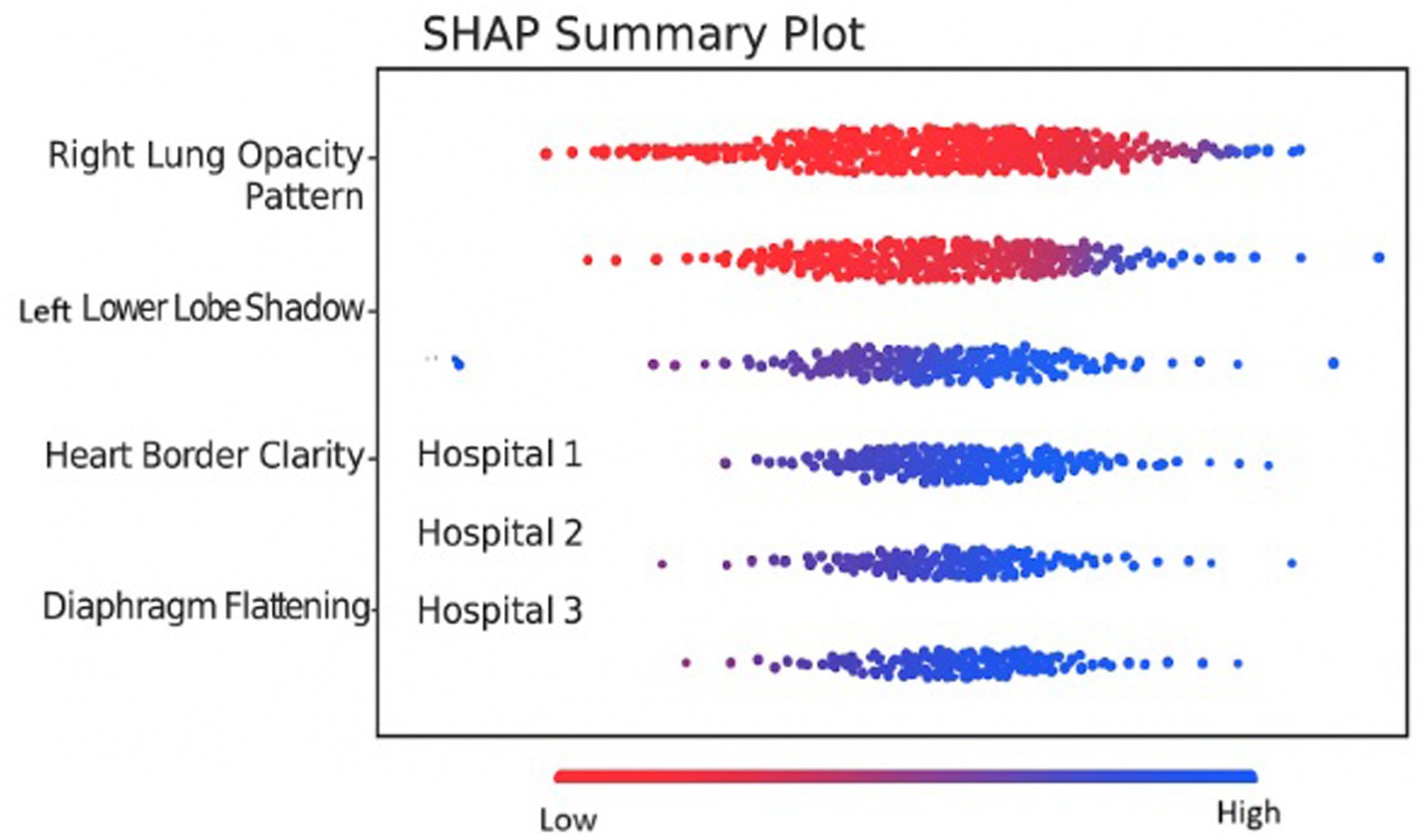
Figure 9 shows the heat maps for each hospital’s model using Grad-CAM. Each column represents participating hospital client nodes (1, 2, 3). The rows show two different input X-ray images. Grad-CAM highlights important regions for prediction in red and yellow. From the figure, we can observe the following variability: Hospital 1 focuses more on both lungs, Hospital 2 emphasizes the central region, and Hospital 3 shows no visible heat map, indicating either low confidence or no activation due to local data variance. Figure 10 illustrates the distributions of the feature importance across hospitals using SHAP (SHapley Additive exPlanations). The X-axis shows the impact of each feature on model output. The Y-axis represents the impact on each features such as pixel groups and lung texture features. Each dot on the graph corresponds to a prediction sample. The coloring indicates feature values, with red representing high values and blue representing low values. The repeated triplets correspond to Hospitals 1, 2, and 3. Hospitals 1 and 2 exhibit a strong focus on Feature 1 and Feature 2, whereas Hospital 3 displays greater variation or noise, which may be attributed to differing data distributions or training instability.
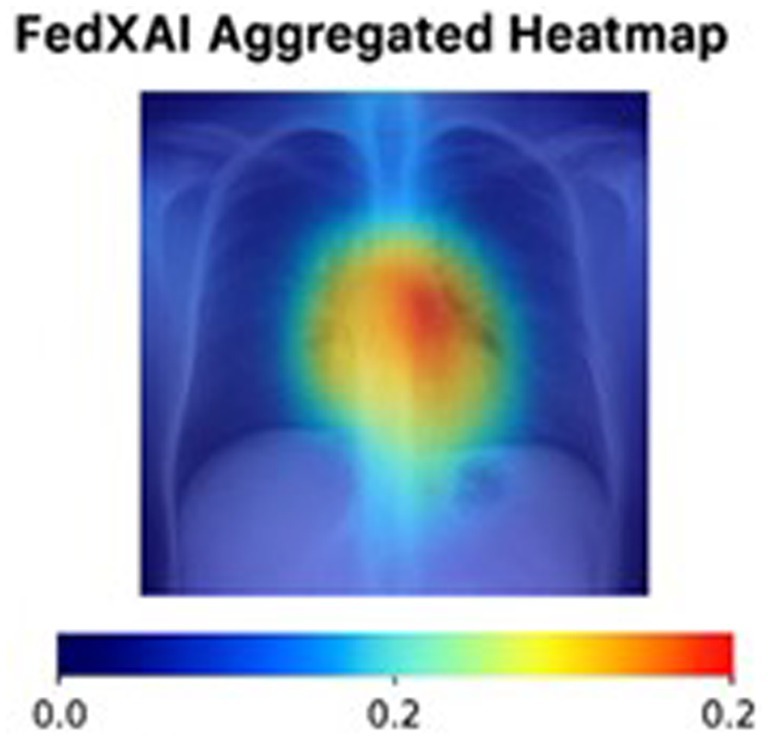
Figure 11 shows the Grad-CAM heatmap generated through FedXAI (federated explainability) which was obtained by averaging the from Grad-CAM outputs from hospital clients. From the figure, it is evident that the Central lung region exhibits the highest relevance. The primary purpose of this demonstration is to highlight the privacy-preserving aggregation of interpretability information, enabling the insights without sharing raw models or data.
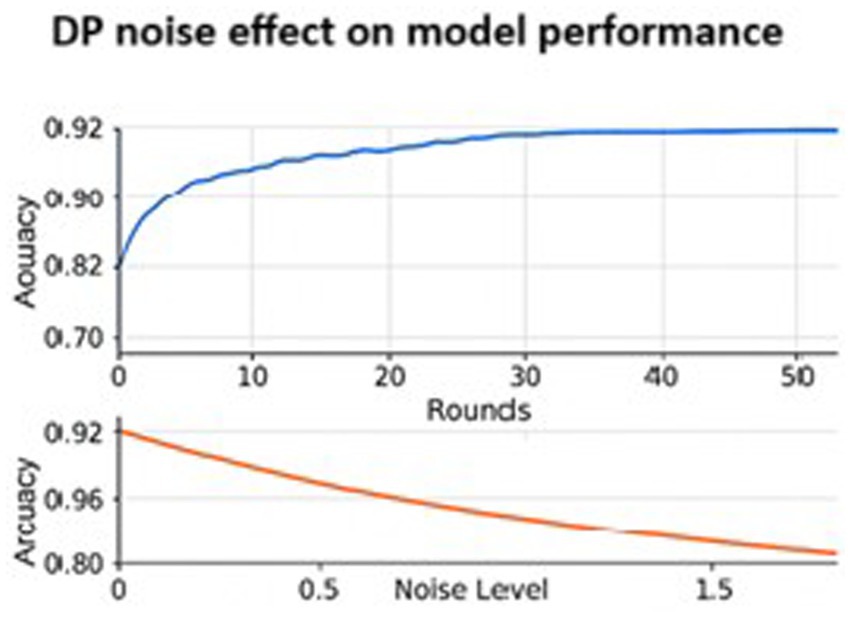
Figure 12 consists of two graphs: the top graph displays Accuracy vs. Rounds and the Bottom graph compares Accuracy vs. DP(Differential Privacy) Noise Level. In the first graph, the blue line illustrates the model performance over training rounds. Accuracy starts at approximately 82%, and plateaus around 91%, indicating that FL with ResNet50 converges fairly well. The second graph has the X-axis representing the noise level added to gradients for DP, and the Y-axis shows the final model accuracy. The orange curve shows accuracy drops as noise increases. The graph emphasizes the trade-off between privacy and performance where increased privacy results in lower accuracy.
6 Conclusion and future work
This research investigates employing the federated learning-XAI based framework for efficient diagnosis of lung diseases. The trained model is deployed in hospitals for real-time privacy-preserving diagnosis. FLEM-XAI uses techniques such as SHAP, Grad-CAM, and FedXAI to promote transparent decision-making. This framework implements Federated Differential Privacy (FedDP) to ensure data security. The FLEM-XAI framework combines Federated Learning (FL) with explainable AI (XAI) techniques such as SHAP, Grad-CAM, and Differential Privacy. This enhances the interpretability of lung disease classification models while ensuring data privacy in medical settings. The feasibility analysis of using the FLEM-XAI model as a replacement for the traditional central server-based learning model has been conducted with COVID-19, tuberculosis, and pneumonia datasets. The performance analysis was carried out using InceptionV3, Conv2D, VGG16, and ResNet50 focusing on metrics such as Precision, Recall, F1 Score, and Accuracy. The FLEM framework was implemented with a simulation. Also, the performance of the proposed FLEM framework was justified by computing the training time, convergence time and the total bandwidth consumption. The testing data accuracy of CNN models such as InceptionV3, Convolution2D, VGG16, and ResNet50 for COVID-19, Pneumonia and Tuberculosis datasets with an increasing number of epochs analysed. Results show that the model resulted in better accuracy with the FLEM environment. The ResNet50 model produced a high average accuracy of 95.5% compared to the other three models at 100 epochs. Also, it is evident from the results that the FLEM framework consumed less bandwidth by sharing only the model update parameters rather than sharing the massive volume of raw data.
Exploring model compression techniques can significantly reduce the bandwidth needed during training. Additionally, examining diverse datasets with different characteristics yields more robust results. Since FLEM involves combining multiple models, improving interpretability is essential. Future work focuses on developing techniques that offer insights into how ensemble decisions are made, thereby fostering user trust in federated environments. In the future, we aim to apply the FLEM-XAI to more real-world applications by utilizing fully homomorphic encryption to safeguard against adversarial attacks within the proposed framework.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: Kaggle, https://www.kaggle.com/datasets/jtiptj/chest-xray-pneumoniacovid19tuberculosis.
Author contributions
SD: Formal analysis, Software, Writing – original draft, Investigation, Data curation, Methodology, Resources, Validation, Writing – review & editing, Project administration, Conceptualization, Supervision. ED: Validation, Software, Resources, Data curation, Writing – review & editing, Investigation. SS: Writing – review & editing, Data curation, Software, Resources. VR: Data curation, Software, Writing – review & editing. VS: Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, A., Abdelsamea, M. M., and Gaber, M. M. (2021). Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 51, 854–864. doi: 10.1007/s10489-020-01829-7
Aggarwal, P., Mishra, N., Fatimah, B., Singh, P., Gupta, A., and Joshi, S. (2022). COVID-19 image classification using deep learning: advances, challenges and opportunities. Comput. Biol. Med. 144:105350. doi: 10.1016/j.compbiomed.2022.105350
Akinwamide, S., Idris-Tajudeen, R., and Akin-Olayemi, T. H. (2024). Prediction of post-covid-19 using supervised machine learning techniques. World J. Adv. Eng. Technol. Sci. 12, 355–369. doi: 10.30574/wjaets.2024.12.2.0297
Alkhanbouli, R., Matar Abdulla Almadhaani, H., Alhosani, F., Simsekler, M. C. E., et al. (2025). The role of explainable artificial intelligence in disease prediction: a systematic literature review and future research directions. BMC Med. Inform. Decis. Mak. 25:110. doi: 10.1186/s12911-025-02944-6
Al-qaness, M. A., Zhu, J., AL-Alimi, D., Dahou, A., Alsamhi, S. H., Abd Elaziz, M., et al. (2024). Chest x-ray images for lung disease detection using deep learning techniques: a comprehensive survey. Arch. Comp. Methods Eng. 31, 3267–3301. doi: 10.1007/s11831-024-10081-y
Apostolopoulos, I. D., Aznaouridis, S. I., and Tzani, M. A. (2020). Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. J. Med. Biol. Eng. 40, 462–469. doi: 10.1007/s40846-020-00529-4
Arun Prakash, J., Asswin, C. R., Ravi, V., Sowmya, V., and Soman, K. P.. (2023). Pediatric pneumonia diagnosis using stacked ensemble learning on multi-model deep CNN architectures. Multimed. Tools Appl. 82, 21311–21351. doi: 10.1007/s11042-022-13844-6
Ashwini, S., Arunkumar, J. R., Prabu, R. T., Singh, N. H., Singh, N. P., et al. (2024). Diagnosis and multi-classification of lung diseases in CXR images using optimized deep convolutional neural network. Soft. Comput. 28, 6219–6233. doi: 10.1007/s00500-023-09480-3
Asswin, C. R., Prakash, J., Dharshan, K., Dora, A., Sowmya, V., Almeshari, M., et al. (2023). Weighted average ensemble approach for pediatric pneumonia diagnosis using channel attention deep CNN architectures. Int. Conf. Mining Intellig. Knowledge Explorat., 13924:250–260. doi: 10.1007/978-3-031-44084-7_24
Bougourzi, F., Contino, R., Distante, C., and Taleb-Ahmed, A. (2021). “CNR-IEMN: a deep learning based approach to recognise COVID-19 from CT-scan,” in ICASSP 2021–2021 IEEE international conference on acoustics, speech and signal processing (ICASSP), pp. 8568–8572.
Chaudhary, S., Sadbhawna, S., Jakhetiya, V., Subudhi, B. N., Baid, U., and Guntuku, S. C. (2021). “Detecting covid-19 and community acquired pneumonia using chest ct scan images with deep learning.” in ICASSP 2021–2021 IEEE international conference on acoustics, speech and signal processing (ICASSP), pp. 8583–8587.
Chowdhury, M. E., Rahman, T., Khandakar, A., Mazhar, R., Kadir, M. A., Mahbub, Z. B., et al. (2020). Can AI help in screening viral and COVID-19 pneumonia? IEEE Access 8, 132665–132676. doi: 10.1109/ACCESS.2020.3010287
Dayan, I., Roth, H. R., Zhong, A., Harouni, A., Gentili, A., Abidin, A. Z., et al. (2021). Federated learning for predicting clinical outcomes in patients with COVID-19. Nat. Med. 27, 1735–1743. doi: 10.1038/s41591-021-01506-3
de Moura, J., Novo, J., and Ortega, M. (2022). Fully automatic deep convolutional approaches for the analysis of COVID-19 using chest X-ray images. Appl. Soft Comput. 115:108190. doi: 10.1016/j.asoc.2021.108190
Deshmukh, M. R., Dhavale, U. S., Dhone, A. B., Hegde, P. V., and Mahalakshmi, B. (2021) “Detection and classification of COVID-19 and other lung diseases from X-ray dataset using deep learning.” in 2021 Asian conference on innovation in technology (ASIANCON), pp. 1–5.
Durga, S., Deepakanmani, S., and Daniel, E. (2025). 2D convolution neural network-based efficient model for brain tumor detection using 5G edge cloud.
Durga, S., Daniel, E., Andrew, J., and Bhat, R. (2024). SmartCardio: advancing cardiac risk prediction through internet of things and edge cloud intelligence. IET Wireless Sensor Syst. 14, 348–362. doi: 10.1049/wss2.12085
Feki, I., Ammar, S., Kessentini, Y., and Muhammad, K. (2021). Federated learning for COVID-19 screening from chest X-ray images. Appl. Soft Comput. 106:107330. doi: 10.1016/j.asoc.2021.107330
Ganeshkumar, M., Ravi, V., Sowmya, V., Gopalakrishnan, E. A., Soman, K. P., and Rupeshkumar, M. (2023). Two-stage deep learning model for automate detection and classification of lung diseases. Soft. Comput. 27, 15563–15579. doi: 10.1007/s00500-023-09167-9
Hu, S., Gao, Y., Niu, Z., Jiang, Y., Li, L., Xiao, X., et al. (2020). Weakly supervised deep learning for covid-19 infection detection and classification from ct images. IEEE Access 8, 118869–118883. doi: 10.1109/ACCESS.2020.3005510
Jasmine Pemeena Priyadarsini, M., Kotecha, K., Rajini, G. K., Hariharan, K., Utkarsh Raj, K., Bhargav Ram, K., et al. (2023). Lung diseases detection using various deep learning algorithms. J. Healthcare Eng. 2023:3563696. doi: 10.1155/2023/3563696
Jin, I. J., Lim, D. Y., and Bang, I. C. (2023). Deep-learning-based system-scale diagnosis of a nuclear power plant with multiple infrared cameras. J. Theor. Appl. Inf. Technol. 100, 493–505. doi: 10.1016/j.compag.2020.105393
Kaggle. (2021). JTIPTJ. Available online at: https://www.kaggle.com/datasets/jtiptj/chest-xray-pneumoniacovid19tuberculosis [Accessed March 1, 2024].
Li, C., Zhang, D., Du, S., and Tian, Z. (2020). Deformation and refined features based lesion detection on chest X-ray. IEEE Access 8, 14675–14689. doi: 10.1109/ACCESS.2020.2963926
Mahbub, M. K., Biswas, M., Gaur, L., Alenezi, F., and Santosh, K. C. (2022). Deep features to detect pulmonary abnormalities in chest X-rays due to infectious diseaseX: Covid-19, pneumonia, and tuberculosis. Inf. Sci. 592, 389–401. doi: 10.1016/j.ins.2022.01.062
Mohale, V. Z., and Obagbuwa, I. C. (2025). A systematic review on the integration of explainable artificial intelligence in intrusion detection systems to enhancing transparency and interpretability in cybersecurity. Front. Artif. Intellig. 8:1526221. doi: 10.3389/frai.2025.1526221
Nasiri, H., and Hasani, S. (2022). Automated detection of COVID-19 cases from chest X-ray images using deep neural network and XGBoost. Radiography 28, 732–738. doi: 10.1016/j.radi.2022.03.011
Ouyang, X., Huo, J., Xia, L., Shan, F., Liu, J., Mo, Z., et al. (2020). Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging 39, 2595–2605. doi: 10.1109/TMI.2020.2995508
Phogat, D., Parasu, D., Prakash, A., and Sowmya, V. (2023). “Selective kernel networks for lung abnormality diagnosis using chest X-rays,” in International Conference on Information, Communication and Computing Technology. pp. 937–950.
Sabry, A. H., Bashi, O. I. D., Ali, N. N., and Al Kubaisi, Y. M. (2024). Lung disease recognition methods using audio-based analysis with machine learning. Heliyon 10:e26218. doi: 10.1016/j.heliyon.2024.e26218
Serte, S., and Demirel, H. (2021). Deep learning for diagnosis of COVID-19 using 3D CT scans. Comput. Biol. Med. 132:104306. doi: 10.1016/j.compbiomed.2021.104306
Keywords: central server based learning model, federated learning, ensemble model, explainable AI, SHapley Additive exPlanations, grad-CAM, DP, lung diseases
Citation: Durga S, Daniel E, Seetha S, Reshma VK and Sachnev V (2025) FLEM-XAI: Federated learning based real time ensemble model with explainable AI framework for an efficient diagnosis of lung diseases. Front. Comput. Sci. 7:1633916. doi: 10.3389/fcomp.2025.1633916
Edited by:
Kannimuthu Subramanian, Karpagam College of Engineering, IndiaReviewed by:
Mohan Sellappa Gounder, Nitte Meenakshi Institute of Technology, IndiaBright Gee Varghese, Maharishi University of Management, United States
Copyright © 2025 Durga, Daniel, Seetha, Reshma and Sachnev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sivan Durga, c19kdXJnYUBjYi5hbXJpdGEuZWR1
†ORCID: Sivan Durga, orcid.org/0000-0003-2376-8365
Esther Daniel, orcid.org/0000-0003-1997-094X
Surleese Seetha, orcid.org/0000-0001-5136-6991
Vijaya Kumar Reshma, orcid.org/0000-0002-0273-0790
Vasily Sachnev, orcid.org/0000-0001-7063-5069
 Sivan Durga
Sivan Durga Esther Daniel
Esther Daniel Surleese Seetha3†
Surleese Seetha3†