- School of Life Sciences and Centre for Cell and Developmental Biology, The Chinese University of Hong Kong, Hong Kong, China
As an essential hormone regulating gonads in vertebrates, the biosynthesis and secretion of follicle-stimulating hormone (FSH) is controlled by a variety of endocrine and paracrine factors in both mammalian and non-mammalian vertebrates. Activin was initially discovered in the ovary for its specific stimulation of FSH secretion by the pituitary cells. Our earlier studies in fish have shown that activin stimulates FSHβ but suppresses LHβ expression in both the goldfish and zebrafish. Further experiments showed that the regulation of FSHβ in fish occurred at the promoter level involving Smads, in particular Smad3. To further understand the mechanisms by which activin/Smad regulates FSHβ transcription, the present study was undertaken to analyze the promoter of goldfish FSHβ gene (fshb) with the aim to identify potential cis-regulatory elements responsible for activin/Smad stimulation. Both serial deletion and site-directed mutagenesis were used, and the promoter activity was tested in the LβT-2 cells, a murine gonadotroph cell line. The reporter constructs of goldfish FSHβ promoter-SEAP (secreted alkaline phosphatase) were co-transfected with an expression plasmid for Smads (2 or 3) followed by measurement of SEAP activity in the medium. Two putative Smad responsive elements were identified in the promoter at distal and proximal regions, respectively. The distal site contained a consensus Smad binding element (AGAC, −1675/−1672) whereas the proximal site (GACCTTGA, −212/−205) was identical to an SF-1 binding site reported in humans, which was preceded by a sequence (AACACTGA) highly conserved between fish and mammals. The proximal site also seemed to be involved in mediating stimulation of FSHβ expression by gonadotropin-releasing hormone and its potential interaction with activin. In conclusion, we have identified two potential cis-regulatory elements in the promoter of goldfish FSHβ that are responsible for activin-induced expression of the gene. Since activin stimulation of FSHβ expression is functionally conserved in fish and mammals, our findings contribute to the understanding of the fundamental mechanisms of this regulation across vertebrates.
Introduction
Follicle-stimulating hormone (FSH) is an essential regulator of gonadal development and function in vertebrates including teleosts. Like other pituitary glycoprotein hormones, FSH is a heterodimeric protein consisting of a hormone-specific β subunit (Fshb, fshb) and a common α subunit (Cga, cga) shared with other pituitary glycoprotein hormones (Pierce and Parsons, 1981). The biosynthesis of FSH in the pituitary is primarily determined by the expression of its β subunit (Shupnik, 1996), which is subject to regulation by a variety of neuroendocrine, endocrine, and paracrine factors including gonadotropin-releasing hormone (GnRH) from the hypothalamus (Ferris and Shupnik, 2006), sex steroids from the gonads (Glidewell-Kenney et al., 2008), and local factors in the pituitary, particularly activin and its binding protein follistatin (Kawakami et al., 2002; Bilezikjian et al., 2004). There are lines of evidence that the effects of GnRH and steroids on FSH biosynthesis may involve interactions with the activin signaling in the pituitary (Melamed, 2010).
Activin is also a dimeric protein (βAβA, βBβB, and βAβB), belonging to the transforming growth factor β (TGF-β) superfamily both structurally and functionally. Activin was first identified in mammalian ovaries as a potent stimulator of pituitary FSH secretion (Ling et al., 1986a; Vale et al., 1986), antagonizing the effect of inhibin. Subsequent studies have provided evidence that activin is also produced locally in the pituitary together with its neutralizing binding protein follistatin in both mammals and fish (Meunier et al., 1988; Roberts et al., 1989; Lau and Ge, 2005; Lin and Ge, 2009) and the pituitary-derived activin plays an important role in controlling FSH biosynthesis and secretion in a paracrine manner (Bilezikjian et al., 2004; Gregory and Kaiser, 2004). In the rat, both in vitro and in vivo studies have demonstrated potent stimulatory effects of activin on FSHβ expression and FSH secretion in the pituitary (Carroll et al., 1989; Woodruff et al., 1993; Lee and Rivier, 1997). In teleosts, activin also stimulates fshb mRNA expression in the goldfish (Yam et al., 1999), tilapia (Yaron et al., 2001), zebrafish (Lin and Ge, 2009), and European eel (Aroua et al., 2011). These data strongly suggest that as a crucial function of activin in reproduction, its stimulation of FSH biosynthesis is well-conserved across vertebrates, indicating the importance of such regulation in vertebrate reproduction.
The regulation of FSHβ mRNA expression by GnRH has been well documented and the effect appears to depend on the frequency of GnRH pulses, with high frequency suppressing its expression and low frequency favoring the expression (Dalkin et al., 1989; Jayes et al., 1997). Interestingly, it has been reported that the regulation of FSH biosynthesis by GnRH involves the activin–follistatin system produced locally in the pituitary. GnRH stimulates FSHβ mRNA expression in a rat pituitary perfusion system and its stimulatory effect is blocked by follistatin (Besecke et al., 1996). Similar result has also been reported in the murine gonadotroph cell line LβT-2 cells (Pernasetti et al., 2001). These findings suggest that the GnRH-regulated FSHβ expression is, at least partially, activin-mediated.
Like other TGF-β superfamily members, activin transduces its signals by interacting with its type I and type II serine/threonine kinase receptors followed by activation of intracellular signaling proteins, the Smads. Activin first binds to its type II receptors, which then recruits a type I receptor to form a type I–type II receptor complex (Mathews, 1994). The activin-receptor complex in turn phosphorylates receptor-regulated Smads (R-Smad), Smad2, or Smad3. The R-Smads then form heteromeric complexes with a common partner Smad4 (Co-Smad), which translocate into the nucleus where they function as transcriptional factors to control the expression of target genes. In addition to these signal transducing Smads, an inhibitory Smad (I-Smad), Smad7, plays an antagonistic role by blocking the activation of these R-Smads (Massague et al., 2005; Ross and Hill, 2008).
Smads exert their transcriptional regulatory activities by interacting with specific DNA elements together with other specific transcription factors (Shi and Massague, 2003). Smad3 and Smad4, but not Smad2, have the ability to bind to the specific DNA sequence (AGAC or GTCT) called Smad binding element (SBE; Dennler et al., 1998; Shi et al., 1998; Zawel et al., 1998; Chai et al., 2003). Smads also interact with various transcription factors to elicit specific gene expression in different cell types, including activating protein-1 (AP-1) transcriptional complex (Jun/Fos; Wang et al., 2008).
Studies using the LβT-2 cells showed that activin stimulated endogenous FSHβ expression in this gonadotroph cell line (Graham et al., 1999) and the stimulation was mediated via both Smad2 and Smad3 (Bernard, 2004). In addition, activin also increased transcriptional activity of mouse, rat, and ovine FSHβ promoters in the LβT-2 cells and the effect was also mediated via Smads (Dupont et al., 2003; Suszko et al., 2003; Lamba et al., 2006). Further experiments showed that it was Smad3 that primarily mediated activin stimulated rat FSHβ promoter activity in the LβT-2 cells (Gregory et al., 2005; Suszko et al., 2005). Promoter analysis revealed an inverted palindrome sequence (GTCTAGAC; −266/−259) in the proximal region (−284/−252) of rat FSHβ promoter (Suszko et al., 2003; Gregory et al., 2005), and the same sequence was also confirmed to bind Smad2/3/4 complex in the promoter of mouse FSHβ gene (−266/−259; Lamba et al., 2006).
Compared to mammalian models, our knowledge on gonadotropin regulation especially FSH expression and secretion in other vertebrates is rather limited. There have been several studies on FSHβ gene and its promoter region in teleosts, providing some clues to the potential mechanisms underlying its regulation at the molecular level (Chong et al., 2004). The genomic structure of FSHβ was first characterized in the goldfish whose genome contains two copies of the gene (gfGTHIβ-1, fshb1 and gfGTHIβ-2, fshb2). Sequence analysis of their promoter regions revealed multiple putative regulatory elements such as steroid responsive elements (estrogen responsive element, ERE; androgen responsive element, ARE), GnRH responsive element (GnRH-RE), and gonadotropin specific element/SF-1 binding element (GSE/SF-1; Sohn et al., 1998). However, none of these potential cis-regulatory elements has been functionally tested. Results from our previous study in the goldfish showed that activin stimulated FSHβ (fshb) promoter activity through the Smad signaling pathway, especially Smad3 (Lau and Ge, 2005); however, the underlying mechanisms for Smad signaling remains largely unknown in this species. This study was undertaken to identify and localize the element(s) responsible for Smad-mediated goldfish fshb transcription. Two potential activin or Smad responsive elements (SRE) were localized at the distal and proximal regions of the promoter, respectively. Our evidence also suggested the existence of additional SRE in the promoter region.
Materials and Methods
Chemicals, Enzymes, and Hormones
All chemicals were purchased from Sigma (St. Louis, MO, USA) or Amersham Biosciences (Piscataway, NJ, USA), and enzymes from Promega (Madison, WI, USA) unless otherwise stated. Recombinant goldfish activin B was generated in our laboratory by an established Chinese hamster ovary (CHO) cell line and partially purified from the medium according to Schmelzer et al. (1990). One unit (U) of activin B is defined as the amount per milliliter to induce a half-maximal differentiation of F5-5 cells in the erythroid differentiation factor assay (ED50; Eto et al., 1987; Schmelzer et al., 1990), which is equivalent to about 7–8 ng/ml recombinant human activin A in the same assay. Recombinant human activin A was provided by Dr. A. F. Parlow through the National Hormone and Pituitary Program (National Institute of Diabetes and Digestive and Kidney Diseases, Torrance, CA, USA). Salmon GnRH (sGnRH) was purchased from Sigma. All PCR primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA, USA).
Construction of Smad Expression Plasmids and SEAP Reporter Plasmids
The expression constructs of Smad2, Smad3, and Smad7 for overexpression have been described in our previous study (Lau and Ge, 2005). The reporter plasmid pSEAP/gfFSHβ(−1744) containing goldfish fshb promoter and the reporter gene SEAP (secreted alkaline phosphatase) was constructed by inserting the fshb promoter region (1744 bp fragment) into the promoter-less reporter plasmid pSEAP2-Enhancer (Clontech, Palo Alto, CA, USA) at the EcoRI and XhoI sites as described previously (Lau and Ge, 2005). As mentioned above, goldfish has two FSHβ genes (fshb1 and fshb2) in the genome (Sohn et al., 1998). In the present study we chose to focus on the promoter of fshb1 because this form is better characterized and its expression seemed to vary more obviously during sexual maturation (Yoshiura et al., 1997; Sohn et al., 1998). The reporter plasmids containing different lengths of fshb promoter were generated by PCR using pSEAP/gfFSHβ(−1744) as the template. All sense primers and antisense primers were designed with XhoI and EcoRI restriction sites added, respectively. PCR-based site-directed mutagenesis was performed to introduce base changes at potential cis-regulatory elements in the promoter fragments by using mutagenic primers. The promoter fragments of different lengths were amplified by PCR on the Thermal Cycler 9600 (Eppendorf, Hamburg, Germany) in 1× Pfu buffer, 0.2 mM dNTPs, 0.2 μM each primer, and 3 U Pfu DNA polymerase in a final volume of 50 μl. All amplified products were digested with XhoI and EcoRI restriction enzymes followed by subcloning into the pSEAP2-Enhancer reporter plasmid at the respective restriction sites.
Cell Culture and Transient Transfection
The mouse gonadotroph cell line, LβT-2 cells, was generously provided by Dr. Pamela Mellon (Department of Reproductive Medicine, University of California, San Diego). The cells were cultured in DMEM medium (Gibco BRL, Gaithersburg, MD, USA) with 10% fetal bovine serum (Hyclone, Logan, UT, USA) at 37°C with 5% CO2. For transient transfection, cells were sub-cultured 24 h before transfection in a 24-well plate and co-transfected with the pSEAP/gfFSHβ reporter plasmids of different promoter lengths, the Smad expression plasmids (Smad2 and Smad3), and the internal control plasmid pSV-β-galactosidase (Promega) using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). pBK-CMV vector (Stratagene, La Jolla, CA, USA) was used to balance the amount of DNA when necessary.
SEAP Reporter Gene Assay
The SEAP activity in the medium of transfected cells was assayed with the Chemiluminescent SEAP Reporter Gene Assay Kit (Roche, Mannheim, Germany) according to the protocol from the manufacturer. Briefly, 100 μl medium was collected from each well and centrifuged to pellet cell debris. Fifty microliters supernatant was then diluted with 150 μl Dilution Buffer in a microtube. After incubation at 65°C for 30 min, the samples were centrifuged for 30 s at room temperature at 14,000 × g before transferring to ice bath. Fifty microliters diluted sample was then transferred to the well of the LumiNunc 96 MicroWell plate (Nalge Nunc International, Rochester, NY, USA) followed by addition of 50 μl Inactivation Buffer. After a 5-min incubation period at room temperature, 50 μl Substrate Reagent (47.5 μl Substrate Buffer + 2.5 μl Alkaline Phosphatase Substrate) was added. The mixture was incubated for 10 min at room temperature with gentle rocking. The light emission was visualized, quantified, and analyzed with the Lumi-Imager F1 (Roche).
β-Galactosidase Reporter Gene Assay
To control transfection efficiency and normalize SEAP assay data, all transfections were performed with pSV-β-galactosidase expression vector (Promega, 500 ng/well) included as the internal control. The activity of pSV-β-galactosidase was assayed with the β-Galactosidase Enzyme Assay System (Promega). Briefly, after medium collection for SEAP assay, the cells were washed twice with ice-cold phosphate-buffered saline (PBS, pH 7.4), and lysed with 100 μl 1× Reporter Lysis Buffer. The lysates were transferred to microtubes, vortexed, and centrifuged at 14,000 × g for 2 min at 4°C. Fifty microliters supernatant of each sample was then transferred to 96-well plate containing 50 μl 2× Assay Buffer (Promega) in each well. After incubation at 37°C for 2 h, 150 μl sodium carbonate (1 M) was added to stop the reaction. The absorbance of the samples was read at 420 nm in the Spectra MAX 250 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Data Analysis
All the experiments were performed at least twice, and all treatments were carried out in triplicate in each experiment. For SEAP assay, the SEAP activity was normalized to the β-galactosidase activity of each sample, and then expressed as the fold change compared to the control. All values are expressed as the mean ± SEM. The data were square-root transformed for normality and variance homogeneity tests. The transformed data were analyzed by one-way ANOVA followed by Newman–Keuls test using GraphPad Prism for Macintosh (GraphPad Software, San Diego, CA, USA). P < 0.05 was considered statistically significant.
Results
Promoter Analysis for Potential Smad Regulatory Element(s) in Goldfish fshb Gene
We previously showed that goldfish fshb promoter strongly responded to activin in the LβT-2 cells (Ge et al., 2003), and co-transfection with Smad expression vectors, especially Smad3, dramatically enhanced the expression level of the reporter, suggesting the presence of Smad regulatory elements in the promoter (Lau and Ge, 2005). To localize the potential regulatory elements, we performed this experiment by examining the activity of goldfish fshb promoter with decreasing size in the LβT-2 cells (a gonadotroph cell line) in the presence of goldfish Smad2 or Smad3. The construct pSEAP/gfFSHβ(−1744) and the promoter-less pSEAP2-Enhancer vector acted as the positive and negative control, respectively. As shown in Figure 1, pSEAP/gfFSHβ(−1744) exhibited the strongest response to goldfish Smad2 and 3. The basal and Smad-stimulated expression of SEAP reporter declined gradually as the length of the promoter decreased. Consistent with the result reported previously (Lau and Ge, 2005), the effect of Smad3 was much higher than that of Smad2 for all sizes of the promoter tested. Although the basal and Smad2/3-induced promoter activity exhibited a general trend of decline with the decreasing size of the promoter, significant drops were noticed at certain locations, including −1744/−1563, −1000/−900, −700/−600, −500/−400 and in particular −300/−200 bp upstream of the potential transcription start site which is designated +1. The predicted transcription start site was defined by 5′-RACE and is located downstream of a TATAA box at −30 (Sohn et al., 1998). The most obvious decrease in activity occurred when the regions −1744/−1563, −700/−600, and −300/−200 were deleted. There is no significant difference between pSEAP/gfFSHβ(−200) and promoter-less pSEAP2-Enhancer vector control (Figure 1).
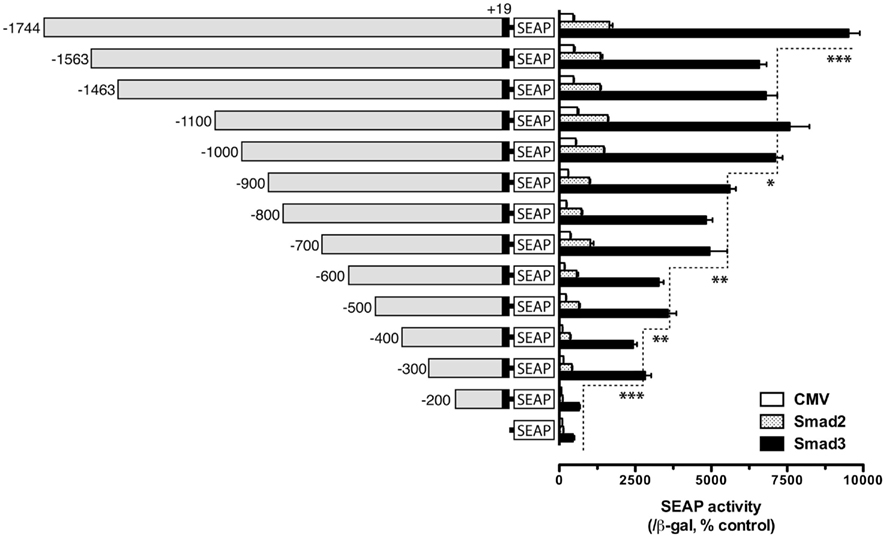
Figure 1. Localization of potential Smad regulatory elements in the goldfish fshb promoter. The reporter constructs of different lengths [pSEAP/gfFSHβ(−200) − (−1744), 300 ng/well] were co-transfected into the LβT-2 cells with the expression plasmids pBK-CMV/Smads (Smad2 or Smad3) or pBK-CMV control (CMV, 300 ng/well) together with 500 ng pSV-β-galactosidase as the internal control. The medium and cells from each well were sampled 48 h after transfection for assaying SEAP and β-galactosidase activities, respectively. The SEAP activity was first normalized by that of β-galactosidase and then expressed as the percentage of the CMV control in the pSEAP2 vector group. Each value represents the mean ± SEM of three replicates (*, **, and *** indicate P < 0.05, 0.01, and 0.001, respectively).
To further define the Smad responsive element(s) in the regions −700/−600 and −300/−200, finer deletions were generated within these regions with 20-bp difference and tested. In these experiments, only Smad3 was used to activate the promoter because of its high potency. The results showed that gradual deletion from −700 to −640 caused no significant change of SEAP activity (−900 was used as the reference); however, the SEAP activity abruptly dropped when the fragment −640/−620 was deleted, suggesting a response element between −620 and −640. Further deletion from −620 to −580 caused no further change of the reporter activity (Figure 2A). For the region −300/−200, 20-bp deletions from −300 to −220 did not affect Smad3-induced promoter activity. However, further deletion from −220 down to −200 nearly abolished the promoter activity (Figure 2B).
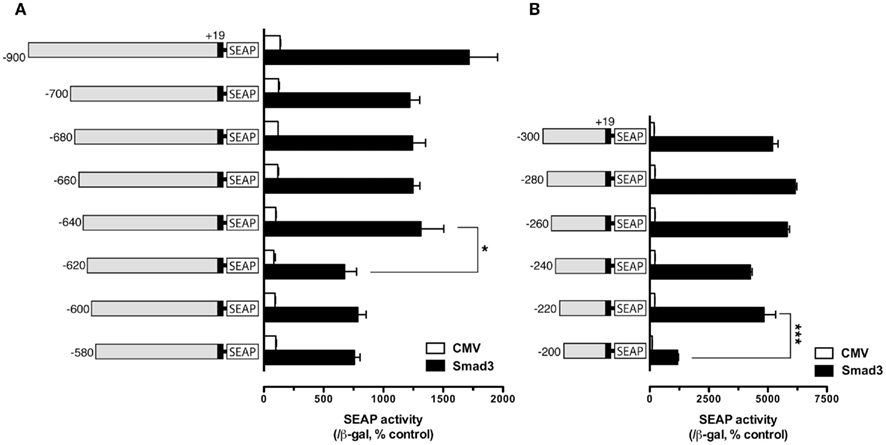
Figure 2. Localization of potential Smad regulatory elements in the −900/−580 region (A) and −300/−200 region (B). The cells in each well were co-transfected with pSEAP/gfFSHβ promoter constructs (300 ng), pBK-CMV/Smad3 (300 ng), and pSV-β-galactosidase (500 ng). The activities of SEAP and β-galactosidase were assayed and analyzed as described in Figure 1. Each value represents the mean ± SEM of three replicates (* and *** indicate P < 0.05 and 0.001, respectively).
Despite its lack of response to Smad3, the proximal region of −200 appeared to be essential for the functionality of the upstream regions. As shown in Figure 3, deletion of the proximal regions completely abolished the activity of the promoter (Figure 3), suggesting that the proximal sequence between −244 and +19 may have essential element(s) for basal transcription activity. Sequence analysis revealed a putative TATA homology element at −30, possibly for initiation of transcription. Therefore, the proximal region of −200/+19 was included in all constructs for characterization or confirmation of putative regulatory regions or elements identified by the deletion experiment described above. Using similar approach, we also analyzed the regions −1000/−900 and −500/−400; however, no distinct sites could be identified over these regions (data not shown).
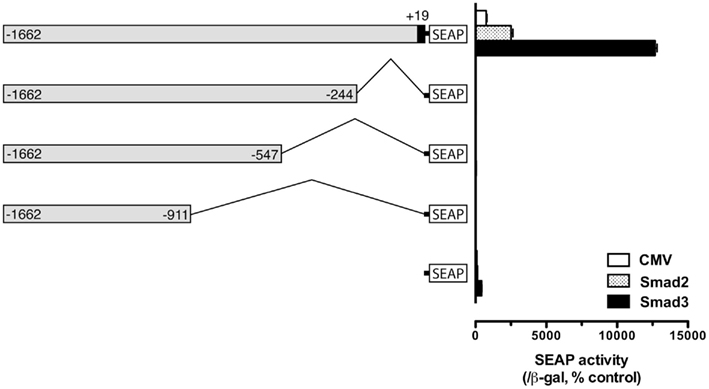
Figure 3. Evidence for an essential element in the proximal region (−244) for fshb promoter activity. The cells in each well were co-transfected with pSEAP/gfFSHβ promoter constructs (300 ng), pBK-CMV/Smad2 or 3 (300 ng), and pSV-β-galactosidase (500 ng). Each value represents the mean ± SEM of three replicates.
Characterization of Smad Response Element(s) in the Distal Region of fshb Promoter
Sequence analysis of the distal region of the goldfish fshb promoter (−1744/−1563) revealed two putative SBEs (AGAC) at −1715/−1712 (site I) and −1675/−1672 bp (site II), respectively (Figure 4A). Interestingly, the two putative SBEs are either near (site I) or overlapping with (site II) an AP-1-like site (site I: 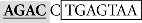 and site II:
and site II: 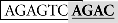 ). To test whether these sites are important for the Smad-mediated fshb expression, we amplified the distal region −1720/−1666 (54 bp) by PCR and placed it upstream of the minimal promoter (−200/+19) in the pSEAP-Enhancer vector, which was then co-transfected with Smad3 into the LβT-2 cells (Figure 4A). The construct pSEAP/gfFSHβ(−1744) and the promoter-less pSEAP2-Enhacer vector were used as the positive and negative control, respectively. The result showed that despite a lower activity than pSEAP/gfFSHβ(−1744), the short 54-bp fragment −1720/−1666 showed a strong response to Smad3, in agreement with the existence of two SBEs (site I and II) in this region. The involvement of these regulatory sites in Smad3-induced promoter activity was confirmed by the evidence that site-directed mutagenesis of these sites resulted in a complete loss of promoter activity. To further demonstrate the role of individual SBE sites, we also mutated each site separately. Interestingly, the removal of SBE and AP-1 motif at site I had no effect on promoter activity whereas the loss of SBE and AP-1 motif at site II completely abolished the promoter activity (Figure 4A), suggesting a role for site II as the functional Smad responsive element in the distal region of fshb promoter.
). To test whether these sites are important for the Smad-mediated fshb expression, we amplified the distal region −1720/−1666 (54 bp) by PCR and placed it upstream of the minimal promoter (−200/+19) in the pSEAP-Enhancer vector, which was then co-transfected with Smad3 into the LβT-2 cells (Figure 4A). The construct pSEAP/gfFSHβ(−1744) and the promoter-less pSEAP2-Enhacer vector were used as the positive and negative control, respectively. The result showed that despite a lower activity than pSEAP/gfFSHβ(−1744), the short 54-bp fragment −1720/−1666 showed a strong response to Smad3, in agreement with the existence of two SBEs (site I and II) in this region. The involvement of these regulatory sites in Smad3-induced promoter activity was confirmed by the evidence that site-directed mutagenesis of these sites resulted in a complete loss of promoter activity. To further demonstrate the role of individual SBE sites, we also mutated each site separately. Interestingly, the removal of SBE and AP-1 motif at site I had no effect on promoter activity whereas the loss of SBE and AP-1 motif at site II completely abolished the promoter activity (Figure 4A), suggesting a role for site II as the functional Smad responsive element in the distal region of fshb promoter.
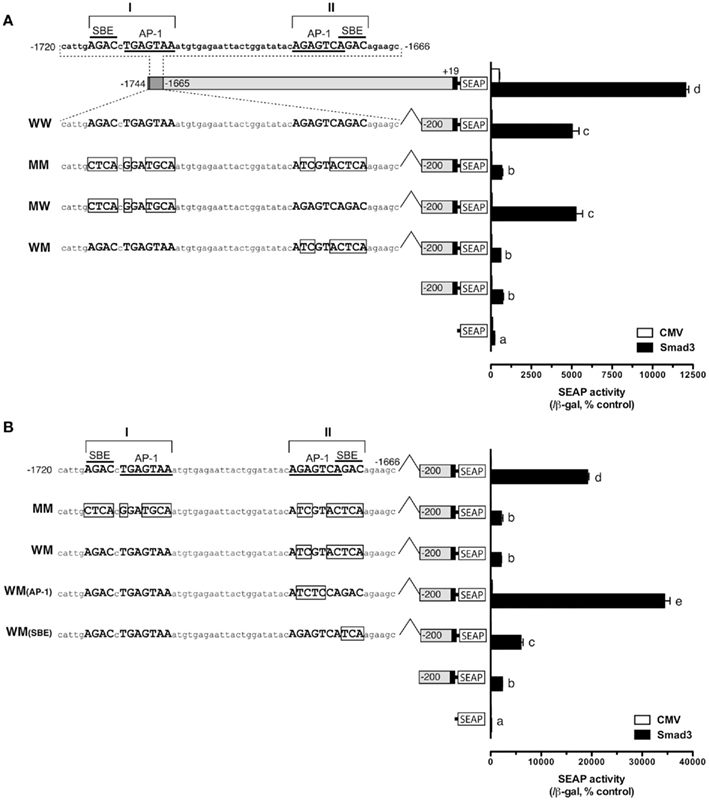
Figure 4. Identification of potential Smad binding element (SBE) in the distal region of fshb promoter (−1720/−1666). (A) Roles of potential SBE/AP-1 sequences at site I and II in the promoter region from −1720 to −1666. The sequence of the region is shown on the top with site I and II each containing an AP-1-like element (underlined) and a SBE (overlined). Mutagenesis of the AP-1-like element and SBE at site I (MW, representing mutation, and wild type for site I and II, respectively) showed no effect on the promoter activity whereas change of sequences at site II site (WM) completely eliminated the promoter activity in the presence of Smad3. (B) Test for roles of AP-1-like element and SBE at site II in promoter activity. Mutation at AP-1-like sequence [WM(AP-1)] had no effect whereas change of SBE sequence [WM(SBE)] completely abolished the promoter activity in the presence of Smad3. The boxed sequences are mutated elements. The cells in each well were transfected with pSEAP/gfFSHβ promoter constructs (300 ng), pBK-CMV/Smad3 (300 ng), and pSV-β-galactosidase (500 ng). Each value represents the mean ± SEM of three replicates. Different letters in the graph indicate statistical significance at P < 0.05.
As shown in Figure 4A, the SBE at −1675/−1672 (site II; italic) overlaps with an AP-1-like site (boxed; 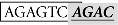 ). To further demonstrate which site is essential for the Smad responsiveness, we prepared two additional mutant constructs with the AP-1 motif and SBE site knocked out separately. The result showed that mutation of the SBE site at −1675/−1672 significantly reduced the Smad3-induced promoter activity. In contrast, instead of reducing transcriptional activity, mutation of the AP-1 site at −1681/−1675 led to an even higher reporter level (Figure 4B).
). To further demonstrate which site is essential for the Smad responsiveness, we prepared two additional mutant constructs with the AP-1 motif and SBE site knocked out separately. The result showed that mutation of the SBE site at −1675/−1672 significantly reduced the Smad3-induced promoter activity. In contrast, instead of reducing transcriptional activity, mutation of the AP-1 site at −1681/−1675 led to an even higher reporter level (Figure 4B).
Characterization of Smad Response Element(s) in the Proximal Region of fshb Promoter
The promoter analysis by serial deletion mutation strongly suggested the existence of a strong Smad responsive element in the proximal region of goldfish fshb promoter between −220 and −200 (AGAACACTGACCTTGAAAAC; Figures 1 and 2B). To further characterize the putative site in this region, we mutated each nucleotide of this region by serial 2-bp mutagenesis (M1 to M10). As shown in Figure 5A, mutation of GACCTT in mutants M5–7 abolished the promoter activity to the level of pSEAP/gfFSHβ(−200) while M8 significantly reduced the activity. No effect was observed with M1–4 and M9–10 as compared to pSEAP/gfFSHβ (−220; Figure 5A). Sequence analysis revealed that the nucleotides mutated in M5–8 at −212/−205 (GACCTTGA) happened to be identical to a steroidogenic factor 1 (SF-1) site reported in the human StAR gene (Sugawara et al., 1997). The same motif is also present in the proximal region of salmon FSHβ gene (−251/−244; Wang et al., 2009). Interestingly, this site in mammals is the location of a highly conserved AP-1 binding site (TGATTCA; Strahl et al., 1997). Despite the difference between the goldfish (SF-1) and mammals (AP-1), this site is preceded and overlapped by a sequence (AACACTGA) that is extremely conserved in FSHβ gene across vertebrates from fish to mammals (Figure 5B).
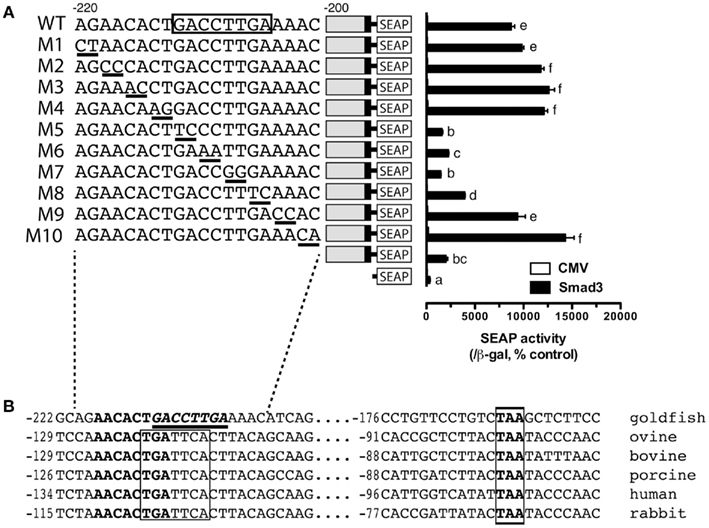
Figure 5. Identification of putative Smad responsive element in the proximal region (−220/−200) of goldfish fshb promoter. (A) Each nucleotide of this region was mutated by serial 2-bp mutagenesis (M1 to M10) and their promoter activity was examined and compared with that of the wild type (WT) in the presence of Smad3. The cells in each well were transfected with pSEAP/gfFSHβ promoter constructs (300 ng), pBK-CMV/Smad3 (300 ng), and pSV-β-galactosidase (500 ng). Each value represents the mean ± SEM of three replicates. Different letters in graph indicate statistical significance at P < 0.05. (B) Comparison of the proximal sequence of goldfish fshb promoter with those of mammals (Strahl et al., 1997). The putative Smad responsive element or SF-1 site (GACCTTGA; underlined) is preceded by and overlaps with a sequence (AACACTGA; bold) fully conserved in the goldfish and mammals. The AP-1 site (TGATTCA) highly conserved in mammals and a TAA motif fully conserved in the goldfish and mammals are boxed.
Interactive Effect of the Distal and Proximal Smad Response Elements
After identification of the distal and proximal SRE (the consensus SBE site at −1675/−1672 and the SF-1 site at −212/−205), we were interested in the relative strength of these two sites and in particular their interactive effect in driving SEAP reporter expression in the presence of Smad3. As described above, both elements could elicit significant response of SEAP expression compared to the minimal pSEAP/gfFSHβ(−200) promoter. When combined together, the two response elements appeared to have an additive to synergistic effect although the level was lower than that of the full-length promoter pSEAP/gfFSHβ(−1744; Figure 6).
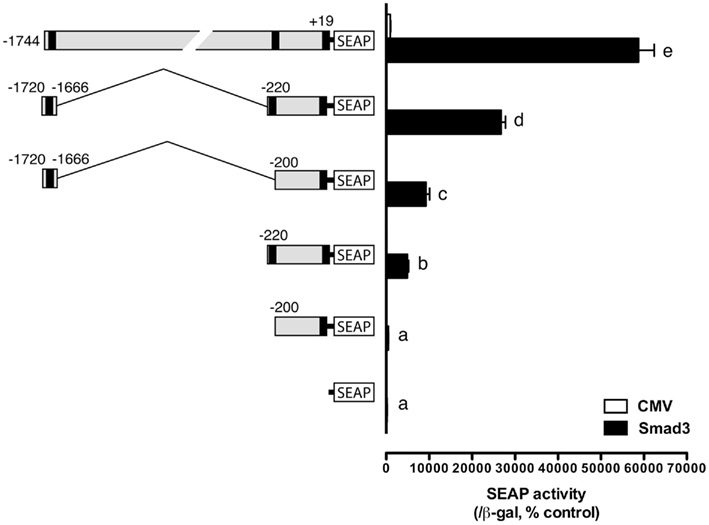
Figure 6. Effects and interactive effects of the proximal and distal Smad responsive elements in fshb promoter activity. The two elements both exhibited promoter activity and their combination produced an even higher synergistic response. The cells in each well were transfected with pSEAP/gfFSHβ promoter constructs (300 ng), pBK-CMV/Smad3 (300 ng), and pSV-β-galactosidase (500 ng). Each value represents the mean ± SEM of three replicates. Different letters in the graph indicate statistical significance at P < 0.05.
Activation of Goldfish fshb Promoter by Activin and GnRH
All the experiments described above involved co-expression of goldfish Smad3, which is the major R-Smad responsible for activin signaling. To demonstrate if the distal and proximal response elements identified also respond to activin, we tested the effects of both human activin A and goldfish activin B. As expected, both activin A (10 ng/ml) and B (4.5 U/ml) significantly stimulated the transcription activity of pSEAP/gfFSHβ(−1744). Activin A and B also elicited significant responses of the two Smad response elements in pSEAP/gfFSHβ(−1720/−1666) and pSEAP/gfFSHβ(−220). Interestingly, GnRH (100 nM) alone also had similar stimulatory effect on the transcription of goldfish fshb gene promoter in the LβT-2 cells. All constructs showed positive response to GnRH treatment and the response levels were generally comparable to those to activins. When applied in combination, GnRH and activin caused even higher responses for all constructs (Figure 7). We also tested the proximal region mutants described in Figure 5 for their response to GnRH. The result showed that all the mutants responded to GnRH in exactly the same way as they did to Smad3. The mutation of the sequence GACCTT (M5–7) completely abolished the effects of Smad3 and GnRH (Figure 8).
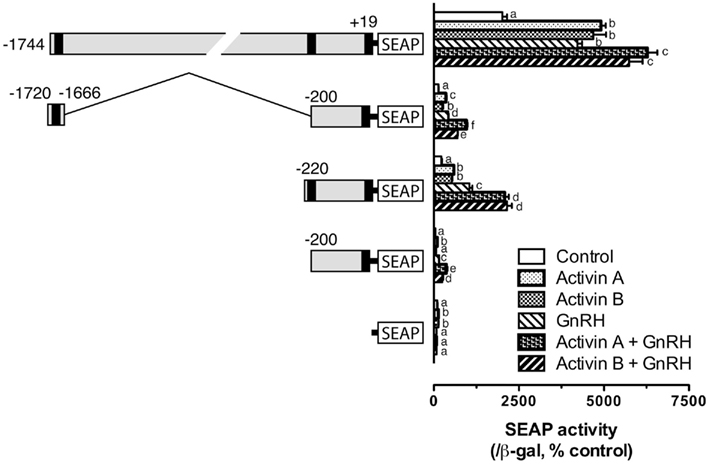
Figure 7. Activin A, activin B, and GnRH effects on goldfish fshb promoter activity in the LβT-2 cells. The cells were transfected with pSEAP/gfFSHβ promoter constructs (300 ng), pBK-CMV/Smad3 (300 ng), and pSV-β-galactosidase (500 ng). After 24-h incubation, the cells were washed and treated with human activin A (10 ng/ml) or goldfish activin B (4.5 U/ml) in the presence or absence of GnRH (100 nM) for 24 h. Each value represents the mean ± SEM of three replicates. Different letters in the graph indicate statistical significance at P < 0.05 in each group.
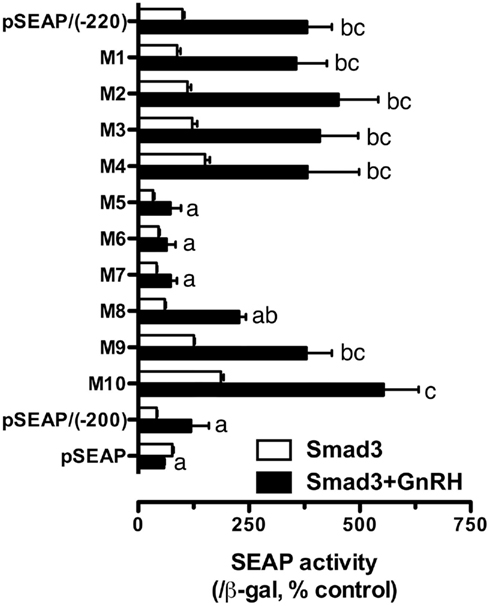
Figure 8. Effect of GnRH on the promoter activity of the mutants described in Figure 5 in the presence of Smad3. The cells were transfected with pSEAP/gfFSHβ promoter constructs (300 ng), pBK-CMV/Smad3 (300 ng), and pSV-β-galactosidase (500 ng). pSEAP/gfFSHβ(−220) and (−200) were included as the positive and negative controls, respectively. Each value represents the mean ± SEM of three replicates. Different letters in the graph indicate statistical significance at P < 0.05 (not shown for Smad3 alone).
Evidence for the Involvement of Activin System in GnRH Signaling in LβT-2 Cells
The experiments described above strongly implicated activin system in GnRH signaling. To provide further evidence for this mechanism, we carried out another experiment in which the LβT-2 cells were co-transfected with pSEAP/gfFSHβ(−220) and goldfish Smad2, Smad3, Smad7 (an inhibitory Smad), or ActRIIA(−) (dominant negative activin type II A receptor) followed by GnRH treatment. As shown in Figure 9, GnRH (100 nM) and TPA (100 nM, a potent PKC activator) significantly increased the promoter activity whereas GF109203x (1 μM, a PKC inhibitor) slightly reduced the Smad-induced expression level. The effects of GnRH and TPA were dramatically increased in the presence of Smads, particularly Smad3; however, their effects were abolished by overexpression of Smad7 or ActRIIA(−) in the cells (Figure 9).
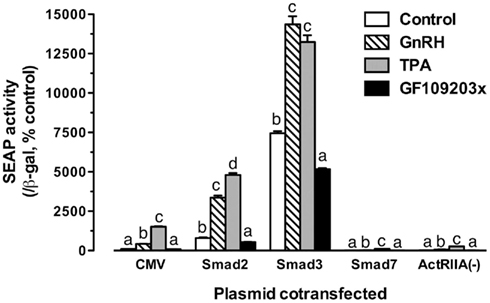
Figure 9. Effects of Smad proteins and the dominant negative activin receptor ActRIIA(−) on the GnRH/TPA-stimulated promoter activity of the proximal region. The cells were transfected with pSEAP/gfFSHβ (−220; 300 ng), pBK-CMV/Smads (2, 3, or 7) or pBK-CMV/ActRIIA(−) (300 ng), and pSV-β-galactosidase (500 ng). After 24-h incubation, the cells were washed and treated with GnRH (100 nM), PKC activator TPA (100 nM), or PKC inhibitor GF109203x (1 μM). Each value represents the mean ± SEM of three replicates. Different letters in the graph indicate statistical significance at P < 0.05 in each group.
Discussion
Activin is a growth and differentiation factor expressed in almost all tissues (Meunier et al., 1988), where it has diverse biological functions (Mather et al., 1997). In the pituitary, activin is a key modulator of FSHβ expression and FSH secretion (Ling et al., 1986a,b; Vale et al., 1986; Weiss et al., 1992, 1995). In the goldfish, our previous results have demonstrated that activin stimulates FSHβ (fshb) expression (Yam et al., 1999; Yuen and Ge, 2004) and its stimulatory effect is exerted at the promoter level (Ge et al., 2003). Similar effect has recently been reported in the zebrafish (Lin and Ge, 2009) and the European eel (Aroua et al., 2011). We have also demonstrated that the effect of activin on goldfish fshb is likely mediated by the Smad signaling pathway, particularly Smad3 (Lau and Ge, 2005). The present study was undertaken to identify the SRE on goldfish fshb promoter using serial deletion and site-directed mutagenesis approaches.
The Smad-induced SEAP activity declined gradually as the length of the promoter was decreased. However, several significant drops in SEAP activity were observed when regions −1744/−1563, −640/−620, and −220/−200 were deleted, suggesting that these regions may contain potential cis-regulatory elements responsible for Smad-induced reporter gene expression. We then focused our analysis on −1744/−1563 and −220/−200 regions because the deletion of these segments produced the most noticeable decline in the promoter activity.
Smad proteins are critical intracellular mediators of activin signaling (Shi and Massague, 2003). The N-terminal MH1 domains of Smad3 and Smad4 are able to bind to specific DNA sequence (AGAC or GTCT) termed SBE to regulate their target gene transcription (Dennler et al., 1998; Shi et al., 1998; Zawel et al., 1998; Chai et al., 2003). One or more SBE sequences have been identified in several activin (Jonk et al., 1998; Nagarajan et al., 1999; Suszko et al., 2003) and TGF-β-responsive promoters (Dennler et al., 1998; Vindevoghel et al., 1998; Hua et al., 1999; von Gersdorff et al., 2000). Sequence analysis of the −1744/−1563 region of goldfish fshb promoter revealed two putative SBEs (AGAC) at −1715/−1712 and −1675/−1672 bp. However, our mutagenesis experiment provided evidence that it was the SBE at −1675/−1672 that was responsible for Smad responsiveness of the −1744/−1563 region whereas the SBE site at −1715/−1712 played no role in mediating Smad-induced promoter activity. The functional SBE site and its adjacent sequence (GTCAGAC; −1678/−1672) was nearly identical to that identified in the proximal region of FSHβ in the mouse and rat (GTCTAGAC; Suszko et al., 2003; Gregory et al., 2005; Lamba et al., 2006). Interestingly, the mutation of the AP-1-like site next to the SBE at −1681/−1675 caused a higher response of the promoter region, suggesting a potential interaction of Smads with AP-1 in regulating fshb promoter activity at this region. This will be an interesting issue to investigate in future studies.
Different from the distal −1744/−1563 region, no consensus SBE element could be found in the proximal −220/−200 region despite its significant response to Smad. This region is interesting in that a previous study has identified a cluster of putative cis-regulatory elements from −154 to −218, including half ARE/ERE and GnRH-RE (Sohn et al., 1998). Furthermore, sequence analysis revealed a putative SF-1 binding site (GACCTTGA) in this region at position −212/−205 (Le Drean et al., 1996; Sohn et al., 1998). Our mutagenesis experiment provided strong evidence for the importance of this site in Smad responsiveness as any sequence change at this site significantly reduced or abolished the Smad-induced transcription activity whereas the bases outside this site had no contribution to the Smad responsiveness. As Smad proteins can also serve as co-activators to interact with other DNA-binding transcription factors to control gene transcription in addition to binding to SBE directly (Chen et al., 1996, 1997; Yeo et al., 1999; Attisano and Wrana, 2000; Wurthner et al., 2001) and SF-1 site is critical for the regulation of FSHβ expression (Ingraham et al., 1994; Brown and Mcneilly, 1997), it is conceivable that Smad proteins may exert their transcriptional effects by associating with SF-1 which binds to this cis-regulatory element.
One interesting finding of the present study was that the SF-1 site (−212/−205) in the proximal region of goldfish FSHβ promoter is absent in the promoter of mammalian FSHβ; instead, the same location is occupied by a highly conserved AP-1 site (TGATTCA) in mammals (Strahl et al., 1997). Both SF-1 site in the goldfish and AP-1 site in mammals are preceded by the same sequence AACACTGA, which is fully conserved in fish and mammals. Interestingly, a recent study on human FSHβ showed that activin and GnRH synergistically stimulated FSHβ expression and their signaling pathways were shown to converge at the conserved high affinity AP-1 site (TGATTCA; −117/−111; Wang et al., 2008). In the present study, we showed that although the AP-1 sequence is replaced by a SF-1 element in the goldfish FSHβ promoter, the same site responded strongly to both Smad3 and GnRH, which agrees with the report in mammals (Wang et al., 2008). The action of GnRH was activin and Smad-dependent as both Smad7 and ActRIIA(−) (dominant negative activin type II A receptor) could abolish the effects of GnRH. The exact mechanism underlying such interaction remains unknown, and it would be an interesting issue to investigate in future studies, especially the role of AP-1 and SF-1. A recent study in the Chinook salmon showed that overexpression of SF-1 in the COS cells could significantly increase the promoter activity of its FSHβ gene (Wang et al., 2009). As TPA mimicked the effect of GnRH and its action could also be abolished by co-expression of Smad7 and ActRIIA(−), it is likely that the activin/Smad pathway is involved downstream of PKC activation by GnRH. One possibility is that GnRH may up-regulate the components of the activin/Smad signal transduction pathway, such as activin subunits, receptors, and/or Smads, which in turn leads to activation of the promoter. This is supported by immunocytochemical studies demonstrating nuclear translocation of cytoplasmic Smad3 in the αT3-1 cells in response to activin and GnRH agonist treatment (Norwitz et al., 2002). Secondly, GnRH may enhance the signaling of SF-1 whose activity of stimulating fshb expression involves cooperation with Smad proteins activated by activin. A blockade of the activin/Smad pathway by Smad7 or ActRIIA(−) would weaken its activity. This is supported by the report in the rat that GnRH stimulated SF-1 gene expression in the pituitary (Haisenleder et al., 1996). A recent study on endogenous FSHβ expression in the LβT-1 suggested that the interaction of activin and GnRH involved the activity of p38 MAPK, which stimulated c-Fos expression and augmented Smad3 phosphorylation (Coss et al., 2007).
Other evidence in mammals also supports the interdependence of GnRH and activin signaling in stimulating FSHβ gene expression. GnRH stimulation of ovine FSHβ promoter was inhibited by follistatin in the LβT-2 cells (Pernasetti et al., 2001), suggesting the involvement of extracellular activin ligand in the action of GnRH. On the other hand, activin increased GnRH signaling by stimulating the synthesis of GnRH receptors (Fernandez-Vazquez et al., 1996), which would sensitize gonadotropes to GnRH. Co-treatment of the LβT-2 cells with activin and GnRH caused a synergistic increase in rat FSHβ promoter activity and the interaction might involve a palindromic Smad binding site (GTCTAGAC) at −266/−259 (Gregory et al., 2005), suggesting the involvement of multiple cis-elements in activin–GnRH interaction.
In conclusion, several potential cis-regulatory elements responsible for activin/Smad-induced fshb expression have been identified in the goldfish fshb promoter, which represents one of the first studies in lower vertebrates. A consensus Smad binding site is located in the distal region of the promoter, which offered a strong response of the promoter to activin/Smad signaling. In the proximal region, it turned out to be a SF-1 site that was responsible for Smad-induced promoter activity, implicating the involvement of SF-1 in the regulation. As activin stimulation of FSH biosynthesis is a well-conserved regulatory mechanism across vertebrates, the present study provides valuable information about the diverse mechanisms by which FSH biosynthesis is controlled and the evolution of such mechanisms in vertebrate history.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Pamela Mellon (Department of Reproductive Medicine, University of California, San Diego) for providing us the LβT-2 cells, and Dr. Makito Kobayashi (International Christian University, Mitaka, Tokyo) for the cloned goldfish FSHβ gene. We also thank Mr. Wai-Kin So for preparing the plasmid pBK-CMV/ActRIIA(−) for expressing the mutant goldfish activin type IIA receptor. The work was substantially supported by RGC/GRF grants from the Research Grants Council of the Hong Kong Special Administrative Region to W. Ge.
References
Aroua, S., Maugars, G., Jeng, S. R., Chang, C. F., Weltzien, F. A., Rousseau, K., and Dufour, S. (2011). Pituitary gonadotropins FSH and LH are oppositely regulated by the activin/follistatin system in a basal teleost, the eel. Gen. Comp. Endocrinol. 175, 82–91.
Attisano, L., and Wrana, J. L. (2000). Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 12, 235–243.
Bernard, D. J. (2004). Both SMAD2 and SMAD3 mediate activin-stimulated expression of the follicle-stimulating hormone β subunit in mouse gonadotrope cells. Mol. Endocrinol. 18, 606–623.
Besecke, L. M., Guendner, M. J., Schneyer, A. L., Bauer-Dantoin, A. C., Jameson, J. L., and Weiss, J. (1996). Gonadotropin-releasing hormone regulates follicle-stimulating hormone-β gene expression through an activin/follistatin autocrine or paracrine loop. Endocrinology 137, 3667–3673.
Bilezikjian, L. M., Blount, A. L., Leal, A. M., Donaldson, C. J., Fischer, W. H., and Vale, W. W. (2004). Autocrine/paracrine regulation of pituitary function by activin, inhibin and follistatin. Mol. Cell. Endocrinol. 225, 29–36.
Brown, P., and Mcneilly, A. S. (1997). Steroidogenic factor-1 (SF-1) and the regulation of expression of luteinising hormone and follicle stimulating hormone b-subunits in the sheep anterior pituitary in vivo. Int. J. Biochem. Cell Biol. 29, 1513–1524.
Carroll, R. S., Corrigan, A. Z., Gharib, S. D., Vale, W., and Chin, W. W. (1989). Inhibin, activin, and follistatin: regulation of follicle-stimulating hormone messenger ribonucleic acid levels. Mol. Endocrinol. 3, 1969–1976.
Chai, J., Wu, J. W., Yan, N., Massague, J., Pavletich, N. P., and Shi, Y. (2003). Features of a Smad3 MH1-DNA Complex: roles of water and zinc in DNA binding. J. Biol. Chem. 278, 20327–20331.
Chen, X., Rubock, M. J., and Whitman, M. (1996). A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383, 691–696.
Chen, X., Weisberg, E., Fridmacher, V., Watanabe, M., Naco, G., and Whitman, M. (1997). Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389, 85–89.
Chong, K. L., Wang, S., and Melamed, P. (2004). Isolation and characterization of the follicle-stimulating hormone β subunit gene and 5′ flanking region of the Chinook salmon. Neuroendocrinology 80, 158–170.
Coss, D., Hand, C. M., Yaphockun, K. K. J., Ely, H. A., and Mellon, P. L. (2007). p38 mitogen-activated protein kinase is critical for synergistic induction of the FSHβ gene by gonadotropin-releasing hormone and activin through augmentation of c-Fos induction and Smad phosphorylation. Mol. Endocrinol. 21, 3071–3086.
Dalkin, A. C., Haisenleder, D. J., Ortolano, G. A., Ellis, T. R., and Marshall, J. C. (1989). The frequency of gonadotropin-releasing-hormone stimulation differentially regulates gonadotropin subunit messenger ribonucleic acid expression. Endocrinology 125, 917–924.
Dennler, S., Itoh, S., Vivien, D., Ten Dijke, P., Huet, S., and Gauthier, J. M. (1998). Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091–3100.
Dupont, J., Mcneilly, J., Vaiman, A., Canepa, S., Combarnous, Y., and Taragnat, C. (2003). Activin signaling pathways in ovine pituitary and LβT2 gonadotrope cells. Biol. Reprod. 68, 1877–1887.
Eto, Y., Tsuji, T., Takezawa, M., Takano, S., Yokogawa, Y., and Shibai, H. (1987). Purification and characterization of erythroid differentiation factor (EDF) isolated from human leukemia cell line THP-1. Biochem. Biophys. Res. Commun. 142, 1095–1103.
Fernandez-Vazquez, G., Kaiser, U. B., Albarracin, C. T., and Chin, W. W. (1996). Transcriptional activation of the gonadotropin-releasing hormone receptor gene by activin A. Mol. Endocrinol. 10, 256–266.
Ferris, H. A., and Shupnik, M. A. (2006). Mechanisms for pulsatile regulation of the gonadotropin subunit genes by GNRH1. Biol. Reprod. 74, 993–998.
Ge, W., Ko, N.-L., Pang, F. Y.-M., Chung, M.-F., Lin, S.-W., Yuen, C.-W., Lau, M.-T., Liu, L., Sohn, Y. C., Kobayashi, M., and Aida, K. (2003). Activin stimulates goldfish FSH biosynthesis by enhancing FSHβ promoter activity. Fish Physiol. Biochem. 28, 65–71.
Glidewell-Kenney, C., Weiss, J., Hurley, L. A., Levine, J. E., and Jameson, J. L. (2008). Estrogen receptor α signaling pathways differentially regulate gonadotropin subunit gene expression and serum follicle-stimulating hormone in the female mouse. Endocrinology 149, 4168–4176.
Graham, K. E., Nusser, K. D., and Low, M. J. (1999). LβT2 gonadotroph cells secrete follicle stimulating hormone (FSH) in response to active A. J. Endocrinol. 162, R1–R5.
Gregory, S. J., and Kaiser, U. B. (2004). Regulation of gonadotropins by inhibin and activin. Semin. Reprod. Med. 22, 253–267.
Gregory, S. J., Lacza, C. T., Detz, A. A., Xu, S., Petrillo, L. A., and Kaiser, U. B. (2005). Synergy between activin A and gonadotropin-releasing hormone in transcriptional activation of the rat follicle-stimulating hormone-β gene. Mol. Endocrinol. 19, 237–254.
Haisenleder, D. J., Yasin, M., Dalkin, A. C., Gilrain, J., and Marshall, J. C. (1996). GnRH regulates steroidogenic factor-1 (SF-1) gene expression in the rat pituitary. Endocrinology 137, 5719–5722.
Hua, X., Miller, Z. A., Wu, G., Shi, Y., and Lodish, H. F. (1999). Specificity in transforming growth factor β-induced transcription of the plasminogen activator inhibitor-1 gene: interactions of promoter DNA, transcription factor muE3, and Smad proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 13130–13135.
Ingraham, H. A., Lala, D. S., Ikeda, Y., Luo, X., Shen, W. H., Nachtigal, M. W., Abbud, R., Nilson, J. H., and Parker, K. L. (1994). The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 8, 2302–2312.
Jayes, F. C., Britt, J. H., and Esbenshade, K. L. (1997). Role of gonadotropin-releasing hormone pulse frequency in differential regulation of gonadotropins in the gilt. Biol. Reprod. 56, 1012–1019.
Jonk, L. J., Itoh, S., Heldin, C. H., Ten Dijke, P., and Kruijer, W. (1998). Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273, 21145–21152.
Kawakami, S., Fujii, Y., Okada, Y., and Winters, S. J. (2002). Paracrine regulation of FSH by follistatin in folliculostellate cell-enriched primate pituitary cell cultures. Endocrinology 143, 2250–2258.
Lamba, P., Santos, M. M., Philips, D. P., and Bernard, D. J. (2006). Acute regulation of murine follicle-stimulating hormone β subunit transcription by activin A. J. Mol. Endocrinol. 36, 201–220.
Lau, M. T., and Ge, W. (2005). Cloning of Smad2, Smad3, Smad4, and Smad7 from the goldfish pituitary and evidence for their involvement in activin regulation of goldfish FSHβ promoter activity. Gen. Comp. Endocrinol. 141, 22–38.
Le Drean, Y., Liu, D., Wong, A. O., Xiong, F., and Hew, C. L. (1996). Steroidogenic factor 1 and estradiol receptor act in synergism to regulate the expression of the salmon gonadotropin II β subunit gene. Mol. Endocrinol. 10, 217–229.
Lee, S., and Rivier, C. (1997). Effect of repeated activin-A treatment on the activity of the hypothalamic-pituitary-gonadal axis of the adult male rat. Biol. Reprod. 56, 969–975.
Lin, S. W., and Ge, W. (2009). Differential regulation of gonadotropins (FSH and LH) and growth hormone (GH) by neuroendocrine, endocrine, and paracrine factors in the zebrafish – an in vitro approach. Gen. Comp. Endocrinol. 160, 183–193.
Ling, N., Ying, S. Y., Ueno, N., Shimasaki, S., Esch, F., Hotta, M., and Guillemin, R. (1986a). A homodimer of the β-subunits of inhibin A stimulates the secretion of pituitary follicle stimulating hormone. Biochem. Biophys. Res. Commun. 138, 1129–1137.
Ling, N., Ying, S. Y., Ueno, N., Shimasaki, S., Esch, F., Hotta, M., and Guillemin, R. (1986b). Pituitary FSH is released by a heterodimer of the β-subunits from the two forms of inhibin. Nature 321, 779–782.
Massague, J., Seoane, J., and Wotton, D. (2005). Smad transcription factors. Genes Dev. 19, 2783–2810.
Mather, J. P., Moore, A., and Li, R. H. (1997). Activins, inhibins, and follistatins: further thoughts on a growing family of regulators. Proc. Soc. Exp. Biol. Med. 215, 209–222.
Mathews, L. S. (1994). Activin receptors and cellular signaling by the receptor serine kinase family. Endocr. Rev. 15, 310–325.
Melamed, P. (2010). Hormonal signaling to follicle stimulating hormone β-subunit gene expression. Mol. Cell. Endocrinol. 314, 204–212.
Meunier, H., Rivier, C., Evans, R. M., and Vale, W. (1988). Gonadal and extragonadal expression of inhibin α, βA, and βB subunits in various tissues predicts diverse functions. Proc. Natl. Acad. Sci. U.S.A. 85, 247–251.
Nagarajan, R. P., Zhang, J., Li, W., and Chen, Y. (1999). Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J. Biol. Chem. 274, 33412–33418.
Norwitz, E. R., Xu, S., Xu, J., Spiryda, L. B., Park, J. S., Jeong, K. H., Mcgee, E. A., and Kaiser, U. B. (2002). Direct binding of AP-1 (Fos/Jun) proteins to a SMAD binding element facilitates both gonadotropin-releasing hormone (GnRH)- and activin-mediated transcriptional activation of the mouse GnRH receptor gene. J. Biol. Chem. 277, 37469–37478.
Pernasetti, F., Vasilyev, V. V., Rosenberg, S. B., Bailey, J. S., Huang, H. J., Miller, W. L., and Mellon, P. L. (2001). Cell-specific transcriptional regulation of follicle-stimulating hormone-β by activin and gonadotropin-releasing hormone in the LβT2 pituitary gonadotrope cell model. Endocrinology 142, 2284–2295.
Pierce, J. G., and Parsons, T. F. (1981). Glycoprotein hormones: structure and function. Annu. Rev. Biochem. 50, 465–495.
Roberts, V., Meunier, H., Vaughan, J., Rivier, J., Rivier, C., Vale, W., and Sawchenko, P. (1989). Production and regulation of inhibin subunits in pituitary gonadotropes. Endocrinology 124, 552–554.
Ross, S., and Hill, C. S. (2008). How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 40, 383–408.
Schmelzer, C. H., Burton, L. E., Tamony, C. M., Schwall, R. H., Mason, A. J., and Liegeois, N. (1990). Purification and characterization of recombinant human activin B. Biochim. Biophys. Acta 1039, 135–141.
Shi, Y., and Massague, J. (2003). Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700.
Shi, Y., Wang, Y. F., Jayaraman, L., Yang, H., Massague, J., and Pavletich, N. P. (1998). Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell 94, 585–594.
Shupnik, M. A. (1996). Gonadotropin gene modulation by steroids and gonadotropin-releasing hormone. Biol. Reprod. 54, 279–286.
Sohn, Y. C., Suetake, H., Yoshiura, Y., Kobayashi, M., and Aida, K. (1998). Structural and expression analyses of gonadotropin Iβ subunit genes in goldfish (Carassius auratus). Gene 222, 257–267.
Strahl, B. D., Huang, H. J., Pedersen, N. R., Wu, J. C., Ghosh, B. R., and Miller, W. L. (1997). Two proximal activating protein-1-binding sites are sufficient to stimulate transcription of the ovine follicle-stimulating hormone-β gene. Endocrinology 138, 2621–2631.
Sugawara, T., Kiriakidou, M., Mcallister, J. M., Holt, J. A., Arakane, F., and Strauss, J. F. III. (1997). Regulation of expression of the steroidogenic acute regulatory protein (StAR) gene: a central role for steroidogenic factor 1. Steroids 62, 5–9.
Suszko, M. I., Balkin, D. M., Chen, Y., and Woodruff, T. K. (2005). Smad3 mediates activin-induced transcription of follicle-stimulating hormone β-subunit gene. Mol. Endocrinol. 19, 1849–1858.
Suszko, M. I., Lo, D. J., Suh, H., Camper, S. A., and Woodruff, T. K. (2003). Regulation of the rat follicle-stimulating hormone β-subunit promoter by activin. Mol. Endocrinol. 17, 318–332.
Vale, W., Rivier, J., Vaughan, J., Mcclintock, R., Corrigan, A., Woo, W., Karr, D., and Spiess, J. (1986). Purification and characterization of an FSH releasing protein from porcine ovarian follicular fluid. Nature 321, 776–779.
Vindevoghel, L., Kon, A., Lechleider, R. J., Uitto, J., Roberts, A. B., and Mauviel, A. (1998). Smad-dependent transcriptional activation of human type VII collagen gene (COL7A1) promoter by transforming growth factor-β. J. Biol. Chem. 273, 13053–13057.
von Gersdorff, G., Susztak, K., Rezvani, F., Bitzer, M., Liang, D., and Bottinger, E. P. (2000). Smad3 and Smad4 mediate transcriptional activation of the human Smad7 promoter by transforming growth factor β. J. Biol. Chem. 275, 11320–11326.
Wang, S., Zhu, Y., and Melamed, P. (2009). The molecular regulation of Chinook salmon gonadotropin β-subunit gene transcription. Gen. Comp. Endocrinol. 161, 34–41.
Wang, Y., Fortin, J., Lamba, P., Bonomi, M., Persani, L., Roberson, M. S., and Bernard, D. J. (2008). Activator protein-1 and smad proteins synergistically regulate human follicle-stimulating hormone β-promoter activity. Endocrinology 149, 5577–5591.
Weiss, J., Guendner, M. J., Halvorson, L. M., and Jameson, J. L. (1995). Transcriptional activation of the follicle-stimulating hormone β- subunit gene by activin. Endocrinology 136, 1885–1891.
Weiss, J., Harris, P. E., Halvorson, L. M., Crowley, W. F. Jr., and Jameson, J. L. (1992). Dynamic regulation of follicle-stimulating hormone-β messenger ribonucleic acid levels by activin and gonadotropin-releasing hormone in perifused rat pituitary cells. Endocrinology 131, 1403–1408.
Woodruff, T. K., Krummen, L. A., Lyon, R. J., Stocks, D. L., and Mather, J. P. (1993). Recombinant human inhibin A and recombinant human activin A regulate pituitary and ovarian function in the adult female rat. Endocrinology 132, 2332–2341.
Wurthner, J. U., Frank, D. B., Felici, A., Green, H. M., Cao, Z., Schneider, M. D., Mcnally, J. G., Lechleider, R. J., and Roberts, A. B. (2001). Transforming growth factor-β receptor-associated protein 1 is a Smad4 chaperone. J. Biol. Chem. 276, 19495–19502.
Yam, K. M., Yoshiura, Y., Kobayashi, M., and Ge, W. (1999). Recombinant goldfish activin B stimulates gonadotropin-Iβ but inhibits gonadotropin-IIβ expression in the goldfish, Carassius auratus. Gen. Comp. Endocrinol. 116, 81–89.
Yaron, Z., Gur, G., Melamed, P., Rosenfeld, H., Levavi-Sivan, B., and Elizur, A. (2001). Regulation of gonadotropin subunit genes in tilapia. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 129, 489–502.
Yeo, C. Y., Chen, X., and Whitman, M. (1999). The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis. J. Biol. Chem. 274, 26584–26590.
Yoshiura, Y., Kobayashi, M., Kato, Y., and Aida, K. (1997). Molecular cloning of the cDNAs encoding two gonadotropin β subunits (GTH-I β and -II β) from the goldfish, Carassius auratus. Gen. Comp. Endocrinol. 105, 379–389.
Yuen, C. W., and Ge, W. (2004). Follistatin suppresses FSHβ but increases LHβ expression in the goldfish – evidence for an activin-mediated autocrine/paracrine system in fish pituitary. Gen. Comp. Endocrinol. 135, 108–115.
Keywords: FSH, promoter, Smad responsive element, activin, GnRH, goldfish
Citation: Lau M-T, Lin S-W and Ge W (2012) Identification of Smad response elements in the promoter of goldfish FSHβ gene and evidence for their mediation of activin and GnRH stimulation of FSHβ expression. Front. Endocrin. 3:47. doi: 10.3389/fendo.2012.00047
Received: 17 November 2011; Accepted: 11 March 2012;
Published online: 27 March 2012.
Edited by:
Vance Trudeau, University of Ottawa, CanadaReviewed by:
José A. Muñoz-Cueto, University of Cadiz, SpainRiccardo Pierantoni, Seconda Università di Napoli, Italy
Copyright: © 2012 Lau, Lin and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Wei Ge, School of Life Sciences, The Chinese University of Hong Kong, Shatin, New Territories, Hong Kong, China. e-mail:d2VpZ2VAY3Voay5lZHUuaGs=

