- 1Department of Animal and Marine Bioresource Sciences, Graduate School of Agriculture, Kyushu University, Fukuoka, Japan
- 2Department of Anatomy and Neurobiology, Kinki University School of Medicine, Osaka, Japan
- 3Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
The peripheral circadian oscillator plays an essential role in synchronizing local physiology to operate in a circadian manner via regulation of the expression of clock-controlled genes. The present study aimed to evaluate the circadian rhythms of clock genes and clock-controlled genes expressed in the rat uterus endometrial stromal cells (UESCs) during the stage of implantation by a DNA microarray. Of 12,252 genes showing significantly expression, 7,235 genes displayed significant alterations. As revealed by the biological pathway analysis using the database for annotation, visualization, and integrated discovery online annotation software, genes were involved in cell cycle, glutathione metabolism, MAPK signaling pathway, fatty acid metabolism, ubiquitin mediated proteolysis, focal adhesion, and PPAR signaling pathway. The clustering of clock genes were mainly divided into four groups: the first group was Rorα, Timeless, Npas2, Bmal1, Id2, and Cry2; the second group Per1, Per2, Per3, Dec1, Tef, and Dbp; the third group Bmal2, Cry1, E4bp4, Rorβ, and Clock; the fourth group Rev-erbα. Eleven implantation-related genes and 24 placenta formation-related genes displayed significant alterations, suggesting that these genes involved in implantation and placenta formation are controlled under circadian clock. Some candidates as clock-controlled genes were evaluated by using RNA interference to Bmal1 mRNA. Down-regulation of Igf1 gene expression was observed by Bmal1 silencing, whereas the expression of Inhβa was significantly increased. During active oscillation of circadian clock, the apoptosis-related genes Fas and Caspase3 remained no significant changes, but they were significantly increased by knockdown of Bmal1 mRNA. These results indicate that clock-controlled genes are up- or down-regulated in rat UESCs during the stage of decidualization. DNA microarray analysis coupled with RNA interference will be helpful to understand the physiological roles of some oscillating genes in blastocyst implantation and placenta formation.
Introduction
Circadian rhythms are primarily synchronized with environmental time by the 24-h period of light-dark cycle. In mammals, the master clock in the suprachiasmatic nucleus coordinates the subsidiary oscillators in the majority of peripheral tissues. However, autonomic circadian oscillators are also functional in peripheral tissues. The peripheral circadian oscillators play critical roles in synchronizing local physiology and metabolism to operate in a circadian manner via regulation of the expression of clock-controlled genes (1). These physiological processes mainly include hormonal secretion, gluconeogenesis, lipogenesis, and bile acid homeostasis (2 –4). In addition, cellular differentiation may cause the suspension of the cyclic expression of clock genes (5, 6). Recent studies suggest that the circadian system is not only required for proper growth control, but also involved in the circadian regulation of cell proliferation and apoptosis. It has been reported that 2–10% of all mammalian genes are controlled under the circadian clock (1, 7, 8). Most of these genes are involved in organ functions and show tissue-specific expression. Only a small set of the clock-controlled genes are expressed in multiple organs. Among them are genes that encode key regulators of cell cycle progression (9, 10).
Several recent studies have demonstrated that circadian clock genes are rhythmically expressed in the uterus (2, 11 –15). In the uterus composed of heterogeneous cell types, ovarian steroids regulate the proliferation and differentiation of uterus endometrial stromal cells (UESCs). In rodents and humans, the UESCs undergo proliferation and differentiation into decidual cells in response to ovarian steroids and blastocyst implantation at the early stage of pregnancy (16 –18). This process ultimately results in the formation of the placenta.
Mice lacking the Clock gene display abnormal estrus cycles and are infertile (19, 20). Furthermore, implantation fails in Bmal1 deficient mice, due to impaired steroidogenesis (21), and mutations of Per1 and Per2 in mice display reproductive deficits in the middle-aged mutant females (22). More recently, we proved the Per2 expression is down-regulated in the UESCs during decidualization that influences the expression of vascular endothelial growth factor A (Vegfa) gene (23). Deregulation of the circadian clock may attenuate or disrupt expression of the clock-controlled genes and can have a profound influence on organ functions. Studies have demonstrated that the circadian clock function is very important for cell cycle, DNA damage response, and tumor suppression in vivo (24 –26).
In the present study, to search the clock-controlled genes expressed during the period of implantation, we analyzed the expression of the clock genes and clock-controlled genes in cultured UESCs prepared from pregnant rats at the stage of implantation using DNA microarray technology. We used transgenic rats constructed with mouse Per2 promoter-destabilized luciferase (Per2-dLuc) reporter gene (27) to precisely adjust the time of gene expression. In addition, several genes of significantly expressed genes including growth factor genes and apoptosis-related genes were analyzed using RNA interference (siRNA) to Bmal1 mRNA to determine whether these were controlled under circadian clockwork.
Materials and Methods
Animals
Mouse Per2 promoter region, assembly by NCBI and the Mouse Genome Sequencing Consortium, was fused to a dLuc reporter gene (27). Per2-dLuc transgenic rats were generated in accordance with the method described in the patent publication number WO/2002/081682 (Y.S. New Technology Institute, Utsunomiya, Japan). Adult females were mated with fertile males, and 12:00 p.m. on the day of finding spermatozoa in the vaginal smear was designated as day 0.5 of gestation. All the experiments were performed under the control of the Guidelines for Animal Experiments in the Faculty of Medicine, Kyushu University, and Law No. 105 and Notification No. 6 of the Government of Japan.
Preparation and Culture of UESCs
The UESCs were isolated from Per2-dLuc transgenic rats on day 4.50 of gestation as reported previously (6, 28, 29). The harvested cells were washed thrice with fresh DMEM/F12, and seeded onto 35 mm collagen-coated dishes at the density of 2 × 105 cells/dish with 2 mL of culture medium (phenol red-free DMEM/F12 supplemented with 10% charcoal-treated FBS and 1× PS). The culture medium was replaced at 15 min after cell seeding to remove epithelial cells. Cells were cultured in a humidified atmosphere of 95% air and 5% CO2 at 37°C for 2 days. Then, cells were cultured in serum-free medium supplemented with 1× antibiotic-antimycotic (AA; Nacalai Tesque, Kyoto), 1× Insulin-Transferrin-Selenium (ITS, Life Technologies, Grand Island, NY, USA), 0.1% bovine serum albumin (BSA, Sigma Chemicals), and 100 nM progesterone (P4, Sigma Chemicals) for additional 2 days prior to other treatments.
Real-Time Monitoring of Per2-dLuc Oscillations
The cultured UESCs were synchronized with 100 nM dexamethasone for 2 h in the serum-free medium containing 1× AA. Then, cells were given the serum-free medium DMEM/F12 supplemented with 15 mM HEPES, 0.1 mM luciferin (Wako, Tokyo), 0.1% BSA, 1× AA, and 1× ITS, and subjected to luminescence determination. Luciferase activity was chronologically monitored at 37°C with a Kronos Dio AB-2550 luminometer (ATTO, Tokyo) interfaced to a computer for continuous data acquisition, as described previously (6, 13, 14). The amplitude and period of Per2-dLuc oscillations were documented by the single Cosinor method using Timing Series Single 6.3 (Expert Soft Tech., Richelieu, France).
Microarray Analysis
RNA samples isolated from cultured UESCs at 30, 36, 42, and 48 h after dexamethasone synchronization were used for microarray analysis using the Whole Rat Genome Microarray 4 × 44 K Ver3.0 (Agilent Technologies, Santa Clara, CA, USA) representing 30,367 probe sets. The preparation of the samples, microarray hybridizations, and bioinformatics analysis were performed by the Cell Innovator at the Kyushu University (Fukuoka, Japan). Bioinformatics analysis was performed using Agilent Future Extraction software (Agilent Technologies). The data were filtered for signal intensity values (p ≤ 0.05, detectable), which allowed removing very low signal values. After this filtering, 12,252 probe sets remained. Probe sets passing the quality control were then analyzed by ANOVA, and p-values of<0.05 were considered significant. The ratio from signal intensity values of four time points was calculated. For the pathway analysis, probe sets were then used for gene ontology analysis using Database for Annotation, Visualization, and Integrated Discovery (DAVID) Bioinformatics Resources (http://david.abcc.ncifcf.gov/) (30). Pathways regulated at p < 0.05 were considered significant.
Bmal1 siRNA Transfection
Three sequences targeting the Bmal1 mRNA and no silencing RNA for rat were purchased from BOVAC Co. (Kurume, Japan). The sequences of RNA oligos used are listed in Table 1. The scrambled RNA for rat was used as no silencing RNA (BOVAC Co.). Both the Bmal1 siRNA and no silencing RNA were used at final concentrations of 25 nM. The cells were maintained with transfection medium for duration of 12 h (31). Then the medium was replaced with DMEM/F12 supplemented with 1× AA, 1× ITS, 0.1% BSA, and 100 nM P4.
RNA Extraction and RT-qPCR
Cultured cells were harvested at the indicated times, and total RNA was isolated using RNeasy Mini kit (Qiagen), according to the manufacturer’s protocol. RNA samples were treated with RNase-free DNase I (Qiagen). The cDNAs were generated by reverse transcription with random primers using a High Capacity Reverse Transcription kit (Applied Biosystems). One microgram of total RNA were reverse transcribed in 20 μL of mixture using MMLV High Performance Reverse Transcriptase (Epicentre Biotechnologies) and Oligo-dT primer according to the manufacture’s protocol. Primer sets used for real-time PCR were listed in Table 2. PCR was performed with a 1:15 dilution of cDNA samples in Master SYBR Green I mixture (Roche Diagnostics) with specific primers (0.25 μM final of each primer) using Mx3000P Real-time QPCR System (Stratagene). Relative quantification of the mRNA levels was performed using the comparative cycle threshold (ΔCt) method. The ΔCt for each sample was normalized to Gapdh and expressed as relative to the control values (23).
Statistical Analyses
All data are presented as means ± SEM of at least three separate experiments, each performed with triplicate samples. The amplitude of Per2-dLuc was documented by Cosinor analysis using Timing Series Single 6.3 (Expert Soft Tech., Richelieu, France). The statistical differences of examined values of target genes in cultured UESCs were determined by Student’s t-test using SigmaPlot software (Ver. 11.2; Systat Software, San Jose, CA, USA). Differences were considered significant at p < 0.05 or less.
Results
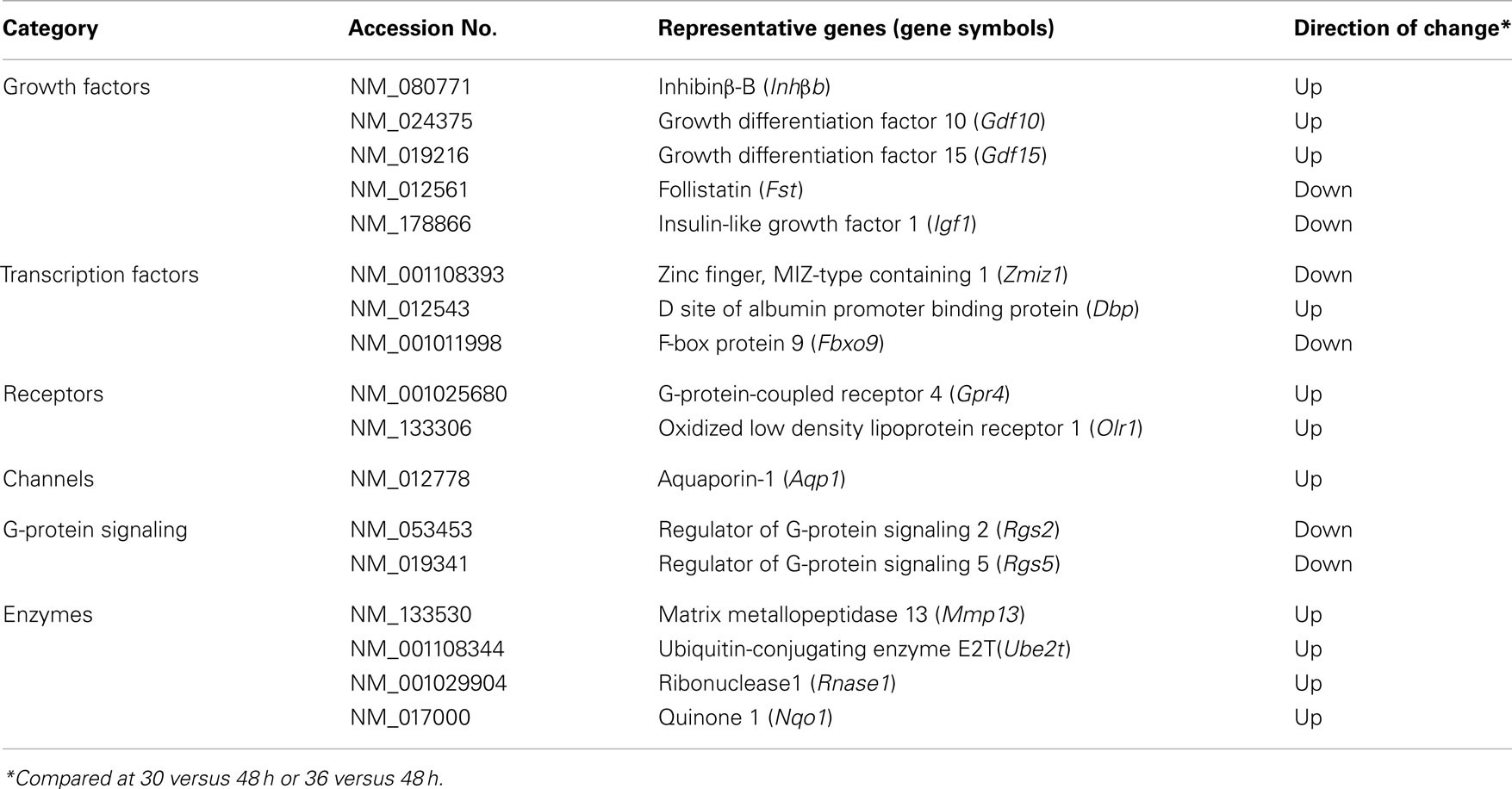
DNA Microarray Analysis of Oscillating Genes Expressed in the Rat UESCs after Dexamethasone Synchronization
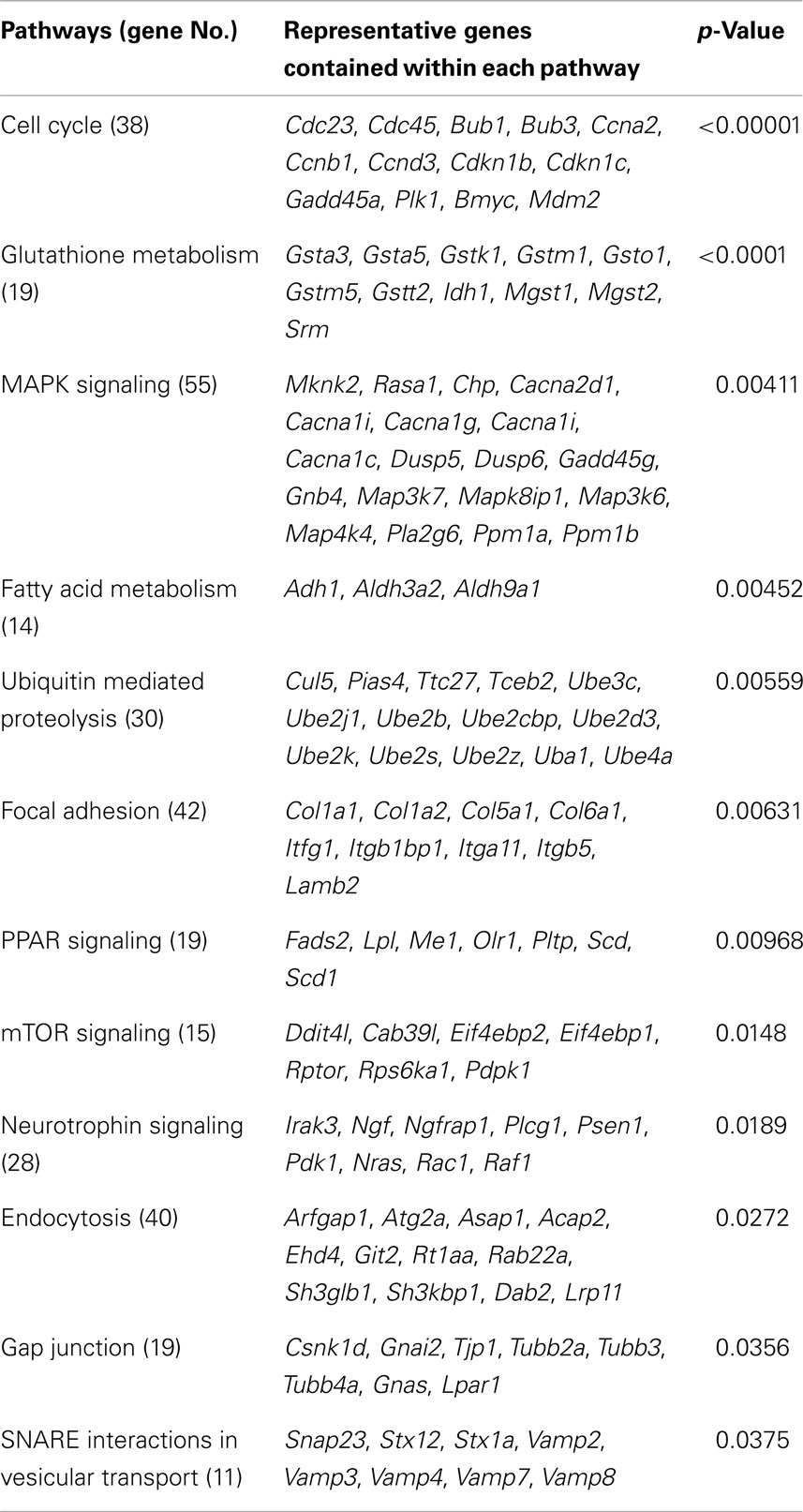
During monitoring of Per2-dLuc activity oscillation, RNA samples were prepared at 30, 36, 42, and 48 h after dexamethasone synchronization and global gene expression patterns were determined using DNA microarray technology (Figure 1). The analysis revealed 7,235 significantly altered genes in the UESCs. The increased expression (357 genes) and decreased expression (202 genes) of genes showing with fold change were obtained from 7,235 significantly altered genes at four time points during oscillation (Table 3). The majority of fold-changed genes were identified in both RNA samples 30 versus 48 h and 36 versus 48 h, including growth factors, transcription factors, receptors, channels, and enzymes (Table 4). For example, the expression of growth factors such as Inhβb, Gdf10, and Gdf15 were increased during the interval, whereas the expression of Igf1 was decreased. To place the microarray results in the cellular context, we performed the biological pathway analysis using the DAVID online annotation software. This analysis revealed that genes were involved in cell cycle, glutathione metabolism, MAPK signaling pathway, fatty acid metabolism, ubiquitin mediated proteolysis, focal adhesion, PPAR signaling pathway, and so on (Table 5). It was identified that the expression of 38 cell cycle related genes were significantly altered during the second to third phase of Per2-dLuc activity oscillation (p < 0.00001).
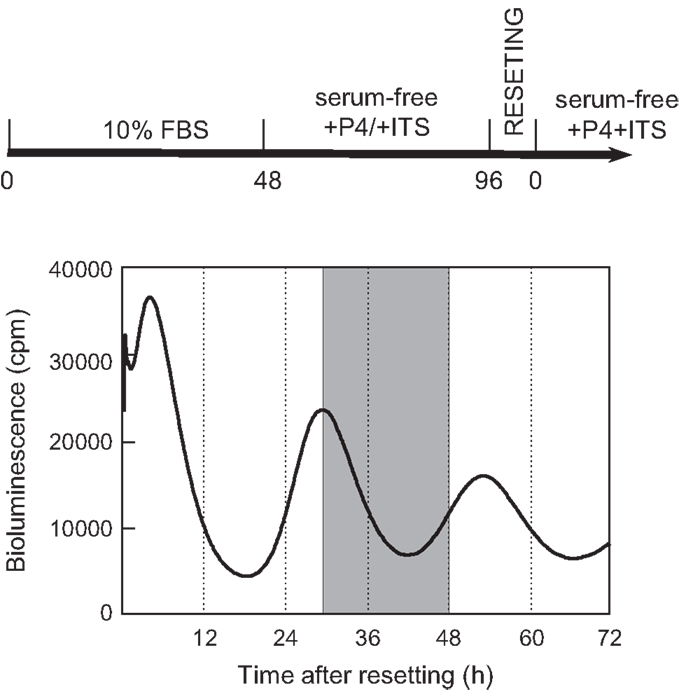
Figure 1. Isolation protocols of total RNA samples from cultured uterus endometrial stromal cells after dexamethasone synchronization. According to the second to the third phases of the Per2-dLuc oscillation, total RNA samples (n = 3) individually isolated from cultured UESCs at 30, 36, 42, and 48 h (shadow area) after dexamethasone synchronization were used for microarray analysis.
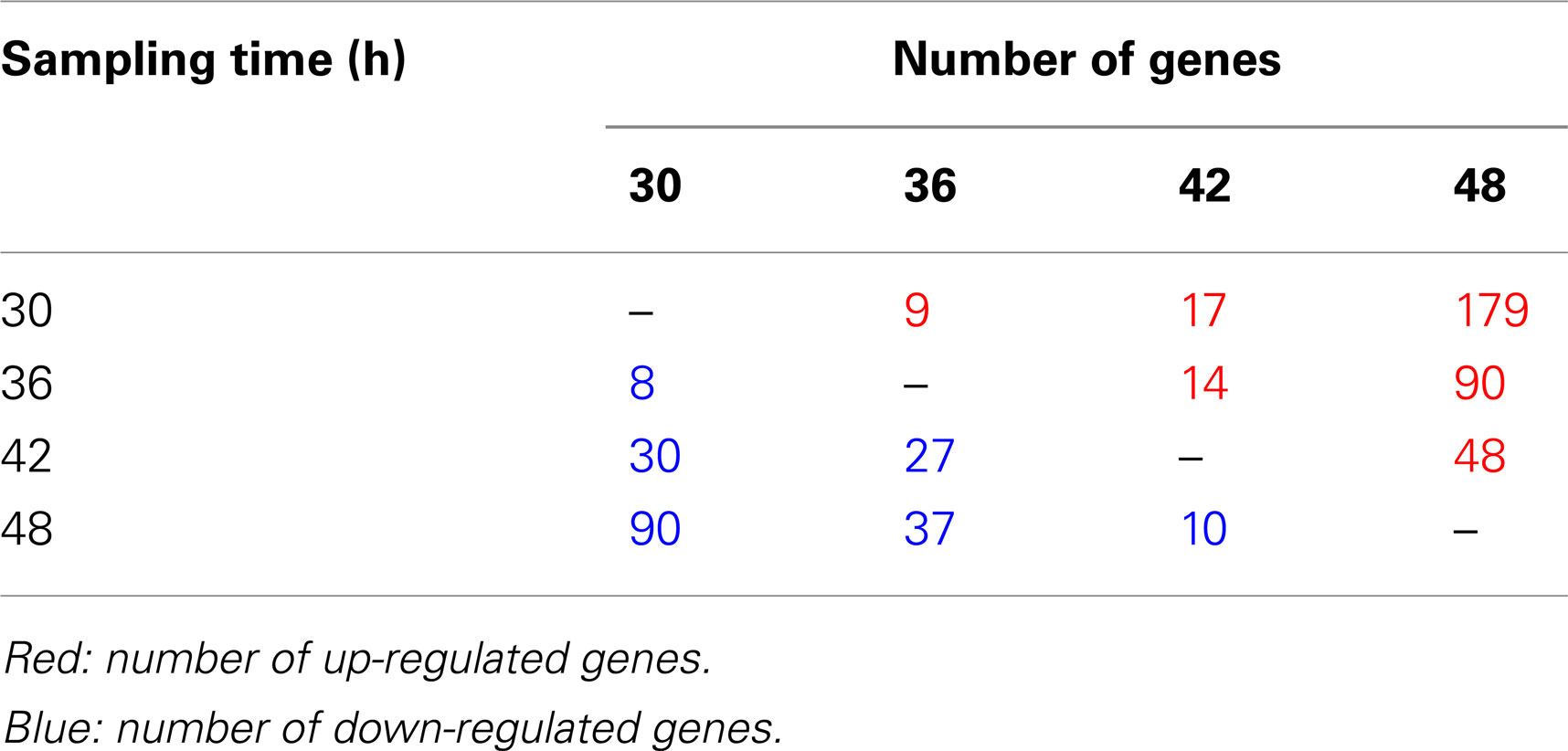
Table 3. Number of altered genes with fold change in the rat UESCs as revealed by microarray analysis.
Functional Molecular Machinery of a Circadian Clock Exists in the Rat UESCs after Dexamethasone Synchronization
In order to determine whether 18 clock genes displayed diurnal rhythmic expression, we performed gene clustering on the microarray results. The gene expression profiles were mainly divided into four groups: the first group was Rorα, Timeless, Npas2, Bmal1 (Arntl), Id2, and Cry2; the second group Per1, Per2, Per3, Dec1 (Bhlhe40), Tef, and Dbp; the third group Bmal2 (Arntl2), Cry1, E4bp4 (Nfil3), Rorβ, and Clock; the fourth group Rev-erbα (Nr1d1) (Figure S1 in Supplementary Material). The Per2 intensity profile was consistent with Per2 circadian oscillation (Figure 2). Profiles of Bmal1 and Rev-erbα expression were obviously anti-phase in the UESCs, because Bmal1 promotes Rev-erbα transcription with Clock and in turn Rev-erbα inhibits Bmal1 transcription through its binding to the RORE in the promoter region. The Per2 and Dbp expression profiles were also out-of-phase with the Bmal1 rhythm. The protein kinase genes such as Csnk1ε and Csnk1δ also displayed the weak rhythmic expression (Figure 2). The expression profiles of core clock genes such as Bmal1, Clock, Per1, Per2, and Rev-erbα were further confirmed by RT-qPCR. As the results are shown in Figure 3, their profiles were very similar to the microarray results. The Cosinor analysis method was used to determine the rhythmic expression of examined genes. Bmal1, Per1, Per2, and Rev-erbα displayed rhythmic expression (p < 0.05).
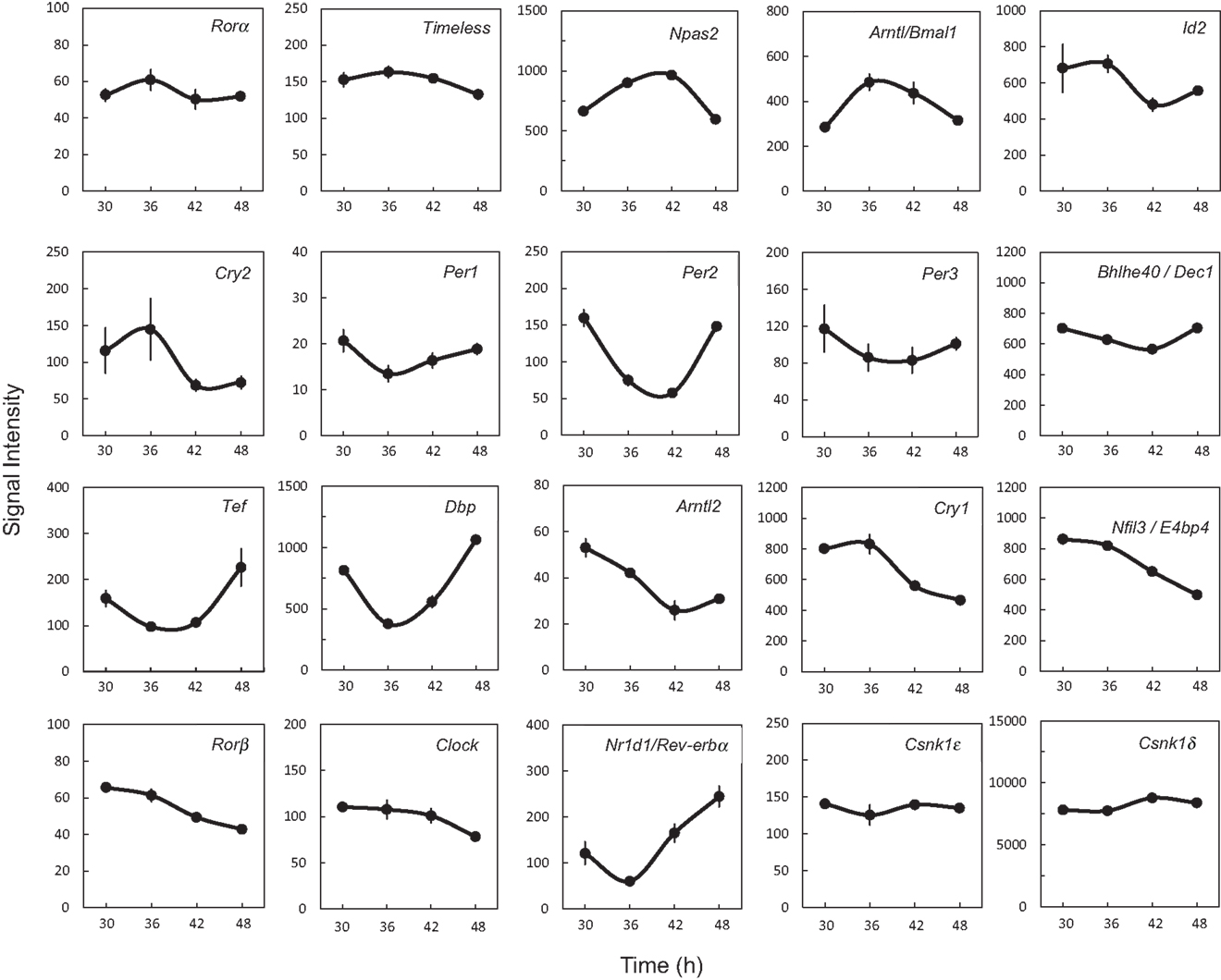
Figure 2. Expression profiles of clock genes and protein kinase genes in the uterus endometrial stromal cells of pregnant rats after dexamethasone synchronization. Each value represents the means ± SEM (n = 3) of signal intensity from the microarray results.
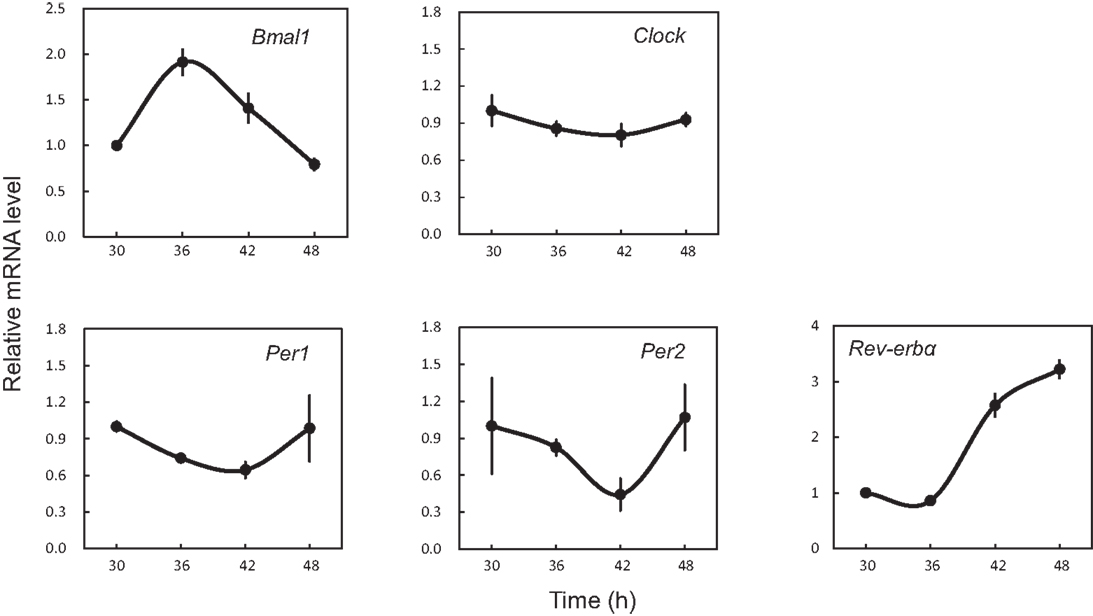
Figure 3. Expression profiles of core clock genes in the uterus endometrial stromal cells after dexamethasone synchronization as revealed by RT-qPCR. According to the Per2-dLuc oscillation as illustrated in Figure 1, total RNA samples were collected at the indicated times after synchronization. qRT-PCR analyses of transcript levels were performed using their specific primers. Gapdh was used as an internal control. Each value represents the mean ± SEM (n = 3).
Significant Alterations of Genes Related to either Implantation or Placenta Formation
In order to determine whether genes involved in implantation or placenta formation displayed significant alteration, we performed gene clustering on the microarray results. As the gene clustering is shown in Figure S2 in Supplementary Material, 11 genes of 33 implantation-related genes displayed significant alterations (p < 0.05). Of 11 significantly altered genes, three genes such as Bsg, Pcsk5, and Klf9 were up-regulated, whereas Tgfβr2, Pparδ, Ptgs2, Grn, and Fkbp4 were significantly down-regulated.
The results of gene clustering for placenta formation are shown in Figure S3 in Supplementary Material. Twenty-four genes of 107 placenta formation-related genes displayed significant alterations (p < 0.05). Of 24 significantly altered genes, nine genes Txnrd1, Pgf, Pkd2, Hif1a, Ccnf, Plac9, Prdx3, Adm, and Pbrm1 were up-regulated, whereas Plac8, Prdm1, Rspo3, Tmed2, Dcn, Epas1, and Stk3 were significantly down-regulated. These results indicate that these genes involved in implantation and placenta formation may be controlled under circadian clock.
Up-Regulation and Down-Regulation of Genes Related to Cell Growth and Apoptosis
Of fold-changed genes (Table 4), we focused on Inhβb and Igf1 that were regulated with opposite directionality. Both Inhβa and Inhβb were high in signal intensity, whereas Inha was low, indicating that activin, but not inhibin, is highly produced in the UESCs. The Fst gene coding the activin-binding protein follistatin was down-regulated. In contrast to Inhβa and Inhβb, Igf1 was down-regulated, whereas its binding proteins Igfbp3 and Ifgbp4 were rather increased (Figure 4). Consequently, the activities of activin and IGF1 may be differentially regulated during circadian rhythms. During active oscillation of circadian clock, however, the apoptosis-related genes Fas and Caspase3 remained no significant changes (Figure 4).
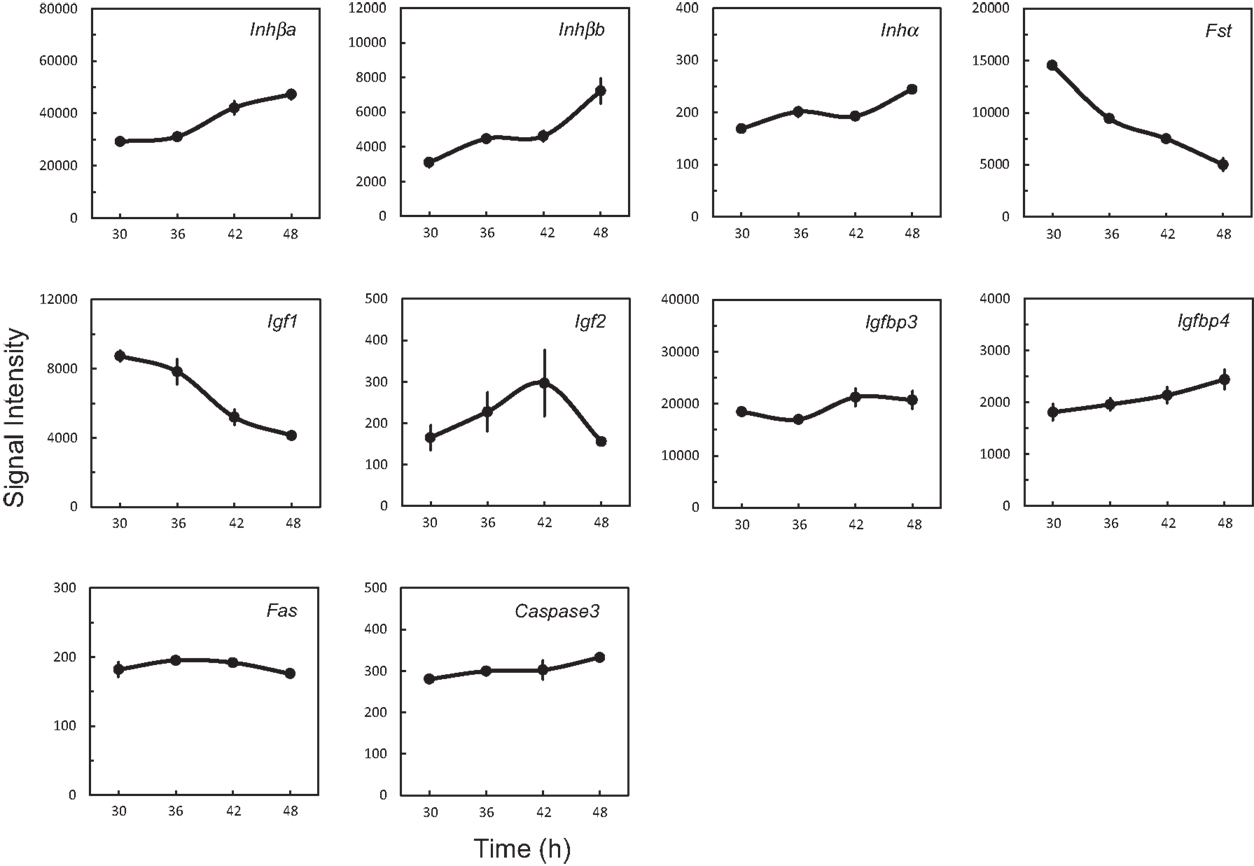
Figure 4. Characteristic expression profiles of genes coding growth factors, their binding proteins, and apoptosis-related proteins. Each value represents the means ± SEM (n = 3) of signal intensity from the microarray results.
Effect of Bmal1 Knockdown on the Expression of Genes Related to Cell Growth and Apoptosis in the Rat UESCs
In order to suppress the cellular clock in the UESCs, Per2-dLuc oscillations were investigated using Bmal1 siRNA (siRNA) or no silencing RNA (CONT). The UESCs transfected with either RNA displayed several Per2-dLuc oscillations. A decline of Per2-dLuc bioluminescence oscillation and significantly decreased amplitude (p < 0.01, versus CONT) were observed in Bmal1 siRNA treated cells (Figure 5A). There was not a significant effect of Bmal1 siRNA treatment on the peak time of the first phase and the cycle time (data not shown). The transcript levels of core clock genes were estimated at the peak time of the first phase. The results are shown in Figure 5B. Bmal1 mRNA expression was inhibited after transfection with Bmal1 siRNA, the Bmal1 transcript level being reduced by 60% compared with that in the CONT group. Rev-erbα and Dbp are regarded as core circadian oscillator genes that fall under the regulation of Bmal1-Clock heterodimer through E-box elements located in their promoter regions. Thus Rev-erbα and Dbp were significantly down-regulated in Bmal1 siRNA cells.
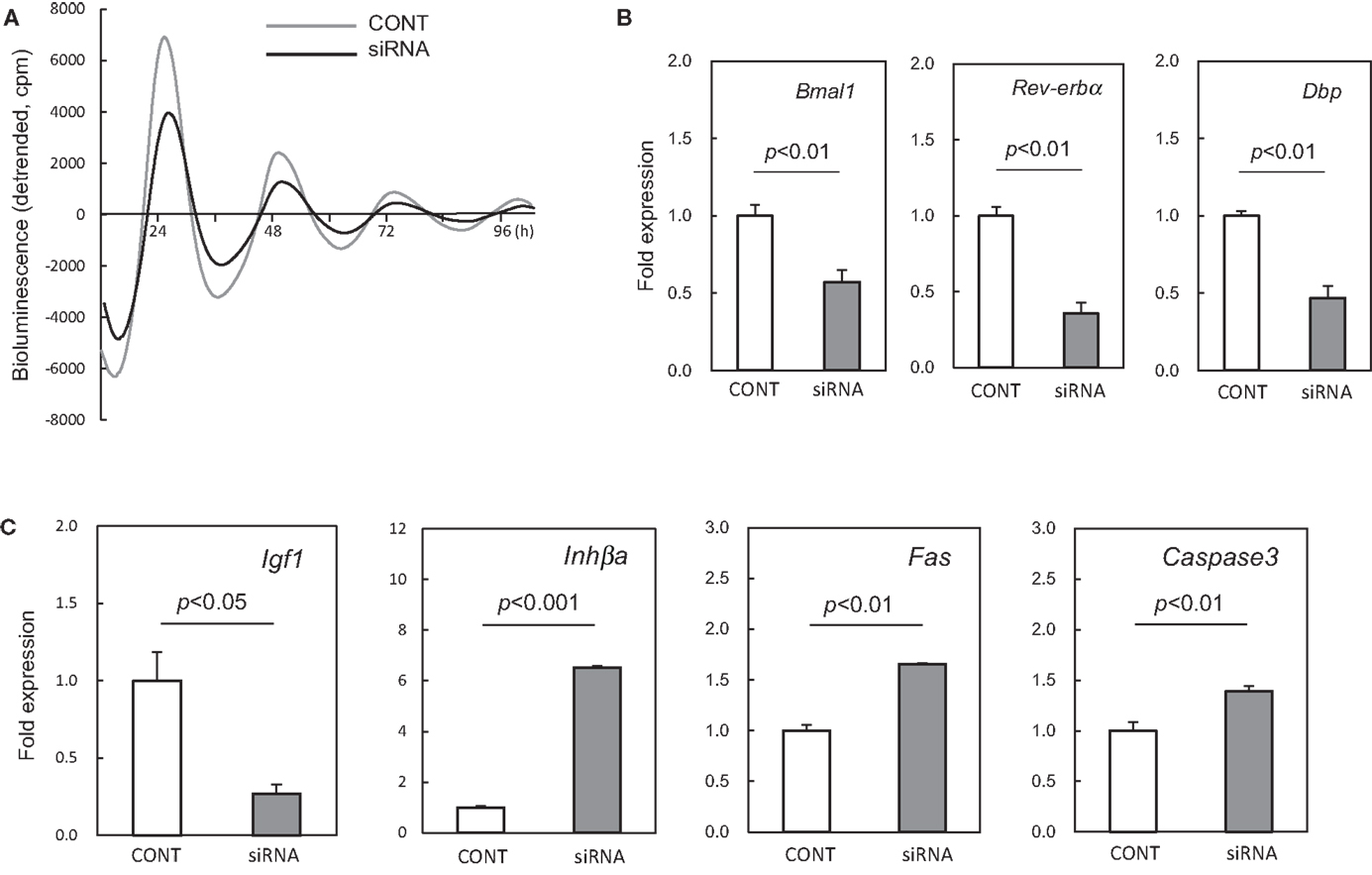
Figure 5. Effect of Bmal1 siRNA treatment on expression of putative clock-controlled genes in cultured uterus endometrial stromal cells. (A) The UESCs were treated with Bmal1 siRNA (siRNA) or no silencing RNA (CONT) according to the indicated protocols. The cells were then synchronized with dexamethasone for bioluminescence determination (time: 0 h), and then monitored Per2-dLuc oscillations. (B) RNA samples were collected 39 h after synchronization, and RT-qPCR analyses of transcript levels were performed using their specific primers. (C) Transcript levels of Igf1, Inhβa, Fas, and Caspase3 were measured in the UESCs treated with siRNA or no silencing RNA. Each value represents the means ± SEM (n = 3).
In addition, we observed that the mRNA expression of several uterus genes (Igf1, Inhβa, Fas, and Caspase3) was differentially affected by the Bmal1 siRNA treatment. The expression of Igf1 gene was significantly down-regulated by Bmal1 silencing (p < 0.05), whereas the Inhβa was up-regulated approximately sevenfold (p < 0.001) (Figure 5C). Although Fas, and Caspase3 did not exhibit cellular circadian rhythms as described above, these genes were significantly up-regulated by Bmal1 silencing (p < 0.01).
Discussion
The present analysis of circadian genes regulated by clockwork system significantly contributes to our understanding of the relationship of cellular circadian clock with implantation and decidualization. Our present study on cultured UESCs of pregnant rats is the first to analyze the global gene expression profiles using DNA microarray technology. We found the specific expression profiles of 18 clock genes and two protein kinase genes. Bmal1 (Arntl) and Rev-erbα (Nr1d1) displayed typically anti-phase expression patterns, showing that the proper range of clock gene oscillation was successfully covered. We identified 7,235 significantly altered genes in the UESCs and 559-fold-changed genes with up or down directionality. The biological pathway analysis using the DAVID online annotation software revealed that many genes involved in cell cycle (38 genes), glutathione metabolism (19 genes), MAPK signaling pathway (55 genes), and so on were significantly expressed with circadian regulation. In addition, some genes involved in implantation or placenta formation displayed significant alteration with up or down directionality: 11 genes involved in implantation and 24 genes involved in placenta formation.
Many genes were listed as the up-regulated genes compared to the RNA sample at 30 h, including Inhβb, Gdf10 (BMP-3), Gdf15 (MIC-1), Dbp, Gpr4, and Olr1. Of 559-fold-changed genes, Gdf10, a member of TGFβ superfamily related to BMP-3 (32), is required for normal decidual development during the post-implantation period (33). Gdf15 (MIC-1) is expressed in the decidual stromal cells and trophoblasts (34). GPR4, a proton-sensing G-protein-coupled receptor, is also expressed in human umbilical vein endothelial cells (35). GPR4 signaling regulates endothelial cell adhesion through the cAMP pathway. It is known that the oxidized low density lipoprotein receptor 1 (Olr1) regulates growth of a variety of cells and is important in inflammation, oxidative stress, and tissue remodeling (36). However, the expression and regulation of Olr1 remain unclear in the uterus. Aquaporin-1 (Aqp1) is proposed as a mediator of estrogen-induced angiogenesis in breast cancer and endometrial cancer (37). The expression of Aqp1 was reported in the pregnant rat uterus (38, 39). Although more than 10 Aqp1 have been identified up to date, only Aqp1 was expressed in the cultured UESCs.
In contrast, the down-regulated representative genes with fold change were also listed, including Fst, Igf1, Zmiz1, Fbxo9, Rgs2, Rgs5, Mt1a, and Mt2A. IGF family genes including Igf1 are differentially expressed throughout gestation, and especially the IGF system contributes phenotypic and functional changes of myometrium smooth muscle cells (40). The overexpression of Zmiz1, recently identified as a candidate oncogene, was reported in human breast, ovarian, and colon cancers (41). Zmiz1 was also reported to function in regulating the activity of the androgen receptor as a co-activator (42). However, the expression and function of Zmiz1 are unclear in the uterus. Fbxo9 was reported to contribute survival of myeloma cells (43). Rgs2 and Rgs5, regulators of G-protein signaling proteins, are abundantly expressed in pregnant human myometrium (44). Rgs2 was reported to up-regulate in response to stress (45). The expression of Mt1a and Mt2A, typical metal response parameters, was reported in the proliferative phase of endometrium (46).
Of up-regulated and down-regulated genes, we focused on the expression of Inhβb, Inhβa, and Igf1 that displayed fold change with up or down directionality. Both the Inhβa and Inhβb genes showed high signal intensities of DNA microarray, whereas Inhα was low. This result indicates the expression of activin. The binding protein of activin, follistatin (Fst), was down-regulated, indicating the relative action of activin is increased. Expression of activin A was increased with reduced stromal cell mitosis, tissue growth, and mitogenic signaling in the decidual endometrium (47). We analyzed using Bmal1 siRNA whether the expression of Inhβa and Igf1 was controlled under circadian clock. Transfection of Bmal1 siRNA into the UESCs induced a significant decline of Per2-dLuc bioluminescence oscillation. However, the transfection did not alter other parameters in the oscillation. The core clock genes Rev-erbα and Dbp as well as Bmal1 were down-regulated. The expression of Igf1 was significantly decreased after Bmal1 silencing, suggesting that Igf1 is positively regulated by circadian clock. Conversely, the expression of Inhβa was enhanced approximately sevenfold after Bmal1 silencing, suggesting that Inhβa is negatively regulated by circadian clock. Although both Inhβa and Igf1 are clock-controlled genes, thus, the two gene are differentially regulated. Rev-erbα is a critical component of circadian clock (48). It acts as a domain clock-controlled gene under the regulation of Bmal1-Clock heterodimer; in turn it has been shown to suppress Bmal1 transcription through binding to RORα response elements (RORE) presenting in Bmal1 promoters (49). Recent studies have shown that Rev-erbα is a transcriptional silencer (49, 50). However, Rev-erbα increases the transcription of Star by binding to its agonist heme (51).
In the uterus endometrial cells of pregnant rats, the early proliferative phase is characterized by tissue remodeling, angiogenesis, and modulation of inflammation; the mid-proliferative phase is characterized not only by proliferation but also marks the onset of expression of genes required for endometrial receptivity. Thus, cell growth and apoptosis support the process of tissue remodeling and implantation. The death receptor Fas/ligand system is a key regulator of apoptotic cell death and irregularity of this signaling pathway has been shown to participate in immune-mediated β-cell apoptosis (52, 53). It is also known that the expression level of the Fas gene is down-regulated in a variety of malignancies including malignant melanoma, adenocarcinoma, and squamous cell carcinoma (54, 55). In the cultured UESCs, however, the expression of Fas and Caspase3 displayed no or small changes. Interestingly, these genes were significantly up-regulated after Bmal1 silencing, similar to Inhβa, suggesting that these apoptosis-related genes are suppressed during active circadian rhythms of clock genes. It is possible that dysfunction of the circadian clockwork during the stage of decidualization is necessary to increase or decrease expression of clock-controlled genes for formation of the placenta.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (B) from the Japan Society for the Promotion of Sciences (JSPS; 22380152) (to Masa-aki Hattori).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Systems_and_Translational_Endocrinology/
10.3389/fendo.2013.00082/abstract
Figure S1. Clustering of clock genes on the microarray results. The expression profiles of clock genes (closed circle) were divided into four groups (1−4). Red, relatively high expression; green, relatively low expression.
Figure S2. Clustering of implantation-related genes on the microarray results. Genes showing with significant alterations (p < 0.05) are listed. Red, relatively high expression; green, relatively low expression.
Figure S3. Clustering of placenta formation-related genes on the microarray results. Genes showing with significant alterations (p < 0.05) are listed. Red, relatively high expression; green, relatively low expression.
References
1. Storch K-F, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, et al. Extensive and divergent circadian gene expression in liver and heart. Nature (2002) 417:78–83. doi:10.1038/nature744
2. Lemos DR, Downs JL, Urbanski HF. Twenty-four-hour rhythmic gene expression in the rhesus macaque adrenal gland. Mol Endocrinol (2006) 20:1164–76. doi:10.1210/me.2005-0361
3. Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet (2006) 40:409–48. doi:10.1146/annurev.genet.40.110405.090603
4. Ma K, Xiao R, Tseng H-T, Shan L, Fu L, Moore DD. Circadian dysregulation disrupts bile acid homeostasis. PLoS One (2009) 4:e6843. doi:10.1371/journal.pone.0006843
5. Alvarez JD, Sehgal A. The thymus is similar to the testis in its pattern of circadian clock gene expression. J Biol Rhythms (2005) 20:111–21. doi:10.1177/0748730404274078
6. He PJ, Hirata M, Yamauchi N, Hashimoto S, Hattori M-A. The disruption of circadian clockwork in differentiating cells from rat reproductive tissues as identified by in vitro real-time monitoring system. J Endocrinol (2007) 193:413–20. doi:10.1677/JOE-07-0044
7. Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell (2001) 107:855–67. doi:10.1016/S0092-8674(01)00610-9
8. Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell (2002) 109:307–20. doi:10.1016/S0092-8674(02)00722-5
9. Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol (2002) 12:551–7. doi:10.1016/S0960-9822(02)00765-0
10. Kornmann B, Preitner N, Rifat D, Fleuury-Olela F, Schibler U. Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res (2001) 29:E51–1. doi:10.1093/nar/29.11.e51
11. Nakamura TJ, Moriya T, Inoue S, Shimazoe T, Watanabe S, Ebihara S, et al. Estrogen differentially regulates expression of Per1 and Per2 genes between central and peripheral clocks and between reproductive and nonreproductive tissues in female rats. J Neurosci Res (2005) 82:622–30. doi:10.1002/jnr.20677
12. Dolatshad H, Campbell EA, O’Hara L, Maywood ES, Hastings MH, Johnson MH. Developmental and reproductive performance in circadian mutant mice. Hum Reprod (2006) 21:68–79. doi:10.1093/humrep/dei313
13. He PJ, Hirata M, Yamauchi N, Hashimoto S, Hattori M-A. Up-regulation of Per1 expression by estradiol and progesterone in the rat uterus. J Endocrinol (2007) 194:511–9. doi:10.1677/JOE-07-0172
14. Hirata M, He PJ, Shibuya N, Uchikawa M, Yamauchi N, Hashimoto S, et al. Progesterone, but not estradiol, synchronizes circadian oscillator in the uterus endometrial stromal cells. Mol Cell Biochem (2009) 324:31–8. doi:10.1007/s11010-008-9981-4
15. Akiyama S, Ohta H, Watanabe S, Moriya T, Hariu A, Nakahata N, et al. The uterus sustains stable biological clock during pregnancy. Tohoku J Exp Med (2010) 221:278–98. doi:10.1620/tjem.221.287
16. Clarke CL, Sutherland RL. Progestin regulation of cellular proliferation. Endocr Rev (1990) 11:266–301. doi:10.1210/edrv-11-2-266
17. Zhang Z, Funk C, Glasser SR, Mulholland J. Progesterone regulation of heparin-binding epidermal growth factor-like growth factor gene expression during sensitization and decidualization in the rat uterus: effects of antiprogestins. Endocrinology (1994) 135:1256–63. doi:10.1210/en.135.3.1256
18. Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, et al. Molecular cues to implantation. Endocr Rev (2004) 25:341–73. doi:10.1210/er.2003-0020
19. Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell (2001) 105:25–42. doi:10.1016/S0092-8674(01)00294-X
20. Miller BH, Olson SL, Turek FW, Levine JE, Horton TH, Takahashi JS. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr Biol (2004) 14:1367–73. doi:10.1016/j.cub.2004.07.055
21. Ratajczak CK, Boehle KL, Muglia LJ. Impaired steroidogenesis and implantation failure in Bmal1−/− mice. Endocrinology (2009) 150:1879–85. doi:10.1210/en.2008-1021
22. Pilorz V, Steinlechner S. Low reproductive success in Per1 and Per2 mutant mouse females due to accelerated ageing? Reproduction (2008) 135:559–68. doi:10.1530/REP-07-0434
23. Uchikawa M, Kawamura M, Yamauchi N, Hattori M-A. Down-regulation of circadian clock gene period 2 in uterine endometrial stromal cells of pregnant rats during decidualization. Chronobiol Int (2011) 28:1–9. doi:10.3109/07420528.2010.522289
24. Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer (2003) 3:350–61. doi:10.1038/nrc1072
25. Lévi F, Okyar A, Dulong S, Innominato PF, Clairambault J. Circadian timing in cancer treatments. Annu Rev Pharmacol Toxicol (2010) 50:377–421. doi:10.1146/annurev.pharmtox.48.113006.094626
26. Sun CM, Huang SF, Zeng JM, Liu DB, Xiao Q, Tian WJ, et al. Per2 inhibits k562 leukemia cell growth in vitro and in vivo through cell cycle arrest and apoptosis induction. Pathol Oncol Res (2010) 16:403–11. doi:10.1007/s12253-009-9227-0
27. Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, et al. A transcription factor response element for gene expression during circadian night. Nature (2002) 418:534–9. doi:10.1038/nature00906
28. Oozono S, Yamauchi N, Nishimura K, Matsumoto K, Watanabe R, Kubota K, et al. Expression of rat uterine serine proteinases homologous to mouse implantation serine proteinase 2. J Exp Zool (2008) 310B:642–9. doi:10.1002/jez.b.21237
29. Matsumoto K, Yamauchi N, Watanabe R, Oozono S, Kubota K, Nishimura K, et al. In vitro decidualization of rat endometrial stromal cells. Cell Tissue Res (2009) 335:575–86. doi:10.1007/s00441-008-0734-1
30. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc (2009) 4:44–57. doi:10.1038/nprot.2008.211
31. Sato F, Kurokawa M, Yamauchi N, Hattori M-A. Gene silencing of myostatin in differentiation of chicken embryonic myoblasts by small interfering RNA. Am J Physiol Cell Physiol (2006) 291:C538–45. doi:10.1152/ajpcell.00543.2005
32. Cunningham NS, Jenkins NA, Gilbert DJ, Copeland NG, Reddi AH, Lee SJ. Growth/differentiation factor-10: a new member of the transforming growth factor-beta superfamily related to bone morphogenetic protein-3. Growth Factors (1995) 12:99–109. doi:10.3109/08977199509028956
33. Rahman MA, Li M, Li P, Wang H, Dey SK, Das SK. Hoxa-10 deficiency alters region-specific gene expression and perturbs differentiation of natural killer cells during decidualization. Dev Biol (2006) 290:105–17. doi:10.1016/j.ydbio.2005.11.016
34. Segerer SE, Rieger L, Kapp M, Dombrowski Y, Müller N, Dietl J, et al. MIC-1 (a multifunctional modulator of dendritic cell phenotype and function) is produced by decidual stromal cells and trophoblasts. Hum Reprod (2012) 27:200–9. doi:10.1093/humrep/der358
35. Chen A, Dong L, Leffler NR, Asch AS, Witte ON, Yang LV. Activation of GPR4 by acidosis increases endothelial cell adhesion through the cAMP/Epac pathway. PLoS One (2011) 6:e27586. doi:10.1371/journal.pone.0027586
36. Wang X, Khaidakov M, Ding Z, Mitra S, Lu J, Dai Y, et al. Lectin-like oxidized low-density lipoprotein receptor-1 (LOX-1) and cardiac fibroblast growth. Hypertension (2012) 60:1437–42. doi:10.1161/HYPERTENSIONAHA.112.200659
37. Zou LB, Shi S, Zhang RJ, Wang TT, Tan YJ, Zhang D, et al. Aquaporin-1 plays a crucial role in estrogen-induced tubulogenesis of vascular endothelial cells. J Clin Endocrinol Metab (2013) 98(4):E672–82. doi:10.1210/jc.2012-4081
38. Lindsay LA, Murphy CR. Aquaporins-1 increases in the rat myometrium during early pregnancy. J Mol Histol (2004) 35:75–9. doi:10.1023/B:HIJO.0000021066.44364.81
39. Lindsay LA, Murphy CR. Redistribution of aquaporins 1 and 5 in the rat uterus is dependent on progesterone: a study with light and electron microscopy. Reproduction (2006) 131:369–78. doi:10.1530/rep.1.00914
40. Shynlova O, Tsui P, Dorogin A, Langille BL, Lye SJ. Insulin-like growth factors and their binding proteins define specific phases of myometrial differentiation during pregnancy in the rat. Biol Reprod (2007) 76:571–8. doi:10.1095/biolreprod.106.056929
41. Rogers LM, Riordan JD, Swick BL, Meyerholz DK, Dupuy AJ. Ectopic expression of Zmiz1 induces cutaneous squamous cell malignancies in a mouse model of cancer. J Invest Dermatol (2013) 133(7):1863–69. doi:10.1038/jid.2013.77
42. Li X, Zhu C, Tu WH, Yang N, Qin H, Sun Z. ZMIZ1 preferably enhances the transcriptional activity of androgen receptor with short polyglutamine tract. PLoS One (2011) 6:e25040. doi:10.1371/journal.pone.0025040
43. Fernández-Sáiz V, Targosz BS, Lemeer S, Eichner R, Langer C, Bullinger L, et al. SCFFbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nat Cell Biol (2013) 15:72–81. doi:10.1038/ncb2651
44. Ladds G, Zervou S, Vatish M, Thornton S, Davey J. Regulators of G protein signalling proteins in the human myometrium. Eur J Pharmacol (2009) 610:23–8. doi:10.1016/j.ejphar.2009.03.042
45. Nguyen CH, Zhao P, Sobiesiak AJ, Chidiac P. RGS2 is a component of the cellular stress response. Biochem Biophys Res Commun (2012) 426:129–34. doi:10.1016/j.bbrc.2012.08.050
46. Petracco RG, Kong A, Grechukhina O, Krikun G, Taylor HS. Global gene expression profiling of proliferative phase endometrium reveals distinct functional subdivisions. Reprod Sci (2012) 19:1138–45. doi:10.1177/1933719112443877
47. Kashiwagi A, DiGirolamo CM, Kanda Y, Niikura Y, Esmon CT, Hansen TR, et al. The postimplantation embryo differentially regulates endometrial gene expression and decidualization. Endocrinology (2007) 148:4173–84. doi:10.1210/en.2007-0268
48. Triqueneaux G, Thenot S, Kakizawa T, Antoch MP, Safi R, Takahashi JS, et al. The orphan receptor Rev-erbα gene is a target of the circadian clock pacemaker. J Mol Endocrinol (2004) 33:585–608. doi:10.1677/jme.1.01554
49. Preitner N, Damiola F, Lopez-Molina L, Zakany L, Duboule D, Albrecht U, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell (2002) 110:251–60. doi:10.1016/S0092-8674(02)00825-5
50. Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms (2005) 20:391–403. doi:10.1177/0748730405277232
51. Chen H, Chu G, Zhao L, Yamauchi N, Shigeyoshi Y, Hashimoto S, et al. Rev-erbα regulates circadian rhythms and StAR expression in rat granulosa cells as identified by the agonist GSK4112. Biochem Biophys Res Commun (2012) 420:374–9. doi:10.1016/j.bbrc.2012.02.164
52. Eizirik DL, Mandrup-Poulsen T. A choice of death – the signal-transduction of immune-mediated β-cell apoptosis. Diabetologia (2001) 44:2115–33. doi:10.1007/s001250100021
53. Lavrik IN, Krammer PH. Regulation of CD95/Fas signaling at the DISC. Cell Death Differ (2012) 19:36–41. doi:10.1038/cdd.2011.155
54. Leithäuser F, Dhein J, Mechtersheimer G, Koretz K, Brüderlein S, Henne C, et al. Constitutive and induced expression of APO-1, a new member of the nerve growth factor/tumor necrosis factor receptor superfamily, in normal and neoplastic cells. Lab Invest (1993) 69:415–29.
Keywords: uterus endometrial stromal cells, clock genes, clock-controlled genes, DNA microarray, implantation, decidualization, Bmal1 siRNA, Per2-dLuc reporter gene
Citation: Tasaki H, Zhao L, Isayama K, Chen H, Yamauchi N, Shigeyoshi Y, Hashimoto S and Hattori MA (2013) Profiling of circadian genes expressed in the uterus endometrial stromal cells of pregnant rats as revealed by DNA microarray coupled with RNA interference. Front. Endocrinol. 4:82. doi: 10.3389/fendo.2013.00082
Received: 29 March 2013; Accepted: 20 June 2013;
Published online: 08 July 2013.
Edited by:
James Olcese, Florida State University College of Medicine, USAReviewed by:
James Olcese, Florida State University College of Medicine, USAPatrick Chappell, Oregon State University, USA
Copyright: © 2013 Tasaki, Zhao, Isayama, Chen, Yamauchi, Shigeyoshi, Hashimoto and Hattori. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Masa-aki Hattori, Department of Animal and Marine Bioresource Sciences, Graduate School of Agriculture, Kyushu University, Hakozaki 6-10-1, Higashi-ku, Fukuoka 812-8581, Japan e-mail:bWhhdHRvcmlAYWdyLmt5dXNodS11LmFjLmpw
†Hirotaka Tasaki and Lijia Zhao have contributed equally to this work.