- 1Department of Clinical Medicine and Surgery, Unesco Chair Health Education and Sustainable Development, Federico II University of Naples, Naples, Italy
- 2Department of Precision Medicine, University of Campania “Luigi Vanvitelli”, Naples, Italy
- 3Department of Clinical Medicine and Surgery, Federico II University of Naples, Naples, Italy
- 4Department of Advanced Medical and Surgical Sciences, University of Campania “Luigi Vanvitelli”, Naples, Italy
Neuroendocrine tumors, or NETs, are cancer originating in neuroendocrine cells. They are mostly found in the gastrointestinal tract or lungs. Functional NETs are characterized by signs and symptoms caused by the oversecretion of hormones and other substances, but most NETs are non-functioning and diagnosis in advanced stages is common. Thus, novel diagnostic and therapeutic strategies are warranted. Epigenetics may contribute to refining the diagnosis, as well as to identify targeted therapy interfering with epigenetic-sensitive pathways. The goal of this review was to discuss the recent advancement in the epigenetic characterization of NETs highlighting their role in clinical findings.
Introduction
Neuroendocrine neoplasms (NENs) are a heterogeneous group of malignancies originating from neuroendocrine cells diffuse throughout the body. The gastroenteropancreatic (GEP) tract and the bronchopulmonary system represent the main site of origin. NENs are mostly sporadic, but in 10–30% they can arise within the context of familial syndromes, mainly multiple endocrine neoplasia type 1 (MEN1) (1). Incidence and prevalence of NENs have markedly increased in the last decades, irrespective of stage and grade (2). Clinical presentation and prognosis of NENs may widely vary. NENs can be functional when they release biologically active hormones that cause distinct clinical syndromes or more often may be non-functional, thus diagnosed incidentally or due to mass effect. Delayed diagnosis is common, as well as the detection of metastases, mainly to the liver, already at diagnosis. Patients with localized disease have a better prognosis, with 5-year survival ranging from 78 to 93%, while in metastatic disease, the 5-year survival is worse (19–38%), although improved over the past years (3). The improvement of survival rates may be the consequence of the availability of effective therapies, as well as earlier and more accurate clinical and pathologic diagnoses with relative downstaging. NENs have usually an indolent course and patients need life-long therapy. Notably, the landscape of the therapeutic options in NENs has considerably expanded in the last decades. The current systemic therapies for locally advanced or metastatic NENs include somatostatin analogs (SSAs), molecular targeted therapy with mTOR inhibitors (Everolimus), or anti-angiogenesis (Sunitinib), peptide receptor radionuclide therapy (PRRT) with either 90Yttrium (90Y) or 177Lutetium (177Lu) and chemotherapies with temozolomide, capecitabine or platinum-based regimens. These options can be used in sequence or association with surgery, locoregional treatments (e.g., radiofrequency ablation, cryoablation, chemoembolization, and radioembolization), and/or other drugs used as supportive therapies (e.g., telotristat, diazoxide and proton pump inhibitors) (4, 5). In this review we will focus on well or moderately differentiated neuroendocrine tumors (NETs), excluding neuroendocrine carcinomas (NEC) for their peculiar pathology and treatment.
Epigenetic Modifications and Neuroendocrine Tumors
Epigenetic changes, such as DNA methylation and histone modification, are critical for regulating genes and non-coding RNA expression. Genomic alterations and gene mutations which are involved in the pathogenesis NETs, as MEN1, VHL-hypoxia-inducible factor, RASSF1A, have a consequence on the aberrant placement of epigenetic markers and related pathways (6–10).
Epigenetic mechanisms can modify gene expression altering DNA methylation status, histones post trascriptional modifications, and influencing the expression of non-coding RNAs. Hypermethylation of a promoter is a mechanism that determined gene silencing, while hypomethylation can lead to chromosomal instability and consequently influences gene expression (9, 10). Histone modifications involves the addition of methyl, acetyl, phosphorylation at different aminoacid residues of histone proteins. These modifications alter chromatin accessibility to transcription factors and lastly gene expression. MicroRNAs (miRNAs) and long noncoding RNAs are other layers of epigenetic regulation. They are small, or long sequences of non-coding RNAs regulating gene expression post-transcriptionally, considered to be a cancer-associated epigenetic mechanism (11).
Methylation Patterns Relevance in the Pathogenesis of NETs and Clinical Findings
The pathogenesis of NETs is further to be elucidated, as in most other solid tumors. Nevertheless, epigenetic studies have improved our knowledge. Pancreatic neuroendocrine tumors (PNETs) account for 1 to 2% of all pancreatic tumors and most of them are sporadic and non-functioning, 5–7% arise within inherited syndromes, including MEN1, Von-Hippel Lindau (VHL) syndrome, neurofibromatosis type 1 (NF1), and tuberous sclerosis. The majority of familial PNETs are caused by germline inactivating mutations in the MEN1 gene, suggesting a key role in PNETs tumorigenesis. MEN1 gene encodes the transcription factor MENIN, ubiquitously expressed, and involved in many biological functions. MENIN, plays an essential role in chromatin remodeling and gene expression recruiting the H3K4me3 histone methyltransferase on mixed-lineage leukemia (MLL1) complex, regulating the expression of the cyclin-dependent kinase inhibitors, and influenced the epigenetic regulation of several genes (12). MEN1 mutations or loss of function deregulated cell growth in 75% cases of PNETs favoring hypermethylation of several tumor suppressor genes including RASSF1A (13), HIC-1, MLH1, CDKN2A, and MGMT (6, 7). Characteristics of the sporadic form of PNETs are mainly gene mutations in DAXX (death-domain-associated protein) or ATRX (alpha thalassemia/mental retardation syndrome X-linked) (12). Both DAXX and ATRX are chromatin remodellers and are involved in the incorporation of the histone variant H3.3 at the telomeres and pericentric heterochromatin necessary (14). Proteins loss, as well as mutations in DAXX or ATRX, are associated with chromosome instability (CIN), reduced genomic H3K9me, and aggressive PNET phenotype (12, 15). Increased risk of PNET was also associated with loss of chromosome 11q containing the genes Men1, but also DNA repair pathway genes as BRCA2 and ATM, and amplification region activating PIK3CA and mTOR pathway. In some cases associated with MENIN loss were also found mutation affecting VHL tumor suppressor gene that determined a constitutive hypoxia transcription factors (HIF) activation and uncontrolled angiogenesis (16, 17), suggesting that MENIN loss or mutation is a key initiator in PNET tumorigenesis (15, 18–21). In pulmonary NET in addition to MEN1 mutations affected also as histone lysine methyltransferase (SETD1B2, SETDB1), histone acetylation modifiers (BRWD3 and HDAC5) and ATP-dependent chromatin remodeling SMARCA1 indicating a key pathogenic role (22). Genomic profile of small intestinal NET (SI-NET) identify two different groups, one characterized by loss of chromosome 18, and another one characterized by the presence of chromosome 18 but with clustered gains on chromosomes 4, 5, 7, 14, and 20 (23). Correlation of loss of chromosome 18 and RASSF1A promoter hyper-methylation and hypo-methylation of long intergenic element 1 (LINE1) and ALU sequences was found in SI-NETs (24) although not associated with grade and tumor size (25).
In hereditary SI-NET causative role was attributed to germline mutations in IPMK (inositol polyphosphate multikinase) p53 activity and MutY DNA glycosylase genes (26) affecting the oxidative pathway. Above mentioned studies emerged that in SI-NET epigenetic machinery is not causative however the uncontrolled pathways of oxidative stress and genomic rearrangement activated several epigenetic modifications (27).
Methylation Patterns Relevance as Differential Diagnostic Biomarkers
The DNA methylation profile of sporadic PNET, VHL and MEN1-related PNETs, and pancreatic islets were analyzed by Illumina array (850k array) with the goal to find novel diagnostic markers. The study identified a distinct cluster of methylation genes associated with VHL, sporadic and MEN1-related PNETs, indicated that mutations in these genes influence the epigenetic pathway and clinical presentation of diseases (28, 29). Differential methylation patterns were also reported among GEP-NETs (24, 30). Indeed, the analysis conducted in 60 tumors selected a pool of 807 genes. These gene sets were able to distinguish NETs in functional GEP-NETs (insulinoma, gastrinoma) and, non-function subtypes underlying the clinical and histological characteristics. Gastrinomas showed hypomethylation of genes including metalloproteinases (MMP1, MMP3, TIMP2, TIMP3), the serpin family (SERPINA5, SERPINB5), and oncogenes (IL2, MCF2, and MOS), whereas hypermethylation was reported for tumor suppressors (SMARCB1, CASP8, and NBL1) (24, 25, 30). Promoter hypermethylation of the IGF2 pathway was characteristic of insulinomas shedding a light on signaling responsible for their differentiation from a common origin (31). A study on SI-NET identified TCEB3C gene hypermethylation to be specific for this histology. Interestingly, treatment of SI-NET cell lines with the de-methylating agent decitabine and the histone methyltransferase inhibitor 3-deazaneaplaoncin A-induced TCEB3C re-expression, confirming an epigenetic regulation of this gene (32). Followed this stem study, Verdugo et al. and then Karpathakis et al. identified hyper-methylation of the gastric inhibitory polypeptide receptor (GIPR) as another specific marker of SI-NETs and reported hyper-methylation in several genes. They selected on chromosome 18 as laminin alpha 3 (LAMA3), serpin peptidase inhibitor clade B member 5 (SERPINB5), and factor receptor superfamily member 11a NFKB activator (RANK or TNSFRSF11A), suggesting that epigenetic silencing could be the possible second step in tumor development upon chromosome 18 loss (33–35).
Methylation Patterns: Relevance in Prognosis and Response to Therapy
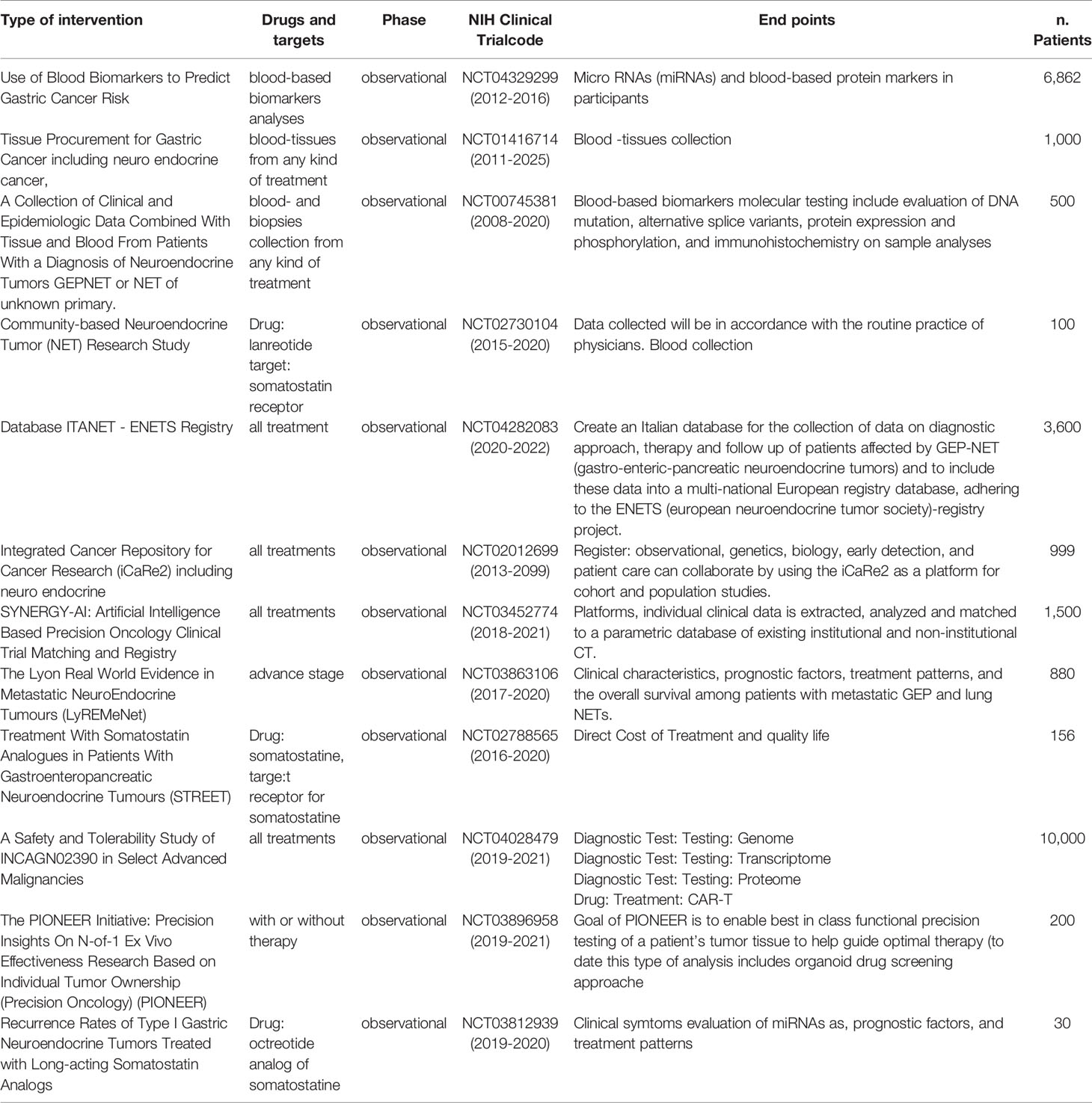
Since epigenetic changes play a key role in the progression of PNETs the finding to select an epigenetic prognostic factor, is crucial (34). In particular, some epigenetic changes area correlated to DAXX or ATRX protein loss because this complex regulates H3K9me and influenced DNA methylase. Indeed, promoter hypermethylation of RASSF1A and p57Kip2 in PNENs was responsible for NAP1L1 overexpression associated with the metastatic phenotype (35–38). Additionally, a peculiar group of PNETs named (CIMP) showed hypermethylation of CpG islands including tumor suppressor genes, such as RASSF1A, hMLH1, and hypomethylation of LINE-1 sequence. These peculiar epigenetic pathways were associated with poor prognosis and advanced stage of PNETs (39). While hypermethylation ofCDKN2A was associated with early tumor recurrence and poor outcomes of GEP-NETs (40). A general decrease in methylation levels was observed in SI-NET metastases compared to the primary tumors. In particular, differential methylation of AXL, CRMP1, FGF5, CXXC5, and APOBEC3C genes were detected in primary tumors compared to metastases (34). However, no validation of these markers was reported in the study population. In a follow-up of primary SI-NET and liver metastases, it was selected a panel of epigenetically dysregulated genes that were progressively methylated or demethylated from the primary tumor to metastases (33, 41), suggesting their potential use as markers. Recently dysregulation of TET1/TET2 enzymes that catalyze DNA demethylation was observed in SI-NETs open a potential novel class of drug treatment (42, 43). Differential methylations of specific gene promoters were also associated with response to therapy. Table 1 shows the most representative observational studies involving epigenetic biomarkers. One example is O6-methylguanine-DNA methyltransferase (MGMT), a DNA repair enzyme removing alkyl groups from an alkylguanine. Retrospective studies have found an association between methylation of MGMT and response to treatment with temozolomide (an alkylating agent) making it a promising marker (44–47) (Table 1). A prospective trial confirmed this correlation (48).
miRNAs Relevance in Differential Diagnosis and Prognosis
MicroRNAs (miRNAs) are small (19–24 nt) regulatory RNA molecules that can also be used to classify cancer because of their abundance, cell-type, and disease-stage specificity which support their possible use to predict clinical outcomes and differential diagnosis. Multiple miRNA profiling studies have been performed on NET pathological types using different RNA isolation, detection, and analysis methods. Although these differences complicate inter-study comparisons, miRNAs still hold much promise as markers. A set of 10 miRNAs (miR-99a, 99b, 100, 125a, 125b-1, 125b-2, 129-2, 130a, 132, and 342) was selected as a potential tool to differentiate pancreatic NEN from pancreatic acinar cell carcinoma (49), while miR-21a was selected as potential biomarker for GEP-NETs (50). Moreover, in another study in insulinomas, miR-204 was the unique miRNA selectively overexpressed while miR-186 showed significantly downregulated in 39 colorectal NET patients (51).
Different sets of miRNAs were identified as predictors of metastases on the base of tissue used as control. Overexpression of miR-21, involved in the regulation of the PI3K/Akt/mTOR pathway, and the Ki-67 proliferation index was significantly associated with liver metastases when pancreatic normal tissue was used as control (52). In contrast proliferation index Ki-67, miR-642, and miR-210 were correlated with metastases of PNETs when pancreatic islets were used as control (49). These data suggest that reference tissue influences the selection of markers. From the comparison of primary tumor and metastasis and then validation in 37 patients, the miRNA-196a was found significantly associated with tumor grade and recurrence (53).
A different approach is the NETest algorithm for the prediction of the clinical status of NETs (54, 55). The test is PCR-based measuring 51 individual circulating genes in 1 ml of blood. An algorithmic analysis provides a numeric score of disease status. It can define the completeness of surgical resection, identify residual disease, monitor disease progression, and determine the efficacy of treatment (56–58). NETest was used to evaluate the alteration in genes during the treatment with SSA, PRRT, and following surgery (59–61). In a Dutch cohort of GEP-NET patients, the NETest had good sensitivity but the specificity was relatively low. Thus, NETest would be less suited for screening but could be valuable for the detection of residual disease after therapy (62). Interestingly, the NETtest was successfully used to evaluate efficacy and response to PRRT in metastatic NETs (63) (Table 1).
Several miRNAs were also associated with tumor progression of SI-NETs. In the miRNAs study performed by Heverhagen (64), the most promising diagnostic miRNA-biomarker was miR-7-5p higher in pathological tissue compared to control and selected miR-885-5p as predictive of rectal NETs metastases (65). In a cohort study, 3 miRNAs 129-5p, 133a, and 143-3p downregulated were associated with the metastatic phenotype of SI-NETs (64, 66–70), while other upregulated were correlated with SSA treatment status or tumor stage (71). In another study, 4 differentially expressed miRNAs (miR-21-5p, miR-22-3p, and miR-150-5p) reached a statistical significance (72) underlying the need to add tissue markers, to discriminate NETs and to confirm the findings in annotated sample sets. Two miRNA profiling studies conducted on SI-NETs (66, 69, 73), compared metastatic tumors to primary malignancy, merging the data from both studies (metastasis vs primary) downregulation of miR-133a and upregulation of miR-183 were associated with poor prognosis and the spread of malignancy.
Role of Long Non-Coding RNAs in NENs Clinical Findings
Long non-coding RNAs (lncRNAs) are non-protein coding RNA transcripts longer than 200 nucleotides that exert multiple types of regulatory functions of all known cellular processes. Increasing evidence supports the role of lncRNAs in NENs development and progression with different mechanisms. In PNETs, tumor hypermethylation and silencing of long noncoding MEG3, determined activation of miR183/BRI3 axis, and cell proliferation due to c-MET oncogene activation (73). The reactivation of MEG3 by demethylating agents suppresses c-MET dependent cell proliferation suggesting that epigenetic targeting of MEG3 may represent an interesting approach in MEN1-PNETs treatment (59).
Moreover, downregulation of noncoding MEG3 and HOX genes has been associated with the development of non-functional pituitary adenomas and parathyroid tumors, respectively (74).
Two other lncRNAs are implicated in the pathogenesis of PNENs, the HOX antisense intergenic RNA chromatin-modifier (HOTAIR) and the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) (75). HOTAIR reprograms neuroendocrine differentiation of prostate cancer (76), and its overexpression increases H3K27me and metastatic potential of breast cancer cells (77). Evidence supports the hypothesis that both lncRNAs through epigenetic modification activate downstream pathways Wnt/β-catenin (78) and ERK/MAPK (79) promoting epithelial-mesenchymal transition (EMT). In contrast, the upregulation of both lncRNAs in primary GEP-NETs was associated with less aggressive disease (80), as well as lncRNA, telomeric repeat-containing RNA (TERRA), is necessary to maintain genome integrity (81).
Epigenetic Modifications Assessed in Liquid Biopsies as Prognostic Markers
Unlike traditional tissue biopsies, liquid biopsies are faster, less invasive, have the potential to reflect all metastatic sites (i.e. tumor heterogeneity), and can indicate therapeutic response or progression through serial sampling. By considering the potential of genomic analysis, liquid biopsies offer a facilitated means of detecting genomic alterations and can be easily repeated over time. Moreover, cancer-specific circulating DNA (ctDNA) methylation can be used to measure circulating tumor DNA, as well as reveal the methylation patterns in the tumor (10).
In metastatic PNET patients, free circulating DNA carrying oncogenic mutations or methylation have been identified by mutation-specific droplet digital PCR (ddPCR) (82). In particular in a prospective trial (“MGMT-NET”), MGMT hypermethylation was also detectable in ctDNA instead of tissue (83, 84).
The Phase II PAZONET study is evaluating the epigenome modification in circulating tumor cells (CTCs), as potential biomarkers of response to therapy. The same goal was also assessed during SSA treatment in association with PRRT (85–87). This novel approach indicates that epigenetic profiling can identify serum biomarkers with prognostic potential (10).
Epigenetic Targeted Agents and Clinical Trials
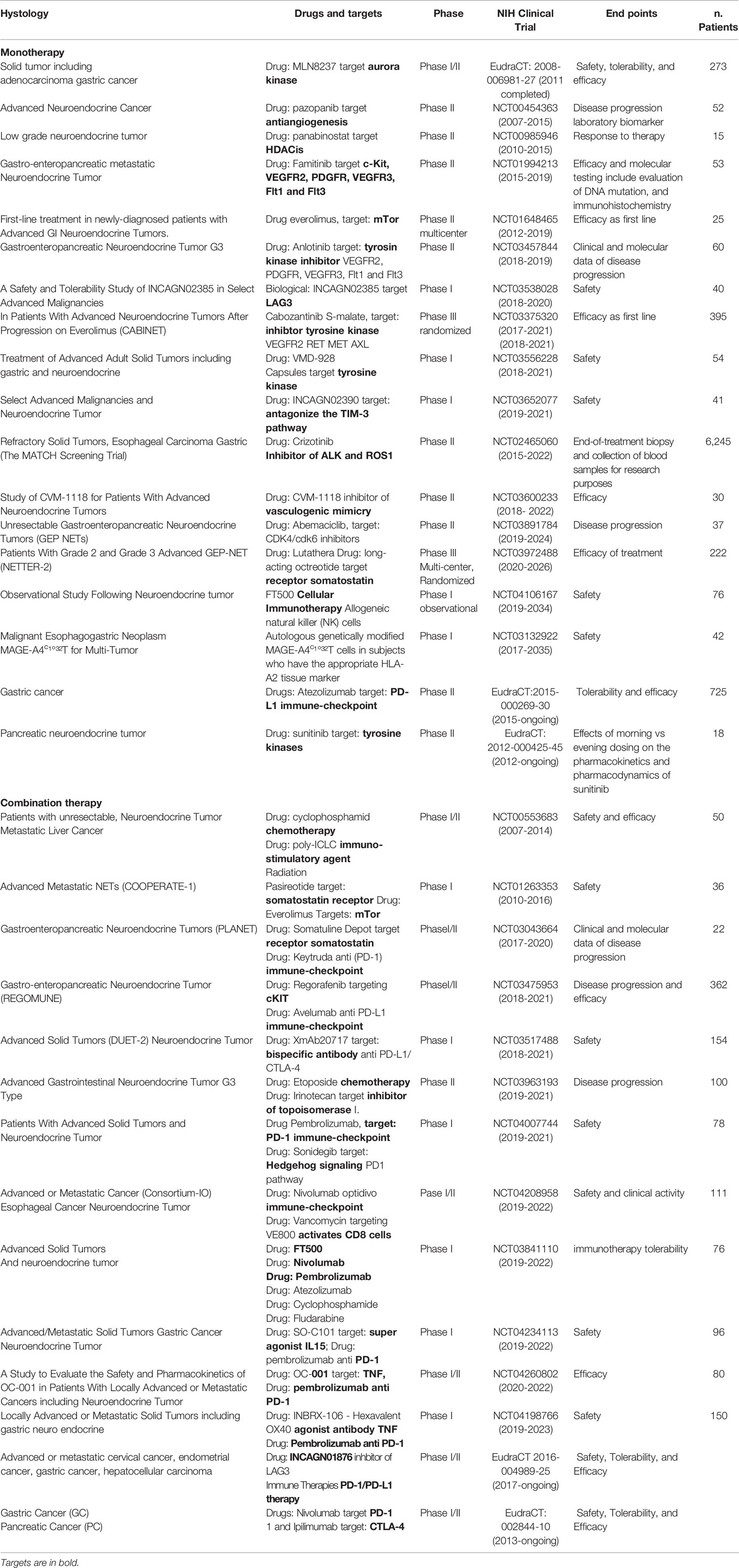
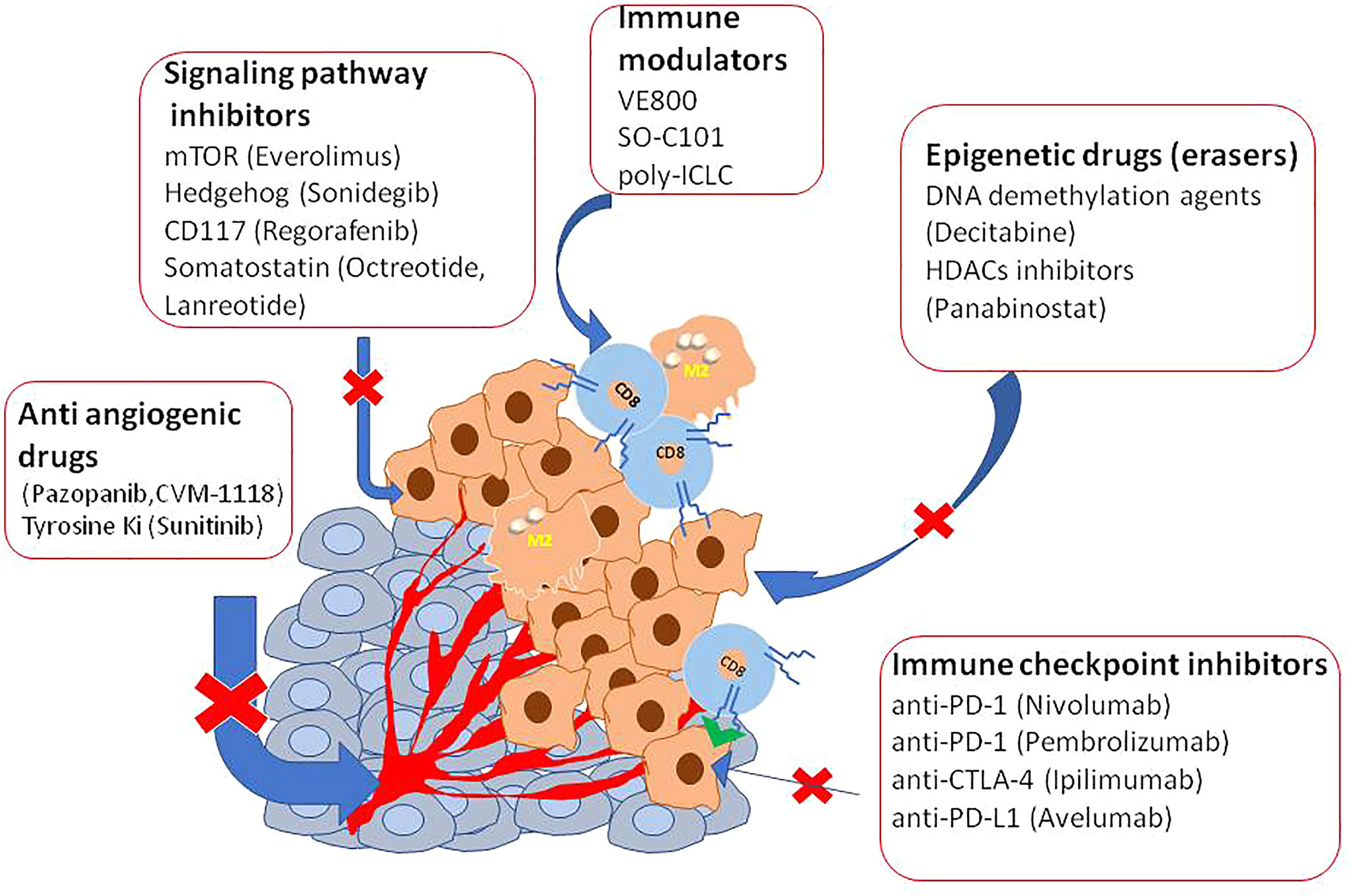
Several clinical studies reported disease control targeting the somatostatin receptor (SSR), overexpressed in 70% of GEP-NETs, supporting the efficacy of both the available SSA octreotide and lanreotid) (88–90). To improve the efficacy and adverse metastatic phenotype, several ongoing trials are evaluating other targets as an inhibitor of angiogenesis, immunotherapy, or combinations of them (Table 2 and Figure 1).
Epigenetics represents a very promising tool in cancer treatment because it can be reverted and epigenetic drugs are in use for the treatment of several cancer types (10, 91).
In vitro studies have already tested DNA methyltransferase inhibitors (DNMTis) because of MEN1 loss increase DNA hypermethylation (92). Promising results in PNET and small intestine cell lines were obtained using inhibitors of DNA methylases and HDAC to reduce cell viability and restoring gene expression (93–97). Interestingly, decitabine increased the expression of SSTR2 and the Ga-DOTATOC uptake also in BON1 tumor-bearing mice, indicating a possible therapy implication (98). However, decitabine has not yet been trialed in humans mainly because this agent targeted the whole methylated genome. Panobinostat, a histone deacetylase inhibitor (HDACi), was used in a completed phase II trial for the treatment of low-grade NENs. Patients showed a high stable disease with the median progression-free survival (PFS) of 9.9 months, and the median overall survival was 47.3 months. However, the low response rates, limitated further investigation (99). Inhibitors of the Bromo and extra terminal domain (BET) protein family, epigenetic readers of histone code, have also tested in experimental models (100). Of particular interest is Rx-001 which acts by blocking both DNMT and HDACs, activity. It showed to induce global epigenetic changes in tumors favoring infiltration of T cells, this histology was correlated with clinical benefit and sensitize tumor microenvironment to chemotherapy (101).
Novel frontier in solid tumor treatment is evaluating a combination of immunotherapy with epigenetic drugs, mainly because some immunosuppressive cancer antigens are regulated by acetylation of their genomic regulative element.
Some trials are testing a combination of agonists of TNF and immunotherapy via checkpoint inhibition (NCT04198766) or antibody with double specificities against PD-L1 and CTLA-4 (NCT03517488). However major interest gained depleting tryptophan enzymes as indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase (ICI). This because tryptophan is able to induce immune suppression within the cancer microenvironment. In tumor cells and nude mice have already targeted tryptophan. The authors by specific inhibitors or by preventing tryptophan promoter acetylation using histone deacetylase inhibitors as BET reported the reduction of immunosuppressive protein expression (10, 102) suggesting a novel therapeutic approach.
Conclusions and the Way Forward
The development of high-throughput techniques and larger datasets (i.e. The Cancer Genome Atlas) have accelerated research even in the field of NENs. Some pioneer studies have used an integrative approach in GEP-NETs (103). EWAS showed that these epigenome profiles can distinguish subtypes with different clinical features (Figure 2). The development of the NETest and liquid biopsy, as well as organoids (104), can be used to predict response to therapy and during the clinical follow-up, although not routinely used. Recently, it was proposed a bioresponsive drug-delivery depot for a combination of epigenetic modulation and immune checkpoint blockade (105). From the analysis of the clinical trials reported in Table 1 and Table 2, it emerges that the evaluation of the epigenetic pathway as a biomarker of response is of most interest in many studies, involving different kinds of therapies, even in combination (10). NCT03475953 and NCT03841110 ongoing trials are evaluating the therapeutic potential of the combination of direct drugs against tyrosine kinases and immune response pathways such as PD-1/PD-L1 and the opportunity to select from patients’ blood epigenetic biomarkers. The major challenge will now be to clinically validate such epigenetic biomarkers, within clinical trials for therapeutics in the new light of precision medicine, as well as network medicine (104, 106–108).
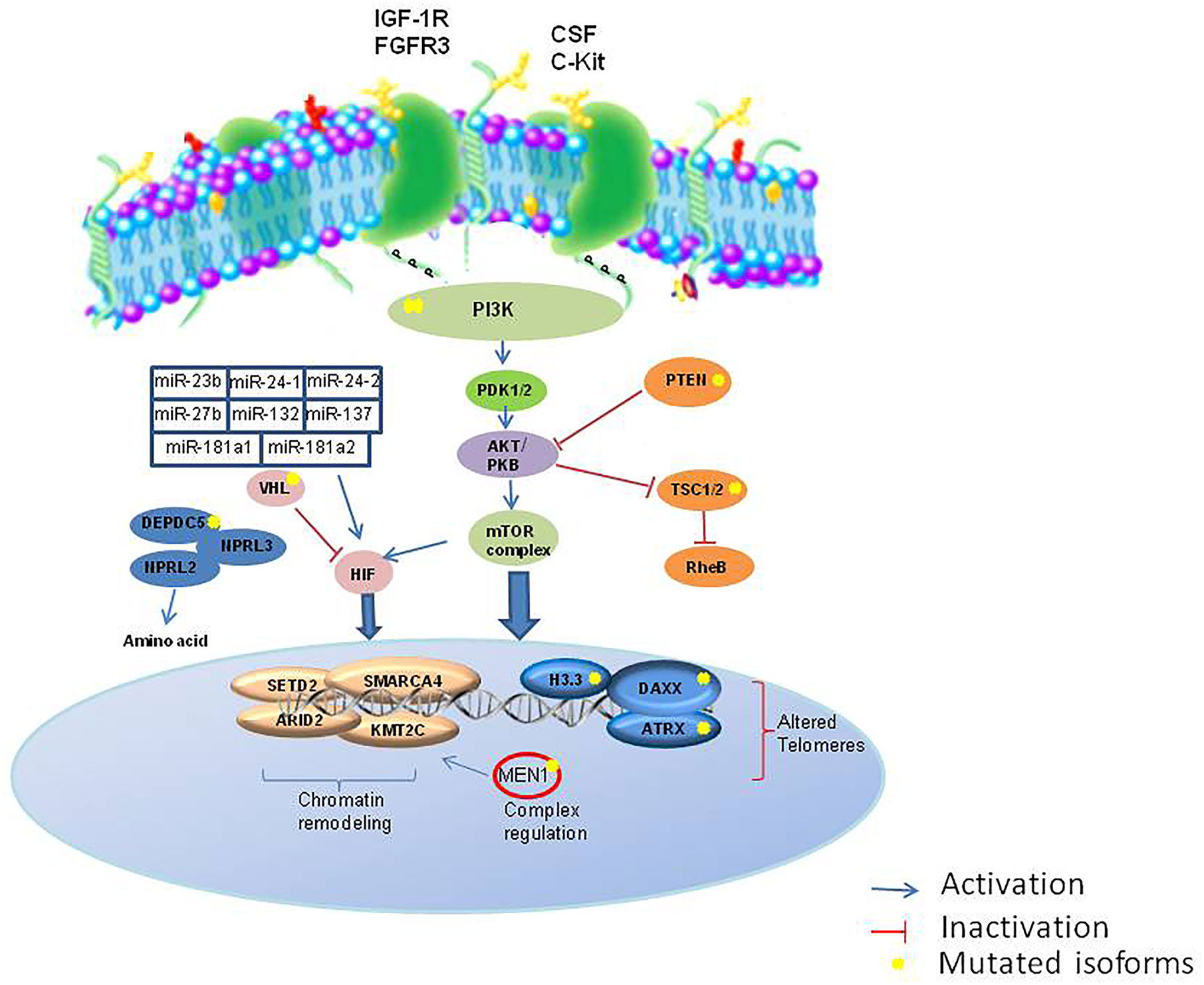
Figure 2 Major epigenetic pathways involved in NETs. IGF1R, insulin growth factor 1 receptor; FGFR, fibroblast growth factor receptor; SCF, colony stimulation factor; c-KIT, c-Kit proto-oncogene; PI3K, phosphatidylinositol 3-kinases; PTEN, Phosphatase and tensin homolog; PDK1/2, protein 3-phosphoinositide-dependent protein kinase-1; TSC1/2, Tuberous sclerosis 1/2; RheB, Ras homolog enriched in brain; HIF, hypoxia factor; RheB, Ras homolog enriched in brain; VHL, Von Hippel-Lindau; DEPDC5, DEP domain containing 5; NPRL3, neuropilin 3.
Author Contributions
AC, FN, and CN contributed to conception and design of the study. FN, RM and CN analyzed the data and wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by POR Campania FESR 2014-2020 “RARE.PLAT.NET” CUP B63D18000380007. This work was supported in part by “Ricerca Finalizzata 2011–12” (project code RF-2011-02349443, PI CN).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
CAPTEM, Capecitabine and temozolomide; cfDNA, Cell-free DNA; CIMP, CpG island methylator phenotype; CNV, Copy number variation; CTC, Circulating tumor cell; ctDNA, Circulating tumor DNA; ddPCR, Droplet digital PCR; GEP, Gastroenteropancreatic; MEN1, Multiple endocrine neoplasia type 1; NEN, Neuroendocrine neoplasm; NET, Neuroendocrine tumor; NEC, Neuroendocrine carcinoma; NF1, Neurofibromatosis type 1; PNEC, Pancreatic neuroendocrine carcinoma; PNEN, Pancreatic neuroendocrine neoplasm; PNET, Pancreatic neuroendocrine tumor; SI-NET, small intestinal neuroendocrine tumors; VHL, Von Hippel Lindau.
References
1. Fraenkel M, Kim M, Faggiano A, de Herder WW, Valk GD. Incidence of gastroenteropancreatic neuroendocrine tumours: a systematic review of the literature. Endocr Relat Cancer (2014) 21(3):R153–63. doi: 10.1530/erc-13-0125
2. Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, et al. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol (2017) 3(10):1335–42. doi: 10.1001/jamaoncol.2017.0589
3. Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, Mares JE, et al. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol (2008) 26(18):3063–72. doi: 10.1200/JCO.2007.15.4377
4. Faggiano A, Lo Calzo F, Pizza G, Modica R, Colao A. The safety of available treatments options for neuroendocrine tumors. Expert Opin Drug Saf (2017) 16(10):1149–61. doi: 10.1080/14740338.2017.1354984
5. Pavel M, Valle JW, Eriksson B, Rinke A, Caplin M, Chen J, et al. ENETS Consensus Guidelines for the Standards of Care in Neuroendocrine Neoplasms: Systemic Therapy - Biotherapy and Novel Targeted Agents. Neuroendocrinology (2017) 105:266–80. doi: 10.1159/000471880
6. House MG, Herman JG, Guo MZ, Hooker CM, Schulick RD, Lillemoe KD, et al. Aberrant hypermethylation of tumor suppressor genes in pancreatic endocrine neoplasms. Ann Surg (2003) 238(3):423–32. doi: 10.1097/01.sla.0000086659.49569.9e
7. Arnold CN, Sosnowski A, Schmitt-Gräff A, Arnold R, Blum HE. Analysis of molecular pathways in sporadic neuroendocrine tumors of the gastro-entero-pancreatic system. Int J Cancer (2007) 120(10):2157–64. doi: 10.1002/ijc.22569
8. Esteller M. Epigenetics in cancer. N Engl J Med (2008) 13;358(11):1148–59. doi: 10.1056/NEJMra072067
9. Shenker N, Flanagan JM. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. Br J Cancer (2012) 106(2):248–53. doi: 10.1038/bjc.2011.550
10. Bates SE. Epigenetic therapies for cancer. N Engl J Med (2020) 383(7):650–63. doi: 10.1056/NEJMra1805035
11. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature (7043) 2005) 435:834–8. doi: 10.1038/nature03702
12. Di Domenico A, Wiedmer T, Marinoni I, Perren A. Genetic and epigenetic drivers of neuroendocrine tumours (NET). Endocr Relat Cancer (2017) 24(9):R315–34. doi: 10.1530/ERC-17-0012
13. Liu L, Broaddus RR, Yao JC, Xie S, White JA, Wu TT, et al. Epigenetic alterations in neuroendocrine tumors: methylation of RAS-association domain family 1, isoform A and p16 genes are associated with metastasis. Mod Pathol (2005) 18(12):1632–40. doi: 10.1038/modpathol.3800490
14. Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA (2010) 107(32):14075–80. doi: 10.1073/pnas.1008850107
15. Pipinikas CP, Dibra H, Karpathakis A, Feber A, Novelli M, Oukrif D, et al. Epigenetic dysregulation and poorer prognosis in DAXX-deficient pancreatic neuroendocrine tumours. Endocr Relat Cancer (2015) 22(3):L13–L8. doi: 10.1530/ERC-15-0108
16. Blansfield JA, Choyke L, Morita SY, Choyke PL, Pingpank JF, Alexander HR, et al. Clinical, genetic and radiographic analysis of 108 patients with von Hippel-Lindau disease (VHL) manifested by pancreatic neuroendocrine neoplasms (PNETs). Surgery (2007) 142(6):814–8. doi: 10.1016/j.surg.2007.09.012
17. Gläsker S, Vergauwen E, Koch CA, Kutikov A, Vortmeyer AO. Von Hippel-Lindau Disease: Current Challenges and Future Prospects. Onco Targets Ther (2020) 13:5669–90. doi: 10.2147/OTT.S190753
18. Iliopoulos O, Levy AP, Jiang C, Kaelin WG Jr, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci U S A (1996) 93(20):10595–9. doi: 10.1073/pnas.93.20.10595
19. Siemeister G, Weindel K, Mohrs K, Barleon B, Martiny-Baron G, Marmé D. Reversion of deregulated expression of vascular endothelial growth factor in human renal carcinoma cells by von Hippel-Lindau tumor suppressor protein. Cancer Res (1996) 56(10):2299–301.
20. Chan CS, Laddha SV, Lewis PW, Koletsky MS, Robzyk K, Da Silva E, et al. ATRX, DAXX or MEN1 mutant pancreatic neuroendocrine tumors are a distinct alpha-cell signature subgroup. Nat Commun (2018) 9(1):4158. doi: 10.1038/s41467-018-06498-2
21. Colao A. Treatment of pituitary tumours in 2014: Advances in therapy of adenomas secreting GH, ACTH and TSH. Nat Rev Endocrinol (2015) 11(2):73–4. doi: 10.1038/nrendo.2014.227
22. Fernandez-Cuesta L, Peifer M, Lu X, Sun R, Ozretic L, Seidel D, et al. Frequent mutations inchromatin-remodelling genes in pulmonary carcinoids. Nat Commun (2014) 5:3518. doi: 10.1038/ncomms4518
23. Andersson E, Sward C, Stenman G, Ahlman H, Nilsson O. High resolution genomic profiling reveals gain of chromosome 14 as a predictor of poor outcome in ileal carcinoids. Endocr Related Cancer (2009) 16(3):953–66. doi: 10.1677/ERC-09-0052
24. Choi IS, Estecio MR, Nagano Y, Kim DH, White JA, Yao JC, et al. Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors). Mod Pathol (2007) 20(7):802–10. doi: 10.1038/modpathol.3800825
25. Stricker I, Tzivras D, Nambiar S, Wulf J, Liffers ST, Vogt M, et al. Site- and grade-specific diversity of LINE1 methylation pattern in gastroenteropancreatic neuroendocrine tumours. Anticancer Res (2012) 32(9):3699–706.
26. Sei Y, Zhao X, Forbes J, Szymczak S, Li Q, Trivedi A, et al. A hereditary form of small intestinal carcinoid associated with a germline mutation in inositol polyphosphate multikinase. Gastroenterology (2015) 149(1):67–78. doi: 10.1053/j.gastro.2015.04.008
27. Dumanski JP, Rasi C, Björklund P, Davies H, Ali AS, Grönberg M, et al. A MUTYH germline mutation is associated with small intestinal neuroendocrine tumors. Endocr Relat Cancer (2017) 24(8):427–43. doi: 10.1530/ERC-17-0196
28. Tirosh A, Mukherjee S, Lack J, Gara SK, Wang S, Quezado MM, et al. Distinct genome-wide methylation patterns in sporadic and hereditary nonfunctioning pancreatic neuroendocrine tumors. Cancer (2019) 125(8):1247–57. doi: 10.1002/cncr.31930
29. Muscogiuri G, Barrea L, Feola T, Gallo M, Messina E, Venneri MA, et al. NIKE (Neuroendocrine Tumors, Innovation in Knowledge and Education) Group. Pancreatic Neuroendocrine Neoplasms: Does Sex Matter? Trends Endocrinol Metab (2020) 31(9):631–41. doi: 10.1016/j.tem.2020.02.010
30. Fotouhi O, Adel Fahmideh M, Kjellman M, Sulaiman L, Höög A, Zedenius J, et al. Global hypomethylation and promoter methylation in small intestinal neuroendocrine tumors: an in vivo and in vitro study. Epigenetics (2014) 9(7):987–97. doi: 10.4161/epi.28936-
31. Dejeux E, Olaso R, Dousset B, Audebourg A, Gut IG, Terris B, et al. Hypermethylation of the IGF2 differentially methylated region 2 is a specific event in insulinomas leading to loss-of-imprinting and overexpression. Endocr Relat Cancer (2009) 16(3):939–52. doi: 10.1677/ERC-08-0331
32. Edfeldt K, Ahmad T, Akerstrom G, Janson ET, Hellman P, Stalberg P, et al. TCEB3C a putative tumor suppressor gene of small intestinal neuroendocrine tumors. Endocr Relat Cancer (2014) 21:275–84. doi: 10.1530/ERC-13-0419-
33. Karpathakis A, Dibra H, Pipinikas C, Feber A, Morris T, Francis J, et al. Prognostic impact of novel molecular subtypes of small intestinal neuroendocrine tumor. Clin Cancer Res (2016) 22(1):250. doi: 10.1158/1078-0432.CCR-15-0373
34. Verdugo AD, Crona J, Starker L, Stalberg P, Akerstrom G, Westin G, et al. Global DNA methylation patterns through an array-based approach in small intestinal neuroendocrine tumors. Endocr Relat Cancer (2014) 21(1):L5–7. doi: 10.1530/erc-13-0481
35. Tirosh A, Kebebew E. Genetic and epigenetic alterations in pancreatic neuroendocrine tumors. J Gastrointest Oncol (2020) (3):567–77. doi: 10.21037/jgo.2020.03.11
36. Attia M, Rachez C, De Pauw A, Avner P, Rogner UC. Nap1l2 promotes histone acetylation activity during neuronal differentiation. Mol Cell Biol (2007) 27:6093–102. doi: 10.1128/mcb.00789-07
37. Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem (2005) 280:1817–25. doi: 10.1074/jbc.M411347200
38. Schimmack S, Taylor A, Lawrence B, Alaimo D, Schmitz-Winnenthal H, Büchler MW, et al. A mechanistic role for the chromatin modulator, NAP1L1, in pancreatic neuroendocrine neoplasm proliferation and metastases. Epigenet Chromatin (2014) 7:15. doi: 10.1186/1756-8935-7-15
39. Stefanoli M, La Rosa S, Sahnane N, Romualdi C, Pastorino R, Marando A, et al. Prognostic relevance of aberrant DNA methylation in G1 and G2 pancreatic neuroendocrine tumors. Neuroendocrinology (2014) 100(1):26–34. doi: 10.1159/000365449
40. Arnold CN, Nagasaka T, Goel A, Scharf I, Grabowski P, Sosnowski A, et al. Molecular characteristics and predictors of survival in patients with malignant neuroendocrine tumors. Int J Cancer (2008) 123(7):1556–64. doi: 10.1002/ijc.23690
41. Karpathakis A, Dibra H, Pipinikas C, Feber A, Morris T, Francis J, et al. Progressive epigenetic dysregulation in neuroendocrine tumour liver metastases. Endocr Relat Cancer (2017) 24(2):L21–l5. doi: 10.1530/erc-16-0419
42. Barazeghi E, Prabhawa S, Norlén O, Hellman P, Stålberg P, Westin G. Decrease of 5-hydroxymethylcytosine and TET1 with nuclear exclusion of TET2 in small intestinal neuroendocrine tumors. BMC Cancer (2018) 18(1):764. doi: 10.1186/s12885-018-4579-z
43. Vitale G, Carra S, Ferraù F, Guadagno E, Faggiano A, Colao A. NIKE. Gastroenteropancreatic neuroendocrine neoplasms and inflammation: A complex cross-talk with relevant clinical implications. Crit Rev Oncol Hematol (2020) 146:102840. doi: 10.1016/j.critrevonc.2019.102840
44. Cros J, Hentic O, Rebours V, Zappa M, Gille N, Theou-Anton N, et al. MGMT expression predicts response to temozolomide in pancreatic neuroendocrine tumors. Endocr Relat Cancer (2016) 23(8):625–33. doi: 10.1530/erc-16-0117
45. Walter T, van Brakel B, Vercherat C, Hervieu V, Forestier J, Chayvialle JA, et al. O6-Methylguanine-DNA methyltransferase status in neuroendocrine tumours: prognostic relevance and association with response to alkylating agents. Br J Cancer (2015) 112(3):523–31. doi: 10.1038/bjc.2014.660
46. Dussol AS, Joly MO, Vercherat C, Forestier J, Hervieu V, Scoazec JY, et al. Gemcitabine and oxaliplatin or alkylating agents for neuroendocrine tumors: Comparison of efficacy and search for predictive factors guiding treatment choice. Cancer (2015) 121(19):3428–34. doi: 10.1002/cncr.29517
47. Campana D, Walter T, Pusceddu S, Gelsomino F, Graillot E, Prinzi N, et al. Correlation between MGMT promoter methylation and response to temozolomide-based therapy in neuroendocrine neoplasms: an observational retrospective multicenter study. Endocrine (2018) 60(3):490–8. doi: 10.1007/s12020-017-1474-3
48. Lemelin A, Barritault M, Hervieu V, Payen L, Peron J, Couvelard A, et al. O6-methylguanine-DNA methyltransferase (MGMT) status in neuroendocrine tumors: a randomized phase II study (MGMT-NET). Dig Liver Dis (2019) 51(4):595–9. doi: 10.1016/j.dld.2019.02.001
49. Thorns C, Schurmann C, Gebauer N, Wallaschofski H, Kumpers C, Bernard V, et al. Global microRNA profiling of pancreatic neuroendocrine neoplasias. Anticancer Res (2014) 34(5):2249–54.
50. Malczewska A, Kidd M, Matar S, Kos-Kudla B, Modlin IM. A comprehensive assessment of the role of miRNAs as biomarkers in gastroenteropancreatic neuroendocrine tumors. Neuroendocrinology (2018) 107(1):73–90. doi: 10.1159/000487326
51. Wang M, Xia X, Chu W, Xia L, Meng T, Liu L, et al. Roles of miR-186 and PTTG1 in colorectal neuroendocrine tumors. Int J Clin Exp Med (2015) 8(12):22149–57.
52. Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol (2006) 24(29):4677–84. doi: 10.1200/jco.2005.05.5194
53. Lee YS, Kim H, Kim HW, Lee J-C, Paik K-H, Kang J, et al. High expression of MicroRNA-196a indicates poor prognosis in resected pancreatic neuroendocrine tumor. Medicine (2015) 94(50):e2224–e. doi: 10.1097/MD.0000000000002224
54. Bodei L, Kidd M, Modlin IM, Prasad V, Severi S, Ambrosini V, et al. Gene transcript analysis blood values correlate with (6)(8)Ga-DOTA-somatostatin analog (SSA) PET/CT imaging in neuroendocrine tumors and can define disease status. Eur J Nucl Med Mol Imaging (2015) 42(9):1341–52. doi: 10.1007/s00259-015-3075-9
55. Kidd M, Drozdov I, Modlin I. Blood and tissue neuroendocrine tumor gene cluster analysis correlate, define hallmarks and predict disease status. Endocr Relat Cancer (2015) 22(4):561–75. doi: 10.1530/erc-15-0092
56. Modlin IM, Drozdov I, Alaimo D, Callahan S, Teixiera N, Bodei L, et al. A multianalyte PCR blood test outperforms single analyte ELISAs (chromogranin A, pancreastatin, neurokinin A) for neuroendocrine tumor detection. Endocr Related Cancer (2014) 21(4):615–28. doi: 10.1530/erc-14-0190
57. Modlin IM, Drozdov I, Kidd M. Gut neuroendocrine tumor blood qPCR fingerprint assay: characteristics and reproducibility. Clin Chem Lab Med (2014) 52(3):419–29. doi: 10.1515/cclm-2013-0496
58. Modlin IM, Kidd M, Bodei L, Drozdov I, Aslanian H. The clinical utility of a novel blood-based multi-transcriptome assay for the diagnosis of neuroendocrine tumors of the gastrointestinal tract. Am J Gastroenterol (2015) 110(8):1223–32. doi: 10.1038/ajg.2015.160
59. Modlin IM, Frilling A, Salem RR, Alaimo D, Drymousis P, Wasan HS, et al. Blood measurement of neuroendocrine gene transcripts defines the effectiveness of operative resection and ablation strategies. Surgery (2016) 159(1):336–47. doi: 10.1016/j.surg.2015.06.056
60. Cwikla JB, Bodei L, Kolasinska-Cwikla A, Sankowski A, Modlin IM, Kidd M. Circulating transcript analysis (NETest) in GEP-NETs treated with somatostatin analogs defines therapy. J Clin Endocrinol Metab (2015) 100(11):E1437–45. doi: 10.1210/jc.2015-2792
61. Bodei L, Kidd M, Modlin IM, Severi S, Drozdov I, Nicolini S, et al. Measurement of circulating transcripts and gene cluster analysis predicts and defines therapeutic efficacy of peptide receptor radionuclide therapy (PRRT) in neuroendocrine tumors. Eur J Nucl Med Mol Imaging (2016) 43(5):839–51. doi: 10.1007/s00259-015-3250-z
62. van Treijen MJC, Korse CM, van Leeuwaarde RS, Saveur LJ, Vriens MR, Verbeek WHM, et al. Blood transcript profiling for the detection of neuroendocrine tumors: results of a large independent validation study. Front Endocrinol (2018) 9:740. doi: 10.3389/fendo.2018.00740
63. Bodei L, Kidd M, Singh A, Wouter A, Zwan W, Severi S, et al. PRRT neuroendocrine tumor response monitored using circulating transcript analysis: the NETest. Eur J Nucl Med Mol Imaging (2020) 47(4):895–906. doi: 10.1007/s00259-019-04601-3
64. Heverhagen AE, Legrand N, Wagner V, Fendrich V, Bartsch DK, Slater EP. Overexpression of MicroRNA miR-7-5p is a potential biomarker in neuroendocrine neoplasms of the small intestine. Neuroendocrinology (2018) 106(4):312–7. doi: 10.1159/000480121
65. Mitsuhashi K, Yamamoto I, Kurihara H, Kanno S, Ito M, Igarashi H, et al. Analysis of the molecular features of rectal carcinoid tumors to identify new biomarkers that predict biological malignancy. Oncotarget (2015) 6(26):22114–25. doi: 10.18632/oncotarget.4294
66. Ruebel K, Leontovich AA, Stilling GA, Zhang S, Righi A, Jin L, et al. MicroRNA expression in ileal carcinoid tumors: downregulation of microRNA-133a with tumor progression. Mod Pathol (2010) 23(3):367–75. doi: 10.1038/modpathol.2009.161
67. Døssing KBV, Binderup T, Kaczkowski B, Jacobsen A, Rossing M, Winther O, et al. Down-regulation of miR-129-5p and the let-7 family in neuroendocrine tumors and metastases leads to up-regulation of their targets Egr1, G3bp1, Hmga2 and Bach1. Genes (2014) 6(1):1–21. doi: 10.3390/genes6010001
68. Miller HC, Frampton AE, Malczewska A, Ottaviani S, Stronach EA, Flora R, et al. MicroRNAs associated with small bowel neuroendocrine tumours and their metastases. Endocr Relat Cancer (2016) 23(9):711–26. doi: 10.1530/erc-16-0044
69. Li SC, Essaghir A, Martijn C, Lloyd RV, Demoulin JB, Oberg K, et al. Global microRNA profiling of well-differentiated small intestinal neuroendocrine tumors. Mod Pathol (2013) 26(5):685–96. doi: 10.1038/modpathol.2012.216
70. Mandal R, Hardin H, Baus R, Rehrauer W, Lloyd RV. Analysis of miR-96 and miR-133a expression in gastrointestinal neuroendocrine neoplasms. Endocr Pathol (2017) 28(4):345–50. doi: 10.1007/s12022-017-9504-5
71. Li SC, Khan M, Caplin M, Meyer T, Öberg K, Giandomenico V. Somatostatin analogs treated small intestinal neuroendocrine tumor patients circulating MicroRNAs. PLoS One (2015) 10(5):e0125553. doi: 10.1371/journal.pone.0125553.152
72. Bowden M, Zhou CW, Zhang S, Brais L, Rossi A, Naudin L, et al. Profiling of metastatic small intestine neuroendocrine tumors reveals characteristic miRNAs detectable in plasma. Oncotarget (2017) 8(33):54331–44. doi: 10.18632/oncotarget.16908
73. Zhang YY, Feng HM. MEG3 Suppresses Human Pancreatic Neuroendocrine Tumor Cells Growth and Metastasis by Down-Regulation of Mir-183. Cell Physiol Biochem (2017) 44(1):345–56. doi: 10.1159/000484906
74. Cheunsuchon P, Zhou Y, Zhang X, Lee H, Chen W, Nakayama Y, et al. Silencing of the imprinted DLK1-MEG3 locus in human clinically nonfunctioning pituitary adenomas. Am J Pathol (2011) 179(4):2120–30. doi: 10.1016/j.ajpath.2011.07.002
75. Ji M, Tang L, Ding R, Shi L, Liu A, Chen D, et al. Long noncoding RNA-mRNA expression profiles and validation in pancreatic neuroendocrine neoplasms. Clin Endocrinol (Oxf) (2020) 92(4):312–22. doi: 10.1111/cen.14156
76. Chang YT, Lin TP, Tang JT, Campbell M, Luo YL, Lu SY, et al. HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Lett (2018) 433:43–52. doi: 10.1016/j.canlet.2018.06.029
77. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature (2010) 464(7291):1071–6. doi: 10.1038/nature08975
78. Wu L, Shi Y, Liu B, Zhao M. Expression of lncRNA-HOTAIR in the serum of patients with lymph node metastasis of papillary thyroid carcinoma and its impact. Oncol Lett (2020) 20(1):907–13. doi: 10.3892/ol.2020.11620
79. Wen D, Liu D, Tang J, Dong L, Liu Y, Tao Z, et al. Malic enzyme 1 induces epithelial-mesenchymal transition and indicates poor prognosis in hepatocellular carcinoma. Tumour Biol (2015) 36(8):6211–21. doi: 10.1007/s13277-015-3306-5
80. Chu YH, Hardin H, Eickhoff J, Lloyd RV. In Situ hybridization analysis of long non-coding RNAs MALAT1 and HOTAIR in gastroenteropancreatic neuroendocrine nNeoplasms. Endocr Pathol (2019) (1):56–63. doi: 10.1007/s12022-018-9564-1
81. Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet (2015) 14(6):143. doi: 10.3389/fgene.2015.00143
82. Boons G, Vandamme T, Peeters M, Beyens M, Driessen A, Janssens K, et al. Cell-free DNA from metastatic pancreatic neuroendocrine tumor patients contains tumor-specific mutations and copy number variations. Front Oncol (2018) 8:467. doi: 10.3389/fonc.2018.00467
83. Garrigou S, Perkins G, Garlan F, Normand C, Didelot A, Le Corre D, et al. A study of hypermethylated circulating tumor DNA as a universal colorectal cancer biomarker. Clin Chem (2016) 62(8):1129–39. doi: 10.1373/clinchem.2015.253609
84. Warton K, Mahon KL, Samimi G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer (2016) 23(3):R157–71. doi: 10.1530/ERC-15-0369
85. Childs A, Vesely C, Ensell L, Lowe H, Luong TV, Caplin ME, et al. Expression of somatostatin receptors 2 and 5 in circulating tumour cells from patients with neuroendocrine tumours. Br J Cancer (2016) 115(12):1540–7. doi: 10.1038/bjc.2016.377
86. Cives M, Quaresmini D, Rizzo FM, Felici C, D’Oronzo S, Simone V, et al. Osteotropism of neuroendocrine tumors: role of the CXCL12/ CXCR4 pathway in promoting EMT in vitro. Oncotarget (2017) 8(14):22534–49. doi: 10.18632/oncotarget.15122
87. Zhu Z, Qiu S, Shao K, Hou Y. Progress and challenges of sequencing and analyzing circulating tumor cells. Cell Biol Toxicol (2018) 34(5):405–15. doi: 10.1007/s10565-017-9418-5
88. Caplin ME, Pavel M, Ćwikła JB, Phan AT, Raderer M, Sedláčková E, et al. CLARINET Investigators. Lanreotide in metastatic enteropancreatic neuroendocrine tumors. N Engl J Med (2014) 371(3):224–33. doi: 10.1056/NEJMoa1316158
89. Rinke A, Krug S. Neuroendocrine tumours - Medical therapy: Biological. Best Pract Res Clin Endocrinol Metab (2016) 30(1):79–91. doi: 10.1016/j.beem.2015.09.004
90. Faggiano A, Modica R, Lo Calzo F, Camera L, Napolitano V, Altieri B, et al. Lanreotide therapy vs active surveillance in MEN1-related pancreatic neuroendocrine tumors < 2 centimeters. J Clin Endocrinol Metab (2020) 105(1):dgz007. doi: 10.1210/clinem/dgz007
91. de Nigris F, Ruosi C, Napoli C. Clinical efficiency of epigenetic drugs therapy in bone malignancies. Bone (2020) 20:115605. doi: 10.1016/j.bone.2020.115605
92. Yuan Z, Gardiner JC, Maggi EC, Adem A, Zhang G, Lee S, et al. Tissue-specific induced DNA methyltransferase 1 (Dnmt1) in endocrine pancreas by RCAS-TVA-based somatic gene transfer system promotes β-cell proliferation. Cancer Gene Ther (2019) 26(3-4):94–102. doi: 10.1038/s41417-018-0046-x
93. Baradari V, Huether A, Hopfner M, Schuppan D, Scherubl H. Antiproliferative and proapoptotic effects of histone deacetylase inhibitors on gastrointestinal neuroendocrine tumor cells. Endocr Related Cancer (2006) 13(4):1237–50. doi: 10.1677/erc.1.01249
94. Zhang HY, Rumilla KM, Jin L, Nakamura N, Stilling GA, Ruebel KH, et al. Association of DNA methylation and epigenetic inactivation of RASSF1A and beta-catenin with metastasis in small bowel carcinoid tumors. Endocrine (2006) 30:299–306. doi: 10.1007/s12020-006-0008-1
95. Habbe N, Bert T, Simon B. Identification of methylation associated gene expression in neuroendocrine pancreatic tumor cells. Pancreatology (2007) 7:352–9. doi: 10.1159/000107270
96. Alexander VM, Roy M, Steffens KA, Kunnimalaiyaan M, Chen H. Azacytidine induces cell cycle arrest and suppression of neuroendocrine markers in carcinoids. Int J Clin Exp Med (2010) 3(2):95–102. doi: 10.1016/j.jss.2009.11.053
97. Arvidsson Y, Johanson V, Pfragner R, Wangberg B, Nilsson O. Cytotoxic effects of valproic acid on neuroendocrine tumour cells. Neuroendocrinology (2016) 103:578–91. doi: 10.1159/000441849
98. Taelman VF, Radojewski P, Marincek N, Ben-Shlomo A, Grotzky A, Olariu CI, et al. Upregulation of key molecules for targeted imaging and therapy. J Nuclear Med (2016) 57:1805–10. doi: 10.2967/jnumed.115.165092
99. Jin N, Lubner SJ, Mulkerin DL, Rajguru S, Carmichael L, Chen H, et al. A phase II trial of a histone deacetylase inhibitor panobinostat in patients with low-grade neuroendocrine tumors. Oncologist (2016) 21(7):785–6. doi: 10.1634/theoncologist.2016-006
100. Lines KE, Stevenson M, Filippakopoulos P, Müller S, Lockstone HE, Wright B, et al. Epigenetic pathway inhibitors represent potential drugs for treating pancreatic and bronchial neuroendocrine tumors. Oncogenesis (2017) 6(5):e332. doi: 10.1038/oncsis.2017.30
101. Abdelfatah E, Kerner Z, Nanda N, Ahuja N. Epigenetic therapy in gastrointestinal cancer: the right combination. Therap Adv Gastroenterol (2006) 9(4):560–79. doi: 10.1177/1756283X16644247
102. Dent P, Booth L, Poklepovic A, Hancock JF. Signaling alterations caused by drugs and autophagy. Cell Signal (2019) 64:109416. doi: 10.1016/j.cellsig.2019.109416
103. Mafficini A, Scarpa A. Genetics and epigenetics of gastroenteropancreatic neuroendocrine neoplasms. Endocr Rev (2019) 40(2):506–36. doi: 10.1210/er.2018-00160
104. Scognamiglio G, De Chiara A, Parafioriti A, Armiraglio E, Fazioli F, Gallo M, et al. Patient-derived organoids as a potential model to predict response to PD-1/PD-L1 checkpoint inhibitors. Br J Cancer (2019) 121(11):979–82. doi: 10.1038/s41416-019-0616-1
105. Ruan H, Hu Q, Wen D, Chen Q, Chen G, Lu Y, et al. A dual-bioresponsive drug-delivery depot for combination of epigenetic modulation and immune checkpoint blockade. Adv Mater (2019) 31(17):e1806957. doi: 10.1002/adma.201806957
106. Colella G, Fazioli F, Gallo M, De Chiara A, Apice G, Ruosi C, et al. Sarcoma spheroids and organoids-promising tools in the era of personalized medicine. Int J Mol Sci (2018) 19(2):615. doi: 10.3390/ijms19020615
107. Horak P, Klink B, Heining C, Gröschel S, Hutter B, Fröhlich M, et al. Precision oncology based on omics data: The NCT Heidelberg experience. Int J Cancer (2017) 141(5):877–86. doi: 10.1002/ijc.30828.108
Keywords: epigenetics, neuroendocrine neoplasms, trials, biomarkers, neuroendocrine, neuroendocrine tumor
Citation: Colao A, de Nigris F, Modica R and Napoli C (2020) Clinical Epigenetics of Neuroendocrine Tumors: The Road Ahead. Front. Endocrinol. 11:604341. doi: 10.3389/fendo.2020.604341
Received: 09 September 2020; Accepted: 12 November 2020;
Published: 15 December 2020.
Edited by:
Antongiulio Faggiano, Sapienza University of Rome, ItalyReviewed by:
Jean-Yves Scoazec, Institut Gustave Roussy, FranceChristian Albert Koch, Fox Chase Cancer Center, United States
Copyright © 2020 Colao, de Nigris, Modica and Napoli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberta Modica, cm9iZXJ0YW1vZGljYUBsaWJlcm8uaXQ=
 Annamaria Colao
Annamaria Colao Filomena de Nigris2
Filomena de Nigris2 Roberta Modica
Roberta Modica Claudio Napoli
Claudio Napoli