- 1Cancer Institute, Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, China
- 2Department of General Surgery, Second Affiliated Hospital School of Medicine, Zhejiang University, Hangzhou, China
- 3Key Laboratory of Respiratory Disease of Zhejiang Province, Department of Respiratory and Critical Care Medicine, Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, China
- 4Zhejiang Provincial Key Laboratory of Pancreatic Disease, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
BRAF mutation accounts for 50% of the PTC (papillary thyroid carcinoma) and is closely associated with high-risk clinicopathological characteristics. Increasing evidence implied that dysregulation of miRNA participated in carcinogenesis and progression of cancer. Clinical data showed the significant up-regulation of miR-222-3p in PTC; however, the role of miR-222-3p and possible relationship with BRAF mutation remained unclear. Here, we identified significant up-regulation of miR-222-3p in PTC tissues harboring BRAFV600E mutation compared with BRAF wild type (BRAFWT) from collected PTC clinical samples. External validation performed with The Cancer Genome Atlas (TCGA) databases was consistent with the above result. Exogenous expression of BRAFV600E oncoprotein increased the expression of miR-222-3p in B-CPAP and TPC-1 cells. The treatment of BRAFV600E and MEK inhibitor, PLX4720 and PD0325901, decreased the expression of miR-222-3p in B-CPAP but not in TPC-1. Inhibition of miR-222-3p significantly suppressed the migration of B-CPAP and induced a mesenchymal-epithelial transition (MET) phenotype via the Snail transcription factor. Immunohistochemistry (IHC) analysis demonstrated the up-regulation of Snail correlated with lymph node metastasis and BRAFV600E mutation in PTC. Besides, in situ hybridization (ISH) and IHC analysis of PTC clinical samples confirmed the correlation between the expression of miR-222-3p and Snail. These results showed miR-222-3p conduced more aggressive clinical manifestation of PTC by promoting Snail-induced EMT.
Introduction
The incidence of PTC has significantly increased in the last decade worldwide. Although most PTCs have a good prognosis, some patients still went through local recurrence or distant metastasis after surgery and radioactive iodine therapy. Treatment of advanced PTC patients characterized by recurrent disease, distant metastasis, and resistance to radioactive iodine, including traditional radiotherapy, chemotherapy, and kinase inhibitors, has limited benefits, but a lot of side effects. These patients showed a poor quality of life and a dismal lifespan (5-year disease-specific survival is ~70%) (1, 2).
miRNA is one endogenous non-coding RNA with a length of ~21 nucleotides, which can promote or inhibit protein expression by combining with target mRNA following the principle of base complementary pairing (3). Aberrant miRNA expression can activate multiple oncogenic pathways in different types of cancers (4). Previous studies showed that miR-222-3p was one of the most consistently overexpressed miRNAs in PTC and an eight to eleven folds increase compared to normal thyroid tissues has been reported (5, 6). Up-regulated miR-222-3p can promote prostate cancer cell cycle transformation, enhance chordoma cell migration and facilitate endometrial carcinoma cell invasion (7–9). In PTC, the up-regulation of miR-222-3p is correlated with high-risk features such as extrathyroidal extension, lymph node metastasis, and recurrence (5, 10, 11). However, the mechanism of the metastasis and invasion conducted by dysregulation of miR-222-3p still needs to be further explored.
There are multiple driver mutations in PTC, including but not limited to BRAF mutation, RAS mutation, TP53 mutation, RET/PTC rearrangement, and NTRK1 rearrangement (12), among which the BRAFV600E mutation accounts for the highest proportion, reaching ~50%. BRAFV600E oncoprotein, caused by a c.1799 T>A transversion, is associated with aggressive clinicopathologic features, such as extrathyroidal invasion, lymph node metastasis, and advanced disease stages (13, 14). Induction of BRAFV600E mutation and simultaneous activation of thyroid-stimulating hormone receptor signaling pathway can transform normal thyroid follicular cells into cancer cells (15). The BRAFV600E oncoprotein could constitutively activate the mitogen-activated protein kinase (MAPK) pathway, thereby enhancing malignancy (16).
Here, by analyzing the data of PTC patients from TCGA, we found that the miR-222-3p expression was related to the BRAFV600E mutation in PTC. We supposed that BRAFV600E could augment the metastasis capability of PTC by upregulating miR-222-3p expression. In this study, we verified the correlation between the BRAFV600E mutation and the up-regulation of miR-222-3p in the tumor samples of PTC patients. By adjusting the expression of BRAFV600E protein in the PTC cell line, the corresponding expression level of miR-222-3p was determined. Furthermore, we tried to elaborate on the metastasis-promoting effect of miR-222-3p through the Snail-induced EMT. The clarification of the correlation between miR-222-3p and BRAFV600E and the metastasis-promoting role of miR-222-3p in PTC may shed more light on the diagnosis and treatment of PTC.
Materials and Methods
Human Thyroid Tissues
70 cases of PTCs were collected by the Department of Pathology of Second Affiliated Hospital of Zhejiang University School of Medicine from 2019 to 2020. All PTC samples were surgically resected and the whole hematoxylin and eosin slides of all samples were reviewed by two senior pathologists. Formalin-fixed paraffin-embedded (FFPE) samples of PTCs were stored at room temperature (51 cases with BRAFV600E mutation and 19 cases with BRAFWT. 45 cases with lymph node metastasis and 25 cases with no metastasis).45 consecutive cases with lymph node metastasis were included in two years to compare the miR-222-3p in PTC tumors with and without lymph node metastasis.
This study was approved by the Human Research Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, China. The IRB protocol number is 2019010.
Cell Culture
B-CPAP (RRID: CVCL_0153) cell line was purchased from the Chinese Academy of Sciences (Shanghai, China). TPC-1 (RRID: CVCL_6298) cell line was purchased from Procell (Wuhan, China). K1 (RRID: CVCL_2537) cell line was purchased from Cellcook (Guangzhou, China). B-CPAP and TPC-1 were cultured in RPMI-1640 medium (Sigma), K1 was cultured in DMEM medium (Gibco). Both media were supplemented with 10% (v/v) fetal bovine serum. All cells were maintained at 5% CO2 and 37°C in a humidified atmosphere.
miRNA Inhibitor Transfection and MAPK Pathway Inhibitor
Cells were seeded in a 12-well plate. After 24h, transfection was performed with Lipofectamine 3000 reagent (Thermo Fisher Scientific), following the manufacturer’s instructions. Transfection experiments were performed using 100nM miR-222-3p inhibitors or negative control (Genepharma). The inhibitor efficiency was greater than ninety-five percent. Sequences of miR-222-3p inhibitor and negative control were as following:
hsa-miR-222-3p inhibitor: 5’-ACCCAGUAGCCAGAUGUAGCU-3’; has-miR-222-3p negative control: 5’-CAGUACUUUUGUGUAGUACAA-3’.
PLX4720 (BRAFV600E inhibitor) and PD0325901 (MEK inhibitor) were purchased from Selleck.
Construction of BRAFV600E Overexpression Plasmid
pcDNA3.1- plasmid was used as the vector plasmid, and Xba I (Takara, TCTAGA) and Hind III (Takara, AAGCTT) were determined as restriction sites. A BRAFV600E point mutation (GTG→GAG) and the amplification primer sequence were shown below:
BRAF forward: 5’-GCTCTAGAATGGCGGCGCTGAGCGGTGG-3’, BRAFV600E reverse: 5’-CTCCATCGAGATTTCTCTGTAGCTAGACCAA-3’; BRAFV600E forward: 5’-TTGGTCTAGCTACAGAGAAATCTCGATGGAG-3’, BRAF reverse: 5’-CCCAAGCTTTCACTTGTCATCGTCGTCCTTGTAATCG TGGACAGGAAACGCACCATATC-3’.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated with RNA-Quick Purification Kit (YiShan Biotech) according to the manufacturer’s instructions. Reverse transcription of mRNA and miRNA (stem-loop primer) was performed following the manufacturer’s protocols of the PrimeScript™ RT reagent kit (Takara). Quantitative real-time PCR was performed following the manufacturer’s protocols of TB Green™ Premix Ex Taq™ II (Takara). Expression of miR-222-3p was normalized to internal control U6. Primer sequences as following: miR-222-3p forward: 5’-GGGAGCTACATCTGGCTA-3’ and reverse: 5’-GTGTCGTGGAGTCGGCAA-3’; U6 forward: 5’-CTCGCTTCGGCAGCACA-3’ and reverse: 5’-AACGCTTCACGAATTTGCGT-3’; miR-222-3p stem-loop primer: 5’- CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGAGACC-3’.
Western Blotting
Cells were lysed with Mammalian Protein Extract Reagent (Thermo Scientific) supplemented with protease inhibitor cocktail (1:100) (Thermo Scientific). Cell lysates were electrophoresed on 10% SDS-PAGE and then transferred to PVDF membranes. The membranes were blocked with 5% milk and then hybridized with a primary antibody at 4°C overnight. The membrane was washed and hybridized with horseradish peroxidase (HRP)-conjugated secondary antibody. Signals were visualized by SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Scientific). Primary antibodies were listed below: anti-E-cadherin (HUABIO, EM0502, 1:1000), anti-Snail (HUABIO, ER1706-22, 1:4000), anti-Zeb2 (HUABIO, ER64964, 1:1000), anti-NF-κB-P65 (HUABIO, ET1603-12, 1:2000), anti-p-P65(S536) (CST, EP2294Y, 1:1000), anti-p-P65(S468) (ABclonal, AP0446, 1:1000), anti-β-actin (HUABIO, M1210-2, 1:5000), anti-DYKDDDDK Tag (HUABIO, M1402-2, 1:2000).
Cell Migration
For transwell migration assay, cell migration ability was measured by transwell permeable chamber with 8.0 μm pore size (Corning Costar, USA). Cell transfected with miR-222-3p inhibitor or negative control were collected after 72 h. 5 × 104 cells were seeded on the upper chamber. 2.5% FBS was used as a chemokine in the lower chambers. After 4-5 h of incubation at cell incubator cells were fixed with 4% paraformaldehyde for 30 min and stained with crystal violet solution for 5 min. Cells were removed from the upper membrane with cotton swabs and migrated cells on the lower membrane were taken photos under a microscope (Random 5 fields, 40 ×).
MiRNA ISH
In brief, FFPE tissue sections were deparaffinized in xylene and rehydrated in a series of decreasing alcohol concentrations. To expose miR-222-3p probes (MiR-222-3p probe sequence: 5’-ACCCAGTAGCCAGATGTAGC-3’), tissue sections were incubated with 15 μg/mL Proteinase K solution at 37°C for 10 min, after a titration experiment that established the optimal unmasking treatment while maintaining tissue morphology. 3% methanol-H2O2 were used to block endogenous peroxidase. Tissues were then pre-hybridized with ISH buffer for 30 min, and then hybridized with locked nucleic acid-based digoxigenin (DIG)-labeled miR binding oligonucleotides at 50°C. Tissues were then washed stringently in decreasing concentrations of saline-sodium citrate (SSC) buffer. Following the stringent wash, tissues were blocked in 2% sheep serum and 1% bovine serum albumin (BSA). Tissues were then incubated with anti-DIG-HRP at 37°C for 40 min. Freshly prepared DAB color developing solution was added and the color development time was controlled under the microscope. The sections were re-dyed by Harris hematoxylin for about 3 minutes and dehydrated. Positive is brownish yellow, and the nucleus is blue.
The scoring system (1-12) is done according to the depth of the color and the proportion of the positive cell. The images corresponding to scores 0, 3, 6, 9, and 12 are shown in Supplementary Figure 1.
IHC
FFPE tissue sections were deparaffinized, rehydrated, antigen retrieved using Proteinase K, and blocked in sheep serum and BSA solution as described above in the ISH section. Snail antigen was retrieved by citrate antigen retrieval solution (Beyotime Biotechnology). Then, tissue sections were incubated with polyclonal rabbit anti-human primary antibody (Snail 1:150, ER1706-22, HUABIO) at RT for 1 h. A DAB Horseradish Peroxidase Color Development Kit (Beyotime Biotechnology) was used for the subsequent steps. Tissue endogenous peroxidase activity was blocked with peroxidase block for 5 min. Thereafter, sections were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 30 min and visualized with diaminobenzidine (DAB) substrate at RT.
Statistical Analysis
A two-tailed t-test was used for continuous variables (GraphPad Prism v9.0 and SPSS v25). Mann-Whitney t-test was used for ranked variables. Ranked correlation analysis was used to analyze the correlation of ISH score (hsa-miR-222-3p) and IHC score (Snail) in 60 PTC samples. Each sample was corresponding to an ISH score and an H-score. The bioinformatics data on the miRNA expression and mutation status of PTC were publicly available from UCSC Xena Browser under GDC TCGA Thyroid Cancer (THCA) datasets. PTC samples in TCGA were divided into two groups (BRAFWT and BRAFV600E mutation) according to BRAF gene status. A two-tailed t-test was used for comparing the miRNA expression. p<0.05 was considered as statistically significant.
Results
The Expression of miR-222-3p in PTCs With or Without BRAFV600E Mutation
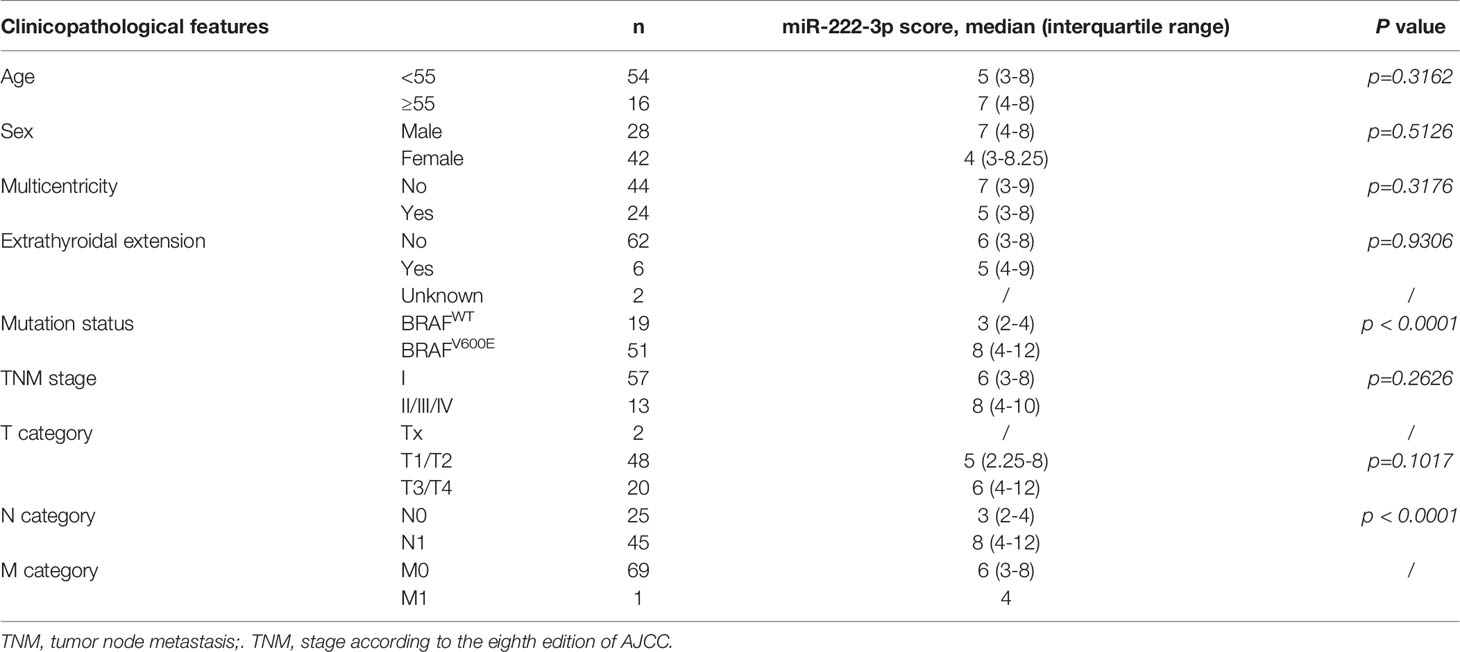
Through differential analysis of miR-222-3p expression in PTC clinical samples from TCGA, we found miR-222-3p had a higher expression level in PTC patients harboring BRAFV600E mutation (n=279) compared with that in PTC patients harboring BRAFWT (n=212) (p<0.001) (Figure 1A). Subsequently, to further verify the clinical correlation of miR-222-3p, we evaluated miR-222-3p expression in PTC clinical samples from our research center through ISH analysis (Table 1). ISH analysis of PTC patients harboring BRAFV600E mutation (n=51) and BRAFWT (n=19) demonstrated that PTC lesions with BRAFV600E mutation showed a higher level of miR-222-3p than that with BRAFWT (p<0.0001) (Figures 1B, C). In addition, we tested the expression of miR-222-3p in PTC cell lines, named TPC-1, B-CPAP, and K1 (TPC-1 cell line harbors BRAFWT, while B-CPAP and K1 cell line harbor BRAFV600E mutation). Through relative quantitative PCR (qPCR), we found the expression of miR-222-3p in B-CPAP (p=0.0154) and K1 (p=0.0035) cell lines was higher than that in TPC-1 (Figure 1D). We found that miR-222-3p was also overexpressed in the PTC cell line harboring BRAFV600E mutation.
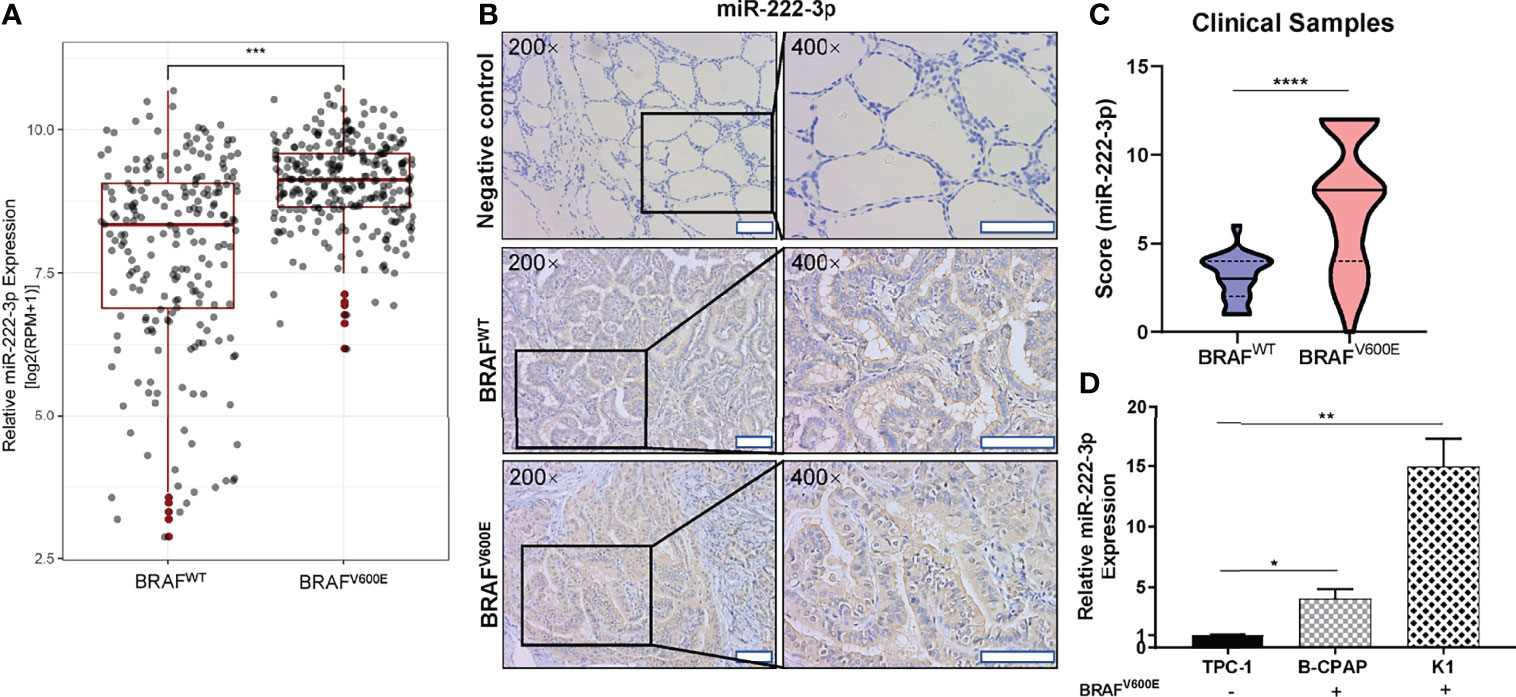
Figure 1 Hsa-miR-222-3p is overexpressed in PTC cells and patients harboring BRAFV600E mutation. (A) Differential analysis of miR-222-3p expression for 491 thyroid cancerous tissues with BRAFWT and BRAFV600E mutation. RPM: reads per million mapped reads. ***p < 0.001. (B) Representative images (200×, 400×) of CISH staining of PTC lesions from clinical patients harboring BRAFWT and BRAFV600E mutation. Negative control from normal thyroid tissues. Scale bar, 100 μM. (C) Mann-Whitney t-test analysis of miR-222-3p ISH score in PTC lesions between BRAFWT and BRAFV600E. The solid line and dotted line respectively represent median and interquartile range, ****p < 0.0001. (D) q-PCR validation of miR-222-3p expression in PTC cell lines including TPC-1, B-CPAP, and K1. Bars represent means ± SD of 3 independent experiments each measured in triplicate. *p < 0.05; **p < 0.01.
BRAFV600E Oncoprotein Promotes miR-222-3p Expression in PTCs
The above experiments showed that miR-222-3p expression in PTC was correlated with BRAFV600E mutation. We speculated that the BRAFV600E oncoprotein might promote miR-222-3p expression in PTC. To verify this hypothesis, we constructed a BRAFV600E-Flag overexpression plasmid (OE) with pcDNA3.1- as a vector. 48 h after transfection, the differential expression of miR-222-3p could not be observed in TPC-1 transfected with OE plasmid (OE group) and empty vector (CTL group), neither was in B-CPAP (Figures 2A–C). However, 72 h after transfection, miR-222-3p expression in the OE group in both TPC-1 (p=0.0069) and B-CPAP (p=0.0385) were significantly increased compared with that in the CTL group (Figures 2B, C). These results confirmed that the BRAFV600E oncoprotein up-regulated the expression of miR-222-3p in PTC cell lines.

Figure 2 BRAFV600E oncoprotein regulates miR-222-3p expression in PTCs. (A) immunoblot of BRAFV600E-Flag protein from extracts of TPC-1 and B-CPAP transfected with OE plasmid or empty vector. (B, C) q-PCR validation of expression level of miR-222-3p in TPC-1 and B-CPAP transfected with OE plasmid or empty vector after 48 h and 72 h. Bars represent means ± SD of 3 independent experiments each measured in triplicate. **p < 0.01; ***p < 0.001. (D) Model of inhibitors suppresses BRAFV600E-MEK-ERK pathway. (E) q-PCR validation of expression level of miR-222-3p in B-CPAP and TPC-1 treated with 10 μM PLX4720 or DMSO after 24 h. Bars represent means ± SD of 3 independent experiments each measured in triplicate. **p < 0.01. (F) q-PCR validation of expression level of miR-222-3p in B-CPAP and TPC-1 treated with 10 μM PD0325901 or DMSO after 24 h. Bars represent means ± SD of 3 independent experiments each measured in triplicate. ***p < 0.001.
To further clarify how the BRAFV600E oncoprotein regulates miR-222-3p, RAF/ERK pathway inhibitors were used to treat PTC cell lines and the expression of miR-222-3p was measured. PLX4720 is a highly selective BRAFV600E inhibitor, and its affinity to BRAFV600E mutant protein is about ten times higher than that to BRAF wild-type protein. PD0325901 is a non-ATP-competitive MEK inhibitor that inhibits the phosphorylation of ERK1 and ERK2 (Figure 2D). We treated TPC-1, K1, and B-CPAP with 10μM PLX4720 and PD0325901, respectively, and measured the expression of miR-222-3p by qPCR after 24 h of treatment. These inhibitors did not affect the expression of miR-222-3p in TPC-1 (Figures 2E, F). For the K1 cell line, we found that 1μM PD0325901 significantly reduced the expression of miR-222-3p by 70% (p=0.0308). However, 1~10μM PLX4720 didn’t inhibit the level of miR-222-3p in K1 (Supplementary Figures 2A, B). After treatment of B-CPAP with PLX4720, the expression of miR-222-3p was reduced by about 50% (p=0.0433), and after treatment with PD0325901, the expression of miR-222-3p in B-CPAP was reduced by about 70% (p=0.0234) (Figures 2E, F). These results showed that the constitutive expression of miR-222-3p in TPC-1, K1, and B-CPAP could be distinct. The RAF/MEK/ERK pathway is responsible for the expression of miR-222-3p in B-CPAP; however, it seems that BRAFV600E inhibitor PLX4720 plays no effect on the activation of the MAPK/ERK pathway and miR-222-3p expression in K1. As for TPC-1, there could be other mechanisms regulating miR-222-3p expression instead of MAPK/ERK pathway.
MiR-222-3p Is Associated With Lymph Node Metastasis in PTCs
Through the above experimental data, the relationship between miR-222-3p expression and BRAFV600E has been clarified. The ISH analysis also showed that PTCs with lymph node metastasis (n=45) had a higher level of miR-222-3p expression (p<0.0001) than those without lymph node metastasis (n=25) (Figures 3A, B). Besides, analysis of the TCGA database implied that the expression of miR-222-3p in PTCs with lymph node metastasis (n=226) was higher than that in PTCs without metastasis (n=231) (p < 0.0001) (Figure 3C). These results indicated that miR-222-3p overexpression might increase the risk of lymph node metastasis. Furthermore, miR-222-3p ISH score and BRAF mutation status were used to predict lymph node metastasis of PTCs. In the ROC curve analysis (Figure 3D), the area under the curve (AUC) was 0.8689, and the optimal cut-off value (0.5859) exhibited a sensitivity and specificity of 0.8667 and 0.76, respectively.
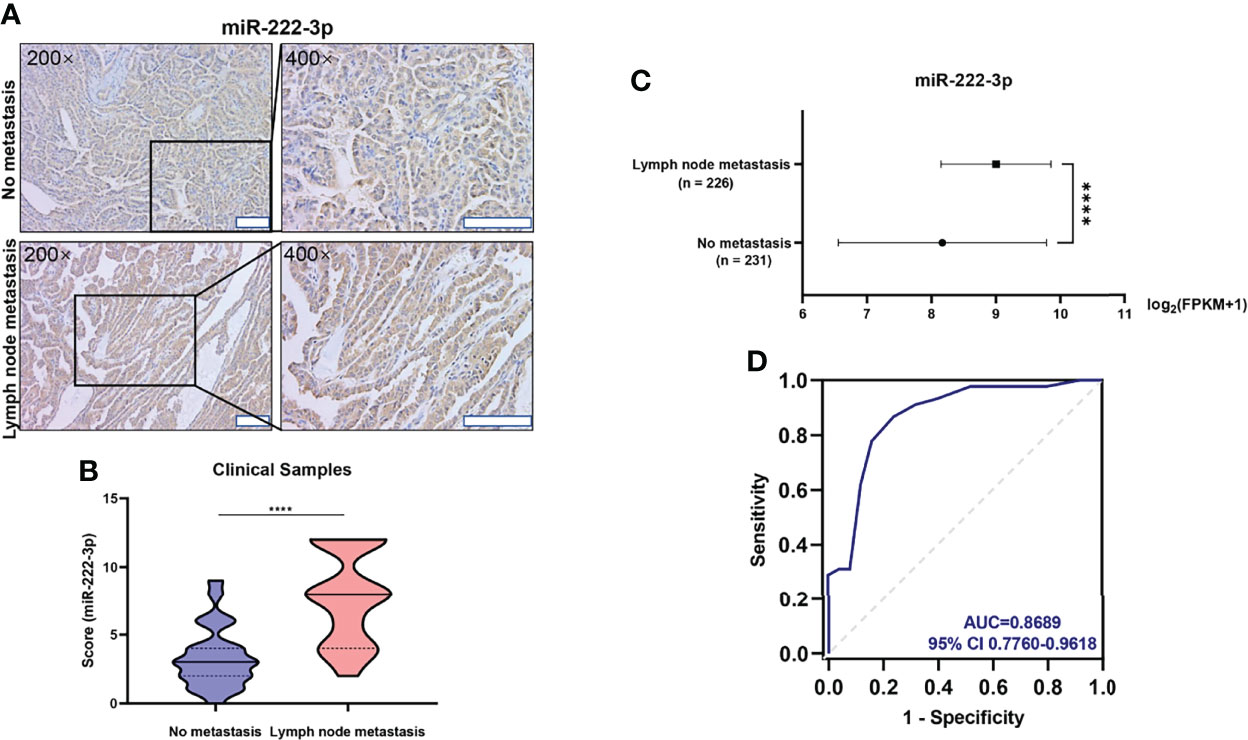
Figure 3 miR-222-3p is correlated with lymph node metastasis in PTCs (A) Representative images (200×, 400×) of CISH staining of PTC lesions from clinical patients with and without lymph node metastasis. Scale bar, 100 μM. (B) Mann-Whitney t-test analysis of miR-222-3p ISH score in PTC lesions between “No metastasis” and “lymph node metastasis” group. The solid line and dotted line respectively represent median and interquartile range, ****p < 0.0001. (C) Differential analysis of miR-222-3p expression between PTCs with lymph node metastasis (n = 226) and without metastasis (n = 231). FPKM: Fragments Per Kilobase per Million, ****p < 0.0001. (D) ROC curve for the prediction of lymph node metastasis in PTCs.
Downregulation of miR-222-3p Suppresses Cell Migration and EMT of PTCs
Next, we examined the role of miR-222-3p in PTC metastasis. The miR-222-3p inhibitor efficiency is greater than ninety-five percent (Figure 4D and Supplementary Figure 2C). Suppression of miR-222-3p in B-CPAP and TPC-1 cells by transfection of inhibitors suppressed cell migration (p=0.0009, p=0.0037) (Figures 4A–C). Nonetheless, the cell migration ability of K1 cell was not influenced by the miR-222-3p inhibitor (Supplementary Figure 2D). Meanwhile, EMT-related markers were measured including E-cadherin, N-cadherin, Twist1, Vimentin, Zeb1, Zeb2, Snail, and Slug through immunoblotting analysis. The decreased expression of Snail and Zeb2 and increased expression of E-cadherin were observed in the B-CPAP inhibitor group (Figure 4E and Supplementary Figure 3A). However, these changes cannot be observed in the TPC-1 cell line (Figure 4F and Supplementary Figure 3B). These results implicated that miR-222-3p influenced the cell migration ability and the underlying mechanism varied from types of cell lines or gene mutation.
Figure 4 Downregulation of miR-222-3p suppresses cell migration and EMT of PTCs. (A) Migration images of representative microscope fields (40x, 100x). (B, C) t-test analysis of the number of B-CPAP and TPC-1 migrating cells of N.C. and IN group. *** p< 0.001,**p < 0.01. (D) q-PCR validation of expression level of miR-222-3p in B-CPAP and TPC-1 transfected with negative control (N.C.) and miR-222-3p inhibitor (IN). ***p < 0.001,****p < 0.0001. (E, F) immunoblot of E-cadherin, Zeb2, and Snail protein from extracts of B-CPAP and TPC-1 transfected N.C. and IN. *p < 0.05.
Expression of Snail Is Correlated With Lymph Node Metastasis in PTCs Harboring BRAFV600E Mutation and miR-222-3p
In vitro experiment, we showed that miR-222-3p regulated the expression of Snail in B-CPAP. We further examined the expression of Snail in 60 clinical PTC samples by IHC. Through rank correlation analysis, we found that, Snail expression was correlated with miR-222-3p in the PTC clinical samples (r=0.33, p=0.0111) (Figure 5D).
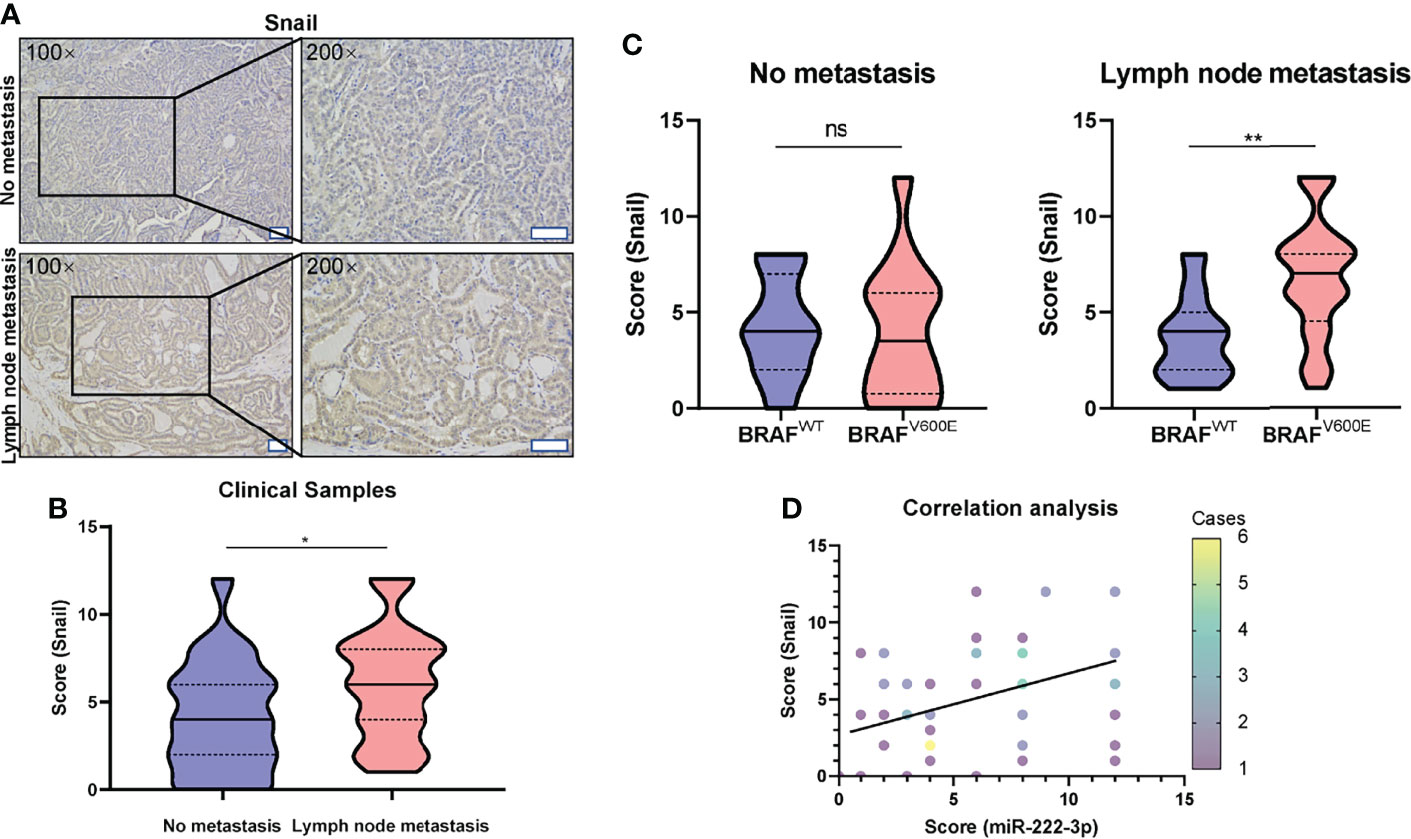
Figure 5 High expression of Snail is correlated with lymph node metastasis of PTC harboring BRAFV600E mutation. (A) Representative images (100×, 200×) of IHC staining of PTC lesions from clinical patients with and without lymph node metastasis. Scale bar, 100 μM. (B) Mann-Whitney t-test analysis of Snail H-score in PTC lesions between “No metastasis” and “lymph node metastasis” group. The solid line and dotted line respectively represent median and interquartile range, *p < 0.05. (C) Mann-Whitney t-test analysis of Snail H-score in PTC lesions with lymph node metastasis (right panel) or without metastasis (left panel) between BRAFWT and BRAFV600E group. The solid line and dotted line respectively represent the median and interquartile range, ns, p > 0.05, **p < 0.01. (D) Rank correlation analysis between the score of miR-222-3p and Snail from PTC clinical patients (Spearman r=0.33, N = 60, p=0.0111). Different color data points represented the different case numbers.
Furthermore, we also analyzed the correlation between Snail expression and PTC lymph node metastasis. The Snail IHC score in the tumor tissue of PTC patients with lymph node metastasis was significantly higher than that without lymph node metastasis (p=0.0219) (Figures 5A, B). Furthermore, in the tumor tissues of PTC patients without lymph node metastasis, BRAFV600E mutation is not significantly related to the Snail IHC score (Figure 5C left); while in the tumor tissues of PTC patients with lymph node metastasis, Snail IHC score in the BRAFV600E mutation group was significantly higher than that in the BRAFWT group (p=0.0072) (Figure 5C right). Consistent with the rank correlation analysis above, PTCs with both lymph node metastasis and BRAFV600E mutation had the highest Snail IHC score and the highest miR-222-3p ISH score among the four groups. Taken together, these results showed that miR-222-3p could promote lymph node metastasis through upregulating Snail in PTCs harboring BRAFV600E mutation.
Discussion
In this study, we clarified the correlation between the BRAFV600E mutation and the overexpression of miR-222-3p in PTC. Meanwhile, we demonstrated that the miR-222-3p could promote the EMT via up-regulation of Snail and lymph node metastasis of PTCs.
Previous studies related to this subject were either focused on the correlation analysis between the BRAFV600E mutation and aggressive clinicopathological characteristics such as recurrence, local extrathyroidal invasion, lymph node metastasis, and distant metastasis (17) or concentrated on the dysregulation of miR-222-3p in PTC patients (10). However, there is a lack of exploration between the BRAFV600E mutation and the high expression of miR-222-3p in PTCs. In the present study, we used the public TCGA database for big data analysis combined with small sample verification to clarify that the BRAFV600E mutation in PTC is related to the upregulation of miR-222-3p expression. Here, the expression level of miR-222-3p in clinical PTC samples was measured via the semi-quantitative CISH because of the small tumor size and the inevitable false positive staining of red blood cells in FISH. By inhibiting BRAFV600E and MEK, or overexpressing exogenous BRAFV600E in PTC cell lines, we confirmed that the expression level of miR-222-3p was indeed regulated by the BRAFV600E mutation and BRAF-MEK-ERK pathway. In another word, miR-222-3p is located downstream of the BRAFV600E/ERK pathway and may act as an intermediate molecule of BRAFV600E oncoprotein to promote the malignant transformation. A similar conclusion was drawn in breast cancer that miR-222-3p was downstream of RAS/RAF/MEK/ERK (18).
Aberrant expression of miR-222-3p played an essential role in various types of cancers. Fan et al. found that decreased miR-222-3p promoted EMT in epithelial ovarian cancer (19). However, up-regulation of miR-222-3p in cancers was likely to be aggressive characteristics including promotion of cell proliferation, inhibition of cell apoptosis, and enhancement of cell migration and invasion (18, 20). Gulluoglu et al. found that increased miR-222-3p promoted chordoma progression by targeting E-cadherin-mediated Zeb1-induced EMT (9). In breast cancer, up-regulation of miR-222-3p promoted Zeb2-induced EMT by targeting transcriptional repressor TRPS1 (18). Here, we found that inhibition of miR-222-3p could suppress EMT-related transcription factors, Snail and Zeb2, and meanwhile suppress cell migration in PTCs. MiR-222-3p could also facilitate or complicate migration and invasion in several types of cancers such as osteosarcoma, gastric cancer, and endometrial carcinoma (7, 21, 22). ISH analysis and IHC analysis of PTC clinical samples also revealed the correlation between miR-222-3p and Snail. Previous studies demonstrated the regulation of BRAFV600E on Snail in thyroid cancer cell lines and the correlation between SNAI1 mRNA expression and lymph node metastasis in PTC patients (23, 24). Here, using IHC analysis of PTC clinical samples, we demonstrated that the expression of Snail in cancerous tissues was related to lymph node metastasis from the protein level, especially in patients harboring BRAFV600E mutation. In summary, our study demonstrated that the regulation of miR-222-3p by BRAFV600E oncoprotein promoted cell migration of PTCs as well as Snail-induced EMT and was correlated with lymph node metastasis of PTC patients.
Aberrant expression of microRNAs in the patients’ body fluids could be a piece of novel evidence to assist diagnosis of thyroid carcinoma and post-operation monitoring. On the basic findings of this topic, more studies needed to be carried out to validate our results. Meanwhile, the molecular mechanisms under how miR-222-3p affect lymph node metastasis of PTC patients also need to be further analyzed. These promising leads will provide a new potential biomarker for both diagnosis and prognosis in the clinic.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Human Research Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, China. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conception and design: TC, YG, and DX. Collection and assembly of the data: DX, ZL, LW, and YG. Language editing and grammar correction: TC. Development of the methodology: WL and XZ. Data analysis and interpretation: all authors. Manuscript writing: all authors. All authors contributed to the article and approved the submitted version.
Funding
The present study was funded by the National Natural Science Foundation of China (grant no. 81500115) and the Science and Technology Department of Zhejiang Province (grant no. 2016C33141).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.843334/full#supplementary-material
Supplementary Figure 1 | The images of the miR-222-3p ISH assay corresponding to scores 0, 3, 6, 9, and 12. Scale bar, 100 μM
Supplementary Figure 2 | The regulation of miR-222-3p in K1 cell line and cell migration experiment. (A) q-PCR validation of expression level of miR-222-3p in K1 treated with 1~10 μM PD0325901 or DMSO after 24 h. Bars represent means ± SD of 3 independent experiments each measured in triplicate. *p < 0.05. (B) q-PCR validation of expression level of miR-222-3p in K1 treated with 1~10 μM PLX4720 or DMSO after 24 h. Bars represent means ± SD of 3 independent experiments each measured in triplicate. (C) q-PCR validation of expression level of miR-222-3p in K1 transfected with N.C and IN. **p < 0.01. (D) Migration images of random microscope fields (200x). There is no significant cell number difference between N.C. and IN in K1.
Supplementary Figure 3 | Statistical analysis graphs of EMT-associated proteins expression between N.C. and IN. (A) Using student’s t-test method to compare the expression level of E-cadherin, Zeb2, and Snail between N.C. and IN in B-CPAP cell line. *p < 0.05. (B) Using student’s t-test method to compare the expression level of E-cadherin, Zeb2, and Snail between N.C. and IN in TPC-1 cell line.
References
1. Nixon IJ, Whitcher MM, Palmer FL, Tuttle RM, Shaha AR, Shah JP, et al. The Impact of Distant Metastases at Presentation on Prognosis in Patients With Differentiated Carcinoma of the Thyroid Gland. Thyroid Off J Am Thyroid Assoc (2012) 22:884–9. doi: 10.1089/thy.2011.0535
2. Wang TS, Sosa JA. Thyroid Surgery for Differentiated Thyroid Cancer - Recent Advances and Future Directions. Nat Rev Endocrinol (2018) 14:670–83. doi: 10.1038/s41574-018-0080-7
3. Krol J, Loedige I, Filipowicz W. The Widespread Regulation of microRNA Biogenesis, Function and Decay. Nat Rev Genet (2010) 11:597–610. doi: 10.1038/nrg2843
4. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in Cancer: Biomarkers, Functions and Therapy. Trends Mol Med (2014) 20:460–9. doi: 10.1016/j.molmed.2014.06.005
5. Wang Z, Zhang H, He L, Dong W, Li J, Shan Z, et al. Association Between the Expression of Four Upregulated miRNAs and Extrathyroidal Invasion in Papillary Thyroid Carcinoma. OncoTarg Ther (2013) 6:281–7. doi: 10.2147/OTT.S43014
6. Kondrotienė A, Daukša A, Pamedytytė D, Kazokaitė M, Žvirblienė A, Daukšienė D, et al. Plasma-Derived miRNA-222 as a Candidate Marker for Papillary Thyroid Cancer. Int J Mol Sci (2020) 21:6445. doi: 10.3390/ijms21176445
7. Liu B, Che Q, Qiu H, Bao W, Chen X, Lu W, et al. Elevated MiR-222-3p Promotes Proliferation and Invasion of Endometrial Carcinoma via Targeting Erα. PloS One (2014) 9:e87563. doi: 10.1371/journal.pone.0087563
8. Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, et al. miR-221 and miR-222 Expression Affects the Proliferation Potential of Human Prostate Carcinoma Cell Lines by Targeting p27Kip1. J Biol Chem (2007) 282:23716–24. doi: 10.1074/jbc.M701805200
9. Gulluoglu S, Tuysuz EC, Kuskucu A, Ture U, Atalay B, Sahin F, et al. The Potential Function of microRNA in Chordomas. Gene (2016) 585:76–83. doi: 10.1016/j.gene.2016.03.032
10. Jiang K, Li G, Chen W, Song L, Wei T, Li Z, et al. Plasma Exosomal miR-146b-5p and miR-222-3p Are Potential Biomarkers for Lymph Node Metastasis in Papillary Thyroid Carcinomas. OncoTarg Ther (2020) 13:1311–9. doi: 10.2147/OTT.S231361
11. Lee JC, Zhao JT, Clifton-Bligh RJ, Gill A, Gundara JS, Ip JC, et al. MicroRNA-222 and microRNA-146b Are Tissue and Circulating Biomarkers of Recurrent Papillary Thyroid Cancer. Cancer (2013) 119:4358–65. doi: 10.1002/cncr.28254
12. Carling T, Udelsman R. Thyroid Cancer. Annu Rev Med (2014) 65:125–37. doi: 10.1146/annurev-med-061512-105739
13. Frasca F, Nucera C, Pellegriti G, Gangemi P, Attard M, Stella M, et al. BRAF(V600E) Mutation and the Biology of Papillary Thyroid Cancer. Endocrine-Related Cancer (2008) 15:191–205. doi: 10.1677/ERC-07-0212
14. Elisei R, Ugolini C, Viola D, Lupi C, Biagini A, Giannini R, et al. BRAF(V600E) Mutation and Outcome of Patients With Papillary Thyroid Carcinoma: A 15-Year Median Follow-Up Study. J Clin Endocrinol Metab (2008) 93:3943–9. doi: 10.1210/jc.2008-0607
15. Franco AT, Malaguarnera R, Refetoff S, Liao X-H, Lundsmith E, Kimura S, et al. Thyrotrophin Receptor Signaling Dependence of Braf-Induced Thyroid Tumor Initiation in Mice. Proc Natl Acad Sci USA (2011) 108:1615–20. doi: 10.1073/pnas.1015557108
16. Caronia LM, Phay JE, Shah MH. Role of BRAF in Thyroid Oncogenesis. Clin Cancer Res Off J Am Assoc Cancer Res (2011) 17:7511–7. doi: 10.1158/1078-0432.CCR-11-1155
17. Moon S, Song YS, Kim YA, Lim JA, Cho SW, Moon JH, et al. Effects of Coexistent BRAF and TERT Promoter Mutations on Poor Clinical Outcomes in Papillary Thyroid Cancer: A Meta-Analysis. Thyroid Off J Am Thyroid Assoc (2017) 27:651–60. doi: 10.1089/thy.2016.0350
18. Stinson S, Lackner MR, Adai AT, Yu N, Kim H-J, O'Brien C, et al. TRPS1 Targeting by miR-221/222 Promotes the Epithelial-to-Mesenchymal Transition in Breast Cancer. Sci Signaling (2011) 4:ra41. doi: 10.1126/scisignal.2001538
19. Fan L, Lei H, Zhang S, Peng Y, Fu C, Shu G, et al. Non-Canonical Signaling Pathway of SNAI2 Induces EMT in Ovarian Cancer Cells by Suppressing miR-222-3p Transcription and Upregulating PDCD10. Theranostics (2020) 10:5895–913. doi: 10.7150/thno.43198
20. Chatterjee A, Jana S, Chatterjee S, Wastall LM, Mandal G, Nargis N, et al. MicroRNA-222 Reprogrammed Cancer-Associated Fibroblasts Enhance Growth and Metastasis of Breast Cancer. Br J Cancer (2019) 121:679–89. doi: 10.1038/s41416-019-0566-7
21. Guo J, Liu Q, Li Z, Guo H, Bai C, Wang F. miR-222-3p Promotes Osteosarcoma Cell Migration and Invasion Through Targeting TIMP3. OncoTarg Ther (2018) 11:8643–53. doi: 10.2147/OTT.S175745
22. Tan X, Tang H, Bi J, Li N, Jia Y. MicroRNA-222-3p Associated With Helicobacter Pylori Targets HIPK2 to Promote Cell Proliferation, Invasion, and Inhibits Apoptosis in Gastric Cancer. J Cell Biochem (2018) 119:5153–62. doi: 10.1002/jcb.26542
23. Baquero P, Sánchez-Hernández I, Jiménez-Mora E, Orgaz JL, Jiménez B, Chiloeches A. (V600E)BRAF Promotes Invasiveness of Thyroid Cancer Cells by Decreasing E-Cadherin Expression Through a Snail-Dependent Mechanism. Cancer Lett (2013) 335:232–41. doi: 10.1016/j.canlet.2013.02.033
Keywords: papillary thyroid carcinoma, lymph node metastasis, miR-222-3p, BRAFV600E, Snail
Citation: Gao Y, Xiang D, Li W, Zheng X, Wang L, Li Z and Chen T (2022) BRAFV600E Mutation-Responsive miRNA-222-3p Promotes Metastasis of Papillary Thyroid Cancer Cells via Snail-Induced EMT. Front. Endocrinol. 13:843334. doi: 10.3389/fendo.2022.843334
Received: 25 December 2021; Accepted: 14 April 2022;
Published: 16 May 2022.
Edited by:
Joachim Feldkamp, Bielefeld University, GermanyReviewed by:
Khawla S. Al-Kuraya, King Faisal Specialist Hospital & Research Centre, Saudi ArabiaLoris Bertazza, University of Padua, Italy
Copyright © 2022 Gao, Xiang, Li, Zheng, Wang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Chen, dGNoZW4yMDEzQHpqdS5lZHUuY24=
†These authors have contributed equally to this work and shared first authorship
 Yuan Gao
Yuan Gao Dapeng Xiang2†
Dapeng Xiang2† Lin Wang
Lin Wang Zhiyu Li
Zhiyu Li Ting Chen
Ting Chen