- Department of Gynecology, Meizhou People’s Hospital, Meizhou, Guangdong, China
Background: Observation studies have confirmed the association between the gut microbiome and reproductive endocrine diseases (REDs), namely, polycystic ovary syndrome (PCOS), endometriosis, and female infertility. However, their association has never been confirmed by a two-sample Mendelian randomization (MR) analysis.
Methods: We conducted a two-sample MR analysis to evaluate the relationship between the gut microbiome and the three aforementioned REDs. In order to get more comprehensive results, two different thresholds were adopted to select instrumental variables (IVs): one was a locus-wide significance threshold (P <1.0×10–5) and the other was a genome-wide significance level (P< 5×10-8). Summary-level statistics for the gut microbiome and REDs were collected from public databases. Inverse-variance weighted (IVW) was the main method used to estimate causality, and sensitivity analyses were conducted to validate the MR results.
Results: At the locus-wide significance level, we identified that the genera Streptococcus (OR=1.52, 95%CI: 1.13-2.06, P=0.006) and RuminococcaceaeUCG005 (OR=1.39, 95%CI: 1.04-1.86, P=0.028) were associated with a high risk of PCOS, while Sellimonas (OR= 0.69, 95%CI: 0.58-0.83, P=0.0001) and RuminococcaceaeUCG011(OR=0.76, 95%CI: 0.60-0.95, P=0.017) were linked to a low PCOS risk. The genus Coprococcus2 (OR=1.20, 95%CI: 1.01-1.43, P=0.039) was correlated with an increased risk of female infertility, while Ruminococcus torques (OR=0.69, 95%CI: 0.54-0.88, P=0.002) were negatively associated with the risk of female infertility. The genera Olsenella (OR= 1.11, 95%CI: 1.01-1.22, P=0.036), Anaerotruncus (OR= 1.25, 95%CI: 1.03-1.53, P=0.025), and Oscillospira (OR= 1.21, 95%CI: 1.01-1.46, P=0.035) were linked to a high risk of endometriosis. However, the results showed that the gut microbiome did not possess a causal link with REDs risk based on the genome-wide significance level. Sensitivity analyses further confirmed the robustness of the MR results.
Conclusion: Our study provides evidence that gut microbiome is closely related with REDs. Subsequent studies should be conducted to promote microbiome-orientated therapeutic strategies for managing REDs.
1 Introduction
The homeostasis of sex hormones plays a significant role in the reproductive endocrine system throughout the lifetime of a female. Disturbance in sex hormones may lead to reproductive endocrine diseases (REDs) such as polycystic ovary syndrome (PCOS), endometriosis, and infertility that have bothered female people of reproductive age for many years. PCOS is one of the most prevalent endocrine and metabolic disorders in reproductive-aged female people. Female people presenting PCOS have a high prevalence of endocrine–metabolic dysfunction, including obesity, insulin resistance, hyperinsulinemia, and dyslipidemia, resulting in a significantly increased risk for mood disorders, type 2 diabetes mellitus, infertility, metabolic disorders, cardiovascular disorders, and the development of cancer (1–3). Infertility is defined as the failure to conceive after 12 months of regular unprotected sexual intercourse. The causes of infertility include male factors, female factors, and unknown factors. It has become a major public health problem affecting 8-12% of reproductive-aged couple (4). Endometriosis is a disease characterized by endometrial tissue outside the uterus, which affects 10% of reproductive-aged female people worldwide and leads to chronic painful symptoms and infertility in severe cases (5). Due to the health, economic, and social burdens caused by these diseases, it is urgent to understand the underlying mechanisms and obtain an adequate treatment for them.
Growing evidence has revealed the relationship between the gut microbiome and REDs. The gut microbiome is considered to be an endocrine organ and plays a major role in the reproductive endocrine system by affecting the fluctuation of sex hormones. The gut microbiome can affect estrogen levels by modulating the secretion of β-glucuronidase. The dysbiosis and reduction of gut microbiota diversity can decrease or increase β-glucuronidase activity and result in the fluctuation of circulating estrogens, which may lead to obesity, metabolic syndrome, cancer, endometrial hyperplasia, endometriosis, PCOS, and infertility (6, 7). The gut microbiome can also affect the level of circulating testosterone. The gut microbiota can synthesize and transform androgens by expressing the enzymes and are involved in the degradation of testosterone via microbial processes (8). For example, Proteobacteria can degrade androgen (9), and Clostridium scindens has a high potential to convert glucocorticoids into androgens (10). A combination of signs and symptoms of hyperandrogenism is a typical feature of PCOS. Previous studies found that the gut microbiome and its metabolites played an important role in the regulation of PCOS-associated ovarian dysfunction and insulin resistance (11). Obesity and PCOS also have a reverse effect on changing the gut microbiome composition, which may disrupt the ovarian function, damage oocyte quality, and cause chronic inflammation, hence further deteriorating fertility (12). Thus, the gut microbiome may have an impact on PCOS pathogenesis through a variety of mechanisms.
The majority of instances of female infertility can be explained in terms of ovulation disorders, uterine or cervical issues, tubal alterations, endometriosis, immune factors, and/or pelvic infections. However, approximately 30% of cases cannot be explained, and these are defined as “unexplained infertility” (13). Growing evidence has confirmed that gut microbiota dysbiosis has an indispensable impact on inflammatory conditions that affect male and female fertility (14, 15). An observational study found that female infertility showed a different bacterial richness and ratio, and an increasing level of inflammation comparing with the fertile group (15). A systematic review demonstrated that many autoantibodies, such as thyroid-related autoantibodies, anti-phospholipid antibodies, and anti-nuclear antibodies, impede the chances of a successful in vitro fertilization cycle (16). Therefore, we concluded that gut microbiome may have a close relationship with female infertility.
The understanding of the etiology of endometriosis is still lacking. Recently, studies have shown that the gut microbiome may be closely associated with the onset and progression of endometriosis due to its influence on the estrogen metabolism and inflammation. The increasing level of circulating estrogen derived by gut microbiome dysbiosis may stimulate the growth and cyclic bleeding of endometriotic lesions (8). Another reason is that dysbiosis in the gut microbiome disrupts the immune function, leading to the elevation of inflammatory cytokines and alteration of immune cell profiles. Over time, a chronic state of inflammation is developed to create an environment conducive to increased adhesion and angiogenesis, which may drive endometriosis onset and progression (17). The gut microbiome also contributes to the chronic pain of endometriosis by regulating microglia, astrocytes, and immune cells, and gut microbiome dysbiosis could lead to incorrect immune responses (18).
These above observations indicate there is a close link between the gut microbiome and the pathogenesis and progression of REDs; however, a Mendelian randomization (MR) analysis about their associations is still lacking. It is necessary to establish a causal relationship analysis to further understand the gut microbiome-derived mechanism and provide new insights into microbiome-orientated therapeutic strategies. Hence, we conducted a two-sample MR analysis to evaluate the relationship between gut microbiome composition and REDs. MR is an effective method to infer causality between exposures and outcomes by using genetic variations strongly associated with exposures as instrumental variables (IVs). MR can be regarded as a natural randomized controlled trial (RCT), which is not easily disturbed by confounding factors and has a high level of evidence.
2 Materials and methods
2.1 Data sources
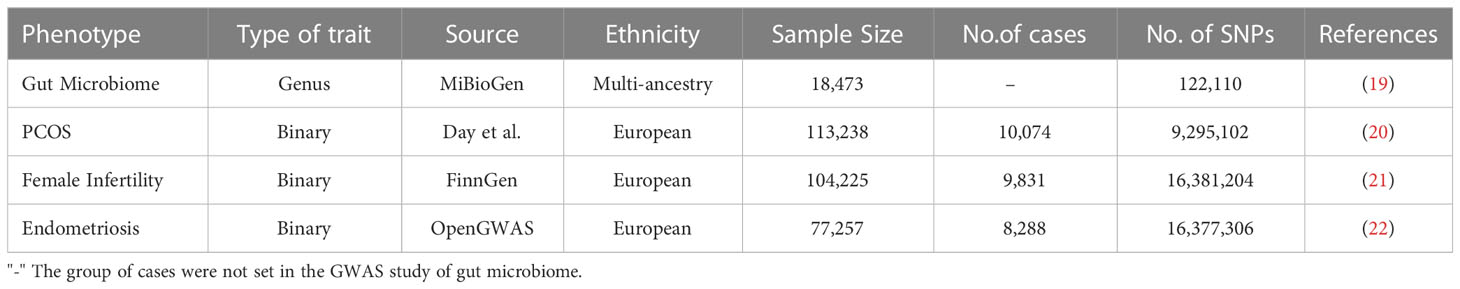
Genome-wide association studies (GWAS) data sources for the gut microbiome and PCOS, pregnancy loss, female infertility, and endometriosis were compiled and made publicly available online (Table 1). Single-nucleotide polymorphisms (SNPs) associated with the composition of the human gut microbiome were selected as IVs. Ethics approval was not required, since the data used were obtained from published studies or public databases.
2.1.1 Gut microbiome
GWAS summary statistics for the human gut microbiome were obtained from the MiBioGen study, which is a large-scale, multiethnic GWAS study recruiting 18, 473 individuals (24 cohorts) from various countries with 122,110 loci of variation (19). Most of the participants had European ancestry (n = 13, 266). A total of 211 taxa were categorized by five biological categories, including 9 phyla, 16 classes, 20 orders, 35 families, and 131 genera. As the genus was the smallest and most precise taxonomic level among all the category criteria, we performed subsequent analyses at the genus level only. Therefore, a total of 119 specific genera were included in the current analysis after removing 12 unknown genera out of 131 genera.
2.1.2 PCOS
GWAS data for PCOS were taken from Apollo (https://doi.org/10.17863/CAM.27720), which includes 10,074 PCOS cases and 103, 164 controls of European ancestry (20). Cases were either diagnosed according to the National Institutes of Health (NIH) or Rotterdam criteria or self‐reported history of PCOS.
2.1.3 Female infertility
Summary-level data for female infertility were also derived from the FinnGen consortium (104,225 female participants recruited, including 9, 831cases and 94,394 controls) (21).
2.1.4 Endometriosis
The GWAS summary datasets for endometriosis were accessed through the OpenGWAS database (22). The diagnostic criterion of endometriosis based on the International Classification of Diseases 10th code and GWAS ID is finn-b-N14_ENDOMETRIOSIS, with 77, 257 female participants recruited, including 8,288 cases and 68, 969 controls.
2.2 Selection of IVs
In this study, several steps were conducted to select eligible SNPs as IVs from the exposure data. First, SNPs strongly associated with the gut microbiome were selected. In order to obtain more comprehensive results, two different thresholds were adopted to select IVs: (1) SNPs at the locus-wide significance threshold (P < 1.0×10–5) were selected as potential IVs; (2) SNPs at the genome-wide significance level (P< 5 × 10-8) were selected as potential IVs. Second, to ensure that IVs used for the gut microbiome were independent, we excluded SNPs that had the linkage disequilibrium (LD) effect (r2 < 0.001, clumping window = 10,000kb). Third, SNPs related to confounders and risk factors for outcome were removed from the analysis by using the online database “PhenoScanner” (http://www.phenoscanner.medschl.cam.ac.uk/) with the filtration of r 2 > 0.8 and p < 1 × 10−5. The IVs of the gut microbiome identified above were extracted from each outcome dataset. Proxy SNPs were not sought by default when specific SNPs were absent in the outcome GWAS. Palindromic SNPs were also excluded. Afterwards, the exposure data and the outcome data were harmonized, which means that the effect of the SNP on the exposure was reconciled with the effect on the outcome in terms of the same allele. The strength of the included IVs was assessed with the F-statistics and R2. R2 reflects the degree to which the IV explains the exposure and is calculated as formula R2 = 2 × EAF × (1 − EAF) ×β2/[2 × EAF × (1 − EAF) × β2 + 2 × EAF × (1 − EAF)×N × se2] (EAF: effect allele frequency, se: the standard error for effect size, β: the effect size, N: the sample size) (23).The F-statistic was calculated by the formula F = R2 × (N − 2)/(1 − R2) (N:the sample size), where weak instrument bias is relatively low with an F-statistic over 10 (24).
2.3 MR analysis
An MR analysis was performed to determine if there is a causal relationship between the gut microbiome and the risk of REDs by using inverse-variance weighted (IVW) as the main method and other methods too, including MR-Egger, weighted median, and weighted mode. IVW was conducted to estimate the causality of each SNP with the assumption of no pleiotropy in these SNPs (25). Comparing with IVW, the MR-Egger not only allows the presence of pleiotropy in > 50% of IVs, but also detects horizontal pleiotropy in term of its intercept with a y-axis (26, 27). There is horizontal pleiotropy when the intercept is not zero. Point estimates from IVW MR are close to that of the MR-Egger when the intercept is close to zero. The weighted median was performed when the presence of pleiotropy was < 50% in IVs (28). The weighted mode method had less power to detect causal effects than the IVW and weighted median methods, but it was larger than that of MR-Egger and presented less bias than the above methods (29).
2.4 Sensitivity analysis
Cochran’s Q test was used to access heterogeneity (30). The MR-Egger regression test was performed to detect pleiotropy. There was horizontal pleiotropy when the intercept was not zero (26). MR-PRESSO was performed to reduce horizontal pleiotropy by detecting and removing final outliers (31). The leave-one-out sensitivity analysis was implemented to validate the robustness of the results by removing a single SNP each time.
A reverse MR analysis was not performed due to the lack of SNPs (related to REDs). All statistical analyses were performed using the package “TwoSampleMR” and “MR-PRESSO” in the R software (Version 4.2.0). Considering multiple-testing correction, FDR correction (Q-value) was performed using the Benjamini–Hochberg method.
3 Results
3.1 Instrumental variables selection
Initially, a total of 7098 SNPs categorized by 119 genera were extracted under the threshold of the locus-wide statistical significance (P< 1× 10-5). There was no genus containing only one SNP for each outcome dataset. In the present study, the F-statistic of IVs were all over 10, indicating no evidence of weak instrument bias. Detailed information including effect allele, other allele, Beta, SE, P-value, and F-statistics in IVs is shown in Supplementary Tables 1A–C. We evaluated the causal effect of each genus on the outcome data.
A total of 396 SNPs were extracted under the threshold of genome-wide statistical significance (P<5×10-8). After a series of quality control steps, a total of 12 independent SNPs were identified as IVs for PCOS, 11 independent SNPs for female infertility, and 11 independent SNPs for endometriosis. There was no weak instrument bias, as the F-statistics of IVs were all greater than 10. Detailed information including effect allele, other allele, Beta, SE, and P-value on IVs is shown in Supplementary Table 4A. Due to the limited number of IVs that met the requirements and each IV representing different genera, we took them as a whole to identify the gut microbiome to estimate its causal effect on outcome data.
3.2 Two-sample MR analysis(locus-wide significance level, P<1×10-5)
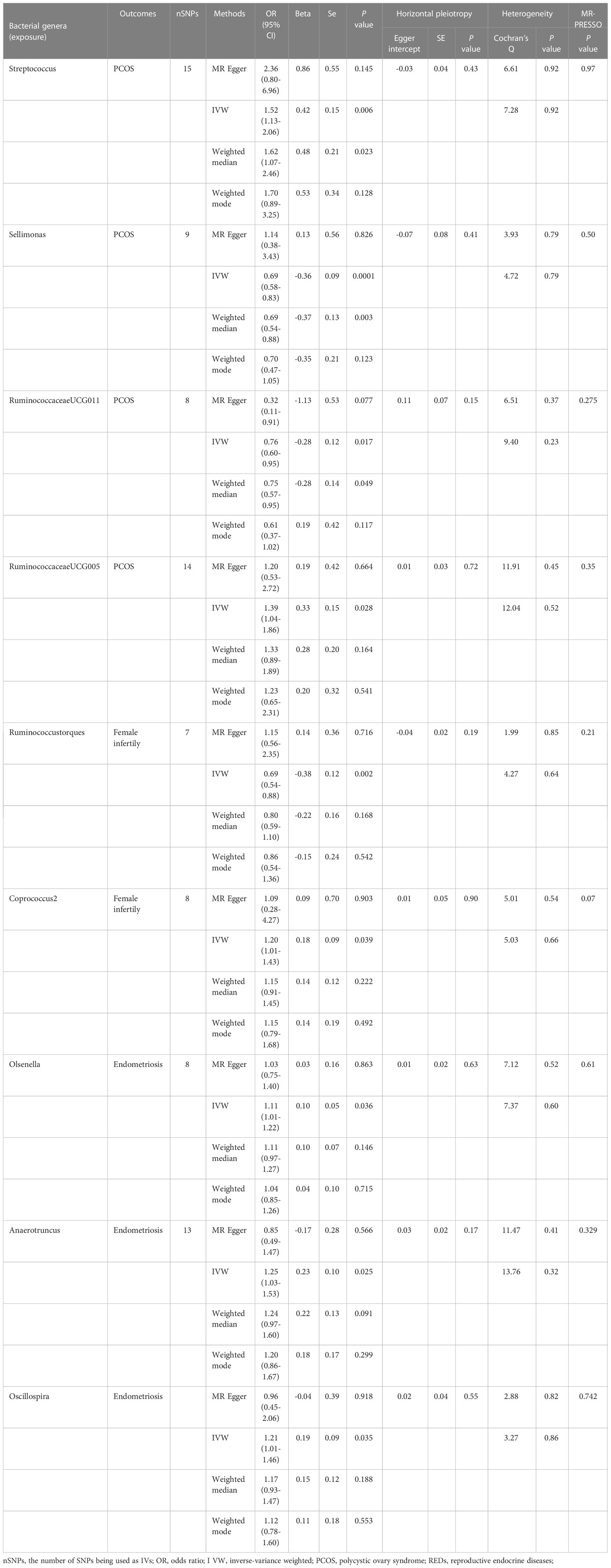
We conducted an MR analysis to evaluate the causal relationship between each genus and PCOS, female infertility, and endometriosis. The comprehensive results are shown in Supplementary Tables 2A–C. Streptococcus, Sellimonas, RuminococcaceaeUCG011, and RuminococcaceaeUCG005 were found to be associated with PCOS when evaluated by IVW. The IVW estimate suggested that the genera Streptococcus (OR:1.52, 95% confidence interval (CI):1.13-2.06, P=0.006) and RuminococcaceaeUCG005(OR:1.39, 95%CI:1.04-1.86, P=0.028) are positively associated with PCOS risk, while Sellimonas (OR:0.69,95%CI: 0.58-0.83, P=0.0001) and Ruminococcaceae UCG011(OR:0.76, 95%CI: 0.60-0.95, P=0.017) are negatively associated with PCOS risk (Table 2). However, only Sellimonas was still significant after FDR correction (Q-value= 0.015) (Supplementary Table 3). Ruminococcus torques and Coprococcus2 were found to be associated with female infertility when evaluated by IVW. The IVW estimate suggests that the genus Coprococcus2 (OR:1.20, 95%CI:1.01-1.43, P=0.039) is associated with an increased risk of female infertility, while Ruminococcus torques (OR:0.69, 95%CI:0.54-0.88, P=0.002) is associated with a decreased risk of female infertility (Table 2). However, no causal association of these genera with female infertility was supported after FDR correction (Supplementary Table 3). Olsenella, Anaerotruncus, and Oscillospira were found to be associated with endometriosis when evaluated by IVW. The IVW estimate suggested that the genera Olsenella (OR:1.11, 95%CI: 1.01-1.22, P=0.036), Anaerotruncus (OR:1.25, 95%CI: 1.03-1.53, P=0.025), and Oscillospira (OR:1.21,95%CI: 1.01-1.46, P=0.035) are associated with an increased risk of endometriosis (Table 2), while these associations were no longer significant after FDR correction (Supplementary Table 3).
The results of Cochran’s Q test evaluated by the IVW test and MR-Egger showed no significant heterogeneity between the gut microbiome and PCOS, female infertility, and endometriosis. There was no evidence of horizontal pleiotropy according to the results of the MR-Egger regression analysis. In addition, MR-PRESSO analysis did not find any significant outliers (Table 2). The leave-one-out results further validated data robustness (Figure 1).
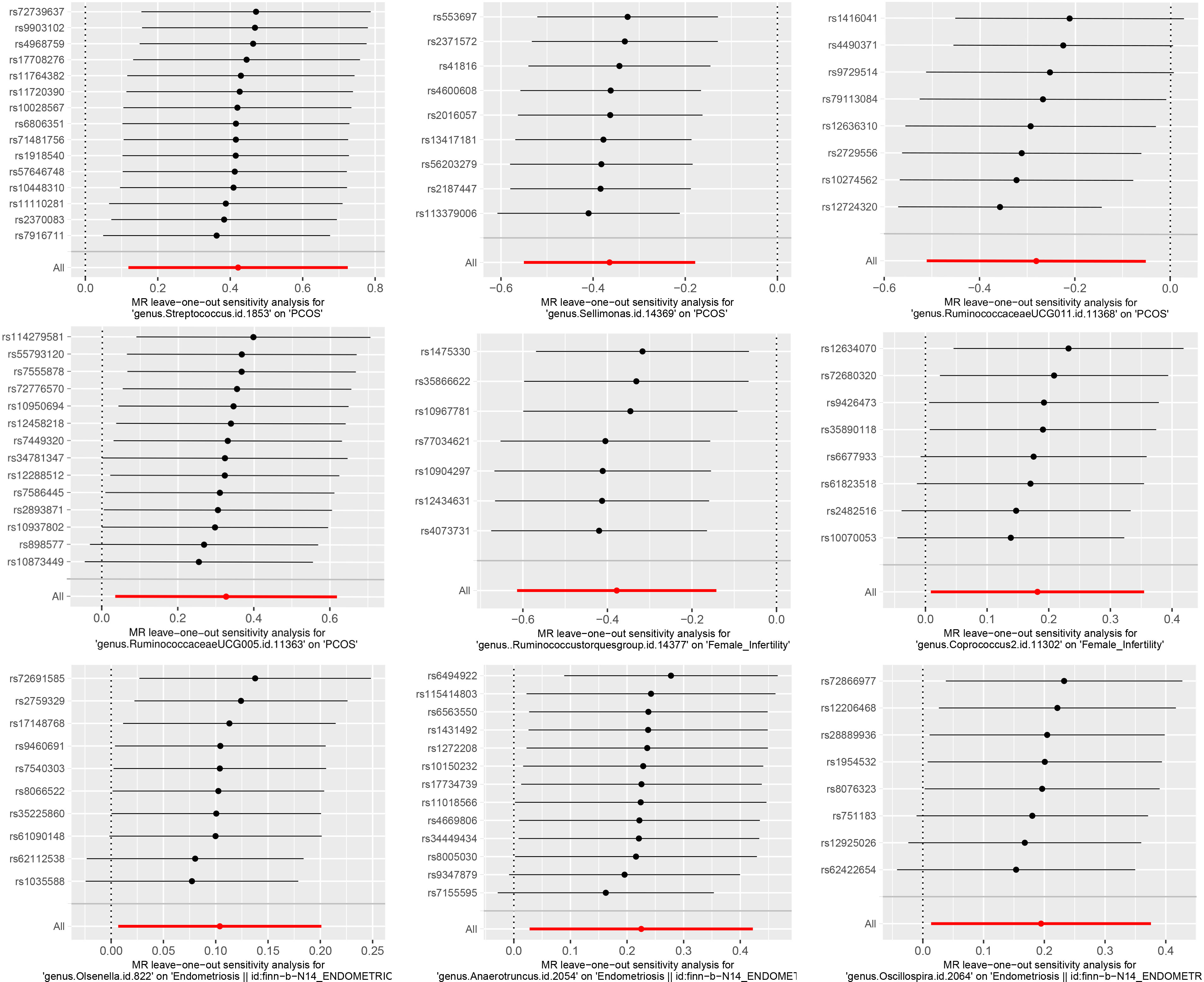
Figure 1 The leave-one-out sensitivity analysis assessed the associations between genera and REDs by removing a single SNP each time (P<1×10-5).
3.3 Two-sample MR analysis(genome-wide significance level, P<5×10-8)
Considering the gut microbiome as a whole, the results of the MR analysis evaluated by IVW (OR = 1.06, 95% CI 0.87-1.30, P =0.58) did not show a significant causal relationship between the gut microbiome and PCOS. The other methods showed directionally consistent results (Supplementary Table 4B). We also could not find a causal link between the gut microbiome and female infertility (IVW: OR = 0.98, 95% CI 0.86-1.13, P =0.82) and endometriosis (IVW: OR = 0.96, 95% CI 0.86-1.09, P =0.56). Cochran’s Q statistics of the IVW test and the MR-Egger regression, respectively, showed no significant heterogeneity between gut the microbiome and PCOS and endometriosis. However, there was heterogeneity between the gut microbiome and female infertility; in this case, we applied the result of the weighted median as main MR result to evaluate the causal association between the gut microbiome and female infertility (OR =1.06, 95% CI 0.93-1.22, P=0.36). The MR-Egger regression results showed that there was no horizontal pleiotropy between the gut microbiome and PCOS, female infertility, and endometriosis. The MR-PRESSO analysis showed that there were no outliers in the analysis (Supplementary Table 4B). Moreover, the leave-one-out results further validated the data robustness (Supplementary Figure 1).
4 Discussion
In the current study, we conducted MR analyses to evaluate the potential causality between the gut microbiota and REDs. Based on the locus-wide significance level, we identified that the genera Streptococcus and RuminococcaceaeUCG005 were associated with a high risk of PCOS, while Sellimonas and RuminococcaceaeUCG011were linked to a low PCOS risk. The genus Coprococcus2 was associated with an increased risk of female infertility, while Ruminococcus torques was negatively associated with the risk of female infertility. Genus Olsenella, Anaerotruncus, and Oscillospira were linked to a high risk of endometriosis. However, the results showed that gut microbiome did not have a causal link with REDs risk based on the genome-wide statistical significance level.
There are many previous studies on the relationship between the gut microbiome and PCOS, with Ruminococcaceae among them (32, 33). RuminococcaceaeUCG005 is a member of the Ruminococcaceae family and is viewed as a harmful bacterium in high-fat diet (HFD)-fed rats, and it correlates with oxidative stress, metabolism genes, and body weight (34). Prior evidence indicated that increased oxidative stress and elevated inflammatory status contribute to the progression of PCOS (35, 36), and weight loss is an important part of PCOS treatment (37, 38), which might partly explain why RuminococcaceaeUCG005 is associated with a high risk of PCOS. Female people presenting PCOS characterized by a combination of signs and symptoms of androgen excess have a high prevalence of obesity, insulin resistance, and dyslipidemia. Streptococcus, considered “bad bacteria”, was previously shown to be associated with obesity and significantly higher in obese and PCOS adults (39–42). Streptococcus was positively correlated with insulin resistance, testosterone, and BMI (33, 43–45). It was also found to be involved with carbohydrate metabolism and positively associated with insulin, connecting peptide, lipopolysaccharide, and pro-inflammatory indicators (42, 46, 47).However, it was negatively correlated with short-chain fatty acids (SCFAs) (42). SCFAs, the microbial fermentation end-products, may help suppress the levels of pro-inflammation cytokines, reduce inflammation in the intestine, and maintain the homeostasis of the intestinal environment (48).SCFAs are associated with insulin releasing and blood glucose levels by supporting the health of beta cells in the pancreas, and they stimulate the secretion of glucagon-like peptide-1 (GLP-1) (49, 50). Additionally, SCFA acetate may help people control weight and support healthy weight maintenance in terms of regulating hormones (such as GLP-1), increasing metabolism, and inhibiting appetite (51). Research showed that acetate could protect ovarian function by supporting normal follicles growth and enhancing circulating 17-β estradiol through the inhibition of histone deacetylase in the rat model of PCOS (52). Additionally, it has been found that probiotics and SCFAs administration, as part of anti-obesity and diabetes interventions, could involve the modification of microbiota, the upregulation of GLP-1 production and related SCFAs, such as acetate, and increasing fasting fat oxidation and resting energy expenditure (53–55). Zhang et al. found that probiotics impact the gut microbiota and sex hormones of PCOS patients by significantly decreasing the levels of luteinizing hormone (LH) and LH/follicle-stimulating hormone (LH/FSH) and markedly increasing SCFAs (56). The above evidence implies probiotics or SCFAs administration can be beneficial as part of the treatment of female disorders. Sellimonas was considered a potential biomarker of gut homeostasis recovery, as studies found that Sellimonas intestinalis was increased in patients with colorectal cancer who recovered their intestinal homeostasis following dysbiosis caused by radical surgery combined with chemotherapy (57)and in patients with liver cirrhosis who underwent therapeutic splenectomy (58). Studies on the association between Sellimonas and PCOS are limited. We speculated Sellimonas may contribute to the recovery of intestinal homeostasis in patients with PCOS. RuminococcaceaeUCG011 was negatively correlated with the serum and hepatic lipid profiles and was significantly increased after hypoglycemic and hypolipidemic intervention in type 2 diabetic mice (59). As Ruminococcaceae UCG011 was shown to be inversely correlated with the serum levels of triglyceride (TG), cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), and the hepatic levels of TC, TG, non-esterified fatty acids (NEFA), and bile acids (BAs) (59), it may show a protective effect in PCOS, as in other metabolic diseases, which is consistent with our findings.
A great deal of evidence supports the role of the gut microbiome in female infertility. Coprococcus2, associated with an increased risk of female infertility in our study, was previously found to be the characteristic genus of obese patients with PCOS (60, 61). Coprococcus2 was also found to be indirectly associated with chronic low-grade systemic inflammation induced by diets and ectopic fat in the Multiethnic Cohort-Adiposity Phenotype Study (62). Increasing evidence suggests that infertility is related to chronic low-grade inflammation characterized by increased inflammatory markers, such as C-reactive protein (CRP), IL-18, TNF-α, and IL-6 (63, 64). Diets with a marked anti-inflammatory signature have been proposed in the nutritional management of infertile patients (65). For the first time, our finding has implied the relationship between Coprococcus2 and female infertility. This may provide new insights into improving fertility through diets that could decrease the abundance of Coprococcus2. Ruminococcus torques, negatively associated with the risk of female infertility in our study, was found to be negatively correlated with pre-pregnancy body weight in a previous study (66). Several studies have shown both underweight (BMI < 19 kg/m2) and overweight (BMI 25–29.9 kg/m2) affect infertility (67, 68). A cohort study with 9232 participants from Denmark found that overweight and obese mothers with a body mass index (BMI) >25 kg/m2 may harm the fertility of fetuses, and especially, sons born to overweight mothers have higher odds of infertility. However, the study did not find an association between maternal overweight and infertility in daughters (69). Tang et al. found that being underweight with BMI < 18.5 kg/m2 is linked to reduced implantation rates, clinical pregnancy rates, and ongoing pregnancy. Rates of miscarriage were markedly increased in the overweight group relative to the normal weight group (70). We speculated that Ruminococcus torques may affect fertility through its impact on pre-pregnancy body weight. Wen at el. found that Ruminococcus torques was generated after a high-cellulose diet in a mouse model of asthma. They speculated Ruminococcus torques is closely correlated with lipid metabolism in vivo (71).Wang et al. discovered that the abundance of Ruminococcus torques was relatively higher after mitigation treatment of colonic inflammation in colitis. Previous studies have revealed the protective role of Ruminococcus torques in other diseases, but further studies need to reveal their protective role in female infertility.
In previous studies, many researchers have revealed the relationship between the gut microbiome and endometriosis. As we know, endometriosis development is influenced by estrogen metabolism and inflammation. The abundance of Olsenella has been found to be associated with the exacerbation of inflammation. The low-abundance and pathogenic bacterium Olsenella proliferates when inflammatory hyperthermia causes host gut microbiota disorders, which further exacerbates the inflammatory response (72), whereas the downregulation of Olsenella can reduce the possibility of inflammation in the rumen epithelium and the organism (73). The abundance of Olsenella is positively correlated with the IL-10 levels (74). IL-10 family proteins, as the main members of Th2 anti-inflammatory cytokines, have been considered a critical factor for the development of endometriosis. Accumulating evidence has demonstrated that IL-10 is sharply increased in the ectopic endometrium and peritoneal fluid of female people with endometriosis, particularly in cases of advanced endometriosis (75–78). Previous research demonstrated that IL-10 from infiltrated plasmacytoid dendritic cells may suppress immunity against endometrial implants to contribute to the development of endometriosis (78) and promote angiogenesis in the early stage of endometriosis (79). Therefore, Olsenella may be involved in the development of endometriosis by modulating the level of IL-10. Moreover, N-acetylserotonin (NAS), an endogenous metabolite, was significantly negatively correlated with Olsenella (72). It has been proposed that the NAS/melatonin ratio is linked to endometriosis pathophysiology. Endometriosis, as an estrogen-dependent condition, is usually mitigated by lowering the estrogen effects. Melatonin inhibits the ERα (80), which modulates the stage transition in endometriosis (81), suggesting that melatonin inhibits ERα-driven pathophysiology in endometriosis. Endometriosis risk is also correlated with CYP1B1 SNPs (82), which increases the backward conversion of melatonin to NAS (83). Further analysis is needed to evaluate the relationships between the NAS/melatonin ratio, Olsenella, and endometriosis. The second genus we found to have a positive association with endometriosis is Oscillospira. Oscillospira was found to associated with adiposity and metabolic dysfunction and gut inflammation and serum triglycerides in mice and humans, respectively (84).Chen et al. discovered that Oscillospira is closely related to human health, because its abundance is positively correlated with high-density lipoprotein, microbial diversity, and sleep time and is inversely correlated with blood pressure, fasting blood glucose, uric acid, triglyceride, and Bristol stool type (85). Jae-Kwon Jo found that Oscillospira was significantly higher in the HFD mice than that those in the control group, producing SCFAs such as acetate, propionate, and butyrate (86). SCFAs butyrate supplement was found to alleviate the symptoms caused by low estrogen, such as excessive osteoclastogenesis and bone loss in estrogen-deficient mice due to ovariectomy (OVX) (87, 88).The association between Oscillospira and host health, SCFAs production, and the estrogen level needs further comprehensive exploration. We speculate that Oscillospira may be involved in endometriosis etiology when gut dysbiosis and estrogen imbalance occur. Anaerotruncus was the last genus we found to positively relate to endometriosis, and it was found to associated with inflammation and obesity in previous studies (89). Li et al. confirmed that the relative abundance of Anaerotruncus decreased when anti-inflammation treatment reduced psoriasis-like inflammation in mice (90). Kong et al. found that Anaerotruncus, as a conditional pathogenic bacterium, increased in a mouse model fed by HFD and high-sucrose diets (HCDs) (91). Anaerotruncus has been positively associated with glucose intolerance and gut permeability and is involved in the pathogenesis of diabetes (92). However, the studies about the mechanism of Anaerotruncus in endometriosis etiology still lack.
Our study has several advantages. To our knowledge, this is the first MR analysis to evaluate the causal relationship between the gut microbiome and REDs. The MR design has the advantage of preventing disturbance from residual confounding and might be more convincing than observational studies. However, as the exact biological function of many genetic variants is still unknown, we cannot completely avoid the impact of horizontal pleiotropy. Thus, the results should be interpreted with caution. Moreover, we analyzed the causal effect of each taxon on REDs primarily from the genus level. This provides new insights for understanding the gut microbiome-derived mechanisms and microbiome-orientated therapeutic strategies. Several limitations should be mentioned. First, our study was unable to count the participants overlapping between the exposure and outcome GWAS, which may lead to an overestimation of the results. We were also unable to identify a reverse causal relationship between them due to the lack of an adequate number of IVs for REDs. Second, our study used gut microbiome data, including multiethnic male and female participants, whereas studies about REDs were conducted on female Europeans, which may have also influenced our results. However, we were unable to avoid this bias by conducting the sex or race subgroup analysis due to the lack of demographic data in the original research. Third, the sample size of each genus from the gut microbiome GWAS was relatively small compared with that for the REDs. Therefore, not enough IVs were identified for certain bacterial features at the genus level. Fourth, multiple statistical corrections are too rigorous and conservative, which may neglect potential genera that have a causal relationship with REDs. Therefore, we did not take the multiple testing results into account. Future studies need to plan to address these limitations.
In conclusion, we comprehensively evaluated the potential association between the gut microbiome and REDs. These strains may provide candidate biomarkers and new insights into the treatment for subsequent studies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YL and LH designed the study. WZ, BW, and TH performed data analysis. YL, RP, HY, and LH structured the manuscript and contributed to the tables, figures, and text editing. All authors contributed to the final manuscript and approved the submitted version.
Acknowledgments
We want to acknowledge the participants and investigators of FinnGen, the UK Biobank study (NealeLab) study, and the MiBioGen consortium for sharing the genetic data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1164186/full#supplementary-material
References
1. Escobar-Morreale HF. Polycystic ovary syndrome: definition, aetiology, diagnosis and treatment. Nat Rev Endocrinol (2018) 14(5):270–84. doi: 10.1038/nrendo.2018.24
2. Yin W, Falconer H, Yin L, Xu L, Ye W. Association between polycystic ovary syndrome and cancer risk. JAMA Oncol (2019) 5(1):106–7. doi: 10.1001/jamaoncol.2018.5188
3. Thong EP, Codner E, Laven JSE, Teede H. Diabetes: a metabolic and reproductive disorder in women. Lancet Diabetes Endocrinol (2020) 8(2):134–49. doi: 10.1016/S2213-8587(19)30345-6
4. Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, Stevens GA. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med (2012) 9(12):e1001356. doi: 10.1371/journal.pmed.1001356
5. Horne AW, Missmer SA. Pathophysiology, diagnosis, and management of endometriosis. BMJ (2022) 379:e070750. doi: 10.1136/bmj-2022-070750
6. Baker JM, Al-Nakkash L, Herbst-Kralovetz MM. Estrogen-gut microbiome axis: physiological and clinical implications. Maturitas (2017) 103:45–53. doi: 10.1016/j.maturitas.2017.06.025
7. Chadchan SB, Singh V, Kommagani R. Female reproductive dysfunctions and the gut microbiota. J Mol Endocrinol (2022) 69(3):R81–94. doi: 10.1530/JME-21-0238
8. Qi X, Yun C, Pang Y, Qiao J. The impact of the gut microbiota on the reproductive and metabolic endocrine system. Gut Microbes (2021) 13(1):1–21. doi: 10.1080/19490976.2021.1894070
9. Yang YY, Pereyra LP, Young RB, Reardon KF, Borch T. Testosterone-mineralizing culture enriched from swine manure: characterization of degradation pathways and microbial community composition. Environ Sci Technol (2011) 45(16):6879–86. doi: 10.1021/es2013648
10. Ridlon JM, Ikegawa S, Alves JM, Zhou B, Kobayashi A, Iida T, et al. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J Lipid Res (2013) 54(9):2437–49. doi: 10.1194/jlr.M038869
11. Qi X, Yun C, Sun L, Xia J, Wu Q, Wang X, et al. Gut microbiota-bile acid-interleukin-22 axis orchestrates polycystic ovary syndrome. Nat Med (2019) 25(8):1225–33. doi: 10.1038/s41591-019-0509-0
12. Snider AP, Wood JR. Obesity induces ovarian inflammation and reduces oocyte quality. Reproduction (2019) 158(3):R79–r90. doi: 10.1530/REP-18-0583
13. Vander Borght M, Wyns C. Fertility and infertility: definition and epidemiology. Clin Biochem (2018) 62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012
14. Lundy SD, Sangwan N, Parekh NV, Selvam MK, Gupta S, McCaffrey P, et al. Functional and taxonomic dysbiosis of the gut, urine, and semen microbiomes in Male infertility. Eur Urol (2021) 79(6):826–36. doi: 10.1016/j.eururo.2021.01.014
15. Azpiroz MA, Orguilia L, Palacio MI, Malpartida A, Mayol S, Mor G, et al. Potential biomarkers of infertility associated with microbiome imbalances. Am J Reprod Immunol (2021) 86(4):e13438. doi: 10.1111/aji.13438
16. Simopoulou M, Sfakianoudis K, Maziotis E, Grigoriadis S, Giannelou P, Rapani A, et al. The impact of autoantibodies on IVF treatment and outcome: a systematic review. Int J Mol Sci (2019) 20(4):892. doi: 10.3390/ijms20040892
17. Jiang I, Yong PJ, Allaire C, Bedaiwy MA. Intricate connections between the microbiota and endometriosis. Int J Mol Sci (2021) 22(11):5644. doi: 10.3390/ijms22115644
18. Ustianowska K, Ustianowski Ł, Machaj F, Gorący A, Rosik J, Szostak B, et al. The role of the human microbiome in the pathogenesis of pain. Int J Mol Sci (2022) 23(21):13267. doi: 10.3390/ijms232113267
19. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-Scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet (2021) 53(2):156–65. doi: 10.1038/s41588-020-00763-1
20. Day F, Karaderi T, Jones MR, Meun C, He C, Drong A, et al. Large-Scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet (2018) 14(12):e1007813. doi: 10.1371/journal.pgen.1007813
21. Kurki MI, Karjalainen J, Palta P, Sipilä T, Kristiansson K, Donner K, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. medRxiv (2022) 2022:2003. doi: 10.1101/2022.03.03.22271360
22. Elsworth B, Lyon M, Alexander T, Liu Y, Matthews P, Hallett J, et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv (2020) 2020:2008. doi: 10.1101/2020.08.10.244293
23. Shim H, Chasman DI, Smith JD, Smith J, Mora S, Ridker PM, et al. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 caucasians. PLoS One (2015) 10(4):e0120758. doi: 10.1371/journal.pone.0120758
24. Burgess S, Thompson SG. Avoiding bias from weak instruments in mendelian randomization studies. Int J Epidemiol (2011) 40(3):755–64. doi: 10.1093/ije/dyr036
25. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol (2013) 37(7):658–65. doi: 10.1002/gepi.21758
26. Burgess S, Thompson SG. Interpreting findings from mendelian randomization using the MR-egger method. Eur J Epidemiol (2017) 32(5):377–89. doi: 10.1007/s10654-017-0255-x
27. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol (2015) 44(2):512–25. doi: 10.1093/ije/dyv080
28. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol (2016) 40(4):304–14. doi: 10.1002/gepi.21965
29. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol (2017) 46(6):1985–98. doi: 10.1093/ije/dyx102
30. Greco MF, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in mendelian randomisation studies with summary data and a continuous outcome. Stat Med (2015) 34(21):2926–40. doi: 10.1002/sim.6522
31. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet (2018) 50(5):693–8. doi: 10.1038/s41588-018-0099-7
32. Eyupoglu ND, Ergunay K, Acikgoz A, Akyon Y, Yilmaz E, Yildiz BO. Gut microbiota and oral contraceptive use in overweight and obese patients with polycystic ovary syndrome. J Clin Endocrinol Metab (2020) 105(12):dgaa600. doi: 10.1210/clinem/dgaa600
33. Liu R, Zhang C, Shi Y, Zhang F, Li L, Wang X, et al. Dysbiosis of gut microbiota associated with clinical parameters in polycystic ovary syndrome. Front Microbiol (2017) 8:324. doi: 10.3389/fmicb.2017.00324
34. Qin S, He Z, Wu Y, Zheng Z, Zhang H, Lv C, et al. Instant dark tea alleviates hyperlipidaemia in high-fat diet-fed rat: from molecular evidence to redox balance and beyond. Front Nutr (2022) 9:819980. doi: 10.3389/fnut.2022.819980
35. Artimani T, Karimi J, Mehdizadeh M, Yavangi M, Khanlarzadeh E, Ghorbani M, et al. Evaluation of pro-oxidant-antioxidant balance (PAB) and its association with inflammatory cytokines in polycystic ovary syndrome (PCOS). Gynecol Endocrinol (2018) 34(2):148–52. doi: 10.1080/09513590.2017.1371691
36. González F. Inflammation in polycystic ovary syndrome: underpinning of insulin resistance and ovarian dysfunction. Steroids (2012) 77(4):300–5. doi: 10.1016/j.steroids.2011.12.003
37. Paoli A, Mancin L, Giacona MC, Bianco A, Caprio M. Effects of a ketogenic diet in overweight women with polycystic ovary syndrome. J Transl Med (2020) 18(1):104. doi: 10.1186/s12967-020-02277-0
38. Li C, Xing C, Zhang J, Zhao H, Shi W, He B. Eight-hour time-restricted feeding improves endocrine and metabolic profiles in women with anovulatory polycystic ovary syndrome. J Transl Med (2021) 19(1):148. doi: 10.1186/s12967-021-02817-2
39. Langley G, Hao Y, Pondo T, Miller L, Petit S, Thomas A, et al. The impact of obesity and diabetes on the risk of disease and death due to invasive group a streptococcus infections in adults. Clin Infect Dis (2016) 62(7):845–52. doi: 10.1093/cid/civ1032
40. Pinart M, Dötsch A, Schlicht K, Laudes M, Bouwman J, Forslund S, et al. Gut microbiome composition in obese and non-obese persons: a systematic review and meta-analysis. Nutrients (2021) 14(1):12. doi: 10.3390/nu14010012
41. Gallè F, Valeriani F, Cattaruzza MS, Gianfranceschi G, Liguori R, Antinozzi M, et al. Mediterranean Diet, physical activity and gut microbiome composition: a cross-sectional study among healthy young Italian adults. Nutrients (2020) 12(7):2164. doi: 10.3390/nu12072164
42. Wang T, Sha L, Li Y, Zhu L, Wang Z, Li K, et al. Dietary α-linolenic acid-rich flaxseed oil exerts beneficial effects on polycystic ovary syndrome through sex steroid hormones-Microbiota-Inflammation axis in rats. Front Endocrinol (Lausanne) (2020) 11:284. doi: 10.3389/fendo.2020.00284
43. Ma M, Su J, Wang Y, Wang L, Li Y, Ding G, et al. Association of body mass index and intestinal (faecal) streptococcus in adults in xining city, China P.R. Benef Microbes (2022) 13(6):465–71. doi: 10.3920/BM2021.0046
44. Zhao F, Dong T, Yuan KY, Wang NJ, Xia FZ, Liu D, et al. Shifts in the bacterial community of supragingival plaque associated with metabolic-associated fatty liver disease. Front Cell Infect Microbiol (2020) 10:581888. doi: 10.3389/fcimb.2020.581888
45. Naderpoor N, Mousa A, Gomez-Arango LF, Barrett HL, Dekker Nitert M, de Courten B. Faecal microbiota are related to insulin sensitivity and secretion in overweight or obese adults. J Clin Med (2019) 8(4):452. doi: 10.3390/jcm8040452
46. Jones RB, Alderete TL, Kim JS, Millstein J, Gilliland FD, Goran MI. High intake of dietary fructose in overweight/obese teenagers associated with depletion of eubacterium and streptococcus in gut microbiome. Gut Microbes (2019) 10(6):712–9. doi: 10.1080/19490976.2019.1592420
47. Liu S, Cao R, Liu L, Lv Y, Qi X, Yuan Z, et al. Correlation between gut microbiota and testosterone in Male patients with type 2 diabetes mellitus. Front Endocrinol (Lausanne) (2022) 13:836485. doi: 10.3389/fendo.2022.836485
48. Gonçalves P, Araújo JR, Di Santo JP. A cross-talk between microbiota-derived short-chain fatty acids and the host mucosal immune system regulates intestinal homeostasis and inflammatory bowel disease. Inflammatory Bowel Diseases (2018) 24(3):558–72. doi: 10.1093/ibd/izx029
49. Pingitore A, Chambers ES, Hill T, Maldonado I, Liu B, Bewick G, et al. The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes Obes Metab (2017) 19(2):257–65. doi: 10.1111/dom.12811
50. He J, Zhang P, Shen L, Niu L, Tan Y, Chen L, et al. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int J Mol Sci (2020) 21(17):6356. doi: 10.3390/ijms21176356
51. Hernández MAG, Canfora EE, Jocken JWE, Blaak EE. The short-chain fatty acid acetate in body weight control and insulin sensitivity. Nutrients (2019) 11(8):1943. doi: 10.3390/nu11081943
52. Olaniyi KS, Bashir AM, Areloegbe SE, Sabinari I, Akintayo C, Oniyide A, et al. Short chain fatty acid, acetate restores ovarian function in experimentally induced PCOS rat model. PLoS One (2022) 17(7):e0272124. doi: 10.1371/journal.pone.0272124
53. Wang Y, Dilidaxi D, Wu Y, Sailike J, Sun X, Nabi XH. Composite probiotics alleviate type 2 diabetes by regulating intestinal microbiota and inducing GLP-1 secretion in db/db mice. BioMed Pharmacother (2020) 125:109914. doi: 10.1016/j.biopha.2020.109914
54. Parascinet O, Mas S, Hang T, Llavero C, Lorenzo Ó. Ruiz-Tovar J. A pilot study: the reduction in fecal acetate in obese patients after probiotic administration and percutaneous electrical neurostimulation. Nutrients (2023) 15(5):1067. doi: 10.3390/nu15051067
55. Canfora EE, van der Beek CM, Jocken JWE, Goossens GH, Holst JJ, Damink SW, et al. Colonic infusions of short-chain fatty acid mixtures promote energy metabolism in overweight/obese men: a randomized crossover trial. Sci Rep (2017) 7(1):2360. doi: 10.1038/s41598-017-02546-x
56. Zhang J, Sun Z, Jiang S, Bai X, Ma C, Peng Q, et al. Probiotic bifidobacterium lactis V9 regulates the secretion of sex hormones in polycystic ovary syndrome patients through the gut-brain axis. mSystems (2019) 4(2):e00017-19. doi: 10.1128/mSystems.00017-19
57. Kong C, Gao R, Yan X, Huang L, He J, Li H, et al. Alterations in intestinal microbiota of colorectal cancer patients receiving radical surgery combined with adjuvant CapeOx therapy. Sci China Life Sci (2019) 62(9):1178–93. doi: 10.1007/s11427-018-9456-x
58. Liu Y, Li J, Jin Y, Zhao L, Zhao F, Feng J, et al. Splenectomy leads to amelioration of altered gut microbiota and metabolome in liver cirrhosis patients. Front Microbiol (2018) 9:963. doi: 10.3389/fmicb.2018.00963
59. Guo WL, Chen M, Pan WL, Zhang Q, Xu J, Lin Y, et al. Hypoglycemic and hypolipidemic mechanism of organic chromium derived from chelation of grifola frondosa polysaccharide-chromium (III) and its modulation of intestinal microflora in high fat-diet and STZ-induced diabetic mice. Int J Biol Macromol (2020) 145:1208–18. doi: 10.1016/j.ijbiomac.2019.09.206
60. Zhou L, Ni Z, Yu J, Cheng W, Cai Z, Yu C. Correlation between fecal metabolomics and gut microbiota in obesity and polycystic ovary syndrome. Front Endocrinol (Lausanne) (2020) 11:628. doi: 10.3389/fendo.2020.00628
61. Zhou L, Ni Z, Cheng W, Yu J, Sun S, Zha D, et al. Characteristic gut microbiota and predicted metabolic functions in women with PCOS. Endocr Connect (2020) 9(1):63–73. doi: 10.1530/EC-19-0522
62. Lozano CP, Wilkens LR, Shvetsov YB, Maskarinec G, Park SY, Shepherd JA, et al. Associations of the dietary inflammatory index with total adiposity and ectopic fat through the gut microbiota, LPS, and c-reactive protein in the multiethnic cohort-adiposity phenotype study. Am J Clin Nutr (2022) 115(5):1344–56. doi: 10.1093/ajcn/nqab398
63. Dull AM, Moga MA, Dimienescu OG, Sechel G, Burtea V, Anastasiu CV. Therapeutic approaches of resveratrol on endometriosis via anti-inflammatory and anti-angiogenic pathways. Molecules (2019) 24(4):667. doi: 10.3390/molecules24040667
64. Rostamtabar M, Esmaeilzadeh S, Tourani M, Rahmani A, Baee M, Shirafkan F, et al. Pathophysiological roles of chronic low-grade inflammation mediators in polycystic ovary syndrome. J Cell Physiol (2021) 236(2):824–38. doi: 10.1002/jcp.29912
65. Fabozzi G, Verdone G, Allori M, Cimadomo D, Tatone C, Stuppia L, et al. Personalized nutrition in the management of female infertility: new insights on chronic low-grade inflammation. Nutrients (2022) 14(9):1918. doi: 10.3390/nu14091918
66. Liang YY, Liu LY, Jia Y, Li Y, Cai JN, Shu Y, et al. Correlation between gut microbiota and glucagon-like peptide-1 in patients with gestational diabetes mellitus. World J Diabetes (2022) 13(10):861–76. doi: 10.4239/wjd.v13.i10.861
67. Mintziori G, Nigdelis MP, Mathew H, Mousiolis A, Goulis DG, Mantzoros CS. The effect of excess body fat on female and male reproduction. Metabolism (2020) 107:154193. doi: 10.1016/j.metabol.2020.154193
68. Boutari C, Pappas PD, Mintziori G, Nigdelis MP, Athanasiadis L, Goulis DG, et al. The effect of underweight on female and male reproduction. Metabolism (2020) 107:154229. doi: 10.1016/j.metabol.2020.154229
69. Arendt LH, Høyer BB, Kreilgaard AF, Bech BH, Toft G, Hougaard KS, et al. Maternal pre-pregnancy overweight and infertility in sons and daughters: a cohort study. Acta Obstet Gynecol Scand (2021) 100(5):843–9. doi: 10.1111/aogs.14045
70. Tang S, Huang J, Lin J, Kuang Y. Adverse effects of pre-pregnancy maternal underweight on pregnancy and perinatal outcomes in a freeze-all policy. BMC Pregnancy Childbirth (2021) 21(1):32. doi: 10.1186/s12884-020-03509-3
71. Wen S, Yuan G, Li C, Xiong Y, Zhong X, Li X. High cellulose dietary intake relieves asthma inflammation through the intestinal microbiome in a mouse model. PloS One (2022) 17(3):e0263762. doi: 10.1371/journal.pone.0263762
72. Gao Y, Liu L, Li C, Liang Y, Lv J, Yang L, et al. Study on the antipyretic and anti-inflammatory mechanism of shuanghuanglian oral liquid based on gut microbiota-host metabolism. Front Pharmacol (2022) 13:843877. doi: 10.3389/fphar.2022.843877
73. Mocanu V, Rajaruban S, Dang J, Kung JY, Deehan EC, Madsen KL. Repeated fecal microbial transplantations and antibiotic pre-treatment are linked to improved clinical response and remission in inflammatory bowel disease: a systematic review and pooled proportion meta-analysis. J Clin Med (2021) 10(5):959. doi: 10.3390/jcm10050959
74. Chen Q, Yin Q, Xie Q, Jiang C, Zhou L, Liu J, et al. 2'-fucosyllactose promotes the production of short-chain fatty acids and improves immune function in human-Microbiota-Associated mice by regulating gut microbiota. J Agric Food Chem (2022) 70(42):13615–25. doi: 10.1021/acs.jafc.2c04410
75. Antsiferova YS, Sotnikova NY, Posiseeva LV, Shor AL. Changes in the T-helper cytokine profile and in lymphocyte activation at the systemic and local levels in women with endometriosis. Fertility Sterility (2005) 84(6):1705–11. doi: 10.1016/j.fertnstert.2005.05.066
76. Podgaec S, Abrao MS, Dias JA Jr., Rizzo LV, de Oliveira RM, Baracat EC. Endometriosis: an inflammatory disease with a Th2 immune response component. Hum Reproduction (2007) 22(5):1373–9. doi: 10.1093/humrep/del516
77. Zhou WJ, Yang HL, Shao J, Mei J, Chang K, Zhu R, et al. Anti-inflammatory cytokines in endometriosis. Cell Mol Life Sci (2019) 76(11):2111–32. doi: 10.1007/s00018-019-03056-x
78. Suen J-L, Chang Y, Chiu P-R, Hsieh T-H, Hsi E, Chen Y-C, et al. Serum level of IL-10 is increased in patients with endometriosis, and IL-10 promotes the growth of lesions in a murine model. Am J Pathol (2014) 184(2):464–71. doi: 10.1016/j.ajpath.2013.10.023
79. Suen JL, Chang Y, Shiu YS, Hsu C-Y, Sharma P, Chiu C-C, et al. IL-10 from plasmacytoid dendritic cells promotes angiogenesis in the early stage of endometriosis. J Pathol (2019) 249(4):485–97. doi: 10.1002/path.5339
80. Chuffa LG, Seiva FR, Fávaro WJ, Teixeira GR, Amorim JP, Mendes LO, et al. Melatonin reduces 17 LH. beta-estradiol and induces differential regulation of sex steroid receptors in reproductive tissues during rat ovulation. Reprod Biol Endocrinol (2011) 9:108. doi: 10.1186/1477-7827-9-108
81. Zhao L, Gu C, Huang K, Fan W, Li L, Ye M, et al. Association between oestrogen receptor alpha (ESR1) gene polymorphisms and endometriosis: a meta-analysis of 24 case-control studies. Reprod BioMed Online (2016) 33(3):335–49. doi: 10.1016/j.rbmo.2016.06.003
82. Tong X, Li Z, Wu Y, Fu X, Zhang Y, Fan H. COMT 158G/A and CYP1B1 432C/G polymorphisms increase the risk of endometriosis and adenomyosis: a meta-analysis. Eur J Obstet Gynecol Reprod Biol (2014) 179:17–21. doi: 10.1016/j.ejogrb.2014.04.039
83. Yu Z, Tian X, Peng Y, Sun Z, Wang C, Tang N, et al. Mitochondrial cytochrome P450 (CYP) 1B1 is responsible for melatonin-induced apoptosis in neural cancer cells. J Pineal Res (2018) 65(1):e12478. doi: 10.1111/jpi.12478
84. Guadagnini D, Rocha GZ, Santos A, Assalin H, Hirabara SM, Curi R, et al. Microbiota determines insulin sensitivity in TLR2-KO mice. Life Sci (2019) 234:116793. doi: 10.1016/j.lfs.2019.116793
85. Chen YR, Zheng HM, Zhang GX, Chen FL, Chen LD, Yang ZC. High oscillospira abundance indicates constipation and low BMI in the guangdong gut microbiome project. Sci Rep (2020) 10(1):9364. doi: 10.1038/s41598-020-66369-z
86. Jo JK, Seo SH, Park SE, Kim HW, Kim EJ, Kim ZS, et al. Gut microbiome and metabolome profiles associated with high-fat diet in mice. Metabolites (2021) 11(8):482. doi: 10.3390/metabo11080482
87. Liu L, Fu Q, Li T, Shao K, Zhu X, Cong Y, et al. Gut microbiota and butyrate contribute to nonalcoholic fatty liver disease in premenopause due to estrogen deficiency. PloS One (2022) 17(2):e0262855. doi: 10.1371/journal.pone.0262855
88. Zhang YW, Cao MM, Li YJ, Lu PP, Dai GC, Zhang M, et al. Fecal microbiota transplantation ameliorates bone loss in mice with ovariectomy-induced osteoporosis via modulating gut microbiota and metabolic function. J Orthop Translat (2022) 37:46–60. doi: 10.1016/j.jot.2022.08.003
89. Valido E, Bertolo A, Fränkl GP, Itodo O, Pinheiro T, Pannek J, et al. Systematic review of the changes in the microbiome following spinal cord injury: animal and human evidence. Spinal Cord (2022) 60(4):288–300. doi: 10.1038/s41393-021-00737-y
90. Li XQ, Chen Y, Dai GC, Zhou BB, Yan XN, Tan RX. Abietic acid ameliorates psoriasis-like inflammation and modulates gut microbiota in mice. J Ethnopharmacol (2021) 272:113934. doi: 10.1016/j.jep.2021.113934
91. Kong C, Gao R, Yan X, Huang L, Qin H. Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition (2019) 60:175–84. doi: 10.1016/j.nut.2018.10.002
Keywords: Mendelian randomization, polycystic ovary syndrome, gut microbiome, endometriosis, female infertility
Citation: Liang Y, Zeng W, Hou T, Yang H, Wu B, Pan R and Huang L (2023) Gut microbiome and reproductive endocrine diseases: a Mendelian randomization study. Front. Endocrinol. 14:1164186. doi: 10.3389/fendo.2023.1164186
Received: 12 February 2023; Accepted: 12 April 2023;
Published: 04 August 2023.
Edited by:
Changyin Zhou, Guangdong Second Provincial General Hospital, ChinaReviewed by:
Oscar Lorenzo, Health Research Institute Foundation Jimenez Diaz (IIS-FJD), SpainMianqun Zhang, Anhui Agricultural University, China
Copyright © 2023 Liang, Zeng, Hou, Yang, Wu, Pan and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lishan Huang, bXpzbGlzaGFuMjAxOUAxNjMuY29t
 Ye Liang
Ye Liang Weihong Zeng
Weihong Zeng