- 1The School of Public Health, Guilin Medical University, Guilin, China
- 2Institute of Biomedical Research, School of Intelligent Medicine and Biotechnology, Guilin Medical University, Guilin, China
- 3Guangxi Key Laboratory of Diabetic Systems Medicine, Guilin Medical University, Guilin, China
Background: Gestational diabetes mellitus (GDM) is an endocrine disorder that occurs easily in women during pregnancy. HLA-DQA1/DQB1 genes play a crucial role in the regulation of the human immune and endocrine systems, potentially influencing the pathogenesis of GDM.
Objective: To explore the associations between single nucleotide polymorphisms (SNPs) in HLA-DQA1/DQB1 genes and the risk of GDM.
Methods: Seven functional SNPs of HLA-DQA1/DQB1 genes were selected and genotyped in 523 GDM patients and 638 normal pregnant women. The odds ratio (OR) and its corresponding 95% confidence interval (CI) were utilized to assess the association between candidate SNPs and the risk of GDM. And then, false positive report probability (FPRP), multifactor dimensionality reduction (MDR) and haplotype analysis were employed to validate the statistically significant associations between studied SNPs and GDM risk.
Results: Compared to those with 0–1 risk genotypes, individuals with 2–7 unfavorable genotypes presented an increased risk of GDM (adjusted OR = 1.54, 95% CI=1.04-2.28, P=0.033). A dose- accumulation effect was detected between the number of unfavorable genotypes and GDM risk (Ptrend=0.024). Furthermore, stratified analysis revealed that the increased GDM risk was more likely to occur in individuals with higher blood pressure and TG, and lower HDL-c levels (P<0.05). Multifactor dimensionality reduction (MDR) analysis revealed that rs9274666 was the best single locus model, whereas the 7-loci model was the best multifactor interaction model for predicting GDM risk (χ²=134.28, P<0.0001). Finally, haplotype analysis revealed that the ACGAGTA and ACGGATA haplotypes were significantly associated with the increased GDM risk. HLA-DQA1/DQB1 SNPs can significantly alter individuals’ genetic susceptibility to GDM.
Conclusions: The genetic variations in the HLA-DQA1 and HLA-DQB1 genes may collectively contribute to the susceptibility to gestational diabetes mellitus. These findings suggest that these genetic markers could be useful for early prediction of GDM, and further validation in larger cohorts is warranted.
1 Introduction
Gestational diabetes mellitus (GDM) is defined as any degree of glucose intolerance first recognized during pregnancy, typically diagnosed through an oral glucose tolerance test during the second trimester. Globally, the incidence of GDM ranges from 2% to 25% (1, 2). As a major complication during pregnancy, GDM not only increases reproductive risk for expectant mothers but also has potential genetic implications for offspring. These genetic effects may increase the risk of chronic conditions such as obesity and diabetes, and lead to abnormal fetal growth patterns, including both excessive and restricted growth (3, 4). Despite its significant impact, the pathogenesis of GDM remains complex and has not yet been fully elucidated.
Clinically, GDM primarily arises from the persistent insulin resistance in pregnant women. Additional risk factors include advanced maternal age, a family history of diabetes, a previous history of GDM, immune factors, overweight or obesity, dietary habits, and physical inactivity (5–7). Given these risk factors, recent research has shifted toward exploring novel contributors to the prevalence of GDM, particularly genetic and epigenetic factors, including gene-environment interactions (7–9). Understanding genetic susceptibility is crucial, as it can lead to early identification and targeted interventions for at-risk populations.
Numerous studies have aimed to clarify the specific impact and risk levels associated with genetic variations in the development of GDM. Single nucleotide polymorphisms (SNPs), which are variations at a single position in a DNA sequence among individuals, provide critical information about genetic predispositions to complex diseases (10). These genetic variations may directly influence GDM risk or interact with clinical traits such as age, pre-pregnancy body mass index (pre-BMI), insulin secretion, pancreatic β-cell function, and blood glucose levels (4, 11–13). To date, genome-wide association studies and candidate gene studies have identified numerous SNPs associated with GDM (10, 14), offering new insights into its pathogenesis.
The human leucocyte antigen (HLA) gene complex plays a pivotal role in regulating the immune system and is characterized by high genetic diversity (15, 16). Various HLA alleles have been linked to susceptibility to different types of diabetes (17, 18). For example, type 1 diabetes mellitus (T1DM) is an autoimmune disorder with a substantial genetic component, where HLA class II gene variants such as DQA1*0301, DQB1*0302, DQA1*0501, DQB1*0201, DQB1*060, DQB1*0302 (DR4), DRB1*03 and DRB1*04 account for 30-50% of its heritability (10, 19, 20). Similarly, specific HLA-DQA1 and HLA-DQB1 genes have been associated with susceptibility to type 2 diabetes mellitus (T2DM) (21, 22). Although studies have explored the relationship between HLA-related genes and genetic susceptibility to GDM, identifying significant polymorphisms such as HLA-DQB10602 are being negatively associated with GDM risk (3, 23), the role of SNPs in the HLA-DQA1 and HLA-DQB1 genes in GDM pathogenesis remains unclear.
In this case-control study involving 523 GDM women and 638 normoglycemic pregnant women, we genotyped seven SNPs in the HLA-DQA1 and HLA-DQB1 genes, which were previously identified via the illumina Asian Screening Array (ASA) BeadChip. Our objective was to investigate the associations between these genetic variants and susceptibility to GDM. These findings may pave the way for genetic screening tools that identify women at higher risk for GDM, contributing to better prevention and management strategies.
2 Materials and methods
2.1 Study subjects
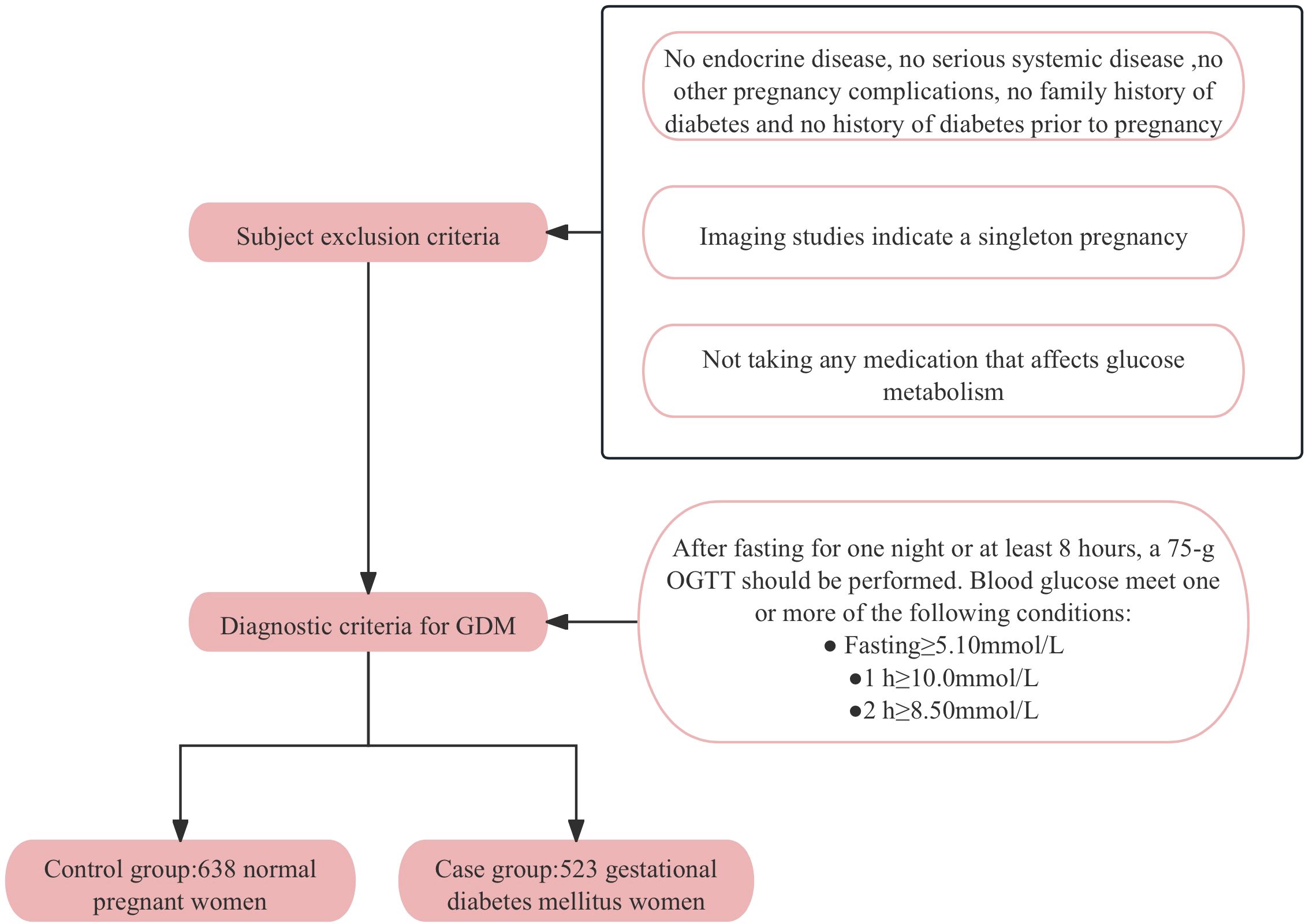
A control group of 638 normoglycemic pregnant women and a case group of 523 GDM women were enrolled at the Affiliated Hospital of Guilin Medical University from September 2014 to April 2016. All participants were genetically unrelated and provided written informed consent. Participants with pre-existing diabetes, chronic diseases, family history of diabetes or pregnancy complications were excluded to ensure a homogenous study population. The diagnostic criteria for GDM followed the standards established by the International Association of Diabetes and Pregnancy Study Groups (IADPSG), which were chosen for their sensitivity and international acceptance. According to these criteria, pregnant women without a previous diagnosis of diabetes underwent a 75-g oral glucose tolerance test (OGTT) after an overnight fast at 24–28 weeks of gestation. A diagnosis of GDM was made if any one or more of the following conditions were met: fasting blood glucose (FBG) ≥ 5.1mmol/L, postprandial 1-hour blood glucose ≥ 10.0mmol/L, or postprandial 2-hour blood glucose ≥ 8.5mmol/L. Figure 1 provides a detailed flowchart of participant recruitment and selection criteria. All participants have signed informed consent forms and the research protocol has been approved by the Ethics Committee of Guilin Medical University (Number: GLMC20131205).
2.2 Asian Screening Array
The DNA extraction kit (Tiangen Biotech) was used to extract DNA from the samples, and then the gene typing module of Genomestudio v2.1 (Illumina) was subsequently used to obtain high-quality data suitable for genome-wide association studies. In the discovery phase, the dataset needs to meet the following criteria: (1) SNP call rate > 95%, in Hardy-Weinberg equilibrium (HWE); (2) sample call rate > 95%; and (3) minimum allele frequency (MAF) < 1%.
2.3 Candidate variant selection
On the basis of the ASA Bead Chip, at the statistical level of 5*10-4, SNPs were screened. In reference to the NCBI (http://www.ncbi.nlm.nih.gov/projects/SNP) and SNP info web server (http://snpinfo.niehs.nih.gov/), potential SNPs are located in the functional regions of the HLA-DQA1 and HLA-DQB1 genes, such as gene regulatory regions, microRNA binding site (MBS), transcription factor binding site (TFBS), splicing site (SS), etc. with minor allele frequency (MAF) greater than 5% in the Chinese Han Beijing population; Finally, seven SNPs, namely, rs1391371, rs9272425, rs9272426, rs9272460, rs9273368, rs9273505 and rs9274666 were selected.
2.4 Clinical and biochemical data
Comprehensive data on age, pre-pregnancy body mass index (pre-BMI), systolic blood pressure (SBP), diastolic blood pressure (DBP), and biochemical parameters were systematically collected from all participants. For biochemical analyses of triglyceride (TG), total cholesterol (TC), and high-density lipoprotein cholesterol (HDL-c) levels, blood samples were collected in no anticoagulant tubes after the participants had fasted for at least 12 hours. Blood samples for fasting blood glucose (FBG), 1-hour OGTT, and 2-hour OGTT were collected in tubes containing sodium fluoride-potassium oxalate as an anticoagulant to inhibit glycolysis. Hemoglobin A1c (HbA1c) testing requires blood samples collected in tubes containing EDTA as an anticoagulant. A minimum of 3 milliliters of blood were collected for each test to ensure sufficient volume for accurate analysis. All biochemical assays were performed in the hospital’s clinical laboratory via standardized procedures and calibrated equipment to ensure the reliability and validity.
2.5 Genomic DNA extraction and genotyping
Genomic DNA was extracted from EDTA-anticoagulated whole blood samples via a commercial DNA extraction kit (Aidlab Biotechnologies Co., Ltd., China) according to the manufacturer’s protocol. The concentration of the extracted DNA was assessed by measuring the absorbance at 260 nm and 280 nm via a UV spectrophotometer, ensuring that the A260/A280 ratio was between 1.80 and 2.00 for optimal quality.
The specific amplification primers used for the selected SNPs are listed in Supplementary Table S1. The total volume of the PCR system was 5 μL, including 1 μL of template DNA (20~100 ng/μL), 1.850 μL of ddH2O, 0.625 μL of 1.25×PCR buffer (15 mmol/L MgCl2), 0.325 μL of 25 mmol/L MgCl2, 0.1 μL of 25 mmol/L dNTP mixture, 1 μL of 0.5 μmol/L primer mixture, and 0.1 μL of 5 U/μL HotStar Taq polymerase. The PCR amplification program was as follows: first, 15 minutes of predenaturation at 94°C was performed, followed by 45 cycles. Each cycle included denaturation at 94°C for 20 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 1 minute. Finally, a final incubation was carried out at 72°C for 3 minutes.
The amplified PCR products were analyzed via the Sequenom MassARRAY platform, which employs matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) for SNP genotyping. The genotype data were processed and analyzed via TYPER 4.0 software. Representative genotyping scatter plots are presented in Supplementary Figure S1, demonstrating the reliability and accuracy of the genotyping.
2.6 Statistical analysis
The changes in the clinical and biochemical test data are represented as the mean (SD), whereas the differences in the demographic variables were evaluated using chi-square tests. The Hardy-Weinberg equilibrium of the genotype distribution was tested using chi-square (χ2) goodness-of-fit test. Binary logistic regression analysis was used to assess the risk of GDM by calculating odds ratios (ORs) and their corresponding 95% confidence intervals (CIs). All statistical analyses were performed using the IBM SPSS Statistics 28 for Windows (IBM Corp, Armonk, NY, USA) with two-tailed tests, and P<0.05 was considered statistically significant.
Multifactor Dimensionality Reduction (MDR) software (version 3.0.2) was used to identifying interaction effects across the studied SNPs and construct the best multifactor model through 1000 permutations under 100-fold cross-validation consistency (CVC) without associations.
Linkage disequilibrium (LD) analysis of each SNP locus and construction of haplotypes were conducted using SHEsis software (http://analysis.bio-x.cn/) (24, 25). SNPs were considered to exhibit strong linkage when r2 > 0.6 and D′ > 0.7, moderate linkage when 0.3≤ r2 ≤ 0.6 and 0.4 ≤ D′ ≤ 0.7, and weak linkage when r2 < 0.3 and D′ < 0.4 (26). Understanding the LD between SNPs allows for the identification of haplotypes that may be associated with GDM risk.
The false positive report probability (FPRP) was estimated to assess the robustness of findings with a statistically significant association with a prior probability set at 0.1 and a relatively strict cutoff value of 0.2 (27).
3 Results
3.1 Characteristics of the subjects
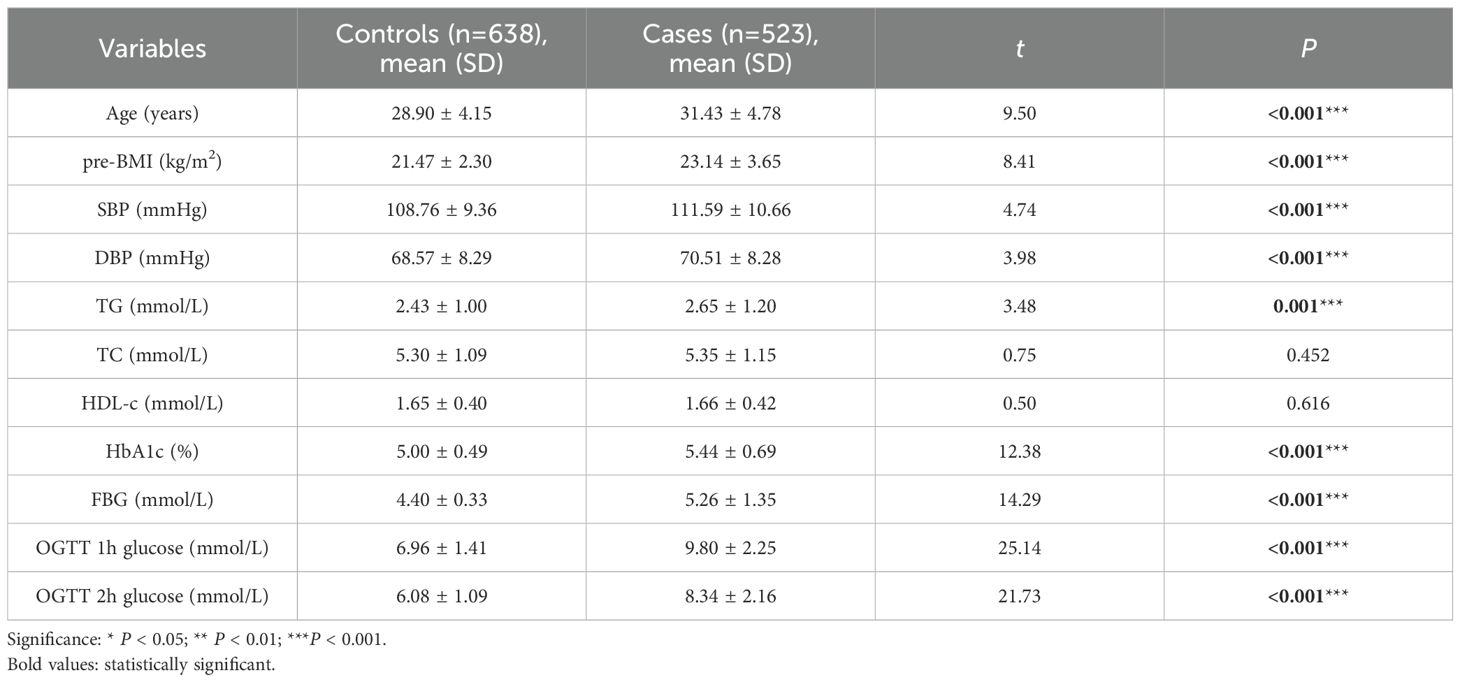
This study included a total of 1161 pregnant women. A comparison between the control group and the GDM group revealed that pregnant women with GDM had significantly higher values for age, pre-BMI, SBP, DBP, TG, HbA1c, FPG, and 1-hour and 2-hour OGTT than those of the normoglycemic pregnant women (P<0.05), as shown in Table 1.
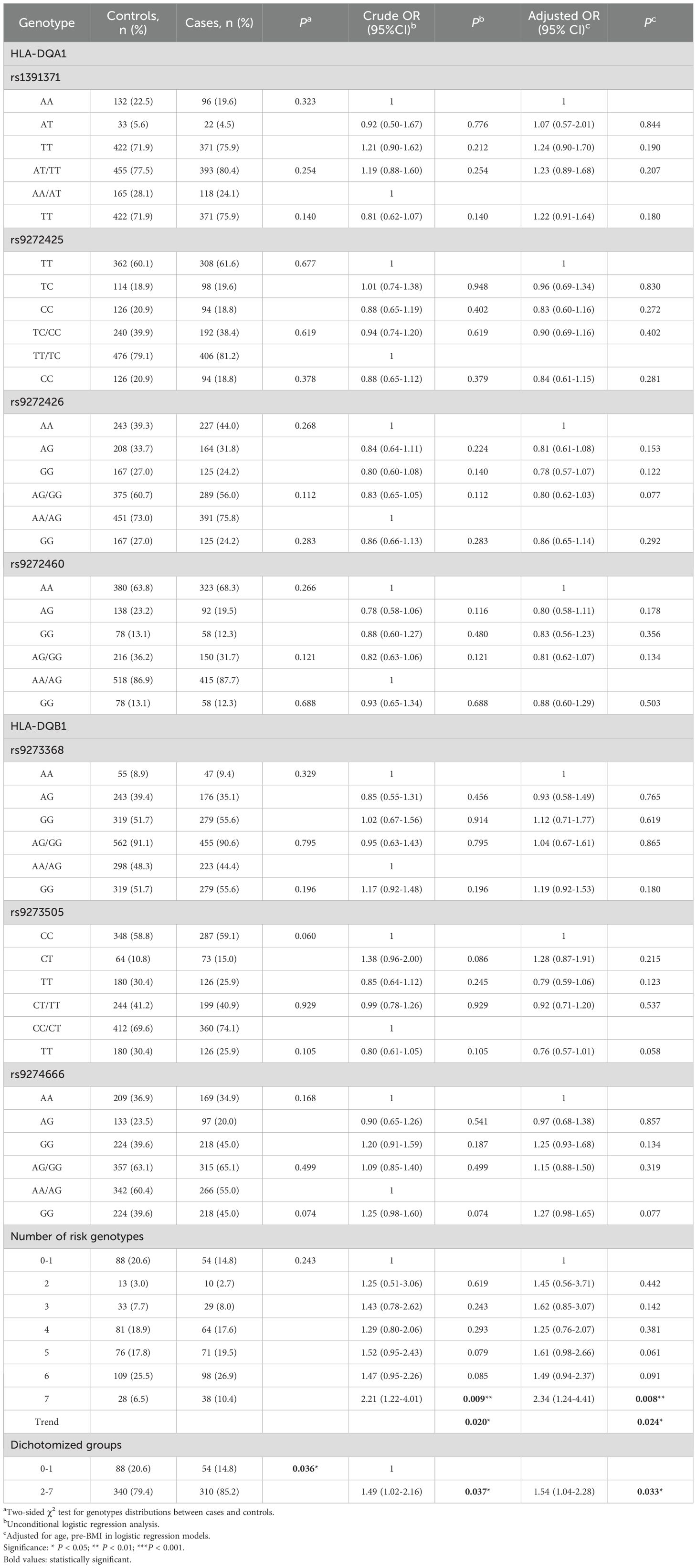
In the term of the association between genetic variant genotype and GDM risk, the genotype frequencies of the four loci of HLA-DQA1 and the three loci of HLA-DQB1 were all in accordance with the Hardy-Weinberg equilibrium as shown in Table 2.
Given the lack of significant associations with individual genotypes, we conducted a combined- genotype effect analysis based on the basis of the number of risk genotypes. All putative risk genotypes, those with an odds ratio (OR) greater than 1.0 (and for OR < 1.0, the reference group was reversed), from the studied SNPs were categorized into new variables according to the number of risk genotypes (28). The risk genotypes used for the calculation were HLA-DQA1 gene rs1391371 AT/TT, rs9272425 TT, rs9272426 AA and rs99272460 AA, and HLA-DQB1 gene rs9273368 GG, rs9273505 CT and rs9274666 AG/GG.
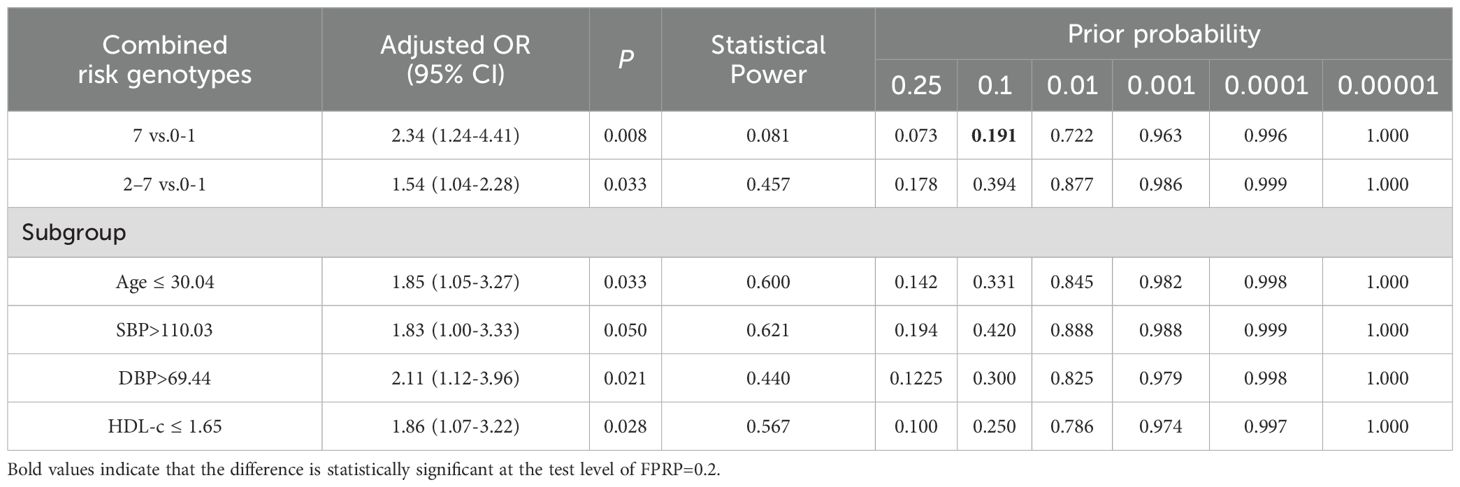
As shown in Table 2, we observed that the likelihood of developing GDM increased with the number of risk genotypes. Compared with individuals with 0–1 risk genotypes, those carrying all 7 unfavorable genotypes had a significantly increased risk of GDM (adjusted OR = 2.34, 95% CI = 1.24-4.41, P=0.008). Additionally, individuals with 2–7 unfavorable genotypes presented increased GDM risk (adjusted OR = 1.54, 95% CI = 1.04-2.28, P= 0.033) compared with those with 0–1 risk genotypes. Notably, this cumulative effect on GDM risk appeared to be dose-dependent with respect to the number of risk genotypes (Ptrend = 0.024).
3.2 Stratified analysis of risk genotypes
We conducted a stratified analysis to assess the association between the number of risk genotypes (2–7 unfavorable genotypes) and GDM risk within subgroups defined by the mean values of clinical variables. As shown in Table 3, the increased GDM risk associated with carrying 2–7 unfavorable genotypes was more pronounced among participants with TG > 2.53 (OR=1.94, 95% CI=1.10-3.42, P=0.022), age ≤ 30.04 (adjusted OR=1.85, 95% CI=1.05-3.27, P=0.033), SBP >110.03 (adjusted OR=1.83, 95% CI=1.00-3.33, P= 0.050), DBP > 69.44 (adjusted OR=2.11, 95% CI=1.12-3.96, P=0.021) and HDL-c ≤ 1.65 (adjusted OR=1.86, 95%CI=1.07-3.22, P=0.028).
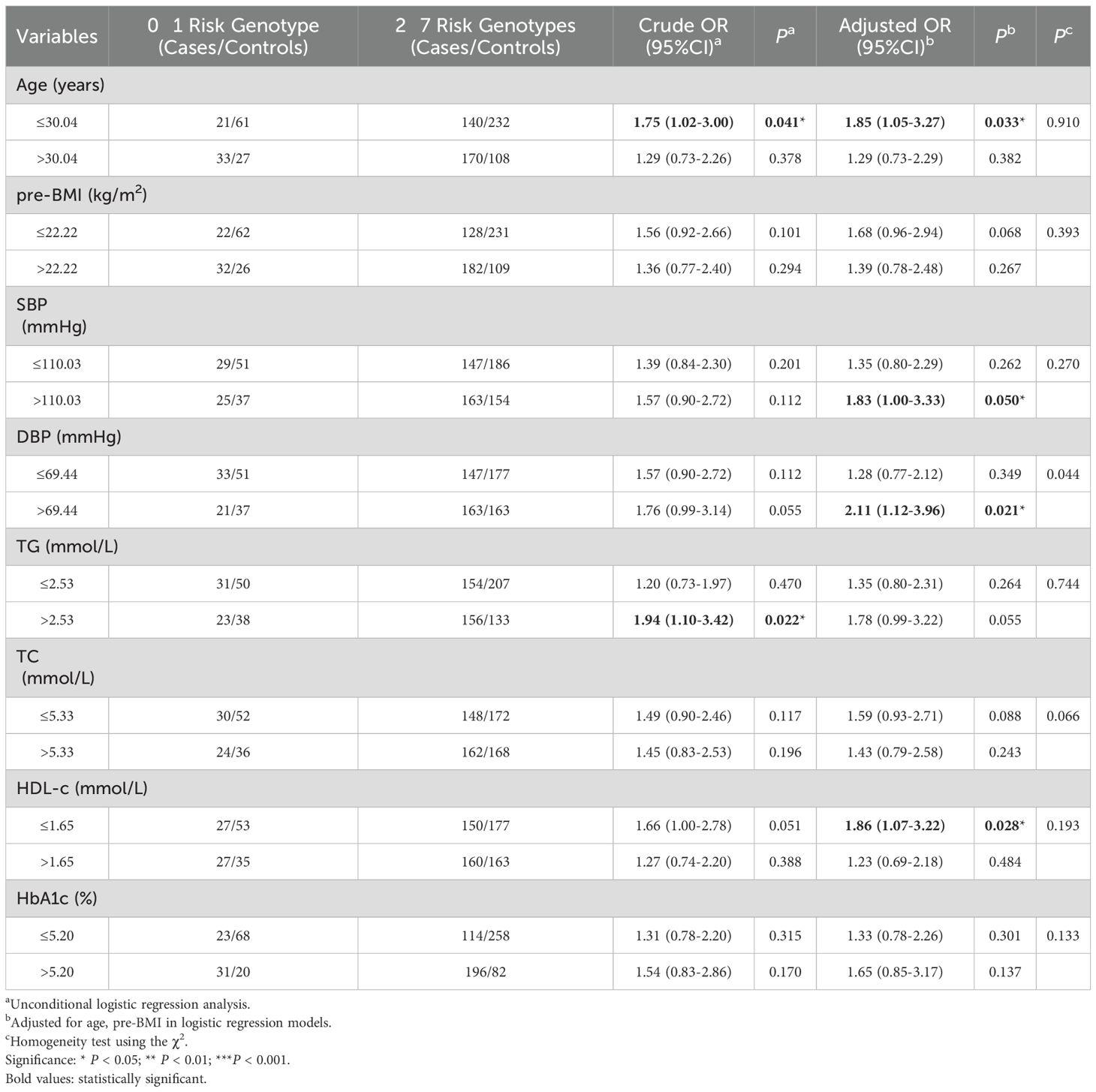
Table 3. Stratification analysis for associations between risk genotypes of HLA-DQA1/DQB1 and GDM risk.
3.3 FPRP analysis
The FPRP test was used to assess whether there was a significant correlation between the accumulation of multiple unfavorable genes in SNPs and the risk of GDM in this study. See Table 4, the FPRP value for the association between seven unfavorable genotypes and GDM risk is 0.191 (less than the threshold value of 0.20), indicating that association is authentic.
3.4 Gene-gene interaction effects on GDM risk according
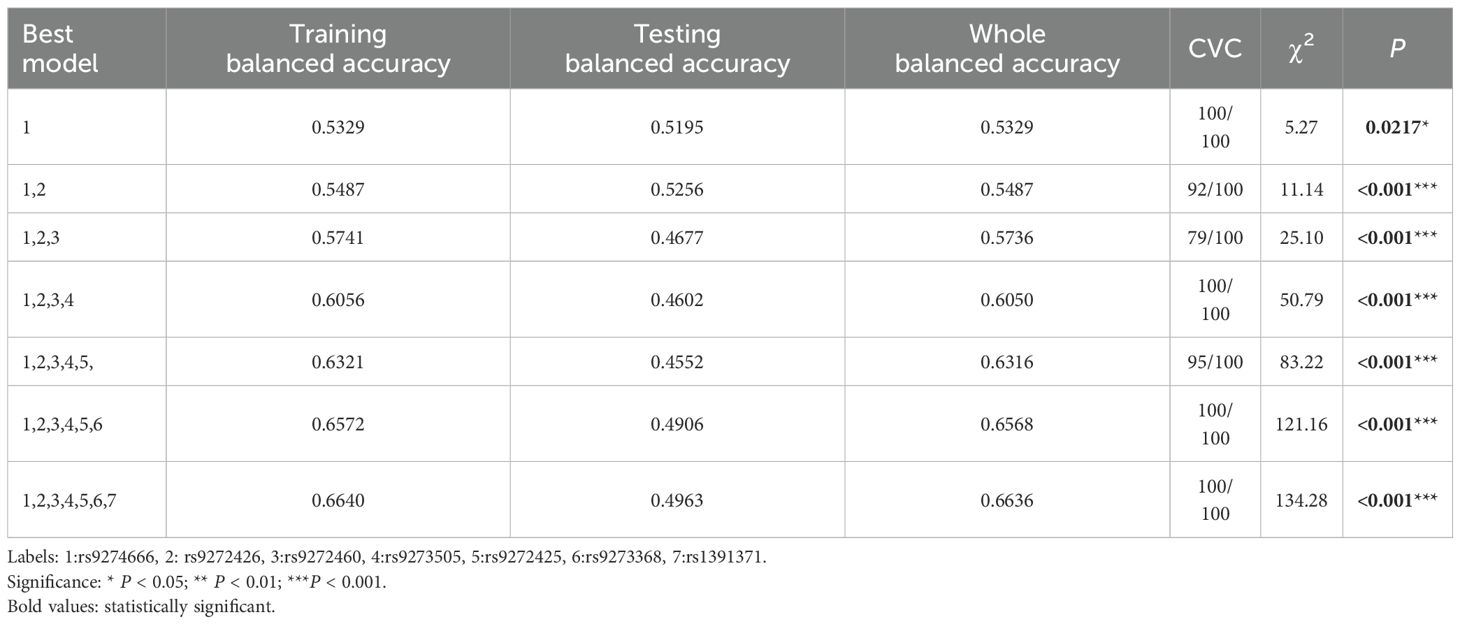
In the MDR analysis, we identified the best single-locus model as rs9274666 in Table 5, which is significantly associated with GDM risk (χ2 = 5.27, P=0.0217) and achieved the highest CVC of 100/100 and a balanced testing accuracy of 0.5195. More interestingly, the model with all seven studied SNPs with a whole balanced accuracy of 0.6636 and a maximum CVC of 100/100 was the best model for evaluate the GDM risk, with a χ2 test of 134.28 and the corresponding P<0.0001.
3.5 LD analysis
For seven SNPs in HLA-DQA1/DQB1, we observed strong LD between rs9272425 and rs1391371 (D′=0.982, r2 = 0.671) in Figure 2. Moderate LD was found between rs9272426 and both rs1391371 (D′=0.954, r2 = 0.458) and rs9272425 (D′=0.929, r2 = 0.521), and between rs9274666 and both rs1391371 (D′=0.932, r2 = 0.322) and rs9272425 (D′=0.954, r2 = 0.491). LD among the other SNPs was relatively weak.
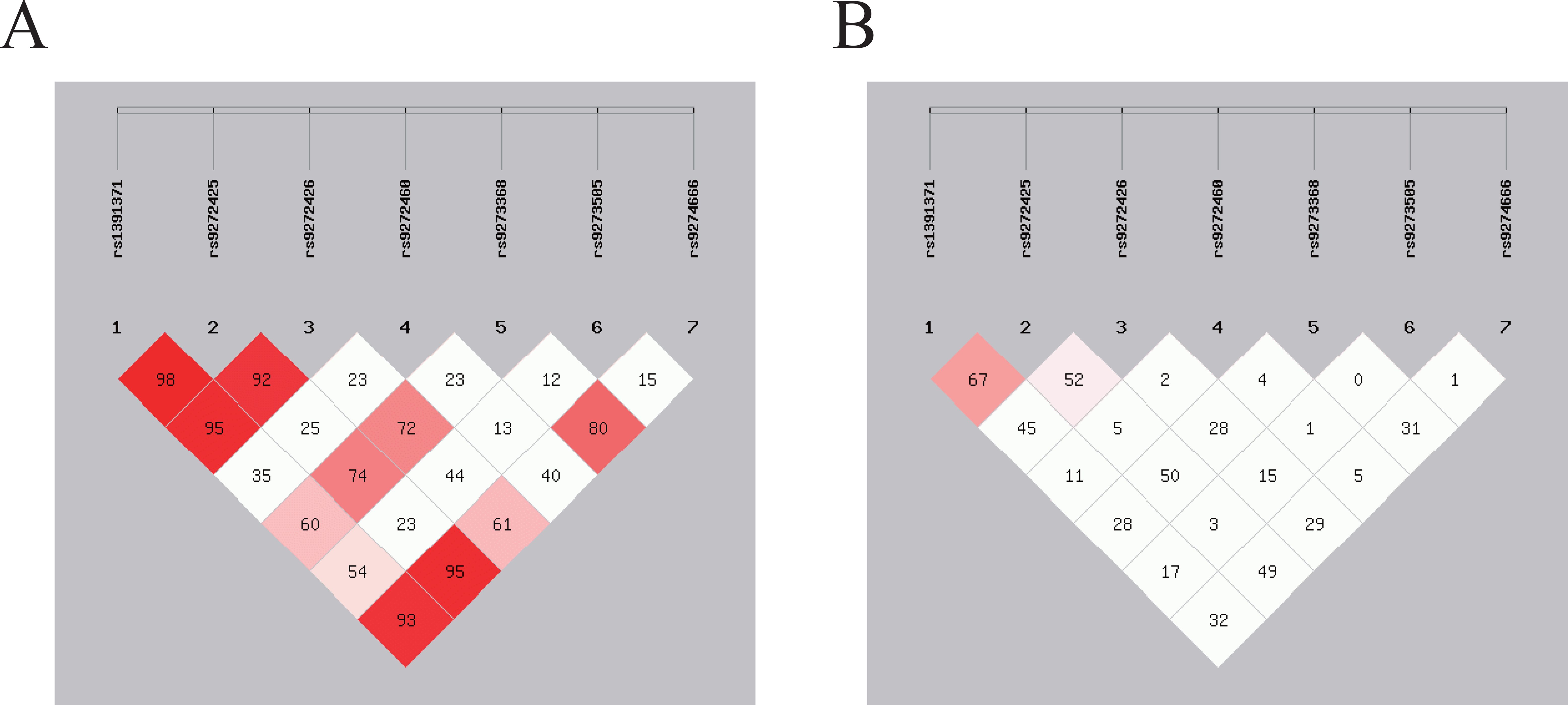
Figure 2. SNPs loci linkage disequilibrium analysis. The strength of LD between SNP pairs, with D’ and r2 values representing the degree of genetic correlation. (A) The D’ value in the box reflects the overall linkage situation of multi-locus chromosome blocks; (B) The r2 value in the box reflects the estimated situation of linkage disequilibrium between two loci.
3.6 Haplotype analysis
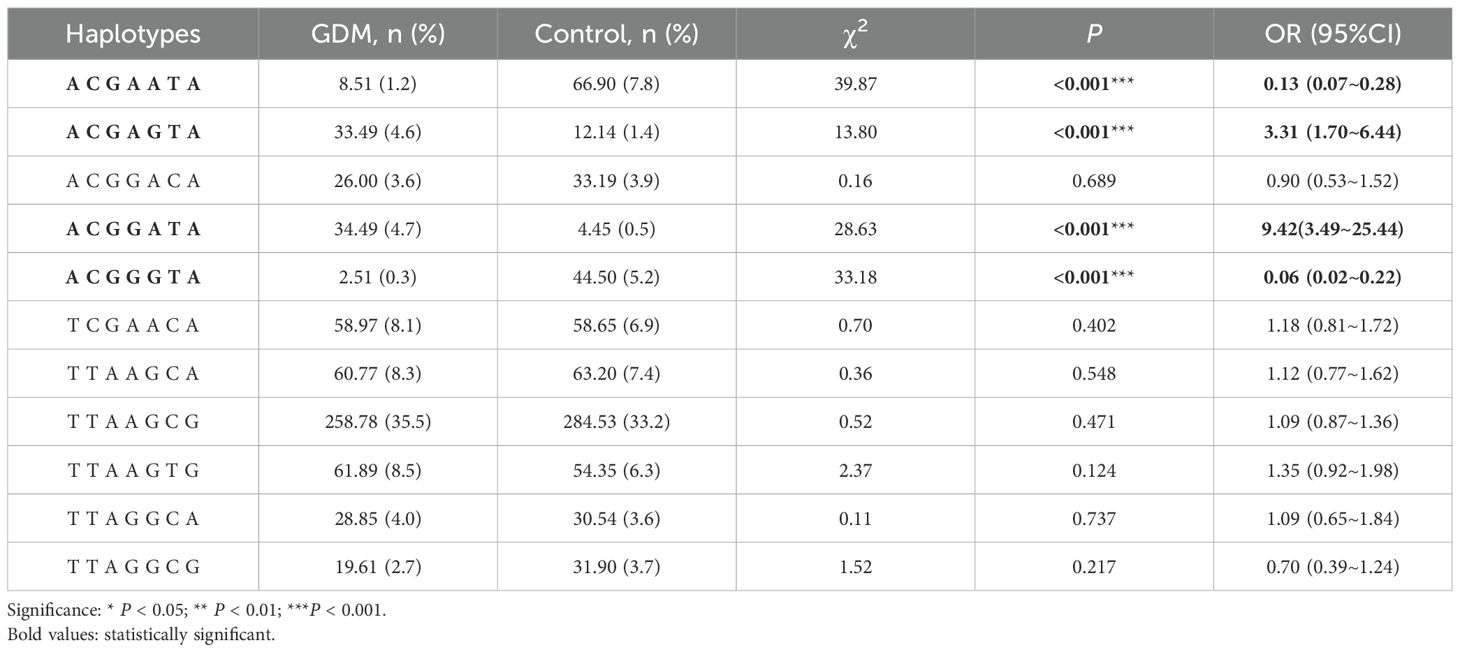
As shown in Table 6, when constructing haplotypes for 7 SNP loci within HLA-DQA1 and HLA -DQB1 for the GDM group and the control group, we found that the haplotype TTAAGCG was the most common, with frequencies of 35.5% in the GDM group and 33.2% in the control group. An increased risk of GDM may be associated with the haplotypes ACGAGTA (OR=3.31, 95% CI=1.70-6.44, P<0.001) and ACGGATA (OR=9.42,95% CI=3.49-25.44, P<0.001); whereas the haplotypes ACGAATA (OR=0.13, 95% CI=0.07-0.28, P<0.001) and ACGGGTA (OR=0.06, 95% CI=0.02-0.22, P<0.001) present a decreased risk effect of GDM.
4 Discussion
The result of the present study suggests that genetic factors, such as SNPs (29), are strongly linked to the onset of GDM. In this study, among the seven selected loci of HLA-DQA1/DQB1 genes (rs1391371, rs9272425, rs9272426, rs9272460, rs9273368, rs9273505, and rs9274666), we also observed that the risk of GDM increased correspondingly with the accumulation of unfavorable genotypes when combined. This cumulative effect was further validated by the results from the multifactor MDR analysis. Identification of these SNPs may enable prediction of GDM risk and allow for early intervention strategies to minimize complications for both the pregnant woman and the newborn (30). These findings are important for the prevention and control of GDM in clinical and public health settings.
Previous studies have shown that HLA class II genes (HLA-DQA1, HLA-DRB1, HLA-DPA1, and HLA-DQB1) and their antigens may play roles in immune regulation during GDM development (31). HLA-DQA1/DQB1, as key molecules within the HLA class II complex, are associated with the pathogenesis of GDM through interactions with regulatory T cells (Tregs) and effector T cells (32). Consistent with this, we established an association between the combined effects of HLA-DQA1/DQB1 genetic variant genotypes and an increased risk of GDM. Using logistic regression, MDR, FPRP and haplotype analysis, we confirmed the joint effects between the studied loci. These results suggest that HLA-DQA1/DQB1 gene variants have significant genetic regulatory effects on the pathogenesis of GDM, potentially altering individuals’ susceptibility to GDM through single-locus effects, multifactor combinations, and gene-environment interactions.
FPRP analysis, plays an important role in molecular epidemiology research, by evaluating the authenticity of research results, helping researchers better understand and control potential false positives in research, and thereby improving the scientific and reliability of research (27). To verify the noteworthiness of the significant associations between the studied variants and GDM risk, a much more stringent FPRP threshold of 0.2 was set, and the 7 risk factors, compared with the 0–1 risk genotype carrying effect, are likely to be authentic. However, the FPRP values detected for other statistically significant associations are much greater than 0.2, indicating that these findings might be possible observations. Thus, the conclusions drawn from here must be considered preliminary and need to be verified in the future.
In exploring gene-environment interactions, we observed that combinations of unfavorable genotypes can significantly increase the risk of GDM in pregnant women under the age of 30.04 years, with SBP exceeding 110.03 mmHg, DBP exceeding 69.44 mmHg, TG greater than 2.53 mmol/L, and HDL-c levels below 1.65 mmol/L. Research has indicated that elevated triglycerides can lead to dyslipidemia, wherein excessive free fatty acids are secreted by abundant adipocytes. This process promotes the overproduction of inflammatory cytokines and oxidative reactions, which can damage pancreatic beta cells or induce insulin resistance, ultimately resulting in abnormal glucose levels. HDL-c, as a crucial lipoprotein, not only influences glucose levels through the aforementioned pathways but also exerts effects through its direct anti-inflammatory properties. Low levels of HDL-c can alter insulin sensitivity and pancreatic insulin secretion (33, 34). Although the complex physiological and pathological mechanisms linking elevated blood pressure to glucose imbalance are not yet clear, studies have shown a close relationship between elevated diastolic blood pressure and insulin resistance and GDM. Researchers thought that the two could interact via the renin-angiotensin-aldosterone and sympathetic nervous systems (35, 36). Lack of gestational weight gain records may affect metabolic assessment. Future studies should incorporate longitudinal anthropometric measurements. These findings provide new insights for further exploration of the relationship between genetic factors and the prediction of GDM and cardiovascular disease. In contrast, while most studies suggest that older age is associated with a greater risk of GDM (37, 38), this research revealed that the combined effect of unfavorable genotypes on GDM risk was more pronounced in individuals under the age of 30.04 years. This could be due to the interaction between genetic susceptibility and early metabolic imbalance, which might lead to the earlier onset of GDM in younger individuals (39). Beyond genetic risk stratification, innovative cardiac imaging modalities such as speckle tracking echocardiography (STE) could provide critical insights for identifying among pregnant women those with increased risk of developing GDM or cardiovascular complications (40, 41). It is likely that several genetic variants may serve as predictors of impaired myocardial deformation indices or subclinical myocardial dysfunction in pregnant women at higher risk of GDM.
Haplotypes can influence disease development by affecting pathways of cell survival, the immune response, and the regulation of cell death (42). Located on chromosome 6, the HLA region comprise an abundant array of immune-related genes and is, to some extent, also influenced by haplotypes (43). We performed LD analysis on seven SNPs within the HLA-DQA1/HLA-DQB1 genes, revealing significant LD among these loci. Haplotype analysis indicated that the ACGAATA and ACGGGTA haplotypes may be associated with a reduced risk of GDM, and the ACGAGTA and ACGGATA haplotypes could increase susceptibility to GDM. Compare the database with previous studies, no correlation was found between these haplotypes and other immune or metabolic diseases. Only cyclin-dependent kinase 5 regulatory subunit related protein 1-like 1 and vascular endothelial growth factor gene haplotypes were found to be associated with the risk of GDM (44, 45). These findings may serve as potential biological markers for predicting GDM risk and enhance our understanding of the genetic basis of this disease.
This study explored the association between HLA-DQA1/DQB1 gene polymorphisms and the risk of GDM, utilizing a relatively large sample size and multiple analytical methods to assess potential interactions. However, there are several limitations in this study. First, as a hospital-based case-control study, there is inherent bias in participant selection and research data recall. Second, although relatively large-size samples were included, however, the lower minor allele frequency of some tested SNPs might limit the statistical power to detect significant associations in some subgroups. Third, the SNPs chosen for this study do not comprehensively cover all variants, which may lead to the omission of some significant variants. Fourth, due to challenges in postpartum follow-up, we were unable to collect reliable postpartum glucose tolerance data. We could not to determine the remission or persistence of GDM, which limits the interpretation of long-term implications of the observed genetic associations. Therefore, more related potential genetic and environmental variables need to be included.
5 Conclusion
In summary, the present study revealed that SNPs in the HLA-DQA1/HLA-DQB1 genes are significantly associated with the risk of GDM via a single locus or joint effects of gene-gene and gene-environment factors. However, larger sample size studies with different populations and related functional experiments are warranted to confirm these positive findings.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Ethics Committee of Guilin Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
LN: Data curation, Formal analysis, Methodology, Software, Writing – original draft, Writing – review & editing. YS: Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. RL: Data curation, Formal analysis, Software, Visualization, Writing – original draft, Writing – review & editing. QL: Software, Visualization, Writing – review & editing. GD: Writing – review & editing, Investigation. XH: Writing – review & editing, Formal analysis. YZ: Investigation, Writing – review & editing. HZ: Investigation, Writing – review & editing. XX: Software, Writing – review & editing. XD: Investigation, Writing – review & editing. JH: Conceptualization, Writing – review & editing. XY: Conceptualization, Data curation, Funding acquisition, Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Guangxi Natural Science Foundation (2025GXNSFHA069270); Guangxi Science and Technology Base and Talent Special Project (AD24010027); Guilin Science Research and Technology Development Plan Project (20230135-2-1); Self-funded research project of The Health Committee of Guangxi (Z-C20241580); Guangxi Young and middle-aged teachers' basic ability improvement project (2020KY12028); Guangxi College Students Innovation Training Program Project (202110601029, 202410601046); Maternal and Child Health Research Project of Guangxi Bagui Scholars (Jun Zhang).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2025.1511561/full#supplementary-material
References
1. Choudhury AA and Devi Rajeswari V. Gestational diabetes mellitus - A metabolic and reproductive disorder. BioMed Pharmacother. (2021) 143:112183. doi: 10.1016/j.biopha.2021.112183
2. Gao C, Sun X, Lu L, Liu F, and Yuan J. Prevalence of gestational diabetes mellitus in mainland China: A systematic review and meta-analysis. J Diabetes Investig. (2019) 10:154–62. doi: 10.1111/jdi.12854
3. American College of Obstetricians and Gynecologists. ACOG practice bulletin no. 190: gestational diabetes mellitus. Obstet Gynecol. (2018) 131:e49–64. doi: 10.1097/aog.0000000000002501
4. Davenport MH, Ruchat SM, Poitras VJ, Jaramillo Garcia A, Gray CE, Barrowman N, et al. Prenatal exercise for the prevention of gestational diabetes mellitus and hypertensive disorders of pregnancy: a systematic review and meta-analysis. Br J Sports Med. (2018) 52:1367–75. doi: 10.1136/bjsports-2018-099355
5. Filardi T, Panimolle F, Crescioli C, Lenzi A, and Morano S. Gestational diabetes mellitus: the impact of carbohydrate quality in diet. Nutrients. (2019) 11:7. doi: 10.3390/nu11071549
6. Martínez-Vizcaíno V, Sanabria-Martínez G, Fernández-Rodríguez R, Cavero-Redondo I, Pascual-Morena C, Álvarez-Bueno C, et al. Exercise during pregnancy for preventing gestational diabetes mellitus and hypertensive disorders: An umbrella review of randomised controlled trials and an updated meta-analysis. Bjog. (2023) 130:264–75. doi: 10.1111/1471-0528.17304
7. Zhang C, Rawal S, and Chong YS. Risk factors for gestational diabetes: is prevention possible? Diabetologia. (2016) 59:1385–90. doi: 10.1007/s00125-016-3979-3
8. Choudhury AA and Rajeswari VD. Polycystic ovary syndrome (PCOS) increases the risk of subsequent gestational diabetes mellitus: A novel therapeutic perspective. Life Sci. (2022) 310:121069. doi: 10.1016/j.lfs.2022.121069
9. Huang X, Liang W, Yang R, Jin L, Zhao K, Chen J, et al. Variations in the LINGO2 and GLIS3 genes and gene-environment interactions increase gestational diabetes mellitus risk in Chinese women. Environ Sci Technol. (2024) 58:11596–605. doi: 10.1021/acs.est.4c03221
10. Pervjakova N, Moen GH, Borges MC, Ferreira T, Cook JP, Allard C, et al. Multi-ancestry genome-wide association study of gestational diabetes mellitus highlights genetic links with type 2 diabetes. Hum Mol Genet. (2022) 31:3377–91. doi: 10.1093/hmg/ddac050
11. McIntyre HD, Catalano P, Zhang C, Desoye G, Mathiesen ER, and Damm P. Gestational diabetes mellitus. Nat Rev Dis Primers. (2019) 5:47. doi: 10.1038/s41572-019-0098-8
12. Wu L, Cui L, Tam WH, Ma RC, and Wang CC. Genetic variants associated with gestational diabetes mellitus: a meta-analysis and subgroup analysis. Sci Rep. (2016) 6:30539. doi: 10.1038/srep30539
13. Zhang C, Bao W, Rong Y, Yang H, Bowers K, Yeung E, et al. Genetic variants and the risk of gestational diabetes mellitus: a systematic review. Hum Reprod Update. (2013) 19:376–90. doi: 10.1093/humupd/dmt013
14. Kwak SH, Kim SH, Cho YM, Go MJ, Cho YS, Choi SH, et al. A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes. (2012) 61:531–41. doi: 10.2337/db11-1034
15. Debebe BJ, Boelen L, Lee JC, Thio CL, Astemborski J, Kirk G, et al. Identifying the immune interactions underlying HLA class I disease associations. Elife. (2020) 9. doi: 10.7554/eLife.54558
16. Schindler E, Dribus M, Duffy BF, Hock K, Farnsworth CW, Gragert L, et al. HLA genetic polymorphism in patients with Coronavirus Disease 2019 in Midwestern United States. Hla. (2021) 98:370–9. doi: 10.1111/tan.14387
17. Ferber KM, Keller E, Albert ED, and Ziegler AG. Predictive value of human leukocyte antigen class II typing for the development of islet autoantibodies and insulin-dependent diabetes postpartum in women with gestational diabetes. J Clin Endocrinol Metab. (1999) 84:2342–8. doi: 10.1210/jcem.84.7.5813
18. Törn C, Gupta M, Sanjeevi CB, Aberg A, Frid A, and Landin-Olsson M. Different HLA-DR-DQ and MHC class I chain-related gene A (MICA) genotypes in autoimmune and nonautoimmune gestational diabetes in a Swedish population. Hum Immunol. (2004) 65:1443–50. doi: 10.1016/j.humimm.2004.09.002
19. Morran MP, Vonberg A, Khadra A, and Pietropaolo M. Immunogenetics of type 1 diabetes mellitus. Mol Aspects Med. (2015) 42:42–60. doi: 10.1016/j.mam.2014.12.004
20. Redondo MJ, Fain PR, and Eisenbarth GS. Genetics of type 1A diabetes. Recent Prog Horm Res. (2001) 56:69–89. doi: 10.1210/rp.56.1.69
21. Jacobi T, Massier L, Klöting N, Horn K, Schuch A, Ahnert P, et al. HLA class II allele analyses implicate common genetic components in type 1 and non-insulin-treated type 2 diabetes. J Clin Endocrinol Metab. (2020) 105. doi: 10.1210/clinem/dgaa027
22. Yang HY, Tai WL, Yuan HY, Xu M, Li J, Ren CF, et al. Study on the association between the polymorphism of HLA-DQA1 alleles and type 2 diabetes in Yunnan Han nationality. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2004) 21:291–3.
23. Papadopoulou A, Lynch KF, Anderberg E, Landin-Olsson M, Hansson I, Agardh CD, et al. HLA-DQB1 genotypes and islet cell autoantibodies against GAD65 and IA-2 in relation to development of diabetes post partum in women with gestational diabetes mellitus. Diabetes Res Clin Pract. (2012) 95:260–4. doi: 10.1016/j.diabres.2011.10.037
24. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. (2009) 19:519–23. doi: 10.1038/cr.2009.33
25. Shi YY and He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. (2005) 15:97–8. doi: 10.1038/sj.cr.7290272
26. Ardlie KG, Kruglyak L, and Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. (2002) 3:299–309. doi: 10.1038/nrg777
27. Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, and Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. (2004) 96:434–42. doi: 10.1093/jnci/djh075
28. Zhu ML, Yu H, Shi TY, He J, Wang MY, Li QX, et al. Polymorphisms in mTORC1 genes modulate risk of esophageal squamous cell carcinoma in eastern Chinese populations. J Thorac Oncol. (2013) 8:788–95. doi: 10.1097/JTO.0b013e31828916c6
29. Arora GP, Åkerlund M, Brøns C, Moen GH, Wasenius NS, Sommer C, et al. Phenotypic and genotypic differences between Indian and Scandinavian women with gestational diabetes mellitus. J Intern Med. (2019) 286:192–206. doi: 10.1111/joim.12903
30. Wu Y, Liu B, Sun Y, Du Y, Santillan MK, Santillan DA, et al. Association of maternal prepregnancy diabetes and gestational diabetes mellitus with congenital anomalies of the newborn. Diabetes Care. (2020) 43:2983–90. doi: 10.2337/dc20-0261
31. Tang W, Wang X, Chen L, Lu Y, and Kang X. Identification of potential gene markers in gestational diabetes mellitus. J Clin Lab Anal. (2022) 36:e24515. doi: 10.1002/jcla.24515
32. Zhang YN, Wu Q, and Deng YH. Phenotypic characterisation of regulatory T cells in patients with gestational diabetes mellitus. Sci Rep. (2024) 14:4881. doi: 10.1038/s41598-023-47638-z
33. Drew BG, Rye KA, Duffy SJ, Barter P, and Kingwell BA. The emerging role of HDL in glucose metabolism. Nat Rev Endocrinol. (2012) 8:237–45. doi: 10.1038/nrendo.2011.235
34. Parhofer KG. Interaction between glucose and lipid metabolism. More than diabetic dyslipidemia. Diabetes Metab J. (2015) 39:353–62. doi: 10.4093/dmj.2015.39.5.353
35. Lailler G, Fosse-Edorh S, Lebreton E, Regnault N, Deneux-Tharaux C, Tsatsaris V, et al. Impact of different types of hypertensive disorders of pregnancy and their duration on incident post-partum risk of diabetes mellitus: Results from the French nationwide study CONCEPTION. Diabetes Metab. (2024) 50:101564. doi: 10.1016/j.diabet.2024.101564
36. Quesada O, Claggett B, Rodriguez F, Cai J, Moncrieft AE, Garcia K, et al. Associations of insulin resistance with systolic and diastolic blood pressure: A study from the HCHS/SOL. Hypertension. (2021) 78:716–25. doi: 10.1161/hypertensionaha.120.16905
37. Gao L, Chen CR, Wang F, Ji Q, Chen KN, Yang Y, et al. Relationship between age of pregnant women with gestational diabetes mellitus and mode of delivery and neonatal Apgar score. World J Diabetes. (2022) 13:776–85. doi: 10.4239/wjd.v13.i9.776
38. Laine MK, Kautiainen H, Gissler M, Raina M, Aahos I, Järvinen K, et al. Gestational diabetes in primiparous women-impact of age and adiposity: a register-based cohort study. Acta Obstet Gynecol Scand. (2018) 97:187–94. doi: 10.1111/aogs.13271
39. Yoon TJ. Early metabolic imbalance in young adults has variable impact on 35-year mortality risk. Am J Prev Cardiol. (2024) 19:100827. doi: 10.1016/j.ajpc.2024.100827
40. Aguilera J, Sanchez Sierra A, Abdel Azim S, Georgiopoulos G, Nicolaides KH, and Charakida M. Maternal cardiac function in gestational diabetes mellitus at 35–36 weeks’ gestation and 6 months postpartum. Ultrasound Obstetrics Gynecology. (2020) 56:247–54. doi: 10.1002/uog.22118
41. Sonaglioni A, Bordoni T, Naselli A, Nicolosi GL, Grasso E, Bianchi S, et al. Influence of gestational diabetes mellitus on subclinical myocardial dysfunction during pregnancy: A systematic review and meta-analysis. Eur J Obstetrics Gynecology Reprod Biol. (2024) 292:17–24. doi: 10.1016/j.ejogrb.2023.11.007
42. Thomsen H, Chattopadhyay S, Weinhold N, Vodicka P, Vodickova L, Hoffmann P, et al. Haplotype analysis identifies functional elements in monoclonal gammopathy of unknown significance. Blood Cancer J. (2024) 14:140. doi: 10.1038/s41408-024-01121-8
43. Evseeva I, Nicodemus KK, Bonilla C, Tonks S, and Bodmer WF. Linkage disequilibrium and age of HLA region SNPs in relation to classic HLA gene alleles within Europe. Eur J Hum Genet. (2010) 18:924–32. doi: 10.1038/ejhg.2010.32
44. Chengjing H, Yan G, Wei L, Bing X, Jing Z, Feng Z, et al. Association of the CDKAL1 gene polymorphism with gestational diabetes mellitus in Chinese women. BMJ Open Diabetes Res Care. (2023) 11:e003164. doi: 10.1136/bmjdrc-2022-003164
Keywords: gestational diabetes mellitus, HLA-DQA1/DQB1, risk, variant, gene-gene interaction
Citation: Nie L, Sun Y, Li R, Liang Q, Deng G, He X, Zeng Y, Zheng H, Xiao X, Ding X, Huang J and Yu X (2025) Genetic variants in HLA-DQA1/DQB1 genes modulate the risk of gestational diabetes mellitus in a southern Chinese population. Front. Endocrinol. 16:1511561. doi: 10.3389/fendo.2025.1511561
Received: 15 October 2024; Accepted: 07 July 2025;
Published: 23 July 2025.
Edited by:
Jan Tesarik, MARGen Clinic, SpainReviewed by:
Claudio Aguayo Tapia, University of Concepcion, ChileAndrea Sonaglioni, IRCCS MultiMedica, Italy
Copyright © 2025 Nie, Sun, Li, Liang, Deng, He, Zeng, Zheng, Xiao, Ding, Huang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Huang, aHVhbmdqaWFuQGdsbWMuZWR1LmNu; Xiangyuan Yu, Z3VpbGlueGlhbmd5dWFuMTIzQDE2My5jb20=
†These authors have contributed equally to this work
 Lijie Nie
Lijie Nie Yan Sun
Yan Sun Ruiqi Li1†
Ruiqi Li1† Qiulian Liang
Qiulian Liang Xiangyuan Yu
Xiangyuan Yu