Abstract
Objective:
This meta-analysis aims to evaluate the diagnostic performance of ultrasound (US)-based artificial intelligence (AI) in assessing cervical lymph node metastasis (CLNM) in patients with papillary thyroid carcinoma (PTC).
Methods:
A comprehensive literature search was conducted in PubMed, Embase, Web of Science, and the Cochrane Library to identify relevant studies published up to November 19, 2024. Studies focused on the diagnostic performance of AI in the detection of CLNM of PTC were included. A bivariate random-effects model was used to calculate the pooled sensitivity and specificity, both with 95% confidence intervals (CI). The I2 statistic was used to assess heterogeneity among studies.
Results:
Among the 593 studies identified, 27 studies were included (involving over 23,170 patients or images). For the internal validation set, the pooled sensitivity, specificity, and AUC for detecting CLNM of PTC were 0.80 (95% CI: 0.75–0.84), 0.83 (95% CI: 0.80–0.87), and 0.89 (95% CI: 0.86–0.91), respectively. For the external validation set, the pooled sensitivity, specificity, and AUC were 0.77 (95% CI: 0.49–0.92), 0.82 (95% CI: 0.75–0.88), and 0.86 (95% CI: 0.83–0.89), respectively. For US physicians, the overall sensitivity, specificity, and AUC for detecting CLNM were 0.51 (95% CI: 0.38–0.64), 0.84 (95% CI: 0.76–0.89), and 0.77 (95% CI: 0.73–0.81), respectively.
Conclusion:
US-based AI demonstrates higher diagnostic performance than US physicians. However, the high heterogeneity among studies and the limited number of externally validated studies constrain the generalizability of these findings, and further research on external validation datasets is needed to confirm the results and assess their practical clinical value.
Systematic review registration:
https://www.crd.york.ac.uk/PROSPERO/view/CRD42024625725, identifier CRD42024625725.
Introduction
Papillary thyroid carcinoma (PTC) is the most common malignant thyroid tumor, with a steadily increasing global incidence, though its mortality rate remains relatively low (1). Approximately 30% to 80% of PTC patients experience lymph node metastasis (LNM), with cervical lymph node metastasis (CLNM) occurring in about 49% of these LNM-positive patients (2, 3). CLNM is a major risk factor for recurrence and reduced survival, often requiring aggressive surgical interventions, such as extensive lymph node dissection, which carry higher risks of complications (4). Accurate and timely detection of CLNM is therefore critical, as it directly influences treatment strategies and improves patient outcomes.
Traditional imaging modalities, including ultrasound (US), computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography-computed tomography (PET-CT), are widely used for evaluating CLNM of PTC (5). Among these, US is the first-line tool due to its non-invasive nature, real-time imaging capabilities, and high spatial resolution (6). However, its diagnostic accuracy is highly operator-dependent, leading to inconsistent results (7). In contrast, CT and MRI offer more detailed anatomical insights but have low sensitivity in identifying small metastatic lymph nodes (<2–3 mm), increasing the risk of missed diagnoses (8, 9). Moreover, these methods often rely on qualitative or semi-quantitative assessments, such as lymph node size and morphology, while neglecting quantitative features like texture, density, and signal intensity, which may be critical for predicting CLNM (10). These limitations highlight the need for more advanced diagnostic tools.
Artificial intelligence (AI) offers promising opportunities to improve the diagnostic performance of US in detecting CLNM. AI algorithms, particularly those based on machine learning and deep learning, can analyze complex imaging data and extract subtle features beyond human perception (11, 12). These algorithms process high-dimensional data and identify patterns that traditional methods may overlook. However, the diagnostic performance of AI remains inconsistent across studies (13, 14), and its comparative performance versus experienced US physicians has not been fully established, raising questions about its integration into routine clinical practice (15).
This meta-analysis aims to systematically evaluate the performance of US-based AI and its relative effectiveness compared to US physicians in detecting CLNM of PTC, providing a comprehensive assessment of its diagnostic capabilities and potential impact on clinical practice.
Methods
The meta-analysis was carried out strictly following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses for Diagnostic Test Accuracy (PRISMA-DTA) guidelines (16). Moreover, the protocol of this study has been registered with the PROSPERO (CRD42024625725).
Search strategy
A comprehensive search across PubMed, Embase, Web of Science, and Cochrane Library, with cutoff date of November 19, 2024. The search strategy included three groups of keywords: the first group related to AI (e.g., artificial intelligence, machine learning, deep learning), the second group related to diseases (e.g., lymphatic metastasis, lymph node metastasis), the third group related to target condition (e.g., thyroid neoplasms, thyroid carcinoma). We employed a combination of Medical Subject Headings (MeSH) and keywords (see Supplementary Table S1). Only studies published in English with full texts were included. Additionally, we manually searched the reference lists of selected studies to identify any potentially missed relevant articles. To ensure no recent studies were overlooked, we repeated the literature search on December 21, 2024.
Inclusion and exclusion criteria
Studies were carefully selected based on the PICOS framework. Population (P): Participants included patients diagnosed with PTC who required evaluation for CLNM. Intervention (I): AI models based on US images. Comparison (C): Either without a control group or compared with experienced ultrasound physicians. Outcome (O): The primary outcomes of interest included sensitivity, specificity, and area under the receiver operating characteristic curve (AUC). Study design (S): Both retrospective and prospective study designs were included.
We excluded animal studies and non-original research articles, including reviews, case reports, conference abstracts, meta-analyses, and letters to the editor. In addition, non-English full-text articles were excluded. Studies that did not meet these criteria were excluded from further analysis.
Quality assessment
We employed a modified version of the Quality Assessment of Diagnostic Performance Studies Revised (QUADAS-2-Revised tool) tool (17) to comprehensively evaluate the methodological quality of included studies. The adaptation involved replacing certain non-relevant criteria with more pertinent standards from the Prediction Model Risk of Bias Assessment tool, accounting for potential sources of bias arising from variations in research design and implementation.
The QUADAS-2-Revised tool assessed four critical domains: participants, index test (AI algorithm), reference standard, and analysis. The detail criteria were shown in Supplementary Table S2. Two independent reviewers systematically evaluated each domain’s risk of bias, with a particular focus on applicability in the first three domains. Divergent assessments were resolved through collaborative discussion.
Data extraction
Two reviewers independently evaluated the eligibility of studies and extracted data. In cases of disagreement, a third reviewer acted as an arbitrator to facilitate consensus. The extracted data included the first author’s name, publication year, country of study origin, study type, AI methods, selected AI algorithms, AI models, and patient-related data.
Since most studies did not report diagnostic contingency tables, we employed two methods to determine the diagnostic 2×2 table: 1) using sensitivity, specificity, the number of true positives determined by the reference standard, and the total number of cases; 2) through receiver operating characteristic (ROC) curve analysis, extracting sensitivity and specificity based on the optimal Youden index.
Outcome measures
The primary outcome measures included sensitivity, specificity, and area under the curve (AUC) for internal validation sets, external validation sets, and radiologists. Sensitivity (also known as recall or true positive rate) measures the probability that the AI model correctly identifies true positive cases, calculated as TP/(TP+FN). Specificity (also known as true negative rate) reflects the probability that the AI model correctly identifies healthy cases, calculated as TN/(TN+FP). AUC represents the area under the ROC curve, serving as a comprehensive measure of the model’s ability to distinguish between positive and negative cases. We extracted AI diagnostic performance data from internal validation sets, external validation sets, and US physicians, including only the models with optimal diagnostic performance (highest AUC values).
Statistical analysis
We summarized the overall sensitivity and specificity of AI analyses predicting CLNM of PTC using a bivariate random effects model for internal validation sets, external validation sets, and clinical diagnoses (18). A forest plot was created to visually represent the pooled sensitivity and specificity. Moreover, a summary receiver operating characteristic (SROC) curve was constructed to illustrate the overall sensitivity and specificity estimates along with their 95% confidence intervals (CI) and prediction intervals. Additionally, a Fagan plot was generated to evaluate the clinical applicability.
Heterogeneity among the included studies was assessed using the I2 statistic, with I2 values of 25%, 50%, and 75% indicating low, moderate, and high heterogeneity, respectively (19). For internal validation sets (greater than 10 studies), meta-regression analysis was conducted when significant heterogeneity was present (I2>50%) to explore potential sources of heterogeneity. The variables for meta-regression included US techniques (B-mode US or multimodal US), AI algorithms, AI models, data analysis types, and the location of CLNM. Furthermore, subgroup analyses were conducted for these variables to assess differences between subgroups. We also used the Z-test to compare the outcome differences between the internal validation sets and US physicians (20). Publication bias was assessed using Deeks’ funnel plot. Statistical analyses were primarily conducted using the Midas and Metadta programs in STATA version 15.1. The risk of bias assessment for study quality was performed using RevMan 5.4 (Cochrane Collaboration). A P-value of <0.05 was defined as statistically significant.
Results
Study selection
The initial database search yielded 593 potentially relevant articles. After removing 103 duplicates, 490 unique articles proceeded to preliminary screening. Following a rigorous application of the inclusion criteria, 446 articles were excluded. After a detailed full-text review, 17 studies were further excluded, including seven studies for not being PTC, three studies due to internal or external validation data being unavailable, and seven studies for being non-US-based AI. Ultimately, 27 studies that met the criteria for evaluating AI diagnostic performance were included in the meta-analysis (2, 13, 21–45). The literature selection method is comprehensively outlined in accordance with the standardized Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram, as shown in Figure 1.
Figure 1

PRISMA flow diagram illustrating the study selection process.
Study description and quality assessment
A total of 27 eligible studies were identified, with the internal validation set comprising all 27 studies and a total of 6,366 patients (range: 50-1,013), while the external validation set included 4 studies with a total of 1,592 patients (range: 95-881). 13 articles provided diagnostic data from US clinicians. One study was prospective, while 26 were retrospective design. Of the studies, 24 used pathology as the gold standard, and three utilized fine needle aspiration (FNA) as the gold standard. The most common modeling methods were logistic regression (LR) (12/27, 44%), convolutional neural network (CNN) (7/27, 26%), and support vector machine (SVM) (2/27, 7%). The characteristics of the studies and patients are summarized in Tables 1 and 2.
Table 1
| Author | Year | Country | Study design | Imaging modality | Location of cervical lymph node metastasis | Analysis | Reference standard | Patients/lesions per set | No. of LNM+ patients/lesions/ | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Training | Internal validation | External validation | |||||||||
| Agyekum et al. (2) | 2022 | China | Retro | B-mode | Central | Patient-based | Pathology | 143 | 62 | NR | Training: 74 Internal validation: 33 |
| Chang et al. (21) | 2023 | China | Retro | B-mode | Central | Patient-based | Pathology | 2114 | 906 | 339 | Training: 1063 Internal validation: 460 External validation:162 |
| Chen et al. (22) | 2021 | China | Retro | B-mode | Central | Patient-based | Pathology | 634 | 272 | NR | Training: 228 Internal validation: 94 |
| Dai et al. (23) | 2023 | China | Retro | CDU&EG | Central | Patient-based | Pathology | 348 | 150 | NR | Training: 167 Internal validation: 74 |
| Gao et al. (13) | 2024 | China | Retro | B-mode | Central | Patient-based | Pathology | 460 | 153 | NR | Training: 228 Internal validation: 76 |
| Guang et al. (24) | 2023 | China | Retro | B-mode | Central& Lateral | Patient-based | Pathology | 196 | 50 | NR | Training: 100 Internal validation: 26 |
| Huang et al. (25) | 2021 | China | Retro | EG&CDU | Central | Patient-based | Pathology | 439 | 220 | NR | Training: 160 Internal validation: 77 |
| Jia et al. (26) | 2024 | China | Retro | SWE&CEUS | Central | Patient-based | Pathology | NR | 126 | NR | Internal validation: 59 |
| Jiang et al. (27) | 2020 | China | Retro | SWE&CDU | Central& Lateral | Patient-based | Pathology | 147 | 90 | NR | Training: 75 Internal validation: 38 |
| Jiang et al. (28) | 2023 | China | Retro | CEUS | NR | Patient-based | Pathology | 148 | 63 | NR | Training: 59 Internal validation: 29 |
| Qian et al. (29) | 2024 | China | Retro | DUV | NR | Patient-based | Pathology | 233 | 78 | NR | Training: 108 Internal validation: 30 |
| Shi et al. (30) | 2022 | China | Retro | B-mode | Central | Patient-based | Pathology | 469 | 118 | NR | Training: 121 Internal validation: 32 |
| Tong et al. (31) | 2022 | China | Retro | B-mode | Central& Lateral | Patient-based | Pathology | 300 | 143 | 277 | Training: 104 Internal validation: 47 External validation:112 |
| Tong et al. (32) | 2021 | China | Retro | B-mode | Lateral | Patient-based | Pathology | 600 | 286 | NR | Training: 55 Internal validation: 31 |
| Wang et al. (33) | 2024 | China | Pro | SWE | NR | Lesion-based | FNA | NR | 84 | NR | Internal validation:36 |
| Wei et al. (34) | 2023 | China | Retro | CEUS | NR | Patient-based | Pathology | 282 | 141 | NR | Training: 138 Internal validation: 68 |
| Wen et al. (35) | 2022 | China | Retro | B-mode | Central | Patient-based | Pathology | 353 | 68 | NR | Training: 185 Internal validation: 35 |
| Wu et al. (36) | 2024 | China | Retro | EG | Central | Patient-based | FNA | 142 | 62 | NR | Training: 75 Internal validation: 27 |
| Park et al. (37) | 2020 | South Korea | Retro | B-mode | Lateral | Patient-based | Pathology | 400 | 368 | NR | Training: 83 Internal validation: 100 |
| Yan et al. (38) | 2023 | China | Retro | B-mode | Central | Lesion-based | Pathology | 212 | 83 | NR | Training: 115 Internal validation: 45 |
| Yao et al. (39) | 2022 | China | Retro | B-mode | NR | Patient-based | Pathology | 5129 | 903 | NR | Training: 2165 Internal validation: 553 |
| Yu et al. (40) | 2020 | China | Retro | B-mode | Central | Patient-based | Pathology | NR | 1013 | 368,513 | Internal validation: 403 External validation: 217,218 |
| Yuan et al. (41) | 2024 | China | Retro | B-mode | Lateral | Lesion-based | FNA | 655 | 206 | NR | Training: 327 Internal validation: 110 |
| Zhang et al. (42) | 2025 | China | Retro | B-mode | Central | Patient-based | Pathology | 340 | 83 | 95 | Training: 185 Internal validation: 47 External validation:47 |
| Zhang et al. (43) | 2023 | China | Retro | CDU | NR | Patient-based | Pathology | 451 | 194 | NR | Training: 67 Internal validation: 35 |
| Zhou et al. (44) | 2022 | China | Retro | B-mode | Central | Patient-based | Pathology | 608 | 326 | NR | Training: 182 Internal validation: 113 |
| Zhu et al. (45) | 2023 | China | Retro | B-mode | Central& Lateral | Lesion-based | Pathology | 282 | 118 | NR | Training: 117 Internal validation: 38 |
Study and patient characteristics of the included studies.
Retro, retrospective; Pro, prospective; NR, not report; FNA, fine needle aspiration; B-mode, B mode ultrasound; CDU, color doppler ultrasound; EG, elastography; CEUS, contrast-enhanced ultrasound; SWE, shear wave elastography; DUV, dynamic ultrasound video.
Table 2
| Author | Year | AI method | Optimal AI Algorithm | AI Mode | Interval validation sets | External validation sets | Ultrasound physician | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FP | FN | TN | TP | FP | FN | TN | TP | FP | FN | TN | |||||
| Agyekum et al. (2) | 2022 | Machine learning | LDA | Ultrasound&clinical model | 20 | 8 | 13 | 21 | NR | NR | NR | NR | 49 | 39 | 49 | 68 |
| Chang et al. (21) | 2023 | Deep learning | CNN | Ultrasound&clinical model | 182 | 104 | 278 | 342 | 59 | 41 | 103 | 136 | 169,59 | 34,15 | 291,103 | 412,162 |
| Chen et al. (22) | 2021 | Deep learning | CNN | Ultrasound-based model | 81 | 33 | 13 | 145 | NR | NR | NR | NR | NR | NR | NR | NR |
| Dai et al. (23) | 2023 | Machine learning | SVM | Ultrasound&clinical model | 59 | 8 | 15 | 68 | NR | NR | NR | NR | NR | NR | NR | NR |
| Gao et al. (13) | 2024 | Deep learning | CNN | Ultrasound&clinical model | 55 | 14 | 21 | 63 | NR | NR | NR | NR | 32 | 23 | 44 | 54 |
| Guang et al. (24) | 2023 | Deep Learning | CNN | Ultrasound-based model | 21 | 4 | 5 | 20 | NR | NR | NR | NR | 61 | 15 | 97 | 135 |
| Huang et al. (25) | 2021 | Machine learning | LR | Ultrasound&clinical model | 60 | 38 | 17 | 105 | NR | NR | NR | NR | NR | NR | NR | NR |
| Jiang et al. (27) | 2020 | Machine learning | LR | Ultrasound&clinical model | 33 | 14 | 5 | 38 | NR | NR | NR | NR | 41 | 19 | 72 | 105 |
| Jiang et al. (28) | 2023 | Machine learning | LR | Ultrasound&clinical model | 24 | 9 | 5 | 25 | NR | NR | NR | NR | NR | NR | NR | NR |
| Qian et al. (29) | 2024 | Deep Learning | CNN | Ultrasound-based model | 26 | 6 | 4 | 42 | NR | NR | NR | NR | NR | NR | NR | NR |
| Jia et al. (26) | 2024 | Machine learning | SVM | Ultrasound-based model | 53 | 18 | 6 | 49 | NR | NR | NR | NR | NR | NR | NR | NR |
| Shi et al. (30) | 2022 | Machine Learning | XGBoost | Ultrasound&clinical model | 28 | 12 | 4 | 74 | NR | NR | NR | NR | NR | NR | NR | NR |
| Tong et al. (31) | 2022 | Machine Learning | LR | Ultrasound&clinical model | 39 | 17 | 8 | 79 | 80 | 21 | 32 | 144 | 23,59 | 9,24 | 24,53 | 87,141 |
| Tong et al. (32) | 2021 | Machine Learning | LR | Ultrasound&clinical model | 25 | 14 | 6 | 241 | NR | NR | NR | NR | 22 | 31 | 9 | 224 |
| Wang et al. | 2024 | Machine Learning | Fisher | Ultrasound-based model | 30 | 8 | 6 | 40 | NR | NR | NR | NR | NR | NR | NR | NR |
| Wei et al. (34) | 2023 | Machine Learning | LR | Ultrasound&clinical model | 52 | 2 | 16 | 71 | NR | NR | NR | NR | 52 | 33 | 16 | 40 |
| Wen et al. (35) | 2022 | Machine Learning | LR | Ultrasound&clinical model | 24 | 8 | 11 | 25 | NR | NR | NR | NR | 7 | 0 | 28 | 33 |
| Wu et al. (36) | 2024 | Machine Learning | LR | Ultrasound&clinical model | 22 | 6 | 5 | 29 | NR | NR | NR | NR | 25 | 15 | 2 | 20 |
| Park et al. (37) | 2020 | Machine Learning | LR | Ultrasound&clinical model | 69 | 126 | 31 | 142 | NR | NR | NR | NR | NR | NR | NR | NR |
| Yan et al. (38) | 2023 | Machine Learning | LR | Ultrasound-based model | 42 | 4 | 3 | 34 | NR | NR | NR | NR | NR | NR | NR | NR |
| Yao et al. (39) | 2022 | Deep Learning | DCNN | Ultrasound&clinical model | 451 | 43 | 102 | 307 | NR | NR | NR | NR | NR | NR | NR | NR |
| Yu et al. (40) | 2020 | Deep Learning | TLR | Ultrasound&clinical model | 379 | 140 | 24 | 470 | 180,207 | 17,74 | 37,11 | 134,221 | NR | NR | NR | NR |
| Yuan et al. (41) | 2024 | Deep Learning | CNN | Ultrasound-based model | 107 | 6 | 14 | 79 | NR | NR | NR | NR | 104 | 16 | 17 | 69 |
| Zhang et al. (42) | 2025 | Deep Learning | CNN | Ultrasound-based model | 37 | 5 | 10 | 31 | 44 | 13 | 3 | 35 | 28 | 17 | 19 | 31 |
| Zhang et al. (43) | 2023 | Machine Learning | LR | Ultrasound&clinical model | 19 | 9 | 16 | 150 | NR | NR | NR | NR | NR | NR | NR | NR |
| Zhou et al. (44) | 2022 | Machine Learning | LR | Ultrasound&clinical model | 92 | 40 | 21 | 173 | NR | NR | NR | NR | 15 | 16 | 98 | 197 |
| Zhu et al. (45) | 2023 | Machine Learning | RF | Ultrasound&clinical model | 26 | 17 | 12 | 63 | NR | NR | NR | NR | NR | NR | NR | NR |
Technical aspects of included studies.
TP, true positive; TN, true negative; FP, false positive; FN, false negative; NR, not report; LDA, linear discriminant analysis; LR, logistic regression; CNN, convolutional neural network; SVM, support vector machine; XGBoost, eXtreme gradient boosting; Fisher, Fisher's stepwise discriminant analysis; DCNN, deep convolutional neural network; TLR, transfer learning radiomics; RF, random forest.
According to the QUADAS-2-Revised tool, the risk of bias for each study is shown in Figure 2. For the bias assessment regarding Patient Selection, 4 studies were rated as “high risk” due to inappropriate exclusion. For the Index Test, 2 studies were rated as “unclear” because it was uncertain whether the AI model provided important training information. Regarding the Reference Standard, 2 studies were rated as “unclear” because it was uncertain whether the pathologists were aware of the pathology results in the final diagnosis. Overall, the quality assessment indicates that the quality of the included studies is acceptable.
Figure 2
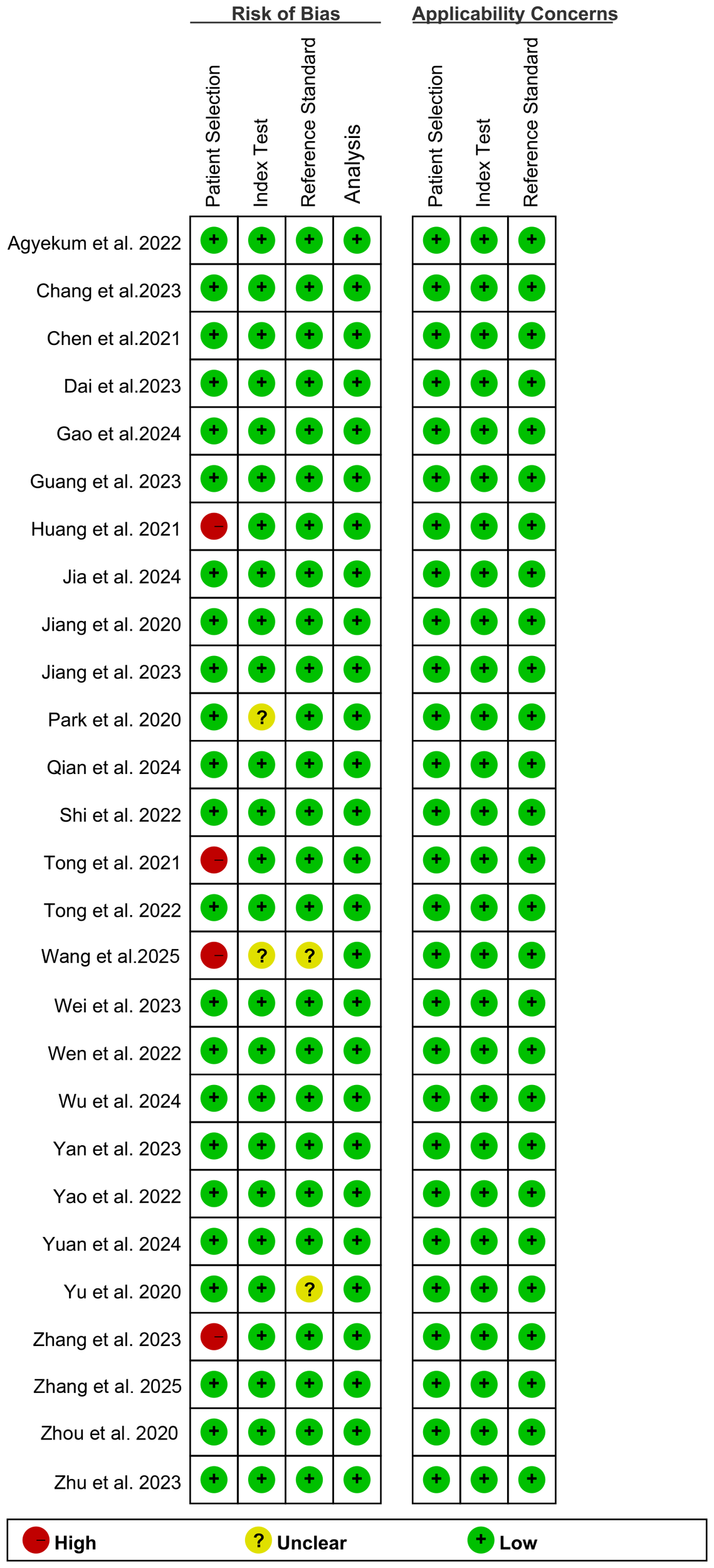
Risk of bias and applicability concerns of the included studies using the Quality Assessment of Diagnostic Performance Studies (QUADAS)-2 Revised tool.
Diagnostic performance of internal validation set for AI and US physicians in predicting CLNM of PTC
For the internal validation set, the sensitivity of AI in detecting CLNM of PTC was 0.80 (95% CI: 0.75-0.84) and the specificity was 0.83 (95% CI: 0.80-0.87) (Figure 3a), with an AUC of 0.89 (95% CI: 0.86-0.91) (Figure 4a). Using a pre-test probability of 20%, the Fagan nomogram indicated a positive likelihood ratio of 55% and a negative likelihood ratio of 6% (Figure 5a). For US physicians, the sensitivity for detecting CLNM of PTC was 0.51 (95% CI: 0.38-0.64) and the specificity was 0.84 (95% CI: 0.76-0.89) (Figure 3b), with an AUC of 0.77 (95% CI: 0.73-0.81) (Figure 4b). Using a 20% pre-test probability, the Fagan nomogram showed a positive likelihood ratio of 44% and a negative likelihood ratio of 13% (Figure 5b). The Z-test indicated that AI had significantly higher sensitivity and AUC values (P < 0.001), while there was no significant difference in specificity (P = 0.79).
Figure 3

Forest plots showing the combined sensitivity and specificity of ultrasonography-based artificial intelligence in patients with cervical lymph node metastasis from papillary thyroid carcinoma: internal validation set (a) and ultrasound physicians (b). Squares represent the sensitivity and specificity in each study, while horizontal bars indicate the 95% confidence intervals.
Figure 4

Summary receiver operating characteristic (SROC) curves for diagnosing cervical lymph node metastasis in papillary thyroid carcinoma: ultrasonography-based artificial intelligence on the internal validation set (a) and ultrasound physicians (b).
Figure 5
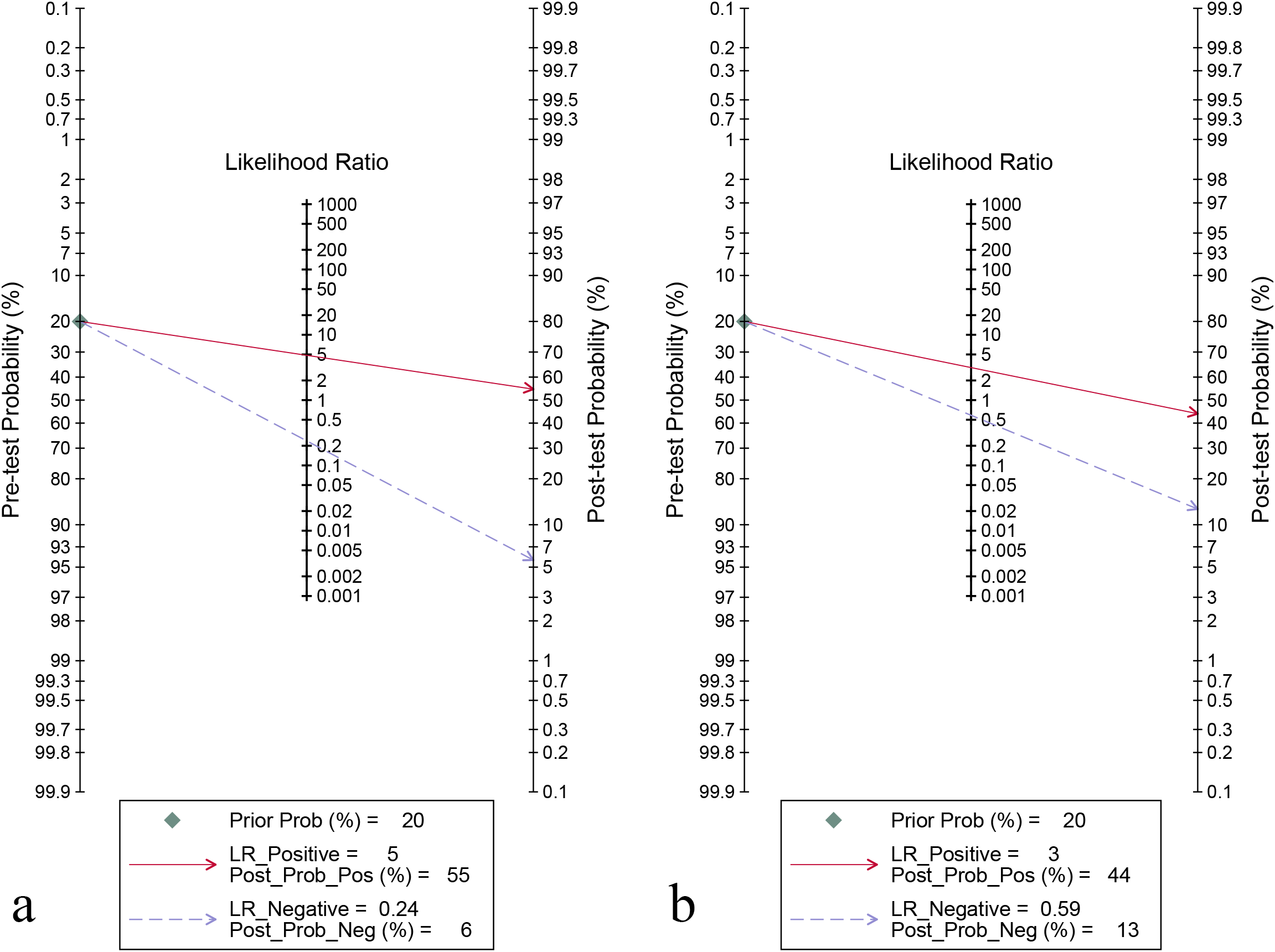
Fagan’s nomogram for diagnosing cervical lymph node metastasis in papillary thyroid carcinoma: ultrasonography-based artificial intelligence on the internal validation set (a) and ultrasound physicians (b).
For the internal validation set, both sensitivity (I2 = 95.21%) and specificity (I2 = 91.33%) exhibited high heterogeneity. Meta-regression analysis indicated that the heterogeneity was primarily attributed to US techniques (sensitivity P < 0.01, specificity P < 0.001), AI methods (sensitivity P < 0.01, specificity P < 0.001), AI models (sensitivity P < 0.05, specificity P < 0.001), and types of data analysis (specificity P < 0.05) (Figure 6).
Figure 6
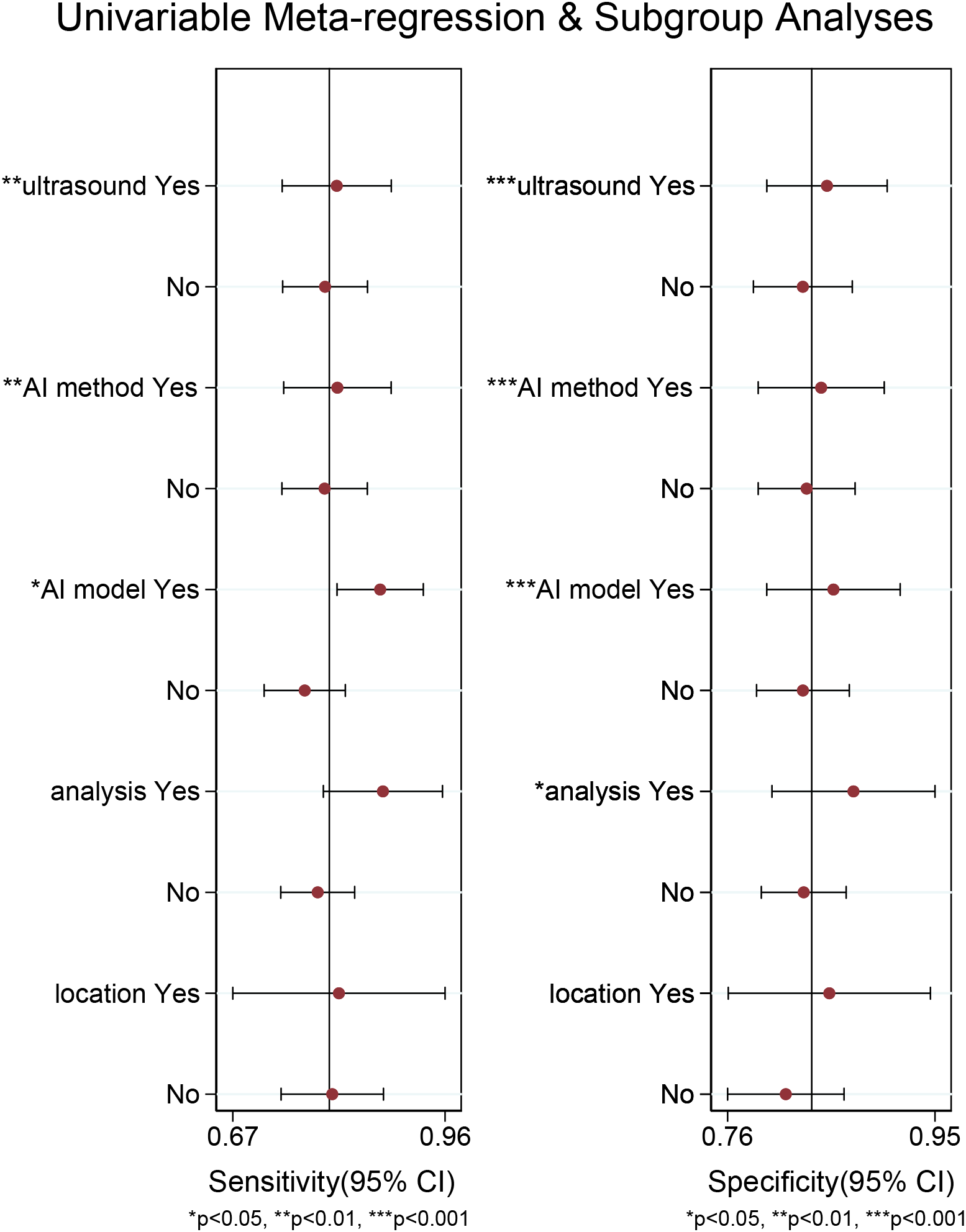
Meta-regression analysis of the internal validation set for diagnosing cervical lymph node metastasis in papillary thyroid carcinoma.
Diagnostic performance of external validation sets for AI in predicting CLNM of PTC
For the external validation set, the sensitivity for detecting CLNM of PTC was 0.77 (95% CI: 0.49-0.92) and the specificity was 0.82 (95% CI: 0.75-0.88) (Supplementary Figure S1), with an AUC of 0.86 (95% CI: 0.83-0.89) (Supplementary Figure S2). Using a pre-test probability of 20%, the Fagan nomogram indicated a positive likelihood ratio of 52% and a negative likelihood ratio of 6% (Supplementary Figure S3).
Diagnostic performance of subgroup analysis for AI in predicting CLNM of PTC
In the subgroups of ultrasound techniques, B-mode US had a sensitivity of 0.81 (95% CI: 0.76-0.86) and Multimodal US 0.78 (95% CI: 0.69-0.85), with no significant difference (P = 0.49). The specificity was 0.82 (95% CI: 0.76-0.86) for B-mode and 0.86 (95% CI: 0.80-0.91) for Multimodal US, also showing no significant difference (P = 0.23) (Table 3).
Table 3
| Subgroup | Studies, n | Sensitivity (95%CI) | Subgroup difference P-value | Specificity (95%CI) | Subgroup difference P-value |
|---|---|---|---|---|---|
| Ultrasound techniques | 0.49 | 0.23 | |||
| B-mode ultrasound | 17 | 0.81 (0.75-0.86) | 0.82 (0.76-0.86) | ||
| Multimodal ultrasound | 10 | 0.78 (0.69-0.85) | 0.86 (0.80-0.91) | ||
| AI method | 0.19 | 0.91 | |||
| Deep learning | 9 | 0.84 (0.76-0.89) | 0.83 (0.76-0.88) | ||
| Machine learning | 18 | 0.78 (0.71 - 0.84) | 0.83 (0.78 - 0.88) | ||
| AI model | <0.001 | 0.93 | |||
| Ultrasound-based model | 8 | 0.88 (0.82-0.92) | 0.83 (0.76-0.89) | ||
| Ultrasound&clinical model | 19 | 0.76 (0.70-0.81) | 0.83 (0.78-0.87) | ||
| Analysis | 0.12 | 0.29 | |||
| Patient-based | 23 | 0.79 (0.73-0.83) | 0.82 (0.78-0.86) | ||
| Lesion-based | 4 | 0.87 (0.77-0.93) | 0.87 (0.78-0.93) | ||
| Location of cervical lymph node metastasis | 0.49 | 0.04 | |||
| Central | 14 | 0.82 (0.76-0.87) | 0.80 (0.74-0.86) | ||
| Lateral | 3 | 0.80 (0.64-0.90) | 0.91 (0.84-0.95) |
Subgroup analysis of cervical lymph node metastasis of papillary thyroid carcinoma of internal validation set.
For AI methods, the sensitivity was 0.84 (95% CI: 0.76-0.89) for deep learning and 0.78 (95% CI: 0.71-0.84) for machine learning, with no significant difference (P = 0.19). Both methods had a specificity of 0.83 (95% CI: 0.76-0.88), with no significant difference (P = 0.91) (Table 3).
Regarding AI models, the sensitivity of the US-based model was 0.88 (95% CI: 0.82-0.92) compared to 0.76 (95% CI: 0.70-0.81) for the US & clinical model, showing a significant difference (P < 0.001). Both models exhibited a specificity of 0.83 (95% CI: 0.76-0.89), with no significant difference (P = 0.93) (Table 3).
For data analysis types, patient-based sensitivity was 0.79 (95% CI: 0.73-0.83) and lesion-based was 0.87 (95% CI: 0.77-0.93), with no significant difference (P = 0.12). Specificity was 0.82 (95% CI: 0.78-0.86) for patient-based and 0.87 (95% CI: 0.78-0.93) for lesion-based, also with no significant difference (P = 0.29) (Table 3).
In terms of CLNM locations, sensitivity was 0.82 (95% CI: 0.76-0.87) for central and 0.80 (95% CI: 0.64-0.90) for lateral locations, showing no significant difference (P = 0.49). However, specificity was 0.80 (95% CI: 0.74-0.86) for central and 0.91 (95% CI: 0.84-0.95) for lateral, indicating a significant difference (P < 0.05) (Table 3).
Publication bias
Deeks’ funnel plot asymmetry test indicated no significant publication bias for the internal validation set of AI and US physicians (P = 0.47, 0.86) (Supplementary Figure S4-S5). For the external validation set, no significant publication bias was observed either (P = 0.49) (Supplementary Figure S6).
Discussion
Our meta-analysis revealed that AI-based ultrasonography demonstrated superior performance compared to human US physicians in detecting CLNM in patients with PTC. Specifically, AI achieved higher sensitivity, specificity, and AUC values. This enhanced diagnostic performance is largely attributable to AI’s ability to process large and complex datasets, extracting subtle, high-dimensional features that may be imperceptible to human observers (46). AI can integrate multiple imaging characteristics—such as texture, density, and signal intensity—into predictive models, thereby improving diagnostic precision (47). Internal validation datasets, which are typically more homogeneous and closely aligned with the training data, tend to yield better algorithm performance due to their consistency in imaging protocols and patient characteristics (48). Conversely, external validation datasets often introduce greater heterogeneity due to the imaging techniques, equipment, and patient populations (48). Interestingly, our findings demonstrate remarkable generalizability of the AI models, with the AUC decreasing only marginally from 0.89 in internal validation to 0.86 in external validation. The lower sensitivity and AUC observed among US physicians underscores the operator-dependent nature of traditional ultrasonography and the inherent limitations of qualitative or semi-quantitative assessments. These findings further highlight the potential of AI to standardize diagnostic processes and improve accuracy in clinical practice.
It’s worth noting that our meta-analysis revealed no statistically significant differences in sensitivity (P = 0.19) or specificity (P = 0.91) between deep learning and machine learning methods. The sensitivity of deep learning and machine learning was 0.84 and 0.78, respectively, while both methods demonstrated a same specificity of 0.83. The comparable diagnostic performance may be explained by their shared reliance on advanced algorithmic frameworks capable of identifying critical imaging features relevant to CLNM prediction (49). Both approaches employ supervised learning techniques to analyze structured imaging data, enabling the detection of patterns such as texture, density, and morphological changes in lymph nodes (50). Deep learning, particularly CNN, has the advantage of automated feature extraction directly from raw data. In contrast, machine learning often relies on handcrafted features derived from expert knowledge (50). However, in this context, the imaging datasets used in the included studies may have been sufficiently optimized, with robust feature engineering for machine learning models, thereby reducing the performance gap between the two methods.
Another finding is that the results demonstrated a statistically significant difference in sensitivity between the US-based model and the US & clinical model for predicting CLNM of PTC patients, with sensitivities of 0.88 and 0.76 (P < 0.001). The higher sensitivity of the US-based model may be attributed to its exclusive reliance on ultrasound imaging features, which are directly associated with structural and morphological changes in lymph nodes, such as size, echogenicity, and vascularity—key indicators for detecting CLNM (51). In contrast, the US & clinical model integrates additional clinical variables, such as patient demographics and laboratory findings, which may not be as strongly correlated with CLNM. These variables could introduce irrelevant or conflicting information, potentially diluting the predictive strength of the imaging features and resulting in lower sensitivity (51).
This meta-analysis also showed no statistically significant difference in sensitivity between the central and lateral locations of CLNM. However, specificity was significantly higher for the lateral lymph nodes (0.91) compared to the central lymph nodes (0.80; P < 0.05). The superior specificity for the lateral location may be attributed to the distinct anatomical and imaging characteristics of lateral lymph nodes. These nodes are typically larger, more superficial, and easier to visualize using ultrasonography (52). They also tend to exhibit clearer morphological changes, such as irregular margins, loss of the hilum, or abnormal vascularity, which facilitate differentiation from benign lymph nodes (52). In contrast, central lymph nodes are situated in a more anatomically complex region, often surrounded by structures such as the thyroid gland, trachea, and blood vessels. This complexity can obscure visualization on ultrasonography and result in overlapping features between metastatic and benign nodes, thereby reducing diagnostic specificity (53).
Previous meta-analyses have provided valuable insights into the diagnostic performance of various imaging modalities for LNM in thyroid cancer. For instance, the 2023 meta-analysis by HajiEsmailPoor et al. evaluated 25 studies assessing the performance of CT, US, and MRI-based radiomics for predicting LNM in PTC (54). Their results indicated that US outperformed CT and MRI, with a sensitivity of 0.77 and a specificity of 0.79. Our study, focusing exclusively on AI-based models using US for predicting CLNM of PTC, revealed even higher diagnostic performance, with pooled sensitivity and specificity of 0.80 and 0.83. This improvement may be attributed to the advanced analytical capabilities of AI, as incorporating more US-based AI studies allows it to extract and analyze subtle imaging features beyond human perception. Furthermore, unlike previous studies, our study is the first meta-analysis to focus on US-based AI models and their relative diagnostic performance compared to US physicians for CLNM of PTC, offering a more targeted and comprehensive result (55).
In comparison to the 2024 meta-analysis by Zhang et al., which examined radiomics-based US models for LNM in thyroid cancer, our study yielded slightly lower diagnostic performance (56). This discrepancy may be explained by differences in study populations, as Zhang et al. included various thyroid cancers (including PTC), while our analysis was restricted to PTC cases. It is important to notethat our study introduced two significant innovations: the first direct comparison of AI models with US physicians, highlighting the potential clinical advantages of AI, and a subgroup analysis evaluating diagnostic performance using internal and external validation datasets. These advancements provide critical evidence for the practical application of AI in clinical settings and address limitations in prior meta-analyses.
This study highlights that significant heterogeneity among the included studies may have impacted the overall sensitivity and specificity of AI in internal test datasets. Meta-regression analysis identified US techniques, AI methods, and AI models as potential sources of heterogeneity affecting sensitivity. The potential source of heterogeneity for specificity were the types of data analysis. Despite this heterogeneity, the findings demonstrate that US-based AI achieves high diagnostic performance for predicting CLNM of PTC across both internal and external validation datasets, surpassing the diagnostic performance of US physicians. This suggests that AI has the potential to alleviate the workload of clinical practitioners, reduce misdiagnoses and missed diagnoses, and prevent adverse outcomes associated with the disease. The integration of US-based AI tools into primary care settings, such as general practice, could support early detection and timely management of PTC. Moreover, US-based AI has the potential to enhance screening efficiency, particularly in resource-constrained or remote areas where access to specialized expertise is limited. In the future, US-based AI systems could serve as valuable tools to assist US physicians in making more accurate diagnoses.
However, while diagnostic performance is crucial, cost-effectiveness is an equally important consideration when introducing new technologies into routine clinical practice. AI’s diagnostic potential raises ethical and operational concerns, including tensions between algorithmic efficiency and clinician autonomy due to opaque “black-box” systems, as well as bias risks from non-representative training data that may worsen health inequities (57). Mitigation strategies could involve adopting explainable AI to clarify decisions, implementing bias-checking validation protocols, and establishing oversight-focused regulatory policies with hybrid human-AI workflows to balance innovation with accountability (58). Notably, this study did not identify any research evaluating the cost-effectiveness of AI in diagnosing CLNM of PTC, underscoring a critical gap that future investigations should address.
The limitations of this study should be acknowledged. First, there is a lack of external validation among the included studies, with only four out of 27 studies performing external validation. External validation is crucial because overfitting is a common issue in AI training (48). Second, most of the included studies were retrospective in design, which may introduce potential biases. Well-designed prospective studies are necessary to confirm the findings of this meta-analysis and ensure their robustness. Third, three studies used non-pathology-based reference standards, which could introduce bias in the evaluation of diagnostic performance. Fourth, this study only included English-language literature, a decision primarily driven by pragmatic considerations of accessibility. However, it may bring potential publication bias. Future research should adopt more standardized and consistent pathology-based reference standards to ensure accuracy and reliability.
Conclusion
US-based AI demonstrates higher diagnostic performance than clinicians. However, the high heterogeneity among studies limits the strength of these findings, necessitating further investigation of external validation datasets to confirm the results and assess their practical clinical value.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
XW: Conceptualization, Formal Analysis, Methodology, Software, Writing – original draft, Writing – review & editing. YQ: Data curation, Formal Analysis, Methodology, Writing – original draft. XZ: Data curation, Formal Analysis, Methodology, Writing – original draft. FL: Data curation, Formal Analysis, Methodology, Writing – original draft. JL: Conceptualization, Data curation, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by “Key Discipline Construction Project of Zunyi Medical University Zhuhai Campus” (No. ZHPY2024-1).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2025.1570811/full#supplementary-material
References
1
ZhangJXuS. High aggressiveness of papillary thyroid cancer: from clinical evidence to regulatory cellular networks. Cell Death Discov. (2024) 10:378. doi: 10.1038/s41420-024-02157-2
2
AgyekumEARenY-ZWangXCranstonSSWangY-GWangJet al. Evaluation of cervical lymph node metastasis in papillary thyroid carcinoma using Clinical-Ultrasound Radiomic Machine Learning-Based model. Cancers. (2022) 14:5266. doi: 10.3390/cancers14215266
3
Popović KrnetaMŠobić ŠaranovićDMijatović TeodorovićLKrajčinovićNAvramovićNBojovićŽet al. Prediction of cervical lymph node metastasis in clinically node-negative T1 and T2 papillary thyroid carcinoma using supervised machine learning approach. J Clin Med. (2023) 12:3641. doi: 10.3390/jcm12113641
4
JiangL-HYinK-XWenQ-LChenCGeM-HTanZ. Predictive risk-scoring model for central lymph node metastasis and predictors of recurrence in papillary thyroid carcinoma. Sci Rep. (2020) 10:710. doi: 10.1038/s41598-019-55991-1
5
SinghNKHageNRamamourthyBNagarajuSKappagantuKM. Nuclear imaging modalities in the diagnosis and management of thyroid cancer. Curr Mol Med. (2024) 24:1091–6. doi: 10.2174/1566524023666230915103723
6
PenetM-FKakkadSPacheco-TorresJBhartiSKrishnamacharyBBhujwallaZM. Chapter 53 - molecular and functional imaging and theranostics of the tumor microenvironment. In: RossBDGambhirSS, editors. Molecular Imaging (Second Edition). San Diego, CA: Academic Press (2021). p. 1007–29.
7
FengJ-WLiuS-QQiG-FYeJHongL-ZWuW-Xet al. Development and validation of clinical-radiomics nomogram for preoperative prediction of central lymph node metastasis in papillary thyroid carcinoma. Acad Radiol. (2024) 31(6):2292–305. doi: 10.1016/j.acra.2023.12.008
8
ChoSSuhCBaekJChungSChoiYLeeJ. Diagnostic performance of MRI to detect metastatic cervical lymph nodes in patients with thyroid cancer: a systematic review and meta-analysis. Clin Radiol. (2020) 75:562.e1–562.e10. doi: 10.1016/j.crad.2020.03.025
9
YangJZhangFQiaoY. Diagnostic accuracy of ultrasound, CT and their combination in detecting cervical lymph node metastasis in patients with papillary thyroid cancer: a systematic review and meta-analysis. BMJ Open. (2022) 12:e051568. doi: 10.1136/bmjopen-2021-051568
10
FanFLiFWangYDaiZLinYLiaoLet al. Integration of ultrasound-based radiomics with clinical features for predicting cervical lymph node metastasis in postoperative patients with differentiated thyroid carcinoma. Endocrine. (2024) 84:999–1012. doi: 10.1007/s12020-023-03644-9
11
SharmaMSavageCNairMLarssonISvedbergPNygrenJM. Artificial intelligence applications in health care practice: scoping review. J Med Internet Res. (2022) 24:e40238. doi: 10.2196/40238
12
TadiboinaSN. The use of AI in advanced medical imaging. J Positive School Psychol. (2022) 6:1939–46.
13
GaoYWangWYangYXuZLinYLangTet al. An integrated model incorporating deep learning, hand-crafted radiomics and clinical and US features to diagnose central lymph node metastasis in patients with papillary thyroid cancer. BMC Cancer. (2024) 24:69. doi: 10.1186/s12885-024-11838-1
14
NamsenaPSongsaengDKeatmaneeCKlabwongSKunapinunASoodchuenSet al. Diagnostic performance of artificial intelligence in interpreting thyroid nodules on ultrasound images: a multicenter retrospective study. Quantitative Imaging Med Surg. (2024) 14:3676. doi: 10.21037/qims-23-1650
15
ShenJZhangCJJiangBChenJSongJLiuZet al. Artificial intelligence versus clinicians in disease diagnosis: systematic review. JMIR Med Inf. (2019) 7:e10010. doi: 10.2196/10010
16
McInnesMDMoherDThombsBDMcGrathTABossuytPMCliffordTet al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. Jama. (2018) 319:388–96. doi: 10.1001/jama.2017.19163
17
QuYYangZSunFZhanS. Risk on bias assessment:(6) a revised tool for the quality assessment on diagnostic accuracy studies (QUADAS-2). Zhonghua Liuxingbingxue Zazhi. (2018) 39:524–31. doi: 10.3760/cma.j.issn.0254-6450.2018.04.028
18
ArendsLHamzaTVan HouwelingenJHeijenbrok-KalMHuninkMStijnenT. Bivariate random effects meta-analysis of ROC curves. Med Decision Making. (2008) 28:621–38. doi: 10.1177/0272989X08319957
19
Huedo-MedinaTBSánchez-MecaJMarín-MartínezFBotellaJ. Assessing heterogeneity in meta-analysis: Q statistic or I² index? psychol Methods. (2006) 11:193. doi: 10.1037/1082-989X.11.2.193
20
YangH-LLiuTWangX-MXuYDengS-M. Diagnosis of bone metastases: a meta-analysis comparing 18 FDG PET, CT, MRI and bone scintigraphy. Eur Radiol. (2011) 21:2604–17. doi: 10.1007/s00330-011-2221-4
21
ChangLZhangYZhuJHuLWangXZhangHet al. An integrated nomogram combining deep learning, clinical characteristics and ultrasound features for predicting central lymph node metastasis in papillary thyroid cancer: A multicenter study. Front Endocrinol. (2023) 14:964074. doi: 10.3389/fendo.2023.964074
22
ChenYWangYCaiZJiangM. Predictions for central lymph node metastasis of papillary thyroid carcinoma via CNN-based fusion modeling of ultrasound images. Traitement Du Signal. (2021) 38:629–38. doi: 10.18280/ts.380310
23
DaiQTaoYLiuDZhaoCSuiDXuJet al. Ultrasound radiomics models based on multimodal imaging feature fusion of papillary thyroid carcinoma for predicting central lymph node metastasis. Front Oncol. (2023) 13:1261080. doi: 10.3389/fonc.2023.1261080
24
GuangYWanFHeWZhangWGanCDongPet al. A model for predicting lymph node metastasis of thyroid carcinoma: a multimodality convolutional neural network study. Quantitative Imaging Med Surg. (2023) 13:8370. doi: 10.21037/qims-23-318
25
HuangCCongSShangSWangMZhengHWuSet al. Web-based ultrasonic nomogram predicts preoperative central lymph node metastasis of cN0 papillary thyroid microcarcinoma. Front Endocrinol. (2021) 12:734900. doi: 10.3389/fendo.2021.734900
26
JiaWCaiYWangSWangJ. Predictive value of an ultrasound-based radiomics model for central lymph node metastasis of papillary thyroid carcinoma. Int J Med Sci. (2024) 21:1701. doi: 10.7150/ijms.95022
27
JiangMLiCTangSLvWYiAWangBet al. Nomogram based on shear-wave elastography radiomics can improve preoperative cervical lymph node staging for papillary thyroid carcinoma. Thyroid. (2020) 30:885–97. doi: 10.1089/thy.2019.0780
28
JiangLZhangZGuoSZhaoYZhouP. Clinical-radiomics nomogram based on contrast-enhanced ultrasound for preoperative prediction of cervical lymph node metastasis in papillary thyroid carcinoma. Cancers. (2023) 15:1613. doi: 10.3390/cancers15051613
29
QianTZhouYYaoJNiCAsifSChenCet al. Deep learning based analysis of dynamic video ultrasonography for predicting cervical lymph node metastasis in papillary thyroid carcinoma. Endocrine. (2024) 87(3):1060–9. doi: 10.1007/s12020-024-04091-w
30
ShiYZouYLiuJWangYChenYSunFet al. Ultrasound-based radiomics XGBoost model to assess the risk of central cervical lymph node metastasis in patients with papillary thyroid carcinoma: Individual application of SHAP. Front Oncol. (2022) 12:897596. doi: 10.3389/fonc.2022.897596
31
TongYZhangJWeiYYuJZhanWXiaHet al. Ultrasound-based radiomics analysis for preoperative prediction of central and lateral cervical lymph node metastasis in papillary thyroid carcinoma: a multi-institutional study. BMC Med Imaging. (2022) 22:82. doi: 10.1186/s12880-022-00809-2
32
TongYLiJHuangYZhouJLiuTGuoYet al. Ultrasound-based radiomic nomogram for predicting lateral cervical lymph node metastasis in papillary thyroid carcinoma. Acad Radiol. (2021) 28:1675–84. doi: 10.1016/j.acra.2020.07.017
33
WangYHanYLiFLinYWangB. Fisher discriminant analysis of multimodal ultrasound in diagnosis of cervical metastatic lymph nodes in papillary thyroid cancer. Korean J Internal Med. (2025) 40:103–14. doi: 10.3904/kjim.2024.122
34
WeiTWeiWMaQShenZLuKZhuX. Development of a clinical-radiomics nomogram that used contrast-enhanced ultrasound images to anticipate the occurrence of preoperative cervical lymph node metastasis in papillary thyroid carcinoma patients. Int J Gen Med. (2023) 16:3921–32. doi: 10.2147/IJGM.S424880
35
WenQWangZTraversoALiuYXuRFengYet al. A radiomics nomogram for the ultrasound-based evaluation of central cervical lymph node metastasis in papillary thyroid carcinoma. Front Endocrinol. (2022) 13:1064434. doi: 10.3389/fendo.2022.1064434
36
WuLZhouYLiLMaWDengHYeX. Application of ultrasound elastography and radiomic for predicting central cervical lymph node metastasis in papillary thyroid microcarcinoma. Front Oncol. (2024), 1354288. doi: 10.3389/fonc.2024.1354288
37
ParkVYHanKKimHJLeeEYoukJHKimE-Ket al. Radiomics signature for prediction of lateral lymph node metastasis in conventional papillary thyroid carcinoma. PloS One. (2020) 15:e0227315. doi: 10.1371/journal.pone.0227315
38
YanXMouXYangYRenJZhouXHuangYet al. Predicting central lymph node metastasis in patients with papillary thyroid carcinoma based on ultrasound radiomic and morphological features analysis. BMC Med Imaging. (2023) 23:111. doi: 10.1186/s12880-023-01085-4
39
YaoJLeiZYueWFengBLiWOuDet al. DeepThy-Net: a multimodal deep learning method for predicting cervical lymph node metastasis in papillary thyroid cancer. Adv Intelligent Syst. (2022) 4:2200100. doi: 10.1002/aisy.202200100
40
YuJDengYLiuTZhouJJiaXXiaoTet al. Lymph node metastasis prediction of papillary thyroid carcinoma based on transfer learning radiomics. Nat Commun. (2020) 11:4807. doi: 10.1038/s41467-020-18497-3
41
YuanYHouSWuXWangYSunYYangZet al. Application of deep-learning to the automatic segmentation and classification of lateral lymph nodes on ultrasound images of papillary thyroid carcinoma. Asian J Surg. (2024) 47(9):3892–8. doi: 10.1016/j.asjsur.2024.02.140
42
ZhangXYZhangDWangZYChenJRenJYMaTet al. Automatic tumor segmentation and lymph node metastasis prediction in papillary thyroid carcinoma using ultrasound keyframes. Med Phys. (2025) 52(1):257–73. doi: 10.1002/mp.17498
43
ZhangMZhangYWeiHYangLLiuRZhangBet al. Ultrasound radiomics nomogram for predicting large-number cervical lymph node metastasis in papillary thyroid carcinoma. Front Oncol. (2023) 13:1159114. doi: 10.3389/fonc.2023.1159114
44
ZhouS-CLiuT-TZhouJHuangY-XGuoYYuJ-Het al. An ultrasound radiomics nomogram for preoperative prediction of central neck lymph node metastasis in papillary thyroid carcinoma. Front Oncol. (2020) 10:1591. doi: 10.3389/fonc.2020.01591
45
ZhuHYuBLiYZhangYJinJAiYet al. Models of ultrasonic radiomics and clinical characters for lymph node metastasis assessment in thyroid cancer: a retrospective study. PeerJ. (2023) 11:e14546. doi: 10.7717/peerj.14546
46
KerJWangLRaoJLimT. Deep learning applications in medical image analysis. IEEE Access. (2017) 6:9375–89. doi: 10.1109/ACCESS.2017.2788044
47
KhanMZGajendranMKLeeYKhanMA. Deep neural architectures for medical image semantic segmentation. IEEE Access. (2021) 9:83002–24. doi: 10.1109/ACCESS.2021.3086530
48
YoussefAPencinaMThakurAZhuTCliftonDShahNH. All models are local: time to replace external validation with recurrent local validation. arXiv preprint, arXiv:2305.03219. (2023). doi: 10.48550/arXiv.2305.03219
49
ZhengBQiuYAghaeiFMirniaharikandeheiSHeidariMDanalaG. Developing global image feature analysis models to predict cancer risk and prognosis. Visual Computing Industry Biomed Art. (2019) 2:1–14. doi: 10.1186/s42492-019-0026-5
50
NayanA-AKijsirikulBIwahoriY. Mediastinal lymph node detection and segmentation using deep learning. IEEE Access. (2022) 10:89289–307. doi: 10.1109/ACCESS.2022.3198996
51
ZhouL-QWuX-LHuangS-YWuG-GYeH-RWeiQet al. Lymph node metastasis prediction from primary breast cancer US images using deep learning. Radiology. (2020) 294:19–28. doi: 10.1148/radiol.2019190372
52
JiangTChenCZhouYCaiSYanYSuiLet al. Deep learning-assisted diagnosis of benign and Malignant parotid tumors based on ultrasound: a retrospective study. BMC Cancer. (2024) 24:510. doi: 10.1186/s12885-024-12277-8
53
AminATRezkKMAttaH. Clinical examination and ultrasonography as predictors of lateral neck lymph nodes metastasis in primary well differentiated thyroid cancer. J Cancer Ther. (2018) 9:55. doi: 10.4236/jct.2018.91007
54
HajiEsmailPoorZKargarZTabnakP. Radiomics diagnostic performance in predicting lymph node metastasis of papillary thyroid carcinoma: a systematic review and meta-analysis. Eur J Radiol. (2023) 168:111129. doi: 10.1016/j.ejrad.2023.111129
55
MarimaRMtshaliNMathabeKBaseraAMkhabeleMBidaMet al. Application of AI in novel biomarkers detection that induces drug resistance, enhance treatment regimens, and advancing precision oncology. In: Artificial intelligence and precision oncology: bridging cancer research and clinical decision support. Cham: Springer (2023). p. 29–48.
56
ZhangSLiuRWangYZhangYLiMWangYet al. Ultrasound-base radiomics for discerning lymph node metastasis in thyroid cancer: A systematic review and meta-analysis. Acad Radiol. (2024) 31(8):3118–30. doi: 10.1016/j.acra.2024.03.012
57
MareyAArjmandPAlerabADSEslamiMJSaadAMSanchezNet al. Explainability, transparency and black box challenges of AI in radiology: Impact on patient care in cardiovascular radiology. Egyptian J Radiol Nucl Med. (2024) 55:183. doi: 10.1186/s43055-024-01356-2
58
ParaRK. The role of explainable AI in bias mitigation for hyper-personalization. J Artif Intell Gen Sci (JAIGS). (2024) 6:625–35. doi: 10.60087/jaigs.v6i1.289
Summary
Keywords
artificial intelligence, ultrasonography, cervical lymph node metastasis, papillary thyroid cancer, meta-analysis
Citation
Wang X, Qi Y, Zhang X, Liu F and Li J (2025) Ultrasound-based artificial intelligence for predicting cervical lymph node metastasis in papillary thyroid cancer: a systematic review and meta-analysis. Front. Endocrinol. 16:1570811. doi: 10.3389/fendo.2025.1570811
Received
04 February 2025
Accepted
19 May 2025
Published
10 June 2025
Volume
16 - 2025
Edited by
Erivelto Martinho Volpi, Hospital Alemão Oswaldo Cruz, Brazil
Reviewed by
Jiayu Ren, Seventh Medical Center of Chinese People’s Liberation Army General Hospital, China
Kathelina Kristollari, Ben-Gurion University of the Negev, Israel
Updates
Copyright
© 2025 Wang, Qi, Zhang, Liu and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Li, lj_070508@163.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.