- 1Molecular Testing Laboratory, Department of Medical Laboratory Sciences, Faculty of Health Sciences, Beirut Arab University, Beirut, Lebanon
- 2Laboratory of Biological Chemistry, Medical School, Aristotle University, Thessaloniki, Greece
- 3College of Medicine, Alfaisal University, Riyadh, Saudi Arabia
Background and objectives: The Coronavirus disease 2019 (COVID-19) pandemic underscored the importance of identifying host factors that influence susceptibility to infection. Vitamin D signaling, mediated via its receptor (VDR), along with innate immune mediators such as antimicrobial peptides (e.g., DEFA1-3) and inflammatory chemokines (e.g., CCL20), plays a critical role in antiviral defense. This study aimed to determine how serum vitamin D status and gene expression of VDR, DEFA1-3, and CCL20 associate with COVID-19 risk in a Lebanese cohort.
Methods: This prospective observational study assessed serum vitamin D concentrations and nasopharyngeal gene expression in Lebanese participants tested for SARS-CoV-2 between January and March 2024. We enrolled 264 patients undergoing RT-qPCR (targeting ORF1, N, and E genes) and quantified serum 25-hydroxyvitamin D [25(OH)D]. In a subset of 70 individuals stratified by COVID-19 status, we measured VDR, DEFA1-3, CCL20, and GAPDH expression by RT-qPCR. Multiple logistic regression and Pearson correlation analyses were performed.
Results: Serum vitamin D levels and CCL20 expression were not significantly associated with COVID-19 status. Elevated VDR expression in nasopharyngeal tissue correlated with lower COVID-19 risk (OR = 0.40, p = 0.05) and inversely with 25(OH)D levels (r = –0.61, p = 0.04). Higher DEFA1–3 expression reduced COVID-19 risk by 81.6% (OR = 0.184, p = 0.012). Among COVID-19 negatives, VDR correlated with CCL20 (r = 0.59, p < 0.01); among positives, VDR correlated with DEFA1-3 (r = 0.45, p < 0.05).
Conclusion: Our findings reveal a complex interplay between systemic vitamin D status, local VDR expression, and innate inflammatory mediators in COVID-19. They support a model in which both micronutrient levels and tissue-specific vitamin D signaling modulate host susceptibility and disease severity.
Introduction
The Coronavirus disease 2019 (COVID-19) pandemic has prompted extensive investigation into host factors that influence susceptibility and disease severity (1). Among these factors, vitamin D has garnered considerable attention due to its immunomodulatory properties (2–6). Early observational studies highlighted a potential protective role for vitamin D, as deficiency was frequently associated with increased disease severity, hospitalization rates, and mortality in COVID-19 patients (7, 8). Vitamin D can impact numerous pathways in the host immune response, promoting an appropriate inflammatory reaction while suppressing an excessive one (5). Vitamin D’s immunomodulatory role is significant in COVID-19, where severe cases involve excessive innate immune activation and lung immunothrombosis (9). Its effects are mediated through the vitamin D receptor (VDR), expressed on macrophages, dendritic cells, T-cells, and respiratory epithelial cells (4, 10, 11). Activation of VDR by active vitamin D modulates gene transcription, enhancing both innate and adaptive immune responses to strengthen antimicrobial defense (4, 9, 10).
While systemic vitamin D status is commonly assessed via circulating serum 25-hydroxyvitamin D [25(OH)D] concentrations, recent research has suggested that local tissue responsiveness—reflected by VDR expression—may be equally, if not more, relevant for immune protection (12). Indeed, local receptor expression levels potentially indicate the ability of tissues to mount effective vitamin D-dependent immune responses better than serum vitamin D concentrations alone. However, the relationship between local VDR expression and systemic vitamin D status remains inadequately explored in the context of viral infections such as COVID-19.
In Lebanon, vitamin D deficiency remains widespread despite plentiful sunshine (13, 14). This is exacerbated by cultural clothing that limits sun exposure and by low dietary intake of vitamin D–rich foods (13, 14). Lebanon’s COVID-19 clinical management protocol, aligned with the World Health Organization’s 2023 guidelines, did not recommend routine vitamin D supplementation as part of standard treatment during the study period.
Innate inflammatory mediators such as the alpha-defensins (DEFA1-3), produced by neutrophils and mucosal cells, exhibit antiviral activity by disrupting viral membranes and blocking entry (15, 16), and are linked to reduced respiratory infections (17, 18). They also maintain immune homeostasis and epithelial integrity during infections (16, 19). Chemokines like CCL20 recruit immune cells to infected tissues (20, 21). Elevated CCL20 levels are associated with severe COVID-19 outcomes, including acute respiratory distress syndrome (ARDS) and multisystem inflammatory syndrome in children (MIS-C), indicating a pathogenic role (22–24).
This prospective observational study aimed to assess how serum 25(OH)D concentrations and nasopharyngeal expression of VDR, DEFA1-3, and CCL20 are associated with COVID-19 status in Lebanese participants tested between January and March 2024.
Materials and methods
Study design and participants
The study was a prospective observational analysis conducted between January and March 2024, involving 264 adult participants who presented for measurement of serum 25(OH)D levels and/or severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) testing. Eligible participants were consecutively enrolled after meeting the inclusion and exclusion criteria at Lebanese Hospital Geitaoui, a tertiary center in Lebanon, during the Omicron BA.5 wave. Serum 25(OH)D levels were measured for all participants during the winter season (year 2024) to minimize the effect of seasonal variation on vitamin D concentrations. Participants were enrolled using a consecutive sampling strategy during the study period. A history of vitamin D supplementation within the past three months was recorded. In a subset of 70 patients, nasopharyngeal tissue was collected for gene expression analysis. Exclusion criteria included chronic autoimmune diseases, active malignancy, uncontrolled diabetes mellitus, chronic renal disease, and acute infections other than COVID-19.
Ethical considerations
This study was conducted in full accordance with ethical guidelines and was approved by the Institutional Review Board (IRB) of Lebanese Hospital Geitaoui-UMC under protocol code 2024-IRB-010. Written informed consent was obtained from the study participants. No personally identifiable information was included in the analyses or subsequent reporting, ensuring that all data were anonymized and handled with the utmost confidentiality.
Data collection and laboratory analyses
Serum vitamin D measurement
Venous blood samples were collected, and total serum 25(OH)D concentrations were quantified using the Roche Elecsys™ Vitamin D Total Assay. Participants were subsequently classified as vitamin D deficient (<20 ng/mL), insufficient (20–30 ng/mL), or sufficient (≥30 ng/mL) based on the Endocrine Society Clinical Practice Guidelines (25). The coefficient of variation for 25(OH)D measurement assays was between 3% to 8%.
RNA extraction and cDNA synthesis
Nasopharyngeal swab samples from a subset of 70 patients were processed for RNA extraction using the ANDiS Viral RNA Auto Extraction & Purification Kit in conjunction with the ANDiS 350 Automated Nucleic Acids Extraction System. In this automated protocol, viral particles were first lysed to release RNA, which was then captured on magnetic beads. Following washes to remove impurities, the RNA was eluted into a clean solution for further analysis. RNA concentration and purity were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific), ensuring A260/A280 ratios between 1.8 and 2.0. The extracted RNA was quantified, stored at –80°C, and subsequently reverse-transcribed into complementary DNA (cDNA) using a commercial reverse transcription kit, following the manufacturer’s instructions.
SARS−CoV−2 detection
SARS-CoV-2 detection was performed using the ANDiS FAST SARS-CoV-2 Detection Kit (3D Biomedicine Science & Technology Co., Limited), targeting the ORF1, N, and E genes. Samples were collected 1–3 days post-symptom onset, between January and March 2024, during which the Omicron BA.5 variant predominated in Lebanon. Although the detection assay targeted conserved SARS-CoV-2 genes (ORF1, N, E), variant-specific genotyping was not performed. Each reaction contained 15 ng of extracted RNA, Positive and negative controls were included to validate the assay. Amplification was conducted on the Bio-Rad CFX96 Real-Time PCR System using the following thermal cycling conditions: reverse transcription at 50°C for 10 minutes, initial denaturation at 95°C for 3 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 30 seconds. Cycle threshold (Ct) values were determined for each gene target, with a Ct ≤40 in at least two of the three targets (ORF1, N, or E) considered positive. All individuals with positive RT-PCR tests also presented with clinical symptoms (fever, cough, myalgia, anosmia), and were considered as having symptomatic COVID-19. As for the control group (non-COVID-19), they were also tested due to the presence of similar respiratory symptoms, but their test results were negative for SARS-CoV-2.
Quantitative real−time PCR for gene expression
RT−qPCR was performed to quantify the expression of VDR and the inflammatory genes DEFA1−3 and CCL20 in nasopharyngeal tissue samples. Gene−specific primers were designed and validated for efficiency and specificity. Each 20 µL reaction mixture contained SYBR Green Supermix, optimized concentrations of forward and reverse primers, and 70 ng of cDNA template, with GAPDH serving as the housekeeping gene for normalization. The thermal cycling protocol commenced with an initial enzyme activation and denaturation step at 95°C for 3 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds and a combined annealing/extension step at 60°C for 30 seconds. All reactions were executed in duplicate to ensure reproducibility and accuracy.
Statistical analysis
Analyses were performed using IBM SPSS Statistics. Continuous variables are expressed as the mean ± standard deviation, and categorical variables are presented as frequencies and percentages. Normality of continuous variables was assessed using the Kolmogorov-Smirnov test prior to applying parametric tests. Independent samples t-tests and Chi-square tests were used to compare continuous and categorical variables, respectively, between patients with and without COVID-19. The Ct values were used solely to define SARS-CoV-2 positivity as a binary variable. Quantitative Ct data were not included in the downstream correlation or regression analyses. The sample size was calculated using G*Power software based on a moderate effect size (Cohen’s d = 0.6), with α = 0.05 and power = 80%, resulting in a minimum of 45 participants per group.
For the entire cohort, a multiple logistic regression analysis was conducted, adjusting for age, sex, and BMI, to identify independent predictors of COVID-19 disease. The results are reported as odds ratios and 95% confidence intervals.
For the subset of 70 patients with gene expression data, separate logistic regression models, adjusted for age, sex, and BMI, were used to evaluate the association between the normalized expression of VDR, DEFA1-3, and CCL20 and COVID-19 status. For each gene, a median value was calculated and further used as a cut-off to classify the gene expression as high or low. Pearson correlation analysis was used to assess the relationship between serum 25(OH)D concentrations and VDR expression, as well as the association between inflammatory biomarker levels and SARS-CoV-2 viral gene expression in COVID-19-positive patients. A two-tailed p-value ≤ 0.05 was considered statistically significant.
Results
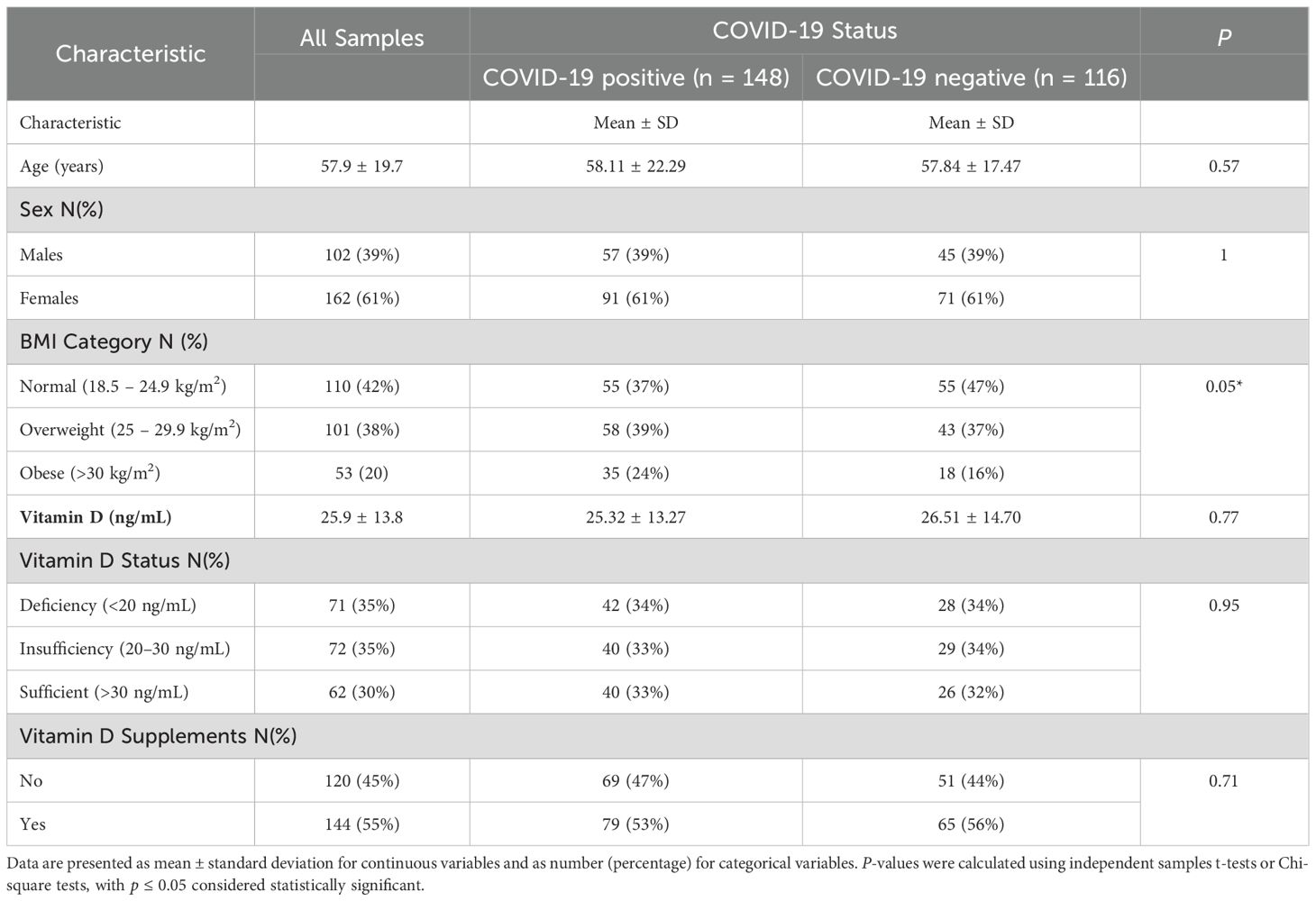
The study cohort consisted of 264 patients, comprising 148 individuals with COVID-19 and 116 individuals without COVID-19 (Table 1). Although the data presentation compares COVID-19-positive and -negative groups, the study was conducted prospectively with no prior matching or retrospective case selection. The mean age was similar between groups (58.11 ± 22.29 years in COVID-positive vs. 57.84 ± 17.47 years in COVID-negative; p = 0.57). Likewise, the sex distribution did not differ significantly between the two groups, with males representing 38.5% of COVID-positive patients and 38.8% of COVID-negative patients (p = 1.00).
Analysis based on BMI categories revealed a significantly lower proportion of COVID-positive patients with a normal BMI (37.2%) compared to those with a negative test result (47.4%; p = 0.05). However, the proportions of overweight and obese participants were comparable between groups. Serum 25(OH)D concentrations did not significantly differ between COVID-positive (25.32 ± 13.27 ng/mL) and COVID-negative patients (26.51 ± 14.70 ng/mL; p = 0.77). Similarly, vitamin D status categories (deficiency, insufficiency, sufficiency) and vitamin D supplement use showed no significant differences (p> 0.05).
Multivariate logistic regression analysis identified age ≥60 years as a significant predictor of increased COVID-19 disease risk (OR = 1.90, 95% CI: 1.02–3.55, p = 0.04). Conversely, sex, BMI categories, and vitamin D status did not independently predict COVID-19 disease (p > 0.05) (Table 2).
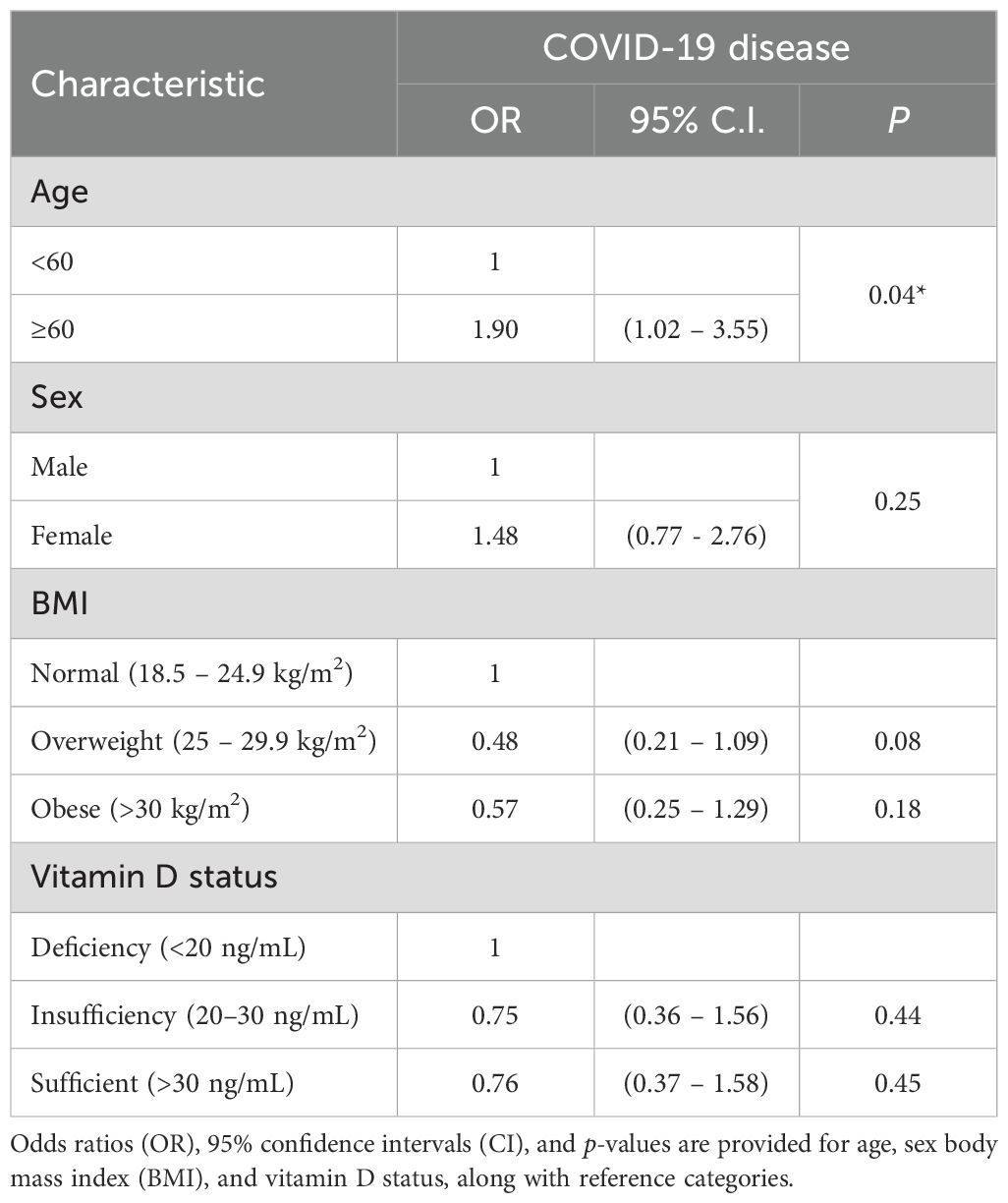
Table 2. Multivariate logistic regression analysis of predictors of COVID-19 disease: clinical parameters.
In the subset of 70 patients evaluated for gene expression (Table 3), higher VDR expression in nasopharyngeal samples was significantly associated with a reduced likelihood of COVID-19 disease (OR = 0.40, 95% CI: 0.15–1.06, p = 0.05). Likewise, elevated DEFA1–3 mRNA expression exhibited strong protective effects, significantly reducing COVID-19 disease risk by 81.6% (OR = 0.184, 95% CI: 0.035–0.97, p = 0.012). Conversely, CCL20 expression did not differ significantly between COVID-positive and COVID-negative patients (p = 0.294).
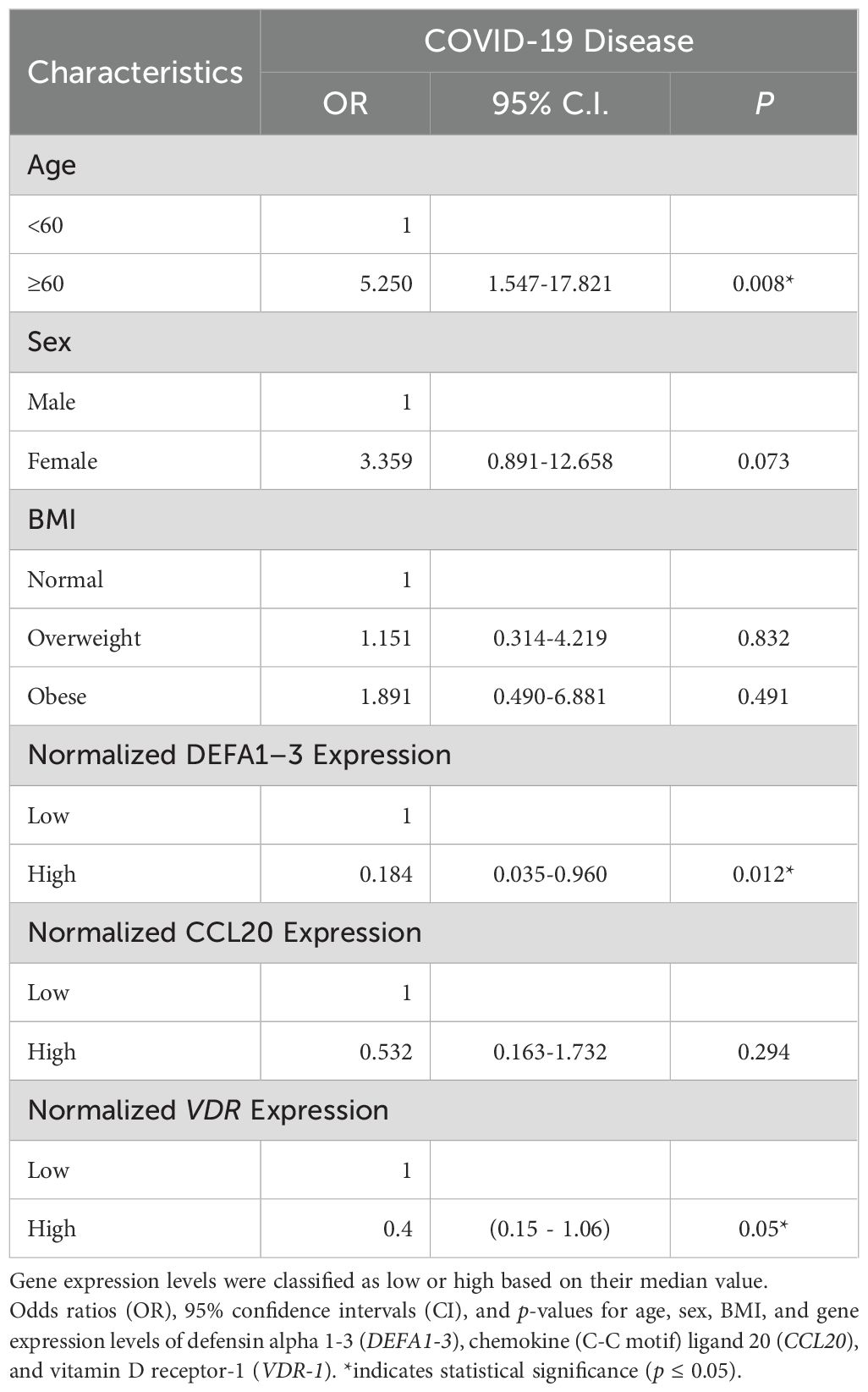
Table 3. Multivariate logistic regression analysis with COVID-19 disease predictors: gene expression data.
The gene expression comparison according to the COVID-19 disease status showed that the normalized VDR, DEFA1–3 and CCL20 expression is significantly higher in the negative group than in the positive group (Figure 1, P<0.05).
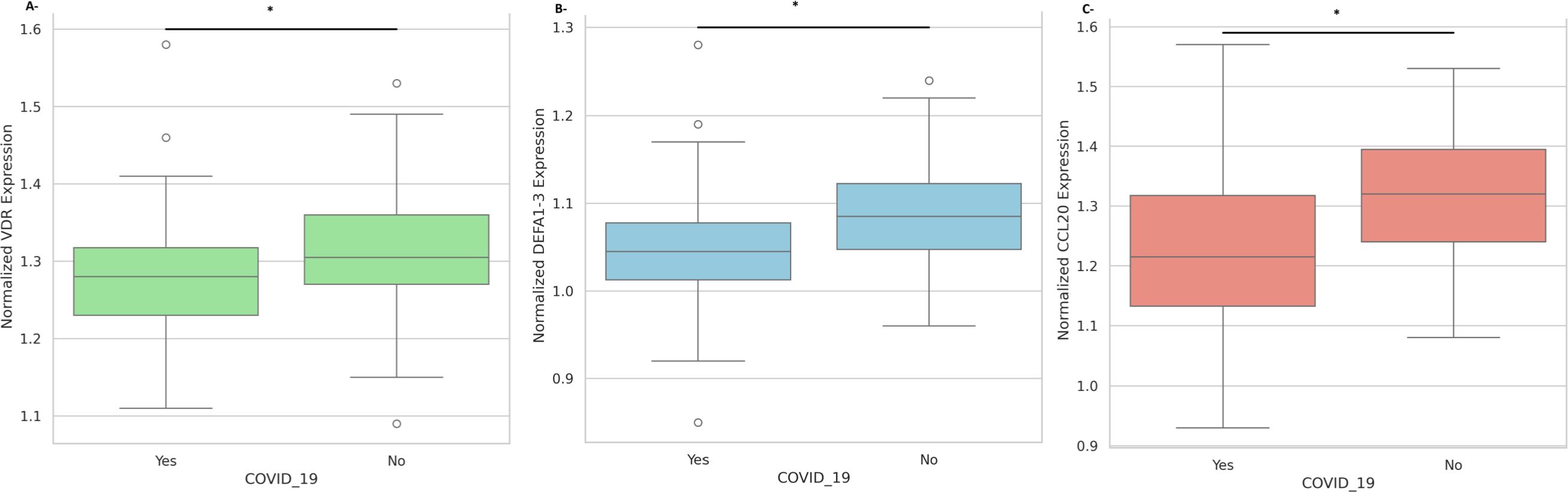
Figure 1. Nasopharyngeal expression of innate immune mediators according to COVID-19 disease status. (A) Normalized expression levels of VDR mRNA in COVID-19 negative versus COVID-19 positive participants. (B) Normalized expression levels of DEFA1–3 mRNA in COVID-19 negative versus COVID-19 positive participants. (C) Normalized expression levels of CCL20 mRNA in COVID-19 negative versus COVID-19 positive participants. All expression values are normalized to GAPDH. Data are presented as mean ± standard deviation. Statistical significance is indicated (*p < 0.05).
Pearson correlation analysis demonstrated a significant inverse relationship between serum 25(OH)D concentrations and VDR expression (r = -0.61, p = 0.04). Interestingly, In the COVID-19 negative group, VDR expression was positively correlated with CCL20 expression (r = 0.59, p < 0.01), while no significant correlation was observed between VDR and DEFA1-3 (r = –0.15) or between CCL20 and DEFA1-3 (r = –0.04). In the COVID-19 positive group, VDR expression showed a significant positive correlation with DEFA1-3 (r = 0.45, p < 0.05). The correlation between CCL20 and DEFA1–3 was weak and nonsignificant (r = 0.16) in the positive group (Table 4). When pooling samples, VDR expression remained significantly correlated with CCL20 (r = 0.45, p < 0.01).
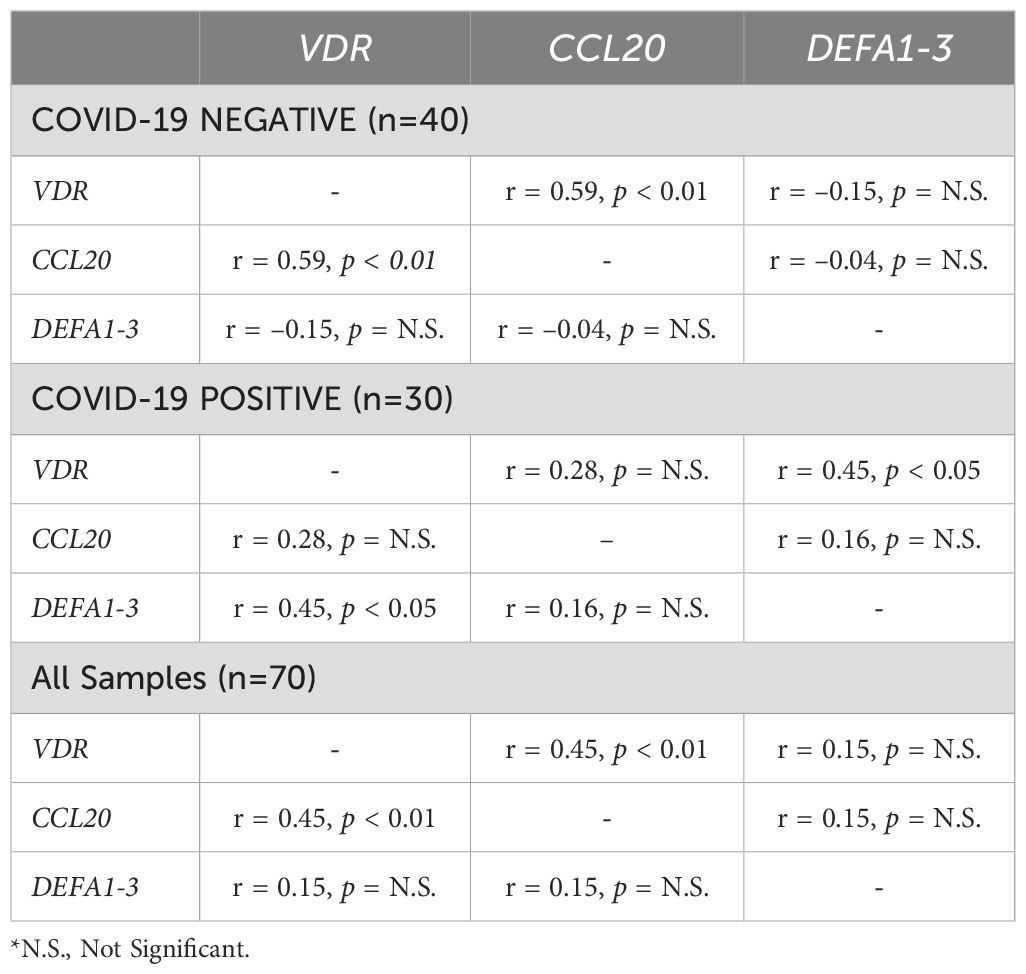
Table 4. Pearson correlation analysis of normalized VDR, CCL20, and DEFA1–3 expression stratified by COVID-19 disease status.
Correlation analysis among COVID-19-positive patients showed no significant associations between DEFA1–3 and CCL20 expression or with SARS-CoV-2 replication genes (ORF1, N, E) (all p >0.05). However, strong correlations were observed among the viral replication genes themselves (ORF1, N, and E; all r >0.99, p <0.001), validating their use as reliable markers of viral replication (Supplementary Table 1).
Discussion
Our integrated analysis, combining clinical data from a cohort with detailed gene expression profiling in a representative subset, provides critical insights into the roles of vitamin D and innate immune responses in COVID-19 susceptibility. Despite similar serum 25(OH)D concentrations between COVID-19-positive and negative patients, this finding suggests that circulating vitamin D concentrations alone may inadequately reflect the full immunomodulatory potential of vitamin D. Rather, local tissue responsiveness—as represented by VDR expression in nasopharyngeal tissues—emerges as a crucial determinant of protective immunity. Higher VDR expression correlated with a significant 60% reduction in COVID-19 disease risk, highlighting the importance of receptor-mediated signaling within local mucosal environments.
Additionally, analysis of inflammatory biomarkers indicated that elevated DEFA1–3 expression significantly reduces COVID-19 susceptibility by 81.6% (OR: 0.184, p = 0.012). DEFA1–3 peptides are known for their potent antiviral activities, including disruption of viral membranes, inhibition of viral entry, and modulation of local immune responses (17, 26). Their protective role was further supported by our finding of higher DEFA1–3 expression in COVID-19-negative individuals, consistent with reduced viral susceptibility. This aligns with observations by Idris et al. (27), who reported significant downregulation of DEFA1–3 during active SARS-CoV-2 infection, suggesting a possible viral evasion mechanism through suppression of host antimicrobial peptides. This suppression may partly explain the increased susceptibility to secondary infections observed in severe COVID-19 cases (27).
However, the role of DEFA1–3 extends beyond direct antiviral activity. Alpha-defensins, including DEFA1-3, have complex functions that involve immune modulation and inflammatory responses. Elevated alpha-defensin levels have been associated with thrombotic complications in COVID-19 through interactions with fibrinogen and interleukin-6, highlighting their dual roles in both protective immunity and pathology (28). DEFA1–3 peptides facilitate neutrophil recruitment and cytokine production, potentially driving beneficial inflammation for pathogen clearance; however, excessive or dysregulated activity could lead to tissue damage and exacerbated pathology (15, 28). This delicate balance underscores the importance of cautious interpretation and further investigation into the role of defensins during SARS-CoV-2 infection.
In our study, DEFA1–3 expression showed no significant correlations with SARS-CoV-2 viral replication genes (ORF1, N, and E), suggesting that defensin activity operates independently of viral replication dynamics and is likely influenced predominantly by host factors (29). Consequently, DEFA1–3 expression may serve as a valuable biomarker for assessing susceptibility to COVID-19. While elevated baseline defensin levels may reflect robust innate immunity, they could also signal immune dysregulation or exhaustion (28). Measuring defensin levels clinically enhances risk assessment and patient management strategies.
In contrast to DEFA1-3, CCL20 expression did not differ significantly between infected and non-infected individuals, suggesting variability in the contribution of inflammatory mediators to COVID-19 susceptibility (22–24). However, our results suggest that the role of CCL20 in disease susceptibility may be context-dependent, warranting further investigation.
The correlation patterns suggest that vitamin D signaling through VDR is functionally linked to immune cell recruitment (via CCL20) under non-infectious conditions. However, this relationship appears to be disrupted during COVID-19 disease, where vitamin D signaling may shift toward enhancing antimicrobial peptide production (DEFA1-3) as part of the host defense mechanism. The loss of VDR-CCL20 correlation during infection could reflect immune system dysregulation or a shift in immune response priorities under infectious stress.
Our study has several limitations. Although our findings highlight reduced 25(OH)D levels in COVID-19 cases compared to controls, clinical severity data were not collected, precluding stratified analysis. Previous reports have shown that vitamin D status may prospectively predict COVID-19 severity and outcomes (30, 31). The study design limits our ability to establish causality and to assess longitudinal changes in vitamin D status, VDR expression, and inflammatory biomarkers. Additionally, although the overall cohort was sizable (n = 264), the subset for gene expression analysis was relatively limited, which may have limited its representativeness and generalizability. Potential confounders, including seasonal variations in vitamin D levels, nutritional status, and other environmental factors, were not fully controlled. Another limitation is the lack of data on participants’ COVID-19 vaccination status, as it was not available at the time of data collection.
Given the high prevalence of vitamin D deficiency in Lebanon and its apparent link to COVID-19 susceptibility, future interventional trials should evaluate the efficacy and optimal dosing of vitamin D supplementation in reducing infection risk and disease severity. We recommend systematic screening for 25(OH)D levels in at-risk populations (e.g., individuals with limited sun exposure) followed by randomized controlled studies to determine whether correcting deficiency can improve clinical outcomes in SARS-CoV-2 infection.
Conclusion
In summary, our findings demonstrate that the protective effects of vitamin D in COVID-19 are more closely associated with local VDR expression and innate antimicrobial pathways, particularly DEFA1-3, than with systemic 25(OH)D concentrations alone (Figure 2). These results support a model in which both micronutrient status and tissue-specific vitamin D signaling modulate host susceptibility and disease severity (Figure 2). Future longitudinal and interventional studies are warranted to validate these associations and explore the therapeutic potential of enhancing local vitamin D responsiveness in respiratory viral infections.
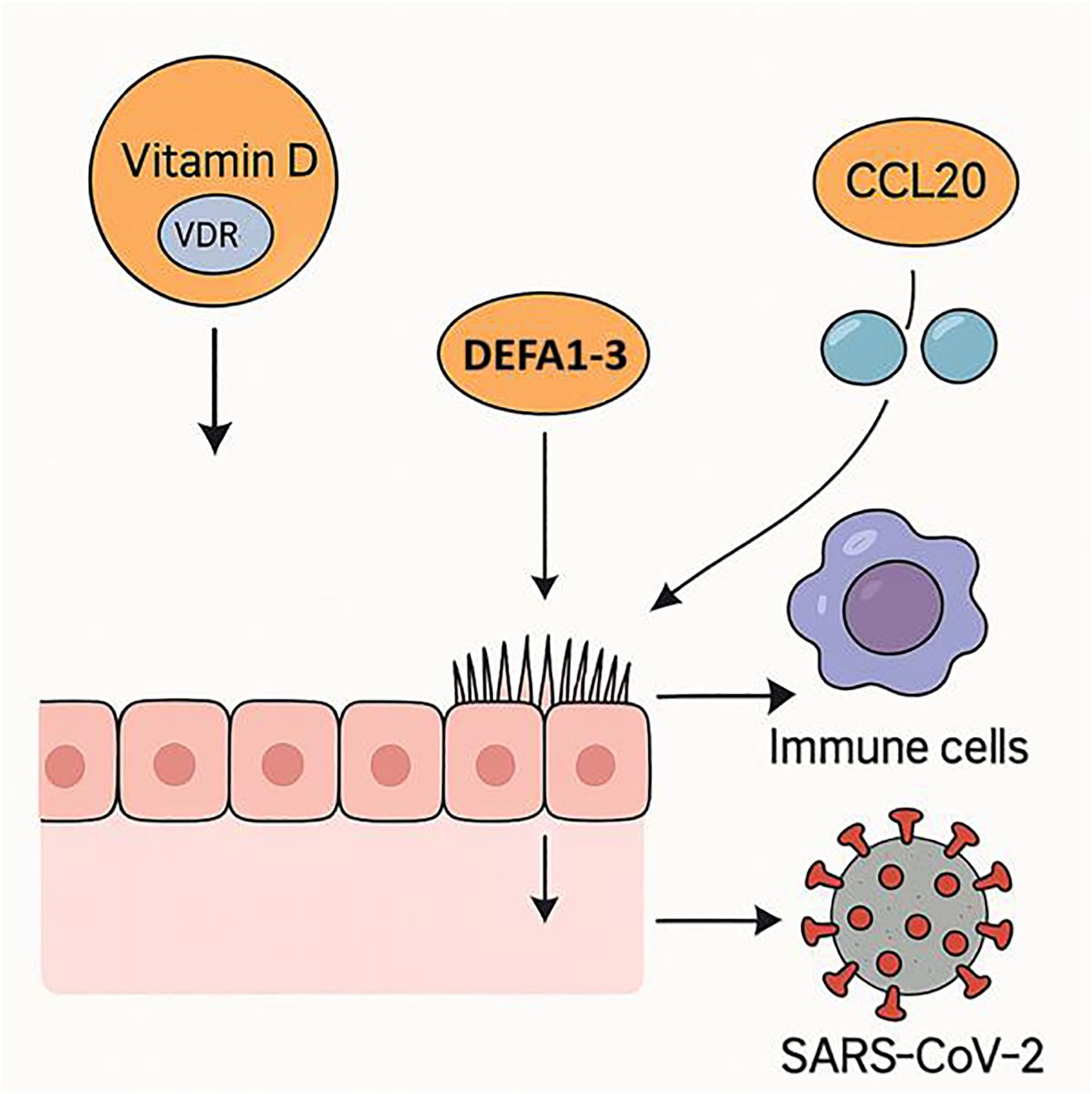
Figure 2. Proposed mechanism of vitamin D in reducing SARS-CoV-2 susceptibility. Vitamin D signaling via VDR in respiratory epithelium enhances antimicrobial defenses through DEFA1–3 upregulation and facilitates immune cell recruitment by modulating CCL20 expression, collectively lowering the risk of SARS-CoV-2 infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
This study was reviewed and approved by the Institutional Review Board (IRB) at Lebanese Hospital Geitaoui-UMC (Protocol code: 2024-IRB-010). All participants provided written informed consent prior to their involvement in the study. The research was conducted following the principles outlined in the Declaration of Helsinki, and all participant data were anonymized and handled confidentially.
Author contributions
FM: Formal Analysis, Investigation, Software, Writing – original draft. DM: Formal Analysis, Investigation, Software, Writing – original draft. ES-S: Conceptualization, Data curation, Project administration, Writing – review & editing. SK: Writing – review & editing. HF: Methodology, Project administration, Resources, Validation, Writing – original draft, Writing – review & editing. SE: Conceptualization, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The Article Processing Charges (APC) were funded by Alfaisal University, Office of Research.
Acknowledgments
The authors wish to thank all the staff at Beirut Arab University and the participating hospital for their support in sample collection and data processing. We also extend our gratitude to the patients who consented to participate in this study. We also thank Dr. Sarah Daher for her valuable assistance in organizing and finalizing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2025.1600623/full#supplementary-material
Abbreviations
25(OH)D, 25-Hydroxyvitamin D; ARDS, Acute Respiratory Distress Syndrome; BMI, Body Mass Index; CCL20, C-C Motif Chemokine Ligand 20; CI, Confidence Interval; COVID-19, Coronavirus Disease 2019; Ct, Cycle Threshold; DEFA1-3, Defensin Alpha 1–3; GAPDH, Glyceraldehyde 3-Phosphate Dehydrogenase; IRB, Institutional Review Board; OR, Odds Ratio; RT-qPCR, Reverse Transcription Quantitative Polymerase Chain Reaction; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; SD, Standard Deviation; VDR, Vitamin D Receptor 1.
References
1. Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, et al. Extrapulmonary manifestations of COVID-19. Nat Med. (2020) 26:1017–32. doi: 10.1038/s41591-020-0968-3
2. Grant WB, Lahore H, McDonnell SL, Baggerly CA, French CB, Aliano JL, et al. Evidence that vitamin D supplementation could reduce risk of influenza and COVID-19 infections and deaths. Nutrients. (2020) 12:988. doi: 10.3390/nu12040988
3. Greiller CL and Martineau AR. Modulation of the immune response to respiratory viruses by vitamin D. Nutrients. (2015) 7:4240–70. doi: 10.3390/nu7064240
4. Aranow C. Vitamin D and the immune system. J Invest Med. (2011) 59:881–6. doi: 10.2310/JIM.0b013e31821b8755
5. Fakhoury HM, Kvietys PR, Shakir I, Shams H, Grant WB, and Alkattan K. Lung-centric inflammation of COVID-19: potential modulation by vitamin D. Nutrients. (2021) 13:2216. doi: 10.3390/nu13072216
6. Hamza FN, Daher S, Fakhoury HM, Grant WB, Kvietys PR, and Al-Kattan K. Immunomodulatory properties of vitamin D in the intestinal and respiratory systems. Nutrients. (2023) 15:1696. doi: 10.3390/nu15071696
7. Wang Z, Joshi A, Leopold K, Jackson S, Christensen S, Nayfeh T, et al. Association of vitamin D deficiency with COVID-19 infection severity: Systematic review and meta-analysis. Clin Endocrinol. (2022) 96:281–7. doi: 10.1111/cen.14540
8. Kaya MO, Pamukçu E, and Yakar B. The role of vitamin D deficiency on COVID-19: a systematic review and meta-analysis of observational studies. Epidemiol Health. (2021) 43:e2021074. doi: 10.4178/epih.e2021074
9. Kvietys PR, Fakhoury HM, Kadan S, Yaqinuddin A, Al-Mutairy E, and Al-Kattan K. COVID-19: lung-centric immunothrombosis. Front Cell Infect Microbiol. (2021) 11:679878. doi: 10.3389/fcimb.2021.679878
10. Haussler MR, Jurutka PW, Mizwicki M, and Norman AW. Vitamin D receptor (VDR)-mediated actions of 1α, 25 (OH) 2vitamin D3: Genomic and non-genomic mechanisms. Best Pract Res Clin Endocrinol Metab. (2011) 25:543–59. doi: 10.1016/j.beem.2011.05.010
11. Wang T-T, Nestel FP, Bourdeau V, Nagai Y, Wang Q, Liao J, et al. Cutting edge: 1, 25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J Immunol. (2004) 173:2909–12. doi: 10.4049/jimmunol.173.5.2909
13. Harkous D, Ghorayeb N, and Gannagé-Yared MH. Prevalence and predictors of vitamin D deficiency in Lebanon: 2016-2022, before and during the COVID-19 outbreak. Endocrine. (2023) 82:654–63. doi: 10.1007/s12020-023-03483-8
14. Arabi A, Chamoun N, Nasrallah MP, and Tamim HM. Vitamin D deficiency in lebanese adults: prevalence and predictors from a cross-sectional community-based study. Int J Endocrinol. (2021) 2021:3170129. doi: 10.1155/2021/3170129
15. Wilson SS, Wiens ME, and Smith JG. Antiviral mechanisms of human defensins. J Mol Biol. (2013) 425:4965–80. doi: 10.1016/j.jmb.2013.09.038
16. Xu C, Wang A, Marin M, Honnen W, Ramasamy S, Porter E, et al. Human defensins inhibit SARS-CoV-2 infection by blocking viral entry. Viruses. (2021) 13:1246. doi: 10.3390/v13071246
17. López-Bermejo A, Chico-Julia B, Castro A, Recasens M, Esteve E, Biarnés J, et al. Alpha defensins 1, 2, and 3: potential roles in dyslipidemia and vascular dysfunction in humans. Arterioscler Thromb Vasc Biol. (2007) 27:1166–71. doi: 10.1161/ATVBAHA.106.138594
18. Shrivastava S, Chelluboina S, Jedge P, Doke P, Palkar S, Mishra AC, et al. Elevated levels of neutrophil activated proteins, alpha-defensins (DEFA1), calprotectin (S100A8/A9) and myeloperoxidase (MPO) are associated with disease severity in COVID-19 patients. Front Cell Infect Microbiol. (2021) 11:751232. doi: 10.3389/fcimb.2021.751232
19. Christensen HM, Frystyk J, Faber J, Schou M, Flyvbjerg A, Hildebrandt P, et al. α-Defensins and outcome in patients with chronic heart failure. Eur J Heart Fail. (2012) 14:387–94. doi: 10.1093/eurjhf/hfs021
20. Lee AY, Eri R, Lyons AB, Grimm MC, and Korner H. CC chemokine ligand 20 and its cognate receptor CCR6 in mucosal T cell immunology and inflammatory bowel disease: odd couple or axis of evil? Front Immunol. (2013) 4:194. doi: 10.3389/fimmu.2013.00194
21. Skovdahl HK, Damås JK, Granlund A, Østvik AE, Doseth B, Bruland T, et al. CC motif ligand 20 (CCL20) and CC motif chemokine receptor 6 (CCR6) in human peripheral blood mononuclear cells: dysregulated in ulcerative colitis and a potential role for CCL20 in IL-1β release. Int J Mol Sci. (2018) 19:3257. doi: 10.3390/ijms19103257
22. Chi Y, Ge Y, Wu B, Zhang W, Wu T, Wen T, et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis. (2020) 222:746–54. doi: 10.1093/infdis/jiaa363
23. Gruber CN, Patel RS, Trachtman R, Lepow L, Amanat F, Krammer F, et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C). Cell. (2020) 183:982–995.e14. doi: 10.1016/j.cell.2020.09.034
24. Hue S, Beldi-Ferchiou A, Bendib I, Surenaud M, Fourati S, Frapard T, et al. Uncontrolled innate and impaired adaptive immune responses in patients with COVID-19 acute respiratory distress syndrome. Am J Respir Crit Care Med. (2020) 202:1509–19. doi: 10.1164/rccm.202005-1885OC
25. Holick MF, Binkley NC, Bischoff-Ferrari HA, Gordon CM, Hanley DA, Heaney RP, et al. Evaluation, treatment, and prevention of vitamin D deficiency: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. (2011) 96:1911–30. doi: 10.1210/jc.2011-0385
26. Kudryashova E, Zani A, Vilmen G, Sharma A, Lu W, Yount JS, et al. Inhibition of SARS-CoV-2 infection by human defensin HNP1 and retrocyclin RC-101. J Mol Biol. (2022) 434:167225. doi: 10.1016/j.jmb.2021.167225
27. Idris MM, Banu S, Siva AB, and Nagaraj R. Down regulation of defensin genes during SARS-CoV-2 infection. Acta Virol. (2022) 66:249–53. doi: 10.4149/av_2022_306
28. Xu D and Lu W. Defensins: a double-edged sword in host immunity. Front Immunol. (2020) 11:3389/fimmu.2020.00764. doi: 10.3389/fimmu.2020.00764
29. Fu J, Zong X, Jin M, Min J, Wang F, and Wang Y. Mechanisms and regulation of defensins in host defense. Signal Transduct Target Ther. (2023) 8:300. doi: 10.1038/s41392-023-01553-x
30. Asla MM, Nawar AA, Elsayed E, Farahat RA, Abdulgadir A, Alsharabasy MA, et al. Vitamin D on COVID-19 patients during the pandemic, 2022. A systematic review and meta-analysis. Curr Res Nutr Food Sci. (2023) 11. doi: 10.12944/CRNFSJ.11.1.3
Keywords: COVID-19, vitamin D, VDR, innate immunity, inflammatory biomarkers
Citation: Missilmani F, Maarabouni D, Salem-Sokhn E, Karras SN, Fakhoury HMA and El Shamieh S (2025) Evaluation of vitamin D status, vitamin D receptor expression, and innate immune mediators in COVID-19. Front. Endocrinol. 16:1600623. doi: 10.3389/fendo.2025.1600623
Received: 26 March 2025; Accepted: 29 July 2025;
Published: 19 August 2025.
Edited by:
Mourad Aribi, University of Abou Bekr Belkaïd, AlgeriaReviewed by:
Luigi Di Filippo, San Raffaele Hospital (IRCCS), ItalySamar Ahmed Amer, Zagazig University, Egypt
Copyright © 2025 Missilmani, Maarabouni, Salem-Sokhn, Karras, Fakhoury and El Shamieh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hana M. A. Fakhoury, aGFuYS5mYWtob3VyeUBnbWFpbC5jb20=; Said El Shamieh, cy5lbHNoYW1pZWhAYmF1LmVkdS5sYg==
†These authors have contributed equally to this work
‡ORCID: Ferdos Missilmani, orcid.org/0009-0002-7620-6016
Dima Maarabouni, orcid.org/0009-0000-6157-6247
Elie Salem-Sokhn, orcid.org/0000-0003-1519-6790
Spyridon N. Karras, orcid.org/0000-0002-4225-2746
Hana M.A. Fakhoury, orcid.org/0000-0001-9974-2108
Said El Shamieh, orcid.org/0000-0002-8522-0445
 Ferdos Missilmani
Ferdos Missilmani Dima Maarabouni
Dima Maarabouni Elie Salem-Sokhn
Elie Salem-Sokhn Spyridon N. Karras
Spyridon N. Karras Hana M. A. Fakhoury
Hana M. A. Fakhoury Said El Shamieh
Said El Shamieh