- Department of Microbiology and Immunology, University of Illinois College of Medicine, Chicago, IL, USA
Introduction
B cell lymphomas represent 95% of all lymphomas diagnosed in the Western world and the majority of these arise from germinal center (GC) B cells (1). Recurrent chromosomal translocations involving Ig loci and proto-oncogenes are a hallmark of many types of B cell lymphoma (2). Three types of breakpoints can be identified in Ig loci. Translocation breakpoints adjacent to the DH or JH gene segments form secondary to V(D)J recombination, a process that occurs in early B cell development. Other translocations are located in rearranged V(D)J exons that have acquired mutations indicating that translocation is a byproduct of somatic hypermutation (SHM) which occurs in GC B cells. A third type of translocation is characterized by breakpoints in the Igh switch regions, a target for double strand DNA breaks (DSBs) during class switch recombination (CSR) that occurs in mature B cells, both inside and outside the GC. Thus, in B lymphocytes, V(D)J joining, CSR, and SHM create obligate single- or double-strand DNA breaks as intermediates for chromosomal translocations (3, 4).
Activation-induced deaminase (AID) is the enzyme that initiates CSR and SHM (5) by inducing the formation of DSBs in switch (S) regions and mutations in V gene exons (6–10). Studies indicate that non-Ig genes are mistargeted by AID (11, 12) and thereby acquire single and double strand DNA breaks at sites coincident with translocation breakpoints (1, 2). Mature B cells are particularly prone to chromosomal translocations that juxtapose Ig genes and proto-oncogenes, including c-myc [Burkitt’s lymphoma (BL)], Bcl-2 (follicular lymphoma), Bcl-6 (diffuse large cell lymphoma), and FGFR (multiple myeloma) and which are characteristic of human B cell malignancies (2). The mouse plasmacytoma (PCT) T(12;15)(Igh-myc) translocation, a direct counterpart of the human BL t(8;14)(q24;q32) translocation, occurs as a dynamic process in mature B cells undergoing CSR and is dependent on the expression of AID (13, 14). Hence, a direct mechanistic link between AID and chromosomal translocations focused to Ig genes has been established.
One of the most puzzling aspects of recurrent chromosomal translocations is that DSBs on two different chromosomes must come into close proximity frequently enough to facilitate the crossover. How do the broken ends located at distal sites in cis or on trans chromosomes come together? Consideration of oncogenic selection, sources of translocation prone DSBs associated with antigen receptor rearrangements in B and T lymphocytes, and the role of DSB persistence in translocations have been recently reviewed [(15, 16) and references therein]. Here we consider the proposition that the spatial organization of mammalian genomes is intrinsically linked to genome stability and modulates the frequency of chromosomal translocations.
A Model for Recurrent Chromosomal Translocations
Two general models have been proposed to explain the non-random nature of higher order spatial genome organization and the correlation with chromosomal translocations (17). The “contact-first” model posits that translocations require pre-existing physical proximity, whereas, the “breakage-first” model postulates that distant DSBs can be juxtaposed, perhaps through DNA repair machinery. These two theories, the dynamic “breakage-first” and the static “contact-first,” differ fundamentally in their requirement for the presence of DSBs and the mobility of the broken ends.
In the contact-first model only limited local positional motion of DSBs is expected. In the breakage-first model, single DSBs are formed and must undergo large scale movement within nuclei to search for appropriate interaction partners. Although evidence for mobility has been found in yeast systems (18–20), the situation in mammalians cells appears different. In mammalian cells, damaged DNA is largely stationary over time (21–23). However, deprotected telomeres as well as joining of broken DNA ends during V(D)J recombination experience higher mobility (24, 25). Accordingly, the VH subdomain of the Igh locus has been described as spatially unstructured (26) although additional studies are required to confirm this conclusion. Nevertheless, the weight of evidence in mammalian systems favors the “contact-first” model in light of the limited spatial mobility of DSBs (27). Comparison of a genomic organization map with sites of chromosomal translocation revealed that the spatial proximity of two DSBs is a dominant factor in determining the translocation landscape genome-wide (28). Therefore, it is useful to examine the disposition of loci within chromatin architecture and how this influences the probability of two DSBs finding each other in nuclear space.
Three Dimensional Organization of the Mammalian Genome
Emerging evidence indicates that a fundamental property of the mammalian nucleus is the non-random organization of the genome in nuclear space (29). Cytogenetic studies reveal that the mammalian nucleus is occupied by non-randomly positioned genes and chromosomes (30). Together these studies have shown that gene activation or silencing is often associated with repositioning of that locus relative to nuclear compartments and other genomic loci. In this regard, it is relevant that in normal B cells, the breakage sites of several common translocations are more frequently found in close spatial proximity in the nucleus than would be expected based on random positioning (31). A similar relationship between translocation frequency and spatial proximity is observed in BL where the myc locus is on average closest to its most frequent translocation partner, Igh (32). The non-random aspect of genome spatial organization in a sub-compartmentalized nuclear space has emerged as a potential contributor to the genesis of chromosomal translocations (23).
The combination of new imaging tools and the comprehensive mapping of long range chromosomal interaction has revealed structural features and biological properties of the three dimensional (3D) genomic organization (33–38). Four features contributing to an ordered 3D organization of eukaryotic genomes have become evident. (1) Individual chromosomes occupy distinct chromosomal territories (CT) with only a limited degree of intermingling (39). (2) The eukaryotic genome is partitioned into functionally distinct euchromatin and heterochromatin (40). (3) Individual genomic loci and elements display preferences for nuclear positioning which correlates well with genomic functions including transcriptional activity and replication timing (39, 41). (4) Distant chromosomal elements associate to form chromatin loops thereby providing a mechanism for long range enhancer function (36, 38, 42). These variables predict that unique and unanticipated spatial genomic relationships may determine unique combinations of chromosomal translocations that may differ in specific tissues and during differentiation.
Chromosomal Looping Interactions Facilitate CSR
The best studied property of chromatin looping is the spatial proximity of genes and their regulatory elements to establish functional states. Of relevance here is the recognition that chromatin looping influences partner selection during V(D)J recombination (43–45), CSR (46, 47), and may drive specific chromosomal translocation events (28, 48, 49). It is of importance to understand the spatial relationships within the Igh locus and how they relate to the preferential expression of Ig gene expression and protect against genome instability. We focus here on CSR because the most prevalent B cell lymphomas arise from GC B cells and are dependent on the expression of AID (1, 13, 14).
Class switch recombination promotes diversification of CH effector function while retaining the original rearranged V(D)J exons. The mouse Igh locus spans 2.9 Mb within which a centromeric 220 kb genomic region contains eight CH genes (encoding μ, δ, γ3, γ1, γ2b, γ2a, ε, and α chains) each paired with repetitive S DNA (with the exception of Cδ) (Figure 1A). CSR is focused on S regions and involves an intra-chromosomal deletional rearrangement (Figure 1B). Germline transcript (GLT) promoters, located upstream of I exon-S-CH regions, focus CSR to specific S regions by differential transcription activation (9, 50). The I-S-CH region genes are embedded between the Eμ intronic and 3′Eα enhancers (51). Chromosome conformation capture (3C) studies reveal that in mature resting B cells the transcriptional enhancer elements, Eμ and 3′Eα, engage in long range chromatin looping interactions (46, 47) (Figure 1C). B cell activation leads to induced recruitment of the GLT promoters to the Eμ:3′Eα complex that in turn facilitates GLT expression and supports S/S synapsis (46).
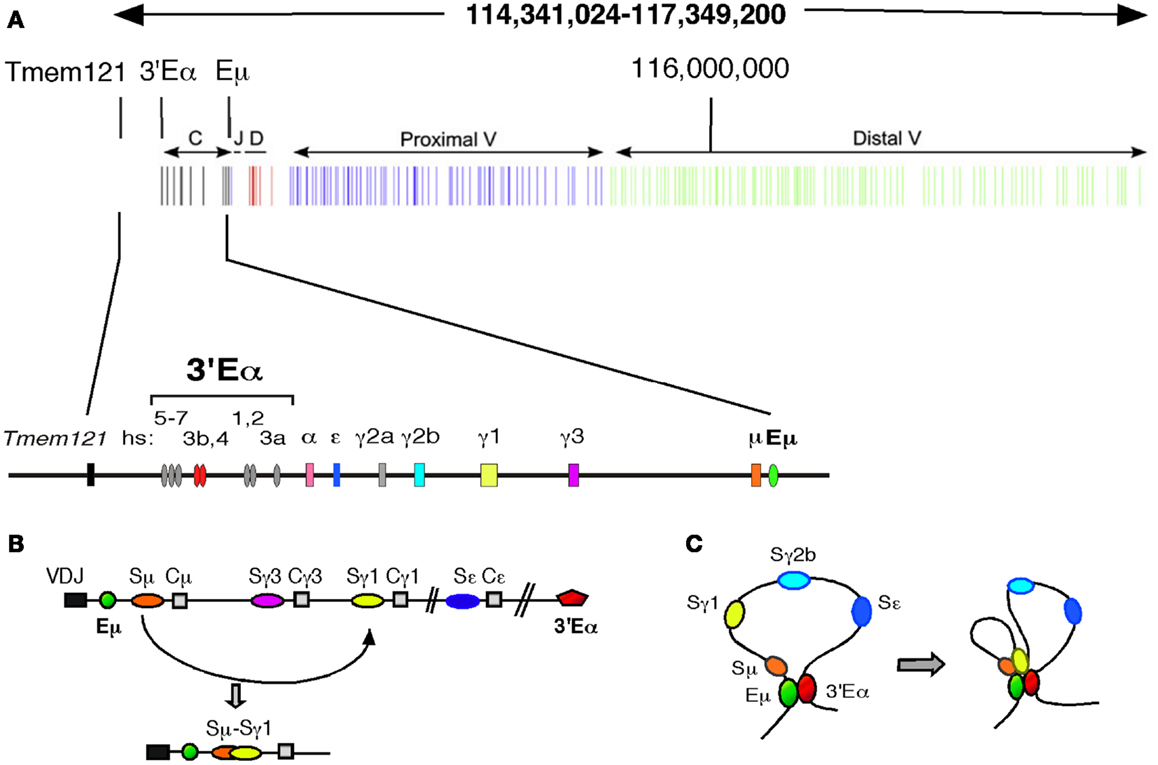
Figure 1. Long range chromatin looping interactions in the Igh locus facilitate CSR in mature B cells. (A) A schematic map, drawn to scale of the 2.9 Mb Igh locus located on chromosome 12 (chr12: 114,341,024–117,349, 200 mm9). The CH, JH, DH, and proximal and distal VH gene segments are indicated. The Igh enhancers, 3′Eα and intronic Eμ bracket the CH region gene cluster (top). A schematic showing an expanded segment of the Igh locus spanning 220 kb and containing the CH region genes (bottom). The orientation of this map follows the chromosomal organization of the Igh locus. (B,C) Diagrams of the Igh CH locus describing CSR are by convention shown with the Eμ enhancer at the 5′ end. (B) CSR promotes diversification of CH effector function while retaining the original V(D)J rearrangement. Within the mouse Igh locus, a 220 kb genomic region contains eight CH genes (encoding μ, δ, γ3, γ1, γ2b, γ2a, ε, and α chains) each paired with repetitive switch (S) DNA (with the exception of Cδ). CSR is focused on S regions and involves an intra-chromosomal deletional rearrangement. Germline transcript (GLT) promoters, located upstream of I exon-S-CH regions, focus CSR to specific S regions by differential transcription activation (50, 67). Prior to CSR and upon GLT expression, S regions become accessible to AID attack. AID initiates a series of events culminating in formation of S region specific double strand breaks (DSBs) at the donor Sμ and a downstream acceptor S region (50). DNA DSBs in transcribed S regions are essential for CSR. Here, Sμ and Sγ1 acquire AID induced DSBs and engage in CSR to form recombinant Sμ/Sγ1 regions. (C) In mature B cells Eμ:3′Eα interactions create a long range chromatin loop encompassing the CH domain of the Igh locus (left). Upon B cell activation with LPS + IL4, long range chromatin interactions directed by the GLT promoters and Igh enhancers creates spatial proximity between Sμ and the downstream Sγ1 region locus (46). This spatial proximity facilitates recombination between the broken S regions and creates a matrix of chromatin contacts, which stabilize the locus during the recombination transaction.
The 3′Eα regulatory region plays a significant role in mediating the spatial structure of the Igh locus during CSR as well as promoting genome stability (52). Targeted deletion of hs3b,4 within 3′Eα abolishes GLT expression and GLT promoter:3′Eα and Eμ:3′Eα looping interactions (46, 53, 54). AID initiates a series of events ending in creation of S region specific DNA DSBs at the donor Sμ and a downstream acceptor S region to create S/S junctions and facilitate CSR (7). S regions targeted by AID for DSB formation are transcriptionally active. Chromatin looping across this region ensures proximity between two S regions targeted for DSB creation and recombination (Figure 1C). Thus, CSR is dependent on 3D chromatin architecture mediated by long range intra-chromosomal interactions between distantly located transcriptional elements that serves to tether broken chromosomal DNA together during the CSR reaction.
Chromosome conformation capture (3C, 4C, 5C, and Hi-C) based studies indicate that the most probable chromatin interactions are the most proximal ones and the probability of contact decreases with distance. Correspondingly, alignment of genomic organization maps with sites of chromosomal translocation generated in Hi-C and 4C studies have shown that translocations are enriched in cis along single chromosomes containing the target DSB and in trans in a manner related to pre-existing spatial proximity (28, 55). The positional immobilization of DSBs in the Igh locus, for example, should render the probability of successful translocation as the product of the frequency of each DSB at the sites of crossover and the frequency with which these sites are synapsed in physical space (28). In B lymphocytes c-myc/Igh translocations occur in trans and may represent a failure of stringent spatial sequestration of AID induced DSBs to within the Igh locus (56, 57).
Dynamic Chromatin Interactions and the Genesis of Chromosomal Translocations
Chromosomal translocation frequency as reported by genome-wide translocation sequencing is determined by the frequency of AID induced DSB at translocation targets, factors that contribute to synapsis of broken loci, and circumvention of DNA repair functions that facilitate intra-chromosomal DSB joining (55–58). Are recurrent chromosomal translocations simply the result of a stochastic process related to the probability of contact between AID induced DSBs? Tagging single loci with Lac operon (LacO) arrays, as well as photobleaching and photoactivation experiments, have shown that interphase chromatin is locally mobile but rarely moves over long distances (59–61). However, lamina associated domains are large genomic regions that are in intermittent molecular contact with the nuclear lamina indicating a dynamic spatial architecture of chromosomes (62). Chromatin looping, clustering, and compartmentalization are dynamic and responsive to developmental and environmental cues. Functionally dynamic chromatin responses include formation of transcription and replication factories, and nuclear relocation of loci during development (63–66). The looping interactions spanning the Igh locus during CSR and in the presence of DSBs may also be dynamic and to some degree transient. In a dynamic chromosomal setting, DSBs present in an Igh locus that lacks Eμ:3′Eα tethering, for example, would be at high risk of re-joining to sites outside the Igh locus along chromosome 12 and at lower frequency to sites on other chromosomes. The dynamism of chromosomal transactions are not yet fully described and represent the next forefront for investigation to appreciate constraints and variables of genome stability and instability.
Author Contributions
Drs. Robert Wuerffel, Satyendra Kumar, Fernando Grigera, and Amy L. Kenter were all involved in developing the ideas regarding long range chromatin interactions and dynamics that are the subject here and all have critiqued and agree to the contents of this piece. Amy L. Kenter wrote the article.
Acknowledgments
This work was supported by the National Institutes of Health RO1AI052400 and R21AI106328 to Amy L. Kenter.
References
1. Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer (2005) 5:251–62. doi: 10.1038/nrc1589
2. Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene (2001) 20:5580–94. doi:10.1038/sj.onc.1204640
3. Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell (2010) 141:27–38. doi:10.1016/j.cell.2010.03.016
4. Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell (2008) 135:1130–42. doi:10.1016/j.cell.2008.10.035
5. Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell (2000) 102:553–63. doi:10.1016/S0092-8674(00)00078-7
6. Chahwan R, Edelmann W, Scharff MD, Roa S. AIDing antibody diversity by error-prone mismatch repair. Semin Immunol (2012) 24:293–300. doi:10.1016/j.smim.2012.05.005
7. Kenter AL. AID targeting is dependent on RNA polymerase II pausing. Semin Immunol (2012) 24:281–6. doi:10.1016/j.smim.2012.06.001
8. Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell (2010) 143:122–33. doi:10.1016/j.cell.2010.09.017
9. Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol (2008) 26:261–92. doi:10.1146/annurev.immunol.26.021607.090248
10. Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol (2008) 26:481–511. doi:10.1146/annurev.immunol.26.021607.090236
11. Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science (1998) 280:1750–2. doi:10.1126/science.280.5370.1750
12. Kotani AI, Okazaki M, Muramatsu M, Kinoshita K, Begum NA, Nakajima T, et al. A target selection of somatic hypermutations is regulated similarly between T and B cells upon activation-induced cytidine deaminase expression. Proc Natl Acad Sci U S A (2005) 102:4506–11. doi:10.1073/pnas.0500830102
13. Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature (2006) 440:105–9. doi:10.1038/nature04495
14. Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell (2004) 118:431–8. doi:10.1016/j.cell.2004.08.006
15. Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell (2013) 152:417–29. doi:10.1016/j.cell.2013.01.007
16. Gostissa M, Alt FW, Chiarle R. Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu Rev Immunol (2011) 29:319–50. doi:10.1146/annurev-immunol-031210-101329
17. Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol (2009) 10:243–54. doi:10.1038/nrm2651
18. Lisby M, Antunez de Mayolo A, Mortensen UH, Rothstein R. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle (2003) 2:479–83. doi:10.4161/cc.2.5.483
19. Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol (2003) 5:572–7. doi:10.1038/ncb997
20. Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr Opin Cell Biol (2004) 16:328–34. doi:10.1016/j.ceb.2004.03.011
21. Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science (1998) 280:590–2. doi:10.1126/science.280.5363.590
22. Kruhlak MJ, Celeste A, Nussenzweig A. Spatio-temporal dynamics of chromatin containing DNA breaks. Cell Cycle (2006) 5:1910–2. doi:10.4161/cc.5.17.3169
23. Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol (2007) 17:80–90. doi:10.1016/j.semcancer.2006.10.008
24. Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature (2008) 456:524–8. doi:10.1038/nature07433
25. Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature (2008) 456:529–33. doi:10.1038/nature07476
26. Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, et al. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity (2013) 39:229–44. doi:10.1016/j.immuni.2013.08.011
27. Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, et al. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol (2007) 9:675–82. doi:10.1038/ncb1591
28. Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell (2012) 148:908–21. doi:10.1016/j.cell.2012.02.002
29. Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet (2007) 8:104–15. doi:10.1038/nrg2041
30. Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature (2007) 445:379–781. doi:10.1038/445379a
31. Neves H, Ramos C, da Silva MG, Parreira A, Parreira L. The nuclear topography of ABL, BCR, PML, and RARalpha genes: evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood (1999) 93:1197–207.
32. Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet (2003) 34:287–91. doi:10.1038/ng1177
34. Osborne CS, Ewels PA, Young AN. Meet the neighbours: tools to dissect nuclear structure and function. Brief Funct Genomics (2011) 10:11–7. doi:10.1093/bfgp/elq034
35. van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol (2010) 28:1089–95. doi:10.1038/nbt.1680
36. Naumova N, Dekker J. Integrating one-dimensional and three-dimensional maps of genomes. J Cell Sci (2010) 123:1979–88. doi:10.1242/jcs.051631
37. Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol (2013) 20:290–9. doi:10.1038/nsmb.2474
38. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature (2012) 485:376–80. doi:10.1038/nature11082
39. Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol (2010) 2:a003889. doi:10.1101/cshperspect.a003889
40. Felsenfeld G, Groudine M. Controlling the double helix. Nature (2003) 421:448–53. doi:10.1038/nature01411
41. Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell (2008) 135:9–13. doi:10.1016/j.cell.2008.09.026
42. Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (2009) 326:289–93. doi:10.1126/science.1181369
43. Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, et al. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell (2011) 147:332–43. doi:10.1016/j.cell.2011.08.049
44. Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature (2011) 477:424–30. doi:10.1038/nature10495
45. Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell (2009) 138:435–48. doi:10.1016/j.cell.2009.07.016
46. Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, et al. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity (2007) 27:711–22. doi:10.1016/j.immuni.2007.09.007
47. Sellars M, Reina-San-Martin B, Kastner P, Chan S. Ikaros controls isotype selection during immunoglobulin class switch recombination. J Exp Med (2009) 206:1073–87. doi:10.1084/jem.20082311
48. Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell (2009) 139:1069–83. doi:10.1016/j.cell.2009.11.030
49. Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science (2009) 326:1230. doi:10.1126/science.1178124
50. Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol (2007) 94:157–214. doi:10.1016/S0065-2776(06)94006-1
51. Perlot T, Alt FW. Cis-regulatory elements and epigenetic changes control genomic rearrangements of the IgH locus. Adv Immunol (2008) 99:1–32. doi:10.1016/S0065-2776(08)00601-9
52. Gostissa M, Yan CT, Bianco JM, Cogne M, Pinaud E, Alt FW. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3′ regulatory region. Nature (2009) 462:803–7. doi:10.1038/nature08633
53. Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, et al. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity (2001) 15:187–99. doi:10.1016/S1074-7613(01)00181-9
54. Pinaud E, Marquet M, Fiancette R, Peron S, Vincent-Fabert C, Denizot Y, et al. The IgH locus 3′ regulatory region: pulling the strings from behind. Adv Immunol (2011) 110:27–70. doi:10.1016/B978-0-12-387663-8.00002-8
55. Rocha PP, Micsinai M, Kim JR, Hewitt SL, Souza PP, Trimarchi T, et al. Close proximity to Igh is a contributing factor to AID-mediated translocations. Mol Cell (2012) 47:873–85. doi:10.1016/j.molcel.2012.06.036
56. Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell (2011) 147:107–19. doi:10.1016/j.cell.2011.07.049
57. Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell (2011) 147:95–106. doi:10.1016/j.cell.2011.07.048
58. Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon KR, Jankovic M, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature (2012) 484:69–74. doi:10.1038/nature10909
59. Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol (2003) 160:685–97. doi:10.1083/jcb.200211103
60. Strickfaden H, Zunhammer A, van Koningsbruggen S, Kohler D, Cremer T. 4D chromatin dynamics in cycling cells: Theodor Boveri’s hypotheses revisited. Nucleus (2010) 1:284–97. doi:10.4161/nucl.1.3.11969
61. Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol (2004) 14:166–72. doi:10.1016/j.cub.2003.12.024
62. Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell (2013) 153:178–92. doi:10.1016/j.cell.2013.02.028
63. Rajapakse I, Perlman MD, Scalzo D, Kooperberg C, Groudine M, Kosak ST. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation. Proc Natl Acad Sci U S A (2009) 106:6679–84. doi:10.1073/pnas.0900986106
64. Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell (2010) 38:603–13. doi:10.1016/j.molcel.2010.03.016
65. Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Curr Opin Genet Dev (2012) 22:110–4. doi:10.1016/j.gde.2012.01.010
Keywords: B cells, Igh locus, chromosomal translocations, AID, genomic structure
Citation: Kenter AL, Wuerffel R, Kumar S and Grigera F (2013) Genomic architecture may influence recurrent chromosomal translocation frequency in the Igh locus. Front. Immunol. 4:500. doi: 10.3389/fimmu.2013.00500
Received: 03 December 2013; Accepted: 18 December 2013;
Published online: 30 December 2013.
Edited by:
Ananda L. Roy, Tufts University School of Medicine, USACopyright: © 2013 Kenter, Wuerffel, Kumar and Grigera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:c3RhcjFAdWljLmVkdQ==
 Amy L. Kenter
Amy L. Kenter Robert Wuerffel
Robert Wuerffel Satyendra Kumar
Satyendra Kumar Fernando Grigera
Fernando Grigera