- Department of Integrative Biology, University of California, Berkeley, Berkeley, CA, United States
The eukaryote immune system evolved and continues to evolve within a microbial world, and as such is critically shaped by—and in some cases even reliant upon—the presence of host-associated microbial species. There are clear examples of adaptations that allow the host to simultaneously tolerate and/or promote growth of symbiotic microbiota while protecting itself against pathogens, but the relationship between immunity and the microbiome reaches far beyond simple recognition and includes complex cross talk between host and microbe as well as direct microbiome-mediated protection against pathogens. Here, we present a broad but brief overview of how the microbiome is controlled by and interacts with diverse immune systems, with the goal of identifying questions that can be better addressed by taking a comparative approach across plants and animals and different types of immunity. As two key examples of such an approach, we focus on data examining the importance of early exposure on microbiome tolerance and immune system development and function, and the importance of transmission among hosts in shaping the potential coevolution between, and long-term stability of, host–microbiome associations. Then, by comparing existing evidence across short-lived plants, mouse model systems and humans, and insects, we highlight areas of microbiome research that are strong in some systems and absent in others with the hope of guiding future research that will allow for broad-scale comparisons moving forward. We argue that such an approach will not only help with identification of generalities in host–microbiome–immune interactions but also improve our understanding of the role of the microbiome in host health.
Introduction
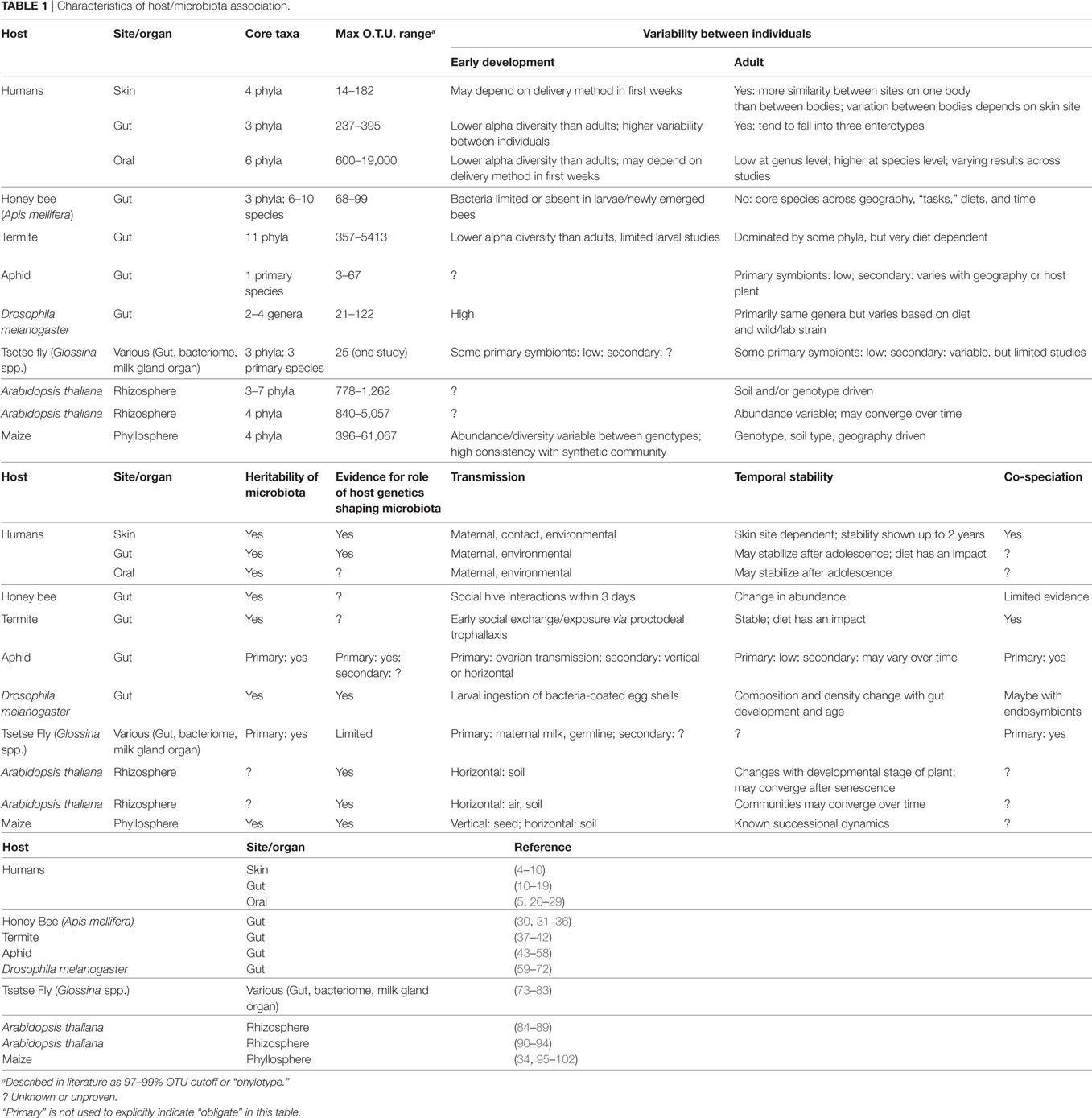
Across kingdoms of life and branches of immunity, there are conserved characteristics in how hosts interact with their microbiome. Plants, mammals, and invertebrates are all able to differentiate between self and non-self, where they tolerate, and in some cases promote, associations with commensal or beneficial microbes while retaining the ability to sense and attack microbial pathogens. In many cases, beneficial microbes can even be considered an extension of the immune system through either competitive exclusion of pathogens or direct inhibition of their growth. Furthermore, non-pathogenic microbiota can both interact with and influence the adaptive and innate immune systems. Across these diverse host systems, the evidence for an interaction between the microbiome and immunity is strong and unsurprising given that eukaryotic evolution has occurred entirely within a microbial world. The topic of immunity is highly complex and may seem inaccessible to those outside the discipline. However, from the perspective of evolutionary ecology, there is much that can be learned about host–microbe adaptation and coevolution through exploring topics in immunity. Therefore, our goal in this perspective piece is to broadly examine the key characteristics of known interplay between host immune systems and symbiotic bacteria across well-studied systems (the more detailed aspects of which, including microbiome variability among individuals, stability over time, mode of transmission, and evidence for host–microbiota co-speciation, we summarize in Table 1). We focus on the bacterial component of the microbiome but recognize the importance of fungal members and viruses, especially bacteriophages, given their known impact on the microbiome [e.g., Ref. (1, 2)] and possible role in host immunity [e.g., Ref. (3)].
The microbiome field is expanding rapidly, and doing so across systems, such as plants, mouse models, humans, and insects. We suggest that taking a broad comparative approach across the diverse mechanisms of immunity and host systems could offer unique insight to how host defenses are shaped by and shape the microbiome. Such an approach can, for example, help identify areas in which research is strong for certain systems but lacking in others. Here, we emphasize areas lacking in plant host systems, but which would likely elucidate important aspects of plant health and resilience against pathogens. Filling in such gaps across systems would allow for more powerful comparative studies and may inform predictions about host–microbe adaptations in light of larger issues such as antibiotic overuse and the spread of agricultural pathogens in a changing climate.
Overview of Host Immune Systems
To begin, we offer a brief description of immunity in mammals, plants, and insects focusing primarily on the aspects of these systems that directly relate to known interactions with the microbiome [thorough and more discipline-specific descriptions of these immune systems exist elsewhere (103–106)]. The adaptive immune system is thought to have arisen in jawed fish ≈500 million years ago (107), whereas the innate immune system likely dates back to early eukaryotic cells themselves (105, 108,). As microbial communities greatly predate the existence of multicellular eukaryotes, both branches of the immune system, therefore, evolved in the presence of microbes, and it follows that tolerance for commensal or mutualistic microbiota (those associated with hosts, but which do not cause disease) must have been a key factor in shaping the evolution of immunity. Innate immunity, found across all kingdoms of life, is largely non-specific and responds broadly to “non-self” cells. Its hallmarks include protective physical barriers and general pattern recognition receptors that sense non-self signals known as microbe-associated or pathogen-associated molecular patterns (MAMPs/PAMPs) and elicit generalized host responses (such as phagocytic ingestion of invading cells in animals or a hypersensitive response in plants). Adaptive immunity is unique to vertebrates and responds to specific pathogens through detection of antigens via somatically generated receptors and specialized white blood cells (B and T cells). Cellular recognition of a specific pathogen leads to clonal expansion of the lymphocyte, resulting in daughter cells that produce the same antigen-specific antibodies. Memory cells are also produced, resulting in specific and long-lasting immunological memory. Other versions of adaptive immunity may exist (discussed below), but broadly speaking, adaptive immune responses are highly specific to particular pathogens or antigens, and the immune response changes over the course of a host’s lifetime.
In many cases in vertebrates, innate immunity is the first line of defense that elicits an adaptive immune response (103), and the two systems work cooperatively to combat infection. In comparison, plants rely on an innate immunity consisting of two primary responses to microbes (106). The first branch of the immune system recognizes MAMPs/PAMPs, such as flagellin and lipopolysaccharides (LPS), through the use of transmembrane pattern recognition receptors and results in pattern-triggered immunity. However, many plant pathogens have evolved to overcome these defenses through the use of effectors. Plants with resistance genes for specific pathogens can detect the effectors through NB-LRR proteins, which represent the second response to microbes: effector-triggered immunity. In addition, plants have physical barriers to infection such as cell wall defenses (109) and can also secrete antimicrobial peptides to ward off infection (110). Insect immunology shares characteristics with both plants and mammals; responses to microbial pathogens are highly diverse among host species, but most are considered innate. Immune responses include production of antimicrobial peptides, pattern recognition receptors, and responding to pathogens via circulating phagocytic cells. Evidence accrued over the last few decades also shows responses reminiscent of adaptive-type immunity, such as immunological memory via virus-derived complementary DNAs that generate systemic immunity (111) and highly specific immune priming both within and across generations (112), but the extent of such adaptive-type immunity and similarity to vertebrate defenses remains an open question in the field (113, 114). Taking into account the type of host immunity is essential when making hypotheses about adaptation and coevolution between host and microbiota. For example, in contrast to adaptive immunity, the innate immune response is a general resistance that can only respond to selection across host generations and not within, an important distinction when considering how plants might adapt in response to microbiota as compared to vertebrates.
As is becoming increasingly evident, the immune system influences both the composition and abundance of non-pathogenic microbiota in addition to its well-studied role in preventing pathogen establishment. In mammals, this is best studied in the gut microbiome, where differentiating between these diverse symbionts and colonizing pathogens is clearly a complex problem. The human immune system maintains a homeostatic relationship with commensal microbiota through mechanisms that include stratification and compartmentalization of the intestine, production of a mucous layer and antimicrobial proteins, and limiting epithelial exposure and immune response (115), and through antibody targeting, which can limit bacterial spread and virulence, among other mechanisms (116). Interactions between the immune system and microbiota in the gut is a heavily studied field (115, 117–121), but we are still learning the ways in which aberrations in cross talk can cause or contribute to conditions, such as inflammatory bowel disease, obesity, and even certain types of cancer (122–126).
In insects, immune system responses also contribute to homeostasis with endosymbionts, reviewed in Ref. (127, 128), and restriction of other commensal bacteria to specific host compartments, as in the gut symbionts of termites (129), bees (32, 33), drosophila (130), and aphids (43), may also help maintain invertebrate symbiotic communities. The plant immune system is also critical in shaping the non-pathogenic microbiome [recently reviewed by Zipfel and Oldroyd (131)]. Two studies in Arabidopsis thaliana demonstrate that disrupting components of the plant immune system, such as the signaling molecules: salicylic acid (SA) and jasmonic acid (JA), influences microbial community composition: the first shows evidence for altered root microbiome communities in plant hosts lacking genes controlling production of SA compared to control plants (132) and the second shows altered microbial communities in plants with mutations in genes controlling ethylene response (another signaling molecule) and cuticle formation (90). Recent work in wheat also demonstrates a role for JA in shaping composition of the microbiome, and again in this case, activation of JA signaling pathways altered microbial diversity and composition of root endophytes (133). However, the importance of resistance genes and diversity, as well as the number of pattern recognition receptors, in shaping the plant microbiome remains an open question.
Importance of Microbiota in Shaping Host Immunity
The interaction between the microbiome and the immune system is far from one-sided, as has been elegantly demonstrated in studies from germ-free mice. Microbiome establishment influences levels of circulating myeloid cells, macrophages in tissues, and proper functioning of innate lymphoid cells, all critical for a healthy immune response (134–136). Furthermore, microbiota is critical in development and function of components of adaptive immunity, such as B and T cell diversity and differentiation (119, 137) and there is evidence from germ-free mice supporting a role in natural killer cell priming and function (137, 138). In insects, microbes also play a role in immune system development. For example, tsetse flies lacking their vertically transmitted symbionts are immunocompromised through both altered expression of immunity-related genes and reduced levels of hemocytes, which play an important role in invertebrate immunity (83, 139–141). Altered gene expression and other physiological effects were also found in axenically raised Drosophila melanogaster (61). In plants, symbiotic bacteria influence host immunity by priming the plant for future exposure to pathogens through the induction of a systemic response, causing broad-range basal levels of protection. A primed plant can respond more rapidly and strongly to pathogen invasion through a variety of mechanisms, including quicker closing of stomata, less sensitivity to bacterial manipulation of defenses, upregulation of defense-related genes, and a stronger SA-related immune responses (142). In some cases, the effects of priming can even be trans-generational through chromatin and histone modification, where the subsequent generation of primed plants exhibits enhanced resistance to bacterial, fungal, and herbivorous pathogens (143–146). Immunological priming by microbiota is also observed in arthropods, where it is often described as functional adaptive immunity, as it can occur within one generation or trans-generationally. Its effects have been observed in bumble bees (147, 148), beetles (149), daphnia (150), moths (151), and many more [summarized by Contreras-Garduño et al. (152)].
Host-associated microbiota can also directly influence host resistance against invading pathogens. Common in insects and also plants and mammals, the microbiome can serve a protective role that is independent of the host immune system through antagonism, competitive exclusion, or physical exclusion of pathogens, collectively referred to as defensive symbiosis (153, 154). For example, the mammalian skin microbiota is known to play a large role in pathogen recognition and infection prevention through amplification of immune responses (155) and production of antimicrobials (156). When germ-free mice were inoculated with gut microbiota from a non-mouse host source, they showed a decreased ability to fight infection against Salmonella, and particular bacterial strains seem to be required for normal adaptive immune response (157). More recently, it has been shown that a mildly pathogenic bacterium of Caenorhabditis elegans can evolve over time to protect its host against the more virulent pathogen, Staphylococcus aureus (158). The importance of such pairwise interactions have been demonstrated many times [reviewed in Ref. (159, 160)], and indeed has motivated many current biocontrol strategies, but an open question in the field is how such microbe-mediated protection might scale up to the whole microbiome level. This leads to the idea that by directly protecting their host against pathogens, microbiota could hinder the evolution of host resistance by relaxing selection on host populations and, therefore, increasing host reliance on the microbiome.
Microbiome Transmission and Timing of Exposure
In mammals, it is clear that early exposure to microbes is crucial to the development of both branches of the immune system (161), influencing not only immune development and response against pathogens but also tolerance to commensal or mutualistic microbiota (162). For example, pregnant female mice treated with antibiotics have been shown to have offspring with not only a depauperate microbiome but also decreased levels of blood neutrophils and precursor cells, resulting in higher susceptibility to infection and increased mortality rates as compared to control mice (163). In line with this, there is increasing evidence for a crucial window of opportunity for exposure to microbiota (135). A study in germ-free mice showed that introducing a healthy microbiome to adult germ-free mice did not restore normal levels of invariant natural killer T cells nor did it lessen the physical effects of induced colitis (164), and altered exposure to bacterial species and their LPS subtypes in human infant guts may have lasting and detrimental effects on development of immunity (165). In the human neonate airway, disruption of microbiome formation as early as the first 2 weeks of life can result in lifelong susceptibility to allergic airway inflammation (166). There are additional documented links between dysbiosis of early-life microbiota and disease or health conditions later in life, reviewed elsewhere (167). Despite the accruing evidence from human and mouse systems, there has been little to no exploration of such a window of opportunity for microbiome–immune system interactions in other systems, such as plants or insects. It also remains unclear whether such early exposure effects should be limited to organisms with adaptive immunity or whether priming of innate immunity at different host developmental stages also affects host–microbiome interactions.
The clear role of early exposure to microbiota, at least in mammals, suggests that it would be advantageous for a community of beneficial microbes to be transmitted vertically from parent to offspring (e.g., through direct contact at birth, seeds, or transovarian) from generation to generation. Vertical transmission in humans may be impacted by delivery mode, as there is good evidence for differences in microbiome composition and diversity between infants delivered via virginal birth versus those delivered via cesarean sections (12, 168), but it remains controversial how long-lived such effects are (4). In insects, symbionts are known to be maintained through both vertical transmission [for example, Buchnera in aphids; (169)] and other transmission mechanisms such as early social interactions [observed in bees; (36)], proctodeal exchange of fluids [e.g., in termites; (170)], or larval consumption of bacteria-coated egg shells [as observed in Drosophila; (59, 70, 171)]. Interestingly, non-social bees (in which early social transmission of symbionts would not occur) do not seem to share the core microbiome that is observed among social bees (33).
Transmission of microbiota in plants can occur vertically through the seeds, or horizontally from the soil and surrounding environment. Plants ranging from trees to grasses are known to harbor bacteria in their seeds, many of which are reported to promote plant health (172–174). Despite this, there is no evidence that plants actively select for transmission of specific microbial communities, and there are no clear examples of adaptations to ensure seed-mediated transmission. Intuitively, vertical transmission of a microbiome or symbionts would allow for maintenance of key members of the microbial community across generations, as beneficial microbes would have primary access to both spatial niches and environmental nutrients provided by seedlings. Interestingly, plants have been shown to have differential onset of resistance to pathogens throughout their life-stages, something described as age-related resistance (ARR) or developmental resistance (175–177). However, much of the work on ARR investigates exposure and resistance to specific pathogens throughout the developmental stage of the plant and does not address if there is a window of opportunity for microbial exposure in general, as observed in mammals.
Conclusion and Opportunities for Advancement in the Field
Unsurprisingly, that the microbiome is both shaped by and shapes the host immune system is a common feature of eukaryotes. However, the mechanisms underlying such cross talk are highly variable. Although we now have a foundation of knowledge demonstrating the microbiome’s role in immune system development and function, key-questions remain unanswered across systems. One specific area for advancement is exploring the importance of both vertical transmission and timing of microbiome exposure across systems with diverse immune mechanisms. For example, despite the known importance of timing of exposure in mouse models and vertical transmission in insects, to our knowledge there are no studies to date that test the importance of timing of non-pathogenic microbial exposure on microbiome establishment or immune function in plants, and few in invertebrates. Would a seedling exposed to beneficial microbes mount as strong of a response as an older plant? And would exposure of otherwise sterile adult plants result in the same successional dynamics of microbiome establishment as has been observed in seedlings of some plant species (93, 178, 179)? Given that we know resistance to pathogens can change throughout the life cycle of a plant, research focused on age-related tolerance and recruitment of beneficial symbionts and plant-growth promoting bacteria has large implications in agricultural practices, such as seed treatment, greenhouse germination, and age-structured planting.
Vertical transmission also ensures stable associations between hosts and their microbiomes over evolutionary time and, therefore, sets the stage for long-term coevolution and even co-speciation. There is good evidence for vertical transmission of microbiota through gametes, secretions, or birth/delivery from across systems, but how this relates to coevolution between microbiota and their hosts remains to be determined. Long-term associations between hosts and microbiota can be uncovered through examination of co-speciation events, and these have been described in insects, such as aphids (51, 52), social bees (31), and termites (40). Furthermore, recent evidence from the hominid phylogeny also strongly supports this phenomenon (180). However, in plant systems, the current evidence is limited to a few pairwise host–symbiont interactions (181, 182). To understand the ways in which microbiota–immunity interactions influence stable association, transmission, and potentially coevolution in organisms such as plants, it may be wise to start by looking for similarities in established examples, such as the reduced genomes of symbionts commonly found in insect symbionts (183), nutritional dependence on symbionts, or physical partitioning of microbiota within the host.
Another area of advancement involves taking into account the whole suite of microbiomes associated with hosts. Despite what we know about spatially distinct microbiota in humans (5) and plants (184, 185), there are still large biases toward the below-ground (rhizosphere) microbiota of plants and the gut microbiota of vertebrates and insects. As more multi-tissue microbiome studies are generated across systems, we will be in a better position to uncover general patterns of potential cross talk among microbiomes within a host, differences in the types of pathogens being protected against across tissues, and perhaps even the role of distinct microbiomes in shaping tissue tropism of pathogens. Furthermore, parallel studies of spatially distinct microbiomes in insects could offer nice insight into, for example, the roles of internally versus externally colonizing microbiota in shaping disease susceptibility, as well as how the host immune response regulates multiple microbiomes simultaneously.
Finally, the field is still limited by challenges in data interpretation for large, complex, and dynamic microbiome systems, explaining many of the open questions regarding heritability, temporal dynamics, and co-speciation (highlighted in Table 1). However, addressing these questions is increasingly feasible through rapidly advancing sequencing and bioinformatics approaches and the compilation of biologically representative synthetic communities. Although we are still some way from having large cross-system comparative microbiome studies, as sequencing costs continue to fall and data standardization across studies becomes more stringent, such meta-analyses will likely uncover larger “rules” of microbiome assembly, diversity, and interplay with host immunity. For example, plant-microbiome literature has forged the way in our understanding of how host genetics versus environment contribute to shaping the adult microbiome [e.g., Ref. (90, 186)], and recent work from humans now raises the question of whether similar rules are true for vertebrates (187). Another, more reductionist, approach for testing fundamental predictions about microbiome establishment genetic underpinning and immune system interactions is using synthetic microbiomes, as has been well-developed in plants (86, 90, 101, 132, 188). For example, a recent study in D. melanogaster explored colonization of gnotobiotic flies with specific strains of bacteria to document how host genotype influences microbial abundance levels (65). Though far from painting a complete picture, approaches such as this may also provide a means to study specific microbial adaptations to the immune systems of hosts across environmental conditions and genotypes. In conclusion, as we accumulate more data across systems, we can take more comparative and/or phylogenetic approaches to better understand the evolution of microbiome–immune system interaction mechanisms and to uncover conserved microbiome-mediated immune functions across systems. Such research has broad application to both human and agricultural health and is critical in light of the emergence of antibiotic and chemical-resistant pathogens and the common use of interventions that disrupt host-microbiome associations across systems.
Author Contributions
NM and BK both contributed to the development of ideas and writing of this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank the editors for the invitation to contribute to this research topic, the reviewers for their helpful feedback, and Callie Cuff for assistance in background research.
References
1. Koskella B. Phage-mediated selection on microbiota of a long-lived host. Curr Biol (2013) 23:1256–60. doi:10.1016/j.cub.2013.05.038
2. Scanlan PD. Bacteria-bacteriophage coevolution in the human gut: implications for microbial diversity and functionality. Trends Microbiol (2017) 25:614–23. doi:10.1016/j.tim.2017.02.012
3. Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A (2013) 110:10771–6. doi:10.1073/pnas.1305923110
4. Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med (2017) 23:314–26. doi:10.1038/nm.4272
5. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science (2009) 326:1694–7. doi:10.1126/science.1177486
6. Gao Z, Tseng C, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A (2007) 104:2927–32. doi:10.1073/pnas.0607077104
7. Grice EA, Kong HH, Renaud G, Young AC; NISC Comparative Sequencing Program, Bouffard GG, et al. A diversity profile of the human skin microbiota. Genome Res (2008) 18:1043–50. doi:10.1101/gr.075549.107
8. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science (2009) 324:1190–2. doi:10.1126/science.1171700
9. Oh J, Byrd AL, Park M; NISC Comparative Sequencing Program, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell (2016) 165:854–66. doi:10.1016/j.cell.2016.04.008
10. Si J, Lee S, Park JM, Sung J, Ko G. Genetic associations and shared environmental effects on the skin microbiome of Korean twins. BMC Genomics (2015) 16:992. doi:10.1186/s12864-015-2131-y
11. Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A (2010) 107:18933–8. doi:10.1073/pnas.1007028107
12. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107:11971–5. doi:10.1073/pnas.1002601107
13. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (2005) 308:1635–8. doi:10.1126/science.1110591
14. Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, de Visser JAGM, de Vos WM. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb Ecol Health Dis (2001) 13:129–34. doi:10.1080/089106001750462669
15. Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science (2006) 312:1355–9. doi:10.1126/science.1124234
16. Greenhalgh K, Meyer KM, Aagaard KM, Wilmes P. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ Microbiol (2016) 18:2103–16. doi:10.1111/1462-2920.13318
17. Hopkins MJ, Sharp R, Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut (2001) 48:198–205. doi:10.1136/gut.48.2.198
18. Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol (2011) 9:279–90. doi:10.1038/nrmicro2540
19. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486:222–7. doi:10.1038/nature11053
20. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol (2005) 43:5721–32. doi:10.1128/JCM.43.11.5721-5732.2005
21. Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J (2010) 4:962–74. doi:10.1038/ismej.2010.30
22. De La Fuente C, Flores S, Moraga M. DNA from human ancient bacteria: a novel source of genetic evidence from archaeological dental calculus. Archaeometry (2013) 55:767–78. doi:10.1111/j.1475-4754.2012.00707.x
23. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, et al. The human oral microbiome. J Bacteriol (2010) 192:5002–17. doi:10.1128/JB.00542-10
24. Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature (2014) 509:357–60. doi:10.1038/nature13178
25. Gomez A, Nelson KE. The oral microbiome of children: development, disease, and implications beyond oral health. Microb Ecol (2017) 73:492–503. doi:10.1007/s00248-016-0854-1
26. Keijser BJF, Zaura E, Huse SM, van der Vossen JMBM, Schuren FHJ, Montijn RC, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res (2008) 87:1016–20. doi:10.1177/154405910808701104
27. Nasidze I, Li J, Quinque D, Tang K, Stoneking M. Global diversity in the human salivary microbiome. Genome Res (2009) 19:636–43. doi:10.1101/gr.084616.108
28. Stahringer SS, Clemente JC, Corley RP, Hewitt J, Knights D, Walters WA, et al. Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome Res (2012) 22:2146–52. doi:10.1101/gr.140608.112
29. Warinner C, Rodrigues JFM, Vyas R, Trachsel C, Shved N, Grossmann J, et al. Pathogens and host immunity in the ancient human oral cavity. Nat Genet (2014) 46:336–44. doi:10.1038/ng.2906
30. Engel P, Kwong WK, McFrederick Q, Anderson KE, Barribeau SM, Chandler JA, et al. The bee microbiome: impact on bee health and model for evolution and ecology of host-microbe interactions. MBio (2016) 7(2):e02164–15.
31. Koch H, Abrol DP, Li J, Schmid-Hempel P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol (2013) 22:2028–44. doi:10.1111/mec.12209
32. Kwong WK, Mancenido AL, Moran NA. Immune system stimulation by the native gut microbiota of honey bees. R Soc Open Sci (2017) 4:170003. doi:10.1098/rsos.170003
33. Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol (2011) 20:619–28. doi:10.1111/j.1365-294X.2010.04959.x
34. Mohr KI, Tebbe CC. Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ Microbiol (2006) 8:258–72. doi:10.1111/j.1462-2920.2005.00893.x
35. Moran NA. Genomics of the honey bee microbiome. Curr Opin Insect Sci (2015) 10:22–8. doi:10.1016/j.cois.2015.04.003
36. Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl Environ Microbiol (2014) 80:7378–87. doi:10.1128/AEM.01861-14
37. Benjamino J, Graf J. Characterization of the core and caste-specific microbiota in the termite, Reticulitermes flavipes. Front Microbiol (2016) 7:171. doi:10.3389/fmicb.2016.00171
38. Dietrich C, Köhler T, Brune A. The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl Environ Microbiol (2014) 80:2261–9. doi:10.1128/AEM.04206-13
39. Diouf M, Roy V, Mora P, Frechault S, Lefebvre T, Hervé V, et al. Profiling the succession of bacterial communities throughout the life stages of a higher termite Nasutitermes arborum (Termitidae, Nasutitermitinae) using 16S rRNA gene pyrosequencing. PLoS One (2015) 10:e0140014. doi:10.1371/journal.pone.0140014
40. Hongoh Y, Deevong P, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, et al. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl Environ Microbiol (2005) 71:6590–9. doi:10.1128/AEM.71.11.6590-6599.2005
41. Tinker KA, Ottesen EA. The core gut microbiome of the American cockroach, Periplaneta americana, is stable and resilient to dietary shifts. Appl Environ Microbiol (2016) 82:6603–10. doi:10.1128/AEM.01837-16
42. Wang Y, Su L, Huang S, Bo C, Yang S, Li Y, et al. Diversity and resilience of the wood-feeding higher termite Mironasutitermes shangchengensis gut microbiota in response to temporal and diet variations. Ecol Evol (2016) 6:8235–42. doi:10.1002/ece3.2497
43. Chen D-Q, Purcell AH. Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol (1997) 34:220–5. doi:10.1007/s002849900172
44. Chong RA, Moran NA. Intraspecific genetic variation in hosts affects regulation of obligate heritable symbionts. Proc Natl Acad Sci U S A (2016) 113:13114–9. doi:10.1073/pnas.1610749113
45. Clark MA, Moran NA, Baumann P, Wernegreen JJ. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution (2000) 54:517–25. doi:10.1111/j.0014-3820.2000.tb00054.x
46. Fukatsu T, Nikoh N, Kawai R, Koga R. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera). Appl Environ Microbiol (2000) 66:2748–58. doi:10.1128/AEM.66.7.2748-2758.2000
47. Funk DJ, Helbling L, Wernegreen JJ, Moran NA. Intraspecific phylogenetic congruence among multiple symbiont genomes. Proc R Soc Lond B Biol Sci (2000) 267:2517–21. doi:10.1098/rspb.2000.1314
48. Gauthier J-P, Outreman Y, Mieuzet L, Simon J-C. Bacterial communities associated with host-adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. PLoS One (2015) 10:e0120664. doi:10.1371/journal.pone.0120664
49. Haynes S, Darby AC, Daniell TJ, Webster G, van Veen FJF, Godfray HCJ, et al. Diversity of bacteria associated with natural aphid populations. Appl Environ Microbiol (2003) 69:7216–23. doi:10.1128/AEM.69.12.7216-7223.2003
50. Macdonald SJ, Thomas GH, Douglas AE. Genetic and metabolic determinants of nutritional phenotype in an insect-bacterial symbiosis. Mol Ecol (2011) 20:2073–84. doi:10.1111/j.1365-294X.2011.05031.x
51. Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc Lond B Biol Sci (1993) 253:167–71. doi:10.1098/rspb.1993.0098
52. Munson MA, Baumann P, Clark MA, Baumann L, Moran NA, Voegtlin DJ, et al. Evidence for the establishment of aphid-Eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol (1991) 173:6321–4. doi:10.1128/jb.173.20.6321-6324.1991
53. Peccoud J, Bonhomme J, Mahéo F, de la Huerta M, Cosson O, Simon J-C. Inheritance patterns of secondary symbionts during sexual reproduction of pea aphid biotypes. Insect Sci (2014) 21:291–300. doi:10.1111/1744-7917.12083
54. Russell JA, Weldon S, Smith AH, Kim KL, Hu Y, Łukasik P, et al. Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol Ecol (2013) 22:2045–59. doi:10.1111/mec.12211
55. Sandström JP, Russell JA, White JP, Moran NA. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol (2001) 10:217–28. doi:10.1046/j.1365-294X.2001.01189.x
56. Smith AH, Łukasik P, O’Connor MP, Lee A, Mayo G, Drott MT, et al. Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol Ecol (2015) 24:1135–49. doi:10.1111/mec.13095
57. Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol Ecol (2002) 11:2123–35. doi:10.1046/j.1365-294X.2002.01606.x
58. Zytynska SE, Weisser WW. The natural occurrence of secondary bacterial symbionts in aphids. Ecol Entomol (2016) 41:13–26. doi:10.1111/een.12281
59. Bakula M. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J Invertebr Pathol (1969) 14:365–74. doi:10.1016/0022-2011(69)90163-3
60. Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes (2012) 3:307–21. doi:10.4161/gmic.19896
61. Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in Drosophila melanogaster host gene expression and gut morphology. MBio (2014) 5:e01117–14. doi:10.1128/mBio.01117-14
62. Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet (2011) 7:e1002272. doi:10.1371/journal.pgen.1002272
63. Chaston JM, Dobson AJ, Newell PD, Douglas AE. Host genetic control of the microbiota mediates the Drosophila nutritional phenotype. Appl Environ Microbiol (2015) 82:671–9. doi:10.1128/AEM.03301-15
64. Corby-Harris V, Pontaroli AC, Shimkets LJ, Bennetzen JL, Habel KE, Promislow DEL. Geographical distribution and diversity of bacteria associated with natural populations of Drosophila melanogaster. Appl Environ Microbiol (2007) 73:3470–9. doi:10.1128/AEM.02120-06
65. Early AM, Shanmugarajah N, Buchon N, Clark AG. Drosophila genotype influences commensal bacterial levels. PLoS One (2017) 12:e0170332. doi:10.1371/journal.pone.0170332
66. Elgart M, Stern S, Salton O, Gnainsky Y, Heifetz Y, Soen Y. Impact of gut microbiota on the fly’s germ line. Nat Commun (2016) 7:11280. doi:10.1038/ncomms11280
67. Ilinsky Y. Coevolution of Drosophila melanogaster mtDNA and Wolbachia genotypes. PLoS One (2013) 8:e54373. doi:10.1371/journal.pone.0054373
68. Mistry R, Kounatidis I, Ligoxygakis P. Interaction between familial transmission and a constitutively active immune system shapes gut microbiota in Drosophila melanogaster. Genetics (2017) 206:889–904. doi:10.1534/genetics.116.190215
69. Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab (2007) 6:144–52. doi:10.1016/j.cmet.2007.06.006
70. Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab (2011) 14:403–14. doi:10.1016/j.cmet.2011.07.012
71. Wong AC-N, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J (2013) 7:1922–32. doi:10.1038/ismej.2013.86
72. Wong CNA, Ng P, Douglas AE. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol (2011) 13:1889–900. doi:10.1111/j.1462-2920.2011.02511.x
73. Aksoy E, Telleria EL, Echodu R, Wu Y, Okedi LM, Weiss BL, et al. Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota. Appl Environ Microbiol (2014) 80:4301–12. doi:10.1128/AEM.00079-14
74. Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S. Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol (2008) 54:1236–42. doi:10.1016/j.jinsphys.2008.06.008
75. Chen X, Li S, Aksoy S. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J Mol Evol (1999) 48:49–58. doi:10.1007/PL00006444
76. Cheng Q, Ruel TD, Zhou W, Moloo SK, Majiwa P, O’neill SL, et al. Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med Vet Entomol (2000) 14:44–50. doi:10.1046/j.1365-2915.2000.00202.x
77. Geiger A, Fardeau M-L, Grebaut P, Vatunga G, Josénando T, Herder S, et al. First isolation of Enterobacter, Enterococcus, and Acinetobacter spp as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Infect Genet Evol (2009) 9:1364–70. doi:10.1016/j.meegid.2009.09.013
78. Geiger A, Fardeau M-L, Njiokou F, Joseph M, Asonganyi T, Ollivier B, et al. Bacterial diversity associated with populations of Glossina spp. from Cameroon and distribution within the campo sleeping sickness focus. Microb Ecol (2011) 62:632–43. doi:10.1007/s00248-011-9830-y
79. Geiger A, Ponton F, Simo G. Adult blood-feeding tsetse flies, trypanosomes, microbiota and the fluctuating environment in sub-Saharan Africa. ISME J (2015) 9:1496–507. doi:10.1038/ismej.2014.236
80. Hamidou Soumana I, Simo G, Njiokou F, Tchicaya B, Abd-Alla AMM, Cuny G, et al. The bacterial flora of tsetse fly midgut and its effect on trypanosome transmission. J Invertebr Pathol (2013) 112:S89–93. doi:10.1016/j.jip.2012.03.029
81. Lindh JM, Lehane MJ. The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek (2011) 99:711–20. doi:10.1007/s10482-010-9546-x
82. Rio RVM, Wu YN, Filardo G, Aksoy S. Dynamics of multiple symbiont density regulation during host development: tsetse fly and its microbial flora. Proc Biol Sci (2006) 273:805–14. doi:10.1098/rspb.2005.3399
83. Wang J, Weiss BL, Aksoy S. Tsetse fly microbiota: form and function. Front Cell Infect Microbiol (2013) 3:69. doi:10.3389/fcimb.2013.00069
84. Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature (2012) 488:91–5. doi:10.1038/nature11336
85. Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by plant development. ISME J (2014) 8:790–803. doi:10.1038/ismej.2013.196
86. Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature (2012) 488:86–90. doi:10.1038/nature11237
87. Micallef SA, Shiaris MP, Colón-Carmona A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot (2009) 60:1729–42. doi:10.1093/jxb/erp053
88. Micallef SA, Channer S, Shiaris MP, Colón-Carmona A. Plant age and genotype impact the progression of bacterial community succession in the Arabidopsis rhizosphere. Plant Signal Behav (2009) 4:777–80. doi:10.4161/psb.4.8.9229
89. Schlaeppi K, Dombrowski N, Oter RG, van Themaat EVL, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci U S A (2014) 111:585–92. doi:10.1073/pnas.1321597111
90. Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet (2014) 10:e1004283. doi:10.1371/journal.pgen.1004283
91. Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, et al. Genome-wide association study of Arabidopsis thaliana’s leaf microbial community. Nat Commun (2014) 5:5320. doi:10.1038/ncomms6320
92. Kniskern JM, Traw MB, Bergelson J. Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol Plant Microbe Interact (2007) 20:1512–22. doi:10.1094/MPMI-20-12-1512
93. Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. Mbio (2014) 5:e00682–13. doi:10.1128/mBio.00682-13
94. Reisberg EE, Hildebrandt U, Riederer M, Hentschel U. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS One (2013) 8:e78613. doi:10.1371/journal.pone.0078613
95. Bouffaud M-L, Kyselková M, Gouesnard B, Grundmann G, Muller D, Moënne-Loccoz Y. Is diversification history of maize influencing selection of soil bacteria by roots? Mol Ecol (2012) 21:195–206. doi:10.1111/j.1365-294X.2011.05359.x
96. Brusetti L, Francia P, Bertolini C, Pagliuca A, Borin S, Sorlini C, et al. Bacterial communities associated with the rhizosphere of transgenic Bt 176 maize (Zea mays) and its non transgenic counterpart. Plant Soil (2005) 266:11–21. doi:10.1007/s11104-005-5399-x
97. Cavaglieri L, Orlando J, Etcheverry M. Rhizosphere microbial community structure at different maize plant growth stages and root locations. Microbiol Res (2009) 164:391–9. doi:10.1016/j.micres.2007.03.006
98. Johnston-Monje D, Raizada MN. Conservation and diversity of seed associated endophytes in zea across boundaries of evolution, ethnography and ecology. PLoS One (2011) 6:e20396. doi:10.1371/journal.pone.0020396
99. Johnston-Monje D, Lundberg DS, Lazarovits G, Reis VM, Raizada MN. Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil (2016) 405:337–55. doi:10.1007/s11104-016-2826-0
100. Li X, Rui J, Mao Y, Yannarell A, Mackie R. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol Biochem (2014) 68:392–401. doi:10.1016/j.soilbio.2013.10.017
101. Niu B, Paulson JN, Zheng X, Kolter R. Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci U S A (2017) 114:E2450–9. doi:10.1073/pnas.1616148114
102. Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci U S A (2013) 110:6548–53. doi:10.1073/pnas.1302837110
103. Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Chapter 24: The adaptive immune system. Molecular Biology of the Cell. 4th ed. New York: Garland Science (2002). Available from: https://www.ncbi.nlm.nih.gov/books/NBK21070/
105. Janeway CA Jr, Travers P, Walport M, Shlomchik MJ. Principles of innate and adaptive immunity. Immunobiology: The Immune System in Health and Disease. 5th ed. New York: Garland Science (2001). Available from: https://www.ncbi.nlm.nih.gov/books/NBK27090/
107. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet (2010) 11:47–59. doi:10.1038/nrg2703
108. Buchmann K. Evolution of innate immunity: clues from invertebrates via fish to mammals. Front Immunol (2014) 5:459. doi:10.3389/fimmu.2014.00459
109. Underwood W. The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci (2012) 3:85. doi:10.3389/fpls.2012.00085
110. Mansfield JW. Antimicrobial compounds and resistance. In: Slusarenko AJ, Fraser RSS, van Loon LC, editors. Mechanisms of Resistance to Plant Diseases. Netherlands: Springer (2000). p. 325–70.
111. Tassetto M, Kunitomi M, Andino R. Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell (2017) 169:314–25.e13. doi:10.1016/j.cell.2017.03.033
112. Dhinaut J, Chogne M, Moret Y. Immune priming specificity within and across generations reveals the range of pathogens affecting evolution of immunity in an insect. J Anim Ecol (2017). doi:10.1111/1365-2656.12661
113. Cooper D, Eleftherianos I. Memory and specificity in the insect immune system: current perspectives and future challenges. Front Immunol (2017) 8:539. doi:10.3389/fimmu.2017.00539
114. Rowley AF, Powell A. Invertebrate immune systems-specific, quasi-specific, or nonspecific? J Immunol (2007) 179:7209–14. doi:10.4049/jimmunol.179.11.7209
115. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science (2012) 336:1268–73. doi:10.1126/science.1223490
116. Kubinak JL, Round JL. Do antibodies select a healthy microbiota? Nat Rev Immunol (2016) 16:767–74. doi:10.1038/nri.2016.114
117. Barroso-Batista J, Demengeot J, Gordo I. Adaptive immunity increases the pace and predictability of evolutionary change in commensal gut bacteria. Nat Commun (2015) 6:8945. doi:10.1038/ncomms9945
118. Feng T, Elson CO. Adaptive immunity in the host-microbiota dialog. Mucosal Immunol (2011) 4:15–21. doi:10.1038/mi.2010.60
119. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature (2016) 535:75–84. doi:10.1038/nature18848
120. Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science (2010) 330:1768–73. doi:10.1126/science.1195568
121. Thaiss CA, Levy M, Suez J, Elinav E. The interplay between the innate immune system and the microbiota. Curr Opin Immunol (2014) 26:41–8. doi:10.1016/j.coi.2013.10.016
122. Alam A, Leoni G, Quiros M, Wu H, Desai C, Nishio H, et al. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol (2016) 1:15021. doi:10.1038/nmicrobiol.2015.21
123. Alam A, Wu H, Matthews J, Saeedi B, Jones R, Neish A. O-010 microbial metabolites establish a gradient of protons, which mediate interkingdom host-microbiome cross-talk and maintain intestinal homeostasis via two proton-sensing GPRs. Inflamm Bowel Dis (2017) 23(Suppl 1):S4. doi:10.1097/01.MIB.0000512520.76586.47
124. Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell (2014) 158:1000–10. doi:10.1016/j.cell.2014.08.006
125. Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci (2017) 74:2959–77. doi:10.1007/s00018-017-2509-x
126. Zeevi D, Korem T, Segal E. Talking about cross-talk: the immune system and the microbiome. Genome Biol (2016) 17:50. doi:10.1186/s13059-016-0921-4
127. Eleftherianos I, Atri J, Accetta J, Castillo J. Endosymbiotic bacteria in insects: guardians of the immune system? Front Physiol (2013) 4:46. doi:10.3389/fphys.2013.00046
128. Ratzka C, Gross R, Feldhaar H. Endosymbiont tolerance and control within insect hosts. Insects (2012) 3:553–72. doi:10.3390/insects3020553
129. Ohkuma M, Brune A. Diversity, structure, and evolution of the termite gut microbial community. In: Bignell DE, Roisin Y, Lo N, editors. Biology of Termites: A Modern Synthesis. Netherlands: Springer (2010). p. 413–38.
130. Marianes A, Spradling AC. Physiological and stem cell compartmentalization within the Drosophila midgut. Elife (2013) 2:e00886. doi:10.7554/eLife.00886
131. Zipfel C, Oldroyd GED. Plant signalling in symbiosis and immunity. Nature (2017) 543:328–36. doi:10.1038/nature22009
132. Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science (2015) 349:860–4. doi:10.1126/science.aaa8764
133. Liu H, Carvalhais LC, Schenk PM, Dennis PG. Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci Rep (2017) 7:41766. doi:10.1038/srep41766
134. Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci (2015) 18:965–77. doi:10.1038/nn.4030
135. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science (2016) 352:539–44. doi:10.1126/science.aad9378
136. Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature (2016) 535:65–74. doi:10.1038/nature18847
137. Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol (2009) 10:83–91. doi:10.1038/ni.1684
138. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity (2012) 37:171–86. doi:10.1016/j.immuni.2012.05.020
139. Benoit JB, Vigneron A, Broderick NA, Wu Y, Sun JS, Carlson JR, et al. Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. eLife (2017) 6:e19535. doi:10.7554/eLife.19535
140. Weiss BL, Wang J, Aksoy S. Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol (2011) 9:e1000619. doi:10.1371/journal.pbio.1000619
141. Weiss BL, Maltz M, Aksoy S. Obligate symbionts activate immune system development in the tsetse fly. J Immunol (2012) 1950(188):3395–403. doi:10.4049/jimmunol.1103691
142. Pastor V, Luna E, Mauch-Mani B, Ton J, Flors V. Primed plants do not forget. Environ Exp Bot (2013) 94:46–56. doi:10.1016/j.envexpbot.2012.02.013
143. Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol (2010) 10:735–44. doi:10.1038/nri2850
144. Luna E, Bruce TJA, Roberts MR, Flors V, Ton J. Next-generation systemic acquired resistance. Plant Physiol (2012) 158:844–53. doi:10.1104/pp.111.187468
145. Rasmann S, De Vos M, Casteel CL, Tian D, Halitschke R, Sun JY, et al. Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol (2012) 158:854–63. doi:10.1104/pp.111.187831
146. Slaughter A, Daniel X, Flors V, Luna E, Hohn B, Mauch-Mani B. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol (2012) 158:835–43. doi:10.1104/pp.111.191593
147. Moret Y, Schmid-Hempel P. Entomology: immune defence in bumble-bee offspring. Nature (2001) 414:506–506. doi:10.1038/35107138
148. Sadd BM, Schmid-Hempel P. Insect immunity shows specificity in protection upon secondary pathogen exposure. Curr Biol (2006) 16:1206–10. doi:10.1016/j.cub.2006.04.047
149. Moret Y, Siva-Jothy MT. Adaptive innate immunity? Responsive-mode prophylaxis in the mealworm beetle, Tenebrio molitor. Proc Biol Sci (2003) 270:2475–80. doi:10.1098/rspb.2003.2511
150. McTaggart SJ, Wilson PJ, Little TJ. Daphnia magna shows reduced infection upon secondary exposure to a pathogen. Biol Lett (2012) 8:972–5. doi:10.1098/rsbl.2012.0581
151. Mikonranta L, Mappes J, Kaukoniitty M, Freitak D. Insect immunity: oral exposure to a bacterial pathogen elicits free radical response and protects from a recurring infection. Front Zool (2014) 11:23. doi:10.1186/1742-9994-11-23
152. Contreras-Garduño J, Lanz-Mendoza H, Franco B, Nava A, Pedraza-Reyes M, Canales-Lazcano J. Insect immune priming: ecology and experimental evidences. Ecol Entomol (2016) 41:351–66. doi:10.1111/een.12300
153. Clay K. Defensive symbiosis: a microbial perspective. Funct Ecol (2014) 28:293–8. doi:10.1111/1365-2435.12258
154. Hussa EA, Goodrich-Blair H. It takes a village: ecological and fitness impacts of multipartite mutualism. Annu Rev Microbiol (2013) 67:161–78. doi:10.1146/annurev-micro-092412-155723
155. Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science (2012) 337:1115–9. doi:10.1126/science.1225152
156. Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol (2012) 12:503–16. doi:10.1038/nri3228
157. Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell (2012) 149:1578–93. doi:10.1016/j.cell.2012.04.037
158. King KC, Brockhurst MA, Vasieva O, Paterson S, Betts A, Ford SA, et al. Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J (2016) 10:1915–24. doi:10.1038/ismej.2015.259
159. May G, Nelson P. Defensive mutualisms: do microbial interactions within hosts drive the evolution of defensive traits? Funct Ecol (2014) 28:356–63. doi:10.1111/1365-2435.12166
160. Oliver KM, Smith AH, Russell JA. Defensive symbiosis in the real world – advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol (2014) 28:341–55. doi:10.1111/1365-2435.12133
161. Kaplan JL, Shi HN, Walker WA. The role of microbes in developmental immunologic programming. Pediatr Res (2011) 69:465–72. doi:10.1203/PDR.0b013e318217638a
162. Kelly D, King T, Aminov R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res (2007) 622:58–69. doi:10.1016/j.mrfmmm.2007.03.011
163. Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O’Leary CE, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med (2014) 20:524–30. doi:10.1038/nm.3542
164. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science (2012) 336:489–93. doi:10.1126/science.1219328
165. Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell (2016) 165:842–53. doi:10.1016/j.cell.2016.04.007
166. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med (2014) 20:642–7. doi:10.1038/nm.3568
167. Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med (2016) 22:713–22. doi:10.1038/nm.4142
168. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med (2016) 8:343ra82. doi:10.1126/scitranslmed.aad7121
169. Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol (1998) 43:17–37. doi:10.1146/annurev.ento.43.1.17
170. Nalepa CA. Origin of termite eusociality: trophallaxis integrates the social, nutritional, and microbial environments. Ecol Entomol (2015) 40:323–35. doi:10.1111/een.12197
171. Wong AC-N, Luo Y, Jing X, Franzenburg S, Bost A, Douglas AE. The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster. Appl Environ Microbiol (2015) 81:6232–40. doi:10.1128/AEM.01442-15
172. Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, et al. Emergence shapes the structure of the seed microbiota. Appl Environ Microbiol (2015) 81:1257–66. doi:10.1128/AEM.03722-14
173. Shade A, Jacques M-A, Barret M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr Opin Microbiol (2017) 37:15–22. doi:10.1016/j.mib.2017.03.010
174. Truyens S, Weyens N, Cuypers A, Vangronsveld J. Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep (2015) 7:40–50. doi:10.1111/1758-2229.12181
175. Develey-Rivière M-P, Galiana E. Resistance to pathogens and host developmental stage: a multifaceted relationship within the plant kingdom. New Phytol (2007) 175:405–16. doi:10.1111/j.1469-8137.2007.02130.x
176. Panter SN, Jones DA. Age-related resistance to plant pathogens. Adv Bot Res (2002) 38:251–80. doi:10.1016/S0065-2296(02)38032-7
177. Whalen MC. Host defence in a developmental context. Mol Plant Pathol (2005) 6:347–60. doi:10.1111/j.1364-3703.2005.00286.x
178. Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS. Seasonal community succession of the phyllosphere microbiome. Mol Plant Microbe Interact (2015) 28:274–85. doi:10.1094/MPMI-10-14-0331-FI
179. Tkacz A, Cheema J, Chandra G, Grant A, Poole PS. Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J (2015) 9:2349–59. doi:10.1038/ismej.2015.41
180. Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, et al. Cospeciation of gut microbiota with hominids. Science (2016) 353:380–2. doi:10.1126/science.aaf3951
181. Schardl CL, Leuchtmann A, Chung K-R, Penny D, Siegel MR. Coevolution by common descent of fungal symbionts (Epichloe spp.) and grass hosts. Mol Biol Evol (1997) 14:133–133. doi:10.1093/oxfordjournals.molbev.a025746
182. Brundrett MC. Coevolution of roots and mycorrhizas of land plants. New Phytol (2002) 154:275–304. doi:10.1046/j.1469-8137.2002.00397.x
183. McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol (2012) 10:13–26. doi:10.1038/nrmicro2670
184. Berg G, Grube M, Schloter M, Smalla K. Unraveling the plant microbiome: looking back and future perspectives. Front Microbiol (2014) 5:148. doi:10.3389/fmicb.2014.00148
185. Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J (2012) 6:1378–90. doi:10.1038/ismej.2011.192
186. Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J (2010) 4:719–28. doi:10.1038/ismej.2010.9
187. Rothschild D, Weissbrod O, Barkan E, Korem T, Zeevi D, Costea PI, et al. Environmental factors dominate over host genetics in shaping human gut microbiota composition. bioRxiv (2017):150540. doi:10.1101/150540
Keywords: timing of exposure, microbiome, defensive symbiont, microbiome transmission, microbiome variation
Citation: Morella NM and Koskella B (2017) The Value of a Comparative Approach to Understand the Complex Interplay between Microbiota and Host Immunity. Front. Immunol. 8:1114. doi: 10.3389/fimmu.2017.01114
Received: 01 May 2017; Accepted: 24 August 2017;
Published: 14 September 2017
Edited by:
Larry J. Dishaw, University of South Florida St. Petersburg, United StatesReviewed by:
Spencer V. Nyholm, University of Connecticut, United StatesSilke Paust, Baylor College of Medicine, United States
Copyright: © 2017 Morella and Koskella. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Norma M. Morella, bW9yZWxsYUBiZXJrZWxleS5lZHU=
 Norma M. Morella
Norma M. Morella Britt Koskella
Britt Koskella