- 1Laboratorio de Terapia Molecular y Celular, Fundación Instituto Leloir, Instituto de Investigaciones Bioquímicas de Buenos Aires (IIBBA-CONICET), Ciudad Autónoma de Buenos Aires, Argentina
- 2Laboratorio Nanomedicina, Gerencia de Desarrollo Tecnológico y Proyectos Especiales, Comisión Nacional de Energía Atómica, Ciudad Autónoma de Buenos Aires, Argentina
Monoclonal antibodies and their fragments have significantly changed the outcome of cancer in the clinic, effectively inhibiting tumor cell proliferation, triggering antibody-dependent immune effector cell activation and complement mediated cell death. Along with a continued expansion in number, diversity, and complexity of validated tumor targets there is an increasing focus on engineering recombinant antibody fragments for lead development. Single-domain antibodies (sdAbs), in particular those engineered from the variable heavy-chain fragment (VHH gene) found in Camelidae heavy-chain antibodies (or IgG2 and IgG3), are the smallest fragments that retain the full antigen-binding capacity of the antibody with advantageous properties as drugs. For similar reasons, growing attention is being paid to the yet smaller variable heavy chain new antigen receptor (VNAR) fragments found in Squalidae. sdAbs have been selected, mostly from immune VHH libraries, to inhibit or modulate enzyme activity, bind soluble factors, internalize cell membrane receptors, or block cytoplasmic targets. This succinct review is a compilation of recent data documenting the application of engineered, recombinant sdAb in the clinic as epitope recognition “modules” to build monomeric, dimeric and multimeric ligands that target, tag and stall solid tumor growth in vivo. Size, affinity, specificity, and the development profile of sdAbs drugs are seemingly consistent with desirable clinical efficacy and safety requirements. But the hepatotoxicity of the tetrameric anti-DR5-VHH drug in patients with pre-existing anti-drug antibodies halted the phase I clinical trial and called for a thorough pre-screening of the immune and poly-specific reactivities of the sdAb leads.
Introduction
The success of monoclonal antibodies (mAbs) in cancer therapy is driven by the overall efficacy of targeted therapies. The rate of approval of recombinant mAbs continues to outperform that of small molecules in all indications and in particular for the treatment of cancer (1, 2). However, a recent rate of advancement of antitumor candidate leads from preclinical to clinical trial was estimated to be only 20% (3). One approach to improving this success rate is to focus early on a set of characteristics termed “developability” based on high-throughput qualification tests applicable to mAb hits for a particular target. Two “developability” issues impacting candidate bioavailability are off-target binding and aggregation that can also result in toxicity and immune-reactivity. A candidate with a favorable profile is more likely to emerge from a large set of hits with a broad epitope coverage, by screening out off-target reactive mAbs (4) and guaranteeing “manufacturability,” or stability and solubility, of the lead candidate early in the pipeline (5–8). Camel and shark serum have provided a source of versatile antibody therapeutics with good “developability” and “manufacturability” prospects (6, 9–11). Most recombinant, variable heavy-chain (or VHH) single domains from homodimeric IgG2 and IgG3 found in camelids and VNAR of the so-called Ig new antigen receptor of sharks display higher solubility (above 1 mg/mL) and rapid refolding after temperature or chemical denaturation in comparison with the heterodimeric VL–VH domains in a Fab fragment (Figure 1A) (12, 13). VHH expression yield, whether in the periplasm of Escherichia coli or the cytoplasm of eukaryotic cells is high. Sequence identity of the VNAR domain with canonical human VH falls as low as 25%, while known camelid VHH domains are distinctly close to human VH3 germline sequences and a source of easily humanized single-domain antibody (sdAb) drugs (10, 14–16). In addition, services such as Hybribody, a platform from Hybrigenics for the selection and validation of antibodies derived from a fully synthetic humanized sdAb library displayed on phage, can supply humanized sdAbs to specific targets (Table 1, item 3) (17). The immunogenicity of humanized sdAbs may be erroneously overlooked yet it is tested in phase I clinical trials (18). The antigen-specific combining sites may be immunogenic providing sufficient justification for the early use of immunogenicity-screening platforms (19). The detection of anti-drug antibodies (ADA) using highly sensitive ELISAs at Ablynx revealed the benefit of mutating sdAb residues in hydrophobic patches at the C-terminus of VH of single-chain variable fragment (scFv) and VHH fragments, shielded by the CH domains in the original structure (20, 21).
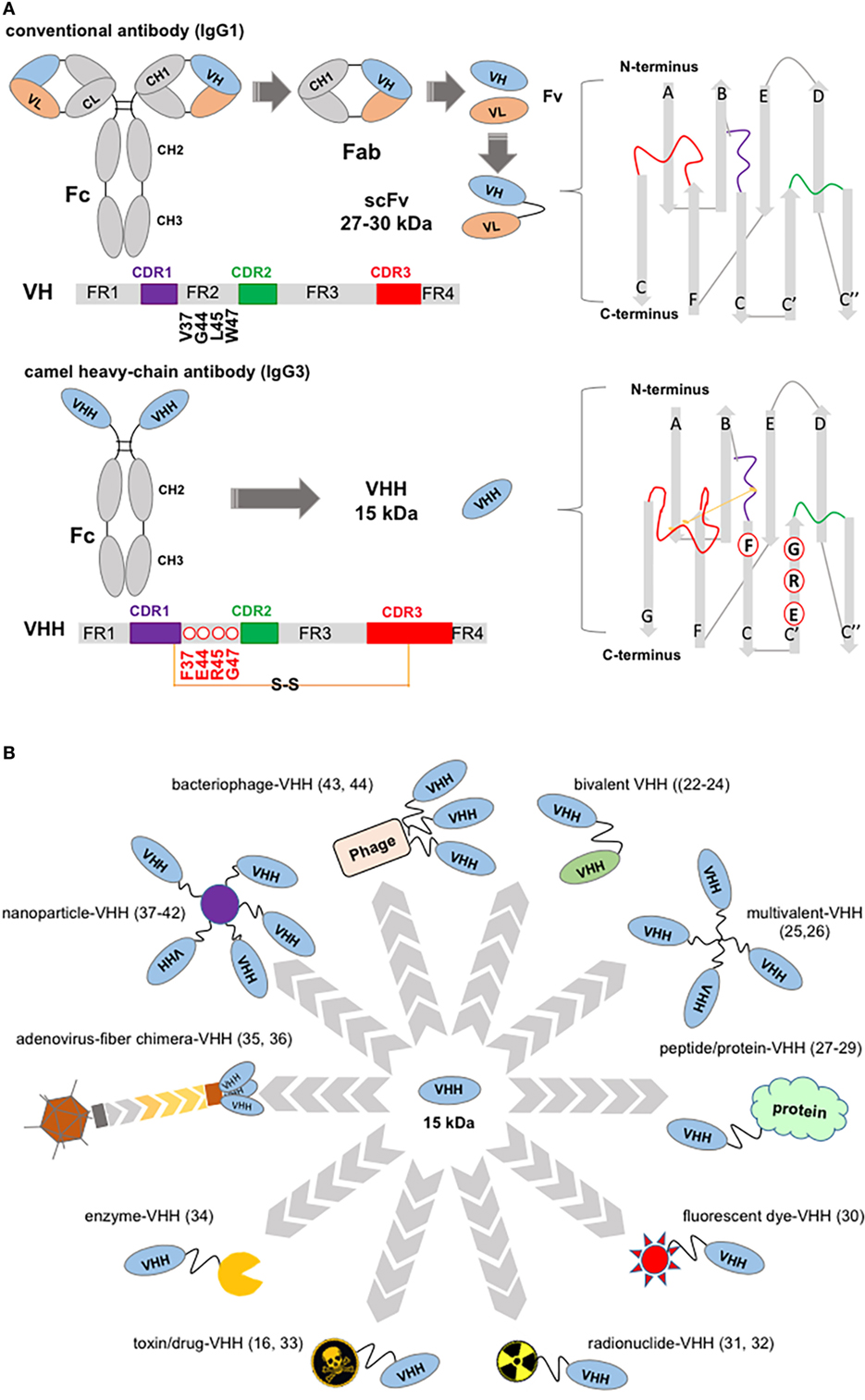
Figure 1. Structure of a “conventional” IgG1 and of a camelid IgG3, showing variable domain differences and illustrations of potential, VHH-based, cancer therapeutics. (A) Schematic of an IgG1 showing canonical hypervariable domains (left top diagram) consisting of two light (L) chains, comprising the VL and CL domains, and two heavy (H) chains composed of the VH, CH1, hinge, and CH2 and CH3 domains; and, below a camelid homodimeric heavy-chain IgG3, a heavy-chain antibody (HCAb) (left bottom diagram) which comprises only H chains; each H chain contains a short VHH hinge, CH2, and CH3 domains. The homodimeric heavy-chain IgG2 (not shown) has longer VHH hinge domains compared to IgG3 and comparable CH2, CH3. The smallest intact functional antigen-binding fragment that can be generated from the immunoglobin G (IgG) canonical variable domains, consists of an oligopeptide linked VH–VL pair known as single-chain variable fragment (top right), while the smallest intact functional antigen-binding fragment of HCAbs is the single-domain VHH (bottom right) known as Nb. VH and VHH bars show framework (FR), complementarity domain regions (CDRs) (color coded), and key residues substitutions. Non-canonical C residues are involved in an inter-CDR disulfide bond in VHH structure. (B) VHH-associated strategies in targeting tumors and tumor accessory cells. Top, clockwise: bivalent bi-specific VHH (22–24); multivalent, high-avidity mono-VHH molecules (25, 26); VHH fusions ranging from vascular penetration peptide-VHH to engineered hu-Fab and albumin-binding domains (27–29); fluorescent dye fusions, for example, one spontaneously crossing the blood–brain barrier (30); radionuclide-VHHs (31, 32); toxin-VHH theragnostics (16, 33); chromogenic enzyme fusions: here an alkaline phosphatase-VHH may be applied in ELISA, dot blot, and transferred protein identification in western blot (34); oncolytic virus (35, 36); VHH decorated nanoparticles for therapeutics delivery and in facilitating photothermal therapy (37–42); bacteriophage engineered to display VHH and deliver targeted therapeutics (43) may also be developed for signal amplification in ELISA assays (44).
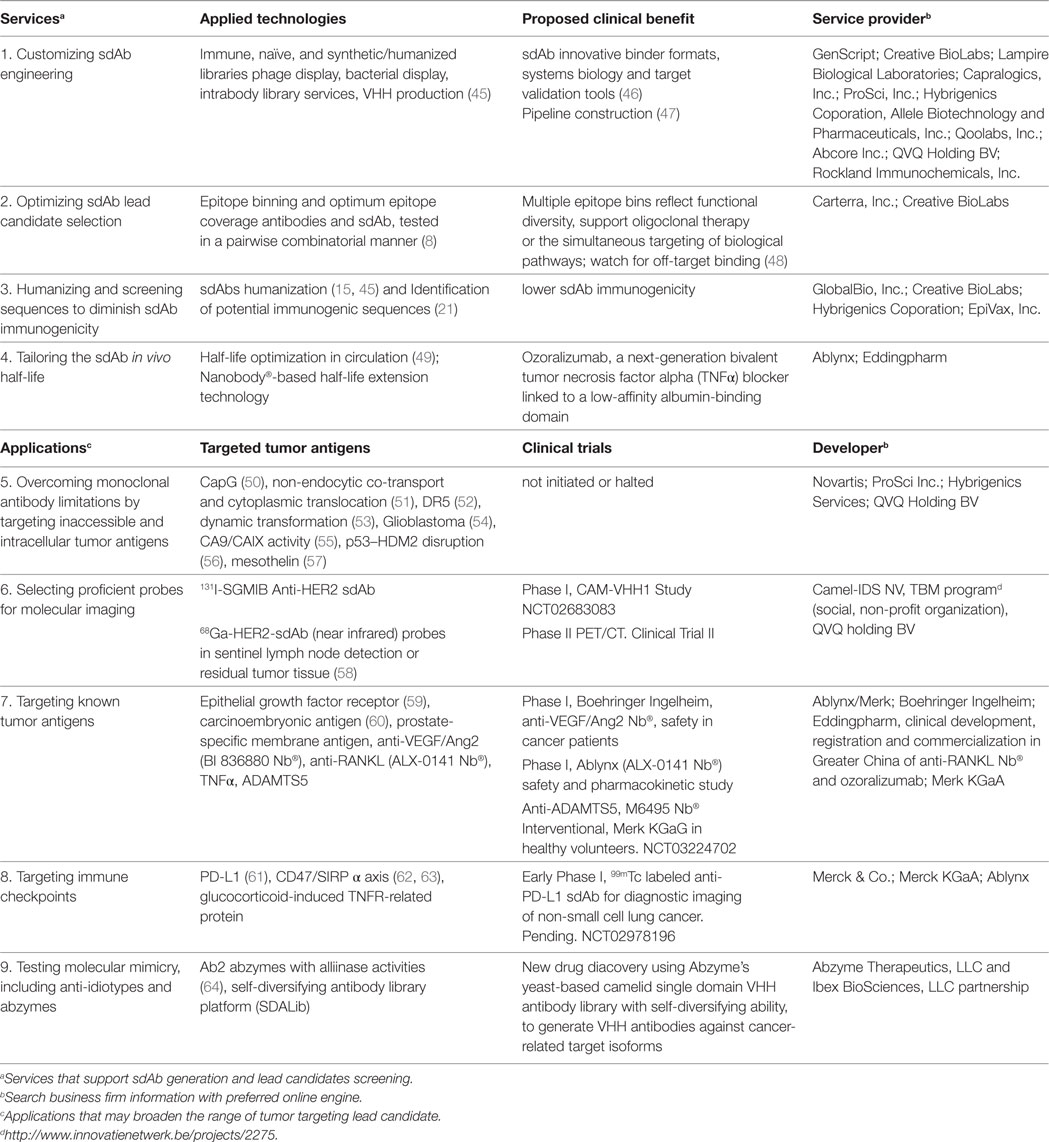
Table 1. Summarized single-domain antibody (sdAb) research and development in cancer diagnostics and therapy.
The VHH repertoire is as complex in sequence diversity as is the IgG1 VH camelid counterpart (65–67). Total peripheral blood lymphocytes and lymph node ribonucleic acid (RNA) from alpaca, llamas, dromedaries, and camels are easily extracted to build recombinant VHH libraries. Typically, a VHH phage display library containing 6 × 107 VHHs clones are generated from 200 µg processed RNA and diverse polymerase chain reaction strategies are available to amplify VHH gene fragments from lymphocyte complementary deoxyribonucleic acid (68, 69). Several reports have confirmed the ease of engineering sdAbs (69, 70) and of selecting specific binders against conformational epitopes in comparison with hit selection of scFv, where library construction shuffles their immune specificity (68, 71, 72).
Two or three VHHs have been combined in a single polypeptide chain to express single, dual, or multimeric specificities without compromising folding or the binding affinities (22, 73). In addition, “self-associating peptide” constructs have been designed to match VHH pairs (69, 74). Concomitantly, the experience gained in site-specific conjugations, in particular those driven by targeted enzymatic reactions, has ensured the preservation of antigen-binding properties of sdAbs (31, 75). The reported affinities of VHH fragments fall in the nanomolar to picomolar range, with binding kinetics comparable to those of conventional antibodies. Selection of stable antigen complexes is often the result of applying selection pressures, such as stringent washing, that enrich a library in VHH with slower off-rates while competitive elution was reported in selecting fragments with novel epitope targeting (70, 76–79). VHH genes are an established source of antibodies, as evidenced by the number of reported co-crystal structures (68, 80–82). Figure 1A highlights hallmark VHH residues and, when present, an inter-CDR disulfide bond in the VHH sequence. Around 10% of HcAbs lack these hydrophobic residues mutation but often show longer CDR3 covering putative VL contacts or a hydrophilic substitution of Trp118. Gonzalez-Sapienza et al. suggested a plausible mechanism of selection of HcAb producing B-cells that supports the emergence of independently folding, soluble VH and VHH domains (72).
Distinctive Properties of sdAbs
The ease of selecting sdAb under denaturing conditions has assisted in the isolation of “superstable” species with improved resistance to proteases that were proposed as antimicrobial therapeutics of oral intake (83, 84). Li et al. have successfully selected VHH expression products with a high isoelectric point (pI) that spontaneously crossed the blood–brain barrier (transcytosis) (30). High-pI sdAb have been found to penetrate cells and bind to intracellular proteins. For instance, a sdAb that bound specifically to the hepatitis C virus (HCV) protease, selected for its ability to penetrate cells (transbodies), interfered with heterologous HCV replication (15). A sdAb-based anti-β-catenin intrabody was expressed and folded in the cytoplasm retaining its ability to bind to β-catenin (85).
The solvent accessible surface (SAS) area of antigen-VHH and VNAR complexes are comparable to antigen–VH–VL complex SAS indicating that complementarity domain region (CDR) loops involved in antigen binding (Figure 1A) contribute similar surface contacts. VHH H1 and H3 loops connecting the β-sheets of the VHH domain are flexible, sometimes longer and packed in a less compact fashion compared to canonical VH of murine and human immunoglobin G (IgGs) (10, 86). Co-crystal structures of enzyme-VHH and -VNAR complexes showed CDRs that often protruded into the active-site cleft and the derived sdAbs were later shown to inhibit catalysis (65, 66, 87, 88). Alternatively, sdAbs have been selected to stabilize “drugable” targets that display multiple conformations (or conformational plasticity) (79, 82). For example, the urokinase-type plasminogen activator (uPA) from the trypsin-like serine protease family, a target involved in metastasis, is known to adopt high and low activity conformations. Selection of sdAbs against mouse uPA yielded both a catalytic-site inhibitor and an allosteric ligand. Crystal structures of the uPA sdAb complexes revealed high and low activity determinants that provided clues of therapeutic value on the regulatory determinants of uPA and of trypsin-like serine proteases in general (89). Table 1 documents the pharmaceutical relevance of sdAbs through the number of research and development companies involved in novel sdAb generation, available contract services, lead candidates under clinical trial, and examples of the sdAbs more recently generated against cancer targets.
sdAbs in Imaging Applications for Cancer Diagnostics
Molecular imaging techniques, of widespread use in the clinic, allow the non-invasive quantitation and visualization of tumors in vivo and sdAbs have become promising, small-sized, high-affinity tracers (58, 90–92) (Figure 1B). Nuclear imaging probes associated to sdAbs have been evaluated in both single-photon emission computed tomography (SPECT) and positron emission tomography (PET) (90, 93) (Table 1, item 6). The most advanced sdAb under clinical evaluation is the 68Ga-labeled anti-HER2 sdAb 2Rs15d probe, developed to screen candidates who qualify for treatment with an anti-HER2 therapeutics. A phase I study resulted in high-quality images without adverse reactions and retained 10% of injected activity in blood after 1 h (94). A phase II trial was launched to correlate tumor uptake with HER2 levels in biopsies of 160 metastatic breast carcinoma patients (Table 1, item 6). In other studies, 2Rs15d labeled with the prosthetic group N-succinimidyl-4-[18F] fluorobenzoate ([18F]-SFB) was validated in preclinical models to advance PET imaging (95). The specific uptake of the sdAb 2Rs15d probe in HER2-positive tumor xenografts showed high tumor-to-blood and tumor-to-muscle ratios, high contrast PET imaging and fast renal clearance (4% intra auricular/g at 3 h post injection.). The lead candidate MSB0010853, a biparatopic sdAb labeled with 89Zr bound efficiently to HER3 kinase, a potential clinical target associated with resistance to epithelial growth factor receptor (EGFR) and HER2 targeted therapies (96, 97).
Organometallic radiopharmaceuticals are also widely used in diagnosis with SPECT imaging. sdAbs that target either EGFR (98), VCAM1, an 8-kDa fragment of gelsolin or carcinoembryonic antigen (CEA) have been conjugated with 99mTc (99). Recently, an anti-PD-L1 sdAb labeled with 99mTc discriminated wild type mice from PD-L1 knock-out mice by SPECT/CT imaging (100). sdAbs used as fluorescence-guided near-infrared wavelength range (NIR) probes are also under preclinical studies addressing sentinel lymph node imaging quality and guiding surgical/endoscopic removal of residual tumor tissue (101). NIR probes, IRDye800CW or IRDye680RD, were conjugated either by lysines or C-terminal cysteine to the 7D12 anti-EGFR sdAb. After IR dye conjugation, comparable specificities and affinities of 7D12 and the conjugate were measured toward EGFR in vitro (58, 102). This study also showed an accumulation of the cysteine-conjugated 7D12 in A431 human tumor xenografts in nude mice or high tumor-to-muscle ratio.
The ultrasound imaging of vessel cell adhesion protein 1 (VCAM1), using specific sdAbs coupled to lipid microbubbles as contrast enhancers, is used to assess potential adhesion sites of melanoma cell extravasation and metastasis (75). Although sdAbs are promising imaging probes renal retention during clearance and toxicity were reported in preclinical studies. Adverse effects were attributed to the polar residue number favoring the interaction with the megalin/cubilin system in the renal tubuli (103). This issue was overcome by mutating positive residues, facilitating filtration at the negatively charged glomerular membrane (104). Toxicity was also controlled by gelofusine or lysine added to the probe (103, 105).
sdAb Against Tumor Targets for Clinical Use
Single-domain antibodies that bind either hepatocyte growth factor, EGFR, bone morphogenetic protein (TGFb superfamily growth factors), HER2, cMET, or VEGFR1, have been shown to efficiently block tumor cell proliferation (81, 106–109). Zhang et al. (61) have recently shown that KN035, an anti-PD-L1 sdAb, can induce T-cell responses and inhibit tumor growth; the KN035 CDRs structure is remarkably similar to that of the VH of Federal Drug Administration-approved Durvalumab (110). Other sdAbs were developed to target uPA, and chemokine receptors such as CXCR4 and CXCR7 (111). More recently, sdAbs targeting antioxidant enzymes such as membrane catalase and superoxide dismutase were selected for their ability to induce reactive oxygen species-dependent cancer cell apoptosis and found to be synergetic to chemotherapy (112).
Single-domain antibodies modules have been engineered into multivalent structures to overcome fast clearance. The anti-DR5 sdAb tetramer showed excellent pharmacokinetics and efficacy in preclinical models, inducing robust antitumor responses and sustained caspase activation in vivo. However, in the phase I trial an unexpected hepatotoxicity which triggered hepatocyte apoptosis, later associated to the immune crosslinking of the tetramer in those patients with pre-existing ADA, prompted its discontinuation (113). A bifunctional sdAb, targeting EGFR and TRAIL, inhibits the growth of different tumor cell types that were not responsive to either EGFR-antagonist or death receptor-agonist monotherapies is a clear step forward of the clinical application of sdAb modules (23). To improve the efficacy of a bifunctional therapeutic, the MaAbNA-PEG2000-ADM chimera consisting of an anti-EGFR1 sdAb linked to two anti-HER2 affibodies was conjugated with Adriamycin (114). The bispecific sdAb chimera recognizing CEA and antigen cluster of differentiation 16 (CD16) (NK-cell marker) was linked to a mutated human IgG1 Fc-fragment that equipped the dimer with an effector function (115). The bispecific antibody HER2-S-Fab, an anti-CD16 sdAb that is linked to a anti-trastuzumab Fab, also exhibited a potent tumor growth inhibition in a human tumor xenografts model (29). A multivalent, sdAb-based, in-tandem trimer was capable of simultaneously binding to CEA, EGFR, and green fluorescence protein with high efficacy for inhibition of human epidermoid carcinoma A431 cell proliferation (26). An interesting approach to increase the half-life of sdAbs without affecting the affinity for its target was the fusion between an anti-TNFα sdAb with an albumin-binding domain derived from Streptococcus zooepidemicus (~39-fold half-life increase with respect to the sdAb alone, Table 1, item 4) (28).
Targeting tumors with ionizing radiation is also a promising area for growth for sdAb therapeutics. The most relevant in vivo study demonstrated that i.v. administration of the sdAb anti-HER2 labeled with 177Lu, a γ-emission radionuclide, completely prevented tumor growth in mice with small HER2-positive tumors (32). The α-emitting radionuclides 213Bi and 211At coupled to sdAbs are tentatively used to treat minimal residual disease and micro-metastasis and their clinical application is being intensely explored (116).
Emerging Drug-Delivery Strategies That Use sdAbs
To improve solid tumor penetration an EGFR-targeted sdAb was fused to an iRGD, a cyclic domain selective of αvβ3 and αvβ5 integrins that carries a CendR motif that binds neuropilin 1 (NRP-1) (117). The efficacy of this construct was measured in BGC-823 multicellular spheroids that overexpress EGFR, NRP-1, and integrins. The anti-EGFRsdAb-iRGD showed better performance in reducing spheroid size than anti-EGFRsdAb or cetuximab. In vivo, anti-EGFRsdAb-iRGD-FITC was shown to bind to αvβ3 and αvβ5 expressed in the tumor vessels, malignant cells, and cancer-associated stromal cells, penetrating further than the anti-EGFR-FITC (27). Recently, anti-EGFRsdAb-iRGD was conjugated to silk fibroin nanoparticles loaded with paclitaxel, resulting in a significant anti-neoplastic activity in EGFR-expressing cells in vitro and in vivo (41).
Single-domain antibody has been successfully used to retarget oncolytic adenovirus to a non-cognate receptor following the incorporation of an anti-CEA sdAb into the adenovirus capsid fiber (Figure 1B). This modification was shown to control viral tropism, entry, and gene transfer specifically in CEA-overexpressing cells (36, 118). sdAb displayed on genetically engineering phage combined with target drugs or imaging probes has recently been proposed for preclinical evaluation (43, 119).
Single-domain antibodies have been used to retarget nanoparticles with particular diagnostic or therapeutic properties (120, 121). Branched gold nanoparticles functionalized with an anti-prostate-specific antigen sdAb were shown to destroy cancer cells in response to laser irradiation in a preclinical model of photothermal therapy (37). Pegylated liposomes, schematized in Figure 1B, may be re-directed away from the reticuloenthoelial system by coupled sdAbs and are under preclinical evaluation as drug nanocarriers (39, 40). A novel potent delivery system based on extracellular vesicles (EVs) has recently been described where an anti-EGFR sdAb was anchored on the surface of EVs via glycosylphosphatidylinositol signal peptides derived from the decay-accelerating factor significantly improving EV targeting (42).
Platforms for the Generation of New sdAbs
Epitope recognition and coverage appear to be dependent on immune-selection pressure of VH and VHH sequences in vivo and by the library diversity (122, 123). To amplify antigenic epitope coverage, naïve and semi-synthetic libraries are being promoted to amplify antigen epitope coverage often limited by B-cell IgG amplification in vivo. Low affinities may be matured or optimized as required. sdAb discovery may now count on high-throughput, high-resolution broad epitope coverage analysis and poly-specificity and affinity screening tools to increase the likelihood of selecting sdAbs with the desired therapeutic functions (Table 1, item 2) as well as to discriminate between functional sdAbs, such as those that can trigger receptor internalization (124) and polyreactive leads (8).
Three novel VHH library presentation and selection platforms have been recently proposed for a high-throughput selection of sdAb to integral membrane tumor antigens, or proteins overexpressed on the surface of whole cells or on virus-like particles (70, 123). Two of the platforms were designed to identify binders to antigen diluted in lysates or in complex mixtures for the discovery of sdAbs that bind critical pathway targets (78, 125). Rosotti et al. reported high throughput, parallel selection and characterization strategies to identify phage-displayed sdAbs against receptors expressed on murine bone marrow-derived dendritic cells (123). As a result of en masse cloning and whole-cell screening, the in vivo biotinylation of selected VHH facilitated the identification of targets. The isolated VHH were effectively mapped, or binned, by epitope, and target coverage was recorded [also see Ref. (126), Table 1, item 2].
Salema and Fernandez optimized the display of VHH on Gram-negative E. coli, and the direct expression of selected VHH clones, by anchoring the expression product on the outer membrane by fusing to the N-terminal, intimin β-domain (Neae) (78, 127, 128). High-affinity clone selection was optimized by magnetic cell sorting on immobilized recombinant biotinylated antigen (MACS) or by flow cytometry on whole cells (CellS) (78).
A third sdAb selection platform was presented by Cavallari using a Gram-positive Staphylococcal (Staphylococcus aureus) display of sdAb (125). Here, VHH clones were engineered with the signal peptide from staphylococcal enterotoxin B, with the sortase A (SrtA) LPXTG motif, to display folded VHH on the surface. Endogenous SrtA covalently, and irreversibly, coupled expressed sdAb on the outer membrane. A nucleophilic attack of the SrtA sdAb-acyl intermediate by polyglycine nucleophile-biotin was used to release and biotinylate selected VHH clones. The major advantages of bacterial display were the efficiency of selection as reflected by a high “hit” frequency, or high frequency of success, in comparison to hit selection by phage display, and minimum avidity. Also attractive is the choice of evaluating selected sdAbs by flow cytometry or in SPR binding assays directly enabling screening sdAbs by epitope and a discrimination of poly-specificity in a high-throughput mode (78, 128).
Concluding Remarks
Single-domain antibodies are soluble, stable, recombinant proteins that fold independently and display an outstanding versatility. The hardware-building concept of “plug and play” appears as an excellent paradigm in which sdAbs are part of a therapeutics generation tool kit that includes engineered recombinant sdAbs, radionuclides, dyes, peptides, proteins, nanostructures, phage, and virus.
Currently, 20–25% of the mAbs in clinical development for cancer and non-cancer indications are recombinant human antibodies derived from phage display libraries or from transgenic mice. Five antibody “fragments” (scFv) were reported in clinical phase 2/3 this past year. These include a human scFv-doxorubicin loaded liposome; two scFv conjugates, a humanized anti-EpCAM scFv-immunotoxin conjugate; and an anti-fibronectin extra-domain B human scFv for cancer indications.
The unexpected toxicity of the anti-DR5 tetramer, TAS266, opened the question of pre-existing immunity against sdAb. This issue has been addressed by developing sensitive immune serum assays and immunogenicity-screening platforms (Table 1, item 3, EpiVax) to identify the safer lead candidates, helping reduce the risk of clinical trial failure of sdAb-based drugs. The promise of recombinant, engineered, antibody-based building modules with optimal efficacy and biovailability may soon translate into tangible cancer drugs.
Author Contributions
MI, LP, SW, OP, and GC: conception, design, and writing of the review manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge Yasmina Abdiche, Ph.D., Ann Rux, Ph.D., and John Rux, Ph.D. for the insightful critique of the manuscript. The authors recognize the assistance of Dr. Monica Perez, Director of the Cardini Library at Fundacion Instituto Leloir for facilitating our searches and in managing our information resources.
Funding
MI, LP, OP, and GC are members of Consejo Nacional de Investigaciones Cientificas y Tecnologicas (CONICET), Argentina. This work was supported by grants from CONICET, Fondo Argentino Sectorial (FONARSEC), Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT), Instituto Nacional del Cáncer, Ministerio de Salud de la Nación Argentina (INC-MSal), and Fundación Florencio Fiorini.
Abbreviations
A431, epidermoid carcinoma cell line; Abzyme, an antibody with catalytic activity, binding a chemical group and stabilizing the transition state of a given reaction; ADA, anti-drug antibody; ADAMTS5, a disintegrin and metalloproteinase with thrombospondin motifs; ADM, Adriamycin; BMCD, bone marrow culture-derived macrophages; BMP, bone morphogenetic protein; CA9/CAIX, carbonic anhydrase IX encoded by the CA9 gene; CD47/SIRP α axis, cluster of differentiation 47 and the myeloid inhibitory immunoreceptor signal regulatory protein α signaling axis; CapG, macrophage capping protein oncogene; CD16, cluster of differentiation 16, a low-affinity Fc receptor; CDR, complementarity determining region or antigen-binding domain; CendR, C-end rule motif R/KXXR/K; CEA, carcinoembryonic antigen; cDNA, complementary deoxyribonucleic acid; cMET, tyrosine-protein kinase Met or hepatocyte growth factor receptor (HGFR); CXCR4, fusin or CD184; CXCR7, atypical chemokine receptor 3 or GPCR 159; DAF, decay-accelerating factor; DR5, death receptor 5 of the TNF receptor superfamily (TNFRSF) 5; EGFR, epidermal growth factos receptor, a membrane tyrosine kinase; EpCAM, epithelial cell adhesion molecule or TROP1; EV, extracellular vesicle; Fab, immunoglobulin antigen-binding fragment composed of one constant and one variable domain of each of the heavy and the light chain; Fc, fragment crystallizable region of Ig; FDA, Federal Drug Administration; FR, framework region is a subdivision of the mAb variable region; FTCI, fluorescein isothiocyanate; GFP, green fluorescence protein; GITR, glucocorticoid-induced TNFR-related protein; GPI-DAF, glycosylphosphatidylinositol-anchored decay-accelerating factor; HcAbs, heavy-chain antibodies; HCV, hepatitis C virus; HER2, human epidermal growth factor receptor 2/neu tyrosine kinase, erbB-2; HER3, human epidermal growth factor receptor tyrosine-protein kinase erbB-3; HGF, hepatocyte growth factor; IA, intra auricular; IgG, immunoglobin G; IgNAR, Ig new antigen receptor; IR, infrared; 131I-SGMIB, iodine-131- labelled N-succinimidyl 4-guanidinomethyl-3-iodobenzoate; iRGF, 9-amino acid cyclic peptide (sequence: CRGDKGPDC) binding tumor cells; i.v., intra venous; mAb, monoclonal antibody; Neae, N-terminal fragment of enterohemorrhagic E. coli intimin; NIR, near infrared; NRP-1, neuropilin 1; PBL, peripheral blood lymphocytes; PCR, polymerase chain reaction; PEG2000, poly(ethylene glycol) methyl ether, average Mw 2,000; PD-L1, Programmed death-ligand 1, CD274; p.i., post injection; pI, isoelectric point; PSA, Prostate-specific antigen; PSMA, prostate-specific membrane antigen; p53-HDM2, functional p53 and human double minute 2 interaction; SAS, solvent accessible surface; scFv, Single-chain variable fragment; sdAb, single-domain antibody fragment; SPECT, single-photon emission computed tomography; SPECT/CT, image fusion for anatomic imaging (CT or MRI) and functional imaging (SPECT) computed tomographies; SrtA, sortase A; RANKL, receptor activator of nuclear factor kappa-B ligand; RNA, ribonucleic acid; TNFα, tumor necrosis factor alpha; uPA, urokinase-type plasminogen activator; VCAM1, vascular cell adhesion protein 1; VEGF/Ang2, vascular endothelial factor/angiopoietin-2; VEGFR1, vascular endothelial growth factor receptor 1; VHH, heavy-chain only antibody fragment or nanobody; VH and VL, variable heavy and light chain domains from conventional IgG structures; VNAR, variable new antigen receptor single domain.
References
1. Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol (2014) 32(1):40–51. doi:10.1038/nbt.2786
2. Reichert JM. Antibodies to watch in 2017. MAbs (2017) 9(2):167–81. doi:10.1080/19420862.2016.1269580
3. Booth B. Human Antibody Discovery: Of Mice and Phage. Forbes (2017). Available from: https://www.forbes.com/sites/brucebooth/2017/05/11/human-antibody-discovery-of-mice-and-phage/-277cbb0c7f26
4. LaBute MX, Zhang X, Lenderman J, Bennion BJ, Wong SE, Lightstone FC. Adverse drug reaction prediction using scores produced by large-scale drug-protein target docking on high-performance computing machines. PLoS One (2014) 9(9):e106298. doi:10.1371/journal.pone.0106298
5. De Genst E, Messer A, Dobson CM. Antibodies and protein misfolding: from structural research tools to therapeutic strategies. Biochim Biophys Acta (2014) 1844(11):1907–19. doi:10.1016/j.bbapap.2014.08.016
6. Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, et al. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci U S A (2017) 114(5):944–9. doi:10.1073/pnas.1616408114
7. Kaelin WG Jr. Common pitfalls in preclinical cancer target validation. Nat Rev Cancer (2017) 17(7):425–40. doi:10.1038/nrc.2017.32
8. Sivasubramanian A, Estep P, Lynaugh H, Yu Y, Miles A, Eckman J, et al. Broad epitope coverage of a human in vitro antibody library. MAbs (2017) 9(1):29–42. doi:10.1080/19420862.2016.1246096
9. Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, et al. Naturally occurring antibodies devoid of light chains. Nature (1993) 363(6428):446–8. doi:10.1038/363446a0
10. Finlay WJ, Almagro JC. Natural and man-made V-gene repertoires for antibody discovery. Front Immunol (2012) 3:342. doi:10.3389/fimmu.2012.00342
11. Ahmadvand D, Rasaee MJ, Rahbarizadeh F, Kontermann RE, Sheikholislami F. Cell selection and characterization of a novel human endothelial cell specific nanobody. Mol Immunol (2009) 46(8–9):1814–23. doi:10.1016/j.molimm.2009.01.021
12. Jespers L, Schon O, Famm K, Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat Biotechnol (2004) 22(9):1161–5. doi:10.1038/nbt1000
13. Feige MJ, Grawert MA, Marcinowski M, Hennig J, Behnke J, Auslander D, et al. The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc Natl Acad Sci U S A (2014) 111(22):8155–60. doi:10.1073/pnas.1321502111
14. Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem (2009) 284(5):3273–84. doi:10.1074/jbc.M806889200
15. Jittavisutthikul S, Thanongsaksrikul J, Thueng-In K, Chulanetra M, Srimanote P, Seesuay W, et al. Humanized-VHH transbodies that inhibit HCV protease and replication. Viruses (2015) 7(4):2030–56. doi:10.3390/v7042030
16. Yu Y, Li J, Zhu X, Tang X, Bao Y, Sun X, et al. Humanized CD7 nanobody-based immunotoxins exhibit promising anti-T-cell acute lymphoblastic leukemia potential. Int J Nanomedicine (2017) 12:1969–83. doi:10.2147/IJN.S127575
18. Brennan FR, Morton LD, Spindeldreher S, Kiessling A, Allenspach R, Hey A, et al. Safety and immunotoxicity assessment of immunomodulatory monoclonal antibodies. MAbs (2010) 2(3):233–55. doi:10.4161/mabs.2.3.11782
19. Wadhwa M, Knezevic I, Kang HN, Thorpe R. Immunogenicity assessment of biotherapeutic products: an overview of assays and their utility. Biologicals (2015) 43(5):298–306. doi:10.1016/j.biologicals.2015.06.004
20. Nieba L, Honegger A, Krebber C, Pluckthun A. Disrupting the hydrophobic patches at the antibody variable/constant domain interface: improved in vivo folding and physical characterization of an engineered scFv fragment. Protein Eng (1997) 10(4):435–44. doi:10.1093/protein/10.4.435
21. Harmsen MM, Ruuls RC, Nijman IJ, Niewold TA, Frenken LG, de Geus B. Llama heavy-chain V regions consist of at least four distinct subfamilies revealing novel sequence features. Mol Immunol (2000) 37(10):579–90. doi:10.1016/S0161-5890(00)00081-X
22. Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs (2017) 9(2):182–212. doi:10.1080/19420862.2016.1268307
23. Zhu Y, Bassoff N, Reinshagen C, Bhere D, Nowicki MO, Lawler SE, et al. Bi-specific molecule against EGFR and death receptors simultaneously targets proliferation and death pathways in tumors. Sci Rep (2017) 7(1):2602. doi:10.1038/s41598-017-02483-9
24. Nosenko MA, Atretkhany KN, Mokhonov VV, Efimov GA, Kruglov AA, Tillib SV, et al. VHH-based bispecific antibodies targeting cytokine production. Front Immunol (2017) 8:1073. doi:10.3389/fimmu.2017.01073
25. Zhu X, Wang L, Liu R, Flutter B, Li S, Ding J, et al. COMBODY: one-domain antibody multimer with improved avidity. Immunol Cell Biol (2010) 88(6):667–75. doi:10.1038/icb.2010.21
26. Alvarez-Cienfuegos A, Nunez-Prado N, Compte M, Cuesta AM, Blanco-Toribio A, Harwood SL, et al. Intramolecular trimerization, a novel strategy for making multispecific antibodies with controlled orientation of the antigen binding domains. Sci Rep (2016) 6:28643. doi:10.1038/srep28643
27. Sha H, Zou Z, Xin K, Bian X, Cai X, Lu W, et al. Tumor-penetrating peptide fused EGFR single-domain antibody enhances cancer drug penetration into 3D multicellular spheroids and facilitates effective gastric cancer therapy. J Control Release (2015) 200:188–200. doi:10.1016/j.jconrel.2014.12.039
28. Cantante C, Lourenco S, Morais M, Leandro J, Gano L, Silva N, et al. Albumin-binding domain from Streptococcus zooepidemicus protein Zag as a novel strategy to improve the half-life of therapeutic proteins. J Biotechnol (2017) 253:23–33. doi:10.1016/j.jbiotec.2017.05.017
29. Li A, Xing J, Li L, Zhou C, Dong B, He P, et al. A single-domain antibody-linked Fab bispecific antibody Her2-S-Fab has potent cytotoxicity against Her2-expressing tumor cells. AMB Express (2016) 6(1):32. doi:10.1186/s13568-016-0201-4
30. Li T, Bourgeois JP, Celli S, Glacial F, Le Sourd AM, Mecheri S, et al. Cell-penetrating anti-GFAP VHH and corresponding fluorescent fusion protein VHH-GFP spontaneously cross the blood-brain barrier and specifically recognize astrocytes: application to brain imaging. FASEB J (2012) 26(10):3969–79. doi:10.1096/fj.11-201384
31. Rashidian M, Wang L, Edens JG, Jacobsen JT, Hossain I, Wang Q, et al. Enzyme-mediated modification of single-domain antibodies for imaging modalities with different characteristics. Angew Chem Int Ed Engl (2016) 55(2):528–33. doi:10.1002/anie.201507596
32. D’Huyvetter M, Vincke C, Xavier C, Aerts A, Impens N, Baatout S, et al. Targeted radionuclide therapy with A 177Lu-labeled anti-HER2 nanobody. Theranostics (2014) 4(7):708–20. doi:10.7150/thno.8156
33. Behdani M, Zeinali S, Karimipour M, Khanahmad H, Schoonooghe S, Aslemarz A, et al. Development of VEGFR2-specific nanobody Pseudomonas exotoxin A conjugated to provide efficient inhibition of tumor cell growth. N Biotechnol (2013) 30(2):205–9. doi:10.1016/j.nbt.2012.09.002
34. Liu X, Xu Y, Wan DB, Xiong YH, He ZY, Wang XX, et al. Development of a nanobody-alkaline phosphatase fusion protein and its application in a highly sensitive direct competitive fluorescence enzyme immunoassay for detection of ochratoxin A in cereal. Anal Chem (2015) 87(2):1387–94. doi:10.1021/ac504305z
35. Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet (2007) 8(8):573–87. doi:10.1038/nrg2141
36. Kaliberov SA, Kaliberova LN, Buggio M, Tremblay JM, Shoemaker CB, Curiel DT. Adenoviral targeting using genetically incorporated camelid single variable domains. Lab Invest (2014) 94(8):893–905. doi:10.1038/labinvest.2014.82
37. Van de Broek B, Devoogdt N, D’Hollander A, Gijs HL, Jans K, Lagae L, et al. Specific cell targeting with nanobody conjugated branched gold nanoparticles for photothermal therapy. ACS Nano (2011) 5(6):4319–28. doi:10.1021/nn1023363
38. Altintas I, Heukers R, van der Meel R, Lacombe M, Amidi M, van Bergen En Henegouwen PM, et al. Nanobody-albumin nanoparticles (NANAPs) for the delivery of a multikinase inhibitor 17864 to EGFR overexpressing tumor cells. J Control Release (2013) 165(2):110–8. doi:10.1016/j.jconrel.2012.11.007
39. Talelli M, Oliveira S, Rijcken CJ, Pieters EH, Etrych T, Ulbrich K, et al. Intrinsically active nanobody-modified polymeric micelles for tumor-targeted combination therapy. Biomaterials (2013) 34(4):1255–60. doi:10.1016/j.biomaterials.2012.09.064
40. Kijanka M, Dorresteijn B, Oliveira S, van Bergen en Henegouwen PM. Nanobody-based cancer therapy of solid tumors. Nanomedicine (Lond) (2015) 10(1):161–74. doi:10.2217/nnm.14.178
41. Bian X, Wu P, Sha H, Qian H, Wang Q, Cheng L, et al. Anti-EGFR-iRGD recombinant protein conjugated silk fibroin nanoparticles for enhanced tumor targeting and antitumor efficiency. Onco Targets Ther (2016) 9:3153–62. doi:10.2147/OTT.S100678
42. Kooijmans SA, Aleza CG, Roffler SR, van Solinge WW, Vader P, Schiffelers RM. Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J Extracell Vesicles (2016) 5:31053. doi:10.3402/jev.v5.31053
43. Pires DP, Cleto S, Sillankorva S, Azeredo J, Lu TK. Genetically engineered phages: a review of advances over the last decade. Microbiol Mol Biol Rev (2016) 80(3):523–43. doi:10.1128/MMBR.00069-15
44. Goldman ER, Anderson GP, Bernstein RD, Swain MD. Amplification of immunoassays using phage-displayed single domain antibodies. J Immunol Methods (2010) 352(1–2):182–5. doi:10.1016/j.jim.2009.10.014
45. Moutel S, Bery N, Bernard V, Keller L, Lemesre E, de Marco A, et al. NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife (2016) 5:e16228. doi:10.7554/eLife.16228
46. Helma J, Cardoso MC, Muyldermans S, Leonhardt H. Nanobodies and recombinant binders in cell biology. J Cell Biol (2015) 209(5):633–44. doi:10.1083/jcb.201409074
47. Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods (2014) 11(12):1253–60. doi:10.1038/nmeth.3170
48. Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu TY, et al. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel (2013) 26(10):663–70. doi:10.1093/protein/gzt047
49. Hoefman S, Ottevaere I, Baumeister J, Sargentini-Maier ML. Pre-clinical intravenous serum pharmacokinetics of albumin binding and non-half-life extended nanobodies® antibodies. Antibodies (2015) 4:141–56. doi:10.3390/antib4030141
50. Van Impe K, Bethuyne J, Cool S, Impens F, Ruano-Gallego D, De Wever O, et al. A nanobody targeting the F-actin capping protein CapG restrains breast cancer metastasis. Breast Cancer Res (2013) 15(6):R116. doi:10.1186/bcr3585
51. Herce HD, Schumacher D, Schneider AFL, Ludwig AK, Mann FA, Fillies M, et al. Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat Chem (2017) 9(8):762–71. doi:10.1038/nchem.2811
52. Huet HA, Growney JD, Johnson JA, Li J, Bilic S, Ostrom L, et al. Multivalent nanobodies targeting death receptor 5 elicit superior tumor cell killing through efficient caspase induction. MAbs (2014) 6(6):1560–70. doi:10.4161/19420862.2014.975099
53. Maier J, Traenkle B, Rothbauer U. Real-time analysis of epithelial-mesenchymal transition using fluorescent single-domain antibodies. Sci Rep (2015) 5:13402. doi:10.1038/srep13402
54. Jovcevska I, Zupanec N, Urlep Z, Vranic A, Matos B, Stokin CL, et al. Differentially expressed proteins in glioblastoma multiforme identified with a nanobody-based anti-proteome approach and confirmed by OncoFinder as possible tumor-class predictive biomarker candidates. Oncotarget (2017) 8(27):44141–58. doi:10.18632/oncotarget.17390
55. van Brussel AS, Adams A, Oliveira S, Dorresteijn B, El Khattabi M, Vermeulen JF, et al. Hypoxia-targeting fluorescent nanobodies for optical molecular imaging of pre-invasive breast cancer. Mol Imaging Biol (2016) 18(4):535–44. doi:10.1007/s11307-015-0909-6
56. Herce HD, Deng W, Helma J, Leonhardt H, Cardoso MC. Visualization and targeted disruption of protein interactions in living cells. Nat Commun (2013) 4:2660. doi:10.1038/ncomms3660
57. Prantner AM, Turini M, Kerfelec B, Joshi S, Baty D, Chames P, et al. Anti-mesothelin nanobodies for both conventional and nanoparticle-based biomedical applications. J Biomed Nanotechnol (2015) 11(7):1201–12. doi:10.1166/jbn.2015.2063
58. Debie P, Van Quathem J, Hansen I, Bala G, Massa S, Devoogdt N, et al. Effect of dye and conjugation chemistry on the biodistribution profile of near-infrared-labeled nanobodies as tracers for image-guided surgery. Mol Pharm (2017) 14(4):1145–53. doi:10.1021/acs.molpharmaceut.6b01053
59. van Driel PB, Boonstra MC, Slooter MD, Heukers R, Stammes MA, Snoeks TJ, et al. EGFR targeted nanobody-photosensitizer conjugates for photodynamic therapy in a pre-clinical model of head and neck cancer. J Control Release (2016) 229:93–105. doi:10.1016/j.jconrel.2016.03.014
60. Wang H, Meng AM, Li SH, Zhou XL. A nanobody targeting carcinoembryonic antigen as a promising molecular probe for non-small cell lung cancer. Mol Med Rep (2017) 16(1):625–30. doi:10.3892/mmr.2017.6677
61. Zhang F, Wei H, Wang X, Bai Y, Wang P, Wu J, et al. Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade. Cell Discov (2017) 3:17004. doi:10.1038/celldisc.2017.4
62. Ingram JR, Blomberg OS, Sockolosky JT, Ali L, Schmidt FI, Pishesha N, et al. Localized CD47 blockade enhances immunotherapy for murine melanoma. Proc Natl Acad Sci U S A (2017) 114(38):10184–89. doi:10.1073/pnas.1710776114
63. Sockolosky JT, Dougan M, Ingram JR, Ho CC, Kauke MJ, Almo SC, et al. Durable antitumor responses to CD47 blockade require adaptive immune stimulation. Proc Natl Acad Sci U S A (2016) 113(19):E2646–54. doi:10.1073/pnas.1604268113
64. Li JW, Xia L, Su Y, Liu H, Xia X, Lu Q, et al. Molecular imprint of enzyme active site by camel nanobodies: rapid and efficient approach to produce abzymes with alliinase activity. J Biol Chem (2012) 287(17):13713–21. doi:10.1074/jbc.M111.336370
65. De Genst E, Handelberg F, Van Meirhaeghe A, Vynck S, Loris R, Wyns L, et al. Chemical basis for the affinity maturation of a camel single domain antibody. J Biol Chem (2004) 279(51):53593–601. doi:10.1074/jbc.M407843200
66. Zielonka S, Empting M, Grzeschik J, Konning D, Barelle CJ, Kolmar H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. MAbs (2015) 7(1):15–25. doi:10.4161/19420862.2015.989032
67. Barelle C, Porter A. VNARs: an ancient and unique repertoire of molecules that deliver small, soluble, stable and high affinity binders of proteins. Antibodies (2015) 4(3):240–58. doi:10.3390/antib4030240
68. Pardon E, Laeremans T, Triest S, Rasmussen SG, Wohlkonig A, Ruf A, et al. A general protocol for the generation of nanobodies for structural biology. Nat Protoc (2014) 9(3):674–93. doi:10.1038/nprot.2014.039
69. Garaicoechea L, Olichon A, Marcoppido G, Wigdorovitz A, Mozgovoj M, Saif L, et al. Llama-derived single-chain antibody fragments directed to rotavirus VP6 protein possess broad neutralizing activity in vitro and confer protection against diarrhea in mice. J Virol (2008) 82(19):9753–64. doi:10.1128/JVI.00436-08
70. Garaicoechea L, Aguilar A, Parra GI, Bok M, Sosnovtsev SV, Canziani G, et al. Llama nanoantibodies with therapeutic potential against human norovirus diarrhea. PLoS One (2015) 10(8):e0133665. doi:10.1371/journal.pone.0133665
71. Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol (2005) 23(9):1105–16. doi:10.1038/nbt1126
72. Gonzalez-Sapienza G, Rossotti MA, Tabares-da Rosa S. Single-domain antibodies as versatile affinity reagents for analytical and diagnostic applications. Front Immunol (2017) 8:977. doi:10.3389/fimmu.2017.00977
73. Efimov GA, Kruglov AA, Khlopchatnikova ZV, Rozov FN, Mokhonov VV, Rose-John S, et al. Cell-type-restricted anti-cytokine therapy: TNF inhibition from one pathogenic source. Proc Natl Acad Sci U S A (2016) 113(11):3006–11. doi:10.1073/pnas.1520175113
74. Wang L, Liu X, Zhu X, Wang W, Liu C, Cui H, et al. Generation of single-domain antibody multimers with three different self-associating peptides. Protein Eng Des Sel (2013) 26(6):417–23. doi:10.1093/protein/gzt011
75. Massa S, Vikani N, Betti C, Ballet S, Vanderhaegen S, Steyaert J, et al. Sortase A-mediated site-specific labeling of camelid single-domain antibody-fragments: a versatile strategy for multiple molecular imaging modalities. Contrast Media Mol Imaging (2016) 11(5):328–39. doi:10.1002/cmmi.1696
76. Maass DR, Sepulveda J, Pernthaner A, Shoemaker CB. Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). J Immunol Methods (2007) 324(1–2):13–25. doi:10.1016/j.jim.2007.04.008
77. Roovers RC, Laeremans T, Huang L, De Taeye S, Verkleij AJ, Revets H, et al. Efficient inhibition of EGFR signaling and of tumour growth by antagonistic anti-EFGR nanobodies. Cancer Immunol Immunother (2007) 56(3):303–17. doi:10.1007/s00262-006-0180-4
78. Salema V, Fernandez LA. High yield purification of nanobodies from the periplasm of E. coli as fusions with the maltose binding protein. Protein Expr Purif (2013) 91(1):42–8. doi:10.1016/j.pep.2013.07.001
79. Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, Garcia KC, et al. Adrenaline-activated structure of beta2-adrenoceptor stabilized by an engineered nanobody. Nature (2013) 502(7472):575–9. doi:10.1038/nature12572
80. Griffin L, Lawson A. Antibody fragments as tools in crystallography. Clin Exp Immunol (2011) 165(3):285–91. doi:10.1111/j.1365-2249.2011.04427.x
81. Schmitz KR, Bagchi A, Roovers RC, van Bergen en Henegouwen PM, Ferguson KM. Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains. Structure (2013) 21(7):1214–24. doi:10.1016/j.str.2013.05.008
82. Manglik A, Kobilka BK, Steyaert J. Nanobodies to study G protein-coupled receptor structure and function. Annu Rev Pharmacol Toxicol (2017) 57:19–37. doi:10.1146/annurev-pharmtox-010716-104710
83. Harmsen MM, van Solt CB, van Zijderveld-van Bemmel AM, Niewold TA, van Zijderveld FG. Selection and optimization of proteolytically sn llama single-domain antibody fragments for oral immunotherapy. Appl Microbiol Biotechnol (2006) 72(3):544–51. doi:10.1007/s00253-005-0300-7
84. Hussack G, Hirama T, Ding W, Mackenzie R, Tanha J. Engineered single-domain antibodies with high protease resistance and thermal stability. PLoS One (2011) 6(11):e28218. doi:10.1371/journal.pone.0028218
85. Newnham LE, Wright MJ, Holdsworth G, Kostarelos K, Robinson MK, Rabbitts TH, et al. Functional inhibition of beta-catenin-mediated Wnt signaling by intracellular VHH antibodies. MAbs (2015) 7(1):180–91. doi:10.4161/19420862.2015.989023
86. Sircar A, Sanni KA, Shi J, Gray JJ. Analysis and modeling of the variable region of camelid single-domain antibodies. J Immunol (2011) 186(11):6357–67. doi:10.4049/jimmunol.1100116
87. Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science (2004) 305(5691):1770–3. doi:10.1126/science.1101148
88. Stanfield RL, Dooley H, Verdino P, Flajnik MF, Wilson IA. Maturation of shark single-domain (IgNAR) antibodies: evidence for induced-fit binding. J Mol Biol (2007) 367(2):358–72. doi:10.1016/j.jmb.2006.12.045
89. Kromann-Hansen T, Louise Lange E, Peter Sorensen H, Hassanzadeh-Ghassabeh G, Huang M, Jensen JK, et al. Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator. Sci Rep (2017) 7(1):3385. doi:10.1038/s41598-017-03457-7
90. Chakravarty R, Goel S, Cai W. Nanobody: the “magic bullet” for molecular imaging? Theranostics (2014) 4(4):386–98. doi:10.7150/thno.8006
91. Van Audenhove I, Gettemans J. Nanobodies as versatile tools to understand, diagnose, visualize and treat cancer. EBioMedicine (2016) 8:40–8. doi:10.1016/j.ebiom.2016.04.028
92. van Driel PB, van der Vorst JR, Verbeek FP, Oliveira S, Snoeks TJ, Keereweer S, et al. Intraoperative fluorescence delineation of head and neck cancer with a fluorescent anti-epidermal growth factor receptor nanobody. Int J Cancer (2014) 134(11):2663–73. doi:10.1002/ijc.28601
93. Siontorou CG. Nanobodies as novel agents for disease diagnosis and therapy. Int J Nanomedicine (2013) 8:4215–27. doi:10.2147/IJN.S39428
94. Keyaerts M, Xavier C, Heemskerk J, Devoogdt N, Everaert H, Ackaert C, et al. Phase I study of 68Ga-HER2-nanobody for PET/CT assessment of HER2 expression in breast carcinoma. J Nucl Med (2016) 57(1):27–33. doi:10.2967/jnumed.115.162024
95. Xavier C, Blykers A, Vaneycken I, D’Huyvetter M, Heemskerk J, Lahoutte T, et al. (18)F-nanobody for PET imaging of HER2 overexpressing tumors. Nucl Med Biol (2016) 43(4):247–52. doi:10.1016/j.nucmedbio.2016.01.002
96. Lipton A, Goodman L, Leitzel K, Cook J, Sperinde J, Haddad M, et al. HER3, p95HER2, and HER2 protein expression levels define multiple subtypes of HER2-positive metastatic breast cancer. Breast Cancer Res Treat (2013) 141(1):43–53. doi:10.1007/s10549-013-2665-0
97. Warnders FJ, Terwisscha van Scheltinga AGT, Knuehl C, van Roy M, de Vries EFJ, Kosterink JGW, et al. Human epidermal growth factor receptor 3-specific tumor uptake and biodistribution of 89Zr-MSB0010853 visualized by real-time and noninvasive PET imaging. J Nucl Med (2017) 58(8):1210–5. doi:10.2967/jnumed.116.181586
98. Piramoon M, Hosseinimehr SJ, Omidfar K, Noaparast Z, Abedi SM. 99m Tc-anti-epidermal growth factor receptor nanobody for tumor imaging. Chem Biol Drug Des (2017) 89(4):498–504. doi:10.1111/cbdd.12871
99. Leung K. 99mTc(CO)3-Anti-Carcinoembryonic Antigen (CEA) Humanized CEA5 Graft Nanobody. Bethesda, MD: Molecular Imaging and Contrast Agent Database (MICAD) (2004).
100. Broos K, Keyaerts M, Lecocq Q, Renmans D, Nguyen T, Escors D, et al. Non-invasive assessment of murine PD-L1 levels in syngeneic tumor models by nuclear imaging with nanobody tracers. Oncotarget (2017) 8(26):41932–46. doi:10.18632/oncotarget.16708
101. Kosaka N, Ogawa M, Choyke PL, Kobayashi H. Clinical implications of near-infrared fluorescence imaging in cancer. Future Oncol (2009) 5(9):1501–11. doi:10.2217/fon.09.109
102. Oliveira S, Heukers R, Sornkom J, Kok RJ, van Bergen En Henegouwen PM. Targeting tumors with nanobodies for cancer imaging and therapy. J Control Release (2013) 172(3):607–17. doi:10.1016/j.jconrel.2013.08.298
103. Gainkam LO, Caveliers V, Devoogdt N, Vanhove C, Xavier C, Boerman O, et al. Localization, mechanism and reduction of renal retention of technetium-99m labeled epidermal growth factor receptor-specific nanobody in mice. Contrast Media Mol Imaging (2011) 6(2):85–92. doi:10.1002/cmmi.408
104. D’Huyvetter M, Xavier C, Caveliers V, Lahoutte T, Muyldermans S, Devoogdt N. Radiolabeled nanobodies as theranostic tools in targeted radionuclide therapy of cancer. Expert Opin Drug Deliv (2014) 11(12):1939–54. doi:10.1517/17425247.2014.941803
105. Chatalic KL, Veldhoven-Zweistra J, Bolkestein M, Hoeben S, Koning GA, Boerman OC, et al. A novel (1)(1)(1)In-labeled anti-prostate-specific membrane antigen nanobody for targeted SPECT/CT imaging of prostate cancer. J Nucl Med (2015) 56(7):1094–9. doi:10.2967/jnumed.115.156729
106. Vosjan MJ, Vercammen J, Kolkman JA, Stigter-van Walsum M, Revets H, van Dongen GA. Nanobodies targeting the hepatocyte growth factor: potential new drugs for molecular cancer therapy. Mol Cancer Ther (2012) 11(4):1017–25. doi:10.1158/1535-7163.MCT-11-0891
107. Calpe S, Wagner K, El Khattabi M, Rutten L, Zimberlin C, Dolk E, et al. Effective inhibition of bone morphogenetic protein function by highly specific llama-derived antibodies. Mol Cancer Ther (2015) 14(11):2527–40. doi:10.1158/1535-7163.MCT-14-0956
108. Slordahl TS, Denayer T, Moen SH, Standal T, Borset M, Ververken C, et al. Anti-c-MET nanobody – a new potential drug in multiple myeloma treatment. Eur J Haematol (2013) 91(5):399–410. doi:10.1111/ejh.12185
109. Heukers R, Altintas I, Raghoenath S, De Zan E, Pepermans R, Roovers RC, et al. Targeting hepatocyte growth factor receptor (Met) positive tumor cells using internalizing nanobody-decorated albumin nanoparticles. Biomaterials (2014) 35(1):601–10. doi:10.1016/j.biomaterials.2013.10.001
110. Tan S, Liu K, Chai Y, Zhang CW, Gao S, Gao GF, et al. Distinct PD-L1 binding characteristics of therapeutic monoclonal antibody durvalumab. Protein Cell (2018) 9(1):135–9. doi:10.1007/s13238-017-0412-8
111. Griffiths K, Dolezal O, Cao B, Nilsson SK, See HB, Pfleger KD, et al. i-bodies, human single domain antibodies that antagonize chemokine receptor CXCR4. J Biol Chem (2016) 291(24):12641–57. doi:10.1074/jbc.M116.721050
112. Bauer G, Motz M. The antitumor effect of single-domain antibodies directed towards membrane-associated catalase and superoxide dismutase. Anticancer Res (2016) 36(11):5945–56. doi:10.21873/anticanres.11182
113. Papadopoulos KP, Isaacs R, Bilic S, Kentsch K, Huet HA, Hofmann M, et al. Unexpected hepatotoxicity in a phase I study of TAS266, a novel tetravalent agonistic nanobody(R) targeting the DR5 receptor. Cancer Chemother Pharmacol (2015) 75(5):887–95. doi:10.1007/s00280-015-2712-0
114. Ding L, Tian C, Feng S, Fida G, Zhang C, Ma Y, et al. Small sized EGFR1 and HER2 specific bifunctional antibody for targeted cancer therapy. Theranostics (2015) 5(4):378–98. doi:10.7150/thno.10084
115. Li J, Zhou C, Dong B, Zhong H, Chen S, Li Q, et al. Single domain antibody-based bispecific antibody induces potent specific anti-tumor activity. Cancer Biol Ther (2016) 17(12):1231–9. doi:10.1080/15384047.2016.1235659
116. Dekempeneer Y, Keyaerts M, Krasniqi A, Puttemans J, Muyldermans S, Lahoutte T, et al. Targeted alpha therapy using short-lived alpha-particles and the promise of nanobodies as targeting vehicle. Expert Opin Biol Ther (2016) 16(8):1035–47. doi:10.1080/14712598.2016.1185412
117. Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci U S A (2009) 106(38):16157–62. doi:10.1073/pnas.0908201106
118. van Erp EA, Kaliberova LN, Kaliberov SA, Curiel DT. Retargeted oncolytic adenovirus displaying a single variable domain of camelid heavy-chain-only antibody in a fiber protein. Mol Ther Oncolytics (2015) 2:15001. doi:10.1038/mto.2015.1
119. Rita Costa A, Milho C, Azeredo J, Pires DP. Synthetic biology to engineer bacteriophage genomes. Methods Mol Biol (2018) 1693:285–300. doi:10.1007/978-1-4939-7395-8_21
120. Hu Y, Liu C, Muyldermans S. Nanobody-based delivery systems for diagnosis and targeted tumor therapy. Front Immunol (1442) 2017:8.
121. Turner KB, Alves NJ, Medintz IL, Walper SA. Improving the targeting of therapeutics with single-domain antibodies. Expert Opin Drug Deliv (2016) 13(4):561–70. doi:10.1517/17425247.2016.1133583
122. Li T, Vandesquille M, Bay S, Dhenain M, Delatour B, Lafaye P. Selection of similar single domain antibodies from two immune VHH libraries obtained from two alpacas by using different selection methods. Immunol Lett (2017) 188:89–95. doi:10.1016/j.imlet.2017.07.001
123. Rossotti M, Tabares S, Alfaya L, Leizagoyen C, Moron G, Gonzalez-Sapienza G. Streamlined method for parallel identification of single domain antibodies to membrane receptors on whole cells. Biochim Biophys Acta (2015) 1850(7):1397–404. doi:10.1016/j.bbagen.2015.03.009
124. Tani H, Osbourn JK, Walker EH, Rush RA, Ferguson IA. A novel in vivo method for isolating antibodies from a phage display library by neuronal retrograde transport selectively yields antibodies against p75(NTR.). MAbs (2013) 5(3):471–8. doi:10.4161/mabs.24112
125. Cavallari M. Rapid and direct VHH and target identification by staphylococcal surface display libraries. Int J Mol Sci (2017) 18(7):E1507. doi:10.3390/ijms18071507
126. Abdiche YN, Yeung AY, Ni I, Stone D, Miles A, Morishige W, et al. Antibodies targeting closely adjacent or minimally overlapping epitopes can displace one another. PLoS One (2017) 12(1):e0169535. doi:10.1371/journal.pone.0169535
127. Salema V, Fernandez LA. Escherichia coli surface display for the selection of nanobodies. Microb Biotechnol (2017) 10(6):1468–84. doi:10.1111/1751-7915.12819
Keywords: camelid heavy-chain antibody, drug-like properties, bioavailability, immunogenicity, broad epitope coverage, poly-specificity
Citation: Iezzi ME, Policastro L, Werbajh S, Podhajcer O and Canziani GA (2018) Single-Domain Antibodies and the Promise of Modular Targeting in Cancer Imaging and Treatment. Front. Immunol. 9:273. doi: 10.3389/fimmu.2018.00273
Received: 31 August 2017; Accepted: 30 January 2018;
Published: 19 February 2018
Edited by:
Leonor Kremer, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Paul Van Bergen En Henegouwen, Utrecht University, NetherlandsAbdul Qader Abbady, Atomic Energy Commission of Syria, Syria
Uener Kolukisaoglu, Universität Tübingen, Germany
Copyright: © 2018 Iezzi, Policastro, Werbajh, Podhajcer and Canziani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Osvaldo Podhajcer, b3BvZGhhamNlckBsZWxvaXIub3JnLmFy;
Gabriela Alicia Canziani, Z2NhbnppYW5pQGxlbG9pci5vcmcuYXI=
 María Elena Iezzi1
María Elena Iezzi1 Santiago Werbajh
Santiago Werbajh Osvaldo Podhajcer
Osvaldo Podhajcer Gabriela Alicia Canziani
Gabriela Alicia Canziani