- Department of Dermatology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States
Tissue-resident memory T cells (TRM) persist in peripheral tissues for long periods of time in the absence of antigenic stimulation. Upon re-encounter with cognate antigen, TRM trigger an immediate immune response at the local tissue microenvironment and provide the first line of host defense. TRM have been reported to play significant roles in host antimicrobial infection, cancer immunotherapy, and pathogenesis of a number of human autoimmune diseases, such as psoriasis, vitiligo, and atopic dermatitis. TRM display a distinct gene transcriptome with unique gene expression profiles related to cellular metabolism that is different from naive T cells (TN), central memory T cells (TCM), and effector memory T cells (TEM). Skin CD8+ TRM upregulate expression of genes associated with lipid uptake and metabolism and utilize mitochondria fatty acid β-oxidation to support their long-term survival (longevity) and function. In this review, we will summarize the recent progresses in the metabolic programming of TRM and will also discuss the potential to target the unique metabolic pathways of TRM to treat TRM-mediated diseases.
Memory T cells mediate immunosurveillance and protect the host through rapid recall responses upon re-exposure to previously encountered pathogens (1). In addition to the two previously identified circulating memory T cells, central memory T cells (TCM), and effector memory T cells (TEM), a new subtype of memory T cells—tissue-resident memory T cells (TRM)—has been identified and characterized (2–4). Unlike TCM and TEM that circulate within blood, TRM reside and remain within epithelial barrier tissues for long periods of time without trafficking back into lymph or blood (5). Upon antigen re-exposure, TRM trigger an immediate immune response and provide the first line of protection against the antigen/pathogen they are specific for (4, 6–11). In addition, TRM create a general antiviral microenvironment at the local tissue site and provide cross-protection against antigenically unrelated pathogens (7, 9). Activation of TRM alters tissue-wide gene expression profiles, induces B cell and circulating memory T cell recruitment through IFN-γ-dependent vascular cell adhesion molecule 1 upregulation, and leads to maturation of local dendritic cells and activation of natural killer cells. These activities support the idea that TRM function as a bridge between the adaptive and innate immune system (7, 9). As many viruses show tissue tropism, TRM also provide protective immune responses for the tissue through which it was previously encountered. TRM specific for HSV are in skin (12–14), TRM specific for rotavirus are in gut (6, 15), and TRM specific for influenza are in lung (16–18). Collectively, we propose that sensitization of relatively small numbers of TRM may lead to an amplified signal to more abundant elements of the innate immune system and trigger an organ-wide antiviral state. The placing of adaptive immune memory cells at the body’s interfaces with the environment, and moreover those specific for a given pathogen, speaks to the elegance of adaptive immune memory.
Upon cognate recognition of antigen via T-cell receptor, naive T cells (TN) undergo extensive clonal expansion and differentiate into several T cell subtypes, including effector T cells (Teff) for immediate pathogen elimination and memory T cells for long-term protection (19). Recent studies showed that T cell activation and differentiation are accompanied with and regulated tightly by metabolic reprogramming, presumably to provide the divergent energetic and functional needs for their development, maintenance, and function (20–23). TN primarily depend on glucose catabolism and oxidative phosphorylation (OXPHOS) to derive energy to support the maintenance of their relatively quiescent state. Teff reprogram their metabolic state to anabolism to enable rapid cell division and cytokine production (24). Teff increase glucose acquisition from blood through upregulating gene expression of glucose transporter-1 (Glut 1) and conduct glycolysis (converting glucose into pyruvate with the production of two molecules of ATP) to meet their energy demand (25). Although glycolysis is less efficient in generating ATP compared to OXPHOS, it is faster and thus rapidly accommodates the increased demand for the energy and biomass formation of Teff. Unlike TN and Teff, TCM utilize endogenously synthesized fatty acids and OXPHOS to support their long-time survival (longevity) and function (26–28). TCM maintain substantial mitochondrial spare respiratory capacity and display increased mitochondrial mass, thus providing metabolic advantage and equipping them for both longevity and the ability of rapid recall upon antigen re-challenge (26). O’Sullivan et al. showed that rather than importing extracellular fatty acids, TCM utilize endogenous fatty acid synthesis and subsequently conduct mitochondrial fatty acid oxidation (FAO) and OXPHOS for their differentiation and maintenance (27). TCM take up extracellular glucose from blood to synthesize fatty acids in the endoplasmic reticulum, a process dependent on lysosomal acid lipase, which is critical in hydrolyzing cholesteryl esters and triglycerides within LDL particles into free cholesterol and free fatty acids (FFAs) (29, 30). Cui et al. additionally showed that interleukin-7, a cytokine critical for TCM differentiation and survival, induced glycerol transport and triacylglycerol synthesis via enhanced gene expression of glycerol channel aquaporin 9, thus providing substrates for mitochondria FAO (28). However, compared to the well-defined metabolic reprogramming of circulating memory T cells, the metabolic programs utilized by TRM to dictate their fate differentiation and sustain their longevity and function, are only beginning to be understood.
TRM Metabolism in Skin
Skin, as the primary interface between the body and outer environment, provides a first line of defense against microbial pathogens, physical damage, and chemical insults. In addition to the role of barrier maintenance and sensing, skin functions as a hotbed of immunological activity (31). It has been shown that healthy skin of an adult human being contains about twice T cells as many as are present in the entire blood volume (1 × 106 T cells/cm2 and an estimated 2 × 1010 T cells in the entire skin surface) (32, 33). T cells contained in human skin are all CD45RO+ memory T cells, co-express skin-homing addressin cutaneous lymphocyte-associated antigen and the chemokine receptor CCR4, and more than half of human skin T cells are resident under resting conditions and do not re-circulate (TRM) (34). A recent study revealed that pathogenic T cell clones persist in “healed” psoriatic lesions as TRM after complete remission using TNFα blocker (35). Studies on vitiligo showed that vitiligo perilesional skin is enriched with a population of CD8+ TRM expressing both CD69 and CD103, in both stable and active disease stages (36, 37). Residing in a nutrient-restricted (particularly glucose) but lipid-rich environments (38, 39), the mechanisms by which skin TRM sustain their longevity and function remained elusive. Using a well-established model of generating CD8+ TRM in skin after cutaneous immunization with Vaccinia virus, we showed that skin CD8+ TRM adapt to utilize lipid metabolism of exogenous FFAs internalized from the surrounding microenvironment to support both their longevity and protective function (Figure 1) (40). CD8+ TRM develop a transcriptional program that features marked overexpression of molecules facilitating exogenous FFAs acquisition and metabolism. Specifically, fatty acid binding proteins 4 and 5 (Fabp4/5), CD36, and lipoprotein lipase (lpl) were in the top 35 most highly overexpressed genes in TRM, as compared to TN, TCM, and TEM. Fabp’s are conventionally thought to function as intracellular chaperones for FFAs, shuttling FFAs from cytoplasm to mitochondria for β-oxidation (41). CD36 is a lipid scavenger receptor that binds to and internalizes FFAs and other lipids (42), and lpl is a lipoprotein lipase that cleaves triglycerides to yield a FFA and diacylglycerol (43). This collection of overexpressed genes involved in lipid uptake and metabolism suggested a special relationship between TRM and lipid metabolism. Further study showed that skin CD8+ TRM upregulated the gene expression of Fabp4/5 in a peroxisome proliferator-activated receptor gamma (PPAR-γ)-dependent manner. When incubated under the presence of exogenous fluorescently conjugated FFAs, skin CD8+ TRM internalized extracellular FFAs much more efficiently compared to other counterparts. Addition of exogenous FFAs induced a significantly higher basal and FCCP-stimulated maximal oxygen-consumption rate in skin CD8+ TRM, which could be blocked by pretreatment with etomoxir, an irreversible inhibitor of mitochondrial carnitine palmitoyltransferase 1, an enzyme that is the rate limiting step for mitochondrial fatty acid β-oxidation and ATP generation (44). Skin CD8+ TRM rendered unable to metabolize exogenous FFAs through mitochondrial β-oxidation, whether through deficiency of Fabp4/5 or pretreatment with etomoxir, cannot persist in skin. By contrast, TCM generated from Fabp4−/−Fabp5−/− mice in parallel have no survival disadvantage. Referring to functionality, skin CD8+ TRM deficient in Fabp4/5 were inferior in clearing viral infection and insufficient to protect host against lethal viral re-challenge. Consistent with data from mice, CD8+ TRM from human skin tissue display higher level of Fabp4/5 expression and internalize more exogenous FFAs compared to other human counterparts, indicating that acquisition of exogenous FFAs for metabolism might represent a conserved feature of TRM across species. Given the dependence of skin CD8+ TRM on lipid metabolism and the increasingly uncovered roles of TRM in skin autoimmune diseases such as psoriasis and vitiligo, it is tempting to speculate a novel and promising treatment strategy for skin immune disorders by blocking critical lipid metabolic pathways in TRM. Still, much remains to be elucidated further for the mechanism of TRM metabolic maintenance and survival. The precise roles of gene upregulation of PPAR-γ in skin CD8+ TRM and the contribution of other lipid metabolism-related genes to the survival and function of skin CD8+ TRM, such as CD36 and lpl, both of which were also upregulated as part of the TRM transcriptional program, remain to be investigated. Overall, a detailed signaling pathway of TRM metabolism, as well as the cross talk among skin tissue microenvironment, TRM metabolism and effector function, will be of great interest and may facilitate the development of efficient treatment strategy for TRM-mediated skin diseases.
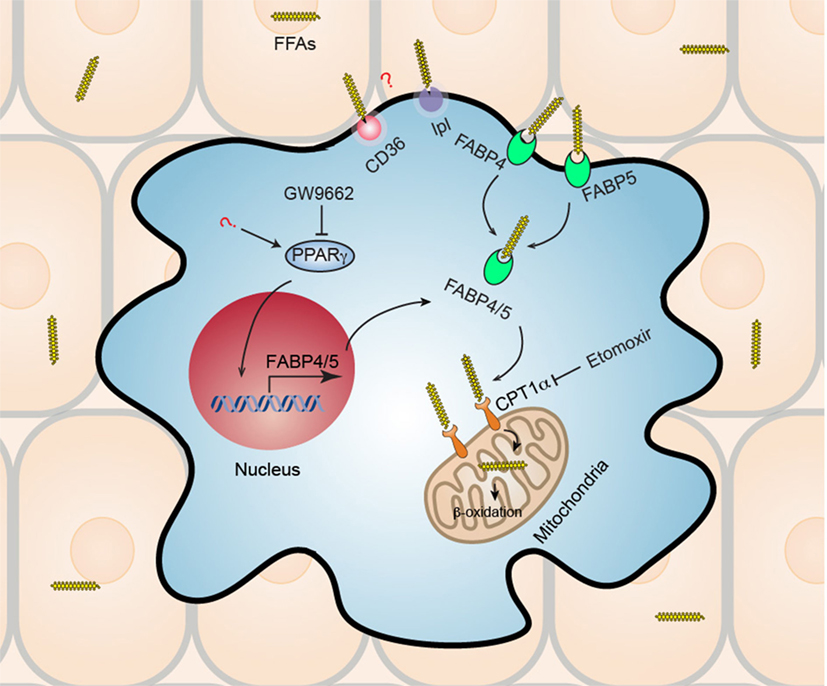
Figure 1. Metabolic reprogramming of skin CD8+ tissue-resident memory T cells (TRM). Skin CD8+ TRM depend on increased uptake of exogenous fatty acid and mitochondrial β-oxidation for their long-term survival (longevity) and function. Skin CD8+ TRM upregulate gene expression of transcription factor peroxisome proliferator-activated receptor gamma (PPAR-γ) and its downstream molecules fatty acid binding proteins 4 and 5 (Fabp4/5), which accounts for the increased uptake of free fatty acids (FFAs) from surrounding tissue environment. Subsequently, Skin CD8+ TRM utilize mitochondrial fatty acid β-oxidation to generate ATP to support their survival and function. Skin CD8+ TRM loss of Fabp4/5 is more prone to cell apoptosis, deficient in long-term survival, and could not protect host efficiently upon viral re-challenge. Treatment with either PPAR-γ inhibitor (GW9662) or with fatty acid mitochondrial β-oxidation inhibitor (etomoxir), results in impaired long-term maintenance of CD8+ TRM in skin. In addition, the roles of CD36 and lipoprotein lipase (lpl), both of which are also upregulated in skin CD8+ TRM and are involved in lipid metabolism, remain unknown and await to be elucidated by future studies.
TRM Metabolism in Tumor Microenvironment (TME)
Solid tumors are infiltrated by heterogeneous immune cell types that work in a coordinated fashion to effect antitumor immunity (45). The presence and abundance of tumor-infiltrating lymphocytes (TILs) in tumors is associated with better clinical outcomes after tumor immunotherapy (46–49). TILs differ from their blood counterparts both in terms of upregulated gene expression of immune checkpoint molecules (PD-1, LAG3, TIGIT, and CTLA-4) and reduced effector functions (tumor immunosuppression) (50). Phenotypic analysis of TILs from melanoma revealed that nearly 60% of CD8+ T cells and 50% of CD4+ T cells are CD45RO+CD69+CCR7–, characteristic of a TRM phenotype (51). Melanoma antigen-specific TRM cells resided predominantly in melanocyte-depleted hair follicles and mediate durable immunity to melanoma (36). Other studies showed that the number of TRM within tumors associates with cytotoxic T cell responses and correlates with a better overall survival in lung cancer, head and neck cancer, and breast cancer (52–54). Local TRM induced via immunization through mucosal vaccine inhibited tumor growth (52, 55). Thus, a TIL TRM phenotype is considered as a new surrogate biomarker for the efficacy of cancer vaccines, and development of vaccine strategies designed to generate TRM against tumor cells has attracted great interest as a potentially significant therapeutic application.
Inside tumors, both tumor cells and TILs compete for the oxygen and nutrients supplied via infiltrating blood vessels. Rapidly growing tumor cells utilize more glucose and glutamine to conduct glycolysis, resulting in a TME of hypoxia and glucose deprivation for TILs (56, 57). This leads to enhanced expression of immune checkpoint molecules and loss of effector functions in TILs, a process that results in T cell exhaustion (51). On the other hand, TILs conduct metabolic reprogramming to adapt to the metabolic constrains within the TME and sustain their maintenance and antitumor function. Using mouse melanoma models, Ying et al. reported that under short-term hypoxia and hypoglycemia, CD8+ T cells decreased transcripts for genes important in glycolysis while increasing transcripts of PPAR-α and downstream molecules involved in FA uptake and mitochondrial FA catabolism (58). Enhanced FA uptake and increased expression of Cpt1a were observed in vaccine-induced CD8+ TILs from late-stage tumors, implying the increasing dependence of TILs on fatty acid metabolism for energy maintenance. Promoting fatty acid catabolism with fenofibrate, a PPAR-α agonist, markedly improves the capacity of CD8+ TILs to delay tumor growth. This enhancement synergizes with PD-1 blockade to efficiently enhance the efficacy of melanoma immunotherapy. With regard to human tumor biology, the same study reported that TILs isolated from human melanoma metastases show evidence of enhanced FA catabolism, which could be fueled by increased level of FAs within tumor intestinal fluid (58). Collectively, these data suggest that TILs in the TME engage in metabolic reprogramming to utilize FAO for their survival and function. The mechanism by which the TME influences TILs metabolic reprogramming, as well as the nutrient source of FAs for fatty acid catabolism, remains to be further investigated. Also, further studies are required to elucidate the mechanisms by which TILs reprogram their metabolism to cope with the TME and how this metabolic switch affect their survival and antitumor function. Since a growing body of studies support the idea that enhancing already present immune responses against tumors leads to considerably long-lasting tumor remissions and delayed tumor metastasis, a better understanding of TIL TRM’s metabolic switch and how to manipulate this process to increase their maintenance and antitumor effector function, may increase the efficacy and improve the outcome of tumor immunotherapy.
TRM Metabolism in White Adipose Tissue (WAT)
White adipose tissue is a storage depot for fat and an endocrine organ that secretes adipokines to regulate whole-body energy homeostasis and metabolism (59). It connects body barrier surfaces and the internal organs, thereby forming a bridge between tissues that are constantly challenged with surrounding microbes and the inner sterile environments. WAT constitutively regulates glucose and lipid homeostasis by sorting and releasing FFAs via lipolysis for usage by other organs (60). Previous work has shed light on cross talks between WAT and immune system in a series of metabolic disorders and inflammatory diseases (61–63). WAT infiltrating lymphocytes are predominantly localized within organized structures referred to as fat-associated lymphoid clusters or milky spots (in the omentum), which can rapidly expand in response to local inflammatory cues (64, 65). Recent studies by Han et al. reveal the residency and occupancy of TRM in WAT and their contribution to immune surveillance and long-term protective memory responses to infection (66). WAT functions as a major hub for adaptive immune memory T cells, predominantly TRM. These adipose TRM express a well-established TRM cell surface marker (CD69) and do not equilibrate between the adipose tissue of conjoined naïve and previously infected mice, confirming the residency of these cells. Transplantation of adipose tissue from previously infected mice was sufficient to protect uninfected mice from lethal pathogen challenge, whereas depletion of T cells abrogated this protective effect, indicating a functional protective role of adipose TRM in systemic pathogen challenge. Following gene expression analysis revealed that adipose TRM cells upregulated genes involved in effector functionalities and lipid metabolism. When incubated ex vivo with fluorescently labeled long chain fatty acid palmitate (Bodipy FL C16), adipose TRM cells displayed high rates of lipid uptake and mitochondrial respiration compared to their counterparts from spleen and small intestine lamina propria (siLP), while no difference could be observed in FFA uptake between siLP TRM and spleen TEM. These data suggest that TRM in WAT might also utilize fatty acid metabolism for their survival and function. To what extent do adipose TRM depend on fatty acid metabolism and the contribution of fatty acid metabolism to their longevity and function remain to be evaluated further. The same study showed that induction of WAT memory responses results in the remodeling of WAT physiology (66). Thus, it would be interesting to investigate the cross-regulation between adipocytes and TRM metabolism within WAT, as well as how to manipulate the regulation of pathways to increase host protection or treat individuals with obesity and metabolic disorders.
Targeting TRM Metabolic Pathways to Treat Associated Autoimmune Diseases
Targeted therapies are increasingly successful at inducing temporary and partial remissions in organ-specific immune mediated autoinflammatory diseases, but it remains nearly impossible to induce durable remission or cure (6). These autoimmune disorders, including diseases of skin (psoriasis, vitiligo, graft vs host disease), GI tract (Crohn’s disease, ulcerative colitis), lung (asthma), joint (rheumatoid arthritis, spondyloarthropathies), CNS (multiple sclerosis), and endocrine system (Type I diabetes), are increasing in incidence and prevalence. Over the past decade, a line of investigation central to the understanding of diseases pathogenesis leads to the discovery of TRM. Increasing evidences from various experimental models and clinical data support a theory that these autoimmune diseases are driven, at least partially, by inappropriate and chronic activation of pathogenic TRM (6). This provides a plausible explanation for the T cell pathogenesis of these diseases and their organ specificity, something that prior explanations of pathogenesis could not adequately explain. This also provides an explanation for the chronicity of these diseases, as TRM are nearly impossible to dislodge from their tissue sites of residence once established. Currently, in clinic these diseases of regional immune hyperactivation (via TRM) are usually being treated with systemic immunomodulation and immunosuppression. After successful therapy is withdrawn, TRM remain in situ and can become reactivated by pathogenic stimuli, thus resulting in disease relapse. Therefore, a therapy that could not only suppress the activity of pathogenic TRM but also dislodge them from their tissue niches has the potential to induce remissions that are much longer and ideally indefinite. The uniqueness of TRM in their dependence on lipid metabolism of FFAs from the external environment makes it a previously unappreciated “Achilles Heel,” and one that could be exploited therapeutically. Indeed, administration in vivo with a pharmacologic mitochondrial β-oxidation, trimetazidine [blocks the long chain 3-ketoacyl CoA thiolase activity (67)], decreased the survival and maintenance of TRM in skin (40). Thus, the likelihood exists that pharmacologic approach targeting the lipid metabolic pathway in TRM could reduce, and theoretically eliminate, the pathogenic TRM that are causative in autoinflammatory disorders of multiple tissues.
Conclusion and Future Perspectives
It has recently become clear that control of metabolism and the adaptive immune system are tightly linked (21, 22, 68). Nutrient availability and cellular metabolism closely regulate the differentiation, survival, and function of immune cells (23). TRM are not simply memory T cells residing in an unexpected location; rather, they are a specific group of memory T cells with unique lineage (40, 69–71). As revealed from gene transcriptional profiling, TRM display a quite distinct transcriptome from those of TCM and TEM, both of which were more similar to that of TN (69, 70, 72). Recent findings have shed light on the role of cellular metabolism in regulating differentiation and memory formation of TCM (26–28). However, it remains unknown how cellular metabolism controls TRM fate decision. Moreover, the focus of previous studies on TRM metabolism is primarily on CD8+ TRM, and little is known about the metabolic reprogramming of CD4+ TRM and their roles in CD4+ TRM differentiation, survival, and function. In addition, attributed to the restricted nutrient availability at specialized tissue sites, more studies will be required to elucidate the metabolic pathways of TRM at other tissue sites such as lung, intestine, and brain. Finally given that generation of long-lived TRM are a goal of efficient vaccination, and considering the dual role of TRM in tumor and autoimmune tissue disorders, a more detailed understanding of the unique metabolic programs intrinsic to TRM, and how these programs might be manipulated to enhance or decrease TRM longevity and function, will be a subject of future study with high clinical relevance and therapeutic significance.
Author Contributions
YP drafted and edited the manuscript; TK edited and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was supported in part by National Institutes of Health Grants R01 AI127654 and R01 AR065807.
References
1. Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol (2004) 22:745–63. doi:10.1146/annurev.immunol.22.012703.104702
2. Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol (2009) 10:524–30. doi:10.1038/ni.1718
3. Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med (2010) 207:553–64. doi:10.1084/jem.20090858
4. Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature (2012) 483:227–31. doi:10.1038/nature10851
5. Mueller SN, Mackay LK. Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol (2016) 16:79–89. doi:10.1038/nri.2015.3
6. Park CO, Kupper TS. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat Med (2015) 21:688–97. doi:10.1038/nm.3883
7. Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song JY, et al. T cell memory. Skin-resident memory CD8(+) T cells trigger a state of tissue-wide pathogen alert. Science (2014) 346:101–5. doi:10.1126/science.1254803
8. Iijima N, Iwasaki A. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science (2014) 346:93–8. doi:10.1126/science.1257530
9. Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science (2014) 346:98–101. doi:10.1126/science.1254536
10. Liu L, Zhong Q, Tian T, Dubin K, Athale SK, Kupper TS. Epidermal injury and infection during poxvirus immunization is crucial for the generation of highly protective T cell-mediated immunity. Nat Med (2010) 16:224–7. doi:10.1038/nm.2078
11. Park CO, Fu X, Jiang X, Pan Y, Teague JE, Collins N, et al. Staged development of long-lived T-cell receptor alphabeta TH17 resident memory T-cell population to Candida albicans after skin infection. J Allergy Clin Immunol (2017). doi:10.1016/j.jaci.2017.09.042
12. Egan KP, Wu S, Wigdahl B, Jennings SR. Immunological control of herpes simplex virus infections. J Neurovirol (2013) 19:328–45. doi:10.1007/s13365-013-0189-3
13. Shin H, Kumamoto Y, Gopinath S, Iwasaki A. CD301b+ dendritic cells stimulate tissue-resident memory CD8+ T cells to protect against genital HSV-2. Nat Commun (2016) 7:13346. doi:10.1038/ncomms13346
14. Khan AA, Srivastava R, Chentoufi AA, Kritzer E, Chilukuri S, Garg S, et al. Bolstering the number and function of HSV-1-specific CD8(+) effector memory T cells and tissue-resident memory T cells in latently infected trigeminal ganglia reduces recurrent ocular herpes infection and disease. J Immunol (2017) 199:186–203. doi:10.4049/jimmunol.1700145
15. Iijima N, Iwasaki A. Tissue instruction for migration and retention of TRM cells. Trends Immunol (2015) 36:556–64. doi:10.1016/j.it.2015.07.002
16. Pizzolla A, Nguyen TH, Sant S, Jaffar J, Loudovaris T, Mannering SI, et al. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J Clin Invest (2018) 128:721–33. doi:10.1172/JCI96957
17. Pizzolla A, Nguyen THO, Smith JM, Brooks AG, Kedzieska K, Heath WR, et al. Resident memory CD8(+) T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci Immunol (2017) 2. doi:10.1126/sciimmunol.aam6970
18. Wilk MM, Misiak A, McManus RM, Allen AC, Lynch MA, Mills KHG. Lung CD4 tissue-resident memory T cells mediate adaptive immunity induced by previous infection of mice with Bordetella pertussis. J Immunol (2017) 199:233–43. doi:10.4049/jimmunol.1602051
19. Santana MA, Esquivel-Guadarrama F. Cell biology of T cell activation and differentiation. Int Rev Cytol (2006) 250:217–74. doi:10.1016/S0074-7696(06)50006-3
20. Slack M, Wang T, Wang R. T cell metabolic reprogramming and plasticity. Mol Immunol (2015) 68:507–12. doi:10.1016/j.molimm.2015.07.036
21. Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic instruction of immunity. Cell (2017) 169:570–86. doi:10.1016/j.cell.2017.04.004
22. Shehata HM, Murphy AJ, Lee MKS, Gardiner CM, Crowe SM, Sanjabi S, et al. Sugar or fat? Metabolic requirements for immunity to viral infections. Front Immunol (2017) 8:1311. doi:10.3389/fimmu.2017.01311
23. Dimeloe S, Burgener AV, Grahlert J, Hess C. T-cell metabolism governing activation, proliferation and differentiation; a modular view. Immunology (2017) 150:35–44. doi:10.1111/imm.12655
24. Adams WC, Chen YH, Kratchmarov R, Yen B, Nish SA, Lin WW, et al. Anabolism-associated mitochondrial stasis driving lymphocyte differentiation over self-renewal. Cell Rep (2016) 17:3142–52. doi:10.1016/j.celrep.2016.11.065
25. Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab (2014) 20:61–72. doi:10.1016/j.cmet.2014.05.004
26. Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature (2009) 460:103–7. doi:10.1038/nature08097
27. O’Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity (2014) 41:75–88. doi:10.1016/j.immuni.2014.06.005
28. Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, et al. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell (2015) 161:750–61. doi:10.1016/j.cell.2015.03.021
29. Burke JA, Schubert WK. Deficient activity of hepatic acid lipase in cholesterol ester storage disease. Science (1972) 176:309–10. doi:10.1126/science.176.4032.309
30. Pagani F, Pariyarath R, Garcia R, Stuani C, Burlina AB, Ruotolo G, et al. New lysosomal acid lipase gene mutants explain the phenotype of Wolman disease and cholesteryl ester storage disease. J Lipid Res (1998) 39:1382–8.
32. Clark RA. Resident memory T cells in human health and disease. Sci Transl Med (2015) 7:269rv1. doi:10.1126/scitranslmed.3010641
33. Clark RA, Chong B, Mirchandani N, Brinster NK, Yamanaka K, Dowgiert RK, et al. The vast majority of CLA+ T cells are resident in normal skin. J Immunol (2006) 176:4431–9. doi:10.4049/jimmunol.176.7.4431
34. Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med (2015) 7:279ra39. doi:10.1126/scitranslmed.3010302
35. Matos TR, O’Malley JT, Lowry EL, Hamm D, Kirsch IR, Robins HS, et al. Clinically resolved psoriatic lesions contain psoriasis-specific IL-17-producing alphabeta T cell clones. J Clin Invest (2017) 127:4031–41. doi:10.1172/JCI93396
36. Malik BT, Byrne KT, Vella JL, Zhang P, Shabaneh TB, Steinberg SM, et al. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci Immunol (2017) 2. doi:10.1126/sciimmunol.aam6346
37. Boniface K, Jacquemin C, Darrigade AS, Dessarthe B, Martins C, Boukhedouni N, et al. Vitiligo skin is imprinted with resident memory CD8 T cells expressing CXCR3. J Invest Dermatol (2018) 138:355–64. doi:10.1016/j.jid.2017.08.038
38. Khnykin D, Miner JH, Jahnsen F. Role of fatty acid transporters in epidermis: implications for health and disease. Dermatoendocrinol (2011) 3:53–61. doi:10.4161/derm.3.2.14816
39. Zhang Y, Li Q, Rao E, Sun Y, Grossmann ME, Morris RJ, et al. Epidermal fatty acid binding protein promotes skin inflammation induced by high-fat diet. Immunity (2015) 42:953–64. doi:10.1016/j.immuni.2015.04.016
40. Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature (2017) 543:252–6. doi:10.1038/nature21379
41. Hotamisligil GS, Bernlohr DA. Metabolic functions of FABPs – mechanisms and therapeutic implications. Nat Rev Endocrinol (2015) 11:592–605. doi:10.1038/nrendo.2015.122
42. Kim TT, Dyck JR. The role of CD36 in the regulation of myocardial lipid metabolism. Biochim Biophys Acta (2016) 1861:1450–60. doi:10.1016/j.bbalip.2016.03.018
43. Hayne CK, Lafferty MJ, Eglinger BJ, Kane JP, Neher SB. Biochemical analysis of the lipoprotein lipase truncation variant, LPL(S447X), reveals increased lipoprotein uptake. Biochemistry (2017) 56:525–33. doi:10.1021/acs.biochem.6b00945
44. Pike LS, Smift AL, Croteau NJ, Ferrick DA, Wu M. Inhibition of fatty acid oxidation by etomoxir impairs NADPH production and increases reactive oxygen species resulting in ATP depletion and cell death in human glioblastoma cells. Biochim Biophys Acta (2011) 1807:726–34. doi:10.1016/j.bbabio.2010.10.022
45. Belli C, Trapani D, Viale G, D’Amico P, Duso BA, Della Vigna P, et al. Targeting the microenvironment in solid tumors. Cancer Treat Rev (2018) 65:22–32. doi:10.1016/j.ctrv.2018.02.004
46. Sharma P, Shen Y, Wen S, Yamada S, Jungbluth AA, Gnjatic S, et al. CD8 tumor-infiltrating lymphocytes are predictive of survival in muscle-invasive urothelial carcinoma. Proc Natl Acad Sci U S A (2007) 104:3967–72. doi:10.1073/pnas.0611618104
47. Baras AS, Drake C, Liu JJ, Gandhi N, Kates M, Hoque MO, et al. The ratio of CD8 to Treg tumor-infiltrating lymphocytes is associated with response to cisplatin-based neoadjuvant chemotherapy in patients with muscle invasive urothelial carcinoma of the bladder. Oncoimmunology (2016) 5:e1134412. doi:10.1080/2162402X.2015.1134412
48. Solinas C, Garaud S, De Silva P, Boisson A, Van den Eynden G, de Wind A, et al. Immune checkpoint molecules on tumor-infiltrating lymphocytes and their association with tertiary lymphoid structures in human breast cancer. Front Immunol (2017) 8:1412. doi:10.3389/fimmu.2017.01412
49. Dunn GP, Dunn IF, Curry WT. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in human glioma. Cancer Immun (2007) 7:12.
50. Boddupalli CS, Bar N, Kadaveru K, Krauthammer M, Pornputtapong N, Mai Z, et al. Interlesional diversity of T cell receptors in melanoma with immune checkpoints enriched in tissue-resident memory T cells. JCI Insight (2016) 1:e88955. doi:10.1172/jci.insight.88955
51. Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol (2015) 15:486–99. doi:10.1038/nri3862
52. Nizard M, Roussel H, Diniz MO, Karaki S, Tran T, Voron T, et al. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat Commun (2017) 8:15221. doi:10.1038/ncomms15221
53. Djenidi F, Adam J, Goubar A, Durgeau A, Meurice G, de Montpreville V, et al. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol (2015) 194:3475–86. doi:10.4049/jimmunol.1402711
54. Ganesan AP, Clarke J, Wood O, Garrido-Martin EM, Chee SJ, Mellows T, et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol (2017) 18:940–50. doi:10.1038/ni.3775
55. Granier C, Blanc C, Karaki S, Tran T, Roussel H, Tartour E. Tissue-resident memory T cells play a key role in the efficacy of cancer vaccines. Oncoimmunology (2017) 6:e1358841. doi:10.1080/2162402X.2017.1358841
56. Anastasiou D. Tumour microenvironment factors shaping the cancer metabolism landscape. Br J Cancer (2017) 116:277–86. doi:10.1038/bjc.2016.412
57. Scharping NE, Delgoffe GM. Tumor microenvironment metabolism: a new checkpoint for anti-tumor immunity. Vaccines (Basel) (2016) 4:E46. doi:10.3390/vaccines4040046
58. Zhang Y, Kurupati R, Liu L, Zhou XY, Zhang G, Hudaihed A, et al. Enhancing CD8(+) T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell (2017) 32:377–91.e9. doi:10.1016/j.ccell.2017.08.004
59. Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell (2014) 156:20–44. doi:10.1016/j.cell.2013.12.012
60. Stern JH, Rutkowski JM, Scherer PE. Adiponectin, leptin, and fatty acids in the maintenance of metabolic homeostasis through adipose tissue crosstalk. Cell Metab (2016) 23:770–84. doi:10.1016/j.cmet.2016.04.011
61. DiSpirito JR, Mathis D. Immunological contributions to adipose tissue homeostasis. Semin Immunol (2015) 27:315–21. doi:10.1016/j.smim.2015.10.005
62. Brestoff JR, Artis D. Immune regulation of metabolic homeostasis in health and disease. Cell (2015) 161:146–60. doi:10.1016/j.cell.2015.02.022
63. Brestoff JR, Kim BS, Saenz SA, Stine RR, Monticelli LA, Sonnenberg GF, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature (2015) 519:242–6. doi:10.1038/nature14115
64. Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature (2010) 463:540–4. doi:10.1038/nature08636
65. Rangel-Moreno J, Moyron-Quiroz JE, Carragher DM, Kusser K, Hartson L, Moquin A, et al. Omental milky spots develop in the absence of lymphoid tissue-inducer cells and support B and T cell responses to peritoneal antigens. Immunity (2009) 30:731–43. doi:10.1016/j.immuni.2009.03.014
66. Han SJ, Glatman Zaretsky A, Andrade-Oliveira V, Collins N, Dzutsev A, Shaik J, et al. White adipose tissue is a reservoir for memory T cells and promotes protective memory responses to infection. Immunity (2017) 47:1154–68.e6. doi:10.1016/j.immuni.2017.11.009
67. Lopatin YM, Rosano GM, Fragasso G, Lopaschuk GD, Seferovic PM, Gowdak LH, et al. Rationale and benefits of trimetazidine by acting on cardiac metabolism in heart failure. Int J Cardiol (2016) 203:909–15. doi:10.1016/j.ijcard.2015.11.060
68. Odegaard JI, Chawla A. The immune system as a sensor of the metabolic state. Immunity (2013) 38:644–54. doi:10.1016/j.immuni.2013.04.001
69. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol (2013) 14:1294–301. doi:10.1038/ni.2744
70. Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, et al. T-box transcription factors combine with the cytokines TGF-beta and IL-15 to control tissue-resident memory T cell fate. Immunity (2015) 43:1101–11. doi:10.1016/j.immuni.2015.11.008
71. Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science (2016) 352:459–63. doi:10.1126/science.aad2035
Keywords: metabolism, longevity, TRM cells, viral vaccines, cancer immunotherapy
Citation: Pan Y and Kupper TS (2018) Metabolic Reprogramming and Longevity of Tissue-Resident Memory T Cells. Front. Immunol. 9:1347. doi: 10.3389/fimmu.2018.01347
Received: 03 April 2018; Accepted: 31 May 2018;
Published: 18 June 2018
Edited by:
Fathia Mami-Chouaib, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Craig Michael Walsh, University of California, Irvine, United StatesLaura K. Mackay, University of Melbourne, Australia
Copyright: © 2018 Pan and Kupper. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas S. Kupper, dGt1cHBlckBid2guaGFydmFyZC5lZHU=
 Youdong Pan
Youdong Pan Thomas S. Kupper
Thomas S. Kupper