- 1Laboratory of Comparative Immunology, Department of Veterinary Medicine, Nihon University, Fujisawa, Japan
- 2Cell Biology and Immunology Group, Wageningen University & Research, Wageningen, Netherlands
- 3Aquaculture and Fisheries Group, Wageningen Institute of Animal Science, Wageningen University & Research, Wageningen, Netherlands
Mammalian granulocyte colony-stimulating factor (G-CSF; CSF3) is a primary cytokine that promotes the development, mobilization, and activation of neutrophils and their precursors. Teleosts have been reported to possess two paralogs as a likely result of the teleost-wide whole genome duplication (WGD) event, but functional divergence of G-CSF paralogs remains poorly understood. Common carp are an allotetraploid species owing to an additional WGD event in the carp lineage and here, we report on genomic synteny, sequence similarity, and phylogeny of four common carp G-CSF paralogs (g-csfa1 and g-csfa2; g-csfb1 and g-csfb2). G-csfa1 and g-csfa2 show differential and relatively high gene expression levels, while g-csfb1 and g-csfb2 show low basal gene expression levels in most tissues. All paralogs are expressed higher in macrophages than in other leukocyte sub-types and are highly up-regulated by treatment of macrophages with mitogens. Recombinant G-CSFa1 and G-CSFb1 both promoted the proliferation of kidney hematopoietic cells, while only G-CSFb1 induced the differentiation of kidney cells along the neutrophil-lineage. Colony-forming unit assays revealed that G-CSFb1 alone stimulates the formation of CFU-G colonies from head- and trunk-kidney whereas the combination of G-CSFa1 and G-CSFb1 stimulates the formation of both CFU-G and CFU-GM colonies. Recombinant G-CSFa1 and G-CSFb1 also exhibit chemotactic activity against kidney neutrophils and up-regulation of cxcr1 mRNA expression was highest in neutrophils after G-CSFb1 stimulation. Furthermore, G-CSFb1 more than G-CSFa1 induced priming of kidney neutrophils through up-regulation of a NADPH-oxidase component p47phox. In vivo administration of G-CSF paralogs increased the number of circulating blood neutrophils of carp. Our findings demonstrate that gene duplications in teleosts can lead to functional divergence between paralogs and shed light on the sub-functionalization of G-CSF paralogs in cyprinid fish.
Introduction
Granulocyte colony-stimulating factor (G-CSF), also called colony-stimulating factor 3 (CSF3), is a primary cytokine that promotes the proliferation, differentiation and survival of neutrophil progenitors and enhances trafficking and immunological functions of mature neutrophils in mammals (1). Human G-CSF is produced mainly by monocytes and macrophages (2), but is also produced by fibroblasts (3), endothelial cells (4), and bone marrow stromal cells (5). Although healthy individuals express low G-CSF protein levels in serum, remarkable elevations of G-CSF production can be induced by several inflammatory stimuli, including increased presence of pro-inflammatory cytokines and LPS during infections (6–8). Effects are mediated by the binding of G-CSF with its cognate receptor G-CSFR on neutrophils and their progenitors, activating downstream signaling cascades and thereby influencing subsequent gene expression and cellular immune responses [reviewed in (1)]. Mice lacking G-CSF/G-CSFR signaling (G-csf-deficient or G-csfr-deficient mice) exhibit a reduction in myeloid progenitors and impaired neutrophil mobilization into the circulation, resulting in chronic neutropenia (9, 10). This suggests that G-CSF is a major regulator of neutrophil development and contributes to the regulation of multipotent hematopoietic progenitors. At the same time, G-CSF also influences the phenotype and function of mature neutrophils and does so by modulating expression of for example chemokine receptors, up-regulating phagocytosis and production of reactive oxygen species (ROS) and enhancing bactericidal activity of neutrophils (11).
G-CSF was first purified and characterized in mice (12), only later followed by studies in non-mammalian vertebrates such as chicken (13), African-clawed frog (14), and a number of teleost fish species including flounder, fugu, green-spotted pufferfish (13), black rockfish (15), and zebrafish (16). Owing to a teleost-specific whole genome duplication (WGD) event (17), teleost can generally be expected to express two G-CSF paralogs, type A (G-CSFa) and type B (G-CSFb), which may not necessarily have the same function. Indeed, zebrafish express an A and B paralog and earlier studies suggest that both G-CSFa and G-CSFb are required for hematopoietic stem cell (HSC) emergence and expansion of primitive and mature neutrophils and macrophages in vivo and in vitro (16). G-csfr morphants were affected on early myeloid cell migration and development, but had functionally normal myeloid cells (18). Zebrafish G-CSFb was involved in neutrophil mobilization toward an injury site (19), but the contribution of G-CSFa remained unclear. Therefore, the exact role of teleost G-CSF paralogs as regulators of diverse markers of neutrophil activation and/or regulators of multipotent hematopoietic progenitor development has remained unresolved.
In this study, we report on the molecular and functional characterization of G-CSF paralogs from the common carp. The close kinship of zebrafish and carp (20) allows for comparative use of genetic information from the well-described zebrafish genome whereas the large size of carp allowed us to perform cell type specific gene expression and ex vivo functional studies on large number of cells. Because common carp is an allotetraploid species owing to an additional WGD event in the carp lineage (21), we report on the cloning and molecular characterization of two type A copies (g-csfa1 and g-csfa2) and two type B copies (g-csfb1 and g-csfb2). For functional characterization we chose to produce recombinant proteins for two G-CSF paralogs particularly highly expressed in macrophages (G-CSFa1 and G-CSFb1) and examined their ability to drive proliferation, differentiation and colony-formation of carp hematopoietic kidney cells along the neutrophil lineage. We also studied the effect of these G-CSF paralogs on neutrophil function including phenotype, chemotaxis and production of ROS. In vivo effects of G-CSF paralogs on circulating blood neutrophils were further investigated. We discuss the functions of teleost G-CSF regarding development, trafficking and activation of neutrophils and discuss the importance of studying paralogs of granulocyte colony-stimulating factor.
Materials and Methods
Animals
Common Carp (Cyprinus carpio L.) were kept at Nihon University (NU) and at Wageningen University (WU). Carp weighing 40–100 g (10 to 15 cm in length) were purchased from commercial farms and reared at NU, Japan. Fish were kept at 23–25°C in a recirculation system with filtered water disinfected by ultraviolet light, fed with pelleted dry food (Hikari, Kyorin CO., LTD., Japan) daily and acclimated to this environment for at least 3 weeks prior to use for all experiments except Figures 2–4. Carp were also bred and reared in the Aquatic Research Facility of WU, the Netherlands. Here, carp were raised at 23°C in recirculating UV-treated tap water, fed pelleted dry food daily (Skretting, Nutreco) and utilized for experiments in Figures 2–4. Since G-CSF paralogs of Asian and European common carp show very high sequence identity (98 to 100%), we combined data from NU and WU. Experiments were performed in accordance with the guidelines of NU and WU and with approval of the animal experimental committee of WU.
Isolation of Carp Tissues and Leukocytes and Purification of Leukocyte Sub-types Such as B Cells, Granulocytes, Macrophages, Thymocytes and Thrombocytes
For tissue and cell isolation, carp were anesthetized with 0.01% Benzocaine (Sigma-Aldrich) or Tricaine Methane Sulfonate (TMS, Crescent Research Chemicals, Phoenix, USA), bled from the caudal vein and euthanized.
Leukocytes were obtained from kidney (head and/or trunk kidney) and spleen. Cell suspensions were obtained by macerating tissues on a sterile mesh in 10 mL of Eagle's minimal essential medium (MEM, Nissui, Tokyo, Japan). Cells were collected by centrifugation at 250 × g for 5 min at 4°C, re-suspended in 5 mL of MEM, layered onto a Percoll (1,075 g/cm3, GE healthcare) and centrifuged at 430 × g for 20 min at 4°C. Cells at the medium/Percoll interface (mononuclear cells) were harvested, washed twice with MEM by centrifugation, re-suspended with E-RDF medium (Kyokuto Pharm. Ind. Co.,Ltd., Tokyo, Japan) containing 20% fetal bovine serum and 2.5% carp serum (E-RDF20/2.5) and passed through 40 μm filter to remove aggregate.
Peripheral blood leukocytes (PBL) were obtained from carp blood. In short, 1 mL of blood was withdrawn from the caudal vein from fish with heparinized syringe, transferred to 9 mL of ice-cold MEM, layered onto a Percoll (1,075) and centrifuged at 430 × g for 20 min at 4°C without brakes. Cells at the medium/Percoll interface were harvested, washed twice with MEM by centrifugation and re-suspended with E-RDF20/2.5.
Kidney neutrophils were isolated as described previously (22) with minor modifications. Briefly, trunk kidney cells were layered onto a Percoll discontinuous gradient (1,080 and 1,100 g/cm3) and centrifuged at 430 × g for 20 min at 4°C. Cells at the 1,080/1,100 interface (neutrophils and erythrocytes) were harvested after complete removal of cells at the upper phase and then washed once. The neutrophil/erythrocyte pellet was treated with 1× red blood cell lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). Cells were washed twice with MEM by centrifugation and re-suspended with appropriate medium for each experiment. The purity of the neutrophils was verified to be >92% by a flow cytometry using a BD FACS Canto (BD Biosciences) and a peroxidase staining according to a DAB oxidization.
Thymocytes (23), macrophages (24), neutrophilic granulocytes (25) were obtained as previously described. Magnetic activated cell sorting (MACS) was used to isolate B cells and thrombocytes from peripheral blood leukocytes (PBLs) using anti-carp IgM [WCI12, (26)] and anti-carp thrombocytes [WCL6, (27)] antibodies and neutrophilic granulocytes from trunk kidney [using monoclonal antibody TCL-BE8; (25)]. The purity of the sorted cells was verified to be >98% by flow cytometry using a BD FACS Canto A (BD Biosciences).
Identification and in silico Analysis of Carp G-CSFs
Genomic loci of carp G-CSF were predicted by the Augustus gene prediction server using information on genes (med24, psmd3, and kpnb1) known to be neighboring G-CSF in several other species. Primers were designed against carp genomic sequences encoding putative carp G-CSFs. The complete list of primers used for PCR, RACE PCR, qRT-PCR and recombinant protein expression are listed in Supplementary Tables S2–S4. PCR reactions were performed using cDNA from carp kidney and heart with PrimeSTAR HS DNA polymerase (Takara, Shiga, Japan). Generated amplicons were gel purified using the FastGene Gel/PCR Extraction kit (Nippon genetics, Tokyo, Japan), ligated into the pMD20-T vector (Takara) using the 10 × A-attachment Mix (Toyobo, Osaka, Japan) and the Ligation high ver. 2 (Toyobo) and transformed into the competent Escherichia coli DH5α. Positive colonies were identified by colony PCR using the vector specific M13 RV and M4 primers, plasmids isolated using the FastGene Plasmid Mini kit (Nippon genetics) and inserts sequenced using a BigDye terminator v3.1 cycle sequencing kit (Applied Biosystems) and an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems). Sequence analysis was performed using the Genetyx version 11 (Genetic Information Processing Software) and sequences aligned and analyzed using BLAST programs.
G-CSF protein sequences from fish, amphibian and mammals were aligned using Clustal Omega software (EMBL-European Bioinformatics Institute). Signal peptide regions of respective G-CSF proteins were estimated using the SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP/) and conserved motifs were predicted using the SMART server (http://smart.embl-heidelberg.de/). Phylogenetic analysis was conducted using the MEGA version 6 software using the neighbor-joining (NJ) method and the Poisson method, and bootstrapped 1,000 times, with values expressed as percentages. The full-length sequences of carp G-CSFa1, G-CSFa2, G-CSFb1, and G-CSFb2 (accession number: MG882495, MG882496, MG882497, and MG882498, respectively) have been submitted to GenBank.
RT-qPCR Analysis of Gene Expression of Carp G-CSF Paralogs in Healthy Carp Tissues, Different Cell Types and Macrophages Stimulated With Mitogens
To investigate basal gene expression levels of G-CSF paralogs, tissues and leukocytes were collected from healthy carp (detailed in the figure legends), then total RNA was isolated using the RNeasy kit (QIAGEN) including on-column DNase treatment according to the manufacturer's instruction and stored at −80°C. cDNA was synthesized with SuperScript III First Strand Synthesis System (Invitrogen) according to manufacturer's instructions. Real-time quantitative PCR (RT-qPCR) was performed with a Rotor-Gene 6000 (Corbett Research) using ABsolute QPCR SYBR Green Mix (Thermo Scientific). Fluorescence data from RT-qPCR experiments were analyzed using Rotor-Gene software v1.7. The take-off value for each sample and the average reaction efficiencies (E) for each primer set were obtained upon Comparative Quantitation Analysis from Rotor Gene Software. The relative expression ratio (R) of target genes were calculated based on the average E and relative to the s11 protein of the 40 s subunit as a reference gene. Take-off values of samples in which genes of interest were non-detectable were given an arbitrary take-off value of 32.
Generation of Recombinant Carp G-CSFa1 and G-CSFb1
The portions of the carp G-CSFa1 and G-CSFb1 sequences corresponding to the mature, signal sequence-cleaved peptides were PCR amplified from carp kidney and heart cDNA using primers designed to meet the requirements of the pET-16b expression vector (Novagen). The resulting PCR products were ligated into pMD20-T vector, digested with two restriction enzymes, NdeI and BamHI, isolated by gel electrophoresis, ligated into the pET-16b which carry an N-terminal 10x His-tag, transformed into competent E. coli DH5α. Plasmids containing the in frame insert of carp G-CSFa1 or G-CSFb1 were transformed into the T7 polymerase expressing Rossetta-gami B (DE3) pLysS E. coli (Novagen), induced with appropriate IPTG and the optimal induction times and temperatures deduced in pilot runs. The bacteria were scaled up accordingly.
Recombinant carp G-CSFa1 and G-CSFb1 were purified from bacterial cells using Ni-NTA agarose (Qiagen, Hilden, Germany) according to the manufacturer's procedure. Briefly, transformed E. coli cells were grown in 100 mL LB medium containing 50 mg/mL ampicillin and 30 mg/mL chloramphenicol to density of OD600 = 0.5 at 37°C, and then cooled on ice. Expression of recombinant G-CSFa1 was induced by addition of 0.5 mM IPTG at 37°C for 4 h, and expression of recombinant G-CSFb1 was induced by addition of 0.25 mM IPTG at 25°C for 8 h. After shaking the cultures, cells were harvested, lysed in the lysis buffer (20 mM sodium phosphate, 500 mM NaCl, 50 mM imidazole, pH 7.4, 0.1% Triton-X and protease inhibitor cocktail) and sonicated. The insoluble materials were removed by centrifugation at 9,600 × g for 20 min and the supernatants were incubated with 500 μL of Ni-NTA agarose slurry at 4°C for 1–2 h with gentle rotation. The resin was then washed with 30 mL of the wash buffer (20 mM sodium phosphate, 500 mM NaCl, 50 mM imidazole, pH 7.4) on columns. Proteins were eluted from the resin using the elution buffer (20 mM sodium phosphate, 500 mM NaCl, 500 mM imidazole, pH 7.4). Subsequently, recombinant G-CSFa1 and G-CSFb1 were purified with gel-filtration to further clarify and simultaneously to determine their molecular weight under a native condition. Gel-filtration was performed using a Sephacryl S-100 column (HR 16/160, GE Healthcare) and the proteins were eluted with 20 mM sodium phosphate buffer containing 300 mM NaCl, pH 7.4 in 0.5 mL/min flow rate. The purified proteins were then passed through Pierce High Capacity Endotoxin Removal Columns (Thermo Fisher Scientific) to remove potential traces of endotoxin, buffer exchanged to 1x PBS with 0.05% BSA, filter sterilized (0.22 μm) and stored at 4°C or −80°C until use. Residual endotoxin was checked to be less than 5 pg/mL according to a Limulus ES-II Single Test (Wako, Osaka, Japan). A Bradford assay (Thermo Fisher Scientific) was performed according to manufactures' directions to determine protein concentration.
Recombinant carp erythropoietin (EPO), kit ligand A (KITLA) and thrombopoietin (TPO) were produced and purified as described previously (28, 29).
Cell Proliferation Assay
Freshly isolated carp head and trunk kidney leukocytes from 4 individuals were adjusted to a concentration of 4–8 × 105 cells/mL in E-RDF20/2.5 medium. Fifty microlitre of this cell suspension was added to each well of a 96-well plate to which 50 μL of treatment in E-RDF20/2.5 medium was added. Treatments consisted of the E-RDF medium (negative control), 25% cell conditioned medium (CCM) derived from the carp kidney leukocyte culture in which macrophages develop [positive control, (30)], a combination of 100 ng/mL TPO and 100 ng/mL KITLA (positive control), recombinant G-CSFa1 or G-CSFb1 at final concentrations of 500, 100, 20, 4, 0.8, 0.16 ng/mL and heat-inactivated (98°C for 30 min) recombinant G-CSFa1 and G-CSFb1. Cell proliferation was determined using the colorimetric MTT assay (Nacalai Tesque, Kyoto, Japan) which was first shown to provide comparable data for different leukocyte cell types (data not shown). Briefly, 10 μL of MTT reagent was added to each well and plates were incubated at 30°C for 5 h to develop a coloration reaction depend on live cell number. One hundred microlitre of solubilization solution (acid-isopropanol) was then added to each well and plates were sealed and kept at 30°C for 12 h. Cell proliferation was determined on days 0, 3, 6, and 9, and plates were read at absorbance of 570 nm and 650 nm as a reference using Multiskan GO microplate reader (Thermo Fisher Scientific). Values obtained at absorbance of 650 nm from each well were subtracted from values obtained at absorbance of 570 nm from each well.
RT-qPCR Analysis of Gene Expressions in Cells Treated With Recombinant G-CSFa1 and/or G-CSFb1
Freshly isolated carp kidney leukocytes were seeded into 24-well plates in 0.5 mL of E-RDF20/2.5 at a concentration of 4 × 105 cells/mL. Cells were either treated with the medium (untreated control), recombinant G-CSFa1 (100 ng/mL final concentration), recombinant G-CSFb1 (100 ng/mL final concentration) or the combination of 100 ng/mL G-CSFa1 and 100 ng/mL G-CSFb1 in the E-RDF20/2.5 for 12 h and 4 days at 30°C. Following incubation, cells were collected, total RNA was isolated using the NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany) and reverse transcribed into cDNA using the Omniscript RT kit (Qiagen) according to manufacturer's instructions. Quantitative PCR was performed for carp transcription factors known to be involved in early hematopoiesis (gata2) (31), myelopoiesis (pu.1, cebpa and irf8) (32–35), erythropoiesis (gata1) (36) and lymphopoiesis (gata3 and pax5) (37, 38) and myeloid cell markers (gcsfr1, gcsfr2, csf1r and mpx) (22, 35) using a Thermal Cycler Dice Real Time System II (Takara). Beta-actin (β-actin) was employed as an endogenous control. Quantitative PCR cycling conditions were 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Data were analyzed using the Thermal Cycler Dice Real Time System software (Takara) and is represented as the average of the four fish (n = 4) with standard deviation.
Likewise, kidney neutrophils from carp were treated with the medium, G-CSFa1 and G-CSFb1 in the E-RDF20/2.5 for 6 h at 30°C, RNA isolated, and cDNA synthesis as described above. Q-PCR was performed for carp chemokine receptors (cxcr1, cxcr2 and cxcr4) (39, 40) and NADPH oxidase components (gp91phox, p22phox, p40phox, p67phox, and p47phox) (41). Beta-actin (β-actin) was employed as an endogenous control. Quantitative PCR cycling conditions were 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Data were analyzed using the Thermal Cycler Dice Real Time System software (Takara) and is represented as the average of the three fish (n = 3) with standard deviation.
Semi-solid Colony-Forming Unit Assay
Freshly isolated leukocytes from carp kidney, spleen or peripheral blood were re-suspended to 2 × 105 cells/mL in E-RDF medium containing 40% FBS and 5% carp serum. Colony-forming unit assay using semi-solid media was carried out as previously described (28) with minor modifications. Briefly, a complete methylcellulose medium was prepared by mixing a 2.0% methylcellulose stock solution with an equal amount of the cell suspension. In some cases (for experiments of colony counting based on the morphology), a complete 0.45% agarose medium was prepared by mixing an 1.8% agarose solution; 2 × E-RDF medium; and the cell suspension in the volume ratio of 1:1:2. Then, 2.4 mL of the cell suspension/complete semi-solid medium was added to a 5 mL tube with a 2.5 mL syringe and 16-gauge blunt-end needle, along with cytokines or PBS. Tubes were tightly capped and the solution gently mixed. One milliliter of the solution (in duplicate) was aliquoted onto a solid E-RDF medium containing 0.45% agarose (Lonza), 20% FBS and 2.5% carp serum in a 35 mm dish or a 6-well plate. Dishes and Plates were incubated at 30°C with an additional 5% CO2 atmosphere and 100% humidity for 7–13 days, followed by examination for colony formation.
The number of progenitor cells in each organ was estimated according to the formula described below.
Characterization of Colony Cells
Cell colonies formed in the methylcellulose media were aspirated by micropipette under a microscope and characterized by morphology, cytochemistry and RT-PCR analyses as previously described (28). In short, the colony cells were re-suspended in MEM, and cyto-centrifuged with a Cytospin (Shandon). Slides were dried, fixed and stained with May-Grünwald Giemsa (MGG, Wako Pure Chemicals, Osaka, Japan) or Peroxidase stain based on the DAB oxidization. For RT-PCR analyses, total RNAs were extracted from each colony cell type using RNeasy Micro Kit (Qiagen) and cDNA was synthesized using Omniscript RT kit (Qiagen). Expression of hematopoietic marker genes was analyzed by PCR using carp specific primers and EmeraldAmp PCR Master Mix (Takara). PCR was conducted as follows: one cycle of 94°C for 1 min, followed by 23 to 38 cycles of denaturation at 98°C for 30 sec, annealing at 58°C for 30 s and elongation at 72°C for 30 s. Colonies treated with recombinant carp erythropoietin (EPO) was utilized for the control group of the erythroid lineage.
Neutrophil Chemotactic Response to Recombinant Carp G-CSFa1 and G-CSFb1
Neutrophils obtained from carp trunk kidney were washed twice in MEM and adjusted to a final concentration of 1 × 106 cells/mL. The chemotaxis assay was performed using blind well chemotaxis chambers (Neuro Probe, Inc.). Two hundred microliters of different concentrations of recombinant carp G-CSFa1 or G-CSFb1 (1, 10, and 100 ng/mL, final concentrations) in the serum free MEM were added to the bottom well of the chemotaxis chambers, and the bottom chamber was separated from the top chamber using 5 μm pore size polycarbonate membrane filters (Neuro Probe, Inc.). To the top chamber, 200 μL of neutrophil suspension was added. Negative controls consisted of medium alone and the positive control was 10 ng/mL of fMLP (Sigma-Aldrich). The chemokinesis control consisted of 100 ng/mL of G-CSFa1 or 100 ng/mL of G-CSFb1 in both the upper and lower chambers of the chemotaxis apparatus.
The incubation period was 1 h after which the cell suspensions were carefully aspirated from the top chamber and the filters removed and applied bottom side up on a slide glass. Filters were stained with MGG. Chemotactic activity was determined by counting the total number of cells found on the underside of the polycarbonate filters in 20 random fields of view (40 × magnification). Technical duplicates were conducted for all treatments (n = 4, biological replicates).
Respiratory Burst Assay
Respiratory burst assay was performed as previously described (42) with minor modifications. The neutrophils harvested from carp kidney were re-suspended in E-RDF20/2.5 medium at a concentration of 2.5 × 106 cells/mL. Four hundred microliters of the cell suspension was added to each 1.5 mL tubes, and cells were treated or untreated with recombinant carp G-CSFa1 (100 ng/mL) or G-CSFb1 (100 ng/mL) at 25°C for 6 h. Following the incubation, DHR123 (Molecular Probes) was added to the cells at a final concentration of 10 μM and incubated for 5 min to allow the cells to take up the DHR123. PMA (Abcam, Cambridge, UK) was then added at a final concentration of 100 ng/ml. The cells were further incubated for 30 min to allow oxidation of the DHR. All samples were appropriately staggered with respect to timing to accommodate the transient state of oxidized DHR fluorescence. Live cells were gated according to the forward scatter and side scatter parameters. DHR fluorescence was detected in the FITC channel, and the mean values of the FITC fluorescence in neutrophils were normalized to untreated controls.
In vivo Effects of G-CSFa1 and G-CSFb1
Carp were bled 200 μL from the caudal vein using a heparinized syringe 2 days before administration of G-CSF paralogs and blood was centrifuged in a capillary glass tube at 1,500× g for 5 min. Leukocytes on top of the erythrocyte layer were obtained, treated with the 1× red blood cell lysis buffer, washed twice with Hanks' balanced salt solution and then used to measure the ratio of neutrophils per total number of leukocytes by flow cytometry analysis, based on forward scatter vs. side scatter parameters. Subsequently, carp were injected intraperitoneal (i.p.) with 100 ng/g body weight of recombinant proteins in 200 μL of 1×PBS, or were injected with 1 × PBS only. After 6, 24 and 48 h, 200 μL of peripheral blood was collected and analyzed as described above. Three fish for each group were examined.
Statistical Analysis
Raw data of technical replicates were first averaged per individual before statistical analysis was performed. Statistical analysis was performed after log-transformation of datasets that were not normally distributed. Subsequently, normality was assumed and statistical significance was tested using an un-paired Student's t-test (independent observations, Figure 3) for one-to-one comparisons and a (repeated-measures) one-way analysis of variance (ANOVA) followed by a Dunnet's post-hoc test (in Figures 4, 5A,D,E, 7, 8). Or, Tukey's HSD (Figure 2) and Dunnet's T3 (in case of unequal variances) (Figure 2) were used for multiple comparisons. A two-way ANOVA was performed followed by Tukey's post-hoc test for multiple comparisons shown in Figures 9, 10. Prism 7 software (GraphPad Software, La Jolla, CA, USA) was used. In absence of sphericity, the Geisser-Greenhouse correction was applied. A value of p ≤ 0.05 was accepted as significant.
Results
Identification of Four Carp G-CSF Paralogs
The presence of four G-CSF paralogs in the genome of common carp was expected as common carp has undergone a WGD event compared to the zebrafish (21), in which two G-CSF paralogs are already present. Referring to carp genome and transcriptome databases (project no. PRJEB7241 and PRJNA73579) at the National Center for Biotechnology Information (NCBI), four putative loci encoding carp G-CSF homologs were found next to conserved genes PSMD3, MED24, or LRRC3 (Figure 1A). Synteny of each paralog is highly conserved with either the zebrafish G-CSFa locus on chromosome 12 or the GCSFb locus on chromosome 19.
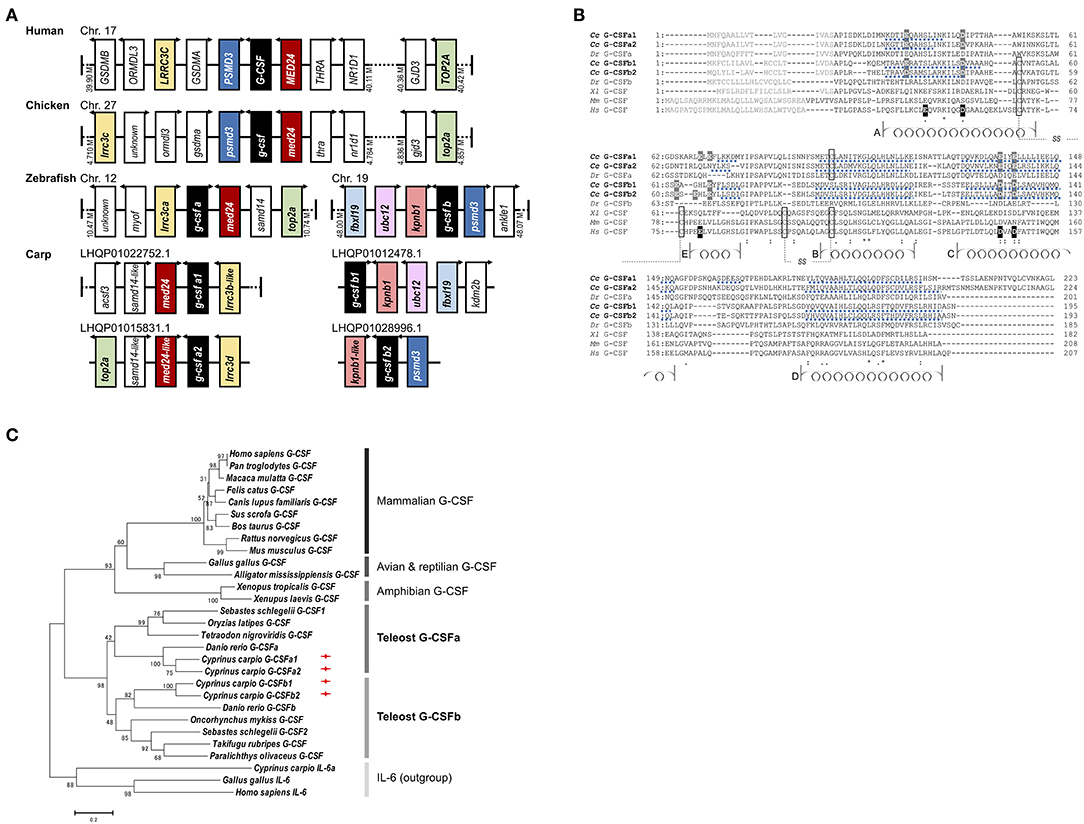
Figure 1. Granulocyte colony-stimulating factor paralogs of common carp. (A) Synteny analysis of G-CSF of human, chicken, zebrafish, and carp. The G-CSF locations in human, chicken, and zebrafish are obtained from respective genome resources at NCBI and carp G-CSF locations are obtained from carp genome and transcriptome projects PRJEB7241 and PRJNA73579 at NCBI. (B) Alignment of vertebrate granulocyte colony-stimulating factor (G-CSF) amino acid sequences. Complete protein sequences of carp Cc G-CSFa1 (Cyprinus carpio, MG882495), carp Cc G-CSFa2 (MG882496), zebrafish Dr G-CSFa (Danio rerio, NP_001138714), carp Cc G-CSFb1 (MG882497), carp Cc G-CSFb2 (MG882498), zebrafish Dr G-CSFb (NP_001137226), african clawed frog Xl G-CSF (Xenopus laevis, Scaffold13265:3008399-3011722), mouse Mm G-CSF (Mus musculus, NP_034101), and human Hs G-CSF (Homo sapiens, NP_000750) were aligned using Clustal Omega. Signal sequences are shown with gray letters and conserved cysteine residues are boxed. In the human G-CSF, helices (A to E) are denoted with coils, and Glutamic acid (E) and Aspartic acid (D) residues representing major interfaces with G-CSF receptor are highlighted in black. In the carp G-CSFs, predicted alpha helical regions are underlined with a broken line, modeled on the structure of human G-CSF, and residues expected to interact with the receptor are highlighted in gray. Amino acids that are conserved in all sequences are denoted with an asterisk (*), strong similarity with a colon (:), and weak similarity with a period (.). (C) Phylogenetic analysis of mammalian, avian, reptilian, amphibian and teleost fish G-CSF proteins. Phylogenetic analysis included carp G-CSFa1 (C. carpio, MG882495); carp G-CSFa2 (MG882496); carp G-CSFb1 (MG882497); carp G-CSFb2 (MG882498); zebrafish G-CSFa (D. rerio, NP_001138714); zebrafish G-CSFb (NP_001137226); rockfish G-CSF1 (S. schlegelii, BAH56611); rockfish G-CSF2 (BAH56612); medaka G-CSF (O. latipes, XP_004080425); green-spotted puffer G-CSF (T. nigroviridis, CAG04394); rainbow trout G-CSF (O. mykiss, CAQ42965); fugu G-CSF (T. rubripes, XP_003965085); flounder G-CSF (P. olivaceus, BAE16320); african clawed frog G-CSF (X. laevis, Scaffold13265:3008399-3011722); tropical clawed frog G-CSF (X. tropicalis, XP_002940261); alligator G-CSF (A. mississippiensis, XP_006270858); chicken G-CSF (G. gallus, NP_990610); human G-CSF (H. sapiens, NP_000750); chimpanzee G-CSF (P. troglodytes, XP_009430519); rhesus monkey G-CSF (M. mulatta, XP_001095097); cat G-CSF (F. catus, NP_001009227); dog G-CSF (C. lupus familiaris, XP_005624600); pig G-CSF (S. scrofa, XP_005653977); cattle G-CSF (B. taurus, NP_776453); rat G-CSF (R. norvegicus, NP_058800); mouse G-CSF (M. musculus, NP_034101); and outgroup including carp interleukin-6a (IL-6a, AGR82313); chicken IL-6 (NP_989959); and human IL-6 (NP_000591). Phylogenetic analysis was conducted using the neighbor joining method in MEGA version 6 and bootstrapped 1,000 times with bootstrap values expressed as percentages.
The complete open reading frames (672, 675, 588, and 582 bp) of four carp paralogs' cDNA transcripts, respectively, encoding 224, 225, 196, and 194 amino acids with 5 exons were obtained (Figure 1B and Supplementary Figures S1A–D). Despite quite low sequence identity and similarity (Supplementary Table S1), carp G-CSF paralogs share a similar predicted structure and one of the copies (G-CSFa1) was predicted to have an additional helical region from Ser160 to Ser164 which is acidic amino acid residue (Asp and Glu)-abundant (Figure 1B and Supplementary Figure S2). Carp G-CSF paralogs all possess the consensus domain of Pfam IL6/GCSF/MGF protein family, whereas the four cysteine residues involved in two disulfide bonds are not conserved. Carp G-CSF copies also share conserved acidic amino acids involved in major ligand-receptor binding demonstrated in human G-CSF, while there is no acidic amino acid residue near the α-helix E in carp G-CSFa2 (Figure 1B). Phylogenetic analysis revealed that all the G-CSFs were found to form a single evolutionary clade outside a related cytokine interleukin-6, suggesting that the G-CSFs are orthologous. Taking into account there may be G-CSF paralogs present in the teleost species shown that have not yet been reported, each of the four carp paralogs did cluster with either teleost G-CSFa or G-CSFb paralogs (Figure 1C). Hence, based on clustering and the conserved synteny, we named the four carp G-CSF paralogs as G-CSFa1, G-CSFa2, and G-CSFb1, G-CSFb2.
Carp g-csf Paralogs Show Differential Expression in Immune Tissues and Cells
Assessment of basal g-csfa1 expression in tissues from healthy carp revealed generally very low expression of g-csfa1 in most tissues, with significantly higher gene expression in spleen, muscle and gill (Figure 2). G-csfa2 was significantly higher expressed in spleen than in gill, brain, thymus, trunk-kidney and head-kidney (Figure 2). Basal expression levels of g-csfb1 and g-csfb2 were generally low or non-detectable in most carp tissues examined (Figure 2).
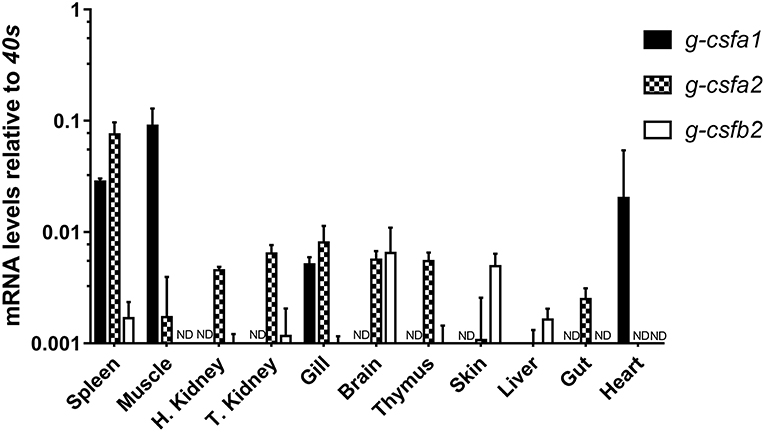
Figure 2. Quantitative mRNA expression analysis of carp g-csf paralogs in carp tissues. Basal gene expression of carp G-CSF paralogs in spleen, muscle, head kidney, trunk kidney, gill, brain, thymus, skin, liver, gut, and heart. Basal expression levels were determined relative to the s11 protein of the 40s subunit (40s) as a reference gene and are presented as mean + standard deviation (n = 3, except thymus, n =2). G-csfb1 expression was non-detectable in all tissues examined (ND indicates “non-detectable”). Significant differences in expression between tissues were determined using one-way ANOVA followed by Tukey's HSD (g-csfa1 and g-csfb2) or Dunnet's T3 post-hoc test for unequal variances (g-csfa2).
At basal levels, g-csfa expression is markedly higher than g-csfb expression in all immune cells examined. Strikingly, all g-csf paralogs were highest expressed at basal level in macrophages, indicating these cells as the major producers of G-CSF, comparable to mammalian macrophages. Within macrophages, g-csfa1 and g-csfb1 were significantly higher expressed compared to their respective counterparts (Figures 3A,B). Remarkably, a clear expression of g-csfa1 was observed also in thrombocytes (Figure 3A).
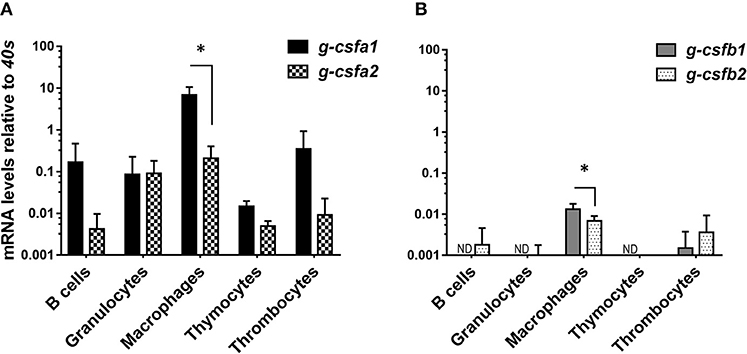
Figure 3. Quantitative mRNA expression analysis of carp g-csf paralogs in immune cells. Basal gene expression of carp g-csfa1 and g-csfa2 (A) and g-csfb1 and g-csfb2 (B) in sorted cells from healthy carp. Basal expression levels were determined relative to the s11 protein of the 40s subunit (40s) as a reference gene and presented as mean + standard deviation (n = 4 except thymocytes and thrombocytes n = 2). Statistical significance within the macrophage group was determined using an un-paired Student's t-test. Asterisks (*) denotes significance (p < 0.05) between indicated genes. ND indicates “non-detectable”.
Expression of Carp g-csf Paralogs Are Immediately Enhanced After Stimulation
In order to determine induction of the different G-CSF paralogs upon antigenic stimulation, we investigated expression levels in freshly isolated kidney leukocytes and head kidney-derived macrophages following the stimulation with LPS, ConA, PMA and poly I:C (only freshly isolated kidney leukocytes). In freshly isolated kidney leukocytes, all paralogs were highly up-regulated after stimulation with LPS and the combination of ConA/PMA at 3 and 6 h but not after stimulation with Poly I:C (Supplementary Figure S3). Likewise, in the cultured macrophages gene expressions of the four paralogs were clearly enhanced by LPS and PMA stimulations (Figure 4). Despite non-detectable g-csfb1 transcripts, its gene expression was induced with LPS stimulation. Interestingly, in macrophages both g-csfa1 and g-csfa2 are relatively high expressed at basal level (Figure 3) and appear to show a relatively small increase upon stimulation with LPS (Figure 4), whereas g-csfb1 and g-csfb2, which are relatively low or non-detectable at basal level (Figure 3), show a large increase in gene expression after LPS stimulation (Figure 4). We could also show that expression in macrophages of interleukin-1 beta, which is a pro-inflammatory cytokine, was significantly up-regulated after LPS stimulation (Figure 4).
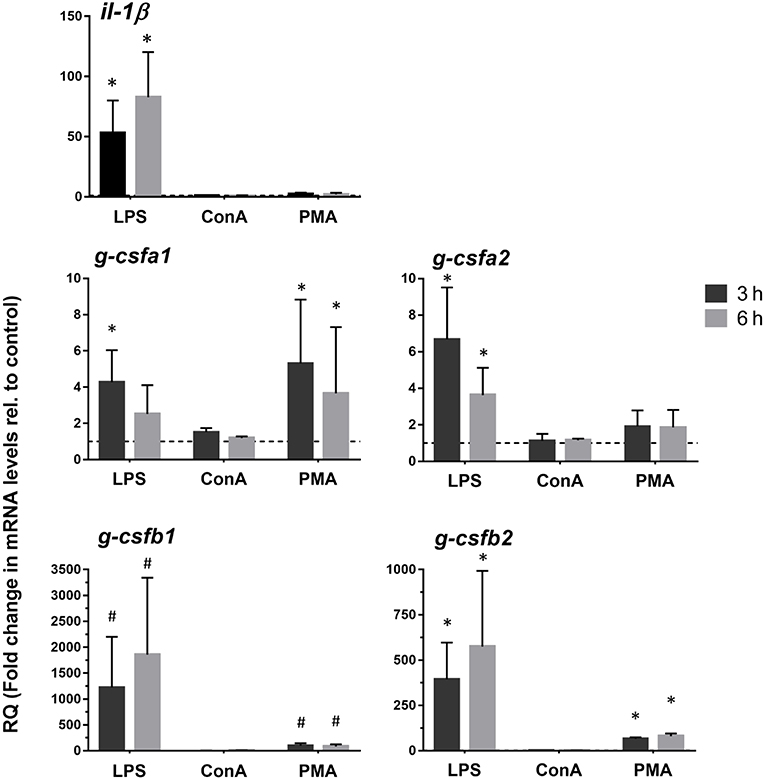
Figure 4. Quantitative expression analysis of carp g-csf paralogs in stimulated macrophages. Gene expression analysis of head kidney-derived macrophages stimulated for 3 or 6 h with 50 μg/mL lipopolysaccharide (LPS), 10 μg/mL Concanavalin A (ConA) or 1 μg/mL phorbol myristate acetate (PMA). Gene expression was normalized relative to the s11 protein of the 40s subunit as a reference gene and expressed relative to unstimulated timepoint controls (dashed line at y = 1). Data are presented as mean + standard deviation (n = 4). Significant differences compared to unstimulated timepoint controls were determined using one-way ANOVA followed by Dunnet's post-hoc test, (p < 0.05) are denoted by asterisks (*). Non-detectable samples were given an arbitrary value of CT = 32. Hash mark (#) indicate significant differences using these arbitrary values.
Recombinant Carp G-CSFa1 and G-CSFb1 Are Monomeric Forms
Based on expression levels in macrophages and the clear induction in stimulated macrophages, we chose to express two copies, G-CSFa1 and G-CSFb1, to investigate their function. Recombinant G-CSFa1 and G-CSFb1 purified using Ni-affinity chromatography were passed through a gel filtration column under a non-denaturing condition to calculate their molecular weights. As a result, the molecular weights of G-CSFa1 and G-CSFb1 were estimated to 25,275 and 22,355, similar to the deduced values based on their primary structures and similar to the result of SDS-PAGE under the denaturing condition, indicating that both recombinants form monomers (Supplementary Figure S4).
Both G-CSFa1 and G-CSFb1 Induce Proliferation of Kidney Leukocytes, but Only G-CSFb1 Induce Differentiation of Cells Along the Neutrophil Lineage
Carp kidney leukocytes treated with the cell conditioned medium containing macrophage growth factor(s) and recombinant TPO plus KITLA exhibited active proliferation, indicating that there are heterogeneous hematopoietic progenitors in the kidney leukocytes (Figure 5A and Supplementary Figure S5). Treatment of carp kidney leukocytes with 0.8, 4, 20, 100, and 500 ng/mL of G-CSFa1 induced a dose-dependent proliferative response, with the highest proliferation observed in cells treated with more than 20 ng/mL of G-CSFa1, whereas heat-inactivated (98°C for 30 min) G-CSFa1 had no effect. Likewise, treatment of kidney leukocytes with 4, 20, 100, and 500 ng/mL of G-CSFb1 induced a dose-dependent proliferative response, with the highest proliferation observed in cells treated with more than 100 ng/mL of G-CSFb1, whereas heat-inactivated G-CSFb1 had no effect (Figure 5A and Supplementary Figure S5). Furthermore, treatment of kidney leukocytes with a combination of 100 ng/mL of G-CSFa1 and 100 ng/mL of G-CSFb1 enhanced the proliferative response compared with those cells treated with G-CSFa1 alone or G-CSFb1 alone (data not shown).
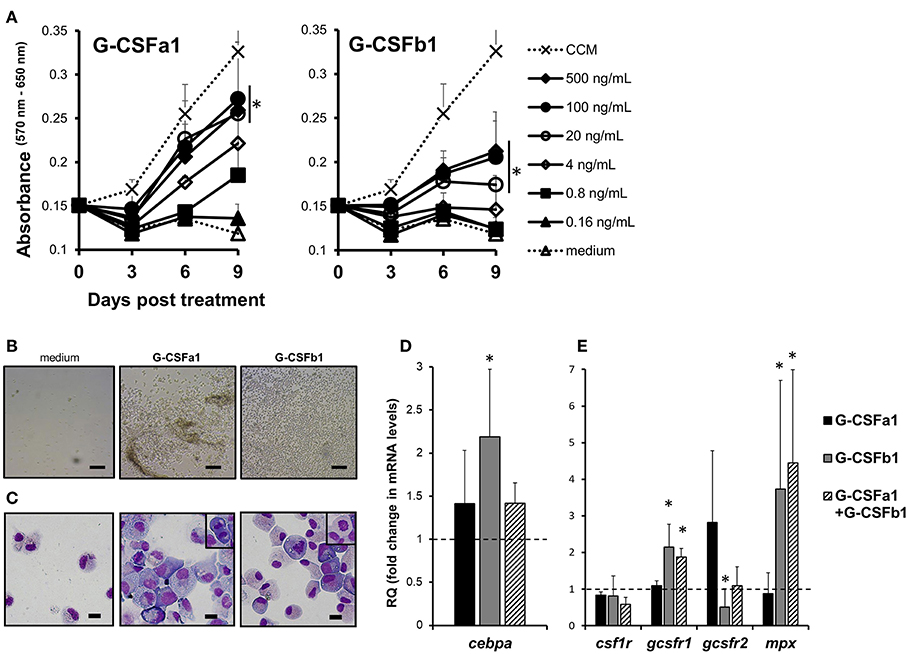
Figure 5. Proliferation and differentiation of carp kidney neutrophilic granulocyte-like cells. (A) Proliferative response of carp kidney leukocytes (20,000 cells) treated with medium alone, cell conditioned medium (CCM) derived from kidney leukocyte cultures in which macrophages develop, or recombinant carp G-CSF paralogs at different doses. Live cells treated with each stimulus were measured with the MTT assay at day 0, 3, 6, and 9 in the culture. Absorbance values at 650 nm were subtracted from experimental absorbance values at 570 nm in each well. Each point in the graphs represents mean + standard deviation (n = 4). Significant differences compared to medium-treated controls in day 9 were determined using one-way ANOVA followed by Dunnet's post-hoc test, (p < 0.05) are denoted by asterisks (*). (B) Photomicrographs of liquid cultures in the absence (medium only) or presence of recombinant G-CSFa1 and G-CSFb1 after 8 days of culture. Scale bars indicate 100 μm. (C) May-Grunwald Giemsa staining of kidney cells after 8 days culture in the absence or presence of G-CSFa1 and G-CSFb1. Mitotic figures were frequently observed (small enclosure). Scale bars indicate 10 μm. (D) Quantitative gene expression analysis of carp transcription factors involved in granulopoiesis (cebpα) in carp kidney leukocytes treated or untreated with G-CSFa1 (100 ng/mL), G-CSFb1 (100 ng/mL) or a combination of G-CSFa1 (100 ng/mL) and G-CSFb1 (100 ng/mL) for 12 h. The mRNA levels were calculated using β-actin as a reference gene. Data were normalized to the control cells (dashed like at y = 1) and mean + standard deviation is shown (n = 4). Significant differences compared to unstimulated controls were determined using one-way ANOVA followed by Dunnet's post-hoc test, (p < 0.05) are denoted by asterisks (*). (E) Quantitative gene expression analysis of myeloid cytokine receptors and myeloperoxidase in carp kidney leukocytes treated or untreated with G-CSFa1, G-CSFb1 or a combination of G-CSFa1 and G-CSFb1 for 4 days. The mRNA levels were calculated using β-actin as a reference gene. Data were normalized to the control cells (dashed line at y = 1) and mean + standard deviation is shown (n = 4). Significant differences compared to unstimulated controls were determined using one-way ANOVA followed by Dunnet's post-hoc test, (p < 0.05) are denoted by asterisks (*).
A lot of growing cells treated with G-CSFa1 adhered onto the plastic and with each other, whereas cells treated with G-CSFb1 exhibited low adhesive property and dispersed (Figure 5B). Morphologically, most cells treated with G-CSFa1 for 8 days were blast-like cells, having a basophilic cytoplasm and round to oval nuclei (Figure 5C). In contrast, the cells treated with G-CSFb1 for 8 days appeared to be at different developmental stages from myeloblast-like to metamyelocyte-like (Figure 5C). Most growing cells with each treatment were ascertained to be myeloid cells by staining with TCL-BE8 monoclonal antibody which mainly binds to carp neutrophils (43) (data not shown).
To characterize the roles of G-CSFa1 and G-CSFb1, we examined the gene expressions of transcription factors (TFs) and cell surface markers involved in the development of various cell lineages in carp kidney leukocytes treated with G-CSFa1, G-CSFb1 and a combination of G-CSFa1 and G-CSFb1. The mRNA levels of the TFs involved in myelopoiesis (pu.1, cebpα and irf8), early hematopoiesis (gata2), erythropoiesis (gata1) and lymphopoiesis (gata3 and pax5) in cells treated with or without G-CSFa1, G-CSFb1 and a combination of them for 12 h were analyzed by quantitative PCR. Kidney cells treated with G-CSFa1 did not undergo any change of TFs mRNA levels. On the other hand, kidney cells treated with G-CSFb1 exhibited a significant up-regulation of cebpα mRNA levels compared to those of the medium-treated controls (Figure 5D), while other TFs tested showed no significant change (data not shown). Cells treated with the combination of G-CSFa1 and G-CSFb1 also showed a moderate up-regulation of cebpα levels compared to those of the controls (Figure 5D). Next, we examined whether G-CSFa1 and G-CSFb1 modulate expression of myeloid cytokine receptors and neutrophil-specific myeloperoxidase in carp kidney cells. The mRNA levels of csf1r, gcsfr1, gcsfr2, and mpx in cells treated with the same treatments for 4 days were analyzed by quantitative PCR. Expression of csf1r, which is the macrophage colony-stimulating factor receptor gene, in the kidney cells was unaffected with any treatment examined (Figure 5E). Gcsfr1 and mpx expression in the kidney leukocytes was up-regulated with the treatment of G-CSFb1 alone and the combination of G-CSFa1 and G-CSFb1, but not with G-CSFa1. On the other hand, gcsfr2 expression in the kidney leukocytes shows a trend toward upregulation with G-CSFa1 treatment but downregulation with G-CSFb1 treatment (Figure 5E).
G-CSFb1 Stimulates Granulocyte Colony Formation and Cooperates With G-CSFa1 to Stimulate Granulocyte/Macrophage Colony Formation
In order to further examine the hematopoietic function G-CSFa1 and G-CSFb1 and identify granulocyte progenitor cells, we used an in vitro methylcellulose/agarose colony assay system. As expected, plating of carp kidney leukocytes (100,000 cells) without addition of cytokine resulted in no colony formation (data not shown). In the presence of G-CSF paralogs, overall two types of colonies appeared (Figure 6A). Surprisingly, when carp kidney leukocytes were cultured with G-CSFa1 alone, few colony formations were observed at any dose (Figure 6B left). On the other hand, in the presence of G-CSFb1, approximately 25 homogeneous colonies were formed after 7 days of the incubation (Figure 6B middle). These colonies consisted of uniform small round cells scattered (type 1, Figure 6A left). When kidney cells were cultured with a combination of G-CSFa1 and G-CSFb1, morphologically two kinds of colonies were observed. One appeared to be similar to the type 1 colonies formed in the presence of G-CSFb1 alone, the other seemed to consist of roughly agminated cells with distinct sizes and shapes (type 2, Figure 6A right). Approximately ten type 1 colonies per 100,000 cells plated were formed at day 5 to 7 in the culture and then gradually disappeared. The peak of type 2 colony formation (about 20 colonies per 100,000 cells plated) was observed after 11 days of cultivation (Figure 6B right). Both type 1 and type 2 colony cells consisted of morphologically neutrophil lineage cells at distinct developmental stages, which are myeloperoxidase-positive and –negative (Figures 6C,D). To characterize colony types, the expression of lineage-associated marker genes was analyzed. Figure 6E shows a typical expression patterns in type 1 and type 2 colonies. Type 1 colonies treated with G-CSFb1 alone or G-CSFa1 plus G-CSFb1 highly expressed g-csfr, cebpα, and mpx mRNAs involved in neutrophil development and slightly expressed csf1r which is the macrophage colony-stimulating factor receptor gene, but did not express other genes examined, indicating that type 1 colonies are derived from the progenitor cells corresponding to mammalian granulocyte colony-forming units (CFU-G). Type 2 colonies treated with the combination of G-CSFa1 and G-CSFb1 highly transcribed not only neutrophil-specific marker genes but also monocyte/macrophage lineage markers csf1r and irf8, suggesting that type 2 colonies are derived from the progenitors corresponding to mammalian granulocyte/macrophage CFU (CFU-GM) (Figure 6E).
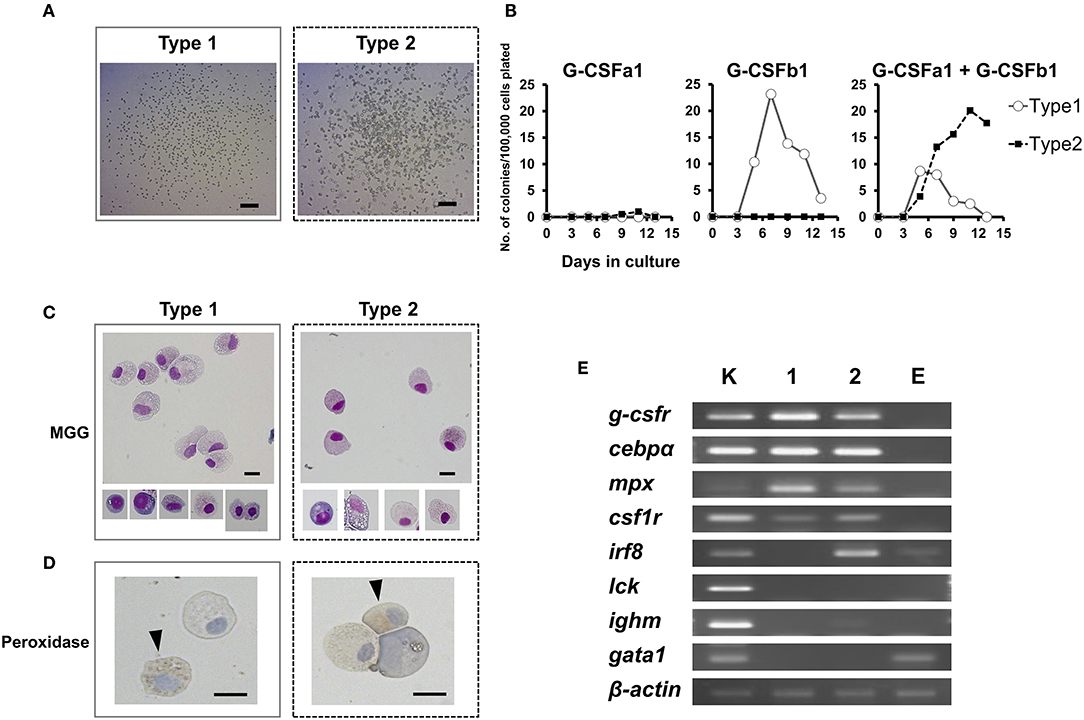
Figure 6. Colony formation of kidney cells in response to recombinant carp G-CSFa1 and G-CSFb1. (A) Colony-formation of kidney cells in response to G-CSF paralogs. Overall two types of colonies (type 1 and type 2) were observed. Bars indicate 200 μm. (B) Colony counts during semi-solid culture of kidney cells (1 × 105) in the presence of 100 ng/mL G-CSFa1 alone, 100 ng/mL G-CSFb1 alone, or a combination of 100 ng/mL G-CSFa1 and 100 ng/mL G-CSFb1. Each point indicates mean colony counts from 4 individual fish under each condition. Cultures scored every 2 days between 3 and 13 days of incubation. (C) May-Grünwald Giemsa (MGG) staining of colony cells (type 1; left and type 2; right). Bars indicate 10 μm. (D) Peroxidase-staining of cells obtained from type 1 colonies (left) and type 2 colonies (right), counterstained with Mayer's Hematoxylin. Arrow heads indicate myeloperoxidase-positive cells. Bars indicate 10 μm. (E) RT-PCR analysis for expression of lineage-associated marker genes in type 1 (lane 1) and type 2 (lane 2) colony cells. cDNA from carp kidney leukocytes was used as a positive control (lane K). cDNA from cells cultured in the presence of 100 ng/mL carp EPO was utilized for the control group of the erythroid lineage (lane E).
Granulocyte/Macrophage Progenitors and Granulocyte Progenitors Are Localized in the Head Kidney and Trunk Kidney but Not in the Spleen of Carp
To assess the contribution of hematopoietic organs to the neutrophil development in common carp, a myeloid colony forming assay was performed. Leukocytes were harvested from head kidney, trunk kidney, spleen and peripheral blood of adult carp (10 to 15 cm in length). Approximately 1 × 105 cells were cultured in the methylcellulose/agarose media in the presence or absence of 100 ng/mL G-CSFa1 plus 100 ng/mL G-CSFb1 and colony counts were performed after 6–11 days in the culture. PBLs and splenocytes did not form any colonies regardless of addition of cytokine or not. Conversely, cells from head kidney and trunk kidney formed about 25 to 40 colonies of each of type 1 and type 2 in the presence of both G-CSFa1 and G-CSFb1. Total number of CFU-G and CFU-GM in each organ was estimated as Table 1.
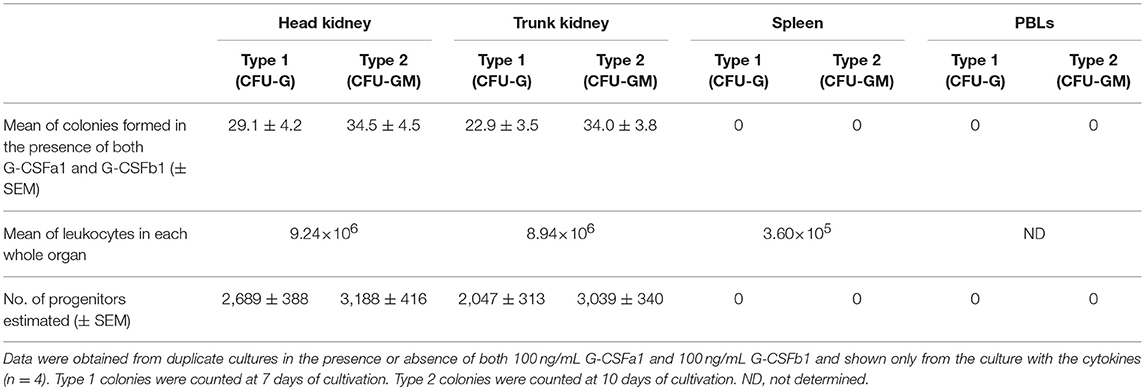
Table 1. The number of type 1 and type 2 colonies formed from 100,000 cells in head kidney, trunk kidney, spleen, and PBLs in the semi-solid culture with the combination of 100 ng/mL G-CSFa1 and 100 ng/mL G-CSFb1.
G-CSFa1 and G-CSFb1 Directly Induce a Chemotactic Response of Kidney Neutrophils and Up-Regulates the Gene Expression of a Chemokine Receptor cxcr1
Following the development of neutrophils at the sites of hematopoiesis, the migration and the recruitment of these cells toward the sites of infection or injury is essential for an efficient inflammatory response. We investigated the chemotactic effect of recombinant G-CSFa1 and G-CSFb1 on kidney neutrophils from normal adult carp employing a blind-well chemotaxis apparatus (Supplementary Figure S6). Neutrophils migrated toward fMLP placed in the bottom chamber (Figure 7), consistent with previous reports (22). In the presence of high doses of G-CSFa1 or G-CSFb1, kidney neutrophils migrated toward the sources (Figure 7). The chemokinesis controls indicated that neutrophil migration was cytokine-gradient dependent, since the migration of neutrophils was similar to the medium control when the recombinants were placed in both upper and lower chemotaxis chamber (Figure 7).
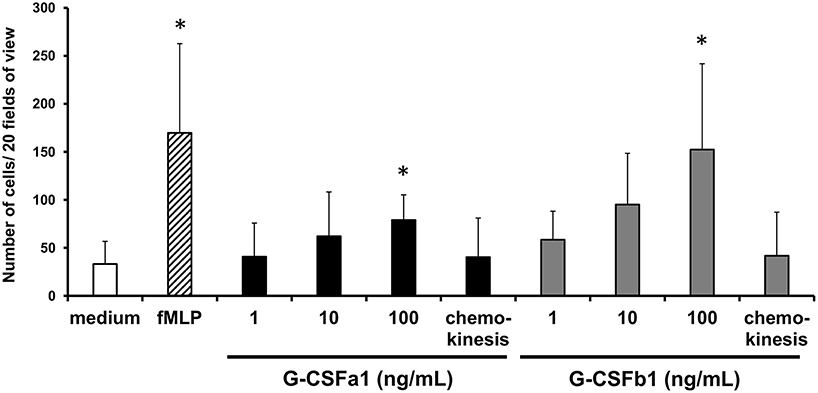
Figure 7. Recombinant G-CSFa1 and G-CSFb1 induces chemotactic response of kidney neutrophils. Chemotactic response of kidney neutrophils after 1 h of incubation with duplicate filters separating cells and cytokines at the concentrations indicated. Cells were stained with MGG and the total number of cells in 20 random fields of view (40× magnification) was determined. Medium and 10 ng/mL fMLP served as negative and positive controls, respectively. Equal concentrations (100 ng/mL) of cytokines in the upper and lower chemotaxis chambers served as chemokinesis control. The data represent mean + standard deviation (n = 4). Significant differences compared to medium control were determined using one-way ANOVA followed by Dunnet's post-hoc test, (p < 0.05) are denoted by asterisks (*).
To assess the ability of G-CSFa1 and G-CSFb1 to modulate the gene expression of chemokine receptors, kidney neutrophils were treated with medium, G-CSFa1 or G-CSFb1 for 6 h. Teleost CXCR1 and CXCR2 are conserved receptors for interleukin-8 (IL-8, also termed CXCL8) and are important for the regulation of neutrophil recruitment and migration to sites of infection and injury (44, 45). Cxcr1 mRNA levels in neutrophils treated with G-CSFa1 and G-CSFb1 were significantly up-regulated compared to the medium control, indicating that both enhances a chemotactic sensibility of neutrophils toward chemotactic mediators such as IL-8 (Figure 8A). Neither cxcr2 mRNA levels in neutrophils treated with G-CSFa1 nor G-CSFb1 were changed compared to the medium control (Figure 8A). The mRNA levels of cxcr4 encoding a receptor for stromal cell-derived factor 1 (SDF-1, also termed CXCL12) in neutrophils were not modulated with the treatment of G-CSFa1 and G-CSFb1 (Figure 8A).
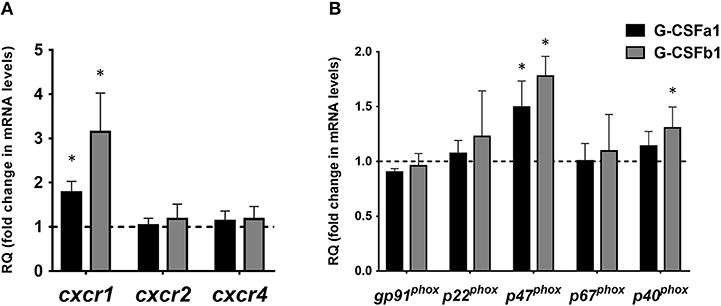
Figure 8. Recombinant G-CSFa1 and G-CSFb1 up-regulates cxcr1 and p47phox mRNA expression levels in carp kidney neutrophils. Quantitative expression analysis of mRNA levels of chemokine receptors (A) and NADPH oxidase components (B) in carp kidney neutrophils treated with the medium, 100 ng/mL G-CSFa1 and 100 ng/mL G-CSFb1 for 6 h. The mRNA levels were calculated using β-actin as a reference gene. Data were normalized to the control cells (dashed line at y = 1) and presented as mean + standard deviation (n = 3). Significant differences compared to unstimulated controls were determined using one-way ANOVA followed by Dunnet's post-hoc test, (p < 0.05) are denoted by asterisks (*).
G-CSFa1 and G-CSFb1 Enhance the Respiratory Burst Capacity in Kidney Neutrophils Through Up-Regulation of a NADPH Oxidase Component p47phox
The respiratory burst in neutrophils is the result of the formation of superoxide anions, in a process catalyzed by NADPH-oxidase (46, 47). Fish NADPH-oxidase components have been shown to have similar modes of activation and functional activities to mammalian counterparts (41, 48). To assess if the NADPH oxidase is induced by G-CSFa1 and G-CSFb1 treatments, we measured the mRNA levels of NADPH oxidase components (gp91phox, p22phox, p47phox, p67phox, and p40phox) in neutrophils treated with G-CSFa1 and G-CSFb1 for 6 h. mRNA levels of p47phox in neutrophils treated with G-CSFa1 and G-CSFb1 and p40phox in neutrophils treated with G-CSFb1 were significantly increased compared to the medium control. In contrast, mRNA levels of other components were not significantly changed (Figure 8B).
Furthermore, in order to investigate whether the treatment of carp kidney neutrophils with G-CSFa1 or G-CSFb1 induces their priming to prepare antimicrobial responses, we measured the respiratory burst in PMA-stimulated neutrophils. Neutrophils were pre-treated with the medium, G-CSFa1 or G-CSFb1 for 6 h. Following these treatments, neutrophils were treated with or without PMA in the presence of DHR123 and then analyzed by flow cytometry. Neither treatment of neutrophils with G-CSFa1 nor G-CSFb1 directly induced the respiratory burst without PMA stimulation (Figure 9). Both G-CSFa1 and G-CSFb1 significantly up-regulated the respiratory burst in PMA-stimulated neutrophils compared to the medium control, while the enhancement of respiratory burst in neutrophils treated with G-CSFb1 was higher than that of G-CSFa1 treated neutrophils (Figure 9), which is consistent with the result of the upregulation of p47phox enhancement (Figure 8B).
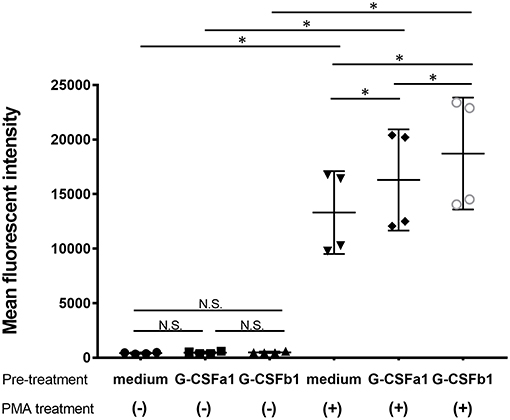
Figure 9. Recombinant G-CSFa1 and G-CSFb1 induces increased respiratory burst capability. Respiratory burst capability of kidney neutrophils after pre-treatment with the medium, 100 ng/mL G-CSFa1 or 100 ng/mL G-CSFb1 for 6 h and subsequently treated with or without 100 ng/mL PMA for 30 min in the presence of DHR123. Mean of DHR123 fluorescence intensity (MFI) in gated neutrophil population was measured by flow cytometry. Data points are presented as mean values of individuals and error bars show standard deviation. Kidney neutrophils were obtained from four fish. Significant differences compared to every other group with two factors of G-CSF pre-treatment and PMA treatment were determined using two-way ANOVA followed by Tukey's post-hoc test, (p < 0.05) are denoted by asterisks (*). N.S. represents ‘not significant’.
In vivo Administration of G-CSFa1 and G-CSFb1 Increases Circulating Neutrophils
Following intraperitoneal (i.p.) injection of PBS and repeated bleeding, the population of peripheral blood neutrophils did not change for 24 h. In contrast, i.p. injection of G-CSFa1 induced a significant increase of peripheral blood neutrophils 6 and 24 h after injection. Likewise, the population of peripheral blood neutrophils was significantly increased after 6 and 24 h of G-CSFb1 injection. At 24 h, G-CSFb1 injection had induced a significantly higher circulating number of neutrophils than injection with G-CSFa1 (Figure 10). However, at 48 h after G-CSFa1 injection, neutrophil numbers no longer were higher than those of unhandled or PBS-injected fish, probably due to the repeated bleedings affecting the peripheral blood neutrophil population of the control groups (Supplementary Figure S7).
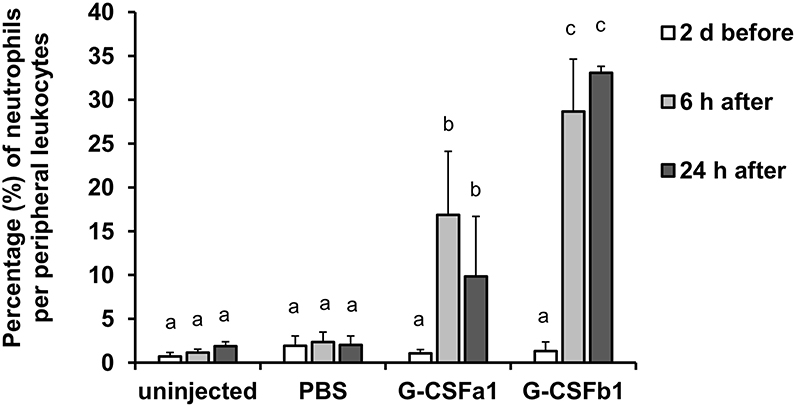
Figure 10. Administration of G-CSFa1 and G-CSFb1 to carp increases circulating blood neutrophil population. Peripheral blood leukocytes were collected from carp 2 days before and 6 and 24 h after intraperitoneal injection of 1 × PBS, recombinant G-CSFa1 and G-CSFb1 or of unhandled carp and analyzed by flow cytometry. Percentage of gated neutrophil population per live peripheral blood leukocytes was measured. Three fish for each group were used and data are presented as mean + standard deviation. Significant differences compared to other treatment groups and other time points were determined using two-way ANOVA followed by Tukey's post-hoc test, (p < 0.05) are denoted by different letters.
Discussion
Here we cloned and functionally characterized carp G-CSF. All four carp g-csf genes contain five exons separated by four introns, retaining the gene structure found in human and mouse G-CSF as well as G-CSF of other teleost species (13). Carp and human G-CSF molecules share a similar structure of a signal peptide and a four-plus-one helical Pfam IL6/GCSF/MGF domain. All teleost fish G-CSF molecules share relatively high homology between each other at the helical regions. They also share acidic residues such as Asp and Glu, involved in the ligand-receptor binding, with mammalian G-CSF (49, 50). The four carp G-CSF paralogs identified in carp may have arisen from an ancestral G-CSF molecule through a series of duplications, including the teleost-specific 3rd WGD event and the carp-specific 4th WGD event (16, 21, 51–53). Overall, despite the overall low homology of teleost fish G-CSF sequences with mammalian G-CSF molecules, our in-silico analyses provide clear evidence that all four paralogs identified in carp are indeed orthologs of mammalian G-CSF.
Carp and other teleost fish G-CSF paralogs share only limited conservation of cysteine residues responsible for disulfide bonds with tetrapod G-CSF. Carp G-CSFa1 and G-CSFb1 express two structural differences: (i) an additional helix enriched with acidic residues in G-CSFa1 and (ii) a location of conserved cysteine residues. These structural differences prompted us to further investigate function of the different paralogs. Where g-csfa1 and g-csfa2 were highly expressed at basal level especially in spleen, g-csfb1 and g-csfb2 basal expression levels were very low in all tissues examined, indicating that g-csf transcription is differentially regulated between paralogs. Similarly, basal g-csfa gene expression was markedly higher than g-csfb expression in all immune cells examined. Macrophages are known to be the major cellular source of mammalian G-CSF (2). Strikingly, G-CSF paralogs were always highest expressed in macrophages of carp.
Basal gene expression levels of g-csfa1 in macrophages were higher than those of g-csfa2 and gene expression levels of g-csfb1 in macrophages were higher than those of g-csfb2, which prompted us to further investigate function of G-CSFa1 and G-CSFb1 by production of these molecules as recombinant proteins. Recombinant proteins were produced in a bacterial expression system with the limitation that proteins are non-glycosylated and could possibly be contaminated with traces derived from bacteria. However, previous studies on mammalian G-CSF reported that glycosylation is not required for its activity and indeed, the non-glycosylated form is utilized in recombinant therapeutics (54). Even though the relative insensitivity to LPS has been reported in fish living in the aquatic environment with high pathogenic pressure (55), the recombinant proteins used in our assays were extensively purified up to the absence of LPS traces. Similar to mammalian G-CSF and zebrafish G-CSF (16, 56), carp G-CSF induced proliferation of hematopoietic cells in a dose-dependent manner, albeit with apparent different activities for the two paralogs studied: G-CSFa1 induced proliferation of blast-like cells adhesive to culture dishes, whereas G-CSFb1 induced proliferation of cells with neutrophil characteristics. Indeed, treatment with G-CSFb1 showed up-regulation of the transcription factor cebpα involved in neutrophil development (34). Also, we investigated at least two carp G-CSF receptor genes (data not shown) and found that only G-CSFb1 enhanced gcsfr1 and myeloperoxidase (mpx) gene expression. Our data indicate that G-CSFb1 and G-CSFR1 are the main players involved with neutrophil development in carp. In zebrafish, both G-CSFa and G-CSFb may bind to the G-CSF receptor, expressed in both neutrophils and macrophages, and promote cell proliferation (16). In contrast to the latter study, carp G-CSFa1 alone did not stimulate colony formation in our semi-solid culture system, in which an agarose layer prevented natural formation of a stromal and an adherent cell layer. This possibly restricted access to spontaneously secreted growth factors from adherent macrophages (57), which are possibly required for colony formation. Further studies would be required to determine if G-CSFa1 directly induces macrophages to produce autocrine growth factors or that G-CSFa1 synergizes with some factors spontaneously secreted from adherent macrophages to synergistically stimulate macrophage development. Meanwhile, carp G-CSFb1 alone did stimulate CFU-G colony formation, whereas the combination of G-CSFa1 and G-CSFb1 stimulated formation of not only CFU-G but also CFU-GM colonies. Our data indicate that carp G-CSFb1 may drive granulopoiesis restricted to neutrophil-lineage development, whereas carp G-CSFa1 may be a cytokine with proliferative effect stimulating CFU-GM or earlier stem/progenitor cells. The functional differences between the G-CSFa1 and G-CSFb1 cytokine preparations make it highly unlikely that the induced cell responses could be due to traces of bacterial contaminations and thus appear cytokine-specific. No matter the indicative differences in biological function between paralogs, carp G-CSFs appears to act as a hematopoietic growth factors.
Mammalian G-CSF is chemo-attractive to neutrophils (58, 59). In zebrafish, G-CSFb but not G-CSFa could be linked to in vivo trafficking of neutrophils to the site of severe injury (19). Our results indicate that carp kidney neutrophils are strongly attracted to G-CSFb1 and are moderately attracted to G-CSFa1, possibly under influence of IL-8 (or CXCL8) (40, 60, 61). Indeed, treatment of carp kidney neutrophil with G-CSF paralogs showed a significant up-regulation of CXCR1 as the IL-8 receptor required for neutrophil recruitment, but not CXCR2 required for neutrophil reverse migration and resolution (45). Unlike mammalian G-CSF, carp G-CSF paralogs did not mediate transcription of CXCR4, important for retention of neutrophils in the hematopoietic tissue in mammalian models (62). In conclusion, carp G-CSFb appears to be the most important G-CSF paralog to induce neutrophil migration.
Once neutrophils receive inflammatory cytokine signals, they become “primed” and capable of promptly and vigorously exerting antimicrobial responses (63). We could not find a significant change of phagocytic activity in neutrophils against beads and zymosan particles following stimulation of G-CSF paralogs for any period tested (data not shown), indicating that neutrophil phagocytosis is regulated by other signals in fish. Although mammalian G-CSF alone is not able to initiate a respiratory burst in naïve neutrophils, pre-incubation with this cytokine primes the cell for an enhanced superoxide anion production following stimulation with physiological stimuli such as fMLP and PMA (11, 64). In fish, following stimulation of phagocytes with inflammatory cytokines, ROS production is activated through at least three sequential steps: (i) activation of protein kinase C (PKC), (ii) phosphorylation of p47phox, and (iii) the production of ROS catalyzed by the NADPH oxidase complex (48). In our hands, expression analysis of NADPH oxidase components in neutrophils treated with carp G-CSF paralogs exhibited up-regulation of especially p47phox, indicating that the priming effect of carp G-CSF paralogs on neutrophils was regulated through the increase of p47phox. Our study provides the first report in teleost fish on the priming effects of G-CSF on neutrophils and analysis of respiratory burst indicated that G-CSFb1 primed neutrophils more effectively than G-CSFa1.
Previous studies in cyprinids showed that circulating blood neutrophils increased in number 6 to 18 h after i.p. injection with killed E. coli or zymosan and then quickly decreased after 24 h, indicating that an intraperitoneal inflammation in fish induces a temporal mobilization of kidney-derived neutrophils into the circulation (42, 65). In our study, administration of G-CSF paralogs increased the number of circulating blood neutrophils 6 and 24 h after i.p. injection, suggesting that also in vivo G-CSFa1 and G-CSFb1 work as chemoattractants and granulopoietic growth factors, in agreement with the in vitro results. However, it remains unclear whether in vivo excess of G-CSF paralogs induce the expression of other inflammatory cytokines and/or chemokines in immune cells. In human clinical medicine, recombinant G-CSF is used as a biophylatic agent to specifically induce granulopoiesis in patients with chemotherapy- and radiation-induced neutropenia to prevent bacterial and fungal infections (66). Further studies will be required to investigate if fish G-CSF paralogs can act as biophylatic agents against infectious diseases in a similar way. Here, functional analyses were limited to G-CSFa1 and G-CSFb1, and we can only speculate that G-CSFa2 and G-CSFb2 could have similar, different, or combinatorial functions in common carp. Additional biochemical investigations involving all native carp G-CSF paralog proteins will be needed to elucidate the full and complicated picture of immune regulation in this polyploid species.
In summary, we identified four carp G-CSF paralogs, studied their gene expression patterns and characterized the functional differences between A and B types of G-CSF on carp hematopoietic cells and neutrophils. We report important differences in their regulation: A type G-CSFs have a relatively high constitutive gene expression and could thus be involved in maintenance of a homeostatic state, whereas B type G-CSFs have a low gene expression and require induction and could thus have a responsive, immunological role associated with a state of infection. In general, G-CSFa1 alone stimulates proliferation of granulocyte/macrophage progenitors, while G-CSFb1 promotes proliferation, differentiation and colony formation of granulocyte/macrophage progenitors and granulocyte progenitors in kidney of carp, similar to the G-CSF mammalian counterpart. G-CSFa1 and G-CSFb1 act as chemo-attractants to neutrophils modulating the expression of the chemokine receptor CXCR1, suggesting a role for G-CSF paralogs in neutrophil trafficking. Both, G-CSFa1 and G-CSFb1 appear to induce neutrophil “priming.” The carp G-CSF paralogs reported herein provide us with valuable tools to further study the immune system of teleost fish.
Author Contributions
FK and KN: conceived and designed the experiments; FK, KN, AW, and EH: performed the experiments; FK, KN, AW, JM, and MO: analyzed the data; FK, KN, AW, GW, and TM: wrote and edited the paper.
Funding
This work was supported in part by a Grant-in Aid for Young Scientists (B) (16K18752) from the Japan Society for the Promotion of Science (JSPS) to FK and a grant of International joint research and training of young researchers for zoonosis control in the globalized world (S1491007) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) Japan. AW was funded by the European Commission under the 8th (H2020) Framework Program for Research and Technological Development of the European Union (PARAFISHCONTROL Grant No. 634429).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors greatly appreciate Dr. Maria Forlenza and Mr. Jules Petit (Wageningen University) for helpful discussion and advice.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00255/full#supplementary-material
Abbreviations
WGD, whole genome duplication; LPS, lipopolysaccharide; ConA, concanavaline A; PMA, phorbol 12-myristate 13-acetate; polyI:C, polyinosinic-polycytidylic acid; IPTG, isopropyl β-D-L-thiogalactopyranoside; MTT, 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyltetrazolium bromide; CFU-G, granulocyte colony-forming unit; CFU-GM, granulocyte/macrophage colony-forming unit; fMLP, N-formyl-methionyl-leucyl-phenylalanine; DHR123, dihydrorhodamine 123; FITC, fluorescein isothiocyanate.
References
1. Panopoulos AD, Watowich SS. Granulocyte colony-stimulating factor: molecular mechanisms of action during steady state and 'emergency' hematopoiesis. Cytokine (2008) 42:277–88. doi: 10.1016/j.cyto.2008.03.002
2. Oster W, Lindemann A, Mertelsmann R, Herrmann F. Production of macrophage-, granulocyte-, granulocyte-macrophage- and multi-colony-stimulating factor by peripheral blood cells. Eur J Immunol. (1989) 19:543–8. doi: 10.1002/eji.1830190320
3. Koeffler HP, Gasson J, Ranyard J, Souza L, Shepard M, Munker R. Recombinant human TNF alpha stimulates production of granulocyte colony-stimulating factor. Blood (1987) 70:55–9.
4. Zsebo KM, Yuschenkoff VN, Schiffer S, Chang D, McCall E, Dinarello CA, et al. Vascular endothelial cells and granulopoiesis: interleukin-1 stimulates release of G-CSF and GM-CSF. Blood (1988) 71:99–103.
5. Rennick D, Yang G, Gemmell L, Lee F. Control of hemopoiesis by a bone marrow stromal cell clone: lipopolysaccharide-and interleukin-1-inducible production of colony-stimulating factors. Blood (1987) 69:682–91.
6. Watari K, Asano S, Shirafuji N, Kodo H, Ozawa K, Takaku F, et al. Serum granulocyte colony-stimulating factor levels in healthy volunteers and patients with various disorders as estimated by enzyme immunoassay. Blood (1989) 73:117–22.
7. Kawakami M, Tsutsumi H, Kumakawa T, Abe H, Hirai M, Kurosawa S, et al. Levels of serum granulocyte colony-stimulating factor in patients with infections. Blood (1990) 76:1962–4.
8. Demetri GD, Griffin JD. Granulocyte colony-stimulating factor and its receptor. Blood (1991) 78:2791–808.
9. Lieschke G, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood (1994) 84:1737–46.
10. Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor–deficient mice. Immunity (1996) 5:491–501. doi: 10.1016/S1074-7613(00)80504-X
11. Spiekermann K, Roesler J, Emmendoerffer A, Elsner J, Welte K. Functional features of neutrophils induced by G-CSF and GM-CSF treatment: differential effects and clinical implications. Leukemia (1997) 11:466–78. doi: 10.1038/sj.leu.2400607
12. Nicola NA, Metcalf D, Matsumoto M, Johnson GR. Purification of a factor inducing differentiation in murine myelomonocytic leukemia cells. Identification as granulocyte colony-stimulating factor. J Biol Chem. (1983) 258:9017–23.
13. Santos MD, Yasuike M, Hirono I, Aoki T. The granulocyte colony-stimulating factors (CSF3s) of fish and chicken. Immunogenetics (2006) 58:422–32. doi: 10.1007/s00251-006-0106-5
14. Yaparla A, Wendel ES, Grayfer L. The unique myelopoiesis strategy of the amphibian Xenopus laevis. Dev Comp Immunol. (2016) 63:136–43. doi: 10.1016/j.dci.2016.05.014
15. Nam BH, An GH, Baeck GW, Kim MC, Kim JW, Park HJ, et al. Molecular cloning and expression of cDNAs for two distinct granulocyte colony stimulating factor genes from black rockfish Sebastes schlegelii. Fish Shellfish Immunol. (2009) 27:360–4. doi: 10.1016/j.fsi.2009.06.005
16. Stachura D, Svoboda O, Campbell C, Espín-Palazón R, Lau R, Zon L, et al. The zebrafish granulocyte colony-stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood (2013) 122:3918–28. doi: 10.1182/blood2012-12-475392
17. Inoue J, Sato Y, Sinclair R, Tsukamoto K, Nishida M. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci USA. (2015) 112:14918–23. doi: 10.1073/pnas.1507669112
18. Liongue C, Hall CJ, O'Connell BA, Crosier P, Ward AC. Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood (2009) 113:2535–46. doi: 10.1182/blood-2008-07-171967
19. Galdames JA, Zuniga-Traslavina C, Reyes AE, Feijoo CG. Gcsf-Chr19 promotes neutrophil migration to damaged tissue through blood vessels in zebrafish. J Immunol. (2014) 193:372–8. doi: 10.4049/jimmunol.1303220
20. Kolder IC, van der Plas-Duivesteijn SJ, Tan G, Wiegertjes GF, Forlenza M, Guler AT, et al. A full-body transcriptome and proteome resource for the European common carp. BMC Genomics (2016) 17:701. doi: 10.1186/s12864-016-3038-y
21. Petit J, David L, Dirks R, Wiegertjes GF. Genomic and transcriptomic approaches to study immunology in cyprinids: what is next? Dev Comp Immunol. (2017) 75:48–62. doi: 10.1016/j.dci.2017.02.022
22. Katzenback BA, Belosevic M. Isolation and functional characterization of neutrophil-like cells, from goldfish (Carassius auratus L.) kidney. Dev Comp Immunol. (2009) 33:601–11. doi: 10.1016/j.dci.2008.10.011
23. Stolte EH, Savelkoul HF, Wiegertjes G, Flik G, Lidy Verburg-van Kemenade BM. Differential expression of two interferon-gamma genes in common carp (Cyprinus carpio L.). Dev Comp Immunol. (2008) 32:1467–81. doi: 10.1016/j.dci.2008.06.012
24. Joerink M, Ribeiro CMS, Stet RJM, Hermsen T, Savelkoul HFJ, Wiegertjes GF. Head kidney-derived macrophages of common carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J Immunol. (2006) 177:61–9. doi: 10.4049/jimmunol.177.1.61
25. Forlenza M, Scharsack JP, Kachamakova NM, Taverne-Thiele AJ, Rombout JH, Wiegertjes GF. Differential contribution of neutrophilic granulocytes and macrophages to nitrosative stress in a host-parasite animal model. Mol Immunol. (2008) 45:3178–89. doi: 10.1016/j.molimm.2008.02.025
26. Secombes C, Van Groningen J, Egberts E. Separation of lymphocyte subpopulations in carp Cyprinus carpio L. by monoclonal antibodies: immunohistochemical studies Immunology (1983) 48:165.
27. Rombout J, Koumans-van Diepen J, Emmer P, Taverne-Thiele J, Taverne N. Characterization of carp thrombocytes with specific monoclonal antibodies. J Fish Biol. (1996) 49:521–31. doi: 10.1111/j.1095-8649.1996.tb00047.x
28. Katakura F, Yabu T, Yamaguchi T, Miyamae J, Shirinashihama Y, Nakanishi T, et al. Exploring erythropoiesis of common carp (Cyprinus carpio) using an in vitro colony assay in the presence of recombinant carp kit ligand A and erythropoietin. Dev Comp Immunol. (2015) 53:13–22. doi: 10.1016/j.dci.2015.06.006
29. Katakura F, Sugie Y, Hayashi K, Nishiya K, Miyamae J, Okano M, et al. Thrombopoietin (TPO) induces thrombocytic colony formation of kidney cells synergistically with kit ligand A and a non-secretory TPO variant exists in common carp. Dev Compar Immunol. (2018) 84:327–36. doi: 10.1016/j.dci.2018.03.005
30. Katakura F, Yamaguchi T, Yoshida M, Moritomo T, Nakanishi T. Demonstration of T cell and macrophage progenitors in carp (Cyprinus carpio) kidney hematopoietic tissues. Development of clonal assay system for carp hematopoietic cells. Dev Compar Immunol. (2010) 34:685–9. doi: 10.1016/j.dci.2010.01.015
31. Tsai F-Y, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood (1997) 89:3636–43.
32. Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. (2003) 4:1029–36. doi: 10.1038/ni973
33. Kurotaki D, Yamamoto M, Nishiyama A, Uno K, Ban T, Ichino M, et al. IRF8 inhibits C/EBPalpha activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat Commun. (2014) 5:4978. doi: 10.1038/ncomms5978
34. Ma O, Hong S, Guo H, Ghiaur G, Friedman AD. Granulopoiesis requires increased C/EBPα compared to monopoiesis, correlated with elevated Cebpa in immature G-CSF receptor versus M-CSF receptor expressing cells. PLoS ONE (2014) 9:e95784. doi: 10.1371/journal.pone.0095784
35. Katzenback BA, Katakura F, Belosevic M. Goldfish (Carassius auratus L.) as a model system to study the growth factors, receptors and transcription factors that govern myelopoiesis in fish. Dev Comp Immunol. (2016) 58:68–85. doi: 10.1016/j.dci.2015.10.024
36. Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell (2005) 8:97–108. doi: 10.1016/j.devcel.2004.11.014
37. Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature (1999) 402:14–20. doi: 10.1038/35005514
38. Hosoya T, Maillard I, Engel JD. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev. (2010) 238:110–25. doi: 10.1111/j.1600-065X.2010.00954.x
39. Fujiki K, Shin D-H, Nakao M, Yano T. Molecular cloning of carp (Cyprinus carpio) CC chemokine, CXC chemokine receptors, allograft inflammatory factor-1, and natural killer cell enhancing factor by use of suppression subtractive hybridization. Immunogenetics (1999) 49:909–14. doi: 10.1007/s002510050573
40. van der Aa LM, Chadzinska M, Tijhaar E, Boudinot P, Verburg-van Kemenade BM. CXCL8 chemokines in teleost fish: two lineages with distinct expression profiles during early phases of inflammation. PLoS ONE (2010) 5:e12384. doi: 10.1371/journal.pone.0012384
41. Mayumi M, Takeda Y, Hoshiko M, Serada K, Murata M, Moritomo T, et al. Characterization of teleost phagocyte NADPH oxidase: molecular cloning and expression analysis of carp (Cyprinus carpio) phagocyte NADPH oxidase. Mol Immunol. (2008) 45:1720–31. doi: 10.1016/j.molimm.2007.09.028
42. Havixbeck JJ, Rieger AM, Wong ME, Hodgkinson JW, Barreda DR. Neutrophil contributions to the induction and regulation of the acute inflammatory response in teleost fish. J Leukoc Biol. (2016) 99:241–52. doi: 10.1189/jlb.3HI0215-064R
43. Nakayasu C, Omori M, Hasegawa S, Kurata O, Okamoto N. Production of a monoclonal antibody for carp (Cyprinus carpio L.) phagocytic cells and separation of the cells. Fish Shellfish Immunol. (1998) 8:91–100. doi: 10.1006/fsim.1997.0125
44. Blaser BW, Moore JL, Hagedorn EJ, Li B, Riquelme R, Lichtig A, et al. CXCR1 remodels the vascular niche to promote hematopoietic stem and progenitor cell engraftment. J Exp Med. (2017) 214:1011–27. doi: 10.1084/jem.20161616
45. Powell D, Tauzin S, Hind LE, Deng Q, Beebe DJ, Huttenlocher A. Chemokine signaling and the regulation of bidirectional leukocyte migration in interstitial tissues. Cell Rep. (2017) 19:1572–85. doi: 10.1016/j.celrep.2017.04.078
46. Babior BM, Kipnes RS, Curnutte JT. Biological defense mechanisms. The production by leukocytes of superoxide, a potential bactericidal agent. J Clin Invest. (1973) 52:741. doi: 10.1172/JCI107236
47. Hampton MB, Kettle AJ, Winterbourn CC. Inside the neutrophil phagosome: oxidants, myeloperoxidase, and bacterial killing. Blood (1998) 92:3007–17.
48. Olavarria VH, Gallardo L, Figueroa JE, Mulero V. Lipopolysaccharide primes the respiratory burst of Atlantic salmon SHK-1 cells through protein kinase C-mediated phosphorylation of p47phox. Dev Comp Immunol. (2010) 34:1242–53. doi: 10.1016/j.dci.2010.07.002
49. Aritomi M, Kunishima N, Okamoto T, Kuroki R, Ota Y, Morikawa K. Atomic structure of the GCSF–receptor complex showing a new cytokine–receptor recognition scheme. Nature (1999) 401:713–7. doi: 10.1038/44394
50. Layton JE, Hall NE, Connell F, Venhorst J, Treutlein HR. Identification of ligand-binding site III on the immunoglobulin-like domain of the granulocyte colony-stimulating factor receptor. J Biol Chem. (2001) 276:36779–87. doi: 10.1074/jbc.M104787200
51. Hultman KA, Bahary N, Zon LI, Johnson SL. Gene Duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. (2007) 3:e17. doi: 10.1371/journal.pgen.0030017
52. Wang T, Hanington PC, Belosevic M, Secombes CJ. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J Immunol. (2008) 181:3310–22. doi: 10.4049/jimmunol.181.5.3310
53. Zou J, Secombes CJ. The function of fish cytokines. Biology (2016) 5:23. doi: 10.3390/biology5020023
54. Bönig H, Silbermann S, Weller S, Kirschke R, Körholz D, Janssen G, et al. Glycosylated vs non-glycosylated granulocyte colony-stimulating factor (G-CSF)–results of a prospective randomised monocentre study. Bone Marrow Transplant. (2001) 28:259. doi: 10.1038/sj.bmt.1703136
55. Novoa B, Bowman T, Zon L, Figueras A. LPS response and tolerance in the zebrafish (Danio rerio). Fish Shellfish Immunol. (2009) 26:326–31. doi: 10.1016/j.fsi.2008.12.004
56. Souza L, Boone T, Gabrilove J, Lai P, Zsebo K, Murdock D, et al. Recombinant human granulocyte colony-stimulating factor: effects on normal and leukemic myeloid cells. Science (1986) 232:61–5. doi: 10.1126/science.2420009
57. Neumann NF, Barreda D, Belosevic M. Production of a macrophage growth factor (s) by a goldfish macrophage cell line and macrophages derived from goldfish kidney leukocytes. Dev Compar Immunol. (1998) 22:417–32. doi: 10.1016/S0145-305X(98)00023-8
58. Wang JM, Chen ZG, Colella S, Bonilla MA, Welte K, Bordignon C, et al. Chemotactic activity of recombinant human granulocyte colony-stimulating factor. Blood (1988) 72:1456–60.
59. Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity (2002) 17:413–23. doi: 10.1016/S1074-7613(02)00424-7
60. de Oliveira S, Reyes-Aldasoro CC, Candel S, Renshaw SA, Mulero V, Calado A. Cxcl8 (IL-8) mediates neutrophil recruitment and behavior in the zebrafish inflammatory response. J Immunol. (2013) 190:4349–59. doi: 10.4049/jimmunol.1203266
61. de Oliveira S, Lopez-Munoz A, Martinez-Navarro FJ, Galindo-Villegas J, Mulero V, Calado A. Cxcl8-l1 and Cxcl8-l2 are required in the zebrafish defense against Salmonella Typhimurium. Dev Comp Immunol. (2015) 49:44–8. doi: 10.1016/j.dci.2014.11.004
62. Summers C, Rankin SM, Condliffe AM, Singh N, Peters AM, Chilvers ER. Neutrophil kinetics in health and disease. Trends Immunol. (2010) 31:318–24. doi: 10.1016/j.it.2010.05.006
63. Hallett MB, Lloyds D. Neutrophil priming: the cellular signals that say ‘amber’but not ‘green’. Immunology Today (1995) 16:264–8.
64. Kitagawa S, Yuo A, Souza LM, Saito M, Miura Y, Takaku F. Recombinant human granulocyte colony-stimulating factor enhances superoxide release in human granulocytes stimulated by the chemotactic peptide. Biochem Biophys Res Commun. (1987) 144:1143–6. doi: 10.1016/0006-291X(87)91430-6
65. Kodama H, Tijiwa K, Moritomo T, Nakanishi T. Granulocyte responses to experimental injection of live and formalin-killed bacteria in carp (Cyprinus carpio). Vet Immunol Immunopathol. (2002) 90:101–5. doi: 10.1016/S0165-2427(02)00230-1
66. Aapro MS, Bohlius J, Cameron DA, Dal Lago L, Donnelly JP, Kearney N, et al. 2010 update of EORTC guidelines for the use of granulocyte-colony stimulating factor to reduce the incidence of chemotherapy-induced febrile neutropenia in adult patients with lymphoproliferative disorders and solid tumours. Eur J Cancer (2011) 47:8–32. doi: 10.1016/j.ejca.2010.10.013
Keywords: teleost, granulocyte colony-stimulating factor, neutrophil, hematopoiesis, cell migration, respiratory burst
Citation: Katakura F, Nishiya K, Wentzel AS, Hino E, Miyamae J, Okano M, Wiegertjes GF and Moritomo T (2019) Paralogs of Common Carp Granulocyte Colony-Stimulating Factor (G-CSF) Have Different Functions Regarding Development, Trafficking and Activation of Neutrophils. Front. Immunol. 10:255. doi: 10.3389/fimmu.2019.00255
Received: 07 September 2018; Accepted: 29 January 2019;
Published: 19 February 2019.
Edited by:
Leon Grayfer, George Washington University, United StatesReviewed by:
James L. Stafford, University of Alberta, CanadaVictoriano Mulero, University of Murcia, Spain
Copyright © 2019 Katakura, Nishiya, Wentzel, Hino, Miyamae, Okano, Wiegertjes and Moritomo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fumihiko Katakura, a2F0YWt1cmEuZnVtaWhpa29Abmlob24tdS5hYy5qcA==
†These authors have contributed equally to this work
 Fumihiko Katakura
Fumihiko Katakura Kohei Nishiya1†
Kohei Nishiya1† Geert F. Wiegertjes
Geert F. Wiegertjes Tadaaki Moritomo
Tadaaki Moritomo