- 1Biology Department, East Tehran Branch, Islamic Azad University, Tehran, Iran
- 2Skin Research Centre, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 3Lübeck Institute of Experimental Dermatology, University of Lübeck, Lübeck, Germany
- 4Department of Dermatology, University of Lübeck, Lübeck, Germany
- 5Institute of Anatomy, University of Lübeck, Lübeck, Germany
Bullous pemphigoid (BP) is a rare autoimmune skin blistering disease, characterized by the presence of autoantibodies against hemidesmosomal autoantigens. Cytokine expression is altered in BP patients, and several of these differently expressed cytokines, including IL-1α, IL-1β, IL-8, and TNF-α, contribute to disease pathogenesis. Since genetic polymorphisms in the genes of these cytokines might be implicated in susceptibility to BP disease, we aimed at testing this implication in susceptibility to BP in an Iranian cohort. Blood samples were collected from the subjects and genomic DNA was extracted. To detect the single nucleotide polymorphisms (SNPs), IL-1α (rs1800587), IL-1β (rs1143627, rs16944, rs1143634), IL-8 (rs4073), and TNF-α (rs1799964, rs1800630, rs1799724, and rs361525) genes were genotyped in BP patients and healthy controls as well as IL-8 (rs4073) in pemphigus vulgaris (PV) patients. Quantitative gene expression was evaluated by RT-PCR analysis. A significant difference was observed in the distribution of genotypes or alleles of IL-8 SNP between the BP patients and controls. The A-allele of IL-8 SNP is significantly more prevalent in the control individuals compared to the BP patient. To further validate this observation, we included PV patients as an additional control. Again, the A-allele of IL-8 SNP is significantly more prevalent in the PV compared to the BP patients. While we observed a trend toward significant differences regarding alleles of TNF-α rs1799724 as well as alleles of TNF-α rs1799964, this difference was, however, not evident after correction for multiple analysis. There was no significant difference in all other studied SNPs. In contrast to IL-1α, IL-1β, and TNF-α, IL-8 gene expression levels were significantly higher in the patients than that of controls. The minor allele in IL-8 SNP might play a protective role in susceptibility to BP in Iranian patients. Although higher expression levels of IL-8 gene was found in the patients compared with healthy controls, these levels, however, suggest no association with the examined polymorphism. Moreover, further investigation revealed an elevation in gene expression between wild and polymorphic genotypes of IL-1α rs1800587 and TNF-α rs361525 in the patient group and these SNPs are therefore associated with altering the levels of gene expression.
Introduction
Bullous pemphigoid (BP) is a rare and difficult-to-treat autoimmune skin blistering disease characterized by the presence of an autoimmune response against type XVII collagen (COL17, BP180) and BP230, and presents with blisters and erythema, and/or urticarial plaques, where itch is clinically a leading symptom (1–4). Binding of autoantibodies to their antigens attenuate skin integrity and initiate a cascade of the inflammatory response (5). This response is mainly mediated by infiltration of inflammatory cells, deposition of complement, and activation of proteases and reactive oxygen species which finally result in blister formation (5). Cytokines, as regulators of inflammatory and immune reactions in the skin, contribute to inflammatory blister induction by recruitment of inflammatory cells as well as activation of resident cells (6).
Humoral immune-mediated mechanisms and cytokine involvement have been described in the pathogenesis of autoimmune blistering disorders (7). High levels of proinflammatory cytokines interleukin (IL) 1 alpha (IL-1α), IL-1β, IL-8, and TNF-α have been detected in the BP patients (8–10). IL-1α and IL-1β are two proinflammatory cytokines belong to the IL-1 family. IL-1 has central roles in infection, inflammation, and disease which the inflammatory process is initiated by IL-1α but multiplied and retained by both forms (11). Tumor necrosis factor- α (TNF- α) is another proinflammatory cytokine and acts as a central and multifunctional mediator of diverse cellular events (12). IL-1 and TNF- α can subsequently induce cytokines involved in leukocyte attraction, including IL-8 (13). IL-8 is a chemoattractant cytokine produced by a variety of cell types which contribute to a variety of proinflammatory activities (14).
Cytokine gene polymorphisms may influence the induction and release of cytokines, contribute to the disease-associated cytokine imbalance, and thereby affect the susceptibility to autoimmune diseases (15, 16). Gene polymorphisms of IL-1α, IL-1β, IL-8, and TNF-α cytokines have substantially been studied in several autoimmune diseases. To date, the contribution of cytokine polymorphisms in BP disease has not been investigated in great detail. One study investigated the cytokine polymorphisms in a Chinese cohort (17). One additional study focused on single polymorphism in TNF-α in Iranian population (18). In the Chinese cohort, a polymorphism in the IL-1β was identified in female patients only. In our study, in Iranian BP patients, we found no significant association of BP with TNF polymorphisms. With the exception of these two studies, no data is available about the association between proinflammatory cytokine polymorphisms and the development of BP. Since these polymorphisms might be associated with susceptibility to the diseases, the present study designed to examine the frequency of different polymorphisms in Iranian BP patients compared with healthy individuals. Due to different gene expression as a result of polymorphism, the RNA expression was evaluated in patients with different allelic variation and furthermore, compared with control subjects.
Materials and Methods
Study Population
Patients with BP disease were selected from Iranian patients attending the Shohada Tajrish, Loghman Hakim, and Razi hospitals in Tehran between 2013 and 2016. The patient group (n = 40) consisted of 12 males and 28 females, with an age ranging from 47 to 100 years with a mean age of 69.9 ± 2.01 years. BP was diagnosed based on clinical presentation, subepidermal blister on skin biopsy, compatible light microscopy findings, and positive direct immunofluorescence microscopy (DIF) data according to investigational assessment guidelines (19). Samples of the patients were collected immediately after diagnosis of BP and the peri-procedural assessment was performed by using the initial data analysis of the patients. Samples with a lack of demographic data or inappropriate processing of initial laboratory specimens which could bias the study are excluded. Relevant information was taken from all study participants such as suffering from autoimmune diseases, past history of BP, skin diseases, and heart diseases using questionnaires. Furthermore, 84 samples from patients with pemphigus vulgaris (PV) were used as control patients. Clinical presentation, as well as direct and indirect immunofluorescence, were performed for the diagnosis of PV. Furthermore, autoantibodies against desmoglein 3 were detected in all PV patients. Local ethics committee (Human Research Ethics Committee of Skin Research Center, Shahid Beheshti University of Medical Sciences) considered and approved the study protocol. Informed patient consent was obtained and the principles outlined in the Declaration of Helsinki was conducted in the study. Unrelated sex and age-matched healthy volunteers were also recruited without any evidence of previous BP. Control individuals (n = 40) included 20 males and 20 females, aged 46 to 92 years with a mean age of 64.7 ± 1.72 years.
Single Nucleotide Polymorphism (SNP) Genotyping and Gene Expression Analysis
Whole blood was taken from the subjects and genomic DNA was extracted using isolation kit (DNGTM—Plus; SinaClon, Iran) according to the manufacturer's protocol. Genetic SNPs of IL-1α (rs1800587), IL-1β (rs1143627, rs16944, rs1143634), IL-8 (rs4073), and TNF-α (rs1799964, rs1800630, rs1799724, and rs361525) genes were analyzed using primers and respective restriction endonucleases by PCR-based restricted fragment length polymorphism (PCR-RFLP) as described previously (20–25).
IL-1α, IL-1β, IL-8, and TNF-α gene expression was evaluated by quantitative RT-PCR, based on RNA extraction and cDNA synthesis from blood leucocytes of all subjects using RNX-Plus kit (SinaClon, Iran) according to the manufacturer's instructions. Expression of candidate genes, as well as housekeeping glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, was analyzed by real-time RT-PCR using SYBR Green assays. Primer sequences and conditions were designed by ABI PCR equipment (Applied Biosystems, USA) except GAPDH which obtained from previously published sequences (26) (Supplementary Table 1).
Statistical Analysis
Statistical analysis was carried out using the SPSS statistical software program or Prism software. Post-hoc power analysis of the present study was performed using G*Power (version 3.1) (http://www.gpower.hhu.de/). Genotype distribution, allele frequencies, and consistency with the Hardy–Weinberg equilibrium was analyzed using Chi-square (χ2) test. Linkage disequilibrium (LD) parameters delta coefficient (D′) and r squared (r2) of the population, as well as haplotype analysis, were calculated by comparisons among the SNPs using the Haploview software version 4.2 and SNPStats online software (http://bioinfo.iconcologia.net/snpstats/start.htm). The χ2 test, analysis of variance (ANOVA), or t-test are used to conduct a possible association between SNPs and clinical-demographic features. Logistic regression analysis was performed to predict the probability occurrence of BP. Shapiro–Wilk test was applied to verify the normality of data distribution and differences between groups, where Mann–Whitney t-test was used to analyze a non-Gaussian distribution. Data were expressed as median with interquartile range and a P < 0.05 was considered statistically significant. The P-value was corrected (Pc) for multiple analysis using the Bonferroni method.
Results
Study Population and Characterization
This study included 40 patients with BP and 40 age- and sex-matched controls. Sample size estimation and the statistical power calculation was conducted based on the reported prevalence of the variants. The sample size of the study is sufficient to reach 70% power at a significance level of 0.05 when considering the medium effect sizes of 0.3. There was no significant difference between patients and controls regarding age and gender (P > 0.05). Frequencies of IL-8 (rs4073) and TNF-α (rs361525) SNPs in both patient and control groups, as well as the distribution of IL-1α (rs1800587), IL-1β (rs1143634), and TNF-α (rs1800630) in control individuals, deviated from Hardy–Weinberg equilibrium. Genotype frequencies of all other polymorphisms were within the equilibrium.
Allele and Genotype Frequencies of Proinflammatory Cytokines
Gene variations of IL-1α, IL-1β, IL-8, and TNF-α were analyzed in BP patients compared to healthy controls to uncover whether these polymorphisms are associated with BP (Table 1). A significant association was found between the genotypes and alleles of our cases and controls in rs4073 of IL-8, where the polymorphic A-allele was significantly more present in the controls (42.5%) than to that of patient individuals (22.5%) (Pc = 0.01). Analysis of logistic regression indicated that the A-allele might be a protective factor for developing BP; the calculated odds ratio (OR) was 0.41 (95% confidence interval (CI): 0.18–0.89). In contrast to IL-8, we observed no significant difference in the distribution of genotypes or alleles of IL-1α and IL-1β SNPs between the BP patients and controls (P > 0.05). In TNF-α rs1799964, the frequency of C-allele was a two-fold increase in the control individuals (30%) compared to the patients (15%), which was significant (P = 0.02). Logistic regression analysis revealed that the minor C-allele at this locus of TNF-α might be protective for developing BP; the calculated odds ratio (OR) was 0.41 (95% confidence interval (CI): 0.18–0.89). However, this difference remains at the border of significance (Pc = 0.05). In TNF-α rs1799724, we found a significant difference in the distribution of genotypes (P = 0.03) and alleles (P = 0.04) as well as genotypes (P = 0.03) in IL-1α rs1800587, but this difference does not achieve the levels of significance after correction for multiple analysis. No significant association was found between our cases and controls in other SNPs of TNF-α (P > 0.05).
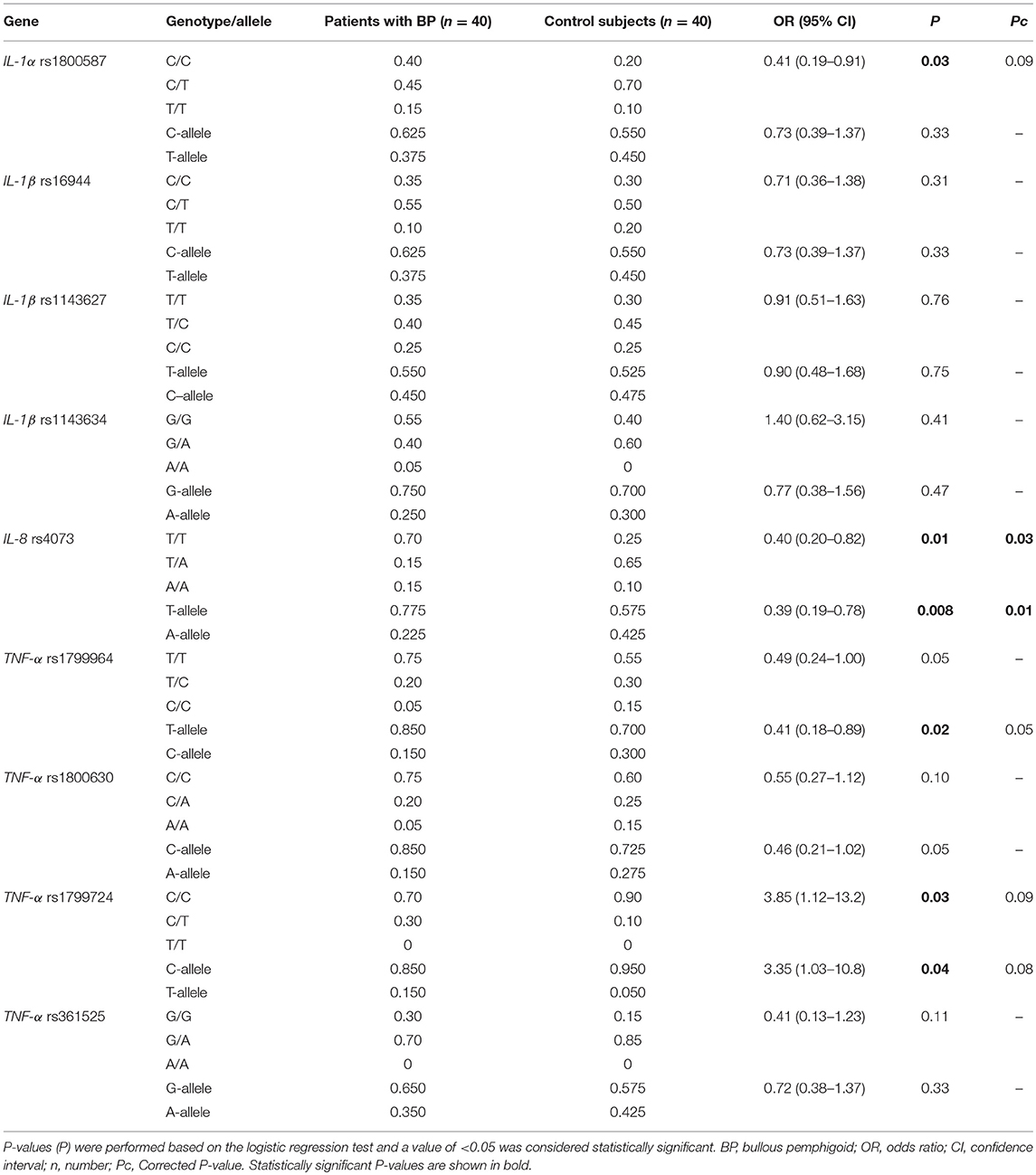
Table 1. Genotype and allele frequencies of IL-1α, IL-1β, IL-8, and TNF-α gene polymorphisms in Iranian patients with BP and respective controls.
Given the significant association between BP patients and respective healthy controls in IL-8 rs4073 and in order to uncover whether the observed association is BP-specific, this polymorphism was likewise analyzed in 84 PV patients (Table 2). The genotypes and alleles were similarly distributed between PV patients and healthy controls; the calculated odds ratio (OR) by logistic regression analysis for A-allele was 1.06 [95% confidence interval (CI): 0.62–1.82]. However, when comparing genotypes and alleles of PV to BP patients, the polymorphic A-allele was significantly more present in PV as compared to BP patients; odds ratio (OR) for A-allele was 0.58 [95% confidence interval (CI): 0.34–0.99]. Although this significance for allele distribution disappears after correction for multiple analysis, it remains significant for genotype analysis.
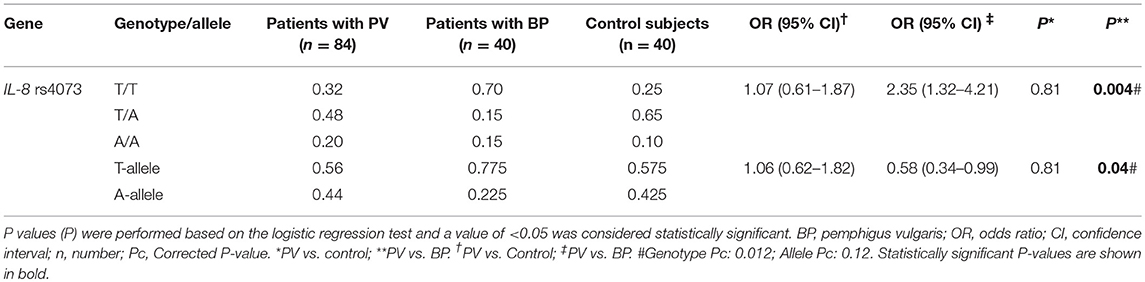
Table 2. Comparison of genotype and allele frequencies of IL-8 rs4073 gene polymorphism in PV Iranian patients with healthy control subjects as well as BP patients.
Linkage Disequilibrium and Haplotype Frequencies
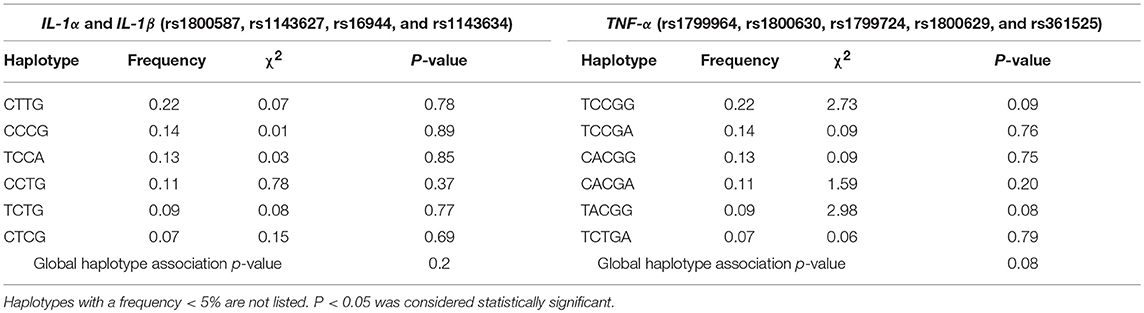
Linkage disequilibrium (LD) was performed in order to investigate the relationship between polymorphisms in IL-1 and TNF-α (Figure 1). The LD analysis of the genotyped SNPs in our samples suggested that two SNPs (rs1800587 and rs1143634) in IL-1 are in strong linkage disequilibrium (D′ = 0.80, LOD = 2.94) (Figure 1, left panel). No correlation was found between polymorphisms in TNF-α (Figure 1, right panel). Haplotype frequencies were compared between IL-1 and TNF-α SNPs and their association with BP which is summarized in Table 3. The P-values for individual and global haplotype score tests did not indicate a significant difference in haplotype frequency profiles between cases and controls (P > 0.05).
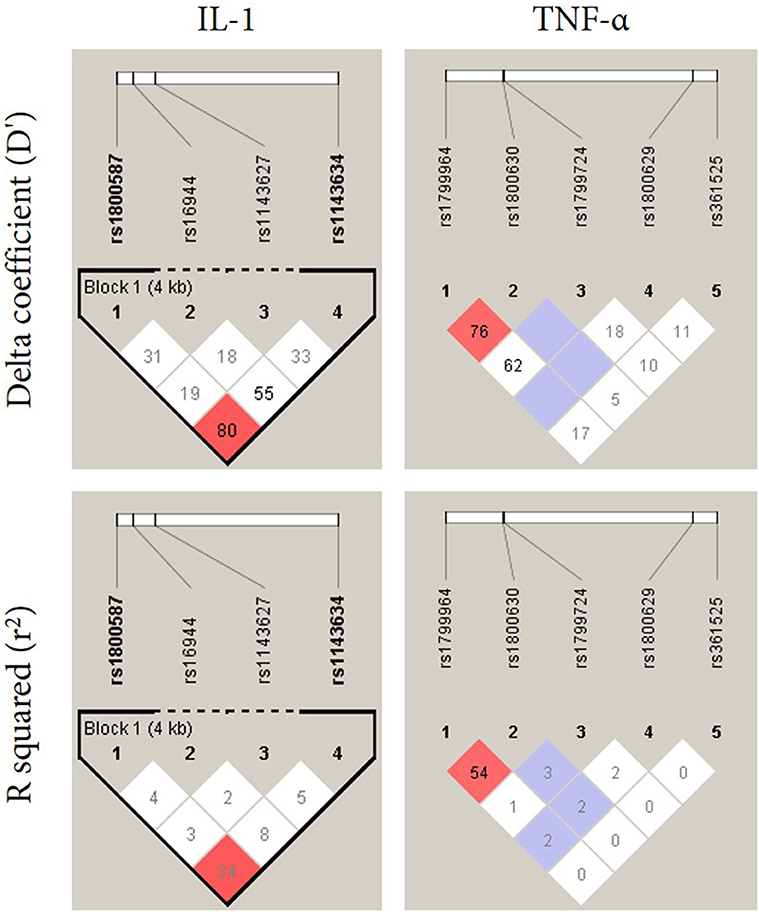
Figure 1. Linkage disequilibrium pattern of the genomic region in chromosome 2 and 6 located between SNPs IL-1α and IL-1β (rs1800587, rs1143627, rs16944, and rs1143634) and TNF-α (rs1799964, rs1800630, rs1799724, rs1800629, and rs361525), respectively.
Genotype Association of BP Patients With Clinical and Demographic Characteristics
Clinical and demographical features were analyzed for a possible association between patients with wildtype and polymorphic genotypes in which the P-values are shown in Table 4. Of note, patients with polymorphic genotypes in TNF-α rs1799964 (T/C+C/C) and rs1800630 (C/A+A/A) SNPs were significantly older than patients with wild-type genotypes (T/T and C/C, respectively) (P = 0.001). Similarly, in these SNPs patients with polymorphic genotypes developed the BP disease in older ages (P = 0.002) and were suffering from heart diseases and focal infection. Furthermore, patients with polymorphic genotypes in TNF-α rs1799724 (C/T +T/T) and rs361525 (G/A+A/A) SNPs had anemia, depression, or other skin diseases. In the case of IL-1β rs1800587, comparison of BP patients carrying wild genotype (C/C) against those with polymorphic genotypes (C/T+T/T) demonstrated that the majority of patients with polymorphic genotypes were suffering heart disorders (P = 0.005). Comparison of cytokine gene polymorphisms in male/female subjects of patients suggested that the polymorphic genotypes of IL-1β rs1143627 and rs1143634, as well as TNF-α rs1799964 and rs1800630, were significantly more frequent in female patients than in male subjects.
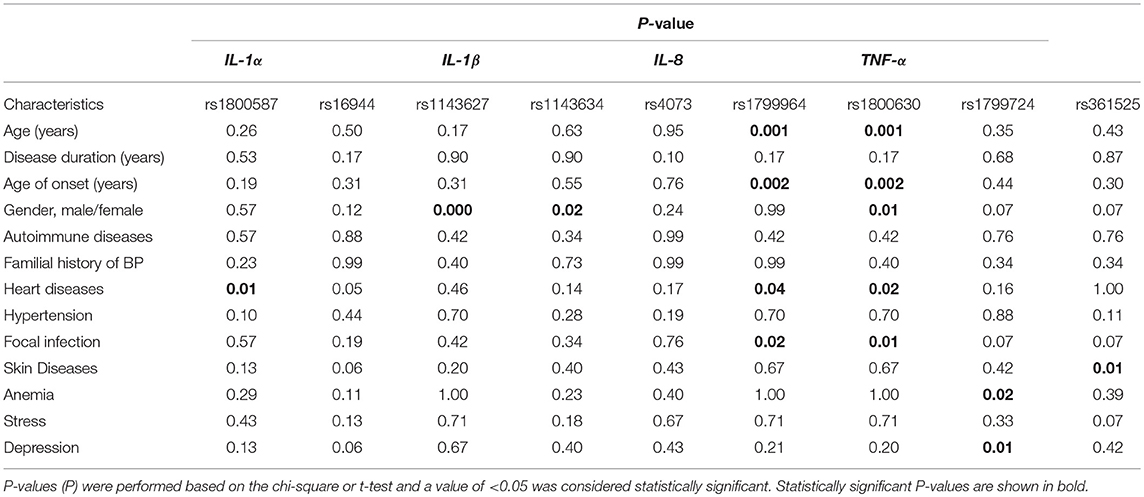
Table 4. Association of clinical characteristics and demographic data in BP patient with polymorphic and wild genotypes in IL-1α, IL-1β, IL-8, and TNF-α gene polymorphisms.
Gene Expression in BP Disease
To investigate whether BP disease influences gene expression, a quantitative analysis of the IL-1α, IL-1β, and IL-8 genes were performed which are shown in Figure 2A. Previously, we could show that the RNA expression levels of TNF-α elevate in circulating leukocytes obtained from patients with BP compared with control subjects (18). Although the gene expression of IL-1β was higher in the controls than in patients, it did not reach the levels of significance (IL-1β/GAPDH: 1.95 [0.42–11.7] vs. 1.13 [0.006–42.5], P = 0.07). The IL-8 gene was expressed significantly higher in the patient subjects than to that of control (IL-8/GAPDH: 3.06 [0.42–10.7] vs. 1.02 [0.001–51.1], P = 0.0005). There is no significant difference between patients and controls in IL-1α gene expression. Further investigation revealed an alteration in gene expression between wild and polymorphic genotypes of IL-1α rs1800587 (P = 0.03) and TNF-α rs361525 (P = 0.01) in the patient group and these SNPs are therefore associated with altering the levels of gene expression (Figure 2B).
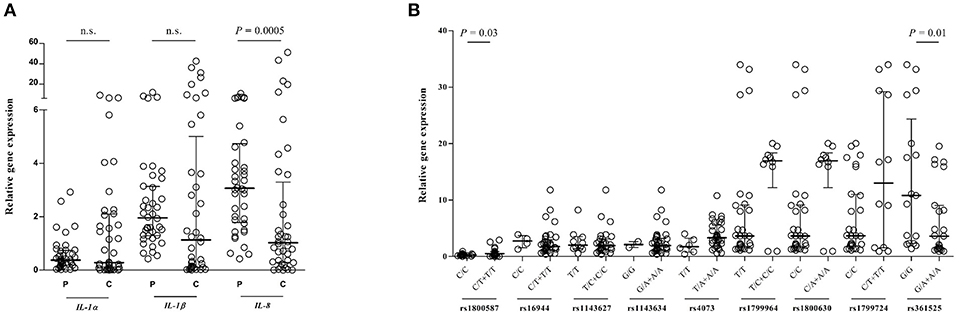
Figure 2. Quantitative real-time RT-PCR analysis of IL-1α, IL-1β, and IL-8 RNA expression of the patients with BP or control individuals. Comparison of candidate gene expressions/GAPDH ratio is shown between patient and control subjects (A) as well as between wild and polymorphic genotypes in the patient group (B), expressed as median with interquartile range and were compared based on the Mann-Whitney U-test. n.s., non-significant.
Discussion
Cytokines such as IL-1, IL-8, and TNF-α are key pro-inflammatory cytokines and play an essential role in inflammatory response through cell activation and triggering a cytokine cascade (27). Despite elevated levels of these cytokines have been demonstrated in BP, the obvious role of cytokines remains controversial. Since altered expression of cytokines could affect the pathogenesis of BP, functional polymorphisms in cytokine genes may alter cytokine production and its biological balance which in terms results in higher susceptibility or severity of BP disease. Lack of sufficient data about any possible effect of gene variations on susceptibility to BP disease in different ethnicity indicates a necessity for further investigations. This is the first study to the best of our knowledge, which investigates the association of alleles and genotypes of proinflammatory susceptible genes IL-1α, IL-1β, IL-8, and TNF-α with the risk of BP as well as the influence of these variations on cytokine gene expression.
IL-1α and IL-1β cytokines stimulate the release of chemotactic cytokines such as IL-8, either directly or in synergy with TNF-α (28, 29) and are involved in the pathologies of several skin diseases including psoriasis, cutaneous lupus erythematosus, atopic dermatitis, and autoimmune blistering diseases (11, 30). IL-1α and IL-1β, mainly produced by the keratinocytes in the skin, as well as TNF-α are key inflammatory cytokines in inflammatory skin diseases such as dermatomyositis and pemphigus (31), as well as experimental pemphigoid disease (32, 33). IL-1β stimulates the inflammatory signals by inducing other cytokines such as TNF-α and IL-8 (13). In our cohort study, we discovered an association between polymorphism of IL-8 cytokine in BP patients compared to the controls. There is disagreement in the results reported for IL-8 rs4073 in Iranian populations with inflammatory diseases. While a study has shown no association between the occurrence of chronic periodontitis (34), another investigation was observed a significant difference for this polymorphism in the same disease (35), which could be due to the diverse ethnicity of the Iranian population. In agreement with the later results of Iranian population, our data indicate that the minor allele is protective for developing the BP disease. Of note, analysis of PV patients revealed that this association is specific for BP because the allele distribution was indicial between healthy controls and PV patients.
However, our observations for IL-8 rs4073 is in contrast to data reported by a publication from China which investigated cytokine gene polymorphisms in a Chinese population (17). In this study, we found similar allele and genotype distribution of all studied SNPs in IL-1α and IL-1β genes. Except for IL-1β (rs1143627) (36), these findings are in line with observations of Iranian investigations on autoimmune diseases (37, 38). Among four studied polymorphisms in TNF-α, our observation revealed that the prevalence of minor allele in rs1799724 is significantly higher in the patients than controls, confirming previously published date from Iran in inflammatory disease (39). However, this significance disappeared after correction for multiple analysis. Similar to reported findings from Iran on autoimmune or inflammatory diseases, no significant differences were observed in our cohort study in all other SNPs in TNF-α (40–42).
There are controversies regarding the concentration of IL-1 in the blister fluid or sera of BP patients (43, 44). Nevertheless, since there is a relationship between IL-1β, IL-8, and TNF-α levels and the number of skin lesions in the patients, they might play a pathogenic role in BP disease (8, 45). Furthermore, blocking of pathogenic effect of rabbit IgG by using neutralizing IL-1 antibodies in an experimental mouse model of BP suggest a critical role of this cytokine in autoimmune blistering diseases (46). Genetic polymorphisms which are located in genes encoding for susceptibility factors may contribute to the gene expression or protein function and in turn to the disease predisposition in certain individuals. Our previous data indicate that the serum or plasma levels of TNF-α are significantly higher in the patients with BP or acute myocardial infarction to that of controls (18, 47). We could also show that the −308A allele is associated with elevated levels of TNF-α mRNA and protein expression in the patients which in turn might be contributed in increased transcription levels of TNF-α gene (47). There is also evidence that the −238G, −863A, and −1031C alleles influence the up-regulation levels of TNF-α expression in individuals (48, 49). In contrast, the −238A and −857T alleles are associated with lower levels or inhibition of TNF-α expression and transcription (50, 51). In other studies, it has been shown a higher serum level of IL-1β in patients with Alzheimer disease who carry −511T/T genotype (52). The SNPs in the IL-8 gene (-251A/T and +781C/T) have been shown to be associated with IL-8 production or protein expression both in vivo and in vitro (53, 54). Since the influence of the BP disease on cytokine gene expression as well as the effect of the SNPs in alteration of gene expression levels are still controversial, a quantitative analysis of the IL-1α, IL-1β, and IL-8 genes was performed. In agreement to previous investigations, the IL-8 gene was expressed significantly higher in the patients than to that of control. However, these expression levels of IL-8 gene suggest no association with the examined polymorphisms and comparable levels were found in wild-type and polymorphic genotypes. In contrast, further investigation revealed that variations of IL-1α rs1800587 and TNF-α rs361525 might contribute to altering the levels of gene expression.
The data presented herein has to be interpreted considering the limitations of our study. First, our study has a power of 70% to detect assumed differences of 30%. Hence, we may have missed significant associations due to the relatively low power of the study. However, nothing had been known about the investigated polymorphisms. Hence, our data now allows for a precise sample size calculation for future studies. In addition, despite this limitation, we identified one significant association. Second, the diagnosis of BP, like in the BLISTER study (55) was based on clinical presentation and DIF. Subsequent serological analysis of the BLISTER Study confirmed BP diagnosis in almost 90% of the cases (56). Hence, a maximum of 10% of our patient population may have had an alternative diagnosis, such as p200 pemphigoid, mucous membrane pemphigoid or epidermolysis bullosa acquisita. Third, genetic association studies cannot provide functional insights into the identified associated polymorphisms. However, these genetic studies may be used as a guide for further studies, addressing the functional impact of the newly identified associated gene polymorphisms.
In conclusion, our findings suggest that the minor allele in IL-8 SNP might play a protective role in susceptibility to BP in Iranian patients. Possible protective influences of IL-8 (rs4073) polymorphism against auto-inflammatory disorders might be helpful toward developing potential therapeutic strategies that intervene with the functions of inflammatory cytokine IL-8 in patients with BP. Given that this association was not observed in PV patients, this would also allow a more specific treatment of different pemphigus and pemphigoid diseases.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
RA, P-ST-P, ZS, HB, MG, SH, AK, FS, and FA participated in the experiments. HM, RL, and MW provided the samples and performed the diagnosis of the disease. P-ST-P and RA designed the study and performed data analysis. RA wrote the manuscript and RL revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the medical and nursing staff of the Skin Research Center, Shohada Tajrish, Loghman Hakim, and Razi hospitals at Tehran, Iran for helpful collaboration. We are also grateful to Claudia Kauderer, Lübeck, for excellent technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00636/full#supplementary-material
Supplementary Table 1. PCR condition and primer pairs for Quantitative RT-PCR of IL-1α, IL-1β, IL-8, TNF-α, and GAPDH gene expression levels.
References
1. Schmidt E, Zillikens D. Pemphigoid diseases. Lancet. (2013) 381:320–32. doi: 10.1016/S0140-6736(12)61140-4
2. Haeberle S, Wei X, Bieber K, Goletz S, Ludwig RJ, Schmidt E, et al. Regulatory T-cell deficiency leads to pathogenic bullous pemphigoid antigen 230 autoantibody and autoimmune bullous disease. J Allergy Clin Immunol. (2018) 142:1831–42.e7. doi: 10.1016/j.jaci.2018.04.006
3. Liu Y, Li L, Xia Y. BP180 is critical in the autoimmunity of bullous pemphigoid. Front Immunol. (2017) 8:1752. doi: 10.3389/fimmu.2017.01752
4. Ludwig RJ, Vanhoorelbeke K, Leypoldt F, Kaya Z, Bieber K, McLachlan SM, et al. Mechanisms of Autoantibody-induced pathology. Front Immunol. (2017) 8:603. doi: 10.3389/fimmu.2017.00603
5. Liu Z. Immunopathology of bullous pemphigoid, an autoimmune and inflammatory skin blistering disease. Keio J Med. (2003) 52:128–33. doi: 10.2302/kjm.52.128
6. Nakashima H, Fujimoto M. Expression of cytokines and chemokines in bullous pemphigoid. Exp Rev Dermatol. (2014) 4:321–7. doi: 10.1586/edm.09.23
7. Didona D, Di Zenzo G. Humoral epitope spreading in autoimmune bullous diseases. Front Immunol. (2018) 9:779. doi: 10.3389/fimmu.2018.00779
8. Ameglio F, D'Auria L, Bonifati C, Ferraro C, Mastroianni A, Giacalone B. Cytokine pattern in blister fluid and serum of patients with bullous pemphigoid: relationships with disease intensity. Br J Dermatol. (1998) 138:611–4. doi: 10.1046/j.1365-2133.1998.02169.x
9. Inaoki M, Takehara K. Increased serum levels of interleukin (IL)-5, IL-6 and IL-8 in bullous pemphigoid. J Dermatol Sci. (1998) 16:152–7. doi: 10.1016/S0923-1811(97)00044-3
10. Rhodes LE, Hashim IA, McLaughlin PJ, Friedmann PS. Blister fluid cytokines in cutaneous inflammatory bullous disorders. Acta Derm Venereol. (1999) 79:288–90. doi: 10.1080/000155599750010689
11. Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. (2011) 117:3720–32. doi: 10.1182/blood-2010-07-273417
12. Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. (2003) 3:745–56. doi: 10.1038/nri1184
13. Larsen C, Zachariae C, Mukaida N, Anderson A, Yamada M, Oppenheim J, et al. Proinflammatory cytokines interleukin 1 and tumor necrosis factor induce cytokines that are chemotactic for neutrophils, T cells and monocytes. Prog Clin Biol Res. (1990) 349:419–31.
14. Skov L, Beurskens FJ, Zachariae CO, Reitamo S, Teeling J, Satijn D, et al. IL-8 as antibody therapeutic target in inflammatory diseases: reduction of clinical activity in palmoplantar pustulosis. J Immunol. (2008) 181:669–79. doi: 10.4049/jimmunol.181.1.669
15. Trajkov D, Mishevska-Perchinkova S, Karadzova-Stojanoska A, Petlichkovski A, Strezova A, Spiroski M. Association of 22 cytokine gene polymorphisms with rheumatoid arthritis in population of ethnic macedonians. Clin Rheumatol. (2009) 28:1291–300. doi: 10.1007/s10067-009-1238-4
16. Tu Y, Fan G, Zeng T, Cai X, Kong W. Association between polymorphism within interleukin related genes and graves' disease: a meta-analysis of 22 case-control studies. Oncotarget. (2017) 8:98993–9002. doi: 10.18632/oncotarget.20114
17. Chang YT, Liu HN, Yu CW, Lin MW, Huang CH, Chen CC, et al. Cytokine gene polymorphisms in bullous pemphigoid in a chinese population. Br J Dermatol. (2006) 154:79–84. doi: 10.1111/j.1365-2133.2005.06938.x
18. Moravvej H, Tabatabaei-Panah PS, Ebrahimi E, Esmaeili N, Ghaderian SMH, Ludwig RJ, et al. TNF-alpha−308G/A gene polymorphism in bullous pemphigoid and alopecia areata. Hum Antibodies. (2018) 26:201–7. doi: 10.3233/HAB-180339
19. Feliciani C, Joly P, Jonkman MF, Zambruno G, Zillikens D, Ioannides D, et al. Management of bullous pemphigoid: the european dermatology forum consensus in collaboration with the european academy of dermatology and venereology. Br J Dermatol. (2015) 172:867–77. doi: 10.1111/bjd.13717
20. Cervin Serrano S, Gonzalez Villareal D, Aguilar-Medina M, Romero-Navarro JG, Romero Quintana JG, Arambula Meraz E, et al. genetic polymorphisms of interleukin-1 alpha and the vitamin d receptor in mexican mestizo patients with intervertebral disc degeneration. Int J Genomics. (2014) 2014:302568. doi: 10.1155/2014/302568
21. Liu H, Mao P, Xie C, Xie W, Wang M, Jiang H. Association between interleukin 8-251 T/A and +781 C/T polymorphisms and glioma risk. Diagn Pathol. (2015)10:138. doi: 10.1186/s13000-015-0378-x
22. Ohashi J, Naka I, Doi A, Patarapotikul J, Hananantachai H, Tangpukdee N, et al. A functional polymorphism in the IL1B gene promoter, IL1B−31C>T, is not associated with cerebral malaria in Thailand. Malaria J. (2005) 4:38. doi: 10.1186/1475-2875-4-38
23. Skoog T, van't Hooft FM, Kallin B, Jovinge S, Boquist S, Nilsson J, et al. A common functional polymorphism (C–>A substitution at position−863) in the promoter region of the tumour necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet. (1999) 8:1443–9.
24. Trevilatto PC, de Souza Pardo AP, Scarel-Caminaga RM, de Brito RB Jr, Alvim-Pereira F, Alvim-Pereira CC, et al. Association of IL1 gene polymorphisms with chronic periodontitis in Brazilians. Arch Oral Biol. (2011) 56:54–62. doi: 10.1016/j.archoralbio.2010.09.004
25. Trujillo-Murillo K, Perez-Ibave D, Munoz-Mejia C, Cordero-Perez P, Munoz-Espinosa L, Martinez-Rodriguez H, et al. Prevalence of the−308 and−238 tumor necrosis factor alpha (TNF-alpha) promoter polymorphisms in mexican chronic hepatitis C patients. Rev Gastroenterol Mex. (2010) 75:7–11.
26. Akatsu T, Nakamura M, Satoh M, Hiramori K. Increased mRNA expression of tumour necrosis factor-alpha and its converting enzyme in circulating leucocytes of patients with acute myocardial infarction. Clin Sci. (2003) 105:39–44. doi: 10.1042/CS20020367
27. Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta. (2014) 1843:2563–82. doi: 10.1016/j.bbamcr.2014.05.014
28. Kemeny L, Ruzicka T, Dobozy A, Michel G. Role of interleukin-8 receptor in skin. Int Arch Allergy Immunol. (1994) 104:317–22. doi: 10.1159/000236686
29. Sanmiguel JC, Olaru F, Li J, Mohr E, Jensen LE. Interleukin-1 regulates keratinocyte expression of T cell targeting chemokines through interleukin-1 receptor associated kinase-1 (IRAK1) dependent and independent pathways. Cell Signal. (2009) 21:685–94. doi: 10.1016/j.cellsig.2009.01.005
30. Ludwig RJ, Schmidt E. Cytokines in autoimmune bullous skin diseases. epiphenomena or contribution to pathogenesis? G Ital Dermatol Venereol. (2009) 144:339–49.
31. Santamaria P. Cytokines and chemokines in autoimmune disease: an overview. Adv Exp Med Biol. (2003) 520:1–7. doi: 10.1007/978-1-4615-0171-8_1
32. Sadeghi H, Lockmann A, Hund AC, Samavedam UK, Pipi E, Vafia K, et al. Caspase-1-independent IL-1 release mediates blister formation in autoantibody-induced tissue injury through modulation of endothelial adhesion molecules. J Immunol. (2015) 194:3656–63. doi: 10.4049/jimmunol.1402688
33. Samavedam UK, Kalies K, Scheller J, Sadeghi H, Gupta Y, Jonkman MF, et al. Recombinant IL-6 treatment protects mice from organ specific autoimmune disease by IL-6 classical signalling-dependent IL-1ra induction. J Autoimmun. (2013) 40:74–85. doi: 10.1016/j.jaut.2012.08.002
34. Khosropanah H, Sarvestani EK, Mahmoodi A, Golshah M. Association of IL-8 (-251 a/t) gene polymorphism with clinical parameters and chronic periodontitis. J Dent. (2013) 10:312–8.
35. Houshmand B, Hajilooi M, Rafiei A, Bidgoli M, Soheilifar S. Evaluation of IL-8 gene polymorphisms in patients with periodontitis in Hamedan, Iran. Dent Res J. (2012) 9:427–32.
36. Mohammadoo-Khorasani M, Salimi S, Tabatabai E, Sandoughi M, Zakeri Z, Farajian-Mashhadi F. Interleukin-1beta (IL-1beta) & IL-4 gene polymorphisms in patients with systemic lupus erythematosus (SLE) & their association with susceptibility to SLE. Indian J Med Res. (2016) 143:591–6. doi: 10.4103/0971-5916.187107
37. Mahmoudi M, Amirzargar AA, Jamshidi AR, Farhadi E, Noori S, Avraee M, et al. Association of IL1R polymorphism with HLA-B27 positive in Iranian patients with ankylosing spondylitis. Eur Cytokine Netw. (2011) 22:175–80. doi: 10.1684/ecn.2011.0293
38. Tahmasebi Z, Akbarian M, Mirkazemi S, Shahlaee A, Alizadeh Z, Amirzargar AA, et al. Interleukin-1 gene cluster and IL-1 receptor polymorphisms in iranian patients with systemic lupus erythematosus. Rheumatol Int. (2013) 33:2591–6. doi: 10.1007/s00296-013-2784-2
39. Abdolmohammadi R, Bonyadi M. Polymorphisms of Promoter Region of TNF-alpha gene in iranian azeri turkish patients with behcet's disease. J Korean Med Sci. (2017) 32:33–7. doi: 10.3346/jkms.2017.32.1.33
40. Ardebili SM, Yeghaneh T, Gharesouran J, Rezazadeh M, Farhoudi M, Ayromlou H, et al. Genetic association of TNF-alpha-308 G/A and−863 C/A polymorphisms with late onset alzheimer's disease in azeri turk population of Iran. J Res Med Sci. (2011) 16:1006–13.
41. Bonyadi M, Abdolmohammadi R, Jahanafrooz Z, Somy MH, Khoshbaten M. TNF-alpha gene polymorphisms in iranian azari turkish patients with inflammatory bowel diseases. Saudi J Gastroenterol. (2014) 20:108–12. doi: 10.4103/1319-3767.126322
42. Hadinedoushan H, Noorbakhsh P, Soleymani-Salehabadi H. Tumor necrosis factor alpha gene polymorphism and association with its serum level in iranian population with rheumatoid arthritis. Arch Rheumatol. (2016) 31:306-13. doi: 10.5606/ArchRheumatol.2016.5907
43. Grando SA, Glukhenky BT, Drannik GN, Kostromin AP, Chernyavsky AI, Barabash TM. Pemphigus and pemphigoid epidermocytes interfere with interleukin cascade reactions. J Clin Lab Immunol. (1990)32:137–41.
44. Schmidt E, Mittnacht A, Schomig H, Dummer R, Brocker EB, Zillikens D. Detection of IL-1 alpha, IL-1 beta and IL-1 receptor antagonist in blister fluid of bullous pemphigoid. J Dermatol Sci. (1996) 11:142–7. doi: 10.1016/0923-1811(95)00435-1
45. Giacalone B, D'Auria L, Ferraro C, Mussi A, Bonifati C, Ameglio F. Bullous pemphigoid blisters of the same duration have similar cytokine concentrations which decrease in older blisters. Br J Dermatol. (1998) 139:158–9. doi: 10.1046/j.1365-2133.1998.02341.x
46. Liu ZG GJ. Zhou, X. Fairley, JA. Diaz, LA. Interleukin-1 and−6 are required for subepidermal blistering in experimental bullous pemphigoid. J Invest Dermatol. (1997) 4:538.
47. Ghaderian SM, Akbarzadeh Najar R, Tabatabaei Panah AS. Tumor necrosis factor-alpha: investigation of gene polymorphism and regulation of TACE-TNF-alpha system in patients with acute myocardial infarction. Mol Biol Rep. (2011) 38:4971–7. doi: 10.1007/s11033-010-0641-x
48. Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, et al. Polymorphism of the 5'-flanking region of the human tumor necrosis factor (TNF)-alpha gene in japanese. Tissue Antigens. (1998) 51:605–12. doi: 10.1111/j.1399-0039.1998.tb03002.x
49. Huizinga TW, Westendorp RG, Bollen EL, Keijsers V, Brinkman BM, Langermans JA, et al. TNF-alpha promoter polymorphisms, production and susceptibility to multiple sclerosis in different groups of patients. J Neuroimmunol. (1997) 72:149–53. doi: 10.1016/S0165-5728(96)00182-8
50. van Heel DA, Udalova IA, De Silva AP, McGovern DP, Kinouchi Y, Hull J, et al. inflammatory bowel disease is associated with a tnf polymorphism that affects an interaction between the OCT1 and NF(-kappa)B transcription factors. Hum Mol Genet. (2002) 11:1281–9. doi: 10.1093/hmg/11.11.1281
51. Kaluza W, Reuss E, Grossmann S, Hug R, Schopf RE, Galle PR, et al. Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphism. J Invest Dermatol. (2000) 114:1180–3. doi: 10.1046/j.1523-1747.2000.00001.x
52. Licastro F, Pedrini S, Ferri C, Casadei V, Govoni M, Pession A, et al. Gene polymorphism affecting alpha1-antichymotrypsin and interleukin-1 plasma levels increases alzheimer's disease risk. Ann Neurol. (2000) 48:388–91. doi: 10.1002/1531-8249(200009)48:3<388::AID-ANA16>3.3.CO;2-7
53. de Oliveira JG, Rossi AF, Nizato DM, Cadamuro AC, Jorge YC, Valsechi MC, et al. Influence of functional polymorphisms in TNF-alpha, IL-8, and IL-10 cytokine genes on mRNA expression levels and risk of gastric cancer. Tumour Biol. (2015) 36:9159–70. doi: 10.1007/s13277-015-3593-x
54. Wacharasint P, Nakada TA, Boyd JH, Russell JA, Walley KR. AA genotype of IL-8−251A/T is associated with low PaO(2)/FiO(2) in critically ill patients and with increased IL-8 expression. Respirology. (2012) 17:1253–60. doi: 10.1111/j.1440-1843.2012.02244.x
55. Williams HC, Wojnarowska F, Kirtschig G, Mason J, Godec TR, Schmidt E, et al. Doxycycline vs. prednisolone as an initial treatment strategy for bullous pemphigoid: a pragmatic, non-inferiority, randomised controlled trial. Lancet. (2017) 389:1630–8. doi: 10.1016/S0140-6736(17)30560-3
Keywords: bullous pemphigoid, autoimmune disease, proinflammatory cytokines, gene polymorphism, gene expression
Citation: Tabatabaei-Panah P-S, Moravvej H, Sadaf Z, Babaei H, Geranmayeh M, Hajmanouchehri S, Karimi A, Sajjadi F, Arghand F, Ludwig RJ, Witte M and Akbarzadeh R (2019) Proinflammatory Cytokine Gene Polymorphisms in Bullous Pemphigoid. Front. Immunol. 10:636. doi: 10.3389/fimmu.2019.00636
Received: 12 November 2018; Accepted: 08 March 2019;
Published: 29 March 2019.
Edited by:
Attila Mócsai, Semmelweis University, HungaryReviewed by:
Kyle T. Amber, University of California, Irvine, United StatesMarian Dmochowski, Poznan University of Medical Sciences, Poland
Cezary Kowalewski, Medical University of Warsaw, Poland
Copyright © 2019 Tabatabaei-Panah, Moravvej, Sadaf, Babaei, Geranmayeh, Hajmanouchehri, Karimi, Sajjadi, Arghand, Ludwig, Witte and Akbarzadeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reza Akbarzadeh, YWtiYXJ6YWRlaEBhbmF0LnVuaS1sdWViZWNrLmRl
 Pardis-Sadat Tabatabaei-Panah1
Pardis-Sadat Tabatabaei-Panah1 Ahmad Karimi
Ahmad Karimi Ralf J. Ludwig
Ralf J. Ludwig Mareike Witte
Mareike Witte Reza Akbarzadeh
Reza Akbarzadeh