- 1Fralin Biomedical Research Institute, Virginia Tech Carilion, Roanoke, VA, United States
- 2Translational Biology, Medicine, and Health Graduate Program, Virginia Tech, Blacksburg, VA, United States
- 3Biomedical and Veterinary Sciences Graduate Program, Virginia-Maryland College of Veterinary Medicine, Virginia Tech, Blacksburg, VA, United States
- 4Department of Biomedical Sciences and Pathobiology, Virginia-Maryland College of Veterinary Medicine, Virginia Tech, Blacksburg, VA, United States
- 5Virginia Tech Carilion School of Medicine, Roanoke, VA, United States
CD4+ T helper cells are capable of differentiating into a number of effector subsets that perform diverse functions during adaptive immune responses. The differentiation of each of these subsets is governed, in large part, by environmental cytokine signals and the subsequent activation of downstream, cell-intrinsic transcription factor networks. Ikaros zinc finger (IkZF) transcription factors are known regulators of immune cell development, including that of CD4+ T cell subsets. Over the past decade, members of the IkZF family have also been implicated in the differentiation and function of individual T helper cell subsets, including T helper 1 (TH1), TH2, TH17, T follicular (TFH), and T regulatory (TREG) cells. Now, an increasing body of literature suggests that the distinct cell-specific cytokine environments responsible for the development of each subset result in differential expression of IkZF factors across T helper populations. Intriguingly, recent studies suggest that IkZF members influence T helper subset differentiation in a feed-forward fashion through the regulation of these same cytokine-signaling pathways. Here, we review the increasingly prominent role for IkZF transcription factors in the differentiation of effector CD4+ T helper cell subsets.
Introduction
The seminal discovery by Mosmann and Coffman that naïve CD4+ T cells could differentiate into either T helper 1 (TH1) or TH2 subsets launched an area of immunological investigation aimed at understanding the mechanisms underlying the functional diversity of CD4+ T helper cell populations (1). In the past three decades, the original TH1 and TH2 dichotomy has been expanded to include additional subsets such as TH17, T follicular helper (TFH), and regulatory T (TREG) cell populations (2–4). The diverse functions performed by these populations permit a highly tailored pathogen-specific immune response to bacterial, viral, and parasitic infections. Conversely, dysregulated T helper cell responses have been implicated in a number of autoimmune disorders, including type 1 diabetes, multiple sclerosis, Crohn's disease, and others (3, 5–7). Thus, due to the importance of these cell populations to human health, extensive efforts have been undertaken to better understand how CD4+ T helper cell subset differentiation is regulated.
Generally, it is recognized that the differentiation of effector CD4+ T cell populations requires three signals. Two of these signals are derived from direct cell-to-cell contact with an antigen-presenting cell (APC), in the form of T cell receptor and co-stimulatory receptor activation (8, 9). Importantly, the third signal, derived from the cytokine environment, drives CD4+ T helper cell subset specification through the activation of cytokine-specific transcription factor networks. Association of cytokines with their specific receptors results in the activation of Janus kinase/Signal Transducer Activator of Transcription (JAK/STAT) pathways, in which JAKs phosphorylate members of the STAT factor family (10). This ultimately leads to dimerization and translocation of STAT factors into the nucleus, where they activate the expression of subset-specific genes including those encoding “lineage-defining” transcription factors, which are required for the differentiation of each T helper cell subset (11, 12).
As with STAT transcription factors, members of the Ikaros Zinc Finger (IkZF) transcription factor family have well-documented roles in the development of immune cell populations (13–15). Ikaros, the founding member of the family, was initially shown to be required for lymphoid cell development, as mice expressing a dominant negative form of Ikaros failed to produce early T and B lymphocyte progenitors, as well as Natural Killer cells (16, 17). In the following decades, four proteins with a high degree of homology to Ikaros were identified and now comprise the IkZF family of transcription factors: Ikaros (encoded by the gene Ikzf1), Helios (Ikzf2), Aiolos (Ikzf3), Eos (Ikzf4), and Pegasus (Ikzf5) (18–22).
Structurally, IkZF family members contain both an N-terminal zinc finger (ZF) DNA-binding domain and a C-terminal ZF protein-protein interaction domain (Figure 1) (23). This distinct structure confers diverse functional capabilities, as IkZF family members can both positively and negatively regulate gene expression through direct interactions with DNA, as well as by forming transcriptional complexes with other proteins. Mechanistically, IkZF factors have been shown to regulate gene expression by (i) remodeling chromatin structure through association with chromatin remodeling complexes such as the nucleosome remodeling deacetylase (NuRD), (ii) interacting with and promoting the activity of the RNA Pol II transcription initiation complex, and (iii) inducing chromosome conformational changes by mediating interactions between distal cis-regulatory regions (14, 15, 24–26).
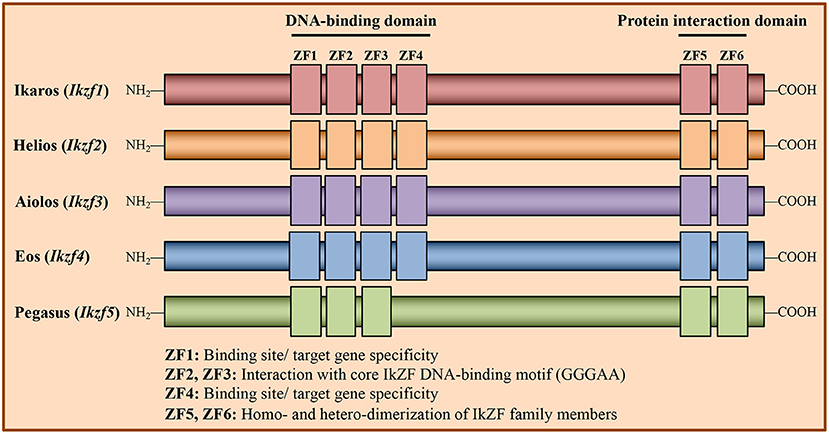
Figure 1. Structure of Ikaros Zinc Finger transcription factor family members. Members of the Ikaros Zinc Finger (IkZF) family of transcription factors are known regulators of hematopoietic cell development, including that of CD4+ T cells. The IkZF family consists of five members: Ikaros (encoded by the gene Ikzf1), Helios (Ikzf2), Aiolos (Ikzf3), Eos (Ikzf4), and Pegasus (Ikzf5). These factors contain N-terminal zinc finger (ZF) domains, which are responsible for mediating direct interactions with DNA, and C-terminal ZFs, which facilitate homo- and heterodimerization between IkZF family members. Of the N-terminal zinc fingers, ZF2 and ZF3 mediate direct interaction with the core IkZF DNA binding motif (GGGAA), while ZF1 and ZF4 regulate factor binding site/target gene specificity.
Recent research efforts have examined potential roles for IkZF family members in regulating the development of effector CD4+ T cell populations (Figure 2). Intriguingly, many of these studies point to mechanisms whereby IkZF factors propagate T helper cell subset differentiation via the modulation of cytokine signaling pathways. Here, we review the literature describing roles for IkZF members in the regulation of CD4+ T cell differentiation, with an emphasis on the interplay that exists between IkZF transcription factors and cell-specific cytokine signaling pathways.
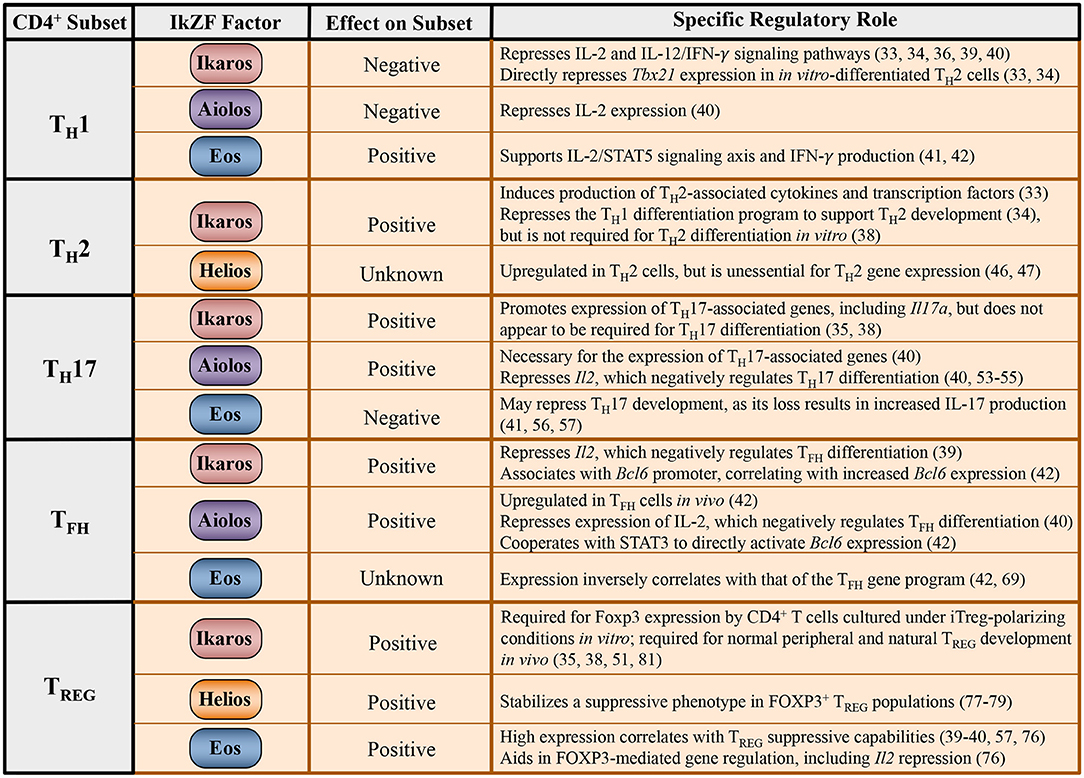
Figure 2. Regulation of CD4+ T cell differentiation programs by IkZF family members. Tabulated summary of the known effects of IkZF family members on expression of the TH1, TH2, TH17, TFH, and TREG gene programs, including associated cytokines and transcription factors. The broad impact of each IkZF factor on the differentiation program of individual T helper cell subsets is also provided, if known.
T Helper 1 (TH1) Cells
The differentiation and function of TH1 cells is critical for effective adaptive immune responses against intracellular pathogens, including viruses, bacteria, and parasites. TH1 differentiation is dependent upon extracellular signals from the cytokine IL-12, which lead to activation of the transcription factor STAT4 (27–29). Upon activation, STAT4 dimerizes and translocates to the nucleus where it directly activates expression of Tbx21, the gene encoding the TH1 lineage-defining transcription factor T-bet. T-bet directly induces the expression of the key TH1 effector cytokine Interferon-gamma (IFN-γ), which functions in a feed-forward fashion, as autocrine IFN-γ signals lead to STAT1 activation and further T-bet expression (30). The production of IFN-γ allows effector TH1 cells to initiate anti-intracellular pathogen responses including increased activation and proliferation of macrophages, as well as the activation of CD8+ T cell populations, which are responsible for the elimination of infected cells. In addition to IL-12 signaling, TH1 cell differentiation is dependent upon autocrine signals from IL-2, the expression of which is induced upon T cell receptor/co-receptor activation. IL-2 signaling results in the activation of STAT5, which, like STAT4, induces expression of genes encoding both transcription factors (including Blimp-1) and cytokine receptors (including components of the IL-12 and IL-2 receptor complexes) that are required for continued commitment to the TH1 lineage (31, 32).
IkZF Factors in TH1 Cell Differentiation and Function
Of the IkZF family members, Ikaros has been most extensively linked to aspects of TH1 cell development. It has been shown in a number of experimental settings that loss of Ikaros expression and/or function results in increased T-bet expression, suggesting that Ikaros plays a role in negatively regulating TH1 differentiation (33–35). Specifically, overexpression of a dominant negative form of Ikaros in TH2 cells resulted in increased T-bet expression, while Ikarosnull in vitro-differentiated TH2 cells expressed increased T-bet as compared to wildtype controls. Supporting a role for Ikaros in the direct regulation of T-bet expression, Ikaros has been shown to directly bind to the Tbx21 promoter in in vitro-polarized TH2 cells (33, 34). However, Ikaros was noticeably absent from the Tbx21 promoter in in vitro differentiated TH1 cells, for which T-bet expression is required (34). Mechanistically, the association of Ikaros with the Tbx21 promoter may be related to alterations in chromatin structure, as another study found increased enrichment of the repressive chromatin mark H3K27me3 at this locus upon Ikaros binding in thymocyte populations (36). However, whether this mechanism is conserved in CD4+ T cell populations is unclear. Regardless, the collective data support a role for Ikaros in the negative regulation of TH1 cell differentiation through direct repression of T-bet expression.
In addition to regulating TH1 differentiation pathways, Ikaros has been shown to negatively regulate expression of the TH1 effector cytokine, IFN-γ. Ikaros enrichment was observed at predicted Ifng regulatory regions in TH2 cells, and the Ifng promoter displayed reduced methylation in TH2 cells expressing a dominant negative form of Ikaros (33, 34). Furthermore, Ikarosnull TH2 cells were shown to exhibit increased IFN-γ production, as well as an increase in both T-bet and STAT1 transcript expression as compared to WT controls (33, 34). In further support of a T-bet-independent role for Ikaros in regulating Ifng expression, it has been shown that overexpression of wildtype Ikaros in Ikarosnull TH2 cells results in reduced IFN-γ production in the absence of a significant impact on T-bet expression (37). Collectively, these data further support a repressive role for Ikaros in both TH1 cell differentiation and function.
It is important to note, however, that all of the above studies utilized germline mutant models to assess the role of Ikaros in regulating T helper cell differentiation programs. Providing further clarity regarding the role of Ikaros in T helper cell differentiation decisions, a recent study assessed the effects of conditional Ikaros knockout exclusively in mature T cell populations on CD4+ T cell differentiation and function (38). Curiously, Ikaros-deficient mature T helper cells exposed to TH1-polarizing conditions did not exhibit increased T-bet or IFN-γ expression as compared to WT. However, Ikaros-deficient TH2 cells displayed increased IFN-γ expression, possibly supporting a role for Ikaros in negatively regulating TH1 gene expression in alternative T helper cell subsets, consistent with previous findings (38).
Illustrating an expanded role for Ikaros in regulating TH1 cytokine signaling pathways, Ikaros has also been shown to directly associate with the Il2 promoter and repress its expression (Figure 3) (39). Loss of Ikaros function was found to result in increased acetylation at the Il2 promoter, which correlated with increased IL-2 production in anergic T helper cells undergoing TCR stimulation. Similarly, Aiolos has also been shown to directly repress IL-2 expression (40). Given the importance of the IL-2/STAT5 pathway to TH1 cell differentiation, these data suggest that Ikaros and Aiolos may also negatively regulate TH1 differentiation by repressing autocrine IL-2 signaling.
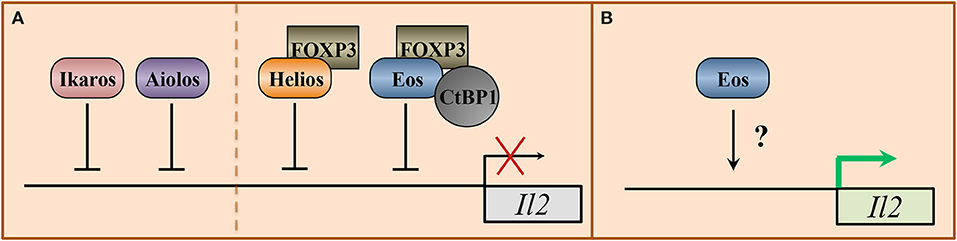
Figure 3. Transcriptional regulation of the interleukin-2 locus by IkZF transcription factors. Signals from the pro-inflammatory cytokine interleukin-2 (IL-2) differentially regulate the expression of T helper cell programs. IL-2 signaling supports the differentiation of TH1, TH2, and TREG cell subsets, but represses the differentiation of TH17 and TFH populations. The Ikaros zinc finger family members Ikaros, Helios, Aiolos, and Eos have all been implicated in regulating IL-2 expression. (A) In anergic CD4+ and TH17 cells, respectively, Ikaros and Aiolos have been shown to directly associate with the Il2 promoter to repress IL-2 expression. For Ikaros, this association correlates with reduced H3 and H4 acetylation. Aiolos association has been linked to a decrease in both acetylation and the positive histone mark H3K4me3 at the Il2 promoter and a concurrent increase in H3K9me3, which is indicative of a transcriptionally inactive locus. In regulatory T (TREG) cells, Eos and Helios have both been implicated in IL-2 repression. Mechanistically, Eos forms a protein complex with the TREG lineage-defining transcription factor FOXP3 and C-terminal binding protein (CtBP) to repress Il2. As with Aiolos, this repression is associated with reduced H3 and H4 acetylation, reduced H3K4me3, and increased H3K9me3 enrichment at the Il2 promoter. Similarly, Helios and FOXP3 are co-enriched at the Il2 locus in TREG cells and correlate with reduced H3 acetylation, consistent with gene repression. (B) In conventional T helper cell populations, in the absence of FOXP3, Eos has been positively linked to the expression of IL-2. However, the mechanism by which Eos influences IL-2 expression is currently unclear.
Unlike Ikaros and Aiolos, the available literature suggests that Eos may function as a positive regulator of the TH1 gene program. Specifically, mixed bone marrow chimera studies utilizing a murine experimental autoimmune encephalomyelitis (EAE) model demonstrated that there was a reduction in IFN-γ production by Eos-deficient vs. WT CD4+ T cells. Curiously, CD4+ T cells in Eos−/− animals displayed no difference in IFN-γ production as compared to those in WT animals during EAE, suggesting that there may be a compensatory mechanism at play in Eos-deficient animals (41). An additional study found that Eos expression correlated with that of TH1 genes at both the transcript and protein level (42). Furthermore, Eos was shown to regulate the expression of both IL-2 and CD25 expression in conventional CD4+ T (TCONV) cell populations (41). Thus, Eos may positively influence TH1 gene expression patterns, at least in part, by promoting the IL-2/STAT5 signaling axis. Additional work will be required to determine the precise role of Eos in promoting TH1 cell differentiation. Collectively, these studies support opposing roles for Ikaros and Aiolos vs. Eos in regulating TH1 cell differentiation and function.
T Helper 2 (TH2) Cells
TH2 responses both provide protection against parasitic infection, such as with helminthic worms, and also assist with tissue repair mechanisms following parasite- and inflammation-induced damage (43). TH2 differentiation is dependent upon both paracrine and autocrine IL-4 signaling, which results in the activation of STAT6. Once activated, STAT6 directly induces the expression of the TH2 lineage-defining transcription factor GATA3 (44). GATA3 then directly activates expression of the TH2 effector cytokines IL-4, IL-5, and IL-13, which recruit and activate additional immune cell types, such as macrophages and eosinophils, and promote IgE production to facilitate parasite clearance. As with TH1 cells, TH2 cell differentiation is also positively regulated by autocrine IL-2/STAT5 signaling, which, in the TH2 context, functions to induce GATA3-indepdendent expression of IL-4, as well as the feed-forward induction of IL-4Rα (45).
IkZF Factors in TH2 Cell Differentiation and Function
Ikaros has been implicated in positively regulating IL-4 production in TH2 cells. Specifically, naïve CD4+ T helper cells isolated from Ikarosnull mice exposed to TH2-polarizing conditions were found to exhibit reduced production of IL-4, and direct association of Ikaros with the Il4 locus was accompanied by increased H3 acetylation (33). Unsurprisingly, this correlated with decreased expression of the TH2 transcription factors GATA3 and cMaf (33). As mentioned previously, Ikaros has also been shown to promote TH2 lineage specification through direct repression of the TH1 lineage-defining transcription factor, T-bet (34). In support of this, in an in vivo experimental setting, mice heterozygous for a dominant negative form of Ikaros displayed an inappropriate TH1 response to Shistosoma mansoni parasitic infection (34). It is important to note that given heterodimeric interactions between IkZF family members, the dominant negative Ikaros protein, which lacks a functional N-terminal DNA binding domain, may disrupt the function of other IkZF factors in this experimental setting. Thus, the precise role of Ikaros in regulating the phenotypes observed in this study is unclear. Additionally, contrary to the above findings, a recent study utilizing a CD4+ specific conditional knockout of Ikaros exclusively in mature T cell populations found that Ikaros-deficient TH2 cells do not display a defect in either Gata3 or IL-4 expression, although they do produce more IFN-γ, as described above (38). Thus, it is possible that Ikaros deficiency during T cell development, vs. the naïve to effector transition, results in an altered phenotype that makes analysis of individual effector T cell populations difficult. Ultimately, these data suggest that Ikaros may support non-TH1 subsets by repressing the Th1 gene program during differentiation.
Similar to Ikaros, Helios expression is upregulated in TH2 cells generated in response to ovalbumin immunization in vivo, coinciding with the expression of GATA3, cMaf, and IL-4 (46). However, unlike Ikaros, Helios does not seem to be required for induction of the TH2 phenotype, as the same study showed that loss of Helios expression had no effect on the expression of TH2-associated cytokines and transcription factors at the transcript level (46). Additionally, no difference in IL-4 production was observed between TH2-polarized WT and Helios-deficient T cells, further establishing Helios as non-essential for TH2 development (47). Thus, although Helios is highly expressed in TH2 cells, there is a lack of evidence to support Helios as a regulator of TH2 development.
In addition to the production of IL-4, TH2 cells mediate their effector functions via secretion of the pro-inflammatory cytokines IL-5 and IL-13 (43). As with the IL-4 studies described above, Ikaros appears to positively mediate TH2-associated functions as exposure of naïve CD4+ T helper cells isolated from Ikarosnull mice to TH2 polarizing conditions resulted in reduced production of IL-5 and IL-13 compared to cells from wild-type mice (33). This is perhaps unsurprising, given the requirement for Ikaros for expression of GATA3, which directly regulates the expression of IL-5 and IL-13. Collectively, the above studies suggest that Ikaros positively regulates TH2 differentiation and function, both via the activation of IL-4 expression and through repression of the opposing TH1 differentiation program.
Finally, as mentioned above, Ikaros has also been implicated in the direct repression of Il2, which is critical for TH2 differentiation, in activated CD4+ T cell populations (39). This is somewhat at odds with the positive role for Ikaros in regulating TH2 development, and suggests that Ikaros may serve differential roles across T helper cell subsets.
T Helper 17 (TH17) Cells
TH17 cells are essential mediators of immunity at mucosal surfaces, and function to eliminate pathogenic extracellular bacteria and fungi (7, 48). A number of different cytokines have been implicated in TH17 cell differentiation, including TGF-β, and IL-6, among others. Signals from these cytokines result in the upregulation of TH17 transcriptional network that includes activation of STAT3, which directly regulates a number of genes required for TH17 differentiation including the TH17 lineage defining transcription factor RORγt (49, 50). Both STAT3 and RORγt are required for the production of the pro-inflammatory TH17 effector cytokines IL-17 and IL-22, which recruit and activate immune cells including neutrophils during the course of infection (2).
IkZF Factors in TH17 Cell Differentiation and Function
Recent work suggests that Ikaros is an important regulator of the TH17 gene program (35). Specifically, loss of Ikaros expression in in vitro-generated TH17 cells has been found to result in decreased expression of TH17 genes including those encoding RORγt and IL-17. Consistent with the known role for Ikaros in modulating the epigenetic landscape, expression of Ikaros in TH17 cells has been shown to correlate with increased enrichment of permissive covalent histone modifications at these loci (35). More recent reports have described a somewhat conflicting role for Ikaros in TH17 function (38, 51). In one study, inhibiting the DNA-binding capability of Ikaros had negligible effects on IL-17 production, while the production of IL-22 was increased (51). One possible explanation for the discrepancy presented by this study is the use of mice expressing a mutant Ikaros protein lacking a functional N-terminal ZF4 domain (Ikzf1Δf4/Δf4), as opposed to cells from Ikzf1Null mice. Zinc fingers 1 and 4 of the N-terminal zinc finger domain of Ikaros are important for binding to specific target genes, while zinc fingers 2 and 3 bind the core consensus sequence GGGAA (14, 15, 52) (Figure 1). Thus, the mutant utilized in the study may have retained some functionality regarding its DNA binding capability, which could explain the lack of an effect on IL-17 production in this setting. Furthermore, when Ikaros was conditionally knocked out in mature T cell populations, the expression of neither Il17 nor Rorc was impacted when Ikaros-deficient CD4+ T cells were exposed to TH17 polarizing conditions. Curiously, these cells exhibited increased expression of a pathogenic TH17 phenotype, including higher levels of T-bet expression and increased IFN-γ production. These data once again support a role for Ikaros in repressing TH1 gene expression during T helper cell differentiation (38).
Similar to Ikaros, recent work has established that Aiolos is necessary for the expression of TH17-associated genes including those that encode both IL-17a and IL-17f (40). Mechanistically, this study also determined that Aiolos aided TH17 lineage commitment, at least in part, through the direct silencing of the Il2 locus, as IL-2/STAT5 signaling negatively regulates TH17 development (40, 53–55). Thus, both Ikaros and Aiolos appear to regulate TH17 differentiation through modulation of TH17 gene expression and via repression of alternative gene programs.
In contrast to Ikaros and Aiolos, there is evidence to suggest that Eos may negatively regulate TH17 differentiation. Specifically, inhibition of Eos expression by the miRNA miR-17 was shown to enhance TH17 cell development (56). Furthermore, Eos-deficient TREG populations were found to gain the ability to produce IL-17 (57). Consistent with this finding, Eos-deficient mice were also found to develop more severe EAE, which correlated with an increased presence of IL-17-producing cells in the central nervous system (CNS) (41).
Taken together, the above findings suggest that Ikaros and Aiolos are positive regulators of TH17 differentiation and function, while Eos appears to functionally antagonize the development of TH17 cell populations.
T Follicular Helper (TFH) Cells
TFH cells play critical roles in the generation of humoral immunity through their specialized ability to provide help to antibody-producing B cells. TFH cells engage in cognate interactions with B cells and produce the cytokine IL-21 to support the formation of germinal centers in secondary lymphoid tissues and B cell production of high-affinity, pathogen-specific antibodies (5, 58). As with TH17 development, TFH cell differentiation can be driven by a number of cytokine signals, with two of the more prominent being IL-6 and IL-21 (59–61). Signals received from these cytokines result in subsequent activation of STAT3, which activates the expression of TFH genes including Bcl6, which encodes the lineage-defining transcription factor for the TFH cell subset (62–65).
IkZF Factors in TFH Cell Differentiation and Function
IL-2 signaling is a potent inhibitor of TFH cell differentiation (32, 66–68). Thus, it is not surprising that as antagonists of IL-2 signaling (39, 40), Ikaros and Aiolos have been implicated as positive regulators of TFH cell differentiation. Surprisingly, however, recent work suggests that Aiolos- and Ikaros-mediated regulation of TFH gene expression patterns may occur independently of this IL-2 dependent repressive mechanism. It was shown that the expression of Aiolos and Ikaros correlates with that of the TFH lineage-defining factor Bcl-6, and that both Aiolos and Ikaros directly associate with the Bcl6 promoter. (42). Furthermore, their enrichment patterns overlapped with those of histone marks consistent with increased chromatin accessibility and gene activation. Intriguingly, it was also shown that Aiolos interacted with STAT3 to form a novel transcriptional complex capable of inducing Bcl-6 expression in CD4+ T cell populations (42). Importantly, this study also found that Aiolos expression was increased in TFH cells generated in vivo in response to both Listeria monocytogenes and influenza infection, further supporting a role for Aiolos in regulating the TFH differentiation program (42).
As observed in other T helper cell populations, Eos appears to oppose the functions of Ikaros and Aiolos in TFH cells as well. Indeed, unlike Ikaros and Aiolos, Eos expression inversely correlates with that of the TFH gene program (42, 69). A recent study utilizing Nr4a-deficient T cells, which display reduced Eos expression, more readily upregulate both TFH gene expression patterns and obtain the ability to support germinal center reactions (69). It is important to note that these cells also exhibit reduced expression of a number of genes in addition to Eos upon Nr4a loss, and thus this phenotype cannot be directly attributed to loss of Eos expression. An additional study demonstrated that Eos-deficient CD4+ T cells are less effective producers of IL-2 as compared to their wild-type counterparts. As IL-2 negatively regulates TFH cell differentiation, these findings suggest that Eos may inhibit TFH differentiation via an IL-2-mediated mechanism (Figure 3) (41). Thus, further work is necessary to determine whether Eos functions to directly repress the TFH gene program or, rather, promotes the expression of alternative effector cell phenotypes.
Regulatory T (TREG) Cells
Unlike the pro-inflammatory effector functions of other T helper cell subsets, the primary role of regulatory T cells is to maintain immune tolerance through a number of suppressive mechanisms, including the secretion of anti-inflammatory cytokines such as IL-10 (70, 71). TREG development is driven by signals propagated through TGF-β engagement with its receptor and the resulting expression of TREG specific transcription factors, including the TREG lineage defining transcription factor Forkhead box P3 (FOXP3), and the IL-2 receptor α chain (CD25) (72–74). This stable expression of CD25, in conjunction with their inability to produce IL-2, allows TREGS to act as “IL-2 sinks,” to restrain pro-inflammatory immune responses. While a number of TREG cell subsets have been identified, much recent work has focused on two major subsets: those that arise from the thymus (tTREGS) and those that are generated in the periphery (pTREGS) (75).
IkZF Factors in TREG Differentiation and Function
Gene expression analysis via microarray studies revealed that Eos is highly expressed in TREG populations and that it functions as a key component of the FOXP3-mediated gene repression complex (76). Mechanistically, it was shown that Eos forms a protein complex with FOXP3 and C-terminal binding protein (CtBP) to promote gene silencing in TREG cells (76). These findings are consistent with another study demonstrating that Eos functions as a co-repressor in cooperation with Foxp3 to maintain the TREG phenotype and suppressive capabilities. Specifically, it was established that Eos downregulation occurs in TREG cells in response to inflammation, permitting their transition to a Foxp3-expressing T helper-like cell phenotype (57). Furthermore, another study found that knockdown of Eos expression resulted in decreased TREG function and a subsequent accentuation of colitis in mice (76). Intriguingly, a conflicting study found that Eos-deficient mice did not exhibit defective TREG development or function, suggesting that another IkZF factor (or factors) may provide a certain level of redundancy (41).
Interestingly, one of the genes targeted by the Eos/FOXP3-repressive complex is the Il2 locus, which appears to be directly regulated by a number of IkZF factors across T helper cell populations (Figure 3) (39, 40, 76). Furthermore, Eos-mediated repression of IL-2 seems to be dependent on the expression and activity of FOXP3, as Eos has also been shown to positively regulate IL-2 production in FOXP3− conventional T helper cell populations, as discussed previously (Figure 3) (41). Similar to Eos, Helios has also been implicated in repressing Il2 expression in TREG populations, supporting the possibility of redundant functions between these factors (Figure 3) (77).
Beyond its role in regulating IL-2 production in TREG cells, several studies have revealed that Helios is also required for the stability of a suppressive phenotype in FOXP3+ TREG populations (77). Consistent with these findings, the expression of pro-inflammatory cytokines, including IFN-γ, TNF-α, and IL-17 by TREG populations is significantly increased in the absence of Helios, and Helios-deficient mice display increased numbers of activated T cells and germinal center B cells, as well as increased production of autoantibodies (78). Curiously, Helios-deficient TREG cells also exhibit reduced STAT5 activation and Foxp3 expression, the latter of which can be rescued upon overexpression of a constitutively active form of STAT5 (78). Furthermore, the production of IL-17 is significantly higher in human FOXP3+Helios− memory TREGS as compared to FOXP3+Helios+ populations, supporting a role for Helios in negatively regulating IL-17 production in memory TREG populations (79). However, it is important to note that some studies suggest that Helios expression does not always negatively correlate with inflammatory cytokine expression in TREG populations. Indeed, Helios expression is consistent between conventional FOXP3+ TREG cells and IL-17-producing TREG cells that co-express FOXP3+ and the TH17 lineage-defining transcription factor RORγt (FOXP3+RORγt+ TREG), indicating that IL-17 production may be unrelated to Helios expression in this population (80). Collectively, while many findings support a role for Helios in promoting TREG suppressive function by repressing effector cytokine production, further work is necessary to establish how Helios functions across diverse TREG subtypes.
A number of studies have implicated Ikaros in the regulation of TREG cell differentiation. To this end, it has been shown that under iTREG-polarizing conditions, Ikaros-deficient CD4+ T cells are unable to upregulate Foxp3 expression (35, 38, 81). Importantly, these studies include cells in which Ikaros had been deleted in the germline (35, 81) and also exclusively in mature T cell populations (38). Additionally, one group observed increased enrichment of the repressive chromatin mark H3K27me2 at the Foxp3 promoter in Ikaros−/− naïve CD4+ T cells as compared to WT (35). In further support of a positive role for Ikaros in TREG differentiation, it has been shown that Ikaros-deficient animals exhibit reduced numbers of peripheral and natural TREG populations as compared to WT controls (81). Another study found that CD4+ T cells expressing a mutant form of Ikaros lacking the N-terminal DNA-binding ZF4 (IkΔZF4) are unable to normally differentiate into iTREG populations (51). Curiously, while mice expressing the IkΔZF4 mutant were found to exhibit increased numbers of total Foxp3+ TREGS in vivo under steady-state conditions, the number of pTREGS was reduced (51). Thus, the authors suggest that Ikaros may differentially regulate different TREG cell compartments. It is important to note that this study found that CD4+ T cells expressing the IkΔZF4 mutant produce significantly higher amounts of IL-21 than their WT counterparts, which negatively regulates iTREG differentiation (51). However, Ikaros-deficient cells were not found to upregulate IL-21 production, indicating that the mechanism underlying the iTREG-deficient phenotypes differs between these two studies (51, 81). Indeed, the authors of the first study found that CD4+ T cells isolated from the spleens of Ikaros-deficient mice exhibit a reduction in the expression of Foxo1, a transcription factor required for the generation of regulatory T cells (81). The different mechanisms observed between these studies may be attributed to use of the IkΔZF4 mutant, which is known to function as a dominant negative isoform. As discussed in previous sections, this may alter the function of other IkZF family members with which Ikaros interacts and may confound interpretation of the data in this system. Collectively, however, the available data support a role for Ikaros in positively regulating iTREG differentiation.
Stat/IkZF Factor Regulatory Modules as Candidate Drivers of T Helper Cell Differentiation
An interesting aspect of IkZF factors as regulators of T helper cell differentiation is that individual family members appear to perform diverse functions across T helper cell subsets. While the entirety of the mechanisms that permit such broad functionality of IkZF family members is currently unclear, one manner by which this differential regulation may be accomplished is through IkZF factor interactions with other transcription factors responsible for regulating individual T helper cell gene programs. As discussed throughout this review, STAT factors have long been established as cytokine-dependent regulators of effector CD4+ T cell differentiation and function (82, 83). Additionally, these factors are known to engage in cooperative mechanisms with other transcription factors to regulate gene expression patterns (84). Thus, STAT factors are prime candidates for potential regulation of IkZF factor function. Indeed, our laboratory recently found that the IkZF factor Aiolos interacts with STAT3 to form a transcriptional complex that induces the expression of Bcl-6, the lineage defining transcription factor for the TFH cell type (42). An interesting feature of this mechanism is that the IL-2/STAT5 signaling axis both inhibits the expression of Aiolos and the activity of STAT3. However, in the absence of IL-2 signaling, increased Aiolos expression and STAT3 activation results in the formation of an Aiolos/STAT3 transcriptional complex capable of inducing a Bcl-6-dependent TFH differentiation program. As detailed in the previous sections, the cytokine-dependent activation of cell-specific STAT factors is accompanied by alterations to expression and activity of different IkZF family members. Given the fact that members of these transcription factor families are both highly conserved and widely expressed across T helper cell subsets, these findings are suggestive of the intriguing possibility that additional IkZF/STAT factor complexes may broadly regulate the differentiation programs of CD4+ T cell populations (Figure 4). Furthermore, IkZF factor expression is not unique to CD4+ T helper cells, as these transcription factors are expressed in cells throughout the immune system (13, 14, 85, 86). Thus, the possibility exists that IkZF/STAT transcriptional complexes may represent a conserved regulatory feature that regulates an array of immune responses. Given the prominent role of STAT factor signaling in human disease pathology, understanding the extent to which they cooperate with IkZF factors to regulate immune cell differentiation is an intriguing area for future research (87).
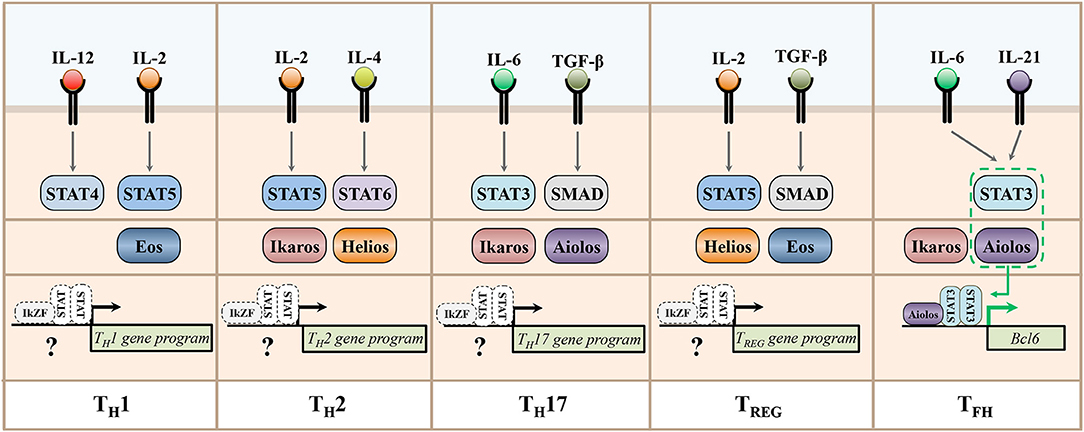
Figure 4. STAT and IkZF regulatory networks underlying the expression of T helper cell gene programs. This schematic depicts representative cytokines implicated in the development of murine T helper cell populations, their downstream STAT signaling pathways, and the Ikaros zinc finger transcription factors recently associated with the differentiation and functions of each subset. A recent study demonstrated that the IkZF factor Aiolos cooperates with STAT3 to induce the expression of Bcl6, the lineage-defining transcription factor for the TFH cell type. The differential expression and/or activity of IkZF and STAT factors across T helper cell subsets is suggestive of the intriguing possibility that additional, cell type-specific STAT/IkZF regulatory modules may regulate T helper cell differentiation decisions across subsets.
Concluding Remarks
In the past decade, IkZF family members have emerged as key regulators of CD4+ T helper subset development and function. Interestingly, many of these studies have identified IkZF factors as regulators of cytokines, cytokine receptors, and other components of cytokine signaling pathways. Likewise, it is becoming increasingly clear that cytokine signals reciprocally regulate the expression and activities of IkZF transcription factors. These points, coupled with the discovery that IkZF factors can engage in cooperative mechanisms with STAT transcription factors, suggest that IkZF factors may continue to emerge as central players in the regulation of T helper cell differentiation. Further work will be required to determine the extent to which IkZF factors may engage in similar mechanisms to regulate the differentiation and function of cells across the immune system. Importantly, cytokine signaling pathways have been popular targets of immunotherapeutic strategies to treat human diseases ranging from cancer to autoimmunity. Therefore, continued study into the role of IkZF factors in the regulation of immune cell differentiation and function will inform the feasibility of targeting these factors in efforts to promote human health.
Author Contributions
The manuscript was jointly conceptualized and written by MP, KR, BS, and KO.
Funding
This work was supported by grants from The National Institutes of Health (R01AI134972) and the Jeffress Memorial Trust to KO.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Oestreich Laboratory for critical review of this manuscript. This review focuses specifically on the role of IkZF factors in regulating T helper cell differentiation. We apologize to any authors whose work we inadvertently failed to mention or those that feel their studies could have been cited here.
References
1. Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. (1986) 136:2348–57.
2. Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. (2009) 27:485–517. doi: 10.1146/annurev.immunol.021908.132710
3. Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. (2008) 8:523–32. doi: 10.1038/nri2343
4. Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. (2015) 15:185–9. doi: 10.1038/nri3803
5. Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. (2014) 41:529–42. doi: 10.1016/j.immuni.2014.10.004
6. Geginat J, Paroni M, Maglie S, Alfen JS, Kastirr I, Gruarin P, et al. Plasticity of human CD4 T cell subsets. Front Immunol. (2014) 5:630. doi: 10.3389/fimmu.2014.00630
7. Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. (2008) 112:1557–69. doi: 10.1182/blood-2008-05-078154
8. June CH, Ledbetter JA, Gillespie MM, Lindsten T, Thompson CB. T-cell proliferation involving the CD28 pathway is associated with cyclosporine-resistant interleukin 2 gene expression. Mol Cell Biol. (1987) 7:4472–81. doi: 10.1128/MCB.7.12.4472
9. Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. (2013) 13:227–42. doi: 10.1038/nri3405
10. O'Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. (2008) 28:477–87. doi: 10.1016/j.immuni.2008.03.002
11. Nakayamada S, Takahashi H, Kanno Y, O'Shea JJ. Helper T cell diversity and plasticity. Curr Opin Immunol. (2012) 24:297–302. doi: 10.1016/j.coi.2012.01.014
12. Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4(+) T cell transcription factors. Nat Rev Immunol. (2012) 12:799–804. doi: 10.1038/nri3321
13. Heizmann B, Kastner P, Chan S. The Ikaros family in lymphocyte development. Curr Opin Immunol. (2017) 51:14–23. doi: 10.1016/j.coi.2017.11.005
14. John LB, Ward AC. The Ikaros gene family: transcriptional regulators of hematopoiesis and immunity. Mol Immunol. (2011) 48:1272–8. doi: 10.1016/j.molimm.2011.03.006
15. Yoshida T, Georgopoulos K. Ikaros fingers on lymphocyte differentiation. Int J Hematol. (2014) 100:220–9. doi: 10.1007/s12185-014-1644-5
16. Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. (1992) 258:808–12. doi: 10.1126/science.1439790
17. Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell. (1994) 79:143–56. doi: 10.1016/0092-8674(94)90407-3
18. Hahm K, Cobb BS, McCarty AS, Brown KE, Klug CA, Lee R, et al. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. (1998) 12:782–96. doi: 10.1101/gad.12.6.782
19. Honma Y, Kiyosawa H, Mori T, Oguri A, Nikaido T, Kanazawa K, et al. Eos: a novel member of the Ikaros gene family expressed predominantly in the developing nervous system. FEBS Lett. (1999) 447:76–80. doi: 10.1016/S0014-5793(99)00265-3
20. Kelley CM, Ikeda T, Koipally J, Avitahl N, Wu L, Georgopoulos K, et al. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr Biol. (1998) 8:508–15. doi: 10.1016/S0960-9822(98)70202-7
21. Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. (1997) 16:2004–13. doi: 10.1093/emboj/16.8.2004
22. Perdomo J, Holmes M, Chong B, Crossley M. Eos and pegasus, two members of the Ikaros family of proteins with distinct DNA binding activities. J Biol Chem. (2000) 275:38347–54. doi: 10.1074/jbc.M005457200
23. Payne MA. Zinc finger structure-function in Ikaros Marvin A Payne. World J Biol Chem. (2011) 2:161–6. doi: 10.4331/wjbc.v2.i6.161
24. Bottardi S, Mavoungou L, Milot E. IKAROS: a multifunctional regulator of the polymerase II transcription cycle. Trends Genet.(2015) 31:500–8. doi: 10.1016/j.tig.2015.05.003
25. Dege C, Hagman J. Mi-2/NuRD chromatin remodeling complexes regulate B and T-lymphocyte development and function. Immunol Rev. (2014) 261:126–40. doi: 10.1111/imr.12209
26. Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. (1999) 10:345–55. doi: 10.1016/S1074-7613(00)80034-5
27. Carter LL, Murphy KM. Lineage-specific requirement for signal transducer and activator of transcription (Stat)4 in interferon gamma production from CD4(+) versus CD8(+) T cells. J Exp Med. (1999) 189:1355–60. doi: 10.1084/jem.189.8.1355
28. Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. (1996) 382:174–7. doi: 10.1038/382174a0
29. Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, et al. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. (1996) 382:171–4. doi: 10.1038/382171a0
30. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. (2000) 100:655–69. doi: 10.1016/S0092-8674(00)80702-3
31. Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. (2011) 12:551–9. doi: 10.1038/ni.2030
32. Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. (2012) 13:405–11. doi: 10.1038/ni.2242
33. Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J Immunol. (2009) 182:741–5. doi: 10.4049/jimmunol.182.2.741
34. Thomas RM, Chen C, Chunder N, Ma L, Taylor J, Pearce EJ, et al. Ikaros silences T-bet expression and interferon-gamma production during T helper 2 differentiation. J Biol Chem. (2010) 285:2545–53. doi: 10.1074/jbc.M109.038794
35. Wong LY, Hatfield JK, Brown MA. Ikaros sets the potential for Th17 lineage gene expression through effects on chromatin state in early T cell development. J Biol Chem. (2013) 288:35170–9. doi: 10.1074/jbc.M113.481440
36. Oravecz A, Apostolov A, Polak K, Jost B, Le Gras S, Chan S, et al. Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat commun. (2015) 6:8823. doi: 10.1038/ncomms9823
37. Umetsu SE, Winandy S. Ikaros is a regulator of Il10 expression in CD4+ T cells. J Immunol. (2009) 183:5518–25. doi: 10.4049/jimmunol.0901284
38. Lyon de Ana C, Arakcheeva K, Agnihotri P, Derosia N, Winandy S. Lack of Ikaros deregulates inflammatory gene programs in T cells. J Immunol. (2019) 202:1112–23. doi: 10.4049/jimmunol.1801270
39. Thomas RM, Chunder N, Chen C, Umetsu SE, Winandy S, Wells AD. Ikaros enforces the costimulatory requirement for IL2 gene expression and is required for anergy induction in CD4+ T lymphocytes. J Immunol. (2007) 179:7305–15. doi: 10.4049/jimmunol.179.11.7305
40. Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol. (2012) 13:770–7. doi: 10.1038/ni.2363
41. Rieder SA, Metidji A, Glass DD, Thornton AM, Ikeda T, Morgan BA, et al. Eos is redundant for regulatory T cell function but plays an important role in IL-2 and Th17 production by CD4+ conventional T cells. J Immunol. (2015) 195:553–63. doi: 10.4049/jimmunol.1500627
42. Read KA, Powell MD, Baker CE, Sreekumar BK, Ringel-Scaia VM, Bachus H, et al. Integrated STAT3 and Ikaros zinc finger transcription factor activities regulate Bcl-6 expression in CD4+ Th cells. J Immunol. (2017) 199:2377–87. doi: 10.4049/jimmunol.1700106
43. Walker JA, McKenzie ANJ. TH2 cell development and function. Nat Rev Immunol. (2018) 18:121–33. doi: 10.1038/nri.2017.118
44. Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. (1997) 89:587–96. doi: 10.1016/S0092-8674(00)80240-8
45. Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol. (2008) 9:1288–96. doi: 10.1038/ni.1656
46. Serre K, Benezech C, Desanti G, Bobat S, Toellner KM, Bird R, et al. Helios is associated with CD4 T cells differentiating to T helper 2 and follicular helper T cells in vivo independently of Foxp3 expression. PLoS ONE. (2011) 6:e20731. doi: 10.1371/journal.pone.0020731
47. Cai Q, Dierich A, Oulad-Abdelghani M, Chan S, Kastner P. Helios deficiency has minimal impact on T cell development and function. J Immunol. (2009) 183:2303–11. doi: 10.4049/jimmunol.0901407
48. Basu R, Hatton RD, Weaver CT. The Th17 family: flexibility follows function. Immunol Rev. (2013) 252:89–103. doi: 10.1111/imr.12035
49. Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. (2010) 32:605–15. doi: 10.1016/j.immuni.2010.05.003
50. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. (2006) 126:1121–33. doi: 10.1016/j.cell.2006.07.035
51. Heller JJ, Schjerven H, Li S, Lee A, Qiu J, Chen ZM, et al. Restriction of IL-22-producing T cell responses and differential regulation of regulatory T cell compartments by zinc finger transcription factor Ikaros. J Immunol. (2014) 193:3934–46. doi: 10.4049/jimmunol.1401234
52. Schjerven H, McLaughlin J, Arenzana TL, Frietze S, Cheng D, Wadsworth SE, et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat Immunol. (2013) 14:1073–83. doi: 10.1038/ni.2707
53. Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. (2007) 26:371–81. doi: 10.1016/j.immuni.2007.02.009
54. Fujimura K, Oyamada A, Iwamoto Y, Yoshikai Y, Yamada H. CD4 T cell-intrinsic IL-2 signaling differentially affects Th1 and Th17 development. J Leukoc Biol. (2013) 94:271–9. doi: 10.1189/jlb.1112581
55. Lim HW, Kang SG, Ryu JK, Schilling B, Fei M, Lee IS, et al. SIRT1 deacetylates RORgammat and enhances Th17 cell generation. J Exp Med. (2015) 212:607–17. doi: 10.1084/jem.20132378
56. Liu SQ, Jiang S, Li C, Zhang B, Li QJ. miR-17-92 cluster targets phosphatase and tensin homology and Ikaros family zinc finger 4 to promote TH17-mediated inflammation. J Biol Chem. (2014) 289:12446–56. doi: 10.1074/jbc.M114.550723
57. Sharma MD, Huang L, Choi JH, Lee EJ, Wilson JM, Lemos H, et al. An inherently bifunctional subset of Foxp3+ T helper cells is controlled by the transcription factor eos. Immunity. (2013) 38:998–1012. doi: 10.1016/j.immuni.2013.01.013
58. Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol. (2016) 34:335–68. doi: 10.1146/annurev-immunol-041015-055605
59. Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, et al. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. (2011) 6:e17739. doi: 10.1371/journal.pone.0017739
60. Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. (2011) 334:825–9. doi: 10.1126/science.1208421
61. Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. (2008) 29:138–49. doi: 10.1016/j.immuni.2008.07.010
62. Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. (2009) 325:1006–10. doi: 10.1126/science.1175870
63. Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, et al. Bcl6 mediates the development of T follicular helper cells. Science. (2009) 325:1001–5. doi: 10.1126/science.1176676
64. Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. (2009) 31:457–68. doi: 10.1016/j.immuni.2009.07.002
65. Ma CS, Avery DT, Chan A, Batten M, Bustamante J, Boisson-Dupuis S, et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. (2012) 119:3997–4008. doi: 10.1182/blood-2011-11-392985
66. Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. (2012) 36:847–56. doi: 10.1016/j.immuni.2012.02.012
67. Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. (2012) 209:243–50. doi: 10.1084/jem.20111174
68. Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem. (2012) 287:11234–9. doi: 10.1074/jbc.M111.324046
69. Sekiya T, Kondo T, Shichita T, Morita R, Ichinose H, Yoshimura A. Suppression of Th2 and Tfh immune reactions by Nr4a receptors in mature T reg cells. J Exp Med. (2015) 212:1623–40. doi: 10.1084/jem.20142088
70. Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. (2012) 30:531–64. doi: 10.1146/annurev.immunol.25.022106.141623
71. Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. (2009) 30:636–45. doi: 10.1016/j.immuni.2009.04.010
72. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. (2003) 4:330–6. doi: 10.1038/ni904
73. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. (2003) 299:1057–61. doi: 10.1126/science.1079490
74. Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J Immunol. (2007) 178:4022–6. doi: 10.4049/jimmunol.178.7.4022
75. Plitas G, Rudensky AY. Regulatory T cells: differentiation and function. Cancer Immunol Res. (2016) 4:721–5. doi: 10.1158/2326-6066.CIR-16-0193
76. Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. (2009) 325:1142–6. doi: 10.1126/science.1176077
77. Baine I, Basu S, Ames R, Sellers RS, Macian F. Helios induces epigenetic silencing of IL2 gene expression in regulatory T cells. J Immunol. (2013) 190:1008–16. doi: 10.4049/jimmunol.1200792
78. Kim HJ, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. (2015) 350:334–9. doi: 10.1126/science.aad0616
79. Raffin C, Pignon P, Celse C, Debien E, Valmori D, Ayyoub M. Human memory Helios- FOXP3+ regulatory T cells (Tregs) encompass induced Tregs that express Aiolos and respond to IL-1beta by downregulating their suppressor functions. J Immunol. (2013) 191:4619–27. doi: 10.4049/jimmunol.1301378
80. Chellappa S, Hugenschmidt H, Hagness M, Line PD, Labori KJ, Wiedswang G, et al. Regulatory T cells that co-express RORgammat and FOXP3 are pro-inflammatory and immunosuppressive and expand in human pancreatic cancer. Oncoimmunology. (2016) 5:e1102828. doi: 10.1080/2162402X.2015.1102828
81. Agnihotri P, Robertson NM, Umetsu SE, Arakcheeva K, Winandy S. Lack of Ikaros cripples expression of Foxo1 and its targets in naive T cells. Immunology. (2017) 152:494–506. doi: 10.1111/imm.12786
82. O'Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. (2011) 11:239–50. doi: 10.1038/nri2958
83. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. (2010) 28:445–89. doi: 10.1146/annurev-immunol-030409-101212
84. Villarino AV, Kanno Y, O'Shea JJ. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol. (2017) 18:374–84. doi: 10.1038/ni.3691
85. Holmes ML, Huntington ND, Thong RP, Brady J, Hayakawa Y, Andoniou CE, et al. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J. (2014) 33:2721–34. doi: 10.15252/embj.201487900
86. Wang JH, Avitahl N, Cariappa A, Friedrich C, Ikeda T, Renold A, et al. Aiolos regulates B cell activation and maturation to effector state. Immunity. (1998) 9:543–53. doi: 10.1016/S1074-7613(00)80637-8
Keywords: CD4+ T helper cells, differentiation, gene regulation, Ikaros zinc finger (IkZF) transcription factors, cytokines
Citation: Powell MD, Read KA, Sreekumar BK and Oestreich KJ (2019) Ikaros Zinc Finger Transcription Factors: Regulators of Cytokine Signaling Pathways and CD4+ T Helper Cell Differentiation. Front. Immunol. 10:1299. doi: 10.3389/fimmu.2019.01299
Received: 28 March 2019; Accepted: 22 May 2019;
Published: 06 June 2019.
Edited by:
Remy Bosselut, National Cancer Institute (NCI), United StatesReviewed by:
Philippe Kastner, Université de Strasbourg, FranceSusan Winandy, Boston University, United States
Copyright © 2019 Powell, Read, Sreekumar and Oestreich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kenneth J. Oestreich, b2VzdHJrQHZ0Yy52dC5lZHU=
†These authors have contributed equally to this work
 Michael D. Powell
Michael D. Powell Kaitlin A. Read
Kaitlin A. Read Bharath K. Sreekumar
Bharath K. Sreekumar Kenneth J. Oestreich
Kenneth J. Oestreich