- 1Duke Molecular Physiology Institute, Duke University School of Medicine, Durham, NC, United States
- 2Division of Rheumatology, Department of Medicine, Duke University School of Medicine, Durham, NC, United States
- 3Laboratory Corporation of America Holdings (LabCorp), Morrisville, NC, United States
Background: MicroRNAs have been implicated in the pathogenesis of rheumatoid arthritis (RA), obesity, and altered metabolism. Although RA is associated with both obesity and altered metabolism, expression of RA-related microRNA in the setting of these cardiometabolic comorbidities is unclear. Our objective was to determine relationships between six RA-related microRNAs and RA disease activity, inflammation, body composition, and metabolic function.
Methods: Expression of plasma miR-21, miR-23b, miR-27a, miR-143, miR-146a, and miR-223 was measured in 48 persons with seropositive and/or erosive RA (mean DAS-28-ESR 3.0, SD 1.4) and 23 age-, sex-, and BMI-matched healthy controls. Disease activity in RA was assessed by DAS-28-ESR. Plasma cytokine concentrations were determined by ELISA. Body composition was assessed using CT scan to determine central and muscle adipose and thigh muscle tissue size and tissue density. Plasma and skeletal muscle acylcarnitine, amino acid, and organic acid metabolites were measured via mass-spectroscopy. Plasma lipoproteins were measured via nuclear magnetic resonance (NMR) spectroscopy. Spearman correlations were used to assess relationships for microRNA with inflammation and cardiometabolic measures. RA and control associations were compared using Fisher transformations.
Results: Among RA subjects, plasma miR-143 was associated with plasma IL-6 and IL-8. No other RA microRNA was positively associated with disease activity or inflammatory markers. In RA, microRNA expression was associated with adiposity, both visceral adiposity (miR-146a, miR-21, miR-23b, and miR-27a) and thigh intra-muscular adiposity (miR-146a and miR-223). RA miR-146a was associated with greater concentrations of cardiometabolic risk markers (plasma short-chain dicarboxyl/hydroxyl acylcarnitines, triglycerides, large VLDL particles, and small HDL particles) and lower concentrations of muscle energy substrates (long-chain acylcarnitines and pyruvate). Despite RA and controls having similar microRNA levels, RA, and controls differed in magnitude and direction for several associations with cytokines and plasma and skeletal muscle metabolic intermediates.
Conclusion: Most microRNAs thought to be associated with RA disease activity and inflammation were more reflective of RA adiposity and impaired metabolism. These associations show that microRNAs in RA may serve as an epigenetic link between RA inflammation and cardiometabolic comorbidities.
Introduction
MicroRNAs (miRNAs) are small, non-coding RNAs, ~22 nucleotides long, that regulate post-transcriptional gene expression (1). miRNAs are synthesized by multiple cells and tissues. While miRNA can be passively released upon injury, active release of miRNA in vesicles or exosomes allows miRNA to communicate in autocrine and paracrine fashions. Upon cellular uptake, miRNAs repress protein synthesis by cleaving or blocking translation of target mRNA (2). Individual miRNAs can have one hundred or more mRNA targets across multiple cells and organs systems, while individual mRNA can be bound and repressed by many miRNAs (3). Altered miRNA expression is associated with many disease states (4) and has been implicated in the pathogenesis of autoimmune disease, including rheumatoid arthritis (RA) (5, 6). Thus, miRNAs have been proposed as both RA biomarkers and therapeutic targets (7, 8).
In addition to autoimmune disease, miRNA contribute to the pathogenesis of sarcopenia (9) and obesity (10). However, evaluation of miRNAs in co-morbid disease states, including RA and its associated comorbidities, has received less attention. Despite revolutionary progress in the management of RA inflammation over the past few decades, patients with RA are still at high risk for sarcopenic obesity—decreased skeletal muscle mass with increased fat mass—which contributes to increased risks of disability, cardiovascular disease (CVD), and mortality (11, 12). RA development, severity, and poor treatment responses are tied to obesity (13). Also, RA is associated with sarcopenia, altered skeletal muscle remodeling, and impaired oxidative metabolism (14, 15). While these metabolic impairments in RA are likely driven in part by epigenetic dysregulation (16), it is unclear whether RA-related miRNAs contribute to the RA comorbidities of obesity and altered metabolism. Additionally, it is unclear how complex associations between miRNAs, RA inflammation, obesity, and metabolism impact the potential for miRNAs to be used as biomarkers and therapeutic targets in RA.
In the present study, we measured plasma expression of six miRNAs proposed to be biomarkers of RA inflammation: miR-21 (17), miR-23b (18), miR-27a (19), miR-143 (20), miR-146a (21), and miR-223 (22). Here, we evaluated relationships between miRNA expression and measures of inflammation, adiposity, and altered metabolism. To better understand RA miRNA specific effects, we then compared RA plasma expression of each miRNA to age-, gender-, and race-matched healthy controls. We hypothesized that some miRNAs would reflect RA disease activity, while others would better reflect obesity and metabolic alterations.
Patients and Methods
Design and Participants
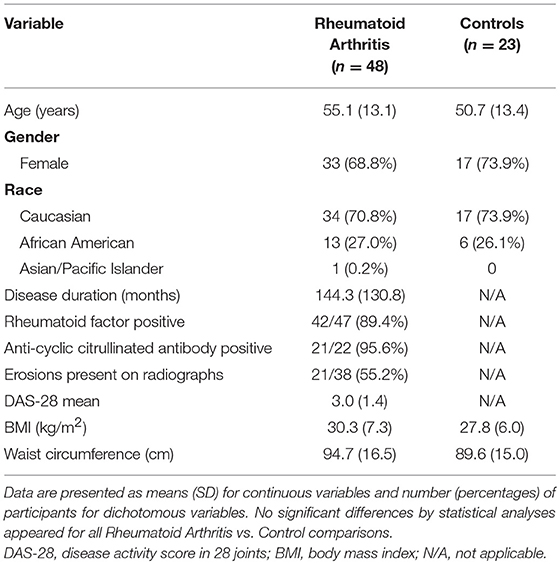
In a cross-sectional design, patients with RA and matched controls were recruited to participate as previously reported (15). RA subjects (n = 48) met American College of Rheumatology 1987 criteria (23); were seropositive (positive rheumatoid factor and/or anti-cyclic citrullinated peptide antibody) or had evidence of erosions on hand or foot imaging; had no medication changes within 3 months of enrollment; and were using ≤ 5 mg prednisone daily. Healthy control subjects (n = 23) without a previous diagnosis of inflammatory arthritis or current joint pain were matched to RA subjects by gender, race, age within 3 years, and body mass index (BMI) within 3 kg/m2. Subjects were excluded with pregnancy, type 2 diabetes mellitus, or known coronary artery disease. This study complied with the Helsinki Declaration and was approved by the Duke University Institutional Review Board.
Outcome Measures
All subjects underwent assessments as previously reported and described (15, 24), which included questionnaires, rheumatologic physical exam, fasting phlebotomy, computed tomography (CT) imaging of abdomen and thigh, and vastus lateralis muscle biopsies. RA disease activity was measured by the Disease Activity Score in 28 joints (DAS28) with erythrocyte sedimentation rate (ESR) (25). Plasma inflammatory marker and cytokine concentrations were determined by immunoassay (24). CT scan analyses were performed to determine central and muscle adipose and thigh muscle tissue size and tissue density (greater tissue density is indicative of less inter-muscular adipose tissue) (24). Standard Bergstrom needle muscle biopsies were performed on the vastus lateralis (26). All plasma and muscle tissue samples were stored at −80°C until analyses.
Analyses of plasma inflammatory markers, plasma, and skeletal muscle metabolic intermediates (acylcarnitines, amino acids, organic acids), and plasma lipoproteins were previously described in detail as follows. Plasma concentrations of inflammatory cytokines interleukin (IL)-1β, IL-6, IL-8, tumor necrosis factor (TNF)-α were measured via enzyme-linked immunosorbent assay (ELISA) (15). Plasma and skeletal muscle acylcarnitines were measured via targeted mass-spectroscopy (15, 27, 28). Skeletal muscle organic acids were measured via gas chromatography–mass spectrometry (GC/MS) (27, 29). Plasma lipoproteins were measured via nuclear magnetic resonance (NMR) spectroscopy (30, 31), while total cholesterol, triglycerides, LDL cholesterol, and HDL cholesterol were measured using standard automated methods.
Plasma miRNA Expression
miRNA was extracted from plasma samples using the miRCURY RNA Isolation kit—Biofluids from Exiqon (Denmark). Using reverse transcription, cDNAs were generated from miRNAs using the Universal cDNA Synthesis kit from Exiqon (No.203301). The miRNA PCR primer set, based on the SYBR Green miRCURY locked nucleic acids (LNA) detection system from Exiqon, was used according to the manufacturer's instructions to profile plasma expression of hsa-miR-21-5p (YP00204230), hsa-miR-23b-3p (YP00204790), hsa-miR-27a-3p (YP00206038), hsa-miR-143-3p (YP00205992), hsa-miR-146a-5p (YP00204688), and hsa-miR-223-3p (YP00205986). Levels of miRNAs were normalized to a reference hsa-miR-191-5p (YP00204306) and reported as delta cycle threshold (ΔCt), where higher values were equal to more abundant expression.
Statistical Analysis
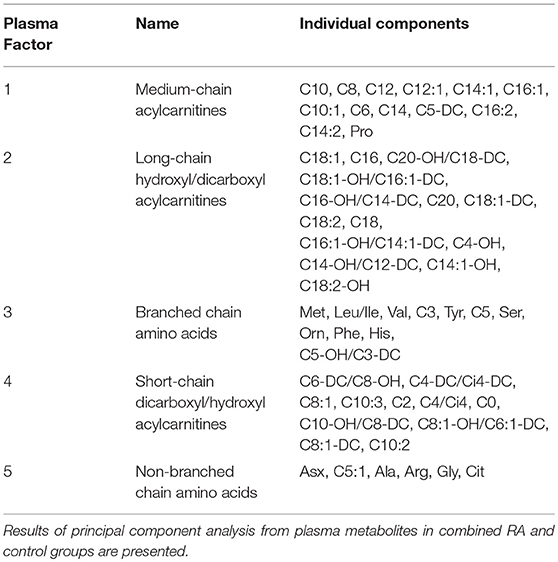
Participant characteristics (Table 1) and plasma miRNAs (Figure 1) were compared in RA vs. control subjects using two sample t-tests or Wilcoxon rank-sum tests dependent on whether data conformed to a normal distribution. Analyses of the metabolomic data of the combined RA and control subjects were performed separately for plasma (Table 2) and skeletal muscle (Table 3). Briefly, metabolic intermediates were standardized and reduced using principal components analysis (PCA) with varimax rotation to five factors, each with an eigenvalue >1.0. For each factor, individual metabolites with a factor load >0.4 were reported as factor components. Factor scores were computed for each individual, and correlations between factor scores and clinical assessments were evaluated using Spearman's rho. Strengths of associations for the two groups were compared with Fisher r-to-z transformations (32). All statistical analyses, besides Fisher transformations, were performed using SAS 9.4 (SAS, Cary, NC). Statistical significance was set at P-value <0.05. Data are available from the corresponding author upon a reasonable request.
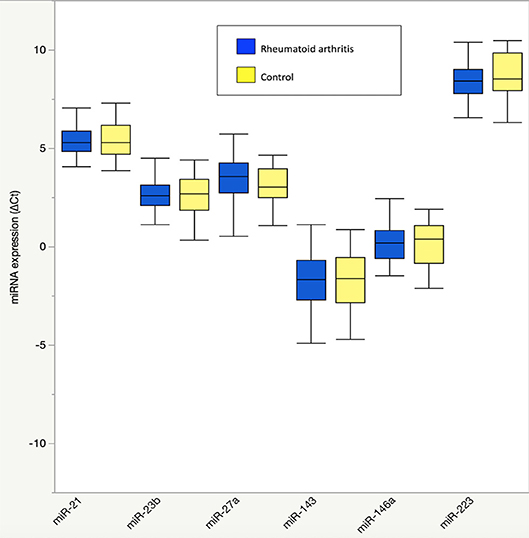
Figure 1. Plasma microRNA expression in rheumatoid arthritis compared to controls. Results are graphically displayed in box plots as medians, inter-quartile ranges (box), and upper and lower 25% of data values, excluding outliers (whiskers) for plasma expression of miR-21, miR-23b, miR-27a, miR-143, miR-146a, and miR-223 in rheumatoid arthritis (n = 48) and age-, gender-, and BMI-matched control subjects (n = 23). Plasma microRNA expression is reported as delta cycle threshold (ΔCt), where higher values are equal to higher expression. No significant group differences were seen in plasma microRNA expression (P > 0.05 for all).
Results
Associations Between MicroRNAs and Markers of RA Disease Activity, Inflammation, Adiposity, and Altered Metabolism in RA
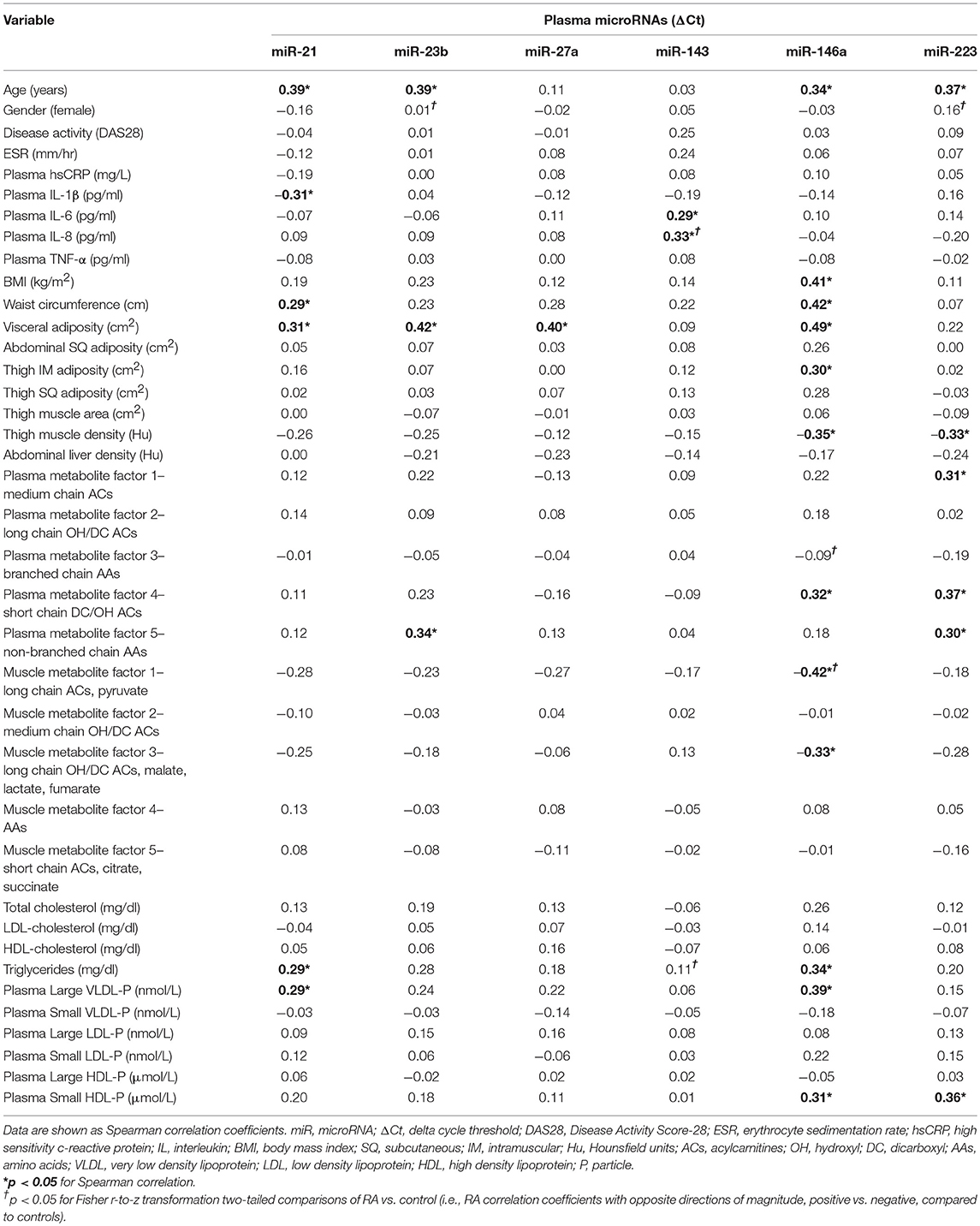
In RA, plasma miR-143 was positively related to RA-related systemic inflammation (plasma IL-6 and IL-8) (Table 4), however no miRNA was significantly associated with disease activity (DAS-28; range 0.6–6.4), ESR, or plasma high sensitivity c-reactive protein. miR-21 was associated with less systemic inflammation, greater adiposity, and an altered, pro-atherogenic plasma lipoprotein profile. miR-146a was associated with greater adiposity, pro-atherogenic lipoproteins, and altered plasma (Table 2) and skeletal muscle metabolic intermediates (Table 3). miR-23b and miR-27a were predominantly associated with adiposity while miR-223 was predominantly associated with increased thigh muscle fat, and altered plasma metabolic and lipoproteins profiles, namely small HDL particles, plasma short, and medium chain acylcarnitines and non-branched chain amino acids.
MicroRNA Associations With Markers of Inflammation and Altered Metabolism Are Different in RA Compared to Controls
There were no differences in RA and control plasma miRNA expression levels (Figure 1), but RA and controls differed in miRNA associations. In controls, miR-143 was negatively correlated with plasma IL-8 (Supplemental Table 1; r = −0.34), which significantly differed from the positive association in RA (r = 0.33; Fisher r-to-z P < 0.05). In controls, miR-146a was positively correlated with plasma branched chain amino acids (Table 2) (Supplemental Table 1; r = 0.45) and skeletal muscle long-chain acylcarnitines and pyruvate (Table 3) (Supplemental Table 1; r = 0.08), differing significantly from the negative associations in RA (r = −0.09 and −0.42, respectively; Fisher r-to-z P < 0.05 for both).
Discussion
In this cohort of established RA, several plasma miRNAs showed unique patterns of association with systemic pro-inflammatory cytokines, adiposity, and impaired metabolism. Among six miRNA selected based on prior associations with RA disease activity and/or inflammation, only miR-143 was reflective of RA systemic inflammation. Rather, plasma miRNAs in our RA cohort associated with measures of adiposity and metabolic alteration. These unique and unexpected associations, along with multiple associations that significantly differed from those of matched controls, highlight the complexity of miRNA functions, especially as they contribute to RA and associated comorbidities.
In contrast to previous reports in the literature (7), we did not find significant associations between inflammation and miR-146a. Inflammation is expected to induce miR-146a expression as part of a feedback mechanism to down-regulate the inflammatory response, including acute inflammation as well as Th1-mediated interferon responses (33–35). We hypothesize that our findings reflect differences in the inflammatory signatures of the RA patients with long-standing disease in our cohort as opposed to those with early, acute inflammatory disease. In our cohort of established RA, miR-146a was instead associated with multiple measures of adiposity as well as plasma short-chain dicarboxyl/hydroxyl acylcarnitines, strong markers of myocardial infarction and coronary artery disease risk (36). Plasma miR-146a was also differentially associated with plasma amino acids, as well as skeletal muscle long-chain acylcarinitines and pyruvate, both key substrates for muscle energy generation, compared to control subjects. These findings are supportive of miR-146a's role in modulating systemic metabolic function, which appears to be altered in RA.
miR-146a mechanistically downregulates TNF-α-induced adipogenesis (37, 38) and oxidative metabolism (39). Our findings suggest that miR-146a expression is appropriate (i.e., increased in response to limit adipogenesis), but is unable to adequately regulate specific metabolic processes due to altered interaction with target mRNAs. Consistent with this hypothesis, miR-146a polymorphisms are not more common in RA (40, 41), but rather RA susceptibility is associated with polymorphisms in known target mRNA binding sites (41).
miRNA alterations likely occur in other RA miRNAs, including miR-143 and miR-223. For example, miR-143 functions to induce inflammation through activation of NF-κB, and other studies show cellular expression of miR-143 is increased in RA synovial tissue (20, 42). In our cohort, miR-143 was positively associated with plasma inflammatory cytokines IL-6 and IL-8 in RA but negatively associated with plasma IL-8 in controls, indicating in RA, the miR-143 pro-inflammatory stimulating response is overactive. In contrast, while miR-223 down-regulates inflammation through multiple mechanisms (43, 44), counterintuitively, in RA, miR-223 expression is increased in multiple sites, including PBMCs, synovial tissue, and plasma (7). In our RA cohort, plasma miR-223 expression did not differ from controls or associate with RA disease activity or inflammation.
miR-223 is also associated with obesity (45) and HDL molecules, which transport miR-223 for lipid metabolic regulatory functions (46). We found RA miR-223 associated with thigh intramuscular fat, small HDL particles, plasma short, and medium chain acylcarnitines and non-branched chain amino acids. Thus, miR-223 alterations may contribute to incomplete systemic beta-oxidation and amino acid catabolism reflecting an obesity-related mitochondrial lipid overload state (47). Further study regarding the effects of these miRNAs on metabolic pathways is warranted.
While this study helps to inform how miRNAs likely exert effects on multiple biologic pathways crucial to RA-associated autoimmunity and impaired metabolism, the findings should be viewed in the context of a few key limitations. First, we did not find any significant differences in expression of candidate plasma miRNA between established RA and control subjects; in contrast, in larger cross-sectional cohorts, there was differential plasma expression of miR-146a and miR-233 between cohorts (7, 48). Whether these findings are the result of the heterogeneity of the RA subjects participating at different investigational sites, low overall RA disease activity, small sample size, or a lack of younger, early RA subjects in our cohort is unclear. Interestingly, previous research shows that epigenetic signatures differ in early vs. longstanding RA (49). Second, the focus of this exploratory study was to identify possible associations between plasma miRNAs and clinical, inflammatory, and metabolic factors to guide further in-depth research; thus, no direct causal pathways were evaluated. Third, we did not choose miRNAs based on microarray studies and thus our analyses were limited to individually measured miRNAs in this study. Finally, we measured miRNA expression only in plasma, and not tissue specific sites, such as immune cells, adipose tissue, or skeletal muscle. We note that quantification of circulating miRNAs may not accurately reflect expression in the tissue (i.e., skeletal muscle) (50). We did not measure the mode of miRNA packaging, either into microvesicles or exosomes, or attached to lipoproteins or other circulating proteins. Knowing the site of miRNA expression, mode of packaging, and target tissue or cellular site of mRNA regulatory function may be helpful in order to better understand these complex miRNA effector pathways.
In conclusion, multiple miRNAs in RA were associated with inflammatory and metabolic pathways in an opposite direction of both expected function and comparative findings in controls. In contrast to previous studies, we found that RA and matched controls had similar amounts of plasma miR-21, miR-23b, miR-27a, miR-143, miR-146a, and miR-223; and only miR-143 was positivity associated with inflammation in RA. Conversely, in RA, miR-146a and miR-223 were prominently associated with age, obesity, plasma and muscle metabolic intermediates, and plasma lipoproteins. Taken together, these findings show in the context of RA, miRNAs influence multiple inflammatory and metabolic pathways across a variety of cells and organ systems. These RA miRNA associations with adipose tissue and metabolic alterations may provide insight into epigenetic connections whereby chronic inflammation leads to common RA comorbidities of obesity and altered metabolism. Further research is needed to clarify the multitude of effects that miRNAs influence in order to utilize miRNAs as diagnostic tools and disease modifying therapies in RA.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
This study was carried out in accordance with the recommendations of the Duke University Medical Center Institutional Review Board with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Duke University Medical Center Institutional Review Board (IRB no. Pro00064057).
Author Contributions
BA, CC, VK, WK, and KH conceived and designed the study and experimental approach. KH performed the skeletal muscle biopsies. CC completed the miRNA analyses. OI and TK completed the metabolomic analyses. MC completed the lipoprotein analyses. BA and KH performed statistical analyses. BA wrote the manuscript. All authors contributed to the writing and approval of the final manuscript.
Funding
This work was supported by NIH/NIAMS K23AR054904 and the Rauch Family Scholarship.
Conflict of Interest Statement
MC was employed by company LabCorp (Morrisville, NC, USA).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the staff members at the Duke Center for Living for their help with recording of data. We appreciate the support of the Division of Rheumatology and Immunology at Duke University. We acknowledge and greatly appreciate all the participants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01475/full#supplementary-material
References
1. Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genome Proteome Bioinformat. (2009) 7:147–54. doi: 10.1016/S1672-0229(08)60044-3
2. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. (2007) 17:118–26. doi: 10.1016/j.tcb.2006.12.007
3. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. (2005) 433:769–73. doi: 10.1038/nature03315
4. Moutinho C, Esteller M. MicroRNAs and epigenetics. Adv Cancer Res. (2017) 135:189–220. doi: 10.1016/bs.acr.2017.06.003
5. Furer V, Greenberg JD, Attur M, Abramson SB, Pillinger MH. The role of microRNA in rheumatoid arthritis and other autoimmune diseases. Clin Immunol. (2010) 136:1–15. doi: 10.1016/j.clim.2010.02.005
6. Chen XM, Huang QC, Yang SL, Chu YL, Yan YH, Han L, et al. Role of micro RNAs in the pathogenesis of rheumatoid arthritis: novel perspectives based on review of the literature. Medicine. (2015) 94:e1326. doi: 10.1097/MD.0000000000001326
7. Churov AV, Oleinik EK, Knip M. MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential. Autoimmun Rev. (2015) 14:1029–37. doi: 10.1016/j.autrev.2015.07.005
8. Jin F, Hu H, Xu M, Zhan S, Wang Y, Zhang H, Chen X. Serum microRNA profiles serve as novel biomarkers for autoimmune diseases. Front Immunol. (2018) 9:2381. doi: 10.3389/fimmu.2018.02381
9. Fan J, Kou X, Yang Y, Chen N. MicroRNA-regulated proinflammatory cytokines in sarcopenia. Mediat Inflamm. (2016) 2016:1438686. doi: 10.1155/2016/1438686
10. Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. (2015) 11:276–88. doi: 10.1038/nrendo.2015.25
11. Biolo G, Cederholm T, Muscaritoli M. Muscle contractile and metabolic dysfunction is a common feature of sarcopenia of aging and chronic diseases: from sarcopenic obesity to cachexia. Clin Nutr. (2014) 33:737–48. doi: 10.1016/j.clnu.2014.03.007
12. Giles JT, Ling SM, Ferrucci L, Bartlett SJ, Andersen RE, Towns M, et al. Abnormal body composition phenotypes in older rheumatoid arthritis patients: association with disease characteristics and pharmacotherapies. Arthritis Rheum. (2008) 59:807–15. doi: 10.1002/art.23719
13. Schulman E, Bartlett SJ, Schieir O, Andersen KM, Boire G, Pope JE, et al. Overweight, obesity, and the likelihood of achieving sustained remission in early rheumatoid arthritis: results from a multicenter prospective cohort study. Arthritis Care Res. (2018) 70:1185–91. doi: 10.1002/acr.23457
14. Andonian BJ, Bartlett DB, Huebner JL, Willis L, Hoselton A, Kraus VB, et al. Effect of high-intensity interval training on markers of muscle remodeling in rheumatoid arthritis. Arthritis Res Ther. (2018) 20:283. doi: 10.1186/s13075-018-1786-6
15. Huffman KM, Jessee R, Andonian B, Davis BN, Narowski R, Huebner JL, et al. Molecular alterations in skeletal muscle in rheumatoid arthritis are related to disease activity, physical inactivity, and disability. Arthritis Res Ther. (2017) 19:12. doi: 10.1186/s13075-016-1215-7
16. Wang Z, Long H, Chang C, Zhao M, Lu Q. Crosstalk between metabolism and epigenetic modifications in autoimmune diseases: a comprehensive overview. Cell Mol Life Sci. (2018) 75:3353–69. doi: 10.1007/s00018-018-2864-2
17. Dong L, Wang X, Tan J, Li H, Qian W, Chen J, et al. Decreased expression of microRNA-21 correlates with the imbalance of Th17 and Treg cells in patients with rheumatoid arthritis. J Cell Mol Med. (2014) 18:2213–24. doi: 10.1111/jcmm.12353
18. Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α. Nat Med. (2012) 18:1077–86. doi: 10.1038/nm.2815
19. Shi DL, Shi GR, Xie J, Du XZ, Yang H. MicroRNA-27a inhibits cell migration and invasion of fibroblast-like synoviocytes by targeting follistatin-like protein 1 in rheumatoid arthritis. Mol Cells. (2016) 39:611–8. doi: 10.14348/molcells.2016.0103
20. Yang Z, Wang J, Pan Z, Zhang Y. miR-143-3p regulates cell proliferation and apoptosis by targeting IGF1R and IGFBP5 and regulating the Ras/p38 MAPK signaling pathway in rheumatoid arthritis. Exp Ther Med. (2018) 15:3781–90. doi: 10.3892/etm.2018.5907
21. Bae SC, Lee YH. MiR-146a levels in rheumatoid arthritis and their correlation with disease activity: a meta-analysis. Int J Rheum Dis. (2018) 21:1335–42. doi: 10.1111/1756-185X.13338
22. Lu MC, Yu CL, Chen HC, Yu HC, Huang HB, Lai NS. Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clin Exp Immunol. (2014) 177:641–51. doi: 10.1111/cei.12374
23. Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. (1988) 31:315–24. doi: 10.1002/art.1780310302
24. AbouAssi H, Tune KN, Gilmore B, Bateman LA, McDaniel G, Muehlbauer M, et al. Adipose depots, not disease-related factors, account for skeletal muscle insulin sensitivity in established and treated rheumatoid arthritis. J Rheumatol. (2014) 41:1974–9. doi: 10.3899/jrheum.140224
25. Prevoo ML, van 't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. (1995) 38:44–8. doi: 10.1002/art.1780380107
26. Bergstrom J. Percutaneous needle biopsy of skeletal muscle in physiological and clinical research. Scand J Clin Lab Invest. (1975) 35:609–16. doi: 10.1080/00365517509095787
27. Huffman KM, Koves TR, Hubal MJ, Abouassi H, Beri N, Bateman LA, et al. Metabolite signatures of exercise training in human skeletal muscle relate to mitochondrial remodelling and cardiometabolic fitness. Diabetologia. (2014) 57:2282–95. doi: 10.1007/s00125-014-3343-4
28. Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. (2009) 9:311–26. doi: 10.1016/j.cmet.2009.02.002
29. Haqq AM, Lien LF, Boan J, Arlotto M, Slentz CA, Muehlbauer MJ, et al. The study of the effects of diet on metabolism and nutrition (STEDMAN) weight loss project: rationale and design. Contemp Clin Trials. (2005) 26:616–25. doi: 10.1016/j.cct.2005.09.003
30. AbouAssi H, Connelly MA, Bateman LA, Tune KN, Huebner JL, Kraus VB, et al. Does a lack of physical activity explain the rheumatoid arthritis lipid profile? Lipids Health Dis. (2017) 16:39. doi: 10.1186/s12944-017-0427-4
31. Jeyarajah EJ, Cromwell WC, Otvos JD. Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clin Lab Med. (2006) 26:847–70. doi: 10.1016/j.cll.2006.07.006
32. VassarStats: website for statistical computation. Vassar College. 1998–2015. Available online at: http://vassarstats.net (accessed March 2019).
33. Saba R, Sorensen DL, Booth SA. MicroRNA-146a: a dominant, negative regulator of the innate immune response. Front Immunol. (2014) 5:578. doi: 10.3389/fimmu.2014.00578
34. Chen Y, Zeng Z, Shen X, Wu Z, Dong Y, Cheng JC. MicroRNA-146a-5p negatively regulates pro-inflammatory cytokine secretion and cell activation in lipopolysaccharide stimulated human hepatic stellate cells through inhibition of toll-like receptor 4 signaling pathways. Int J Mol Sci. (2016) 17:1076. doi: 10.3390/ijms17071076
35. Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. (2010) 142:914–29. doi: 10.1016/j.cell.2010.08.012
36. Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. (2010) 3:207–14. doi: 10.1161/CIRCGENETICS.109.852814
37. Roos J, Enlund E, Funcke JB, Tews D, Holzmann K, Debatin KM, et al. miR-146a-mediated suppression of the inflammatory response in human adipocytes. Sci Rep. (2016) 6:38339. doi: 10.1038/srep38339
38. Wu D, Xi QY, Cheng X, Dong T, Zhu XT, Shu G, et al. miR-146a-5p inhibits TNF-α-induced adipogenesis via targeting insulin receptor in primary porcine adipocytes. J Lipid Res. (2016) 57:1360–72. doi: 10.1194/jlr.M062497
39. Heggermont WA, Papageorgiou AP, Quaegebeur A, Deckx S, Carai P, Verhesen W, et al. Inhibition of MicroRNA-146a and overexpression of its target dihydrolipoyl succinyltransferase protect against pressure overload-induced cardiac hypertrophy and dysfunction. Circulation. (2017) 136:747–61. doi: 10.1161/CIRCULATIONAHA.116.024171
40. Yang B, Zhang JL, Shi YY, Li DD, Chen J, Huang ZC, et al. Association study of single nucleotide polymorphisms in pre-miRNA and rheumatoid arthritis in a Han Chinese population. Mol Biol Rep. (2011) 38:4913–9. doi: 10.1007/s11033-010-0633-x
41. Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. A polymorphism in the 3'-UTR of interleukin-1 receptor-associated kinase (IRAK1), a target gene of miR-146a, is associated with rheumatoid arthritis susceptibility. Joint Bone Spine. (2010) 77:411–3. doi: 10.1016/j.jbspin.2010.05.013
42. Yu B, Zhao Y, Zhang H, Xie D, Nie W, Shi K. Inhibition of microRNA-143-3p attenuates myocardial hypertrophy by inhibiting inflammatory response. Cell Biol Int. (2018) 42:1584–93. doi: 10.1002/cbin.11053
43. Yuan X, Berg N, Lee JW, Le TT, Neudecker V, Jing N, et al. MicroRNA miR-223 as regulator of innate immunity. J Leukoc Biol. (2018) 104:515–24. doi: 10.1002/JLB.3MR0218-079R
44. Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol. (2012) 189:4175–81. doi: 10.4049/jimmunol.1201516
45. Wen D, Qiao P, Wang L. Circulating microRNA-223 as a potential biomarker for obesity. Obes Res Clin Pract. (2015) 9:398–404. doi: 10.1016/j.orcp.2015.01.006
46. Tabet F, Cuesta Torres LF, Ong KL, Shrestha S, Choteau SA, et al. High-density lipoprotein-associated miR-223 is altered after diet-induced weight loss in overweight and obese males. PLoS ONE. (2016) 11:e0151061. doi: 10.1371/journal.pone.0151061
47. Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. (2008) 7:45–56. doi: 10.1016/j.cmet.2007.10.013
48. Ormseth MJ, Solus JF, Vickers KC, Oeser AM, Raggi P, Stein CM. Utility of select plasma MicroRNA for disease and cardiovascular risk assessment in patients with rheumatoid arthritis. J Rheumatol. (2015) 42:1746–51. doi: 10.3899/jrheum.150232
49. Ai R, Whitaker JW, Boyle DL, Tak PP, Gerlag DM, Wang W, et al. DNA methylome signature in synoviocytes from patients with early rheumatoid arthritis compared to synoviocytes from patients with longstanding rheumatoid arthritis. Arthritis Rheumatol. (2015) 67:1978–80. doi: 10.1002/art.39123
Keywords: rheumatoid arthritis, microRNA, metabolomics, skeletal muscle, obesity, lipoproteins, disease activity
Citation: Andonian BJ, Chou CH, Ilkayeva OR, Koves TR, Connelly MA, Kraus WE, Kraus VB and Huffman KM (2019) Plasma MicroRNAs in Established Rheumatoid Arthritis Relate to Adiposity and Altered Plasma and Skeletal Muscle Cytokine and Metabolic Profiles. Front. Immunol. 10:1475. doi: 10.3389/fimmu.2019.01475
Received: 05 April 2019; Accepted: 13 June 2019;
Published: 27 June 2019.
Edited by:
Marzia Dolcino, University of Verona, ItalyReviewed by:
Nicoletta Ronda, University of Parma, ItalyAntonio Puccetti, University of Genoa, Italy
Copyright © 2019 Andonian, Chou, Ilkayeva, Koves, Connelly, Kraus, Kraus and Huffman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian J. Andonian, YnJpYW4uYW5kb25pYW5AZHVrZS5lZHU=
 Brian J. Andonian
Brian J. Andonian Ching-Heng Chou1
Ching-Heng Chou1 William E. Kraus
William E. Kraus