- 1Department of Pediatrics, Center for Host Defense, Inflammation, and Lung Disease Research, The Pennsylvania State University College of Medicine, Hershey, PA, United States
- 2Departments of Pharmacology and Biochemistry and Molecular Biology, Institute for Personalized Medicine, The Pennsylvania State University College of Medicine, Hershey, PA, United States
- 3Department of Obstetrics and Gynecology, The Pennsylvania State University College of Medicine, Hershey, PA, United States
In humans there are two surfactant protein A (SP-A) functional genes SFTPA1 and SFTPA2 encoding innate immune molecules, SP-A1 and SP-A2, respectively, with numerous genetic variants each. SP-A interacts and regulates many of the functions of alveolar macrophages (AM). It is shown that SP-A variants differ in their ability to regulate the AM miRNome in response to oxidative stress (OxS). Because humans have both SP-A gene products, we were interested to determine the combined effect of co-expressed SP-A1/SP-A2 (co-ex) in response to ozone (O3) induced OxS on AM miRNome. Human transgenic (hTG) mice, carrying both SP-A1/SP-A2 (6A2/1A0, co-ex) and SP-A- KO were utilized. The hTG and KO mice were exposed to filtered air (FA) or O3 and miRNA levels were measured after AM isolation with or without normalization to KO. We found: (i) The AM miRNome of co-ex males and females in response to OxS to be largely downregulated after normalization to KO, but after Bonferroni multiple comparison analysis only in females the AM miRNome remained significantly different compared to control (FA); (ii) The targets of the significantly changed miRNAs were downregulated in females and upregulated in males; (iii) Several of the validated mRNA targets were involved in pro-inflammatory response, anti-apoptosis, cell cycle, cellular growth and proliferation; (iv) The AM of SP-A2 male, shown, previously to have major effect on the male AM miRNome in response to OxS, shared similarities with the co-ex, namely in pathways involved in the pro-inflammatory response and anti-apoptosis but also exhibited differences with the cell-cycle, growth, and proliferation pathway being involved in co-ex and ROS homeostasis in SP-A2 male. We speculate that the presence of both gene products vs. single gene products differentially impact the AM responses in males and females in response to OxS.
Introduction
Surfactant protein A (SP-A) plays important role in lung innate immunity and surfactant-related functions under basal conditions (1–5) and in response to various insults such as infection and oxidative stress (6–10). The human SP-A locus consists of two functional genes, SFTPA1 and SFTPA2, and one pseudogene (11, 12). The functional genes encode human SP-A1 and SP-A2 proteins, respectively, and each gene has been shown to have several genetic and splice variants (13, 14).
Human SP-A is expressed in alveolar epithelial type II cells (15) and in other tissues (16–19). It has been reported that human SP-A exists as octadecamer with six trimers (20), and that SP-A trimers have two SP-A1 molecules, and one SP-A2 molecule in the ratio of 2:1 (21). Previous studies from our group and others have shown that the ratio of the proposed model at mRNA and protein levels varies (22, 23). In bronchoalveolar lavage (BAL) fluid the ratio between SP-A1 to total SP-A varies as a function of age and health status (23, 24). The SP-A1 and SP-A2 mRNA content was found to vary in explant cultures under different conditions (25–27). Moreover, more SP-A2 mRNA than SP-A1 was observed in lung tissues of adults, whereas more SP-A1 mRNA transcripts were detected in neonates (28).
Several studies demonstrated that single gene products, SP-A1 and SP-A2, exhibit both qualitative (i.e., functional, biochemical and/or structural) (29–43), and quantitative (regulatory) differences (23, 25–27, 44–46). For example, SP-A1 and SP-A2 variants have been shown to differ in their ability to modulate the proteomic expression profile of AM and the AM actin cytoskeleton (47–49). The proteome profile of AM from KO mice, after treatment with exogenous SP-A1 or SP-A2 resulted in significant changes in proteins involved in the oxidative stress response pathway, with females being more responsive to SP-A1 and males to SP-A2 (48), as well as the single cell analysis revealed sex- and age-related differences in alveolar macrophage phenotypes from KO mice in response to SP-A1 and SP-A2 proteins (49). Moreover, sex differences have been observed between SP-A1 and SP-A2 and among variants in survival and lung function mechanics in response to bacterial infection (42, 43), and SP-A1 compared to SP-A2 exhibits a higher efficiency in pulmonary surfactant reorganization (50). The major contributor for at least some of these differences appears to be amino acid 85 of the precursor molecule; where SP-A1 has a cysteine and SP-A2 has an arginine (14). This Cys/Arg is a key difference between SP-A1 and SP-A2. This single amino acid change has a major impact on SP-A oligomerization, lipopolysaccharide (LPS) aggregation, and phagocytosis (38). The replacement of cysteine of SP-A1 with arginine or the arginine of SP-A2 with cysteine resulted in a reversal pattern of SP-A oligomerization and functional activity of both mutants of SP-A1 and SP-A2 (38). Thus, structural differences due to Cys85 and other amino acids may underlie differences in function observed between SP-A1 and SP-A2.
Ozone (O3), a major component of air pollution and a strong oxidizing agent known to cause toxicity in the lower airways, has significant effects on innate host defense and lung function (51). The O3 exposure can cause, edema, contributing to lung injury, and pulmonary surfactant derangement (52). A significant difference in survival has been observed with females being more affected than males in several lung diseases (53–56). In our animal studies, we observed significant differences in survival after infection and O3 exposure, with females being more susceptible to oxidative stress than males (7, 9) and sex hormones have been implicated in the observed differences in survival (57) but the mechanism underlying these differences is unknown. Moreover, during pneumonie infection and bacterial clearance, the ability to limit bacterial dissemination, and the phagocytic activity of alveolar macrophages may play an important role in the differential outcome in survival between males and females in the presence or absence of oxidative stress (7, 9, 58). Previously, we observed significant changes in AM miRNome of SP-A2 males but not in SP-A2 females or in SP-A1 males and females in response to OxS (41).
In the present study, we investigated the hypothesis that male and female mice expressing both SP-A1/SP-A2 gene products (co-ex) differentially regulate the AM miRNome in response to ozone-induced oxidative stress and that this differs from that previously observed in SP-A single gene variants (41). Toward this co-ex male and female mice were exposed to filtered air (FA) or O3 and the expression levels of 307 miRNAs was measured with or without normalizing to miRNAs identified from KO under the same conditions. We found significant differences in the AM miRNome of co-ex in terms of sex, exposure, with or without normalization to KO. Comparison of the co-ex miRNome to that of hTG mice carrying SP-A2 variant showed that the pathways involved in AM SP-A2 share some similarities to that of co-ex, but also exhibit differences.
Methods
Animals
Humanized transgenic (hTG) mice carrying both gene variants, SP-A1/SP-A2 (6A2/1A0, co-ex), as well as SP-A knockout (KO) mice were used in this study. They were 12 weeks old. hTG mice were generated on the C57BL/6J SP-A (KO) background (59). The animals used in this study were raised and maintained in a pathogen-free environment, at the Penn State College of Medicine animal facility as described previously (43). Both males and females were used. The females were synchronized with regard to the estrous cycle as described previously (43). A total of 44 mice (32 for miRNA analysis and 12 for qRT-PCR analysis) were used in the present study. All the procedures were approved by The Penn State Hershey Medical Center Institutional Animal Care and Use Committee (IACUC).
Filtered Air (FA) and Ozone (O3) Exposure
The animals were exposed to FA or O3 in parallel as described previously (60). A group of 4 animals (males, females) were exposed to FA or O3 for 3 h, and alveolar macrophages (AM) were isolated after 4 h of recovery as described (61).
RNA Preparation, Library Construction, and Sequencing
Total RNA was extracted from AMs using mirVana kit (#AM1560, Ambion, Waltham, MA). The extracted RNAs were quantified and quality checked using a BioAnalyzer RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA). Small RNA-seq libraries were generated by NEXTflex Small RNA Library Prep Kit v3 for Illumina (BioO Scientific, Austin, TX), followed by deep sequencing on an Illumina HiSeq 2500 as per the manufacturer's instructions. Briefly, 1-2 ng of total RNA was ligated with chemically modified 3′- and 5′- adapters that can specifically bind to mature micro RNAs, followed by reverse transcription and PCR amplification. Unique index sequence tags were introduced during PCR to enable multiplexed sequencing. Each library was assessed for the presence of desired micro RNA population and approximate library quantity by Bioanalyzer High Sensitivity DNA Kit (Agilent Technologies). Pooled libraries were denatured and loaded onto a TruSeq Rapid flow cell on an Illumina HiSeq 2500 and run for 50 cycles using a single-read recipe according to the manufacturer's instructions. De-multiplexed sequencing reads passed the default purify filtering of Illumina CASAVA pipeline (released version 1.8) and were quality trimmed/filtered using The FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit). The filtered reads were further trimmed with both 5′ and 3′ adapter sequences and subjected to Chimira suite to align and count miRNA expression (62). The differentially expressed miRNAs (DEG) between FA to O3, males and females were identified by using the edgeR (63) and the TCC v1.14.0 R package (64) with false discovery rate (FDR) adjusted p-value of 0.1 as a significance cutoff.
miRNA Data Analysis
We successfully identified 307 miRNAs with good correlation between mice (3 out of 4, Supplementary Table 1). We used two different parameters (methods) to analyze the identified miRNA expression (fold change). (a) The expression levels (fold change) of miRNAs from co-ex (FA to O3 for males and females) and KO (FA to O3 for males and females) were analyzed and differentially expressed miRNAs in co-ex and KO males and females were identified in response to FA or O3. (b) The changes in miRNA expression in co-ex were calculated by normalizing to KO, i.e., the level of expression of each individual experimental miRNA in co-ex males and females exposed to FA or O3 was divided by the corresponding miRNA in the KO. Next, the differentially expressed miRNAs between co-ex males and females were determined by dividing a specific individual male miRNA by the corresponding female miRNA (Supplementary Table 1).
Gene Expression Analysis
The expression levels of CCND1, CCND2, CCNE1, CDK7, IL-6, IL-10, TLR2, TLR3, STAT3, MYD88, IL-4, IL2RG, EGR2, PTEN, TNFSF12, MDTH, JUN, E2F3, BCL2, TNF, CDK2, SMAD2, MMP2, ARG1, AKT1, PPARA, and MYC genes at mRNA level in the female and male co-ex and KO AM, were validated by qRT-PCR as described previously (41). The RT2 qPCR Primer assays were purchased from Qiagen. The AM cell samples [3 animals/treatment (FA or O3)] were analyzed in triplicates/animal and quantified relative to GAPDH mRNA.
Statistical Analysis
Statistical differences between miRNA expression levels (fold change) in FA vs. O3 and male vs. female were evaluated by two-tailed t-test and nonparametric Mann-Whitney test. For multiple comparison analysis one-way analysis of variance (ANOVA) was employed followed by Bonferroni multiple comparisons. The p-values <0.05 were considered to be significant. All the data points are means ± standard deviation, and analyses were performed using Graph-Pad Prism software version 5.0 (Graph-Pad Software, San Diego, USA).
Results
We have previously studied the AM miRNome in hTG mice carrying either SP-A1 or SP-A2 variants using the qRT-PCR method (41). In this study, because humans have both SP-A gene products, we were interested to a) determine the combined effect of SP-A1/SP-A2 (6A2/1A0, co-ex) on the AM miRNome in response to oxidative stress with or without normalization to KO in males and females; b) identify sex, treatment, and gene (co-ex) impact on the AM miRNome; c) use differentially expressed miRNAs of ≥2 fold in co-ex in response to O3 exposure in Ingenuity Pathway Analysis (IPA) to identify biological functions and regulatory network targets of the identified and differentially expressed miRNAs after normalized to KO and Bonferroni multiple corrections; d) compare the co-ex miRNome (present study) and its targets to that of hTG mice carrying SP-A2 (41) variant under the same condition.
Effect of SP-A1/SP-A2 (6A2/1A0, co-ex) on AM miRNome Regulation
Without Normalization to KO
A total of 307 miRNAs from AMs of co-ex and KO in response to FA or O3 were identified from males and females (Supplementary Table 1). We observed significant differences (p < 0.05) in the expression of AM miRNAs in response to FA or O3 in co-ex females and KO males and females (Figures 1A,C,D). No significant differences were observed in co-ex males, after exposure to FA or O3 (Figure 1B). One-way ANOVA and Bonferroni multiple comparison analysis was performed to find the effect of treatment (FA, and O3) and sex (males, and females) as well as the interaction between the two parameters (sex and treatment). In co-ex, there was a significant difference between FA-exposed males and females (Figure 1E) but no significant difference was observed after O3 exposure between males and females (Figure 1E). However, females (but not males) showed a significant difference between FA or O3 exposure (Figure 1E). Whereas, in KO (i.e., in the absence of SP-A altogether), there was a significant difference between male and female after O3 exposure, no significant difference was observed after FA exposure (Figure 1F), indicating a role of SP-A in O3 exposure between sexes. However, similar to what was shown for the co-ex (Figure 1E), the KO females (but not the males) showed a significant difference after FA or O3 exposures (Figure 1F). Moreover, the miRNAs identified from KO females exposed to O3, showed a significant difference compared to KO males exposed to FA (Figure 1F).
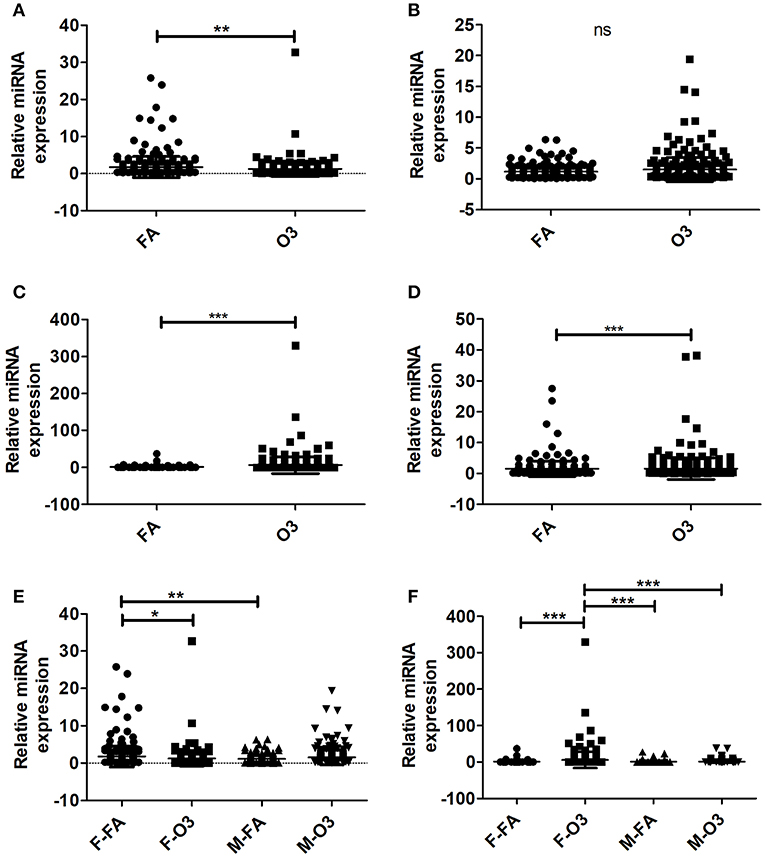
Figure 1. Regulation of the AM miRNome in co-ex and KO males (M) and females (F) after Filter air (FA) and Ozone (O3) exposure. Comparisons between miRNAs identified after FA or O3 exposure in co-ex females (A), males (B), and KO females (C), males (D). Significant differences observed between treatment in co-ex females, KO males and females (A,C,D; P < 0.05). (E,F) Depict the Bonferroni multiple comparisons of the miRNAs identified after FA or O3 exposure in co-ex and KO, respectively. A significant difference is observed in co-ex and KO females as a function of treatment (E,F). Significant differences (P < 0.05) in miRNA regulation were observed between sexes in co-ex (F-FA vs. M-FA, E), and in KO (F-O3 vs. M-O3, F-O3 vs. M-FA, F). ns, not significant. *p < 0.05, **p < 0.001, **p < 0.0001.
Normalization to KO
By normalizing the expression of miRNAs in co-ex to KO, i.e., the level of expression of each individual experimental miRNA (i.e., in SP-A1/SP-A2, co-ex) was divided by the level of the corresponding miRNA in the KO (Supplementary Table 1), we found significant differences in the differential expression of miRNAs in both males and females after FA or O3 exposure (Figures 2A,B). The one-way ANOVA and Bonferroni multiple comparison analysis resulted in similar observations as those observed in Figure 1E in the absence of KO normalization. Significant differences in miRNAs differentially expressed were observed in females between FA to O3 exposure and FA-exposed females to FA-exposed males, with no significant difference observed of differentially expressed miRNAs after O3 exposure between males and females (Figure 2C). Unlike in Figure 1E, FA-exposed females differed significantly from O3-exposed males (Figure 2C).
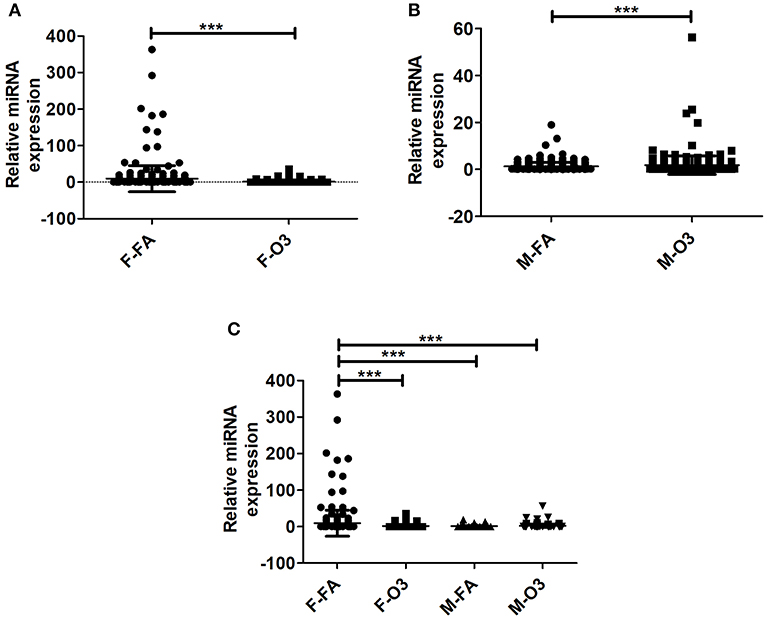
Figure 2. Effect of normalization on the regulation of miRNAs. The miRNAs identified from co-ex were normalized to same miRNAs identified in KO males and females after FA or O3 exposure. (A,B) Depict the differences in miRNA regulation in females and males after FA or O3. Significant differences (P < 0.05) were observed in both sexes after FA or O3 exposure (A,B). (C) Depicts the Bonferroni multiple comparisons of the miRNAs identified after FA or O3 exposure in co-ex. A significant difference (P < 0.05) was observed between co-ex FA or O3 exposed females and between sexes in co-ex (F-FA vs. M-FA, and F-FA vs. M-O3) (C). ***p < 0.0001.
The observations made with or without normalization to KO indicate that the AM miRNome of hTG mice carrying both SP-A1/SP-A2 (6A2/1A0, co-ex), exhibit no differences in miRNA expression between sexes in response to oxidative stress (O3 exposure), but sex differences are observed in controls (i.e., after FA exposure; Figures 1E, 2C). Female co-ex exhibited significant differences between FA or O3 exposure. The presence of the two genes may play a protective role in the outcome of miRNA expression in response to oxidative stress, especially in males.
Regulation of miRNAs That Changed ≥2 Fold in Response to FA, O3, and Sex
We further looked into the AM miRNAs, whose expression compared to control miRNAs is altered ≥2 fold in response to FA or O3 from co-ex and KO males and females, and compared them between males and females.
Without Normalization to KO
First we compared AM miRNAs of FA vs. O3 exposed animals. We found that in co-ex females, 49 miRNAs were changed after FA and 36 miRNAs after O3 exposure, whereas in males, 31 miRNAs were changed in FA and 45 in O3 exposure. The same comparison in KO AM miRNome showed that in females 45 miRNAs were changed ≥2 fold in FA and 94 miRNAs in O3, while in males 34 miRNAs were changed ≥2 fold in FA and 41 miRNAs in O3 (Figure 3A, Supplementary Table 1). In both co-ex and KO, all the miRNAs that had ≥2 fold are specific either to FA or O3 exposure, with no miRNAs in common between FA or O3 in either males or females.
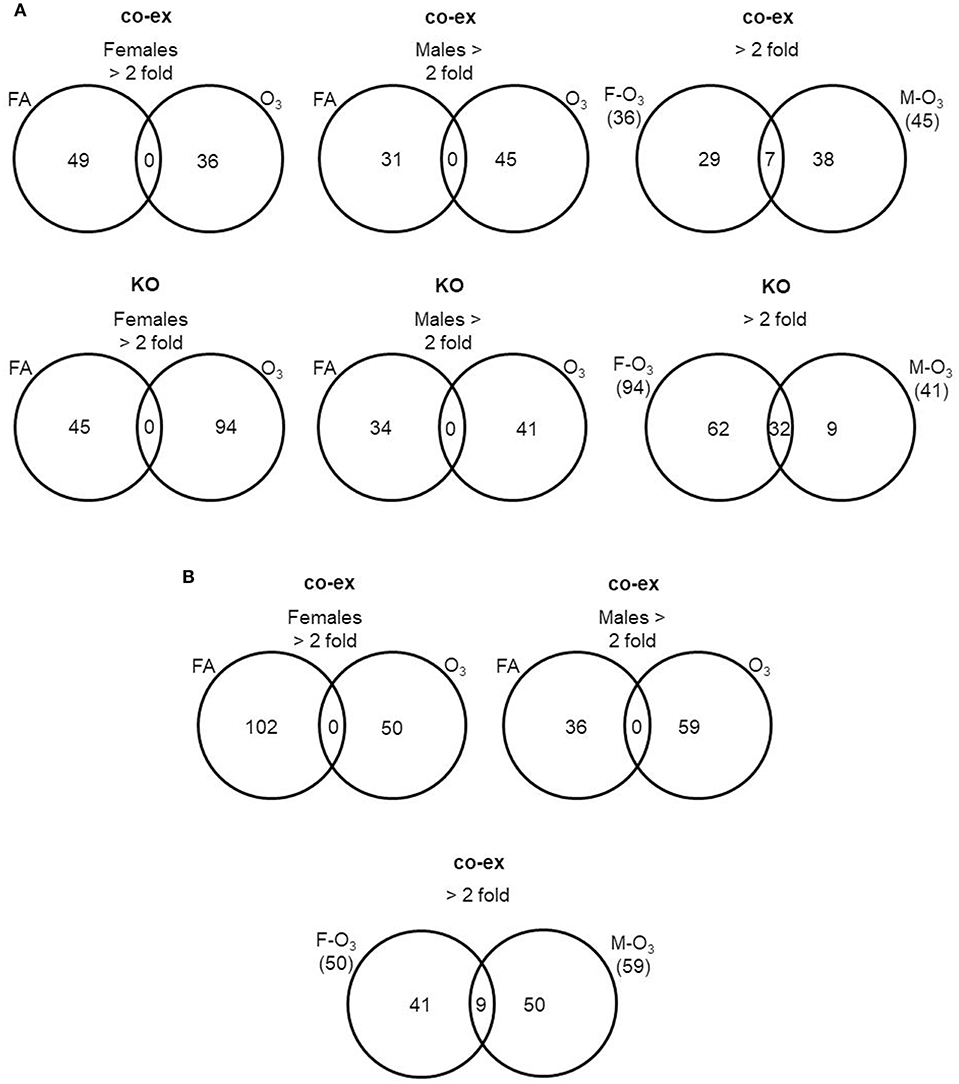
Figure 3. Comparison of miRNAs affected by FA or O3 in co-ex females and males. A. The Venn diagrams show miRNAs with significant changes ≥2 fold in response to FA or O3 in AM of males and female co-ex and KO. Out of the 307 miRNAs identified after FA or O3, 49 miRNAs had ≥2 fold and 36 miRNAs were ≥2 fold after FA or O3 exposure respectively, in co-ex females and in co-ex males 31 miRNAs had ≥2 fold and 45 miRNAs were ≥2 fold after FA or O3 exposure (A). A similar comparison in KO resulted in 45 miRNAs with ≥2 fold and 94 miRNAs with ≥2 fold after FA or O3 exposure, respectively, in KO females, and in KO males 34 miRNAs and 41 miRNAs with ≥2 fold after FA or O3 exposure, respectively. In both co-ex and KO there were no differentially regulated miRNAs found in common after FA or O3 exposure (A). A comparison of differentially regulated miRNAs after O3 exposure in co-ex between females and males, identified 36 miRNAs ≥2 fold in females and of these 29 were specific to females. In males 45 miRNAs ≥2 fold were identified and of these 38 were specific to males. Seven miRNAs were identified to be in common between females and males after O3 exposure (A). Whereas, in KO of the 94 miRNAs ≥2 fold in females, 62 were specific to females, and of the 41 miRNAs ≥2 fold in males, 9 were specific to males. Thirty-two miRNAs were identified to be in common between females and males after O3 exposure (A). (B) The Venn diagrams show the number of miRNAs identified from co-ex after normalization to the same miRNAs identified in KO males and females after FA or O3 exposure. Out of the 307 miRNAs identified after FA or O3, 102 miRNAs ≥2 fold and 50 miRNAs ≥2 fold were observed after FA or O3 exposure, respectively, in co-ex females. In co-ex males, 36 miRNAs ≥2 fold and 59 miRNAs ≥2 fold were identified after FA or O3 exposure, respectively. In both co-ex females and males, there were no differentially regulated miRNAs found in common after FA or O3 exposure. A comparison of differentially regulated miRNAs after O3 exposure in co-ex between females and males, identified 50 miRNAs ≥2 fold in females and of these, 41 were specific to females. In males of the 59 miRNAs ≥2 fold, 50 were specific to males and 9 miRNAs were identified to be in common between females and males after O3 exposure.
Next, we compared males vs. females (co-ex, KO) in response to oxidative stress. In co-ex females, 36 miRNAs were differentially expressed (≥2 fold) compared to 45 miRNAs in males, with 7 miRNAs (≥2 fold) being in common in both males and females. Of these, 29 miRNAs are unique to females and 38 are unique to males (Figure 3A). In KO females after O3 exposure, 94 miRNAs had ≥2 fold expression compared to 41 miRNAs in males. Of these 32 miRNAs were found in common in both sexes (Figure 3A), and 62 miRNAs are unique to females and 9 to males (Figure 3A). Of the seven miRNAs found to be in common between co-ex male and females after O3 exposure only one miRNA is included in the 32 miRNAs found in common between KO males and females (Supplementary Table 1). Of interest the co-ex had a significantly lower number of shared miRNAs between the sexes compared to KO (7 vs. 32), indicating the major effect of sex on co-ex (Supplementary Table 1).
With Normalization to KO
By normalizing the miRNAs identified in co-ex to KO, we found 102 miRNAs to be differentially expressed ≥2 fold in females after FA exposure compared to 50 miRNAs after O3 exposure (Figure 3B). In the case of males, 36 miRNAs are differentially expressed after FA exposure and 59 miRNAs had ≥2 fold after O3 exposure. In both co-ex males and females, all the miRNAs with ≥2 fold are specific to either FA or O3. A comparison of miRNAs from males and females after O3 exposure showed 50 miRNAs ≥2 fold in females vs. 59 miRNAs in males (Figure 3B). Of these, 41 are unique to females and 50 to males, with 9 being found in common. These data indicate that when the expression of the AM miRNome in co-ex, after normalization to KO is compared in males and females, following exposure to FA or O3, the observed changes in miRNA levels are either specific to males or females. However, when the miRNome between males and females co-ex normalized KO is compared after oxidative stress there are few miRNAs that are found in common in co-ex males and females (Supplementary Table 1).
Ingenuity Pathway Analysis (IPA)
We performed IPA to further understand the role of significantly changed miRNAs in co-ex males and females under the studied conditions. After Bonferroni correction, expression of miRNAs in co-ex females exhibited significant differences in response to OxS, but not in males (Figure 2C). However, the IPA analyses before and after the Bonferroni correction were identical because the miRNA data input in IPA was same. We were able to identify biological functions and regulatory network targets of these differentially expressed miRNAs. The targets shown in Figure 4 are involved in anti-apoptosis, cell cycle, cellular growth and proliferation, as well as pro-inflammatory response are affected by differential expression of miRNAs in response to O3 exposure (Supplementary Table 1). These targets include CCND1, CCND2, CCNE1, CDK7, IL-6, IL-10, TLR2, TLR3, STAT3, TNFSF12, MYD88, IL-4, IL2RG, EGR2, PTEN, MDTH, JUN, E2F3, BCL2, TNF, CDK2, MYC, SMAD2, MMP2, ARG1, AKT1, and PPARA mRNAs. The miRNAs that were changed significantly in co-ex females and males in response to O3 exposure and their targets are listed in Table 1.
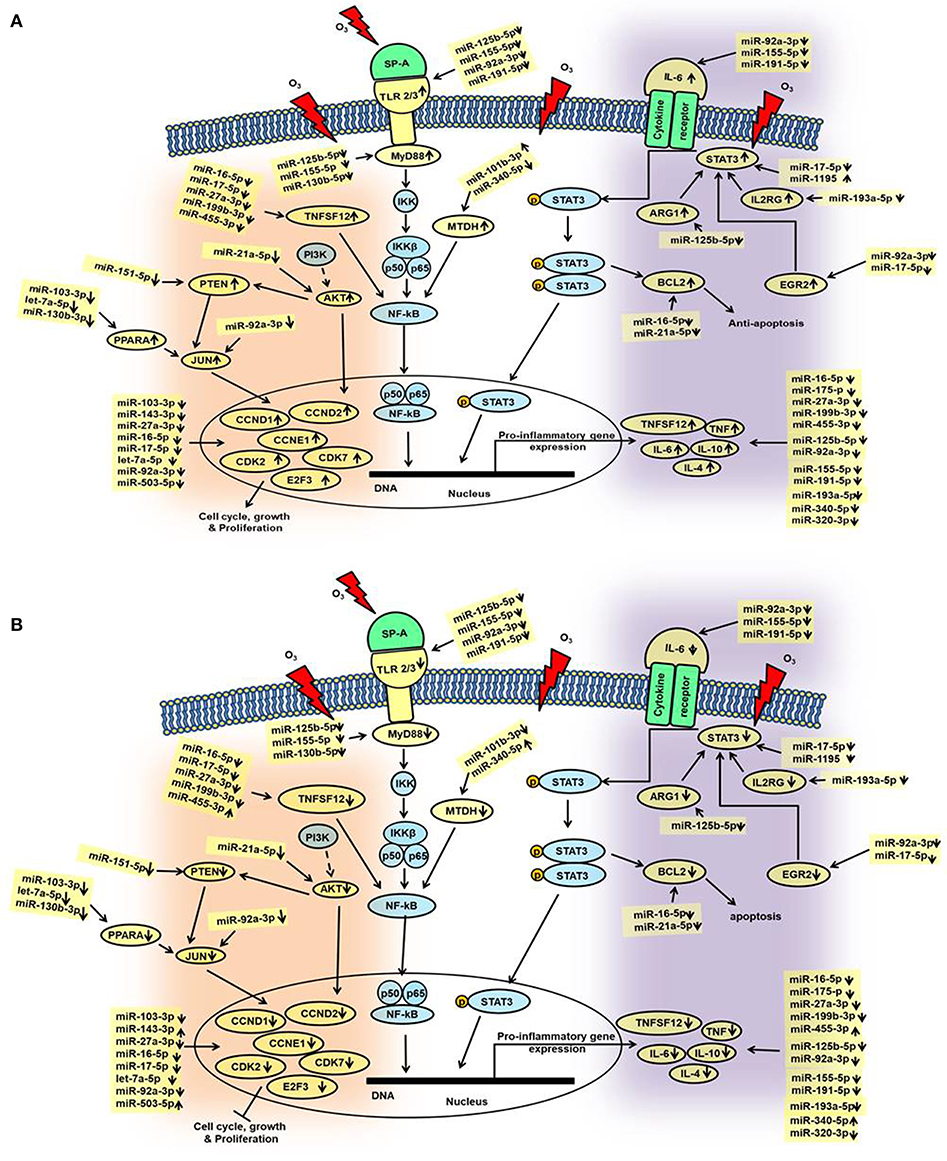
Figure 4. Schematic representation of the identified miRNAs in co-ex AM and their targets in response to OxS. This diagram depicts the significantly changed miRNAs and their targets in the various pathways. These include pathways of cell cycle, and cellular growth and proliferation as well as, pathways of pro-inflammatory response and anti-apoptosis, (A) males and (B) females. The miRNAs and genes studied in the present study are highlighted with yellow. Up (↑) and down (↓) arrows indicate increase or decrease.
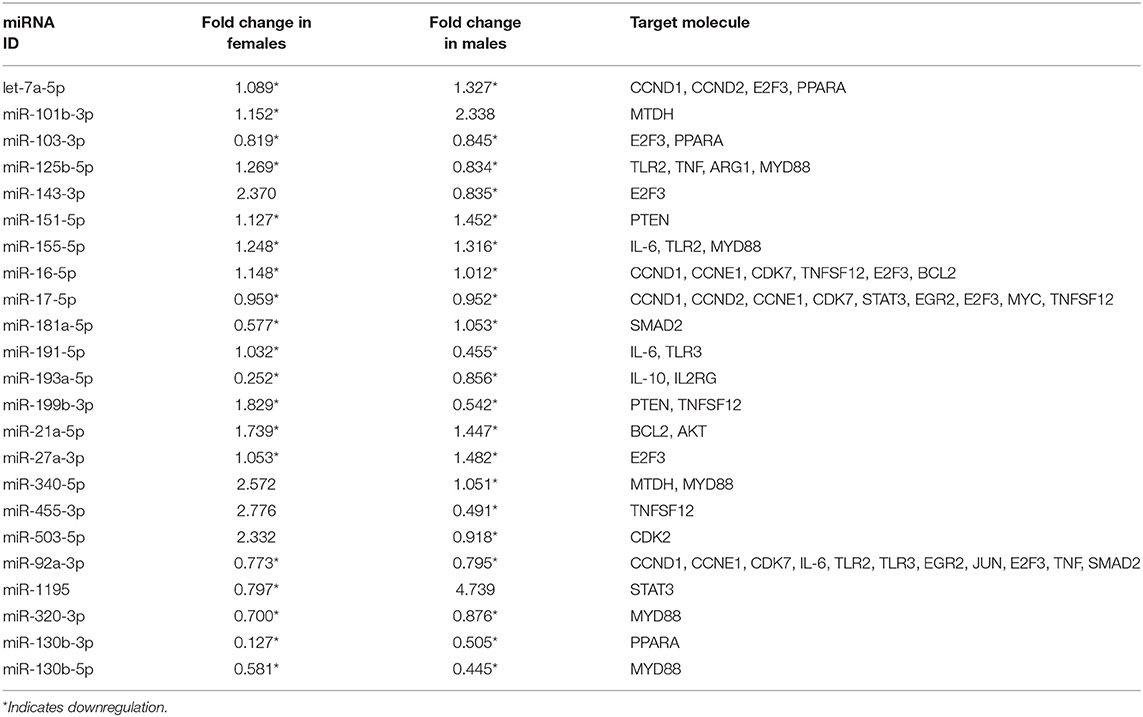
Table 1. Expression levels of co-ex AM miRNAs (males and females) in response to OxS and its mRNA targets identified by IPA analysis.
In general, we observed largely a downregulation of miRNAs in both males and females and upregulation of their targets in co-ex males but not in females (Figures 4A,B). For example, a significant downregulation of miR-191-5p, miR-155-5p, and miR-92a-3p expression was observed in response to O3 in both co-ex males and females but their target IL-6 mRNA is upregulated in males but downregulated in females. Though most of the miRNAs are downregulated in both males and females, the expression of miR-340-5p, miR455-3p, miR-143-3p, and miR-503-5p in females, and miR1195 and miR101b-3p in males is upregulated (Figures 4A,B).
Validation of miRNA Target Genes
To measure the expression levels of target genes by differentially expressed miRNA in response to FA or O3, we performed qRT-PCR analysis on AM cell samples isolated from co-ex and KO males and females after FA or O3 exposure (Figure 5).
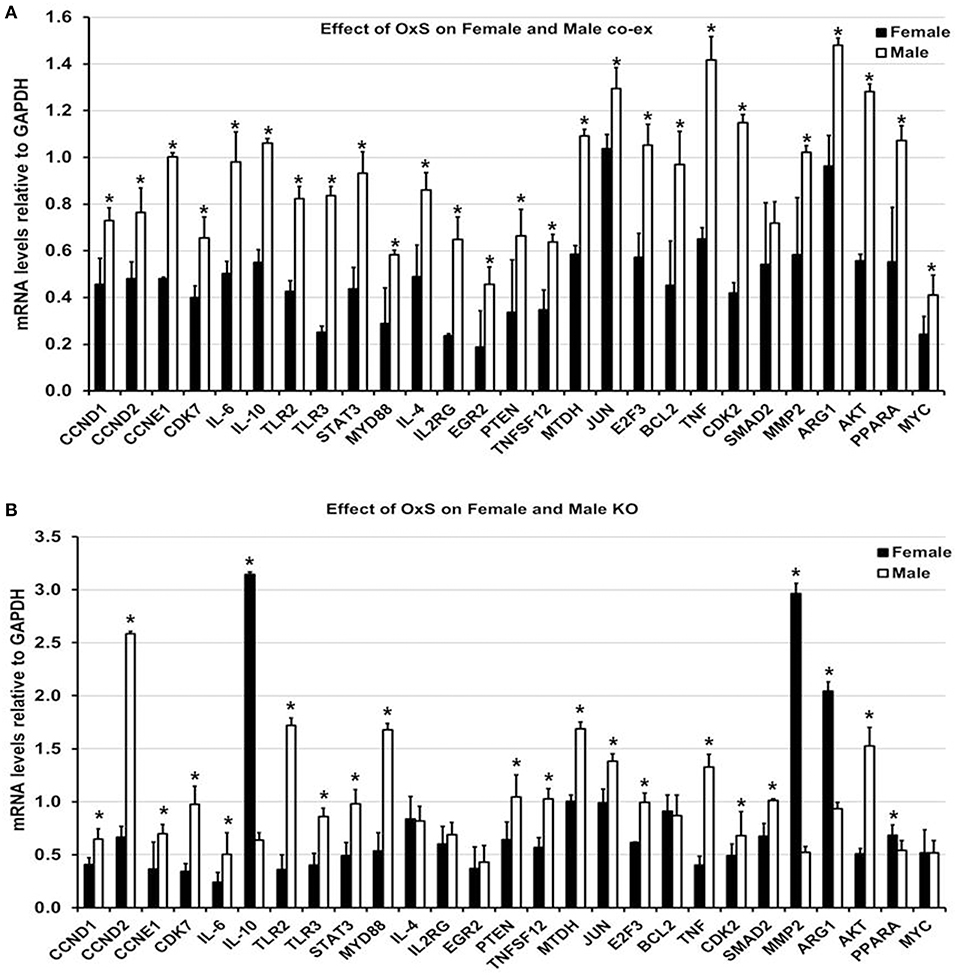
Figure 5. Effect of OxS on mRNA targets of co-ex and KO. (A) The expression of level of CCND1, CCND2, CCNE1, CDK7, IL-6, IL-10, TLR2, TLR3, STAT3, MYD88, IL-4, IL2RG, EGR2, PTEN, TNFSF12, MTDH, JUN, E2F3, BCL2, TNF, CDK2, MMP2, ARG1, AKT1, PPARA, and MYC, were significantly upregulated in co-ex males compared to females. The level of SMAD2 did not change between males and females. The expression levels were normalized to GAPDH and significant differences (P < 0.05) between sexes in co-ex are noted by an asterisk (*). (B) Similar analysis of the above genes was performed in KO males and females. Significant upregulation was observed for CCND1, CCND2, CCNE1, CDK7, IL-6, TLR2, TLR3, STAT3, MYD88, PTEN, TNFSF12, MTDH, JUN, E2F3, TNF, CDK2, SMAD2, and AKT1 in KO males, whereas the expression levels of IL-10, MMP2, ARG1, and PPARA were significantly upregulated in KO females. The expression levels were normalized to GAPDH and the significant differences (P < 0.05) between sexes in KO are noted by an asterisk (*).
In response to OxS, the expression level of all target genes studied, with the exception of SMAD2 that remained similar in both sexes under OxS, was significantly upregulated in co-ex males compared to females (Figure 5A, Supplementary Table 2). These included CCND1, CCND2, CCNE1, CDK7, IL-6, IL-10, TLR2, TLR3, STAT3, MYD88, IL-4, IL2RG, EGR2, PTEN, TNFSF12, MTDH, JUN, E2F3, BCL2, TNF, CDK2, MMP2, ARG1, AKT1, PPARA, and MYC. A similar analysis in KO males and females resulted in significant upregulation of CCND1, CCND2, CCNE1, CDK7, IL-6, TLR2, TLR3, STAT3, MYD88, PTEN, TNFSF12, MTDH, JUN, E2F3, TNF, CDK2, SMAD2, and AKT1 in KO males (Figure 5B, Supplementary Table 2), whereas the expression level of IL-10, MMP2, ARG1, and PPARA was significantly upregulated in KO females (Figure 5B). The levels of IL-4, IL2RG, EGR2, BCL2, and MYC remained similar in both sexes under OxS (Figure 5B).
In Summary the collective information of the significantly changed miRNAs and their targets indicate sex specific differences. Females (unlike males) largely failed to upregulate the target genes as expected when regulatory miRNAs are downregulated.
Discussion
Surfactant protein-A (SP-A), is a key molecule in the lung innate immunity and surfactant related functions. The human SP-A locus consists of two functional genes, SFTPA1 and SFTPA2 (11, 12) and encodes two functional proteins, SP-A1 and SP-A2, respectively, and each is identified with several genetics variants (13, 14). Recently, it has been shown that SP-A1 and SP-A2 variants differ in their ability to regulate the AM miRNome in response to ozone (O3)-induced oxidative stress (OxS) (41) as well as in lung function mechanics and survival in response to bacterial infection (42, 43). Because humans express both SP-A gene products, we wished to investigate the combined effect of co-expressed SP-A1/SP-A2 (co-ex) in response to O3 induced OxS on AM miRNome. Toward this, human transgenic (hTG) mice, carrying both SP-A1/SP-A2 (6A2/1A0, co-ex) and SP-A-KO were exposed to filtered air (FA) and O3 and miRNA levels were measured after AM isolation with or without normalization to KO. The observation made include, (i) Significant differences in AM miRNome of co-ex in terms of sex, exposure, with or without normalization to KO, and after Bonferroni multiple comparison analysis; (ii) After normalization with KO, both males and females showed significant differences in response to OxS; (iii) The AM miRNome of females was largely down regulated significantly in response to OxS compared to control (FA) in all comparisons made including the multiple comparison analysis; (iv) The miRNA targets were largely downregulated in females and upregulated in males; (v) Several of the mRNA targets identified of the significantly altered miRNAs in females were involved in pro-inflammatory response, anti-apoptosis, cell cycle, cellular growth and proliferation; (vi) The AM of the SP-A2 male (41) shares similarities with the co-ex, as well as differences.
We studied the AM miRNome in co-ex male and female mice that express human SP-A1/SP-A2 (6A2/1A0, co-ex) after O3 exposure and compared it to that of KO mice lacking SP-A. In response to OxS, AM miRNome changes were observed in both males and females (with or without normalization to KO), although after multiple comparison analysis, AM miRNAs significantly changed only in females. Furthermore, IPA of the differentially expressed co-ex AM miRNome data identified several miRNA targets involved in the pro-inflammatory response, anti-apoptosis, cell cycle, and cellular growth and proliferation, as shown in Figure 4 and discussed below.
Pro-inflammatory Responses
The expression of miR-191-5p, miR-155-5p and miR-92a-3p were decreased significantly in response to O3 in both co-ex males and females (Figures 4A,B). These miRNAs target pro- and anti-inflammatory IL-6 cytokine (41, 65–67). The IL-6 level decreased significantly in co-ex females, whereas this increased significantly in co-ex males (Figures 4A,B) compared to the control mRNA (GAPDH). The latter is consistent with our previous observation with SP-A2 males in response to OxS (41).
IL-6 plays a crucial role in the activation of STAT3 (68–70). In response to OxS, STAT3 gets phosphorylated which results in the activation of genes involved in inflammation and injury (71). miR-17-5p, and miR-1195 that were significantly altered in response to OxS, are known to interact with STAT3 (Figures 4A,B). In co-ex females and males miR-17-5p is downregulated whereas miR-1195 is downregulated in co-ex females but upregulated in co-ex males. Although, the downregulation of these miRNAs in females should have resulted in an increase in the expression of STAT3, the opposite was observed, indicating that either the regulation of STAT3 by these (and perhaps other) miRNAs is dysfunctional or mechanisms other than miRNAs are involved. The downregulation of STAT3 in females was associated with decreased levels of TNF, TNFSF12, IL-6, IL-10, and IL-4 (Figure 4B). In contrast, in male co-ex, although miR-17-5p is decreased, and miR-1195 is increased, STAT3, which is target for both, is increased, as well as the levels of the pro-inflammatory cytokines were increased as one may expect. Also in males the levels of a number of target genes (ARG1, EGR2, and IL2RG) that contribute, via STAT3, to the upregulation of pro-inflammatory cytokines were upregulated. Although, the details of the underlying mechanisms are unknown currently, these data show a disconnect between miRNA expression and target gene expression in females. The emerging picture is that pro-inflammatory cytokines are downregulated in females and upregulated in males (Figure 4A).
Toll like receptors (TLRs) are a family of membrane bound proteins that recognize pathogen-associated molecular patterns and mediate innate immune response (72). SP-A differentially regulates TLR expression (73). We found four co-ex regulated miRNAs (miR-92a-3p, miR-125b-5p, miR-155-5p, and miR-191-5p) that target TLR2, and TLR3. The expression of these miRNAs is significantly downregulated in our study in both males and females, and this is associated with increased mRNA levels of TLR2 and TLR3 in co-ex males but a decrease in co-ex females. Both miR-125b-5p and miR-155-5p are shown in several studies to regulate TLRs (74–77). TLR2 engages the ubiquitous intracellular adaptor MyD88 (myeloid differentiation primary response 88) and TLR3 engages TRIF (TIR-domain containing adaptor protein). In the current study, the level of miR-125b-5p, miR155-5p and miR130b-5 targeting MyD88 are downregulated and the mRNA level of MyD88 is upregulated in co-ex males but decreased in co-ex females. The involvement of TLR2, TLR3, and activation of MyD88 in co-ex males may result in the recruitment of other genes involved in the activation of NF-kB (78, 79), and this may result in the transcription of pro-inflammatory genes, such as TNF, TNFSF12, IL-10, and IL-4, as observed in the present study (Figures 4A,B). Previous studies have provided evidence that SP-A activates NF-kB (80) either through accumulation of inhibitory IkBa (81, 82) or via direct interaction with TLR2 and TLR4 (73, 80, 83–85) or SIRPα and CD91 (86). It has also been shown that, SP-A is unable to activate NF-kB in response to O3 as assessed by the lack of changes in the nuclear p65 subunit and the cytoplasmic IkBa levels as it would have been expected in the classical NF-kB pathway (87). Moreover, it has been shown that decreased levels of MTDH expression attenuate NF-kB signaling (88) and that TNFSF12 regulates NF-kB activity (89). Upregulation of TNFSF12 and MTDH as it occurs in co-ex males in the current study may alter NF-kB signaling, enhance its translocation to the nucleus to facilitate the transcription of pro-inflammatory genes in co-ex males but not in females. The present data support the idea that NF-kB and STAT3-mediated pathways play a role in the pro-inflammatory gene expression in co-ex males, and that these pathways in females are compromised in response to OxS.
Anti-apoptosis, Cell Cycle, Growth, and Proliferation
The expression of two miRNAs miR-16-5p and miR-21a-5p (TargetScan) that bind BCL2 (90–93) was significantly downregulated in both males and females in response to OxS and the BCL2 mRNA levels were increased in co-ex males but decreased in females. An increase in BCL2 is likely to result in the inhibition of apoptosis and cell proliferation in males, but not in co-ex females (Figures 4A,B). These findings together indicate that OxS differentially affects anti-apoptotic pathways in co-ex males and females, and that females seem to have a disconnect between miRNA expression and target gene expression.
A number of miRNAs whose expression was for the most part downregulated significantly in response to OxS were predicted to target CCND1, CCND2, CCNE1, CDK2, CDK7, and E2F3 (Figures 4A,B). miR-16-5p and miR-17-5p, predicted to bind CCND1, CCND2, CCNE1, and E2F3 mRNAs (TargetScan), have been shown in several studies that these genes are regulated by these two miRNAs (94–97). The expression of all the target genes followed a similar pattern as described above, there was an increase in males and decrease in females. These genes are involved in cell cycle, and growth and proliferation, indicating that ozone differentially affects expression of molecules involved in cell cycle and proliferation pathways in co-ex male and female mice. Of interest significant differences in survival have been observed with females being more affected than males in several lung diseases (53–56). In our animal studies, we observed a significant difference in survival after infection and O3 exposure. Females were more susceptible to oxidative stress than males and exhibited lower survival (7, 9). Sex hormones were shown to play a role in the observed survival differences (57).
AKT, PPARA, PTEN, and JUN involved in the MAPK pathway were significantly upregulated in co-ex males but not in females in response to OxS. The miRNAs that targeted these genes, miR-21-5p, miR-103-3p, let-7a-5p, miR-130b-3p, miR-151-5p, and miR-92-3p, were significantly downregulated in co-ex males and females (Figures 4A,B). The upregulation of MAPK pathway genes has the potential to regulate the genes involved in cell cycle, growth and proliferation (as shown in Figure 4), as well as pro-inflammatory and anti-apoptotic genes (not shown) in co-ex males but not in co-ex females in response to OxS.
We have previously studied the effect of a single, SP-A1 or SP-A2 gene, on the AM miRNome after OxS, and found that SP-A2 (but not SP-A1) had a significant impact on AM in males (41). To gain further insight into the contributions of SP-A2 vs. the co-ex on AM miRNome, we compared the data from two studies. This comparison is shown diagrammatical in Figure 6. A number of observations are readily evident. Broadly these show (1) the miRNAs that changed (≥2 fold) significantly in response to OxS were decreased in male SP-A2 and co-ex male and female after normalization to KO. (2) In males (SP-A2 & co-ex) the expression of all validated target genes (except SMAD2), of the significant miRNAs, identified by IPA, is increased but decreased in co-ex females. (3) The common pathways include pro-inflammatory response, and anti-apoptosis. However the SP-A2 males include ROS-homeostasis processes and the co-ex include cell cycle, growth, and proliferation processes. These data show that SP-A2 alone or in combination with SP-A1 may regulate the expression of pro-inflammatory genes via the STAT3-NF-kB pathway and anti-apoptotic genes in response to OxS (Figure 4A). Furthermore, this indicated that these pathways may in part (if not in their entirety) be regulated or driven by SP-A2 and that the presence of SP-A1 does not negatively affect this. However, although SP-A1 by itself did not have any significant effect on AM, in presence of SP-A2 is shown to regulate genes involved in cell cycle, growth and proliferation, whereas SP-A2 alone regulates genes involved in homeostasis of ROS (41).
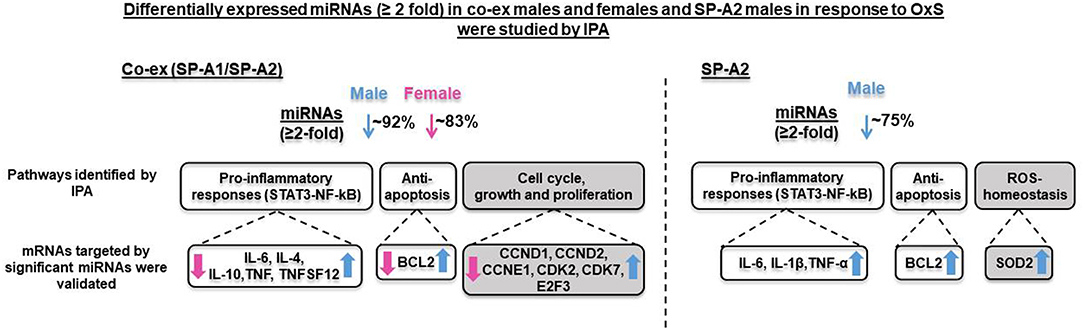
Figure 6. Overview of the effect of both gene (SP-A1/SP-A2) product and of single gene product (SP-A2) on the regulation AM miRNome on both males and females, and males respectively. The diagram depicts comparison of miRNAs, target genes, and pathways after OxS in AM from mice carrying both (co-ex) SP-A1/SP-A2 variants (male and female, left, current study) and from mice carrying a single SP-A2 (male) gene variant (right) (41). The SP-A2 work has been published (41) and is used here for comparison reasons only. The significantly regulated miRNAs (23 and 11) in co-ex male and female and in SP-A2 male, respectively, in response to OxS were largely decreased (~92% in co-ex male, ~83% in co-ex female, and ~75% in SP-A2 male) with the exception of few miRNAs that were increased (see Figure 4). All the mRNA targets were upregulated in co-ex male and in SP-A2 and are shown in blue arrow (on the right). The mRNA targets in co-ex females were downregulated and are shown in pink arrow (on the left). The pathways that differ in SP-A2 and co-ex in response to OxS are shown in gray background and those that are similar between co-ex males and SP-A2 male are shown in white background.
Although the data of this study are largely in line with our previous observations, the current study has a few limitations: (a) the study is carried out at single time point, (b) validation analysis was performed only for genes which are targeted by significantly changed miRNAs in co-ex males and females after normalizing to KO, and we did not look at the protein levels of the targeted mRNAs, (c) we did not study the molecular mechanisms of the identified pathways, (d) we did not differentiate the impact of varying amount SP-A1 and SP-A2 on miRNA expression in response to OxS. It has been shown that the ratio of SP-A1 to SP-A differs in lung diseases (23, 24), and this may have functional consequences given the varying activities of SP-A1 and SP-A2, (e) we did not study the impact of gonadectomy on the expression of AM miRNAs from co-ex and KO males and females. However, we have previously studied the effect of a single, SP-A1, or SP-A2 gene, on the AM miRNome after OxS, and found that SP-A2 (but not SP-A1) had a significant impact on AM in males (41). In this study, we observed that the regulation of the miRNome of the SP-A2 male mice compared to that of female mice in response to OxS is significantly altered after gonadectomy (41). It has also been shown that different stages of the estrous cycle have significant impact on the lung miRNA expression (98). In addition, a role of sex hormones on survival after Klebsiella pneumoniae infected wildtype (SP-A) mice with or without exposure to ozone has been observed (57). This study indicated that (1) after removal of gonadal hormones, differences in survival in animals after infection, and oxidative stress are minimized in males, and eliminated in females. (2) Treatment of gonadectomized females with DHT and males with E2 resulted in a similar kind of survival compared to the intact male and female animals, respectively. This further supports a role of DHT and E2 in survival after infection and oxidative stress. Based on these observations, we speculate that in co-ex males and females sex hormones play a significant role in the regulation of AM miRNome. However, the result of this study advances our knowledge of the differential impact of co-expressed SP-A1/SP-A2 and sex on the AM miRNome.
We postulate that in males in response to OxS, SP-A2 via its activity in ROS-homeostasis provides some protection from the injurious ROS in its microenvironment. Whereas, the co-ex males via cell cycle, growth, and proliferation process may promote cellular recovery, perhaps a more sustained recovery. We further postulate that in co-ex females the disconnect between the downregulation of miRNAs and the expression of their target genes is responsible or contributes to the reduced ability in females to enhance phagocytosis by AM as well as to the poorer survival we observed in females after OxS and infection (9). Although the details of the underlying mechanisms are unknown, the AM miRNome appears to play a significant role in OxS.
In summary, SP-A1/SP-A2 (6A2/1A0, co-ex) regulate miRNAs that play a role in pathways involved in inflammatory responses, anti-apoptosis, cell cycle, growth, and proliferation. Both gene products are needed to alleviate the deleterious effects of OxS in males and promote cellular recovery. However, in females even in the presence of both SP-A1 and SP-A2 genes, expression of target genes to mitigate the OxS injury is lacking, indicating that other hormone dependent mechanisms are involved. Because the innate immune molecules, SP-A1 and SP-A2 appear to play a differential role in the outcome of males and females after OxS, the potential impact on health of innate immune genetics should be considered separately in males and females.
Data Availability
The datasets generated for this study are included in the manuscript and the Supplementary Files, and has been deposited in the Gene Expression Omnibus repository GSE135233 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE135233).
Ethics Statement
All protocols used in this study was evaluated and approved by the Pennsylvania State University College of Medicine Institutional Animal Care and Use Committee and Confirmed to the guidelines of the National Institute of Health on the care and use of laboratory animals.
Author Contributions
NT performed experiments, run statistics, analyzed and synthesized the data, contributed to the manuscript writing. YK performed RNA sequencing analysis. CG contributed to the manuscript writing. XZ performed mouse line maintenance, breeding. JF designed the study and provided oversight to the entire project, involved in data analysis, integration, and writing of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by CHILD fund, Department of Pediatrics, College of Medicine at Pennsylvania State University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.01960/full#supplementary-material
Supplementary Table 1. Identification of miRNAs from SP-A1/SP-A2 (co-ex), KO males and females in response to Filtered air or Ozone.
Supplementary Table 2. Expression levels of target genes by differentially expressed miRNAs in response to FA or Ozone by qRT-PCR.
Abbreviations
AKT1, AKT Serine/Threonine Kinase 1; ARG1, Arginase 1; AM, Alveolar macrophages; ANOVA, analysis of variance; BAL, bronchoalveolar lavage; BCL2, B-cell lymphoma 2; CCND1, Cyclin D1; CCND2, Cyclin D2; CCNE1, Cyclin E1; CDK2, Cyclin-dependent kinase 2; CDK7, Cyclin-dependent kinase 7; E2F3, E2F transcription factor 3; EGR2, Early growth response 2; FA, Filtered air; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; hTG, Humanized transgenic; IkBa, NFKB Inhibitor Alpha; IL4, Interleukin 4; IL6, Interleukin 6; IL10, Interleukin 10; IL2RG, Interleukin 2 receptor subunit gamma; IPA, Ingenuity Pathway Analysis; JUN, Jun proto-oncogene; KO, knock-out; LPS, lipopolysaccharide; MAPK, Mitogen-activated protein kinases; MDTH, Metadherin; miRNAs, microRNAs; MMP2, Matrix metallopeptidase 2; MYC, MYC proto-oncogene; MYD88, Myeloid differentiation primary response 88; NF-kB, Nuclear factor kappa-light-chain-enhancer of activated B cells; O3, ozone; OxS, oxidative stress; PPARA, Peroxisome proliferator activated receptor alpha; PTEN, Phosphatase and tensin homolog; ROS, reactive oxygen species; SFTPA1, gene encoding SP-A1; SFTPA2, gene encoding SP-A2; SMAD2, SMAD family member 2; SP-A, surfactant protein A; STAT3, Signal transducer and activator of transcription 2; TLR2, Toll-like receptor 2; TLR3, Toll-like receptor 3; TNF, Tumor necrosis factor; TNFSF12, TNF super family member 12; TRIF, TIR-domain containing adaptor protein.
References
1. Johansson J, Curstedt T. Molecular structures and interactions of pulmonary surfactant components. Eur J Biochem. (1997) 244:675–93. doi: 10.1111/j.1432-1033.1997.00675.x
2. Batenburg JJ, Haagsman HP. The lipids of pulmonary surfactant: dynamics and interactions with proteins. Prog Lipid Res. (1998) 37:235–76. doi: 10.1016/S0163-7827(98)00011-3
3. Phelps DS. Surfactant regulation of host defense function in the lung: a question of balance. Pediatr Pathol Mol Med. (2001) 20:269–92. doi: 10.1080/15513810109168822
4. Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol. (2005) 5:58–68. doi: 10.1038/nri1528
5. Bates SR, Dodia C, Tao JQ, Fisher AB. Surfactant protein-A plays an important role in lung surfactant clearance: evidence using the surfactant protein-A gene-targeted mouse. Am J Physiol Lung Cell Mol Physiol. (2008) 294:L325–33. doi: 10.1152/ajplung.00341.2007
6. Mason RJ, Greene K, Voelker DR. Surfactant protein A and surfactant protein D in health and disease. Am J Physiol. (1998) 275(1 Pt 1):L1–13. doi: 10.1152/ajplung.1998.275.1.L1
7. Mikerov AN, Gan X, Umstead TM, Miller L, Chinchilli VM, Phelps DS, et al. Sex differences in the impact of ozone on survival and alveolar macrophage function of mice after Klebsiella pneumoniae infection. Respir Res. (2008) 9:24. doi: 10.1186/1465-9921-9-24
8. Mikerov AN, Haque R, Gan X, Guo X, Phelps DS, Floros J. Ablation of SP-A has a negative impact on the susceptibility of mice to Klebsiella pneumoniae infection after ozone exposure: sex differences. Respir Res. (2008) 9:77. doi: 10.1186/1465-9921-9-77
9. Mikerov AN, Hu S, Durrani F, Gan X, Wang G, Umstead TM, et al. Impact of sex and ozone exposure on the course of pneumonia in wild type and SP-A (-/-) mice. Microb Pathog. (2012) 52:239–49. doi: 10.1016/j.micpath.2012.01.005
10. Mikerov AN, Phelps DS, Gan X, Umstead TM, Haque R, Wang G, et al. Effect of ozone exposure and infection on bronchoalveolar lavage: sex differences in response patterns. Toxicol Lett. (2014) 230:333–44. doi: 10.1016/j.toxlet.2014.04.008
11. Floros J, Hoover RR. Genetics of the hydrophilic surfactant proteins A and D. Biochim Biophys Acta. (1998) 1408:312–22. doi: 10.1016/S0925-4439(98)00077-5
12. Hoover RR, Floros J. Organization of the human SP-A and SP-D loci at 10q22-q23. Physical and radiation hybrid mapping reveal gene order and orientation. Am J Respir Cell Mol Biol. (1998) 18:353–62. doi: 10.1165/ajrcmb.18.3.3035
13. Karinch AM, Floros J. 5' splicing and allelic variants of the human pulmonary surfactant protein A genes. Am J Respir Cell Mol Biol. (1995) 12:77–88. doi: 10.1165/ajrcmb.12.1.7811473
14. DiAngelo S, Lin Z, Wang G, Phillips S, Ramet M, Luo J, et al. Novel, non-radioactive, simple and multiplex PCR-cRFLP methods for genotyping human SP-A and SP-D marker alleles. Dis Markers. (1999) 15:269–81. doi: 10.1155/1999/961430
15. Phelps DS, Floros J. Localization of surfactant protein synthesis in human lung by in situ hybridization. Am Rev Respir Dis. (1988) 137:939–42. doi: 10.1164/ajrccm/137.4.939
16. Goss KL, Kumar AR, Snyder JM. SP-A2 gene expression in human fetal lung airways. Am J Respir Cell Mol Biol. (1998) 19:613–21. doi: 10.1165/ajrcmb.19.4.3155
17. Saitoh H, Okayama H, Shimura S, Fushimi T, Masuda T, Shirato K. Surfactant protein A2 gene expression by human airway submucosal gland cells. Am J Respir Cell Mol Biol. (1998) 19:202–9. doi: 10.1165/ajrcmb.19.2.3239
18. Lin Z, deMello D, Phelps DS, Koltun WA, Page M, Floros J. Both human SP-A1 and Sp-A2 genes are expressed in small and large intestine. Pediatr Pathol Mol Med. (2001) 20:367–86. doi: 10.1080/15513810109168621
19. MacNeill C, Umstead TM, Phelps DS, Lin Z, Floros J, Shearer DA, et al. Surfactant protein A, an innate immune factor, is expressed in the vaginal mucosa and is present in vaginal lavage fluid. Immunology. (2004) 111:91–9. doi: 10.1111/j.1365-2567.2004.01782.x
20. Voss T, Eistetter H, Schafer KP, Engel J. Macromolecular organization of natural and recombinant lung surfactant protein SP 28-36. Structural homology with the complement factor C1q. J Mol Biol. (1988) 201:219–27. doi: 10.1016/0022-2836(88)90448-2
21. Voss T, Melchers K, Scheirle G, Schafer KP. Structural comparison of recombinant pulmonary surfactant protein SP-A derived from two human coding sequences: implications for the chain composition of natural human SP-A. Am J Respir Cell Mol Biol. (1991) 4:88–94. doi: 10.1165/ajrcmb/4.1.88
22. Karinch AM, deMello DE, Floros J. Effect of genotype on the levels of surfactant protein A mRNA and on the SP-A2 splice variants in adult humans. Biochem J. (1997) 321(Pt 1):39–47. doi: 10.1042/bj3210039
23. Tagaram HR, Wang G, Umstead TM, Mikerov AN, Thomas NJ, Graff GR, et al. Characterization of a human surfactant protein A1 (SP-A1) gene-specific antibody; SP-A1 content variation among individuals of varying age and pulmonary health. Am J Physiol Lung Cell Mol Physiol. (2007) 292:L1052–63. doi: 10.1152/ajplung.00249.2006
24. Wang Y, Voelker DR, Lugogo NL, Wang G, Floros J, Ingram JL, et al. Surfactant protein A is defective in abrogating inflammation in asthma. Am J Physiol Lung Cell Mol Physiol. (2011) 301:L598–606. doi: 10.1152/ajplung.00381.2010
25. Karinch AM, Deiter G, Ballard PL, Floros J. Regulation of expression of human SP-A1 and SP-A2 genes in fetal lung explant culture. Biochim Biophys Acta. (1998) 1398:192–202. doi: 10.1016/S0167-4781(98)00047-5
26. Kumar AR, Snyder JM. Differential regulation of SP-A1 and SP-A2 genes by cAMP, glucocorticoids, and insulin. Am J Physiol. (1998) 274(2 Pt 1):L177–185. doi: 10.1152/ajplung.1998.274.2.L177
27. Scavo LM, Ertsey R, Gao BQ. Human surfactant proteins A1 and A2 are differentially regulated during development and by soluble factors. Am J Physiol. (1998) 275(4 Pt 1):L653–69. doi: 10.1152/ajplung.1998.275.4.L653
28. McCormick SM, Mendelson CR. Human SP-A1 and SP-A2 genes are differentially regulated during development and by cAMP and glucocorticoids. Am J Physiol. (1994) 266(4 Pt 1):L367–74. doi: 10.1152/ajplung.1994.266.4.L367
29. Wang G, Phelps DS, Umstead TM, Floros J. Human SP-A protein variants derived from one or both genes stimulate TNF-alpha production in the THP-1 cell line. Am J Physiol Lung Cell Mol Physiol. (2000) 278:L946–54. doi: 10.1152/ajplung.2000.278.5.L946
30. Garcia-Verdugo I, Wang G, Floros J, Casals C. Structural analysis and lipid-binding properties of recombinant human surfactant protein a derived from one or both genes. Biochemistry. (2002) 41:14041–53. doi: 10.1021/bi026540l
31. Wang G, Umstead TM, Phelps DS, Al-Mondhiry H, Floros J. The effect of ozone exposure on the ability of human surfactant protein a variants to stimulate cytokine production. Environ Health Perspect. (2002) 110:79–84. doi: 10.1289/ehp.0211079
32. Oberley RE, Snyder JM. Recombinant human SP-A1 and SP-A2 proteins have different carbohydrate-binding characteristics. Am J Physiol Lung Cell Mol Physiol. (2003) 284:L871–881. doi: 10.1152/ajplung.00241.2002
33. Selman M, Lin HM, Montano M, Jenkins AL, Estrada A, Lin Z, et al. Surfactant protein A and B genetic variants predispose to idiopathic pulmonary fibrosis. Hum Genet. (2003) 113:542–50. doi: 10.1007/s00439-003-1015-4
34. Huang W, Wang G, Phelps DS, Al-Mondhiry H, Floros J. Human SP-A genetic variants and bleomycin-induced cytokine production by THP-1 cells: effect of ozone-induced SP-A oxidation. Am J Physiol Lung Cell Mol Physiol. (2004) 286:L546–53. doi: 10.1152/ajplung.00267.2003
35. Wang G, Bates-Kenney SR, Tao JQ, Phelps DS, Floros J. Differences in biochemical properties and in biological function between human SP-A1 and SP-A2 variants, and the impact of ozone-induced oxidation. Biochemistry. (2004) 43:4227–39. doi: 10.1021/bi036023i
36. Mikerov AN, Umstead TM, Huang W, Liu W, Phelps DS, Floros J. SP-A1 and SP-A2 variants differentially enhance association of Pseudomonas aeruginosa with rat alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. (2005) 288:L150–158. doi: 10.1152/ajplung.00135.2004
37. Mikerov AN, Wang G, Umstead TM, Zacharatos M, Thomas NJ, Phelps DS, et al. Surfactant protein A2 (SP-A2) variants expressed in CHO cells stimulate phagocytosis of Pseudomonas aeruginosa more than do SP-A1 variants. Infect Immun. (2007) 75:1403–12. doi: 10.1128/IAI.01341-06
38. Wang G, Myers C, Mikerov A, Floros J. Effect of cysteine 85 on biochemical properties and biological function of human surfactant protein A variants. Biochemistry. (2007) 46:8425–35. doi: 10.1021/bi7004569
39. Wang G, Taneva S, Keough KM, Floros J. Differential effects of human SP-A1 and SP-A2 variants on phospholipid monolayers containing surfactant protein B. Biochim Biophys Acta. (2007) 1768:2060–9. doi: 10.1016/j.bbamem.2007.06.025
40. Mikerov AN, Umstead TM, Gan X, Huang W, Guo X, Wang G, et al. Impact of ozone exposure on the phagocytic activity of human surfactant protein A (SP-A) and SP-A variants. Am J Physiol Lung Cell Mol Physiol. (2008) 294:L121–30. doi: 10.1152/ajplung.00288.2007
41. Noutsios GT, Thorenoor N, Zhang X, Phelps DS, Umstead TM, Durrani F, et al. SP-A2 contributes to miRNA-mediated sex differences in response to oxidative stress: pro-inflammatory, anti-apoptotic, and anti-oxidant pathways are involved. Biol Sex Differ. (2017) 8:37. doi: 10.1186/s13293-017-0158-2
42. Thorenoor N, Umstead TM, Zhang X, Phelps DS, Floros J. Survival of surfactant protein-A1 and SP-A2 transgenic mice after Klebsiella pneumoniae infection, exhibits sex-, gene-, and variant specific differences; treatment with surfactant protein improves survival. Front Immunol. (2018) 9:2404. doi: 10.3389/fimmu.2018.02404
43. Thorenoor N, Zhang X, Umstead TM, Scott Halstead E, Phelps DS, Floros J. Differential effects of innate immune variants of surfactant protein-A1 (SFTPA1) and SP-A2 (SFTPA2) in airway function after Klebsiella pneumoniae infection and sex differences. Respir Res. (2018) 19:23. doi: 10.1186/s12931-018-0723-1
44. Wang G, Guo X, Floros J. Human SP-A 3'-UTR variants mediate differential gene expression in basal levels and in response to dexamethasone. Am J Physiol Lung Cell Mol Physiol. (2003) 284:L738–748. doi: 10.1152/ajplung.00375.2002
45. Silveyra P, Raval M, Simmons B, Diangelo S, Wang G, Floros J. The untranslated exon B of human surfactant protein A2 mRNAs is an enhancer for transcription and translation. Am J Physiol Lung Cell Mol Physiol. (2011) 301:L795–803. doi: 10.1152/ajplung.00439.2010
46. Silveyra P, DiAngelo SL, Floros J. An 11-nt sequence polymorphism at the 3'UTR of human SFTPA1 and SFTPA2 gene variants differentially affect gene expression levels and miRNA regulation in cell culture. Am J Physiol Lung Cell Mol Physiol. (2014) 307:L106–19. doi: 10.1152/ajplung.00313.2013
47. Phelps DS, Umstead TM, Silveyra P, Hu S, Wang G, Floros J. Differences in the alveolar macrophage proteome in transgenic mice expressing human SP-A1 and SP-A2. J Proteom Genom Res. (2013) 1:2–26. doi: 10.14302/issn.2326-0793.jpgr-12-207
48. Phelps DS, Umstead TM, Floros J. Sex differences in the acute in vivo effects of different human SP-A variants on the mouse alveolar macrophage proteome. J Proteomics. (2014) 108:427–44. doi: 10.1016/j.jprot.2014.06.007
49. Tsotakos N, Phelps DS, Yengo CM, Chinchilli VM, Floros J. Single-cell analysis reveals differential regulation of the alveolar macrophage actin cytoskeleton by surfactant proteins A1 and A2: implications of sex and aging. Biol Sex Differ. (2016) 7:18. doi: 10.1186/s13293-016-0071-0
50. Lopez-Rodriguez E, Pascual A, Arroyo R, Floros J, Perez-Gil J. Human pulmonary surfactant protein SP-A1 provides maximal efficiency of lung interfacial films. Biophys J. (2016) 111:524–36. doi: 10.1016/j.bpj.2016.06.025
51. Al-Hegelan M, Tighe RM, Castillo C, Hollingsworth JW. Ambient ozone and pulmonary innate immunity. Immunol Res. (2011) 49:173–91. doi: 10.1007/s12026-010-8180-z
52. Putman E, van Golde LM, Haagsman HP. Toxic oxidant species and their impact on the pulmonary surfactant system. Lung. (1997) 175:75–103. doi: 10.1007/PL00007561
54. Mudway IS, Kelly FJ. An investigation of inhaled ozone dose and the magnitude of airway inflammation in healthy adults. Am J Respir Crit Care Med. (2004) 169:1089–95. doi: 10.1164/rccm.200309-1325PP
55. Wong CM, Thach TQ, Chau PY, Chan EK, Chung RY, Ou CQ, et al. Part 4. Interaction between air pollution and respiratory viruses: time-series study of daily mortality and hospital admissions in Hong Kong. Res Rep Health Eff Inst. (2010) 283–62.
56. Tam A, Churg A, Wright JL, Zhou S, Kirby M, Coxson HO, et al. Sex differences in airway remodeling in a mouse model of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2016) 193:825–34. doi: 10.1164/rccm.201503-0487OC
57. Durrani F, Phelps DS, Weisz J, Silveyra P, Hu S, Mikerov AN, et al. Gonadal hormones and oxidative stress interaction differentially affects survival of male and female mice after lung Klebsiella pneumoniae infection. Exp Lung Res. (2012) 38:165–72. doi: 10.3109/01902148.2011.654045
58. Mikerov AN, Cooper TK, Wang G, Hu S, Umstead TM, Phelps DS, et al. Histopathologic evaluation of lung and extrapulmonary tissues show sex differences in Klebsiella pneumoniae - infected mice under different exposure conditions. Int J Physiol Pathophysiol Pharmacol. (2011) 3:176–90.
59. Wang G, Guo X, Diangelo S, Thomas NJ, Floros J. Humanized SFTPA1 and SFTPA2 transgenic mice reveal functional divergence of SP-A1 and SP-A2: formation of tubular myelin in vivo requires both gene products. J Biol Chem. (2010) 285:11998–2010. doi: 10.1074/jbc.M109.046243
60. Haque R, Umstead TM, Ponnuru P, Guo X, Hawgood S, Phelps DS, et al. Role of surfactant protein-A (SP-A) in lung injury in response to acute ozone exposure of SP-A deficient mice. Toxicol Appl Pharmacol. (2007) 220:72–82. doi: 10.1016/j.taap.2006.12.017
61. Phelps DS, Umstead TM, Floros J. Sex differences in the response of the alveolar macrophage proteome to treatment with exogenous surfactant protein-A. Proteome Sci. (2012) 10:44. doi: 10.1186/1477-5956-10-44
62. Vitsios DM, Enright AJ. Chimira: analysis of small RNA sequencing data and microRNA modifications. Bioinformatics. (2015) 31:3365–7. doi: 10.1093/bioinformatics/btv380
63. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
64. Sun J, Nishiyama T, Shimizu K, Kadota K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. (2013) 14:219. doi: 10.1186/1471-2105-14-219
65. Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta. (2011) 1813:878–88. doi: 10.1016/j.bbamcr.2011.01.034
66. Tufekci KU, Oner MG, Genc S, Genc K. MicroRNAs and multiple sclerosis. Autoimmune Dis. (2010) 2011:807426. doi: 10.4061/2011/807426
67. Migita K, Iwanaga N, Izumi Y, Kawahara C, Kumagai K, Nakamura T, et al. TNF-α-induced miR-155 regulates IL-6 signaling in rheumatoid synovial fibroblasts. BMC Res Notes. (2017) 10:403. doi: 10.1186/s13104-017-2715-5
68. Zhang Z, Jones S, Hagood JS, Fuentes NL, Fuller GM. STAT3 acts as a co-activator of glucocorticoid receptor signaling. J Biol Chem. (1997) 272:30607–10. doi: 10.1074/jbc.272.49.30607
69. Nguyen VA, Gao B. Cross-talk between alpha(1B)-adrenergic receptor (alpha(1B)AR) and interleukin-6 (IL-6) signaling pathways. Activation of alpha(1b)AR inhibits il-6-activated STAT3 in hepatic cells by a p42/44 mitogen-activated protein kinase-dependent mechanism. J Biol Chem. (1999) 274:35492–8. doi: 10.1074/jbc.274.50.35492
70. Miller AM, Wang H, Park O, Horiguchi N, Lafdil F, Mukhopadhyay P, et al. Anti-inflammatory and anti-apoptotic roles of endothelial cell STAT3 in alcoholic liver injury. Alcohol Clin Exp Res. (2010) 34:719–25. doi: 10.1111/j.1530-0277.2009.01141.x
71. Carballo M, Conde M, El Bekay R, Martin-Nieto J, Camacho MJ, Monteseirin J, et al. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J Biol Chem. (1999) 274:17580–6. doi: 10.1074/jbc.274.25.17580
72. Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. (2004) 4:499–511. doi: 10.1038/nri1391
73. Henning LN, Azad AK, Parsa KV, Crowther JE, Tridandapani S, Schlesinger LS. Pulmonary surfactant protein A regulates TLR expression and activity in human macrophages. J Immunol. (2008) 180:7847–58. doi: 10.4049/jimmunol.180.12.7847
74. Ruckerl D, Jenkins SJ, Laqtom NN, Gallagher IJ, Sutherland TE, Duncan S, et al. Induction of IL-4Rα-dependent microRNAs identifies PI3K/Akt signaling as essential for IL-4-driven murine macrophage proliferation in vivo. Blood. (2012) 120:2307–16. doi: 10.1182/blood-2012-02-408252
75. Elton TS, Selemon H, Elton SM, Parinandi NL. Regulation of the MIR155 host gene in physiological and pathological processes. Gene. (2013) 532:1–12. doi: 10.1016/j.gene.2012.12.009
76. Hu X, Ye J, Qin A, Zou H, Shao H, Qian K. Both microRNA-155 and virus-encoded MiR-155 ortholog regulate TLR3 expression. PLoS ONE. (2015) 10:e0126012. doi: 10.1371/journal.pone.0126012
77. Arboleda JF, Fernandez GJ, Urcuqui-Inchima S. Vitamin D-mediated attenuation of miR-155 in human macrophages infected with dengue virus: implications for the cytokine response. Infect Genet Evol. (2019) 69:12–21. doi: 10.1016/j.meegid.2018.12.033
78. Tak PP, Firestein GS. NF-κB: a key role in inflammatory diseases. J Clin Invest. (2001) 107:7–11. doi: 10.1172/JCI11830
79. Lim R, Barker G, Lappas M. TLR2, TLR3 and TLR5 regulation of pro-inflammatory and pro-labour mediators in human primary myometrial cells. J Reprod Immunol. (2017) 122:28–36. doi: 10.1016/j.jri.2017.08.004
80. Koptides M, Umstead TM, Floros J, Phelps DS. Surfactant protein A activates NF-kappa B in the THP-1 monocytic cell line. Am J Physiol. (1997) 273(2 Pt 1):L382–8. doi: 10.1152/ajplung.1997.273.2.L382
81. Wu Y, Adam S, Hamann L, Heine H, Ulmer AJ, Buwitt-Beckmann U, et al. Accumulation of inhibitory κB-α as a mechanism contributing to the anti-inflammatory effects of surfactant protein-A. Am J Respir Cell Mol Biol. (2004) 31:587–94. doi: 10.1165/rcmb.2004-0003OC
82. Moulakakis C, Adam S, Seitzer U, Schromm AB, Leitges M, Stamme C. Surfactant protein A activation of atypical protein kinase C zeta in IκB-α-dependent anti-inflammatory immune regulation. J Immunol. (2007) 179:4480–91. doi: 10.4049/jimmunol.179.7.4480
83. Guillot L, Balloy V, McCormack FX, Golenbock DT, Chignard M, Si-Tahar M. Cutting edge: the immunostimulatory activity of the lung surfactant protein-A involves Toll-like receptor 4. J Immunol. (2002) 168:5989–92. doi: 10.4049/jimmunol.168.12.5989
84. Konishi M, Nishitani C, Mitsuzawa H, Shimizu T, Sano H, Harimaya A, et al. Alloiococcus otitidis is a ligand for collectins and Toll-like receptor 2, and its phagocytosis is enhanced by collectins. Eur J Immunol. (2006) 36:1527–36. doi: 10.1002/eji.200535542
85. Yamada C, Sano H, Shimizu T, Mitsuzawa H, Nishitani C, Himi T, et al. Surfactant protein A directly interacts with TLR4 and MD-2 and regulates inflammatory cellular response. Importance of supratrimeric oligomerization. J Biol Chem. (2006) 281:21771–80. doi: 10.1074/jbc.M513041200
86. Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, et al. By binding SIRPalpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. (2003) 115:13–23. doi: 10.1016/S0092-8674(03)00758-X
87. Janic B, Umstead TM, Phelps DS, Floros J. Modulatory effects of ozone on THP-1 cells in response to SP-A stimulation. Am J Physiol Lung Cell Mol Physiol. (2005) 288:L317–325. doi: 10.1152/ajplung.00125.2004
88. Jiao Y, Yang H, Qian J, Gong Y, Liu H, Wu S, et al. miR36645P suppresses the proliferation and metastasis of gastric cancer by attenuating the NFκB signaling pathway through targeting MTDH. Int J Oncol. (2019) 54:845–58. doi: 10.3892/ijo.2019.4680
89. Zhu C, Zhang L, Liu Z, Li C, Bai Y. TWEAK/Fn14 interaction induces proliferation and migration in human airway smooth muscle cells via activating the NF-κB pathway. J Cell Biochem. (2018) 119:3528–36. doi: 10.1002/jcb.26525
90. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. (2005) 102:13944–9. doi: 10.1073/pnas.0506654102
91. Shen J, Wan R, Hu G, Yang L, Xiong J, Wang F, et al. miR-15b and miR-16 induce the apoptosis of rat activated pancreatic stellate cells by targeting Bcl-2 in vitro. Pancreatology. (2012) 12:91–9. doi: 10.1016/j.pan.2012.02.008
92. Jia X, Ouyang H, Abdalla BA, Xu H, Nie Q, Zhang X. miR-16 controls myoblast proliferation and apoptosis through directly suppressing Bcl2 and FOXO1 activities. Biochim Biophys Acta Gene Regul Mech. (2017) 1860:674–84. doi: 10.1016/j.bbagrm.2017.02.010
93. Yang Y, Hu Z, Du X, Davies H, Huo X, Fang M. miR-16 and fluoxetine both reverse autophagic and apoptotic change in chronic unpredictable mild stress model rats. Front Neurosci. (2017) 11:428. doi: 10.3389/fnins.2017.00428
94. Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. (2008) 182:509–17. doi: 10.1083/jcb.200801079
95. Attar M, Arefian E, Nabiuni M, Adegani FJ, Bakhtiari SH, Karimi Z, et al. MicroRNA 17-92 expressed by a transposone-based vector changes expression level of cell-cycle-related genes. Cell Biol Int. (2012) 36:1005–12. doi: 10.1042/CBI20110089
96. Shu J, Xia Z, Li L, Liang ET, Slipek N, Shen D, et al. Dose-dependent differential mRNA target selection and regulation by let-7a-7f and miR-17-92 cluster microRNAs. RNA Biol. (2012) 9:1275–87. doi: 10.4161/rna.21998
97. Wang F, Mao A, Tang J, Zhang Q, Yan J, Wang Y, et al. microRNA-16-5p enhances radiosensitivity through modulating Cyclin D1/E1-pRb-E2F1 pathway in prostate cancer cells. J Cell Physiol. (2018) 234:13182–90. doi: 10.1002/jcp.27989
Keywords: surfactant protein A, alveolar macrophages, miRNA, surfactant protein A1/A2, sex differences, oxidative stress
Citation: Thorenoor N, Kawasawa YI, Gandhi CK, Zhang X and Floros J (2019) Differential Impact of Co-expressed SP-A1/SP-A2 Protein on AM miRNome; Sex Differences. Front. Immunol. 10:1960. doi: 10.3389/fimmu.2019.01960
Received: 01 May 2019; Accepted: 02 August 2019;
Published: 16 August 2019.
Edited by:
Francesca Granucci, University of Milano-Bicocca, ItalyReviewed by:
Eswari Dodagatta-Marri, University of California, San Francisco, United StatesTaruna Madan, National Institute for Research in Reproductive Health (ICMR), India
Copyright © 2019 Thorenoor, Kawasawa, Gandhi, Zhang and Floros. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanna Floros, anhmMTlAcHN1LmVkdQ==; amZsb3Jvc0BwZW5uc3RhdGVoZWFsdGgucHN1LmVkdQ==
 Nithyananda Thorenoor
Nithyananda Thorenoor Yuka Imamura Kawasawa
Yuka Imamura Kawasawa Chintan K. Gandhi
Chintan K. Gandhi Xuesheng Zhang1
Xuesheng Zhang1