Abstract
There is a developing interest in how immune genes may function in other physiological roles, and how traditionally non-immune peptides may, in fact, be active in immune contexts. In the absence of infection, the induction of the immune response is costly, and there are well-characterized trade-offs between immune defense and fitness. The agents behind these fitness costs are less understood. Here we implicate antimicrobial peptides (AMPs) as particularly costly effectors of immunity using an evolutionary framework. We describe the independent loss of AMPs in multiple lineages of Diptera (true flies), tying these observations back to life history. We then focus on the intriguing case of the glycine-rich AMP, Diptericin, and find several instances of loss, pseudogenization, and segregating null alleles. We suggest that Diptericin may be a particularly toxic component of the Dipteran immune response lost in flies either with reduced pathogen pressure or other environmental factors. As Diptericins have recently been described to have neurological roles, these findings parallel a developing interest in AMPs as potentially harmful neuropeptides, and AMPs in other roles beyond immunity.
Introduction
The innate immune system plays a vital role in host defense against pathogens. This is particularly true in invertebrates, which lack an adaptive immune system. Antimicrobial peptides (AMPs) are one of the main effector molecules of innate immunity in many organisms and, as such, they represent the front lines in the coevolutionary struggle between host and pathogen. AMPs are often cationic, amphipathic peptides that defend their hosts against infection by disrupting the cell membranes of invading microbes (1). However, the dose makes the poison, and AMPs can also be toxic to eukaryotic host cells under certain conditions. This suggests that host immunity needs to strike a delicate balance: AMPs need to be potent enough to quickly inhibit pathogenic microbes, but not so potent that they upset the balance of the microbiota or damage host tissue.
Indeed, many pathologies in humans have been observed when this balance is perturbed. These include chronic inflammatory skin or bowel diseases (2–4), and pulmonary infections including cystic fibrosis wherein reduced levels of β-Defensins and the cathelicidin LL-37 are associated with increased risk of infection (5–9). The cathelicidin LL-37 is implicated in autoimmune reactions because it can be toxic to white blood cells (10), induce inflammation in the nervous system (11), or even damage host tissues during anti-cancer responses (12). Recent studies have also suggested the Alzheimer's peptide Amyloid-beta is an AMP in the nervous system, and that Alzheimer's may in part be an infectious disease (13–15). These observations of AMPs as toxic agents are further supported by reduced lifespan in Drosophila fruit flies ubiquitously expressing AMPs in the brain (16), or systemically (17). During aging, Drosophila NF-κB signaling is also implicated in neurodegeneration with AMPs as prime suspects (18). Thus, AMP dysregulation can impose a significant threat to organismal health.
Insects, and particularly Drosophila melanogaster, have been integral to unraveling the innate immune response, including the regulation of AMPs by the Toll and Imd NF-κB signaling pathways (19). Thus, far seven AMP gene families have been described in Drosophila: Defensin, Cecropin, Attacin, Diptericin, Drosocin, Metchnikowin, and Drosomycin. Another class of AMP-like effectors called the Bomanins are also essential for Toll-mediated defense, however their antimicrobial properties await functional clarification (20, 21). Drosophila AMP evolution is shaped both by balancing and diversifying selection at the sequence level (22, 23). Following a duplication event and subsequent speciation, Drosophila Diptericins rapidly diverged into distinct Diptericin clades (24). In contrast, balancing selection seems to maintain a stable polymorphism amongst alleles that provide either moderate or poor protection against systemic infection with Providencia rettgeri (P. rettgeri) (25). Why selection should favor Diptericin alleles that result in loss of immune competence is unclear. One possibility is that the immune-poor Diptericin allele is selected for through trade-offs between poor immune defense when infected and higher fitness when uninfected. Indeed, rare Diptericin null alleles are observed in North American populations (25), and patterns of duplication and loss in Diptericin and other Drosophila AMPs have resulted in copy number variations amongst species (24, 26–28).
As AMP dysregulation can affect health, copy number variation may impose a significant challenge for the maintenance of optimal gene expression (29). Yet perhaps the most overt patterns in AMP evolution are duplication events affecting copy number, which is widespread in both humans and fruit flies (30–34). Therefore, conflict between maintenance of healthy expression levels and improved immune competence may drive patterns of AMP gain/loss or changes in expression patterns. In this model, we expect that AMPs are evolutionary liabilities in the absence of infection, and that host ecology and associated pathogen pressure will drive the evolution of AMP content both at the level of broad AMP gain/loss, and also of AMP expression: species with strong pathogen pressures would evolve to increase potential AMP production, while species whose ecologies involve less exposure to pathogens would be expected to reduce their AMP complement.
While characterizing pathogen pressure in different animal hosts is exceedingly difficult, we can use host ecology as a proxy for infectious pressures. The use of sterile food resources (such as plant sap) reduces the opportunities for microbes to inoculate hosts. There are several insects that spend large portions of their lives (larval, adult, or both) feeding on sterile or near sterile food resources—likely reducing the evolutionary benefits of AMPs and/or AMP induction. The pea aphid (which feeds on sterile plant phloem) is one such insect that has lost not just effectors, but also an entire NF-κB immune signaling pathway (35). Loss of immune signaling is also observed in plant-feeding Tetranychus mites (36, 37), as well as bed bugs (38) and body lice (39), suggesting blood-feeding may be a similarly clean feeding ecology. It should be noted that in some cases these hosts have intimate associations with endosymbionts, microbes that supplement nutrition or protect against infection. One argument to explain loss of immune signaling is that it is a direct consequence of endosymbiont presence to avoid negative consequences associated with chronic activation of the host immune response (40). However, cereal weevils live in sterile environments and have nutritional endosymbionts, but they instead utilize AMPs to regulate their symbiont populations (41). Thus, what factors of sterile lifestyles and/or endosymbionts promote immune gene loss remains poorly resolved.
As AMP copy number evolves rapidly, we suspected AMP evolution might respond to shifts in host ecology before entire immune pathways are affected. To test this, we surveyed Diptera (true flies) for AMP presence or absence and interpret this in the context of host ecology. Diptera are an extremely diverse lineage with equally diverse and unique ecologies and life histories, and boast numerous sequenced genomes and transcriptomes. We probed these diverse flies for classic AMP families described in Drosophila and other insects to better understand the forces driving AMP gain or loss. We further analyzed Drosophila copy number and sequence variation in conserved AMPs, tying these results back to life history. Globally, we describe a pattern suggesting AMPs are lost in Diptera lineages with more sterile life histories, with striking parallels to loss of immune signaling in other arthropods with sterile food resources. We also focus on Diptericin, which we suggest is a particularly costly AMP, describing distinct evolutionary patterns across ecologically diverse Drosophila and within D. melanogaster.
Results
Some AMP Families Are Absent in Diptera Living in More Sterile Environments
Diptera diverged from other related holometabolous insects about 272 mya [timetree.org; Kumar et al. (42)] and diversified into extremely broad ecological habitats. We surveyed 31 Dipteran genomes as well as diverse Drosophila species for the presence of eight AMP/AMP-like families either described in Drosophila (Bomanins, Drosocin, Drosomycin, Metchnikowin) or characterized more broadly across Dipterans and other insects (Diptericins, Cecropins, Attacins, Defensins). We also annotated the feeding ecologies of these diverse flies to better understand which lineages may have reduced pathogen pressure owing to food resource use (Supplementary Data File 1). We performed an iterative reciprocal BLAST search using known AMPs against genomic or transcriptomic sequence. We found that Drosocin, Metchnikowin, Bomanins, and Drosomycin are restricted to Drosophila and their close relatives (Figure 1). Using a lenient E-value threshold, we were able to recover Metchnikowin from diverse mushroom-feeding Drosophila and perhaps other flies, and confirmed their identities by alignment and reciprocal BLAST (Figure S1), improving on previous annotations of immune genes in this lineage (24). The other AMP families show a broader taxonomic distribution (Figure 1).
Figure 1
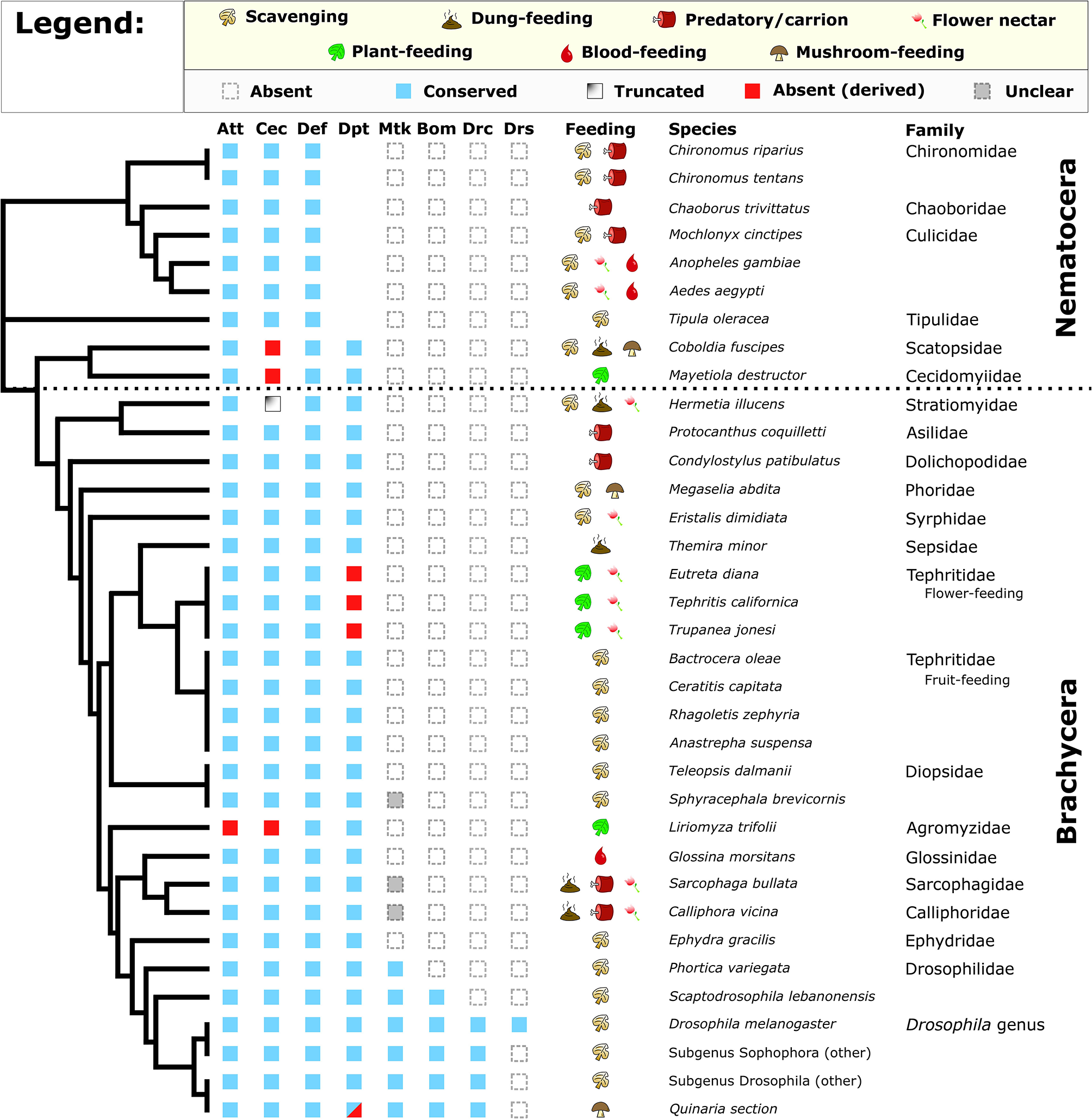
Conservation of Drosophila AMP families in diverse Diptera. Broadly, Attacin, Cecropin, Defensin, and Diptericin are conserved in most flies. However, Cecropins are absent or truncated in select lineages, while flower-feeding Tephritids and Liriomyza trifolii lack subsets of AMPs. We also recover a pattern of Diptericin loss in members of the Quinaria section of Drosophila (shown later). Full annotation of larval and adult feeding ecologies is given in Supplementary Data File 1. Cladogram adapted from Vicoso and Bachtrog (43).
One striking pattern is the absence of Cecropin in two Nematocerans: the plant-feeding Hessian fly Mayetiola destructor and the oyster mushroom pest Coboldia fuscipes. The Coboldia genome is a small, well-assembled genome (~100 Mbp, scaffold N50 = 242 Kbp) (43, 44), and Cecropins throughout Diptera share similar motifs from the N terminus to C terminus. As such, we interpret this absence of Cecropin in both M. destructor (plant sap-feeding) and C. fuscipes (scavenger-feeding) as a likely true loss of Cecropin in this basal lineage. We also found a partial Cecropin sequence truncated by a premature stop codon in Hermetia illucens (scavenger-feeding). Finally, we did not recover Cecropin from the genome of the leafminer Liriomyza trifolii, an independent transcriptome of L. trifolii pupae (a life stage when AMPs are often highly upregulated), or from a sequence read archive (SRA) file (GenBank accession: DRX064600) of the related Liriomyza chinensis. We see no immediate pattern in feeding ecology or life history that predicts Cecropin loss, but we also failed to recover an Attacin from Liriomyza, suggesting Liriomyza has lost AMPs from two gene families (Cecropins and Attacins). Called “leafminers,” Liriomyza larvae feed internally in the leaves of plants, an environment protected from external microbes by the immune system of the host plant; a more sterile food resource than most Dipterans. Moreover, we also failed to recover Diptericin in three flower-feeding Tephritid species. Like the leafminers, these flower-feeding flies live in a protected environment owing to larval development inside budding flower inflorescences (45).
Within the genus Drosophila, we observed two unique instances of AMP gain/loss we note separately. First, the genome of the cosmopolitan fly Drosophila busckii encodes no less than nine intact Diptericin genes, and we further recovered three pseudogenes in the D. busckii Diptericin gene region (Figure S2); for context, other Drosophila typically have only 2–3 Diptericin genes (24). Drosophila busckii is a cosmopolitan generalist species in common association with D. melanogaster, however D. busckii arrives later to rotting fruits and compost relative to D. melanogaster (46, 47). To favor the retention of so many Diptericin copies suggests the Diptericin response is highly important for D. busckii ecology. Second, we found that one paralog of the Attacin A/B duplication event has been lost in Drosophila sechellia, a species closely-related to D. melanogaster. Drosophila sechellia is famous for its unique ecology, feeding on toxic morinda fruit that repels other flies (48). Beyond losing this Attacin paralog, D. sechellia also lacks the ability to encapsulate and kill invading parasitoid wasps, associated with loss of function in immune genes involved in the melanization and stress responses (49). It seems likely that the toxins in morinda fruit would repel parasites such as wasps, reducing infectious pressure on D. sechellia. Thus, this ecology—already associated with loss of immune genes—may have additionally promoted loss of an Attacin as well.
Overall, we observe numerous instances of AMP loss across the Diptera phylogeny. The loss of Cecropins in ecologically diverse lineages is puzzling. For the mushroom pest C. fuscipes and Hessian fly M. destructor, either scavenging (C. fuscipes) or sap-feeding (M. destructor) could reflect an ancestral ecology promoting Cecropin loss. These two last shared a common ancestor ~250 mya [Timetree; (42)], and transitions from generalist to specialist ecologies, and back again, have been inferred in Drosophila (50). However, more strikingly we observe AMP gene family loss in all three strictly plant-feeding fly lineages assessed (the Hessian fly M. destructor, Liriomyza leafminers, and flower-feeding Tephritids), reminiscent of immune gene loss in sap-feeding Pea Aphids.
Parallel Loss of Diptericins in Lineages With Divergent Ecology
In our screen of AMP conservation in Diptera, we were intrigued by the loss of Diptericin in some Tephritid fruit flies and some Drosophila; Diptericin was previously shown to have rare null alleles segregating in a North American D. melanogaster population (25). While assembly quality was variable amongst the Tephritid genomes, the absence of Diptericin in three independent flower-feeding Tephritid species, but presence in all screened fruit-feeding Tephritid species suggests Diptericin is lost in the flower-feeding Tephritid lineage. Diptericin is an AMP that has attracted a great deal of attention as the canonical readout of Imd signaling in D. melanogaster (19), and for its highly specific interaction with Providencia rettgeri bacteria (25, 51). Interestingly, the Diptericin sequence retained in the fruit-feeding Tephritids has converged on a Drosophilid DptB-like sequence (Figure 2A). Furthermore, it was previously reported that DptB was pseudogenized in the mushroom-feeding Drosophila species D. guttifera and likely also in Drosophila neotestacea (D. neotestacea) (24). However, when we screened the recently-sequenced mushroom-feeding Drosophila innubila genome, we recovered intact coding sequence for DptB. It is possible that the intact DptB sequence in D. innubila could reflect that DptB in mushroom-feeding flies was initially pseudogenized not due to loss of coding sequence, but rather due to mutations affecting gene expression. We therefore performed qPCR following infection to determine the expression profile of Diptericins amongst mushroom-feeding flies and included outgroup Drosophila to inform our interpretations.
Figure 2
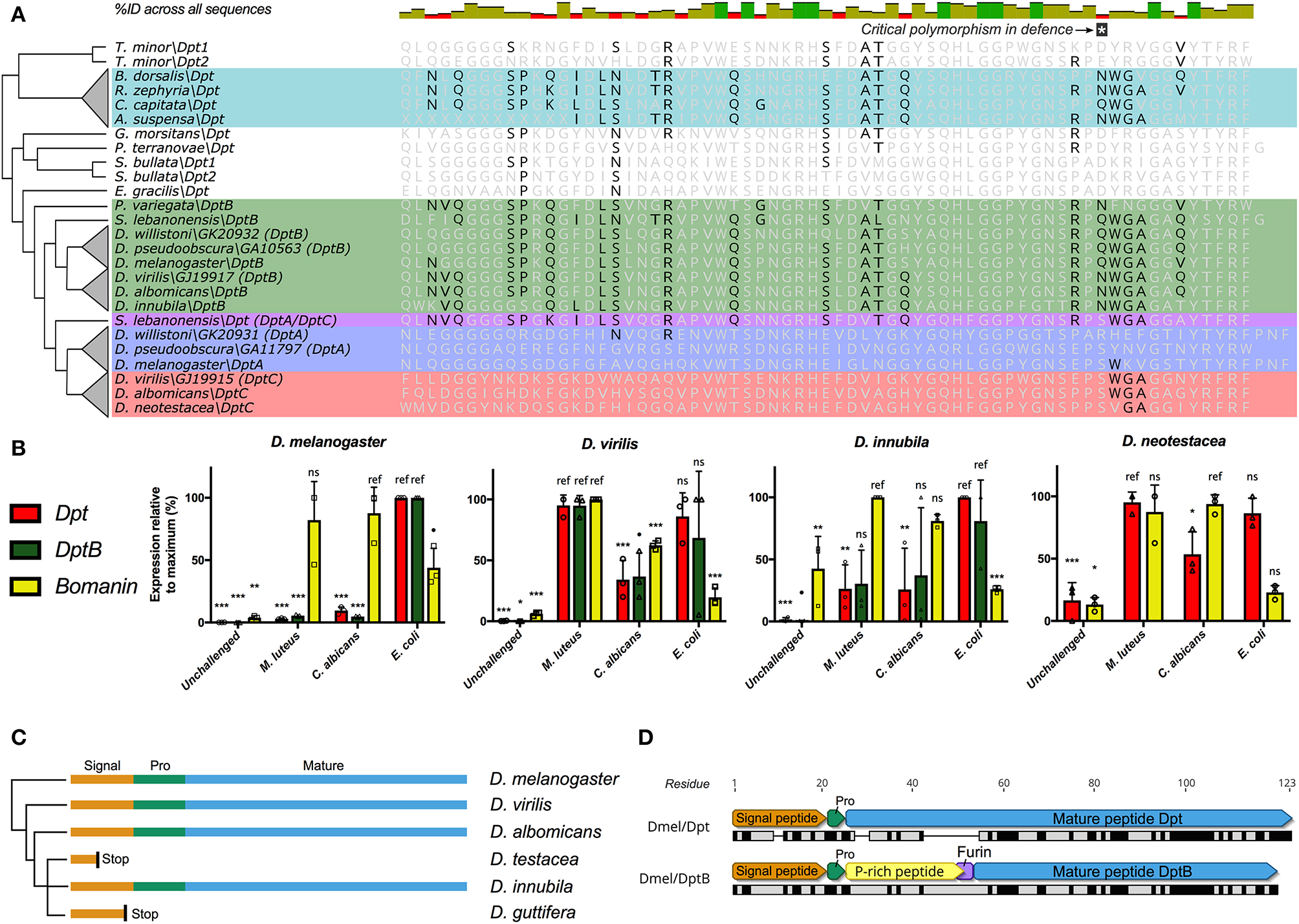
Diptericin evolution in Tephritids and Drosophilids. (A) The Diptericin C-terminal sequence of fruit-feeding Tephritids has converged on DptB-like residues (also see Figure S3). Major Diptericin clades: Blue, DptA; Green, DptB; Red, DptC; Teal, Tephritid Dpts, and S. lebanonensis Dpt in purple is a direct outgroup to the DptA and DptC clades (collectively referred to as “Dpt” in B). The polymorphism critical for defense (25) is indicated, and Tephritid Diptericins have converged on the Q/N polymorphism found in the Drosophila DptB clade. (B)Diptericin expression in select lineages of Drosophila. Expression is normalized to the maximum for each gene within each species, and statistical comparisons are done with reference to the treatment that induced expression most (see Materials and Methods). (C)DptB has been pseudogenized in two independent lineages of mushroom-feeding Drosophila: D. testacea and D. guttifera. Signal, Signal peptide; Pro, propeptide; Mature, Mature peptide. (D) The mature peptides of D. melanogaster Dpt and DptB differ markedly in mature structure, including a critical furin cleavage site in DptB that likely leads to two mature peptides. At the protein sequence level Dpt has only 41.7% pairwise identity to DptB (black bars = identical sites).
We used a Bomanin (Bom791) as a positive control for infections more specific to the Toll pathway in D. melanogaster. We further intended to use Dpt as a specific readout of Imd signaling, and as an independent control gene for assessing DptB expression. First, we found that DptB is strongly induced in D. innubila, suggesting it is not pseudogenized as in sister lineages. However, we found that Dpt expression is highly variable across Drosophila species (Figure 2B). Dpt is more specifically induced by Gram-negative bacterial challenge in D. melanogaster, and indeed we see this pattern in the outgroup flies Drosophila pseudoobscura (D. pseudoobscura) and Drosophila immigrans (D. immigrans) (Supplementary Data File 1), and also broadly in D. innubila. However, Dpt and DptB are similarly induced by either Gram-negative or Gram-positive bacterial challenge in D. virilis, and the same is true for Dpt in both Drosophila testacea (D. testacea) (not shown), and D. neotestacea (Figure 2B, and see Materials and methods). Using Sanger sequencing, we additionally confirmed that DptB is pseudogenized in D. testacea by a premature stop codon, supporting its absence in the D. neotestacea transcriptome (GenBank accession: MN311476). The mutation affecting DptB in the Testacea group is distinct from the mutation in the Quinaria group species D. guttifera (Figure 2C), suggesting independent loss events.
Thus, the pseudogenization of DptB-like genes in both flower-feeding Tephritids and two lineages of mushroom-feeding flies reflects that first there was convergent evolution toward DptB-like sequence in both Tephritids and Drosophilids. Thereafter, subsequent independent losses of DptB-like Diptericins occurred in lineages with ecologies that diverged from fruit-feeding. This pattern suggests DptB may be attuned to fruit-feeding ecology, but not as useful in other ecological niches.
We can only speculate on how evolution shapes patterns of DptB gain/loss: in D. melanogaster, functional characterization of DptB was long-ignored in favor of its more potently-induced paralog Dpt. However, recent studies have revealed that the two Diptericins have markedly different activities in immunity and host physiology. First, Unckless et al. (25) showed that a specific serine allele in Dpt confers defense against P. rettgeri, however no DptB gene in any fly encodes a serine at this site. Alternately, Barajas-Azpeleta et al. (52) found that DptB, but not Dpt, is required for long-term memory formation. There are a number of overt structure and sequence differences amongst Dpt and DptB (Figure 2D). First, Dpt encodes an 83-residue mature peptide with a proline-rich domain tailed by a glycine-rich Attacin-family domain. This 83-residue mature peptide is secreted following cleavage of the Dpt signal peptide and propeptide. On the other hand, DptB encodes a furin cleavage site (RVRR) between its proline-rich and glycine-rich domains, similar to other AMPs of the Attacin gene family (53); In Attacin C, this furin cleavage results in two secreted peptides, a proline-rich AMP (called MPAC) and a separate glycine-rich AMP (54). Furthermore, amongst the many sequence differences between Dpt and DptB (see Figure 2A) is the aforementioned serine residue of Dpt that confers defense against P. rettgeri bacteria. In Dpt genes, this residue is polymorphic (S/R/Q/N) in D. melanogaster and close relatives (25). However, DptB encodes a polymorphism for only Q/N at this site, including in convergent DptB-like Diptericins of Tephritid fruit flies. Globally, in D. melanogaster, Dpt appears to be key in mediating defense against P. rettgeri bacteria, while DptB is uniquely required for memory formation. Accordingly, these two Diptericins have overt differences in mature peptide products. Here we implicate host ecology as a likely determinant of Diptericin evolution, and suggest that these overt differences may have ecology-dependent effects on fitness leading to distinct patterns of gain/loss.
Null Alleles of Diptericin Are Segregating in Wild Populations of D. melanogaster
Our findings on Diptericin evolution coupled with recent descriptions of distinct Diptericin activities uniquely position this AMP family for providing insight into how conflicting roles in immunity and physiology can shape AMP evolution. Unckless et al. (25) reported the maintenance of two alternate alleles (serine/arginine) at residue 92 of the full length Dpt protein (residue 69 of the mature peptide) in wild populations of D. melanogaster and D. simulans. Providencia rettgeri is a natural pathogen of D. melanogaster (55), yet the Dpt arginine allele is maintained by balancing selection in the wild despite being associated with poor immune defense against P. rettgeri infection. Additionally, Unckless et al. (25) reported a rare null mutation in a D. melanogaster North American population (DGRP) (56) resulting in a premature stop codon affecting ~1% of the reference strains. Surprisingly, when we investigated a set of African populations (DPGP) (57), we found multiple independent null mutations segregating in different sub-populations throughout the Africa sampling range (Figure 3A). Even more surprising, the prevalence of these null mutations reaches over 20% in some populations and appears to follow a latitudinal cline (Figure 3B). As such, selective pressure on Dpt may follow a clinal gradient in Africa, favoring Dpt loss in southern African populations. Note that the cline crosses the equator and so may not be driven by climate alone. We also recover a similar, though not significant, trend for null alleles segregating in North American collections (Figure 3C).
Figure 3
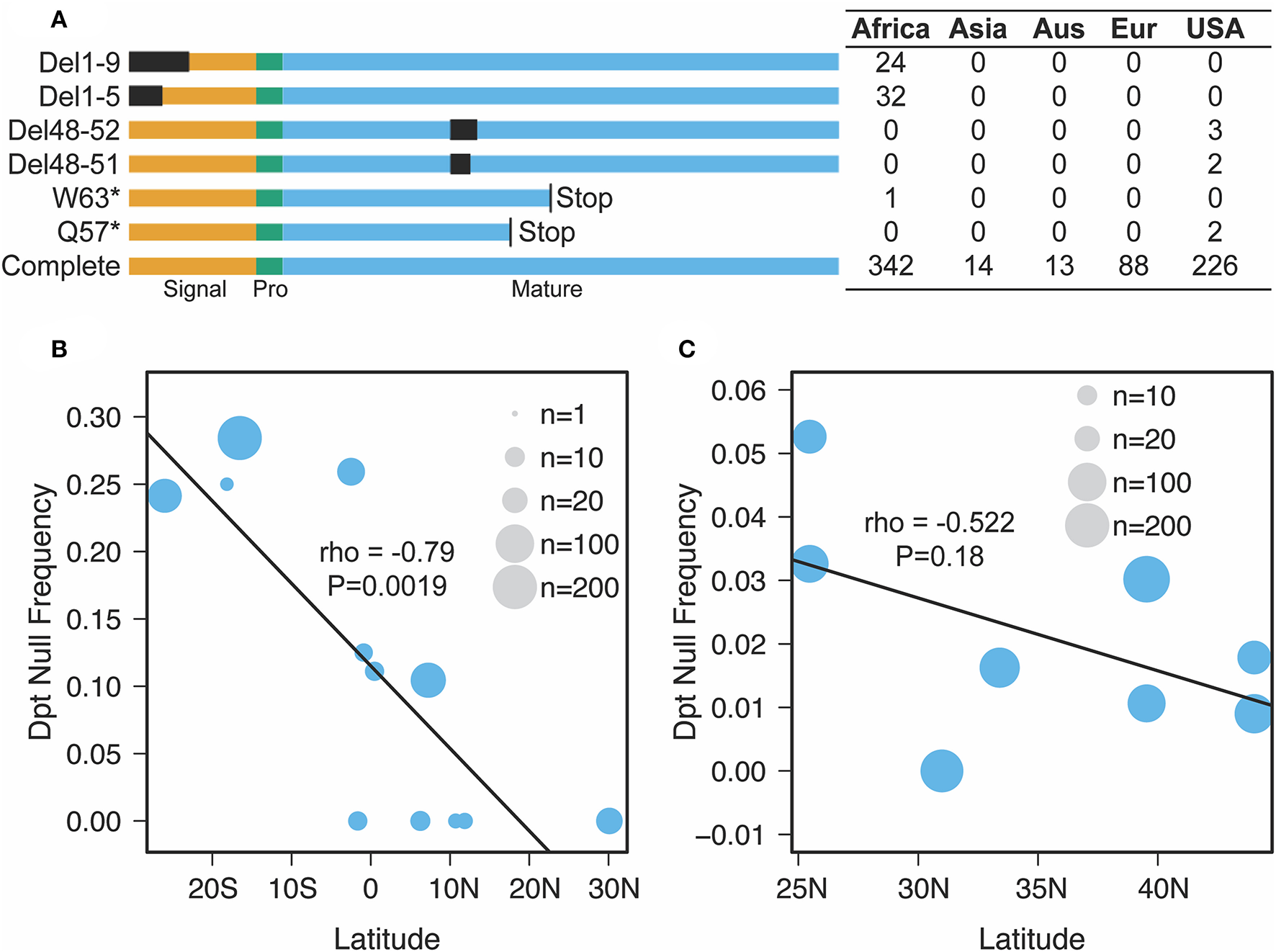
Nature and geographic distribution of Dpt null alleles. (A) At least six unique null alleles segregate in D. melanogaster Dpt worldwide. Del1-9 and Del1-5 are deletions removing the first 9 or 5 amino acids, Del 48-52 and Del 48-51 are in frame deletions that remove amino acids in the mature peptide and W63* and Q57* are premature stop codons. Signal, Pro and Mature correspond to the signal peptide, propeptide and mature peptide of the protein. The box to the right denotes the counts of each allele in each population (Aus, Australia; Eur, Europe, USA, DGRP population only). (B,C) The correlation between latitude and null frequency in African (B) and North American (C) populations (data from pooled sequencing of populations along a cline). The size of the circle represents the number of individuals sequenced (B) or reads mapped (C) in each population.
These data suggest that despite the described importance of Dpt in defense against the ecologically relevant pathogen P. rettgeri, null alleles associated with extremely poor immune defense are actively segregating in wild D. melanogaster. This suggests that the evolutionary forces behind Diptericin loss are not entirely passive. Taken together with the loss of DptB in other flies, instead this implicates active selection on Diptericins as peptides deleterious for fitness in alternate ecological conditions.
Discussion
AMPs must maintain a fine balance: being potent enough that they can kill harmful pathogens but not so harmful that they damage host tissues directly or by damaging beneficial components of the host's microbiome. It stands to reason that host ecology drives pathogen pressure and therefore might indirectly shape the complement of AMPs in a given host. In our survey of AMPs across Diptera and within Drosophila presented above, we find some support for an adaptive loss of AMPs in hosts associated with more sterile habitats. There is increasing awareness that these classic immune molecules can play diverse physiological roles, and that evolution may be shaping AMP copy number and sequence due to selection on non-immune functions. When considering the internal plant parasites of the Tephritid family and the leaf miner, clear parallels can be drawn regarding sterile food resource use and other fluid-feeding arthropods that have similarly lost or re-organized their immunity genes, namely: aphids, some mites, bed bugs, and body lice (35–39). It may also be that these plant-parasitic flies have yet-uncharacterized bacterial endosymbionts that impose selection against certain AMPs, enabling their specialist lifestyle. We also describe multiple incidents of Diptericin loss in Drosophila: DptB in some mushroom-feeding flies, a lineage with a specialist ecology whose microbiota differs drastically from D. melanogaster (58), while Dpt null alleles are segregating in wild D. melanogaster populations.
The central question then becomes: why should immune-inducible AMPs, antimicrobial agents required for competent host defense, be lost so readily? We can think of two evolutionary scenarios that would lead to the loss of AMPs. First, when infection pressure is low, relaxed constraint on protein sequence and/or expression could lead to the accumulation of mutations that compromise protein function and eventually lead to pseudogenization and loss. This represents a neutral process where genetic drift is the force removing AMPs. The second evolutionary scenario is that AMPs are costly in the absence of infection, so when infection pressure is low, mutations that compromise function (indels, premature stop codons, cis-regulatory mutations) are actually selected for. If periods of low infection pressure persist long enough, those mutations can become fixed, and gene function is lost.
Several lines of evidence support the second, “selective loss” scenario. First, if relaxed constraint in the absence of infection drove AMP loss, we would expect the loss of AMPs to be somewhat random. However, we see convergent loss of DptB-like genes in independent lineages with divergent ecologies, and Dpt null alleles segregating in wild populations of D. melanogaster. Of course, if AMPs are specific to a small suite of pathogens (e.g., Dpt and P. rettgeri), perhaps those pathogens are relatively absent in some natural populations compared to others. This would also lead to increased loss of AMPs, but via neutral processes. In the case of Dpt, however, Providencia rettgeri is distributed worldwide including throughout Africa (59, 60). Therefore, it is unlikely to be completely absent from African populations of D. melanogaster where null alleles are common. Instead, the specific loss of Diptericins in Drosophila might reflect a deleterious consequence when dysregulated in non-immune tissues. For example, AMP expression increases dramatically in the head tissue of aging flies (18). This explanation seems more likely, as non-cell autonomous DptB is known to affect memory formation in D. melanogaster (52), evidence of direct Diptericin impact on brain function. A second line of support for the “selective loss” scenario is the null allele cline observed in African populations alongside a parallel (though not significant) cline in North America. Such parallel clines are often used as evidence for selection acting on alleles (61–63). While neutral processes could lead to clines in null alleles as well, with the null allele spreading from an initial source population, the likelihood of this happening in parallel on two continents is small. Finally, there is growing evidence that several AMPs may inflict damage on host tissues. For instance, the cathelicidin LL-37 is toxic to leukocytes (64), and constitutive expression of AMPs reduces lifespan in Drosophila (17). These direct observations of deleterious effects strongly undermine the idea that neutral processes are driving the loss of AMPs, instead suggesting these molecules impose a significant effect on host fitness.
One exciting explanation for AMP fitness costs is the idea that AMPs are dysregulated through aging, leading to chronic inflammatory responses and eventually cell death. Additionally, the idea that AMPs may be active in the nervous system is an attractive recent hypothesis that demands more consideration (65, 66), particularly to understand the roles these short peptides play in neuronal homeostasis (67). For instance, while implicated in Alzheimer's disease for decades, the specific nature of how Amyloid-beta contributes to dementia remains unclear (68). Understanding its role in immunity may shed light on the cause and progression of amyloid plaques (13), and reveal the true culprit(s) behind Alzheimer's progression; an interesting recent study found that Amyloid-beta binds to the human cathelicidin LL-37, forming heterodimers that reduce the toxicity of LL-37 to host cells (69). Alongside evidence that Alzheimer's may be an infectious disease (14), dysregulation of AMPs in the nervous system upon chronic infection could lead to host cell toxicity. Appreciating the role of AMPs in the nervous system, particularly during infection, may lead to breakthroughs in treating neurodegenerative disorders such as Alzheimer's disease or Parkinson's disease.
If indeed AMPs are deleterious in non-immune contexts, this may promote balancing selection in populations with dynamic immune pressures. Beyond AMPs, trade-offs between immunity and fitness are well-documented, implying that an immune system is advantageous only in the context of immune challenge, but otherwise is detrimental to reproductive fitness (70–73). As the front line of innate immunity, AMPs should be primary actors on this evolutionary stage, and selection for or against immunity genes should therefore act strongly on context-dependent AMPs. Recent studies report that both balancing and diversifying selection has shaped the Drosophila AMP arsenal (22–25), revising how we view AMPs as actors in host-pathogen interactions (28). If balancing selection is driven by trade-offs between alleles that provide increased resistance during infection but are costly when hosts are uninfected, this could explain the dynamic patterns of AMP gain and loss described here. By characterizing e.g. Diptericin loss throughout Diptera, we provide the beginnings of an immunological fossil record with extinct (pseudogenes) and extant Diptericin gene copies in different lineages. The observations of other AMP gene losses throughout Diptera extend this fossil record back in time, describing lineages with different stages of loss stemming from an ancestral immune-competent fly to derived lineages lacking subsets of certain AMPs.
Globally, we highlight how host ecology predicts AMP loss, and follow the evolution of AMP lineages throughout Diptera. We describe that selection on the innate immune system can act swiftly and directly on AMPs, implicating some AMPs as deleterious molecules in the absence of microbial challenges. These results could relate to the newly discovered role of AMP-like peptides in neurodegenerative diseases and autoimmune disorders. If so, our findings offer evolutionary signatures supporting the notion that trade-offs between immunity and fitness are mediated by costs related to the maintenance of autotoxic host AMPs.
Materials and Methods
Survey of AMP Families in Published Diptera Genomes and Transcriptomes
We first conducted a thorough literature review to annotate the life histories of diverse Diptera. We then searched for Drosophila AMP families present in other Diptera using an iterative step-wise tBLASTn approach followed by manual curation; of note, we used an extremely lenient E-value for the shortest peptides (e.g., E <100 for Mtk, 26 residues long), followed by manual curation. In brief, we collected AMP genes from sequenced Drosophila and then BLASTed all available orthologs against outgroup genomes from Vicoso and Bachtrog (43). We collected all confirmed outgroup orthologs and re-performed this BLAST against any species where no match was found, until we ceased to recover new orthologs. To verify any patterns of loss we observed (e.g., Dpt loss in Tephritid species), we further searched for outgroup sequence data (genomes, transcriptomes, or raw SRAs) to include in our analyses as independent databases. For some orthologs, only a partial sequence was recovered on a scaffold assembled with many gaps (NNNs). If sequence similarity was highly conserved we annotated these AMPs as “present” but do not include them in phylogenetic analyses as their information content was poor. All sequence databases used in this study are included in Supplementary Data File 1. Sanger sequencing results are deposited in GenBank under accessions: MN311474-MN311476.
To investigate sequence similarity and validate curated orthologs, we aligned sequences using MAFFT and performed phylogenetic analysis using Neighbor-joining (1000 bootstraps) and/or Maximum likelihood (100 or 500 bootstraps) methods implemented in Geneious R10 and the PhyML webserver (74). For Diptericin evolution in Figure S3, sequences were also codon-aligned using MAFFT.
Fly Stocks and Strain Information
The following strains were used in this study for gene expression analysis and Sanger sequencing: D. melanogaster (DrosDel iso w1118), D. pseudoobscura, D. virilis, D. immigrans, D. innubila, D. testacea, and D. neotestacea. Drosophila pseudoobscura and D. immigrans were generously provided by Ben Longdon and correspond to strains used in Duxbury et al. (75). Drosophila innubila used in this study is the same as the genome-sequenced strain from Hill et al. (76). Drosophila virilis was a gift from Richard Benton corresponding to Sackton et al. (28). Steve Perlman kindly provided Testacea group flies. The D. testacea strain used corresponds to the wild-type D. testacea described in Keais et al. (77) cleared of Wolbachia symbionts by the Perlman lab. The D. neotestacea strain is the same as used in Hanson et al. (24). Drosophila melanogaster, D. pseudoobscura, and D. virilis were reared on standard food medium for D. melanogaster and reared at 25°C. Drosophila immigrans, D. innubila, and D. neotestacea were reared on Nutri FlyTM instant formulation supplemented with a piece of Agaricus bisporus mushroom, and reared at 22°C. All species were kept at 22°C during the course of infection. All flies used in this study were previously shown to be negative for Wolbachia, a common endosymbiont of Drosophila.
Gene Expression Analysis
Infections, RNA extraction, and cDNA synthesis were performed as previously described (51). Pooled samples of 6 flies (3 males and 3 females) were used for each replicate experiment, and three repeats were performed (18 total flies per treatment per species). Flies were frozen either 6 hpi (Unchallenged, Escherichia coli (E. coli)] or 24hpi (M. luteus, C. albicans) at −20°C in TRIzol. Quantitative PCR was performed on a LightCycler 480 (Roche) in 96-well plates using Applied Biosystems PowerUP Master Mix. Error bars represent one standard deviation from the mean. Statistical analysis was performed using One-way ANOVA with Holm's-Sidak post-hoc comparisons to the treatment that induced expression most in each species for each gene (marked as “ref”); e.g., the E. coli treatment was the point of comparison for Dpt, and C. albicans for Bomanin in D. melanogaster. P-values are reported as: not significant = ns, <0.1 = •, <0.05 = * <0.01 = ** and <0.001 = ***.
We note the pattern of Dpt induction we observed conflicts with a previous report that Dpt is not inducible in D. neotestacea (24), which is likely explained by measuring alternate Dpt isoforms. Primers used in this study, additional expression data for AMPs in different species, and D. neotestacea Dpt primer comparisons suggesting alternate Dpt isoforms can be found in Supplementary Data File 1.
Diptericin Evolution in Drosophila
We identified segregating putative null alleles in Drosophila melanogaster populations by visually inspecting alignments of re-sequenced individual inbred lines (Figures 3A,B) or pool-seq alignments (North American populations in Figure 3C) (57, 78). We found three classes of putative null alleles: (a) premature stop codons, (b) deletions that disrupt core parts of the transcript (i.e., the start codon), and (c) deletions that are in frame but were associated with reduced immune defense against P. rettgeri in prior studies (79). Thus, while the counts in Figure 3A for the USA represent individual inbred lines, those in Figure 3C represent the proportion of reads at a given site carrying the particular null allele.
Statements
Data availability statement
The datasets generated for this study can be found in the GenBank database, accession numbers MN311474-MN311476.
Author contributions
MH performed live specimen experiments. MH and RU screened and analyzed data. MH, BL, and RU wrote the manuscript.
Funding
This work was supported by the National Institutes of Health Grants R00GM114714 and R01AI139154 to RU.
Acknowledgments
We would like to thank Florent Masson, Joanne Chapman, Tom Hill and members of the Unckless lab for useful comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.02620/full#supplementary-material
Figure S1Alignment of Metchnikowin (Mtk) and Mtk-like sequences. We recovered Mtk genes from mushroom-feeding flies (brown highlight) that retain the major portion of the Mtk mature peptide. We also recovered a clear Mtk ortholog in the outgroup Drosophilid P. variegata (Pvar\Mtk), which resembles Mtk-like sequence in the Brachyceran fly Solenopsis brevicornis (S. brevicornis) (Sbre\Mtk-like). Other Mtk-like sequences from Brachycerans are also shown, and the full open reading frame is shown for all sequences.
Figure S2The Drosophila busckii Diptericin gene region encodes 6 copies of the subgenus Drosophila-restricted Dpt (DptC clade, see Figure S3) and 3 copies of DptB. There are an additional 3 Diptericin pseudogenes apparent in the gene region.
Figure S3Maximum likelihood tree showing that Tephritid Diptericins paraphyletically cluster within the Drosophila DptB clade, though bootstraps for exact sorting are poor (as expected of paraphyletic clustering); 100 bootstraps, where bootstraps <30 are not shown. Major Dpt clades are highlighted as follows: Blue, DptA; Red, DptC; Green, DptB; Teal, Tephritid Dpts.
Supplementary Data File 1Annotated life histories: Annotations of feeding ecology in larvae and adults of Diptera in this study. Used in Figure 1. Datasets used in this study: Datasets and their accesions and quality scores for diverse Diptera used in this study. Used in Figure 1. Primers used in this study: Primers used in qPCR analysis or Sanger sequencing. qPCR reactions were run with a 60°C annealing and extension phase. Used in Figure 2. qPCR data from alt. species: qPCR data from initial explorations using Drosophila pseudoobscura, Drosophila immigrans, and Drosophila testacea (1 Experiment). Remarked as “not shown” in manuscript. Dneo DptC primer comparison: Comparison of Dneo\Dpt induction as measured by primers from Hamilton et al. (2014) and universal Testacea group Dpt primers from this study. Revises interpretation of Hamilton et al. (2014) and (24).
References
1.
ZhangLGalloRL. Antimicrobial peptides. Curr Biol. (2016) 26:R14–9. 10.1016/j.cub.2015.11.017
2.
MarcinkiewiczMMajewskiS. The role of antimicrobial peptides in chronic inflammatory skin diseases. Postep Dermatologii i Alergol. (2016) 33:6–12. 10.5114/pdia.2015.48066
3.
AntoniLNudingSWellerDGersemannMOttGWehkampJStangeEF. Human colonic mucus is a reservoir for antimicrobial peptides. J Crohn's Colitis. (2013) 7:e652–64. 10.1016/j.crohns.2013.05.006
4.
MukherjeeSHooperL V. Antimicrobial defense of the intestine. Immunity. (2015) 42:28–39. 10.1016/j.immuni.2014.12.028
5.
DalcinDUlanovaM. The role of human beta-defensin-2 in Pseudomonas aeruginosa pulmonary infection in cystic fibrosis patients. Infect Dis Ther. (2013) 2:159–66. 10.1007/s40121-013-0015-5
6.
GoldmanMJAndersonGMStolzenbergEDKariUPZasloffMWilsonJM. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. (1997) 88:553–60. 10.1016/S0092-8674(00)81895-4
7.
BergssonGReevesEPMcNallyPChotirmallSHGreeneCMGreallyPet al. LL-37 complexation with glycosaminoglycans in cystic fibrosis lungs inhibits antimicrobial activity, which can be restored by hypertonic saline. J Immunol. (2009) 183:543–51. 10.4049/jimmunol.0803959
8.
BuckiRByfieldFJJanmeyPA. Release of the antimicrobial peptide LL-37 from DNA/F-actin bundles in cystic fibrosis sputum. Eur Respir J. (2007) 29:624–32. 10.1183/09031936.00080806
9.
SinganayagamAGlanvilleNCuthbertsonLBartlettNWFinneyLJTurekEet al. Inhaled corticosteroid suppression of cathelicidin drives dysbiosis and bacterial infection in chronic obstructive pulmonary disease. Sci Transl Med. (2019) 11:eaav3879. 10.1126/scitranslmed.aav3879
10.
BjörstadÅAskariehGBrownKLChristensonKForemanHÖnnheimKet al. The host defense peptide LL-37 selectively permeabilizes apoptotic leukocytes. Antimicrob Agents Chemother. (2009) 53:1027–38. 10.1128/AAC.01310-08
11.
LeeMShiXBarronAEMcGeerEMcGeerPL. Human antimicrobial peptide LL-37 induces glial-mediated neuroinflammation. Biochem Pharmacol. (2015) 94:130–41. 10.1016/j.bcp.2015.02.003
12.
PiktelENiemirowiczKWnorowskaUWatekMWollnyTGłuszekKet al. The role of cathelicidin LL-37 in cancer development. Arch Immunol Ther Exp. (2016) 64:33–46. 10.1007/s00005-015-0359-5
13.
GosztylaMLBrothersHMRobinsonSR. Alzheimer's Amyloid-β is an antimicrobial peptide: a review of the evidence. J Alzheimer's Dis. (2018) 62:1495–506. 10.3233/JAD-171133
14.
DominySSLynchCErminiFBenedykMMarczykAKonradiAet al. Porphyromonas gingivalis in Alzheimer's disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. (2019) 5:eaau3333. 10.1126/sciadv.aau3333
15.
SosciaSJKirbyJEWashicoskyKJTuckerSMIngelssonMHymanBet al. The Alzheimer's disease-associated amyloid β-protein is an antimicrobial peptide. PLoS ONE. (2010) 5:e9505. 10.1371/journal.pone.0009505
16.
CaoYChtarbanovaSPetersenAJGanetzkyB. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc Natl Acad Sci USA. (2013) 110:E1752–60. 10.1073/pnas.1306220110
17.
BadinlooMNguyenESuhWAlzahraniFCastellanosJKlichkoVIet al. Overexpression of antimicrobial peptides contributes to aging through cytotoxic effects in Drosophila tissues. Arch Insect Biochem Physiol. (2018) 98:e21464. 10.1002/arch.21464
18.
KounatidisIChtarbanovaSCaoYHayneMJayanthDGanetzkyBet al. NF-κB immunity in the brain determines fly lifespan in healthy aging and age-related neurodegeneration. Cell Rep. (2017) 19:836–48. 10.1016/j.celrep.2017.04.007
19.
LemaitreBHoffmannJ. The host defense of drosophila melanogaster. Annu Rev Immunol. (2007) 25:697–743. 10.1146/annurev.immunol.25.022106.141615
20.
ClemmonsAWLindsaySAWassermanSA. An effector peptide family required for drosophila toll-mediated immunity. PLoS Pathog. (2015) 11:e1004876. 10.1371/journal.ppat.1004876
21.
LindsaySALinSJHWassermanSA. Short-form bomanins mediate humoral immunity in Drosophila. J Innate Immun. (2018) 10:306–14. 10.1159/000489831
22.
UncklessRLLazzaroBP. The potential for adaptive maintenance of diversity in insect antimicrobial peptides. Philos Trans R Soc B Biol Sci. (2016) 371:20150291. 10.1098/rstb.2015.0291
23.
ChapmanJRHillTUncklessRL. Balancing selection drives maintenance of genetic variation in Drosophila antimicrobial peptides. Genome Biol Evol. (2019) 11:2691–701. 10.1093/gbe/evz191
24.
HansonMAHamiltonPTPerlmanSJ. Immune genes and divergent antimicrobial peptides in flies of the subgenus Drosophila. BMC Evol Biol. (2016) 16:228. 10.1186/s12862-016-0805-y
25.
UncklessRLHowickVMLazzaroBP. Convergent balancing selection on an antimicrobial peptide in Drosophila. Curr Biol. (2016) 26:257–62. 10.1016/j.cub.2015.11.063
26.
QuesadaHRamos-OnsinsSEAguadM. Birth-and-death evolution of the Cecropin multigene family in Drosophila. J Mol Evol. (2005) 60:1–11. 10.1007/s00239-004-0053-4
27.
JigginsFMKimKW. The evolution of antifungal peptides in Drosophila. Genetics. (2005) 171:1847–59. 10.1534/genetics.105.045435
28.
SacktonTBLazzaroBPSchlenkeTAEvansJDHultmarkDClarkAG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. (2007) 39:1461–8. 10.1038/ng.2007.60
29.
ZhouJLemosBDopmanEBHartlDL. Copy-number variation: the balance between gene dosage and expression in Drosophila melanogaster. Genome Biol Evol. (2011) 3:1014–24. 10.1093/gbe/evr023
30.
HolloxEJArmourJALBarberJCK. Extensive normal copy number variation of a β-defensin antimicrobial-gene cluster. Am J Hum Genet. (2003) 73:591–600. 10.1086/378157
31.
LinzmeierRMGanzT. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in α- and β-defensin regions at 8p22-p23. Genomics. (2005) 86:423–30. 10.1016/j.ygeno.2005.06.003
32.
MachadoLROttoliniB. An evolutionary history of defensins: a role for copy number variation in maximizing host innate and adaptive immune responses. Front Immunol. (2015) 6:115. 10.3389/fimmu.2015.00115
33.
ChenJHuddlestonJBuckleyRMMaligMLawhonSDSkowLCet al. Bovine NK-lysin: copy number variation and functional diversification. Proc Natl Acad Sci USA. (2015) 112:E7223–9. 10.1073/pnas.1519374113
34.
SacktonTB. Comparative genomics and transcriptomics of host-pathogen interactions in insects: evolutionary insights and future directions. Curr Opin Insect Sci. (2018) 31:106–13. 10.1016/j.cois.2018.12.007
35.
GerardoNMAltincicekBAnselmeCAtamianHBarribeauSMde VosMet al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. (2010) 11:R21. 10.1186/gb-2010-11-2-r21
36.
PalmerWJJigginsFM. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol Biol Evol. (2015) 32:2111–29. 10.1093/molbev/msv093
37.
Santos-MatosGWybouwNMartinsNEZéléFRigaMLeitãoABet al. Tetranychus urticae mites do not mount an induced immune response against bacteria. Proc Biol Sci. (2017) 284:20170401. 10.1098/rspb.2017.0401
38.
BenoitJBAdelmanZNReinhardtKDolanAPoelchauMJenningsECet al. Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat Commun. (2016) 7:10165. 10.1038/ncomms10165
39.
SuttonGGStrausbergRKirknessEFHaasBJSunWBraigHRet al. Genome Sequences of the Human Body Louse and Its Primary Endosymbiont Provide Insights into the Permanent Parasitic Lifestyle. (2010). Available online at: http://www.pnas.org.ezproxy.york.ac.uk/content/107/27/12168.full
40.
DouglasAEBouvaineSRussellRR. How the insect immune system interacts with an obligate symbiotic bacterium. Proc R Soc B. (2011) 278:333–8. 10.1098/rspb.2010.1563
41.
HeddiAZaidman-RémyA. Endosymbiosis as a source of immune innovation. Comptes Rendus. (2018) 341:290–6. 10.1016/j.crvi.2018.03.005
42.
KumarSStecherGSuleskiMHedgesSB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. (2017) 34:1812–9. 10.1093/molbev/msx116
43.
VicosoBBachtrogD. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. (2015) 13:e1002078. 10.1371/journal.pbio.1002078
44.
Schmidt-OttURafiqiAMSanderKJohnstonJS. Extremely small genomes in two unrelated dipteran insects with shared early developmental traits. Dev Genes Evol. (2009) 219:207–10. 10.1007/s00427-009-0281-0
45.
AlbrectsenBR. Flowering phenology and seed predation by a tephritid fly: Escape of seeds in time and space. Ecoscience. (2000) 7:433–8. 10.1080/11956860.2000.11682614
46.
AtkinsonWShorrocksB. Breeding site specificity in the domestic species of Drosophila. Oecologia. (1977) 29:223–32. 10.1007/BF00345697
47.
MarkowTA. The natural history of model organisms: the secret lives of Drosophila flies. Elife. (2015) 4:e06793. 10.7554/eLife.06793
48.
HigaIFuyamaY. Genetics of food preference in Drosophila sechellia. Genetica. (2006) 88:129–36. 10.1007/BF02424469
49.
Salazar-JaramilloLPaspatiAVan De ZandeLVermeulenCJSchwanderTet al. Evolution of a cellular immune response in Drosophila: a phenotypic and genomic comparative analysis. Genome Biol Evol. (2014) 6:273–89. 10.1093/gbe/evu012
50.
Scott ChialvoCHWhiteBEReedLKDyerKA. A phylogenetic examination of host use evolution in the quinaria and testacea groups of Drosophila. Mol Phylogenet Evol. (2019) 130:233–43. 10.1016/j.ympev.2018.10.027
51.
HansonMADostálováACeroniCPoidevinMKondoSLemaitreB. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. Elife. (2019) 8:e44341. 10.7554/eLife.44341
52.
Barajas-azpeletaRWuJGillJWelteR. Antimicrobial peptides modulate long-term memory. PLoS Genet. (2018) 14:e1007440. 10.1371/journal.pgen.1007440
53.
HedengrenMBorgeKHultmarkD. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem Biophys Res Commun. (2000) 279:574–81. 10.1006/bbrc.2000.3988
54.
RabelDCharletMEhret-SabatierLCavicchioliLCudicMOtvosLet al. Primary structure and in vitro antibacterial properties of the Drosophila melanogaster attacin C Pro-domain. J Biol Chem. (2004) 279:14853–9. 10.1074/jbc.M313608200
55.
JunejaPLazzaroBP. Providencia sneebia sp. nov. and Providencia burhodogranariea sp. nov., isolated from wild Drosophila melanogaster. Int J Syst Evol Microbiol. (2009) 59:1108–11. 10.1099/ijs.0.000117-0
56.
MackayTFCRichardsSStoneEABarbadillaAAyrolesJFZhuDet al. The Drosophila melanogaster Genetic Reference Panel. Nature. (2012) 482:173–8. 10.1038/nature10811
57.
LackJBLangeJDTangADCorbett-DetigRBPoolJE. A Thousand fly genomes: an expanded Drosophila genome nexus. Mol Biol Evol. (2016) 33:3308–13. 10.1093/molbev/msw195
58.
MartinsonVGDouglasAEJaenikeJ. Community structure of the gut microbiota in sympatric species of wild Drosophila. Ecol Lett. (2017) 20:629–39. 10.1111/ele.12761
59.
YohMMatsuyamaJOhnishiMTakagiKMiyagiHMoriKet al. Importance of Providencia species as a major cause of travellers' diarrhoea. J Med Microbiol. (2005) 54:1077–82. 10.1099/jmm.0.45846-0
60.
TshisevheVSLekalakalaMRTshumaNJanse van RensburgSMbelleN. Outbreak of carbapenem-resistant Providencia rettgeri in a tertiary hospital. S Afr Med J. (2016) 107:31–3. 10.7196/SAMJ.2016.v107.i1.12002
61.
LinnenCRHoekstraHE. Measuring natural selection on genotypes and phenotypes in the wild. In: Cold Spring Harbor Symposia on Quantitative Biology, Vol. 74. Cold Spring Harbor Laboratory Press (2009) p. 155–68. 10.1101/sqb.2009.74.045
62.
YangYEderyI. Parallel clinal variation in the mid-day siesta of Drosophila melanogaster implicates continent-specific targets of natural selection. PLoS Genet. (2018) 14:e1007612. 10.1371/journal.pgen.1007612
63.
AdrionJRHahnMWCooperBS. Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. (2015) 31:434–44. 10.1016/j.tig.2015.05.006
64.
WehkampJSchmidMStangeEF. Defensins and other antimicrobial peptides in inflammatory bowel disease. Curr Opin Gastroenterol. (2007) 23:370–8. 10.1097/MOG.0b013e328136c580
65.
AugustyniakDNowakJ T. Lundy F. Direct and indirect antimicrobial activities of neuropeptides and their therapeutic potential. Curr Protein Pept Sci. (2013) 13:723–38. 10.2174/138920312804871139
66.
SchluesenerHJ. Antimicrobial peptides in the brain neuropeptides and amyloid. Front Biosci. (2012) S4:1375–80. 10.2741/s339
67.
BrogdenKAGuthmillerJMSalzetMZasloffM. The nervous system and innate immunity: the neuropeptide connection. Nat Immunol. (2005) 6:558–64. 10.1038/ni1209
68.
NelsonPTAlafuzoffIBigioEHBourasCBraakHCairnsNJet al. Correlation of alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J Neuropathol Exp Neurol. (2012) 71:362–81. 10.1097/NEN.0b013e31825018f7
69.
De LorenziEChiariMColomboRCretichMSolaLVannaRet al. Evidence that the human innate immune peptide LL-37 may be a binding partner of amyloid-β and inhibitor of fibril assembly. J Alzheimer's Dis. (2017) 59:1213–26. 10.3233/JAD-170223
70.
SchwenkeRALazzaroBPWolfnerMF. Reproduction–immunity trade-offs in insects. Annu Rev Entomol. (2015) 61:239–56. 10.1146/annurev-ento-010715-023924
71.
LochmillerRLDeerenbergC. Trade-offs in evolutionary immunology: just what is the cost of immunity?Oikos. (2000) 88:87–98. 10.1034/j.1600-0706.2000.880110.x
72.
SchlenkeTAMoralesJGovindSClarkAG. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog. (2007) 3:1486–501. 10.1371/journal.ppat.0030158
73.
RolffJ. Bateman's principle and immunity. Proc R Soc B Biol Sci. (2002) 269:867–72. 10.1098/rspb.2002.1959
74.
GuindonSDufayardJFLefortVAnisimovaMHordijkWGascuelO. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. (2010) 59:307–21. 10.1093/sysbio/syq010
75.
DuxburyEMDayJPMaria VespasianiDThüringerYTolosanaISmithSCet al. Host-pathogen coevolution increases genetic variation in susceptibility to infection. Elife. (2019) 8:e46440. 10.7554/eLife.46440
76.
HillTKosevaBSUncklessRL. The genome of Drosophila innubila reveals lineage-specific patterns of selection in immune genes. Mol Biol Evol. (2019) 36:1405–17. 10.1101/383877
77.
KeaisGLHansonMAGowenBEPerlmanSJ. X chromosome drive in a widespread Palearctic woodland fly, Drosophila testacea. J Evol Biol. (2017) 30:1185–94. 10.1111/jeb.13089
78.
BerglandAOBehrmanELO'BrienKRSchmidtPSPetrovDA. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. (2014) 10: e1004775. 10.1371/journal.pgen.1004775
79.
UncklessRLRottschaeferSMLazzaroBP. The complex contributions of genetics and nutrition to immunity in Drosophila melanogaster. PLoS Genet. (2015) 11:e1005030. 10.1371/journal.pgen.1005030
Summary
Keywords
innate immunity, antimicrobial peptide (AMP), molecular evolution, population genetics, diptericin, drosophila, diptera, attacin
Citation
Hanson MA, Lemaitre B and Unckless RL (2019) Dynamic Evolution of Antimicrobial Peptides Underscores Trade-Offs Between Immunity and Ecological Fitness. Front. Immunol. 10:2620. doi: 10.3389/fimmu.2019.02620
Received
21 August 2019
Accepted
22 October 2019
Published
08 November 2019
Volume
10 - 2019
Edited by
Charles Lee Bevins, University of California, Davis, United States
Reviewed by
Abdelaziz Heddi, Institut National des Sciences Appliquées de Lyon (INSA Lyon), France; Neal Silverman, Worcester Foundation for Biomedical Research, United States
Updates
Copyright
© 2019 Hanson, Lemaitre and Unckless.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark A. Hanson mark.hanson@epfl.chRobert L. Unckless unckless@ku.edu
This article was submitted to Microbial Immunology, a section of the journal Frontiers in Immunology
†Mark A. Hanson orcid.org/0000-0002-6125-3672
Bruno Lemaitre orcid.org/0000-0001-7970-1667
Robert L. Unckless orcid.org/0000-0001-8586-7137
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.