- 1Pediatric Gastroenterology Unit, Edmond and Lily Safra Children's Hospital, Sheba Medical Centre, Ramat Gan, Israel
- 2Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel
- 3Pediatric Department A, Edmond and Lily Safra Children's Hospital, Sheba Medical Centre, Ramat Gan, Israel
- 4Immunology Service, Edmond and Lily Safra Children's Hospital, Sheba Medical Centre, Ramat Gan, Israel
- 5Jeffrey Modell Foundation Center, Edmond and Lily Safra Children's Hospital, Sheba Medical Centre, Ramat Gan, Israel
- 6Pediatric Gastroenterology Unit, Soroka University Medical Center, Ben Gurion University of the Negev, Be'er Sheva, Israel
- 7Department of Gastroenterology, Great Ormond Street Hospital, London, United Kingdom
- 8Translational Gastroenterology Unit, Nuffield Department of Experimental Medicine, John Radcliffe Hospital, University of Oxford, Oxford, United Kingdom
- 9Department of Pediatrics, University of Oxford, Oxford, United Kingdom
- 10NIHR Oxford Biomedical Research Centre, University of Oxford, Oxford, United Kingdom
- 11Division of Gastroenterology, Hepatology and Nutrition, Boston Children's Hospital, Boston, MA, United States
- 12Harvard Medical School, Boston, MA, United States
Patients with loss-of-function mutations in IL10 or IL10 receptor (IL10R) genes develop severe, medical-refractory, infantile-onset inflammatory bowel disease (IBD). We have previously reported significant alterations in innate and adaptive immune responses in these patients. Next generation sequencing platforms enable a comprehensive assessment of T cell receptor (TCR) and B cell receptor (BCR) repertoire patterns. We aimed to characterize TCR and BCR features in peripheral blood of patients with deleterious IL10 signaling defects. DNA was isolated from blood of seven patients with IL10R mutations and one with an IL10 mutation, along with eight controls, and subjected to next generation sequencing of TRB and IgH loci. A significant increase in clonality was observed in both TCR and BCR repertoires in circulating lymphocytes of IL10/IL10R-deficient patients, but to a much greater extent in T cells. Furthermore, short CDR3β length and altered hydrophobicity were demonstrated in T cells of patients, but not in B cells, secondary to lower rates of insertions of nucleotides, but not deletions, at the V-, D-, or J-junctions. We were unable to observe specific T or B clones that were limited only to the patients or among controls. Moreover, the expanded T cells clones were unique to each patient. In conclusion, next generation sequencing of the TCR and BCR is a powerful tool for characterizing the adaptive immune cell phenotype and function in immune-mediated disorders. The oligoclonality observed among IL10/IL10R-deficient patients may suggest specialization of unique clones that likely have a role in mediating tissue damage. Nevertheless, the lack of shared clones between patients provides another piece of evidence that the adaptive immune response in IBD is not triggered against common antigens. Additional studies are required to define the specific antigens that interact with the expanded IL10/IL10R-deficient clones.
Introduction
Inflammatory bowel disease (IBD), such as Crohn's disease (CD), and ulcerative colitis (UC), are chronic, relapsing-remitting inflammatory disorders of the gastrointestinal tract. These diseases develop in genetically susceptible subjects due to a dysregulated immune response to microbial dysbiosis and/or environmental changes (1). Most cases of IBD present between the ages of 20–40 years, and in up to 20% disease manifests in childhood, mainly in adolescent years. However, in 1–2% these disorders present in the first 5–6 years of life, defined as very early-onset IBD (VEO-IBD) (2), whereas infantile-onset IBD is defined when manifestations develop before the age of 2 years.
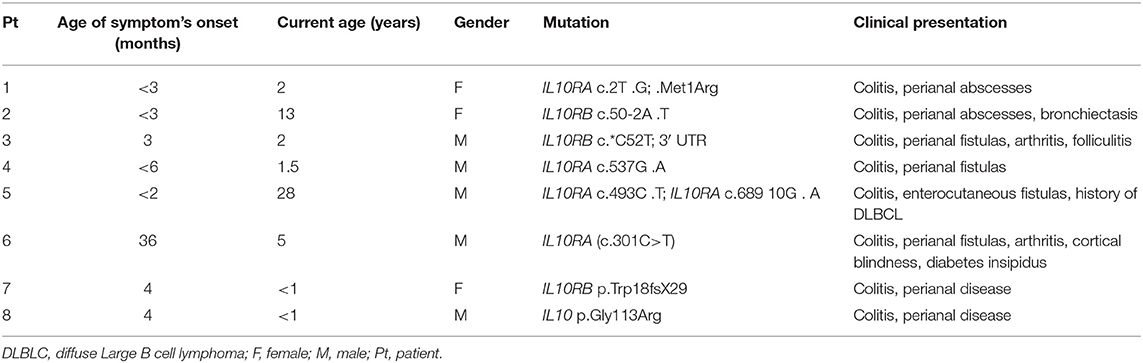
Extensive use of advanced sequencing platforms in the last decade has facilitated identification of more than 50 different monogenic disorders that directly cause IBD (3). Most of these patients develop IBD in the first years of life (3). Patients with deleterious mutations in IL10 and the IL10 receptor (IL10R) were first reported in 2009 and 2010 (4–6). These patients develop colitis and perianal disease in the first months of life, along with arthritis and folliculitis. In addition, they possess an increased risk of developing diffuse large B cell lymphoma (7, 8). The gastrointestinal disease is severe and refractory to multiple immunosuppressive medications, including steroids, thiopurines, and TNFα antagonists (5, 9). We have reported in two patients that anakinra, an IL1 receptor antagonist, was effective in ameliorating colitis (10) and was used as a bridge to hematopoietic stem cell transplantation (HSCT), which is the treatment of choice in these conditions (5, 9).
Our previous studies have elucidated important roles of IL10 in regulating different immune responses (11). IL10 is a key anti-inflammatory cytokine secreted by immune and non-immune cells. It signals through the IL10 receptor (IL10R), which is a heterotetramer composed of two sub-units of IL10Ra (IL10R1 in humans) that bind IL10 and two sub-units of IL10Rb (IL10R2) that stabilize this complex. Ligation of IL10 to its receptor leads to phosphorylation of STAT3, which translocates to the nucleus and initiates an anti-inflammatory transcriptional program (11). In innate cells, we have shown that IL10 regulates macrophage development and canonical and non-canonical inflammasome-mediated IL1β production (10, 12). In T cells we demonstrated that loss of IL10R-dependent signals enhance a TH17 phenotype, but is not required for regulatory T cells (Tregs)-mediated suppression (13). There is paucity of data on IL10R regulation of B cell responses in both mice and humans.
T and B cells express unique antigen-specific receptors that are formed following a complex rearrangement of genetic segments (14). This process involves recombination of V, D, and J gene segments, accompanied by a deletion as well as insertion of random nucleotides, generating millions of T and B cell clones that bind unique antigens. Next generation sequencing (NGS) platforms allow detailed assessment of the T and B cell receptor (TCR and BCR) repertoires at the nucleotide level (15, 16). Such a strategy permits an in-depth overview of adaptive immune cell composition and allows determining whether specific diseases are associated with expansion of distinct clones. By applying NGS on the TRB region on rectal biopsies, we have recently reported that patients with UC demonstrate oligoclonal expansion of specific clonotypes in the rectum, while the blood was relatively polyclonal (17). NGS of the TRB and IgH loci has also been used to characterize adaptive immune function in other immune-mediated disorders, such as juvenile idiopathic arthritis (18, 19), malignancies (20), and primary immunodeficiencies (21, 22).
Given the unique clinical presentation of patients with deleterious IL10/IL10R mutations, and the known alterations in adaptive immune function in these patients, we aimed to characterize T and B cell repertoire patterns in patients with IL10 signaling defects. We hypothesized that these patients possess unique T and B cell repertoire patterns.
Methods
DNA Isolation
The study was approved at each site by the local ethics committees. Informed written consent was obtained from each participating subject. Genomic DNA was extracted from blood using a commercially available kit (Wizard kit, Promega), according to the manufacturer's instructions. Up to 2 μg DNA were used for TRB and IgH repertoire library generation by Adaptive Biotechnologies platform (Seattle, USA), with a minimum input of 0.7 μg DNA. Amplified sequences were subjected to high-throughput sequencing using Illumina technology. To ensure equal depth of sequencing among the different samples, we used the Survey level (up to 500,000 reads per sample). Further details for the pipeline utilized for corrections of PCR clustering and sequencing errors can be found in previous reports (15, 23).
TREC and KREC Analysis
T-cell receptor excision circle (TREC) and kappa-deleting recombination excision circle (KREC) copy numbers were determined as described previously (24). Briefly, 50 ng genomic DNA per sample was used in quantitative real-time polymerase chain reaction (qRT–PCR). A standard curve was constructed using serial dilutions containing 103-106 copies of a known TREC or KREC plasmid. Patient and control samples were tested in triplicate, and the number of TREC or KREC copies in a given sample were calculated by comparing the obtained cycle threshold (Ct) value of the sample to the standard curve using a quantification algorithm.
TCRβ and IgH Repertoire Analysis
ImmunoSeq software was used for determination of productive clonality, clonal sharing, CDR3β, and CDR3H length, and percent productiveness. Percentage of T cells was calculated using housekeeping genes as controls to measure the total number of nucleated cells. Clonality is calculated as 1-normalized Shannon's entropy. This measures how evenly receptor sequences are distributed amongst a set of T or B cells, with values ranging from 0 to 1. Values near 1 represent samples with one or a few predominant clones (monoclonal or oligoclonal samples) dominating the observed repertoire. Clonality values near 0 represent polyclonal samples. Shannon's H, which measures the overall diversity in a given population, and takes into account the number of unique sequences (richness of the repertoire) and how evenly the sequences are distributed, was calculated using the following formulas:
R = Total templates
i = Unique rearrangements
pi = Proportion of the total sequences belonging to the “i”th unique rearrangement
Graphical presentation of the repertoire was presented using hierarchical tree maps using the Treemap software (www.treemap.com). Hydrophobicity scale was determined using a modified version of the Kyte-Doolittle index (25) of average hydrophobicity as previously described (26). Briefly, for each unique CDR, the amino acids composition at consequential positions was determined and value of hydrophobicity assigned. Average hydrophobicity at each position was calculated for all CDRs. Our calculation examined clones with length of 14 amino acids for T cells, and 18 amino acids for B cells; as these were found to be the average lengths of the CDR3β and CDR3H regions, respectively. Unless stated otherwise, all analyses were performed solely on rearrangements (defined as a single, unique nucleotide sequence) which were productive. Analysis for CDR3 length and hydrophobicity were performed on unique rearrangements (1 rearrangement per clone), in order to exclude effects of the expanded clones.
Statistical Analysis
Values are expressed as mean ± standard error of the mean (SEM). Unpaired Student's t-test was used to test for statistical significance. Significance was determined if P ≤ 0.05 (with P-value summaries as: *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001). For differential V-, D-, and J- gene usage, Bonferoni's adjustment for multiple comparisons was performed.
Results
Clinical and Genetic Characteristics of the Patients
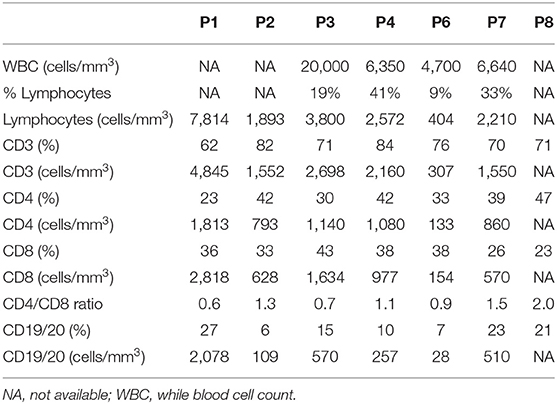
Genomic DNA was isolated from peripheral blood of seven IL10R-deficient patients and one IL10-deficient patient. All patients presented with severe colitis and perianal disease within the first months of life, expect for Patient 6 who had an atypical course and developed associated symptoms at age 3 years (Table 1). All patients were refractory to multiple immunosuppressive medications (data not shown). One patient (P5) developed diffuse large B cell lymphoma at the age of 19 years, 9 years before he was identified with IL10R deficiency. Deleterious variants in the IL10RA, IL10RB, or IL10 were identified in all patients, and presented in Table 1. As controls we obtained blood samples from eight pediatric patients referred for evaluation of abdominal pain or diarrhea, but without a diagnosis of IBD, celiac disease, or another immune-mediated disorder. Although the mean age at the time of repertoire analysis was not statistically different between the groups (10.5 ± 1.9 vs. 7.0 ± 3.3 years among controls and patients, respectively; P = 0.38), four patients were 2 years of age or younger, compared to none among controls. Basic lymphocyte immunophenotyping for seven patients, performed as part of clinical care, is presented in Table 2.
TREC and KREC Analysis in IL10/IL10R Deficient Patients
TREC are small circular DNA segments that are created in T cells during TCR rearrangement in the thymus, and thus can reflect thymic output (27). Similarly, KREC reflects B cell output from the bone marrow. TREC values were significantly lower in IL10/IL10R-deficient patients vs. controls (436 ± 158 vs. 2,302 ± 412 copies/50 ng DNA; P ≤ 0.01, Supplementary Figure 1A). Specifically, two of the older subjects (aged 13 and 30 years) in the patient's group exhibited low levels (64 and 149 copies/50 ng DNA), which may be in part to decreasing TREC values with age (28). Nevertheless, even when these two samples were excluded from analysis, TREC levels were significantly lower in patients in comparison with controls (data not shown). In B cells, KREC levels were comparable between groups (Supplementary Figure 1B). These data may suggest a defect in thymic output of T cells, but not of bone-marrow derived B cells, in patients with IL10 signaling defects.
Alterations of TCR Repertoire Features in Blood of Patients With IL10/IL10R Mutations
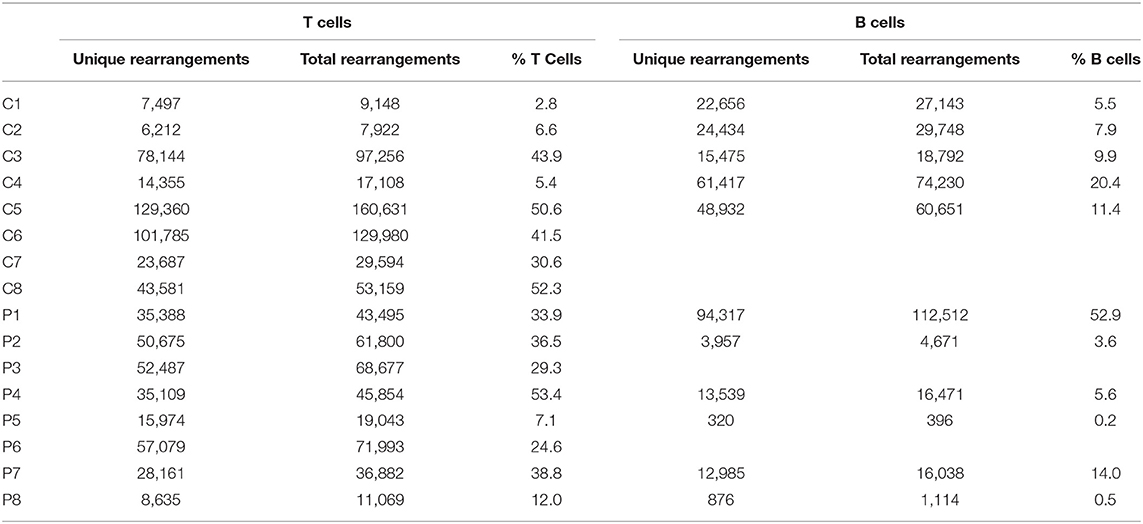
The number of total TCR rearrangements among patients and controls was comparable (44,852 ± 7,866 vs. 63,100 ± 20,893; P = 0.43); as were the unique rearrangements (35,439 ± 6,184 vs. 50,578 ± 16,635; P = 0, see Table 3). The calculated percentage of circulating T cells, based on NGS data, was also similar between groups (29.5 ± 5.3% vs. 29.2 ± 7.5% among patients and controls, respectively; P = 0.98). Next, we examined differential TRBV and TRBJ gene usage among unique rearrangements of IL10/IL10R-deficient patients, in comparison to controls. TRBV and TRBJ gene usage was similar between the groups, and specific patterns of gene usage could not be observed (Figures 1A,B).
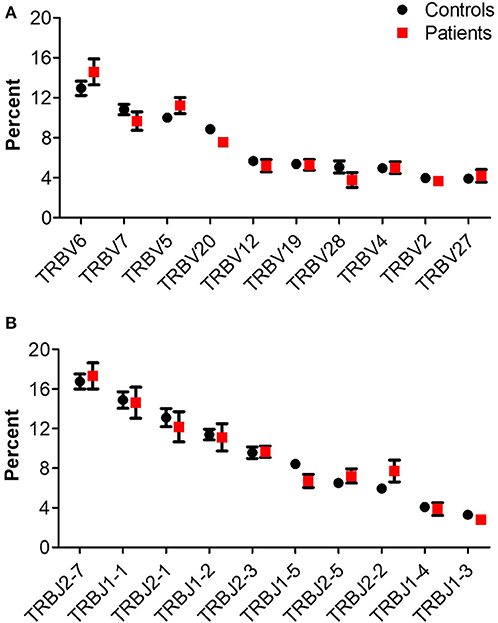
Figure 1. TRB gene usage in patients with IL10/IL10R defects and controls. Comparison of usage of the 10 most frequently used (A) TRBV and (B) TRBJ genes between control and patient groups.
For a broad overview of the overall repertoire, we performed Treemap analysis. In these images, each square represents a specific amino acid sequence, and the size correlates with its frequency. Marked clonal expansion was observed in patients with IL10 signaling defects, compared with controls (Figure 2A; Supplementary Figure 2). In Table 4 we present the three most expanded clones in each of the patients, and their respective gene family. The cumulative frequency of the top 100 most frequent clones was significantly higher among patients vs. controls (24.2 ± 4.2 vs. 14.1 ± 1.5%; P ≤ 0.001, Figure 2B). Furthermore, the calculated clonality was significantly higher in the blood of IL10/IL10R-deficient patients (Figure 2C) and inversely, Shannon's H diversity index was significantly lower (Figure 2D), compared with controls.
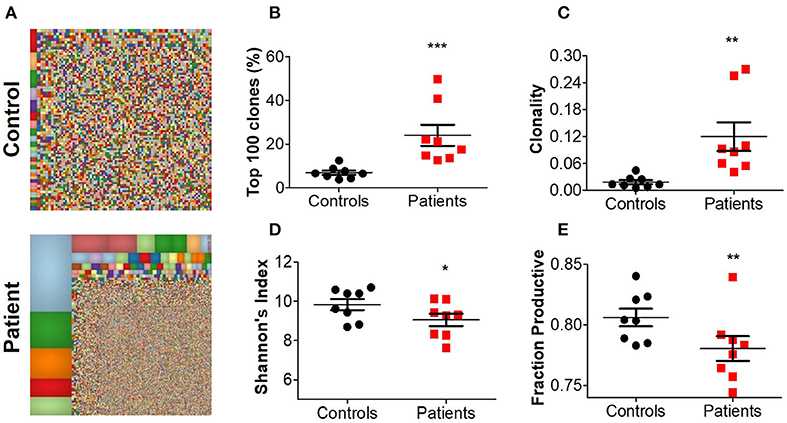
Figure 2. Aberrant TCR repertoire features in blood from patients with IL10 signaling defects. (A) Representative Treemaps of entire TCR repertoire in a control (C1) and an IL10R-deficient patient (P2). (B) Cumulative percentage of the 100 most abundant clones from the total repertoire, (C) calculated clonality, (D) Shannon's diversity index, and (E) fraction productiveness in control and patient groups. Bars represent mean ± SEM.
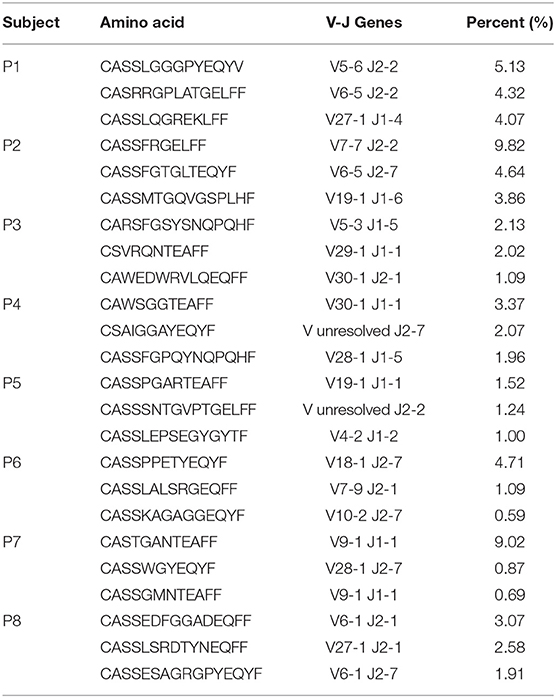
Table 4. The amino-acid sequence, and corresponding TRBV-TRBJ pair, of the three most expanded T cell clones for each of the patients.
Next, we assessed the fraction of productive clones in each subject. Since a T-cell undergoes random somatic rearrangement, the outcome can be a non-productive allele (for example, generation of stop codon, or out-of-frame sequence). Calculation of the fraction of productive TCR sequences in each sample showed significantly lower productiveness among patients, compared with controls (78.0 ± 1.0 vs. 80.6 ± 0.7%; P ≤ 0.01, Figure 2E). Overall, these results suggest that patients with IL10/IL10R mutations exhibit clonal expansion of specific T cell clones, which may have a role in mediating the clinical phenotype.
Next, we assessed different features of the CDR3β, which is the most variable region of the TCR and thus can affect its antigenic binding affinity. The length of the CDR3β loop itself is important in determining T cell specificity, as longer CDR3β regions have a greater potential for sequence variation and can potentially reach into narrow antigenic pockets (29). The mean CDR3β length in peripheral blood of the patients was significantly lower compared to controls (43.4 ± 0.08 vs. 43.8 ± 0.07 nucleotides, respectively; P ≤ 0.001, Figure 3A). This statistically significant difference was maintained at the amino acid level as well (14.48 ± 0.02 vs. 14.59 ± 0.03 in patients and controls, respectively; P ≤ 0.01). A sub-analysis revealed similar results when comparing only productive clones (43.4 ± 0.06 vs. 43.8 ± 0.06 nucleotides in patients and controls, respectively; P ≤ 0.01), while the length of non-productive clones was comparable between the groups (Figure 3B). These differences in CDR3β length can be partially explained by a significantly lower rate of random insertions of nucleotides, but not deletions, at the V-, D-, or J-junctions, among patients with IL10/IL10R defects (Figure 3C). Interestingly, the length of the non-productive clones was significantly higher compared with the productive clones in both groups (Figure 3B).
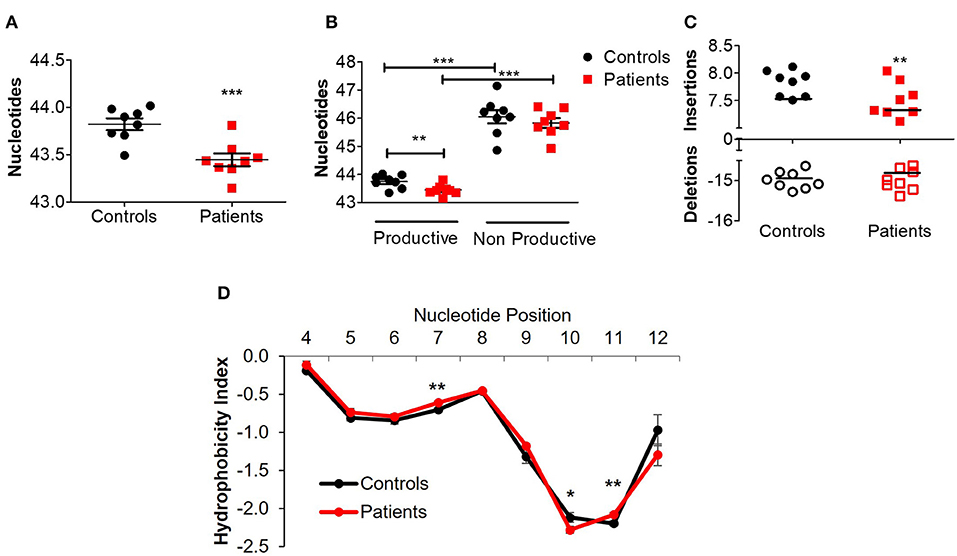
Figure 3. Aberrant TCRβ length and hydrophobicity in T cell lymphocytes of patients with IL10/IL10R defects. (A) Mean CDR3β length in circulating lymphocytes of controls and patients. (B) Sub-analysis of CDR3β length according to productive and non-productive clones. (C) Analysis of the number of nucleotide insertions and deletions in total clones. (D) Hydrophobicity index of the CDR3β region. Position signifies successive amino acid location at the CDR3β region. Bars represent mean ± SEM.
Another important biochemical property of the TCR is its hydrophobicity, which could promote development of self-reactive clones (30) and affects antigenic recognition (31), since intrinsic hydrophobicity of specific residues influences binding properties. As shown in Figure 3D, for CDR3β with length of 14 amino acids (the most frequent length), significant differences in hydrophobicity were evident between patients and controls at residue positions 7, 10, and 11. Residues 7 and 11 were significantly more hydrophobic in patients, while residue 10 was significantly less hydrophobic. Examination of hydrophobicity for CDR3β with lengths of either 13 or 15 amino acids, showed no differences between patients and controls (data not shown). Overall, these results may suggest biochemical property differences of the CDR3β in T cells from patients with IL10 signaling defects, which can consequently affect their function.
BCR Repertoire of IL10/IL10R-Deficient Patients
NGS of the IgH genetic segments was performed on blood samples of 6 patients with IL10/IL10R deficiency and five controls. One of the patients (P1), a 2 years old female with an IL10RA mutation with severe colitis and perianal disease, but without any signs of lymphoma, displayed a very high percentage of B cells (52.9%) and total rearrangements (112,512). These values were markedly elevated compared with the other patients or controls examined, and therefore were excluded from analysis of frequency and number of rearrangements. The frequency of B cells was slightly decreased in the patient group (4.8 ± 2.5 vs. 11.0 ± 2.5% in patients and controls, respectively; P = 0.12). This is also reflected by their lower numbers of total (7,738 ± 3,552 vs. 42,113 ± 10,715 rearrangements among patients and controls, respectively; P = 0.02) and unique (6,335 ± 2,896 vs. 34,583 ± 8,764 rearrangements among patients and controls, respectively; P = 0.02, see Table 3). We next looked at gene usage among unique rearrangements and found that frequency of IGHV, IGHJ, and IGHD gene groups were overall similar between groups (Figures 4A–C).
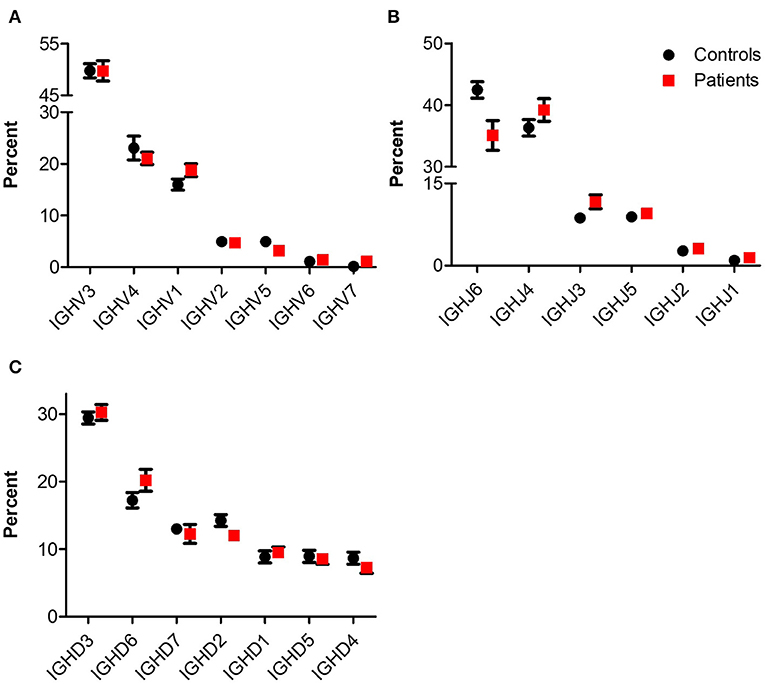
Figure 4. IgH gene usage in patients with IL10/IL10R defects. Comparison of usage of (A) IGHV, (B) IGHJ, and (C) IGHD genes between control and patient groups.
Treemap images demonstrated the lower number of rearrangements among patients (Figure 5A; Supplementary Figure 3). Comparable cumulative frequencies of the top 100 most common clones were observed in both groups, despite a trend for a higher frequency among patients (9.5 ± 5.1 vs. 1.1 ± 0.1%. P = 0.08; Figure 5B). The calculated clonality was significantly higher (though still much lower than for T cells, Figure 5C), while Shannon's diversity index was significantly lower (Figure 5D), in patients compared with controls. No difference in productiveness was found between the groups (Figure 5E). Last, the mean CDR3H length (Figure 6A) and hydrophobicity of CDR3H with length of 18 amino acids (Figure 6B) were comparable between groups. Collectively, these results suggest that although some changes in IgH repertoire features were observed in patients with IL10/IL10R defects in the blood, these were much less prominent compared to alterations in TCRβ repertoire.
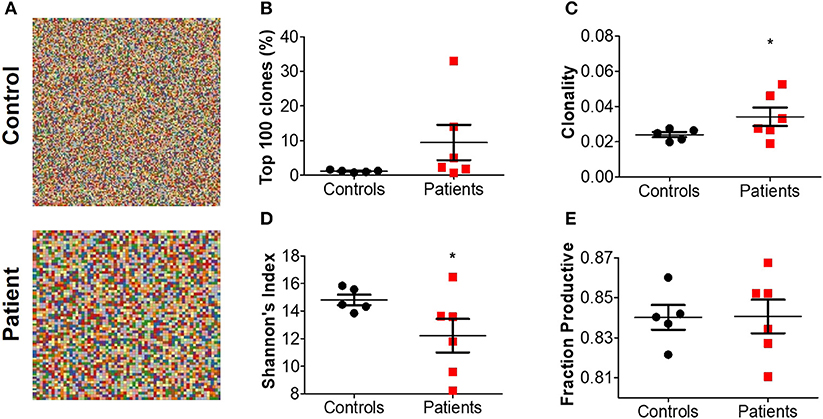
Figure 5. Increased clonality of BCR repertoire in blood from patients with IL10 signaling defects. (A) Representative Treemaps of entire BCR repertoire in a representative control (C1) and IL10R-deficient patient (P2). (B) Cumulative percentage of the 100 most abundant clones from the total repertoire, (C) calculated clonality, (D) Shannon's diversity index, and (E) fraction productiveness in control and patient groups. Bars represent mean ± SEM.
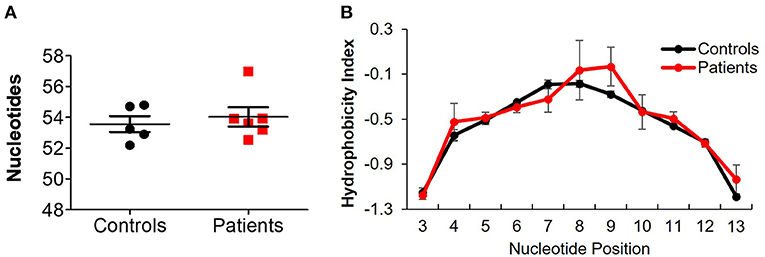
Figure 6. Comparable IgH length and hydrophobicity between controls and IL10/IL10R-deficient patients. (A) CDR3H length in blood of patients and controls. (B) Hydrophobicity index of the CDR3H region. Position signifies successive amino acid location at the CDR3β region. Bars represent mean ± SEM.
Degree of Clonal Sharing Between Controls and Between Patients
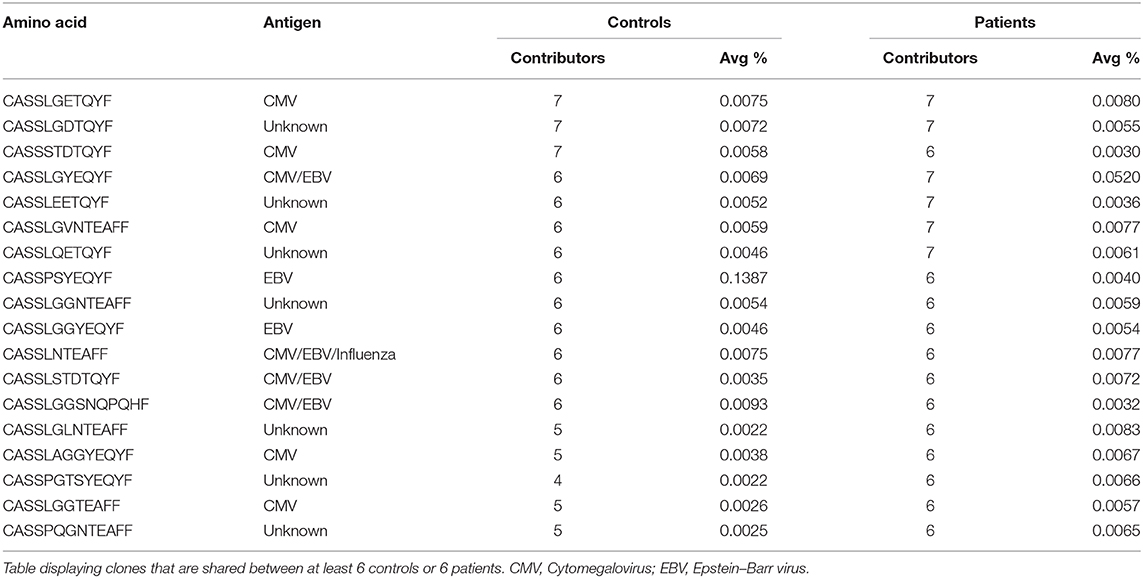
Our previous studies among patients with UC demonstrated sharing of specific T cell clones in the gut (but not in the blood), although they were also identified in healthy subjects, but to a lesser extent (17). Such sharing may point to unique clones that have an important role in the pathogenesis of this disorder. In contrast to UC patients, in our IL10/IL10R-deficient patient cohort, there were no unique blood-borne T or B cell clones shared exclusively either among the IL10/IL10R-deficient patients or among the controls. Interestingly, the 10 most expanded T cell clones in each IL10/IL10R-deficient patient were unique, and not identified in other patients, or publicly available databases, such as VDJdb. We did identify public T cell clones (most of them directed against viral antigens) shared between controls and patients, although their frequency was very low (Table 5).
We next calculated the Jaccard index, reflecting degree of clonal sharing between any two individuals (Figures 7A,B). Overall, clonal sharing in the blood was low in both groups, reflecting the heterogenic circulating repertoire. The mean degree of T cell sharing was comparable between groups (2.11 ± 0.25 vs. 1.70 ± 0.24% among patients and controls, respectively; P = 0.12, see Figure 7A). However, sharing of clones was lower in B cells of patients vs. controls (0.03 ± 0.01 vs. 0.14 ± 0.02%, P ≤ 0.001; Figure 7B). Looking at the nucleotide level, nearly no sharing was identified in both groups for T and B cells (data not shown).
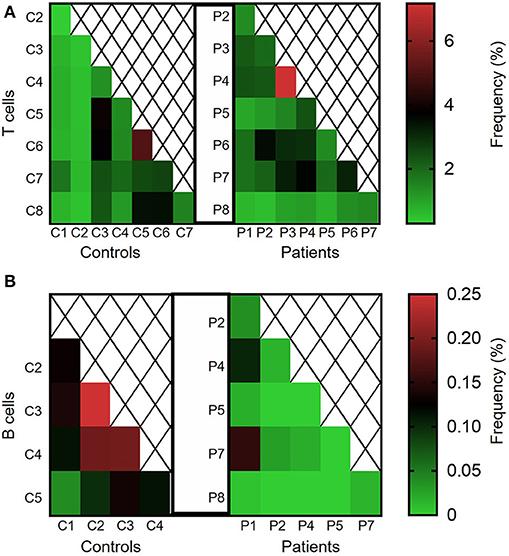
Figure 7. Clonal sharing between controls and between different IL10/IL10R-deficient patients. Degree of clonal sharing is presented in heat maps images, depicting percentage of the (A) TCR and (B) BCR repertoires shared between any two individuals.
Discussion
Patients with rare deleterious mutations in the IL10 axis develop a unique infantile-onset form of IBD. As such, we and others have previously characterized various dysregulated immune responses in these patients in both innate and adaptive immunity (10, 12, 32–36). We were able to show that loss of IL10 signaling does not impair the suppressive capacity of Tregs, but leads to enhanced generation of TH17 cells and upregulation of IL1-mediated responses (13). Our current study provides another aspect of adaptive immunity in these patients.
Using NGS of the TCRβ and IgH, we were able to deeply characterize T and B cell repertoire features in a relatively large cohort of rare patients with IL10 and IL10R mutations. One of the key findings in our study is marked clonal expansion, mainly in T cells, and to a lesser extent in B cells, in these patients. Upregulation of specific clones is observed in various immune-mediated disorders, primary immunodeficiencies, and malignancies. In our previous study, we have shown that patients with UC also exhibit an oligoclonal TCRβ repertoire; nevertheless, this was observed only in the inflamed colon, but not in the blood (17). This makes our findings on IL10/IL10R-deficient patients even more striking, since in circulating blood there are high frequencies of naïve T cells that likely have a polyclonal distribution. It will be interesting to look in these patients at repertoire features of specific immune subsets, including effector memory, which acquire a TH17 phenotype, and Tregs, which seem to function normally based on our previous observations (13).
A skewed TCRβ repertoire in the blood was previously reported in different primary immunodeficiencies, including Ras-associated lymphoproliferative disease (RALD) (24) and wiskott-Aldrich syndrome (WAS) (18) which demonstrated significant restriction of the TCRβ repertoire. These disorders are associated with recurrent infections and result from mutations in genes critical for normal immune development and function. Patients with deleterious IL10/IL10R mutations are generally not more susceptible to infections, but are still considered to have an immunodeficiency. These abnormal repertoire features, which are not seen in the blood of patients with UC (17) or CD (37), are another reflection of the important role of IL10 in mediating immune responses.
Another altered TCRβ repertoire feature we show in patients with IL10 signaling defects is a significantly shorter CDR3β length and changes in hydrophobicity. Abnormalities in CDR3β length were previously described in primary immunodeficiencies, including in patients with WAS and recombination-activating genes (RAG) mutations, where a correlation between length and severity of phenotype was demonstrated (18, 38). Alterations in distribution of the CDR3β length were also described in autoimmune disorders, including insulin-dependent diabetes mellitus (39), and multiple sclerosis (40). Although the differences in CDR3β appear minor, they might still have functional consequences. For example, in insulin-dependent diabetes mellitus a slight, yet significant decrease in CDR3β length may be associated an increased potential for self-recognition (39). The decreased CDR3β length we describe was demonstrated only in productive clones of patients with IL10 defects (representing >75% of the total repertoire). If shorter CDR3β length were genetically determined in the patients (due to IL10 regulation of thymic selection, for example), we would expect to detect shorter CDRs in the non-productive clones as well, implying that other triggers may be responsible for the reduced CDR3β lengths. Thus, together with the aberrant hydrophobicity of the CDR3β, as hydrophobic residues are thought to form high affinity binding “hotspots” to antigen (41, 42), the characterization of the CDR3β suggests a defect in proper antigen recognition in the TCR repertoire of IL10/IL10R-deficient patients.
Findings of the IgH repertoire features were relatively comparable between groups, despite increased clonality among IL10/IL10R-deficient patients. A trend toward the focus on B cell immune repertoire is important, given the high risk of these patients to develop EBV-negative diffuse large B cell lymphoma (7, 8). The mechanisms leading to lymphoma in these conditions are unclear but might be related to impaired immune surveillance by cytotoxic CD8+ T cells (43). One patient in our cohort had a history of lymphoma, 9 years before the diagnosis of IL10R deficiency was made, and blood was obtained for the current study. While we could not identify a specific clone that was markedly upregulated in the blood of these patients, clonal expansion might occur at other sites or at later stages. In the IL10/IL10R-deficient patients lymphoma developed in the intestine, liver and/or spleen (7, 8), and therefore unique B cell signatures might only be identified at that specific sites and not in the blood.
Clonal sharing of specific T and B cell clonotypes was very low in our cohort, both between patients and between controls. In addition, we were unable to identify unique sequences limited to one of the groups (patients or controls). Although our sample size was relatively small, this may suggest that IL10/IL10R deficiency is not triggered by common antigens, and are in line with microbiome studies showing large heterogeneity among IBD patients (44). Nevertheless, sharing of clones might be evident at mucosal surfaces. Additional studies are required to identify specific microbial triggers that stimulate T cells in these patients and lead to clonal expansion.
Our study has several limitations. First, we studied repertoire features of total T and B lymphocytes. While this provides a broad and important overview, it fails to capture changes is specific subsets, especially those that are less frequent, such as Tregs. Analysis of these characteristics in sorted immune populations or in single-cell RNAseq coupled to TCRβ repertoire should provide more specific information. Second, usage of RNA for immune repertoire studies, instead of DNA, would enable detection of somatic hypermutations and isotype switching in B cells. In addition, usage of primers for longer reads within the entire V-region would also allow analysis of somatic hypermutations in B cells. Third, comparisons were performed between patients and controls without IBD or other immune-mediated disorder. While the patients' repertoire features were significantly different from those we reported for UC patients (17), a comparison to a group of patients with a systemic inflammatory phenotype, including those with CD would be valuable, but this was beyond the scope of this study. Another potential limitation is the age discrepancy between control and patient groups, as four of the patients were 2 years old or younger. Britanova et al. reported a decrease in TCR repertoire diversity with age; nevertheless, the youngest subject in that study was 6 years of age, and most participants were adult (45). We have recently reported that the intestinal B and T cell receptor repertoire among infants is diverse, similar to pediatric adolescent subjects (46). Although we did not assess repertoire of circulating blood-borne lymphocytes, we can speculate it is also diverse in the first year of life. Finally, immune repertoire assessment of intestinal samples and analysis of autologous blood-intestinal clones of these IL10/IL10R-deficient patients would be interesting; however, obtaining research biopsies from these young, sick and rare patients is challenging.
In conclusion, we reveal that both the T and B cell receptor repertoires are skewed in patients lacking intact IL10 signaling and are characterized by enhanced clonality, and alterations in several repertoire features, mostly in T cells. Given the severe and unique inflammatory phenotype of these patients, additional studies are required to elucidate the dysfunctional adaptive arm. Last, this study is another proof-of-concept that NGS is a powerful tool for in-depth analysis of adaptive immune function.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by Sheba Medical Center, Boston Children's Hospital and NIHR Oxford Biomedical Research Center. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
LW designed the study, conducted the analysis, and wrote the manuscript. YL, ER, AL, BY, and GL contributed to data analysis. NS, HU, BW, RS, and SS contributed to acquisition of samples. DS designed the study, conducted the analysis, and wrote the manuscript.
Funding
This study was funded by a Specific Defect Research Program grant of the Jeffrey Modell Foundation. DS, HU, and SS are supported by the Leona M. and Harry B. Helmsley Charitable Trust. HU was also supported by the Oxford Biomedical Research Center (BRC). Finally, SS is also supported by the Wolpow Chair in IBD Research and Treatment and the Translational Research Program (Boston Children's Hospital).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the patients and their families that participated in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.00109/full#supplementary-material
References
1. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. (2011) 474:307–17. doi: 10.1038/nature10209
2. Moran CJ, Klein C, Muise AM, Snapper SB. Very early-onset inflammatory bowel disease: gaining insight through focused discovery. Inflamm Bowel Dis. (2015) 21:1166–75. doi: 10.1097/MIB.0000000000000329
3. Uhlig HH, Schwerd T, Koletzko S, Shah N, Kammermeier J, Elkadri A, et al. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology. (2014) 147:990–1007 e3. doi: 10.1053/j.gastro.2014.07.023
4. Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. (2009) 361:2033–45. doi: 10.1056/NEJMoa0907206
5. Kotlarz D, Beier R, Murugan D, Diestelhorst J, Jensen O, Boztug K, et al. Loss of interleukin-10 signaling and infantile inflammatory bowel disease: implications for diagnosis and therapy. Gastroenterology. (2012) 143:347–55. doi: 10.1053/j.gastro.2012.04.045
6. Glocker EO, Frede N, Perro M, Sebire N, Elawad M, Shah N, et al. Infant colitis–it's in the genes. Lancet. (2010) 376:1272. doi: 10.1016/S0140-6736(10)61008-2
7. Neven B, Mamessier E, Bruneau J, Kaltenbach S, Kotlarz D, Suarez F, et al. A Mendelian predisposition to B-cell lymphoma caused by IL-10R deficiency. Blood. (2013) 122:3713–22. doi: 10.1182/blood-2013-06-508267
8. Shouval DS, Ebens CL, Murchie R, McCann K, Rabah R, Klein C, et al. Large B-cell lymphoma in an adolescent patient with interleukin-10 receptor deficiency and history of infantile inflammatory bowel disease. J Pediatric Gastroenterol Nutr. (2016) 63:e15–7. doi: 10.1097/MPG.0000000000000532
9. Engelhardt KR, Shah N, Faizura-Yeop I, Kocacik Uygun DF, Frede N, Muise AM, et al. Clinical outcome in IL-10- and IL-10 receptor-deficient patients with or without hematopoietic stem cell transplantation. J Allergy Clin Immunol. (2013) 131:825–30. doi: 10.1016/j.jaci.2012.09.025
10. Shouval DS, Biswas A, Kang YH, Griffith AE, Konnikova L, Mascanfroni ID, et al. Interleukin 1beta mediates intestinal inflammation in mice and patients with interleukin 10 receptor deficiency. Gastroenterology. (2016) 151:1100–4. doi: 10.1053/j.gastro.2016.08.055
11. Shouval DS, Ouahed J, Biswas A, Goettel JA, Horwitz BH, Klein C, et al. Interleukin 10 receptor signaling: master regulator of intestinal mucosal homeostasis in mice and humans. Adv Immunol. (2014) 122:177–210. doi: 10.1016/B978-0-12-800267-4.00005-5
12. Shouval DS, Biswas A, Goettel JA, McCann K, Conaway E, Redhu NS, et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. (2014) 40:706–19. doi: 10.1016/j.immuni.2014.03.011
13. Shouval DS, Konnikova L, Griffith AE, Wall SM, Biswas A, Werner L, et al. Enhanced TH17 responses in patients with IL10 receptor deficiency and infantile-onset IBD. Inflamm Bowel Dis. (2017) 23:1950–61. doi: 10.1097/MIB.0000000000001270
14. Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. (2002) 109 (Suppl):S45–55. doi: 10.1016/S0092-8674(02)00675-X
15. Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. (2013) 4:2680. doi: 10.1038/ncomms3680
16. Six A, Mariotti-Ferrandiz ME, Chaara W, Magadan S, Pham HP, Lefranc MP, et al. The past, present, and future of immune repertoire biology - the rise of next-generation repertoire analysis. Front Immunol. (2013) 4:413. doi: 10.3389/fimmu.2013.00413
17. Werner L, Nunberg MY, Rechavi E, Lev A, Braun T, Haberman Y, et al. Altered T cell receptor beta repertoire patterns in pediatric ulcerative colitis. Clin Exp Immunol. (2019) 196:1–11. doi: 10.1111/cei.13247
18. O'Connell AE, Volpi S, Dobbs K, Fiorini C, Tsitsikov E, de Boer H, et al. Next generation sequencing reveals skewing of the T and B cell receptor repertoires in patients with wiskott-Aldrich syndrome. Front Immunol. (2014) 5:340. doi: 10.3389/fimmu.2014.00340
19. Henderson LA, Volpi S, Frugoni F, Janssen E, Kim S, Sundel RP, et al. Next-generation sequencing reveals restriction and clonotypic expansion of treg cells in juvenile idiopathic arthritis. Arthr Rheum. (2016) 68:1758–68. doi: 10.1002/art.39606
20. Ye B, Smerin D, Gao Q, Kang C, Xiong X. High-throughput sequencing of the immune repertoire in oncology: applications for clinical diagnosis, monitoring, and immunotherapies. Cancer Lett. (2018) 416:42–56. doi: 10.1016/j.canlet.2017.12.017
21. Ghraichy M, Galson JD, Kelly DF, Trück J. B-cell receptor repertoire sequencing in patients with primary immunodeficiency: a review. Immunology. (2018) 153:145–60. doi: 10.1111/imm.12865
22. Wong GK, Heather JM, Barmettler S, Cobbold M. Immune dysregulation in immunodeficiency disorders: the role of T-cell receptor sequencing. J Autoimmun. (2017) 80:1–9. doi: 10.1016/j.jaut.2017.04.002
23. Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. (2009) 114:4099–107. doi: 10.1182/blood-2009-04-217604
24. Levy-Mendelovich S, Lev A, Rechavi E, Barel O, Golan H, Bielorai B, et al. T and B cell clonal expansion in Ras-associated lymphoproliferative disease (RALD) as revealed by next-generation sequencing. Clin Exp Immunol. (2017) 189:310–7. doi: 10.1111/cei.12986
25. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. (1982) 157:105–32. doi: 10.1016/0022-2836(82)90515-0
26. Rechavi E, Lev A, Lee YN, Simon AJ, Yinon Y, Lipitz S, et al. Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Sci Transl Med. (2015) 7:276ra25. doi: 10.1126/scitranslmed.aaa0072
27. Hazenberg MD, Verschuren MC, Hamann D, Miedema F, van Dongen JJ. T cell receptor excision circles as markers for recent thymic emigrants: basic aspects, technical approach, and guidelines for interpretation. J Mol Med. (2001) 79:631–40. doi: 10.1007/s001090100271
28. Cho S, Ge J, Seo SB, Kim K, Lee HY, Lee SD. Age estimation via quantification of signal-joint T cell receptor excision circles in Koreans. Leg Med. (2014) 16:135–8. doi: 10.1016/j.legalmed.2014.01.009
29. Hou X, Wang M, Lu C, Xie Q, Cui G, Chen J, et al. Analysis of the repertoire features of TCR beta chain CDR3 in human by high-throughput sequencing. Cell Physiol Biochem. (2016) 39:651–67. doi: 10.1159/000445656
30. Stadinski BD, Shekhar K, Gómez-Touriño I, Jung J, Sasaki K, Sewell AK, et al. Hydrophobic CDR3 residues promote the development of self-reactive T cells. Nat Immunol. (2016) 17:946–55. doi: 10.1038/ni.3491
31. Chowell D, Krishna S, Becker PD, Cocita C, Shu J, Tan X, et al. TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc Natl Acad Sci USA. (2015) 112:E1754–62. doi: 10.1073/pnas.1500973112
32. Zigmond E, Bernshtein B, Friedlander G, Walker CR, Yona S, Kim KW, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. (2014) 40:720–33. doi: 10.1016/j.immuni.2014.03.012
33. Li B, Gurung P, Malireddi RK, Vogel P, Kanneganti TD, Geiger TL. IL-10 engages macrophages to shift Th17 cytokine dependency and pathogenicity during T-cell-mediated colitis. Nat Commun. (2015) 6:6131. doi: 10.1038/ncomms7131
34. Li B, Alli R, Vogel P, Geiger TL. IL-10 modulates DSS-induced colitis through a macrophage-ROS-NO axis. Mucosal Immunol. (2014) 7:869–78. doi: 10.1038/mi.2013.103
35. Girard-Madoux MJ, Ober-Blöbaum JL, Costes LM, Kel JM, Lindenbergh-Kortleve DJ, Brouwers-Haspels I, et al. IL-10 control of CD11c+ myeloid cells is essential to maintain immune homeostasis in the small and large intestine. Oncotarget. (2016) 7:32015–30. doi: 10.18632/oncotarget.8337
36. Veenbergen S, Li P, Raatgeep HC, Lindenbergh-Kortleve DJ, Simons-Oosterhuis Y, Farrel A, et al. IL-10 signaling in dendritic cells controls IL-1beta-mediated IFNgamma secretion by human CD4(+) T cells: relevance to inflammatory bowel disease. Muc Immunol. (2019) 12:1201–11. doi: 10.1038/s41385-019-0194-9
37. Chapman CG, Yamaguchi R, Tamura K, Weidner J, Imoto S, Kwon J, et al. Characterization of T-cell receptor repertoire in inflamed tissues of patients with crohn's disease through deep sequencing. Inflamm Bowel Dis. (2016) 22:1275–85. doi: 10.1097/MIB.0000000000000752
38. Lee YN, Frugoni F, Dobbs K, Tirosh I, Du L, Ververs FA, et al. Characterization of T and B cell repertoire diversity in patients with RAG deficiency. Sci Immunol. (2016) 1:eaah6109. doi: 10.1126/sciimmunol.aah6109
39. Gomez-Tourino I, Kamra Y, Baptista R, Lorenc A, Peakman M. T cell receptor beta-chains display abnormal shortening and repertoire sharing in type 1 diabetes. Nat Commun. (2017) 8:1792. doi: 10.1038/s41467-017-01925-2
40. Laplaud DA, Ruiz C, Wiertlewski S, Brouard S, Berthelot L, Guillet M, et al. Blood T-cell receptor beta chain transcriptome in multiple sclerosis. Characterization of the T cells with altered CDR3 length distribution. Brain. (2004) 127:981–95. doi: 10.1093/brain/awh119
41. Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. (2005) 437:640–7. doi: 10.1038/nature04162
42. Baker BM, Gagnon SJ, Biddison WE, Wiley DC. Conversion of a T cell antagonist into an agonist by repairing a defect in the TCR/peptide/MHC interface: implications for TCR signaling. Immunity. (2000) 13:475–84. doi: 10.1016/S1074-7613(00)00047-9
43. Mumm JB, Emmerich J, Zhang X, Chan I, Wu L, Mauze S, et al. IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer Cell. (2011) 20:781–96. doi: 10.1016/j.ccr.2011.11.003
44. Sun Y, Li L, Xia Y, Li W, Wang K, Wang L, et al. The gut microbiota heterogeneity and assembly changes associated with the IBD. Scient Rep. (2019) 9:440. doi: 10.1038/s41598-018-37143-z
45. Britanova OV, Putintseva EV, Shugay M, Merzlyak EM, Turchaninova MA, Staroverov DB, et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J Immunol. (2014) 192:2689–98. doi: 10.4049/jimmunol.1302064
Keywords: IBD, VEOIBD, NGS, T cell receptor repertoire, B cell receptor repertoire, IL10, IL10R, adaptive immunity
Citation: Werner L, Lee YN, Rechavi E, Lev A, Yerushalmi B, Ling G, Shah N, Uhlig HH, Weiss B, Somech R, Snapper SB and Shouval DS (2020) Alterations in T and B Cell Receptor Repertoires Patterns in Patients With IL10 Signaling Defects and History of Infantile-Onset IBD. Front. Immunol. 11:109. doi: 10.3389/fimmu.2020.00109
Received: 03 October 2019; Accepted: 15 January 2020;
Published: 06 February 2020.
Edited by:
Yu Lung Lau, The University of Hong Kong, Hong KongReviewed by:
Xiao Liu, Beijing Genomics Institute (BGI), ChinaHanna IJspeert, Erasmus University Rotterdam, Netherlands
Katherine J. L. Jackson, Garvan Institute of Medical Research, Australia
Copyright © 2020 Werner, Lee, Rechavi, Lev, Yerushalmi, Ling, Shah, Uhlig, Weiss, Somech, Snapper and Shouval. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dror S. Shouval, ZHJvci5zaG91dmFsQGdtYWlsLmNvbQ==
 Lael Werner
Lael Werner Yu Nee Lee
Yu Nee Lee Erez Rechavi2,3,4,5
Erez Rechavi2,3,4,5 Baruch Yerushalmi
Baruch Yerushalmi Batia Weiss
Batia Weiss Raz Somech
Raz Somech Scott B. Snapper
Scott B. Snapper Dror S. Shouval
Dror S. Shouval