- The Jenner Institute, University of Oxford, Oxford, United Kingdom
Plasmodium spp.-infected mosquitos inject sporozoites into the skin of a mammalian host during a blood meal. These enter the host's circulatory system and establish an infection in the liver. After a silent metamorphosis, merozoites invade the blood leading to the symptomatic and transmissible stages of malaria. The silent pre-erythrocytic malaria stage represents a bottleneck in the disease which is ideal to block progression to clinical malaria, through chemotherapeutic and immunoprophylactic interventions. RTS,S/AS01, the only malaria vaccine close to licensure, although with poor efficacy, blocks the sporozoite invasion mainly through the action of antibodies against the CSP protein, a major component of the pellicle of the sporozoite. Strikingly, sterile protection against malaria can be obtained through immunization with radiation-attenuated sporozoites, genetically attenuated sporozoites or through chemoprophylaxis with infectious sporozoites in animals and humans, but the deployability of sporozoite-based live vaccines pose tremendous challenges. The protection induced by sporozoites occurs in the pre-erythrocytic stages and is mediated mainly by antibodies against the sporozoite and CD8+ T cells against peptides presented by MHC class I molecules in infected hepatocytes. Thus, the identification of malaria antigens expressed in the sporozoite and liver-stage may provide new vaccine candidates to be included, alone or in combination, as recombinant protein-based, virus-like particles or sub-unit virally-vectored vaccines. Here I review the efforts being made to identify Plasmodium falciparum antigens expressed during liver-stage with focus on the development of parasite, hepatocyte, mouse models, and resulting rate of infection in order to identify new vaccine candidates and to improve the efficacy of the current vaccines. Finally, I propose new approaches for the identification of liver-stage antigens based on immunopeptidomics.
Introduction
According to the latest WHO report, around 219 million clinical episodes of malaria were reported in 91 countries, most of these occurring in sub-Saharan African countries, representing a decrease of 1 million cases over the previous year. 435,000 deaths were registered in 2017 due to malaria, identical to the numbers of reported deaths in 2015. Dramatically, most of these deaths are African children under 5 years of age (1). Increasing parasite drug resistance and mosquito insecticide resistance threatens to lead to worse control and higher mortality in the coming years (2). Therefore, the control and eventual eradication of this disease relies on the development of a highly effective malaria vaccine. The vaccine RTS,S/AS01, the first ever malaria vaccine to enter Phase III clinical trials and the first human parasitic vaccine ever created, shows modest efficacy, short durability and needs to be administrated in a four-dose schedule for maximum efficacy (3). Nevertheless, RTS,S/AS01 brings hope for the development of more efficacious vaccines. Currently there are about 20 vaccines in clinical trials based on whole organisms or very well-known antigens, reviewed elsewhere (4, 5). The Malaria Vaccine Technology Roadmap proposes two main objectives for the development of new malaria vaccines by 2030: (1) vaccines with protective efficacy of at least 75% against clinical malaria and, (2) vaccines that reduce transmission of the parasite to reduce the incidence of human malaria infection (6). Vaccination with the pre-erythrocytic malaria vaccine RTS,S induces high levels of antibodies (7), CD4+ T cells (8, 9), and CD8+ T cells (9) specific for the circumsporozoite protein (CSP). To improve the efficacy of pre-erythrocytic malaria vaccines, however, there is need to identify new vaccine candidate antigens, especially antigens able to induce strong CD8+ T cell responses.
It has been 50 years since Ruth Nussenzweig's seminal paper describing that radiation-attenuated sporozoites induce sterile protection (10), remain in the liver (11) and require CD8+ T-cell responses for protection (12) against the CSP protein (12), and a single epitope, known as Pb9, can induce protection in mice (13, 14). In the 1990's, our group has developed a CSP P. berghei vaccine that can induce sterile protection in mice, based in a Modified Vaccinia Virus viral vector (13). Protection can be induced with peptides presented by MHC Class I molecules in the mouse liver, mediated by CD8+ T cells, using viral vectors against a single antigen (13). In humans, there is an important role of CD8+ T cells in inducing protection against pre-erythrocytic malaria. It has been shown in naturally exposed individuals or volunteers vaccinated with radiation-attenuated sporozoites (RAS) that these responses are against CSP and other antigens, which have not been extensively characterized (15, 16). Moreover, it has been shown in clinical trials with ChAd63-MVA expressing ME-TRAP that CD8+ T cells correlate with protection (17). Thus, the identification of antigens presented by MHC-I molecules in infected hepatocytes may provide the yet unidentified antigens required for full protection. This review focuses on the work that has been done to identify liver-stage protective antigens from P. falciparum, and proposes new strategies to complement or improve current methods of antigen discovery for malaria vaccines.
Biology of Plasmodium
Plasmodium sp. are polymorphic, obligate intracellular parasites with a complex life cycle that has both an asexual and a sexual stage and parasitize two very different hosts: an invertebrate host, mainly Anopheles mosquitos, and a vertebrate mammalian host. Five species P. falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi are recognized as responsible of natural infection in humans, although infections in other primate species have been reported with these pathogens (18).
Malaria is transmitted by different species of Plasmodium-infected Anopheles female mosquitos (18). The infected female mosquito injects approximately one hundred sporozoites into the skin of a mammalian host whilst taking a blood meal (19). The sporozoites search for capillaries in the skin and, within minutes, enter the host's circulatory system, eventually infecting cells in the liver (20, 21). In the liver, the sporozoites traverse several hepatocytes before establishing a successful infection within a hepatocyte (22, 23), residing in a parasitophorous vacuole with specialized functions (22, 24). The hepatic infection is asymptomatic and takes about 7 days to complete for P. falciparum. P. vivax and P. ovale can establish a latent form, the hypnozoite, that persists in the liver and may cause relapses by invading the bloodstream months or years later (18). In the hepatocytes, parasites undergo asexual schizogony to form tens of thousands of merozoites (18). The parasites inside the hepatocyte are known as exo-erythrocytic forms (EEFs). The pre-erythrocytic stage or exo-erythrocytic cycle includes both the sporozoite invasion of the mammalian host and the liver-stage. This stage represents a bottleneck in the disease which is ideal to block progression to clinical malaria, through chemotherapeutic and immunoprophylactic interventions.
Following merozoite egress from the infected liver cell, the parasites escape into the blood circulation to infect erythrocytes, where they asexually replicate exponentially. This constitutes the symptomatic and well-studied blood-stage malaria or erythrocytic cycle (18). Once merozoites invade erythrocytes, they become trophozoites that develop into schizonts which eventually rupture the host cell and re-infect new erythrocytes. Alternatively, some trophozoites develop into gametocytes. If a female mosquito takes a blood meal from an infected mammalian host, containing at least one female and one male gametocyte, sexual development of the parasite resumes (18). Inside the mosquito, the sexual stage or sporogonic cycle occurs, forming gametes, which develop into ookinetes that cross the mosquitoes' mid gut wall to become oocysts. Inside oocysts, thousands of sporozoites are formed that eventually migrate to the salivary glands (18, 25).
Major Achievements Leading to the Identification of Pre-Erythrocytic Proteins
Antigen Identification Based on Genomic, Transcriptomic, and Proteomic Studies
In 2002, P. falciparum genome was sequenced, which constituted a landmark in the field and the beginning of the post-genomic era. P. falciparum nuclear genome consists of an unusually high A-T content, 22.8 megabases long, composed of 14 chromosomes encoding 5,268 genes (26). Surprisingly, only 733 (14% of total) genes encoding enzymes and transporters were identified but a large proportion of genes were thought to be involved in host–parasite interactions and immune evasion (26). Also, many proteins seem to be targeted to the apicoplast, which is an organelle homologous to the chloroplasts of plants and algae, with a role in the anabolism of fatty acids, isoprenoids and haeme. The apicoplast is present in many organisms from the phylum apicomplexa and is essential for parasite survival (26). In the same year, the first large-scale, high-throughput proteomics study on four stages of the parasite (sporozoites, merozoites, trophozoites and gametocytes) was published revealing 2,415 proteins in total and 1,049 in the sporozoite but the liver stage was not surveyed (27) (Table 1). Interestingly, the antigenically variant proteins (var and rif) known to be expressed on the surface of infected erythrocytes were also detected in sporozoites (27), which are thought to be involved in immune escape and suggests a promiscuity in gene expression between stages. Moreover, chromosomal clusters of proteins expressed simultaneously were also identified (27), suggesting some form of gene expression regulation. Both studies were performed with P. falciparum clone 3D7. In a back-to-back publication, another large-scale proteomics study revealed 1,289 proteins in three stages of the parasite (asexual blood stages, gametocytes and gametes), using P. falciparum isolate NF54 (34) (Table 1). The pre-erythrocytic stages were not included in this study. The combined protein identifications of these two studies represent 52% of the predicted gene products (2,731) (26), and in both studies infected hepatocytes were not included. The genome sequencing and the first two large-scale proteomics studies brought new hope for the identification of new targets for therapies as well as new vaccine candidates. Regrettably, liver-stage proteins were not identified, thus urging for the identification of antigens expressed at this stage. However, in an integrated study that combined both transcriptome and proteome analysis, Kappe produced P. yoelii expressing GFP which allowed the isolation of liver-stage-infected hepatocytes, through cell sorting. This allowed performing gene and protein expression directly in purified infected hepatocytes, which resulted in the identification of 1,985 genes expressed during liver-stage using P. yoelii microarrays. This revealed interesting genes such as the fatty acid synthesis II (FASII) and other apicoplast pathways as potential druggable targets for malaria prophylaxis (38).
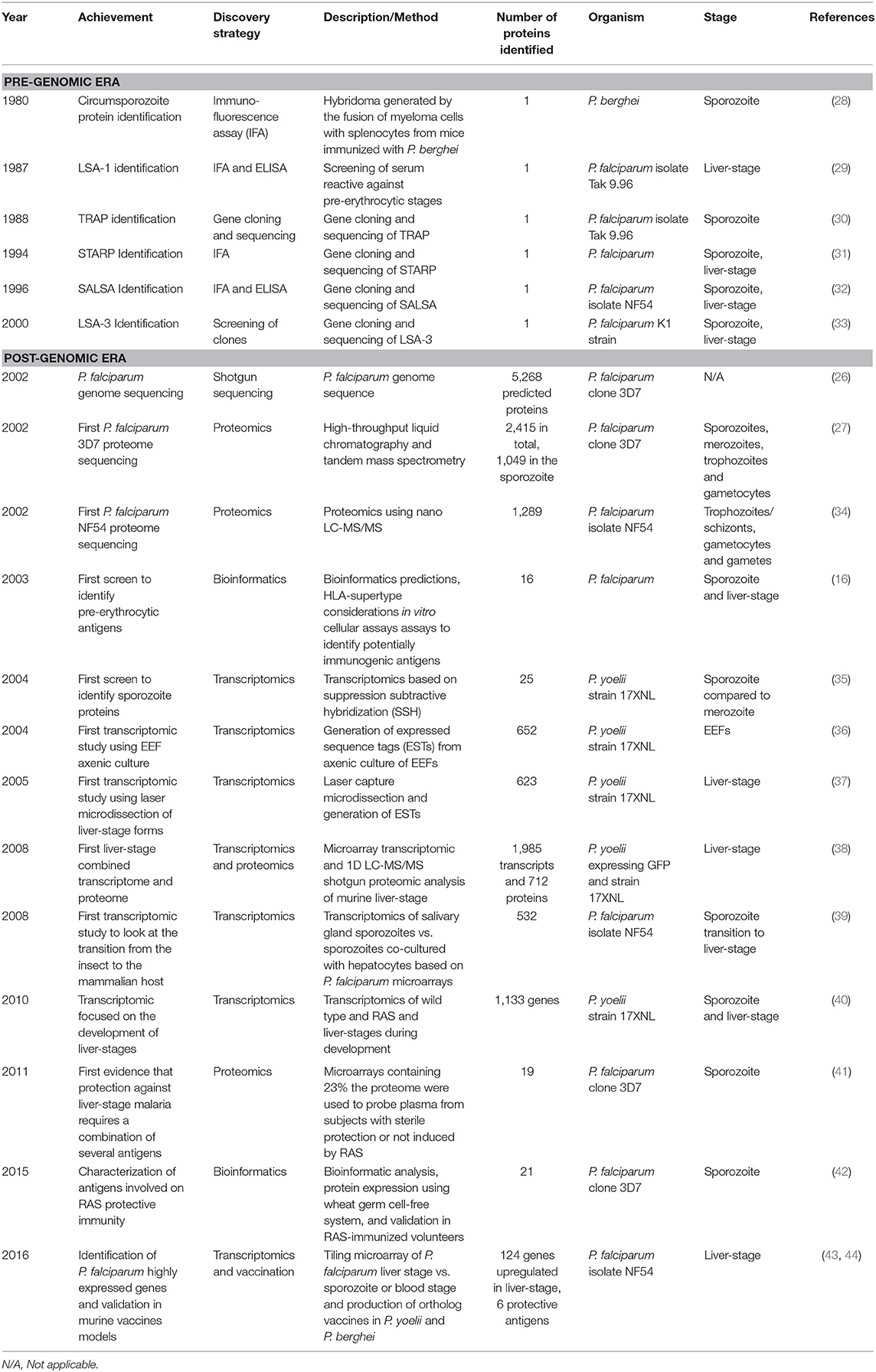
Table 1. Major achievements leading to the identification of pre-erythrocytic proteins in chronological order.
Antigen Identification Based on Clinical and Challenge Studies
Remarkably, sterile immunity to a challenge with infectious sporozoites can be obtained through immunization with RAS (10, 45), through genetically attenuated parasites (GAP) (46, 47), and through chemoprophylaxis with infectious sporozoites (CPS) (48, 49), both in animals and humans. The protection induced occurs in the pre-erythrocytic stages (11), and is mediated by CD8+ T cells (50). Benefiting from this fact and from the genome and proteome studies of 2002, Doolan has identified 16 pre-erythrocytic antigenic proteins recognized by volunteers immunized with radiation-attenuated P. falciparum sporozoites, using bioinformatics predictions, HLA analysis, and in vitro cellular assays (16).
Also, benefitting from the genome sequencing, transcriptomic studies were performed in an attempt to identify new pre-erythrocytic antigens. By using suppression subtractive hybridization (SSH) of P. yoelii sporozoites and comparing them to merozoites, Kappe has identified 25 genes expressed in pre-erythrocytic stage, including the well-known CSP and TRAP (35). Although some interesting antigens were identified, this study may have excluded many antigens that are co-expressed in both sporozoite and merozoite stages.
In an attempt to characterize the transcriptome of liver-stage malaria, axenically cultured EEFs of P. yoelii were produced, which resulted in the identification of 652 unique transcripts based on 1,453 expressed sequence tags from cultured EEFs (36). Even though a number of transcripts were identified, axenic cultures do not mimic the natural hepatocyte infection, thus making this system poorly suited for the identification of liver-stage expressed antigens. To look into in vivo expression of parasite mRNA during liver-stage, Aguiar used laser capture microdissection to produce enriched samples of parasite mRNA for the construction of a liver-stage cDNA library, resulting in the expression of 623 unique P. yoelii genes (37).
Dominique Mazier hypothesized that P. falciparum sporozoites would undergo changes in gene expression during the transition from the insect to the mammalian host, in order to be prepared for the liver-stage. P. falciparum sporozoites were co-cultured in vitro with primary human hepatocytes and maintained at 37°C, to mimic this transition, which resulted in the identification of 532 up-regulated genes, suggesting that the salivary gland sporozoites are indeed activated for hepatocyte invasion upon contact with these cells at 37°C (39). Many interesting antigens were identified in this study, and later evaluated as pre-erythrocytic vaccine candidates (51).
In another study, RNA was collected from P. yoelii wild type, RAS and of developmental liver-stage samples obtained by laser microdissection at 24 and 48 h post-infection. Transcriptional analysis made with microarrays on these samples identified 1,133 genes significantly differentially expressed compared to blood stages (40).
Doolan et al. have produced microarrays containing 23% of P. falciparum proteome and used them to probe plasma from subjects with sterile protection or no protection after experimental immunization with RAS. Nineteen pre-erythrocytic stage antigens were strongly associated with sporozoite-induced protective immunity, 16 of which were novel. This study revealed that sterile protection against malaria requires a combination of several antigens, and the authors suggested that a malaria vaccine should be multivalent in order to improve its efficacy (41).
Another study based on bioinformatic analysis and expression databases produced a list of 27 recombinant proteins using wheat germ cell-free protein expression system. Twenty-one proteins were recognized by plasma and 20 by PBMCs from RAS-immunized volunteers (42).
Using tiling microarrays, Duffy and colleagues identified 124 P. falciparum genes expressed in liver-stage compared to sporozoite or blood-stage. Produced 21 of vaccines with orthologs in P. yoelii and P. berghei, six being protective and two offered improved protection when in combination with CSP, compared to CSP alone (43, 44). Many other studies followed these with the purpose of identifying liver-stage antigens, to be used as vaccines or drug targets and to understand the pathophysiology of liver-stage malaria and host-pathogen interactions.
Liver-Stage Antigens
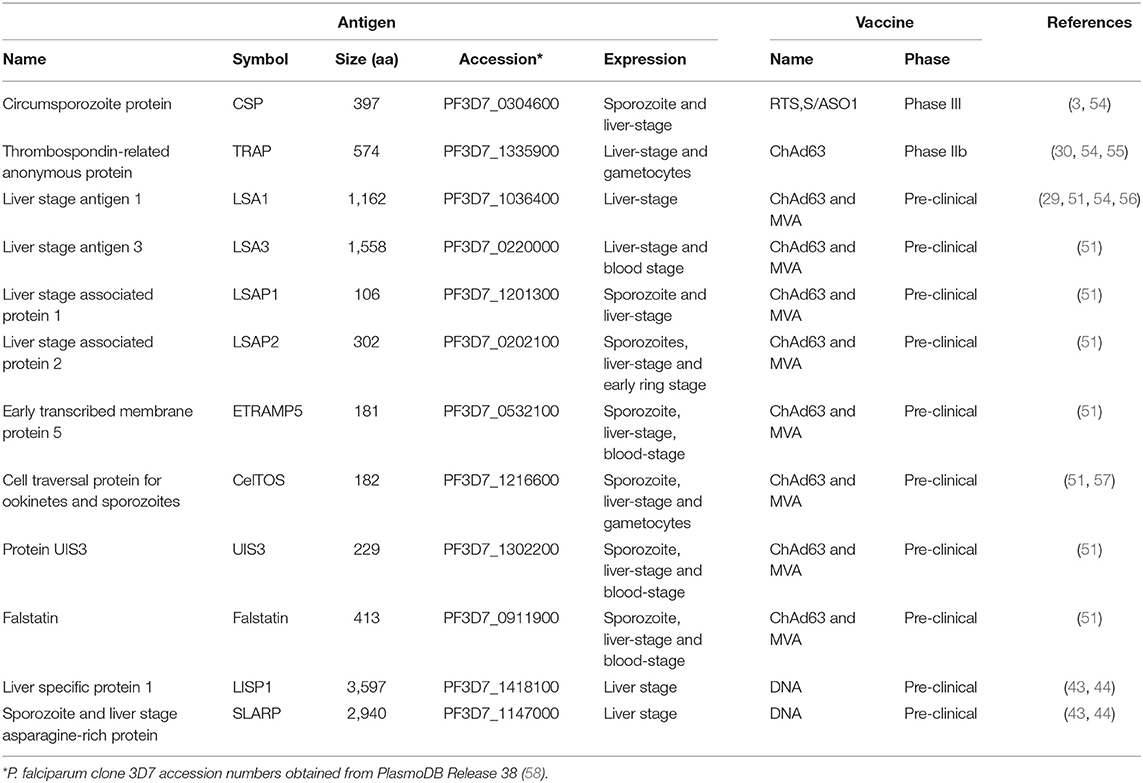
Sterile protection against malaria can be obtained through immunization with RAS. Based on this observation, many studies were performed to identify antigens expressed during liver-stage that would be associated with protection. CSP was the first antigen to be described conferring protection at the pre-erythrocytic level. CSP is predominantly expressed in the sporozoite and early liver forms (52). Due to incomplete protection provided by CSP, comparing to immunizations with radiation-attenuated sporozoites, several studies attempted to identify other antigens expressed at this stage, resulting in the identification of LSA-1 (29), STARP (31), SALSA (32), LSA-3 (33) and others recently described in a comprehensive review (53).
The first pre-erythrocytic antigen identified was the circumsporozoite protein (CSP), in 1980 (Tables 1, 2). An hybridoma resulting from the fusion of myeloma cells with splenocytes from mice immunized with P. berghei was generated and its antibodies recognized the surface of P. berghei sporozoites (28) (Table 2). RTS,S/AS01 is a recombinant protein vaccine containing part of the CSP antigen, including 19 NANP repeats and the carboxyl terminus, expressed in virus-like particle hepatitis B surface antigen and formulated with AS01 adjuvant (3, 59).
The first liver-stage antigen identified was the Liver Stage-Specific-Antigen-1 (LSA-1). The authors searched for sera from patients that were restricted to pre-erythrocytic stages. One such individual, living in a malaria-endemic area and undergoing continuous drug prophylaxis for 26 years, had high antibody titers against sporozoites and liver-stage, yet negative for blood-stage. Serum was used to screen against a clinical strain P. falciparum genomic expression library, which led to the identification of clones expressing LSA-1. One clone was sequenced and contained a DNA fragment of 196 bp composed of a 51 bp repeat sequence, encoding the 17 amino-acid sequence (EQQSDLEQERLAKEKLQ) recognized in ELISA by affinity-purified human antibodies (29) (Table 2). The pre-erythrocytic antigens LSA3, LSAP1, LSAP2, ETRAMP5, UIS3, and Falstatin, used in pre-clinical and clinical vaccine development were identified in the abovementioned Mazier's sporozoite screen (Table 2).
Tools to Study Liver-Stage Malaria Biology
Although many attempts to characterize liver-stage proteins have been performed in the last 50 years, the difficulties in performing these studies stem from three main reasons: first, it is difficult, laborious and complex to obtain large numbers of P. falciparum sporozoites required for hepatocyte infection experiments; secondly, primary hepatocytes are poorly suited for research because they show great phenotypic variability across donors and hepatoma cell lines are not sufficiently metabolically mature when compared to primary human hepatocytes; thirdly, the sporozoite infectivity, i.e., the percentage of infected hepatocytes, is traditionally very low. This will be discussed in greater detail below.
Parasites
During transmission, sporozoites are injected with mosquito saliva into the skin. The current methods for obtaining viable sporozoites for liver stage studies include the well-known hand dissection of mosquitoes, followed by grinding the salivary glands with a pestle to release sporozoites (60), passing the glands through a needle and syringe (61), or purifying the sporozoites through a density gradient (62), which may produce a heterogeneous mix of sporozoites, mosquito debris, salivary glands, and mosquito saliva. Indeed, an infected Anopheles mosquito saliva protein that is associated with saliva sporozoites was recently identified by mass spectrometry, with similarity to the human gamma interferon inducible thiol reductase (GILT), and has a negative impact on the speed and cell traversal activity of Plasmodium (63).
Animal models have been used for more than half a century to ascertain the efficacy (10) and immunogenicity (64) of vaccines against malaria. However, malaria mouse models do not allow the successful infection of the human parasite P. falciparum. The rodent malaria parasites (RMP) P. yoelii, P. berghei and P. chabaudi, have been extensively used as models for the human disease. RMPs have more than 90% homology with primate parasites such as P. falciparum (65). However, some P. falciparum vaccine candidates don't have orthologs in the murine malaria parasites, as is the case of LSA1 (29), some antigens are present in both but not with complete overlapping sequences, as is the case of CSP (30), and their expression might be species specific, as is often the case with many genes (66). Furthermore, it has been suggested that transcriptional evolution in Plasmodium species may be under different selection pressures, due to host specific variations (66). To overcome this, transgenic parasites have been developed to express P. falciparum antigens, so that these antigens can be assessed for immunogenicity in murine models. Moreover, transgenic RPMs that express unique P. falciparum antigens can be used to assess the efficacy of vaccines containing the same antigens, in mice. For a recent review on this subject please refer to Longley et al. (67).
RMPs expressing P. falciparum genes can be produced by genetic manipulation based on the insertion of exogenous DNA through homologous recombination mechanisms and transfection methods (68). P. falciparum genes can be either introduced to replace a given RMP endogenous gene or to introduce an additional gene (67). Recently, CRISPR/Cas9 technology has emerged as an improvement in the generation of transgenic parasites (69).
New tools to study pre-erythrocytic malaria are urgently needed. These include Plasmodium parasites expressing reporter molecules throughout the entire life cycle, but especially detectable in the liver-stage. Reporter molecules sensitive to changes in the pH, oxidative stress or other intracellular conditions would be of particular interest to pinpoint metabolic changes in the parasitized cell, which could provide additional information on the host-pathogen interactions during infection. Of particular interest is the recent P. berghei reporter parasite line that reveals membrane dynamics by GFP-tagging of a non-essential protein localized in the plasma membrane, and its trafficking in living parasites through the entire life, cycle using live-cell microscopy can be followed (70).
Recently, new P. falciparum clinical isolates NF135 and NF166 were identified by Sauerwein, that present a significantly higher infectivity on human primary hepatocytes in vitro, around 3%, and showed faster egress to the blood compared to NF54, which correlated directly with the magnitude of the first wave of blood-stage parasites to emerge from the liver in vivo, and correlated inversely with the pre-patent period in controlled human malaria infection (CHMI) subjects (71).
Even though some progress has been achieved by the introduction of reporter genes, development of transgenic strains and characterization of new clinical isolates, the difficulties of producing pre-erythrocytic forms in high numbers, hinders the identification of liver-stage antigens.
Hepatocytes
The liver, the largest human organ, is the target of the sporozoites upon invasion. At the cellular level, the architecture of the liver is organized in hexagonal lobules composed of several cell types from the most abundant hepatocytes, to endothelial cells, cholangiocytes, stellate cells, küppfer cells, dendritic cells and resident lymphocytes. These are supplied by an intricate network of bile canaliculi and sinusoids, leading to bile ducts and central and portal branch veins, respectively, which in turn, combined with the hepatic branch artery, constitute the portal triad. This highly organized organ, with a complex three-dimensional architecture, is involved in crucial functions of metabolism, storage, and detoxification as well as endocrine and exocrine functions (72–74).
Early studies in pre-erythrocytic malaria were performed in microscopic observations of liver sections (75), and later in monolayers of hepatoma cell lines (76). Currently, primary cells from human, non-human primate, murine and rabbit cells have been extensively used, as well as hepatoma cell lines (77). More recently, induced pluripotent stem cells-derived hepatocyte-like cells (iHLCs), have been shown to support pre-erythrocytic malaria using P. berghei, P. yoelii, P. falciparum, and P. vivax (78). This variety of cultivable cell types promoted a rapid improvement in the understanding of the biology of EEFs. However, primary hepatocytes fail to proliferate in vitro in contrast to their natural regenerative ability in vivo. Furthermore, although primary hepatocytes are metabolically competent, they are phenotypically unstable, exhibit great variability between batches and possess limited ability to proliferate ex vivo (79–81). In contrast, hepatoma cell lines proliferate generously, but lack several maturity markers, with especial relevance to a reduced Cytochrome P450 gene expression (79, 80), making them poorly suited for the complete understanding of liver-stage malaria infection. Several hepatoma cell lines are commercially available both from murine or primate origin. For a review on this subject please read the useful Prudêncio' s toolbox article describing all host-parasite combinations of Plasmodium hepatic infection models, in vivo, ex vivo, and in vitro (77).
Models of two-dimensional (2D) monolayer cultures in a supportive matrix (76, 82, 83) or 2D co-cultures of primary hepatocytes surrounded by stromal cells (61), have been used to study pre-erythrocytic malaria. The latter were improvements on the monolayers, but still lack the three-dimensional architecture of an organ. To overcome that, 3D models have been developed, containing several cell types organized in a three-dimensional structure. A further improvement, these co-cultured cells were organized in a functional matrix to mimic the organ, known as organoids. Although mimicking somehow the three-dimensional architecture of the organ, are not able to reproduce the physical dynamics of the blood and duct vessels and shear stress typical of the liver. To include the complex fluid dynamics in these models, new models of liver-on-a-chip engineered liver platforms have been designed to mimic the hepatic fluid dynamics environment in the liver (72). These models were originally developed with the purpose of drug testing and cell differentiation studies may be now employed for pre-erythrocytic malaria studies.
To reduce the gap between in vitro liver-mimicking models and in vivo human livers, humanized mice were developed. These are immunodeficient mice xenographed with human primary hepatocytes and constitute the closest in vivo models of human liver developed so far and were received both with enthusiasm and frustration.
Mouse Models
Mice have been instrumental in our understanding of malaria pathogenesis and have driven most of the progress made in the field. The initial studies using mouse models were on inbred A/J mice (10, 11, 84, 85), BALB/c (28, 86), C57BL/6 (87, 88), as well as outbred Theiler's Original (T.O.) mice (89). Experiments in mice revealed that CD8+ T cells are required for protection against a RAS challenge, while CD4+ T cells are dispensable, by depleting CD8+ and CD4+ T cells respectively, in immunocompetent mice (84). Additionally, IFN-γ and antibodies were also shown to be required for inhibition of the development of EEFs during protection induced by vaccination with RAS (84).
Naturally immunocompromised mice, as well as laboratory produced knock-out and transgenic mouse models have been used to improve our understanding of the disease. Congenitally athymic mice were used to demonstrate the T cell requirement for protection induced with RAS against a lethal challenge of P. berghei sporozoites (85). Beta 2-microglobulin knockout mice were instrumental in showing the requirement of MHC-I presentation to CD8+ T cells in inducing protection mediated by RAS (88). Transgenic mice were generated to express a T cell receptor specific for the epitope (SYVPSAEQI) from the circumsporozoite protein of P. yoelii. This peptide was recognized by transgenic CD8+ T cells but not CD4+ T cells, was able to inhibit parasite development (90) and its protection was independent of IFN-γ production (87). Even though a great diversity of mouse model exists, mice may not recapitulate the pathophysiology of human severe malaria, as in the case of human cerebral malaria (91). Therefore better models of disease are required.
Humanized mouse models consist of chimeric mice containing human cells, and have been developed in an attempt to provide in vivo animal models to study human disease. The first liver humanized mouse developed was based on a strain of immunocompromised SCID mice homozygous for the Alb-uPA transgene, which causes liver injury. The Alb-uPA transgenic mouse has an accelerated hepatocyte death and, as this strain is immunocompromised, human hepatocytes were successfully transplanted and repopulated the mouse liver (92). Moreover, these animals were successfully infected with hepatitis C virus (92), and later with P. falciparum (93).
Another model, the FRG mouse that lacks the fumarylacetoacetate hydrolase (Fah−/−) which causes liver injury, and immunocompromised (with a Rag2−/− and Il2rγ−/− background), can harbor up to 90% of human hepatocytes in the liver (94), and when crossed with NOD mice (non-obese diabetic mice that tolerate human hematopoietic cells), and transplanted with human hepatocytes and O+ human red blood cells, supports the transition of liver-stage to blood-stage malaria, after P. falciparum infection (95).
Recently a double engraftment of TK-NOG mice by human primary hepatocytes and red blood cells was developed (96). For the first time, this mouse model allows the complete hepatic development of P. falciparum, and the transition to erythrocytic stages, including the appearance of mature gametocytes. Even though the human hepatocytes that repopulate the mouse liver account for only 60–80% of total hepatocytes, this mouse model closely mimics the physiological complexity and specificity of an in vivo infection in the human environment.
Another recent model consists in engineered artificial human livers, implantable in mice without the requirement of liver injury. Composed of macroporous PEG cryogels, the human ectopic artificial liver (HEAL) is amenable to liver stage Plasmodium infection in vitro and in vivo. With intraperitoneal implantation and support infection with both liver stage rodent and human Plasmodium parasites in vivo (97).
Although these mouse models are attractive and offer the possibility of studying pre-erythrocytic malaria in vivo with P. falciparum and other human pathogenic Plasmodium parasites, these animals are immunodeficient thus not allowing the study of vaccine efficacy and immunogenicity, due to a constitutive lack of immune response.
Another humanized mouse model, the DRAG (HLA-DR4, Rag1−/−, Il2rγ−/−, NOD) mice, were generated by transplantation of HLA-II-matched human Hematopoietic stem cells (HSC) and reconstituted human hepatocytes, küpffer cells, liver endothelial cells, and erythrocytes, allowed the full vertebrate life cycle of P. falciparum and developed functional human T and B cells (98).
Even though none of these mouse models has served to identify antigens expressed by the liver stages of P. falciparum, they have been instrumental to study the dynamics of host-parasite interactions, to dissect the immune response against RMPs and vaccine efficacy and immunogenicity evaluation. For a recent review liver-stage response against malaria antigens using mouse models, please see (99).
Infectivity
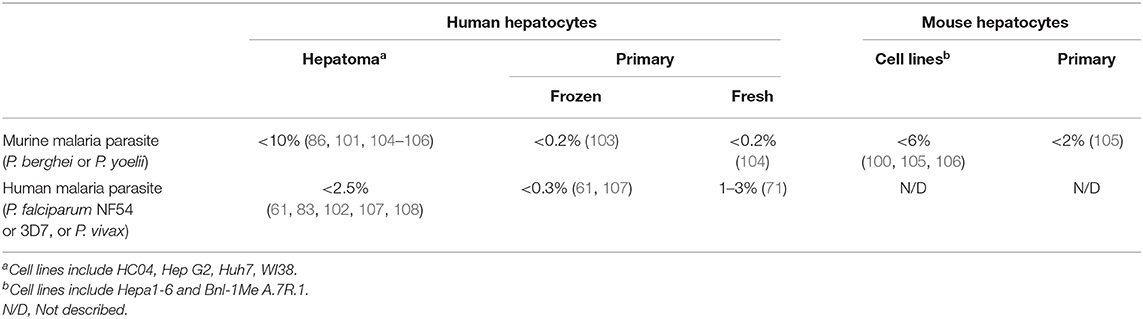
Infectivity, or rate of infection of sporozoites in hepatocytes, can be determined semi-quantitatively by fluorescence microscopy, quantitatively by flow cytometry, and indirectly by RT-PCR. For both fluorescence microscopy and flow cytometry, the parasite has to be labeled with a fluorophore to be detected. For RT-PCR, a standard curve with a known copy number of Plasmodium genomes has to be produced, which serves as a ruler to measure the amount of parasites in a given sample. Then, if the total number of hepatocytes is known, a rate of infection can be estimated by RT-PCR. Usually, these three methods correlate relatively well. It has been reported that <5% cells from hepatoma cell lines are infected with murine parasites in most publications (100, 101), however, infectivity's of nearly 10% could be reached when small numbers of hepatocytes were infected with P. berghei (86). In several reports, <2.5% of hepatoma cell lines were infected with P. falciparum (61, 102). In frozen human primary hepatocytes, 0.2% or less of cells were infected with murine parasites (103) and lower than 0.3% were infected with P. falciparum (61). In freshly harvested human primary hepatocytes, between 1 and 3% of cells were infected with P. falciparum (71), in contrast to 0.2% when infected with RMPs. Primary mouse hepatocytes and mouse cell lines, have had infectivity's lower than 6 and 2%, respectively, using RMPs, based on immunofluorescence microscopy or flow cytometry (Table 3). The lower infectivity's observed are a typical feature of malaria liver-stage infection and depend not only on the origin of the host cells and strain of parasite, but also on the expression of CD81 receptor, which has been described as essential for primary human hepatocyte invasion (107, 109). The infectivity may vary due to the expression levels of EhpA2 (110) and the class B, type I scavenger receptor (SR-BI) (111). Additionally, primary hepatocytes in humanized mouse models, as well as obtained from genetic altered mice may display different levels of infectivity. Primary hepatocytes originating from other animal models such as old and new world monkeys, rabbits and rats, may offer different infectivity's in vitro. Also, the treatment of hepatocytes with diverse compounds may change the rate of infection. Interestingly, it has been reported a staggering 10–20% rate of infection in irradiated HepG2 cells infected with P. berghei (89). Large scale antigen discovery experiments using current methods depend on large number of infected cells, therefore, it is extremely laborious and time-consuming to obtain sufficient number infected hepatocytes in vivo, ex vivo, and in vitro. Thus, the identification of new antigens has been hindered.
Antigen Identification for Vaccine Development
On the pre-genomic era, vaccine candidate identification was relatively empiric. Surprisingly, most of the current vaccine candidates were identified before the publication of P. falciparum genome, and they were mostly based on the screening of cDNA libraries for functional antibodies (112). The post-genomic era, initiated with the publication of P. falciparum genome (26), followed by the first transcriptomics (113, 114) and proteomic studies (27, 34), alone or in combination (38, 115), provided the tools for the systematic interrogation of the biology of the parasite at every stage. A consequence of the genome sequencing, allowed the combining data from genomics, transcriptomics and proteomics studies, with an unprecedented depth allowing the identification of new vaccine candidates, promoting the exponential creation and development of new bioinformatics tools, with or without experimental validation, opening new avenues for the discovery and functional analysis of new vaccine antigens at a faster pace and larger scale (116). This led to development of a more rational approach to identify vaccine candidates termed reverse vaccinology.
Reverse Vaccinology
Reverse vaccinology consists of the integration of whole genome sequencing data from a pathogen and the identification of vaccine candidates using bioinformatics as was employed for the first time by Rappuoli et al. on a virulent strain of Neisseria meningitidis serogroup B. Bioinformatics analysis were performed to shortlist 350 candidate antigens, followed by expression and purification of selected antigens in E. coli, and its immunization in mice. Sera from immunized mice were used to identify seven antigens that were positive all three assays: ELISA against whole cell MenB, FACS to detect proteins at the surface of MenB and MenB bactericidal activity (117). This strategy allows for the reduction of number of candidate antigens and has been extensively applied on number of subsequent studies identifying vaccine antigens against group B streptococcus (118), Chlamydia pneumoniae (119), Streptococcus pneumoniae (120), Bacillus anthracis (121), Porphyromonas gingivalis (122), among other bacterial pathogens with relatively small genomes. Reverse vaccinology in parasitology, and more specifically in malaria has been relatively successful in identifying some potential antigens for vaccine development. For example, Mu et al. searched for polymorphisms in ~65% of P. falciparum genes and identified several polymorphic loci. From a list of 56 antigens, half of those being already known, some were confirmed as potential vaccine candidates, using human immune sera (123), one of which the Apical membrane antigen 1 (AMA1), a well-known blood-stage vaccine candidate (124). Reverse vaccinology has been applied to malaria, particularly in the identification of the transmission blocking vaccine candidates, discussed in a recent review (125).
New Challenges in Antigen Identification—Immunopeptidomics
Large scale proteomic studies on parasite forms are a rapid and sensitive manner to discover new vaccine candidates. However, from the 5,300 predicted proteins and 3,000 described so far, the identification of the most immunogenic antigens is not a trivial task, not only due to the large number of potential candidates but also because strong immune responses may not correlate with protection. During liver stage malaria, infected hepatocytes present pathogen peptides through MHC Class I molecules from some of the many hundreds to thousands of genes expressed by the liver-stage parasite. Which genes are most abundantly presented as peptides on HLA class I molecules on the surface of hepatocytes that can be targeted by protective CD8+ T cells, is still largely unknown.
The peptides presented by MHC molecules are designated as the immunopeptidome (126). MHC-I derived peptides are typically 9–12 amino acids long and, based on the current methods, the minimal requirements for the identification of MHC-I derived peptides using mass spectrometry requires a sample containing 5 × 108 cells expressing ~2 × 105 MHC molecules per cell (126). Additionally, for the identification of Plasmodium peptides in a sample containing infected cells, the requirements may be more stringent as P. falciparum and P. berghei may interfere with MHC-I antigen presentation at the late time-points of liver-stage (104).
In the proteomics studies presented on Table 1, tryptic peptides were produced for the mass spectrometry analysis. Trypsin cleaves peptides at the C-terminal lysine and arginine residues (127). Enquiring peptides with Lys or Arg residues at the C terminus, although facilitates the identification of proteins, the majority of tryptic peptides (56%) are too small (≤6 residues) and thus not identified by mass spectrometry (127). An important advantage of immunopeptidomics is that no digestion protocol is used on the pathogen peptides, thus the identification of non-tryptic peptides that is, of native peptides (≥7 residues), can be very challenging although possible, however with a relatively low identification success rate of 10% compared to 50% for trypsinized peptides (126).
Moreover, as the rate of infection of hepatocytes infected with Plasmodium spp. is extremely low, arguably 5% (see Table 3), the mass spectrometry identification of pathogen peptides would require a higher number in the order of 1 × 1010 infected hepatocytes at a 5% rate of infection, which makes extremely laborious and complex to produce samples this size.
There is hope as mass spectrometry has been successful in the identification of MHC-II derived peptides from Leishmania major, from infected bone marrow-derived dendritic cells. Synthetic peptides were able to activate CD4+ T cells, and vaccination with the main synthetic peptide from Glycosomal phosphoenolpyruvate carboxykinase (PEPCK) protein, recombinant protein or DNA vaccines induced protection in mice. Although the protection was relatively long and protected from a L. donovani challenge as well, sterile protection was not achieved (128). More recently, a survey on the MHC-II immunopeptidome was conducted on murine blood-stage malaria, generating a list of 14 MHC-II ligands presented by cDC1 dendritic cells, which included the blood-stage vaccine candidates AMA1 and MSP1 (129).
The identification of antigens as vaccine candidates against malaria based on immunopeptidomics is in its infancy and major achievements should be produced in the upcoming years.
Discussion
The development of a useful malaria vaccine is a major priority for tropical infectious diseases: mortality from malaria exceeds half a million each year and there are hundreds of millions of cases. Control is threatened by increasing drug and insecticide resistance. A highly effective malaria vaccine could be extremely valuable for disease control but this has been very difficult to develop. However, there are increasing signs that success is possible and maybe even in sight. RTS,S/AS01, a sporozoite vaccine with modest efficacy may be licensed in the next 5 years. A liver-stage vaccine recently showed 67% efficacy in preventing malaria infection in Kenyan adults (55) and intravenous cryopreserved sporozoites can provide high level efficacy in challenge studies (130).
However, a cost-effective vaccine with high level durable efficacy in young infants remains elusive. The genome of P. falciparum is yielding new antigens that show promise such as PfRH5 (131) at the blood-stage, PfLSA1 (51), PfLSAP2 (51) and PfCelTOS (51, 57) at the pre-erythrocytic stage. New antigens are urgently needed to achieve the highest possible protection efficacy against malaria. Most likely, a combination of antigens expressed in every stage of the parasite's life cycle would improve the efficacy of a malaria vaccine.
Viral vector vaccines have the capacity to induce strong protective T cell responses against pathogens (25). For more than 20 years our group has been developing vaccine strategies with Adenovirus and MVA as antigens delivery strategies for vaccination, both in pre-clinical and clinical trials, with more than 1,500 people vaccinated in several countries. This viral vector vaccine approach, has been extensively reported to be safe, well-tolerated and relatively easy to deploy in remote settings (132). Viral vectored vaccines expressing new liver-stage antigens, alone or in combination, are expected to provide new and exciting antigens in the near future.
The identification of new liver-stage sub-unit vaccine candidates against malaria requires both the exploration of the genomics, transcriptomics and proteomics tools already developed, as well as new tools, based in three main pillars.
First, sporozoites: parasite strains with higher infectivity, such as NF135 and NF166 that improve the rate of infection of hepatocytes (71), may have promising consequences for the field. New genetic manipulation tools could facilitate the generation of diverse genetic attenuated strains or transgenic strains able to express multiple antigens simultaneously under different promoters. A good example was the generation of a resource developed by Billiker and colleagues that consists in a large-scale reverse genetic screening method based on barcoded vectors inserted into the P. berghei genome, which can be measured through barcode sequencing. This resource can provide the identification of essential genes involved in the parasite development. Additionally, this allows the study of the function of each barcoded KO genes, phenotyping each of these mutants, which can provide in depth information on the function of each gene, as the authors showed by studying the function of a set of promiscuous kinases (133). Improved methods to dissect and obtain pure sporozoites should also be developed to promote better yields of infected hepatocytes. Alternatively, improvements in the development of axenic cultures could offer an abundant source of sporozoites.
Second, hepatocytes: improved methods for culturing primary hepatocytes as well as the generation of fully matured and differentiated hepatocytes derived from stem cells (79), would be advantageous for the improvement of liver-stage infection experiments, especially to reduce the extensive variability observed as a consequence of the diversity of primary cells. Further progress, taking advantage of stem cell models for infection in combination with humanized mice or 3D organoids, could provide new models for infection. Humanized mice are the first in vivo model of a human liver, reason for great excitement in the field, but these mice are hard to maintain, and suffer from the inherent genetic variability of the primary human hepatocytes, which limit large-scale experiments. Improvements in the scalability and engraftment of these models could produce models with several humanized tissues to further explore the interaction between the liver, blood and with other tissues as well. Additionally, the development of new delivery methods of sub-unit vaccines specifically designed to target the liver could provide more robust protection and a better understanding of the protection mechanisms at the site of infection, in vivo.
Third, antigen identification: although there is currently a fair number of transcripts and proteins known to be expressed in the liver-stage, not all highly expressed proteins might be presented by MHC-I molecules, hence, not being recognized by CD8+ T cells. The pathogen peptides that are processed and presented via MHC-I, by the infected hepatocytes are largely unknown. Thus, identifying these peptides can provide and prioritize new vaccine candidates. To identify pathogen-specific MHC-associated peptides, a large number of infected cells are required, which depend on three key aspects: (i) the difficulty in obtaining large number of infectious sporozoites, (ii) the difficulty in obtaining a reliable source of human hepatocytes and (iii) the low yield of infection, or low infectivity. A combination of improvements in these three key aspects, with the current methods for antigen identification by mass spectrometry, would offer new vaccine candidates.
Other promising strategies for identifying liver-stage antigens have been described. Among these, bioinformatic analysis of MHC binding predictions to identify and select T cell epitopes as new antigen vaccine candidates (134). Also, in silico approaches, consisting of variations of reverse vaccinology combined with immunological analysis. Based on samples collected in CHMI or natural exposure to malaria, large-scale screens have been performed to identify targets able to induce T cell or antibody responses (135).
Although significant progress has been achieved in the last 50 years, culminating with the implementation of the only malaria vaccine in Phase III clinical trials RTS,S/AS01, exciting progress is expected to happen in the coming years, to improve the efficacy of current vaccines toward the final objective of malaria eradication.
Author Contributions
PB conceived and wrote the article.
Funding
This review received funding from the European Union's Horizon 2020 Research and Innovation Programme under grant agreement no. 733273.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to acknowledge Prof. Adrian V. S. Hill and Dr. Alexandra J. Spencer for reviewing this manuscript, the research group members, reviewers and collaborators whose work and ideas have contributed to this review.
References
2. Knox TB, Juma EO, Ochomo EO, Pates Jamet H, Ndungo L, Chege P, et al. An online tool for mapping insecticide resistance in major Anopheles vectors of human malaria parasites and review of resistance status for the Afrotropical region. Parasit Vectors. (2014) 7:76. doi: 10.1186/1756-3305-7-76
3. RTS, S Clinical Trials Partnership. Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: final results of a phase 3, individually randomised, controlled trial. Lancet. (2015) 386, 31–45. doi: 10.1016/S0140-6736(15)60721-8
4. Coelho CH, Doritchamou JYA, Zaidi I, Duffy PE. Advances in malaria vaccine development: report from the 2017 malaria vaccine symposium. NPJ Vaccines. (2017) 2:34. doi: 10.1038/s41541-017-0035-3
5. Halbroth BR, Draper SJ. Recent developments in malaria vaccinology. Adv Parasitol. (2015) 88:1–49. doi: 10.1016/bs.apar.2015.03.001
6. Moorthy VS, Newman RD, Okwo-Bele JM. Malaria vaccine technology roadmap. Lancet. (2013) 382:1700–1. doi: 10.1016/S0140-6736(13)62238-2
7. Stoute JA, Slaoui M, Heppner DG, Momin P, Kester KE, Desmons P, et al. A preliminary evaluation of a recombinant circumsporozoite protein vaccine against Plasmodium falciparum malaria. RTS,S Malaria Vaccine Evaluation Group. N Engl J Med. (1997) 336:86–91. doi: 10.1056/NEJM199701093360202
8. Lumsden JM, Schwenk RJ, Rein LE, Moris P, Janssens M, Ofori-Anyinam O, et al. Protective immunity induced with the RTS,S/AS vaccine is associated with IL-2 and TNF-alpha producing effector and central memory CD4 T cells. PLoS ONE. (2011) 6:e20775. doi: 10.1371/journal.pone.0020775
9. Sun P, Schwenk R, White K, Stoute JA, Cohen J, Ballou WR, et al. Protective immunity induced with malaria vaccine, RTS,S, is linked to Plasmodium falciparum circumsporozoite protein-specific CD4+ and CD8+ T cells producing IFN-gamma. J Immunol. (2003) 171:6961–7. doi: 10.4049/jimmunol.171.12.6961
10. Nussenzweig RS, Vanderberg J, Most H, Orton C. Protective immunity produced by the injection of x-irradiated sporozoites of Plasmodium berghei. Nature. (1967) 216:160–2. doi: 10.1038/216160a0
11. Vanderberg JP, Nussenzweig RS, Most H, Orton CG. Protective immunity produced by the injection of x-irradiated sporozoites of Plasmodium berghei. II Effects of radiation on sporozoites. J Parasitol. (1968) 54:1175–80. doi: 10.2307/3276987
12. Weiss WR, Sedegah M, Beaudoin RL, Miller LH, Good MF. CD8+ T cells (cytotoxic/suppressors) are required for protection in mice immunized with malaria sporozoites. Proc Natl Acad Sci USA. (1988) 85:573–6. doi: 10.1073/pnas.85.2.573
13. Plebanski M, Gilbert SC, Schneider J, Hannan CM, Layton G, Blanchard T, et al. Protection from Plasmodium berghei infection by priming and boosting T cells to a single class I-restricted epitope with recombinant carriers suitable for human use. Eur J Immunol. (1998) 28:4345–55.
14. Romero P, Maryanski JL, Corradin G, Nussenzweig RS, Nussenzweig V, Zavala F. Cloned cytotoxic T cells recognize an epitope in the circumsporozoite protein and protect against malaria. Nature. (1989) 341:323–6. doi: 10.1038/341323a0
15. Doolan DL, Hoffman SL, Southwood S, Wentworth PA, Sidney J, Chesnut RW, et al. Degenerate cytotoxic T cell epitopes from P. falciparum restricted by multiple HLA-A and HLA-B supertype alleles. Immunity. (1997) 7:97–112. doi: 10.1016/S1074-7613(00)80513-0
16. Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci USA. (2003) 100:9952–7. doi: 10.1073/pnas.1633254100
17. Ewer KJ, O'Hara GA, Duncan CJ, Collins KA, Sheehy SH, Reyes-Sandoval A, et al. Protective CD8+ T-cell immunity to human malaria induced by chimpanzee adenovirus-MVA immunisation. Nat Commun. 4:2836. doi: 10.1038/ncomms3836
18. Antinori S, Galimberti L, Milazzo L, Corbellino M. Biology of human malaria plasmodia including Plasmodium knowlesi. Mediterr J Hematol Infect Dis. (2012) 4:e2012013. doi: 10.4084/mjhid.2012.013
19. Medica DL, Sinnis P. Quantitative dynamics of Plasmodium yoelii sporozoite transmission by infected anopheline mosquitoes. Infect Immun. (2005) 73:4363–9. doi: 10.1128/IAI.73.7.4363-4369.2005
20. Hopp CS, Chiou K, Ragheb DR, Salman AM, Khan SM, Liu AJ, et al. Longitudinal analysis of Plasmodium sporozoite motility in the dermis reveals component of blood vessel recognition. eLife. (2015) 4:e07789. doi: 10.7554/eLife.07789
21. Shortt HE, Garnham PCC, Covell G, Shute PG. The pre-erythrocytic stage of human malaria, Plasmodium vivax. Br Med J. (1948) 1:547. doi: 10.1136/bmj.1.4550.547
22. Mota MM, Hafalla JCR, Rodriguez A. Migration through host cells activates Plasmodium sporozoites for infection. Nat Med. (2002) 8:1318–22. doi: 10.1038/nm785
23. Mota MM, Pradel G, Vanderberg JP, Hafalla JC, Frevert U, Nussenzweig RS, et al. Migration of Plasmodium sporozoites through cells before infection. Science. (2001) 291:141–4. doi: 10.1126/science.291.5501.141
24. Spielmann T, Montagna GN, Hecht L, Matuschewski K. Molecular make-up of the Plasmodium parasitophorous vacuolar membrane. Int J Med Microbiol. (2012) 302:179–86. doi: 10.1016/j.ijmm.2012.07.011
25. Hill AV. Vaccines against malaria. Philos Trans R Soc Lond B Biol Sci. (2011) 366:2806–14. doi: 10.1098/rstb.2011.0091
26. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. (2002) 419:498–511. doi: 10.1038/nature01097
27. Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. (2002) 419:520–6. doi: 10.1038/nature01107
28. Yoshida N, Nussenzweig RS, Potocnjak P, Nussenzweig V, Aikawa M. Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science. (1980) 207:71–3. doi: 10.1126/science.6985745
29. Guerin-Marchand C, Druilhe P, Galey B, Londono A, Patarapotikul J, Beaudoin RL, et al. A liver-stage-specific antigen of Plasmodium falciparum characterized by gene cloning. Nature. (1987) 329:164–7. doi: 10.1038/329164a0
30. Robson KJ, Hall JR, Jennings MW, Harris TJ, Marsh K, Newbold CI, et al. A highly conserved amino-acid sequence in thrombospondin, properdin and in proteins from sporozoites and blood stages of a human malaria parasite. Nature. (1988) 335:79–82. doi: 10.1038/335079a0
31. Fidock DA, Bottius E, Brahimi K, Moelans II, Aikawa M, Konings RN, et al. Cloning and characterization of a novel Plasmodium falciparum sporozoite surface antigen, STARP. Mol Biochem Parasitol. (1994) 64:219–32. doi: 10.1016/0166-6851(94)00012-3
32. Bottius E, BenMohamed L, Brahimi K, Gras H, Lepers JP, Raharimalala L, et al. A novel Plasmodium falciparum sporozoite and liver stage antigen (SALSA) defines major B, T helper, and CTL epitopes. J Immunol. (1996) 156:2874–84.
33. Daubersies P, Thomas AW, Millet P, Brahimi K, Langermans JA, Ollomo B, et al. Protection against Plasmodium falciparum malaria in chimpanzees by immunization with the conserved pre-erythrocytic liver-stage antigen 3. Nat Med. (2000) 6:1258–63. doi: 10.1038/81366
34. Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, Sauerwein RW, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. (2002) 419:537–42. doi: 10.1038/nature01111
35. Kaiser K, Matuschewski K, Camargo N, Ross J, Kappe SH. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol Microbiol. (2004) 51:1221–32. doi: 10.1046/j.1365-2958.2003.03909.x
36. Wang Q, Brown S, Roos DS, Nussenzweig V, Bhanot P. Transcriptome of axenic liver stages of Plasmodium yoelii. Mol Biochem Parasitol. (2004) 137:161–8. doi: 10.1016/j.molbiopara.2004.06.001
37. Sacci JB Jr, Ribeiro JM, Huang F, Alam U, Russell JA, Blair PL, et al. Transcriptional analysis of in vivo Plasmodium yoelii liver stage gene expression. Mol. Biochem. Parasitol. (2005) 142, 177–183. doi: 10.1016/j.molbiopara.2005.03.018
38. Tarun AS, Peng X, Dumpit RF, Ogata Y, Silva-Rivera H, Camargo N, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci U.S.A. (2008) 105:305–10. doi: 10.1073/pnas.0710780104
39. Siau A, Silvie O, Franetich JF, Yalaoui S, Marinach C, Hannoun L, et al. Temperature shift and host cell contact up-regulate sporozoite expression of Plasmodium falciparum genes involved in hepatocyte infection. PLoS Pathog. (2008) 4:e1000121. doi: 10.1371/journal.ppat.1000121
40. Williams CT, Azad AF. Transcriptional analysis of the pre-erythrocytic stages of the rodent malaria parasite, Plasmodium yoelii. PLoS ONE. (2010) 5:e10267. doi: 10.1371/journal.pone.0010267
41. Trieu A, Kayala MA, Burk C, Molina DM, Freilich DA, Richie TL, et al. Sterile protective immunity to malaria is associated with a panel of novel P. falciparum antigens. Mol. Cell. Proteomics. (2011) 10:M111.007948. doi: 10.1074/mcp.M111.007948
42. Aguiar JC, Bolton J, Wanga J, Sacci JB, Iriko H, Mazeika JK, et al. Discovery of novel Plasmodium falciparum pre-erythrocytic antigens for vaccine development. PLoS ONE. (2015) 10:e0136109. doi: 10.1371/journal.pone.0136109
43. Speake C, Pichugin A, Sahu T, Malkov V, Morrison R, Pei Y, et al. Correction: identification of novel pre-erythrocytic malaria antigen candidates for combination vaccines with circumsporozoite protein. PLoS ONE. (2016) 11:e0165489. doi: 10.1371/journal.pone.0165489
44. Speake C, Pichugin A, Sahu T, Malkov V, Morrison R, Pei Y, et al. Identification of novel pre-erythrocytic malaria antigen candidates for combination vaccines with circumsporozoite protein. PLoS ONE. (2016) 11:e0159449. doi: 10.1371/journal.pone.0159449
45. Clyde DF, Most H, McCarthy VC, Vanderberg JP. Immunization of man against sporozite-induced falciparum malaria. Am J Med Sci. (1973) 266:169–77. doi: 10.1097/00000441-197309000-00002
46. Mueller AK, Labaied M, Kappe SH, Matuschewski K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature. (2005) 433:164–7. doi: 10.1038/nature03188
47. Spring M, Murphy J, Nielsen R, Dowler M, Bennett JW, Zarling S, et al. First-in-human evaluation of genetically attenuated Plasmodium falciparum sporozoites administered by bite of Anopheles mosquitoes to adult volunteers. Vaccine. (2013) 31:4975–83. doi: 10.1016/j.vaccine.2013.08.007
48. Belnoue E, Costa FT, Frankenberg T, Vigario AM, Voza T, Leroy N, et al. Protective T cell immunity against malaria liver stage after vaccination with live sporozoites under chloroquine treatment. J Immunol. (2004) 172:2487–95. doi: 10.4049/jimmunol.172.4.2487
49. Roestenberg M, McCall M, Hopman J, Wiersma J, Luty AJ, van Gemert GJ, et al. Protection against a malaria challenge by sporozoite inoculation. N Engl J Med. (2009) 361:468–77. doi: 10.1056/NEJMoa0805832
50. Schofield L, Villaquiran J, Ferreira A, Schellekens H, Nussenzweig R, Nussenzweig V. Gamma interferon, CD8+ T cells and antibodies required for immunity to malaria sporozoites. Nature. (1987) 330:664–6. doi: 10.1038/330664a0
51. Longley RJ, Salman AM, Cottingham MG, Ewer K, Janse CJ, Khan SM, et al. Comparative assessment of vaccine vectors encoding ten malaria antigens identifies two protective liver-stage candidates. Sci Rep. (2015) 5:11820. doi: 10.1038/srep11820
52. Singh AP, Buscaglia CA, Wang Q, Levay A, Nussenzweig DR, Walker JR, et al. Plasmodium circumsporozoite protein promotes the development of the liver stages of the parasite. Cell. (2007) 131:492–504. doi: 10.1016/j.cell.2007.09.013
53. Duffy PE, Sahu T, Akue A, Milman N, Anderson C. Pre-erythrocytic malaria vaccines: identifying the targets. Expert Rev Vaccines. (2012) 11:1261–80. doi: 10.1586/erv.12.92
54. Hill AV, Elvin J, Willis AC, Aidoo M, Allsopp CE, Gotch FM, et al. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature. (1992) 360:434–9. doi: 10.1038/360434a0
55. Ogwang C, Kimani D, Edwards NJ, Roberts R, Mwacharo J, Bowyer G, et al. Prime-boost vaccination with chimpanzee adenovirus and modified vaccinia Ankara encoding TRAP provides partial protection against Plasmodium falciparum infection in Kenyan adults. Sci. Transl. Med. (2015) 7:286re285. doi: 10.1126/scitranslmed.aaa2373
56. Cummings JF, Spring MD, Schwenk RJ, Ockenhouse CF, Kester KE, Polhemus ME, et al. Recombinant liver stage antigen-1 (LSA-1) formulated with AS01 or AS02 is safe, elicits high titer antibody and induces IFN-gamma/IL-2 CD4+ T cells but does not protect against experimental Plasmodium falciparum infection. Vaccine. (2010) 28:5135–44. doi: 10.1016/j.vaccine.2009.08.046
57. Bergmann-Leitner ES, Mease RM, De La Vega P, Savranskaya T, Polhemus M, Ockenhouse C, et al. Immunization with pre-erythrocytic antigen CelTOS from Plasmodium falciparum elicits cross-species protection against heterologous challenge with Plasmodium berghei. PLoS ONE. (2010) 5:e12294. doi: 10.1371/journal.pone.0012294
58. Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. (2009) 37:D539–543. doi: 10.1093/nar/gkn814
59. Gordon DM, McGovern TW, Krzych U, Cohen JC, Schneider I, LaChance R, et al. Safety, immunogenicity, and efficacy of a recombinantly produced Plasmodium falciparum circumsporozoite protein-hepatitis B surface antigen subunit vaccine. J Infect Dis. (1995) 171:1576–85. doi: 10.1093/infdis/171.6.1576
60. Sinnis P, De La Vega P, Coppi A, Krzych U, Mota MM. Quantification of sporozoite invasion, migration, and development by microscopy and flow cytometry. Methods Mol Biol. (2013) 923:385–400. doi: 10.1007/978-1-62703-026-7_27
61. March S, Ng S, Velmurugan S, Galstian A, Shan J, Logan DJ, et al. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host Microbe. (2013) 14:104–15. doi: 10.1016/j.chom.2013.06.005
62. Kennedy M, Fishbaugher ME, Vaughan AM, Patrapuvich R, Boonhok R, Yimamnuaychok N, et al. A rapid and scalable density gradient purification method for Plasmodium sporozoites. Malar J. (2012) 11:421. doi: 10.1186/1475-2875-11-421
63. Schleicher TR, Yang J, Freudzon M, Rembisz A, Craft S, Hamilton M, et al. A mosquito salivary gland protein partially inhibits Plasmodium sporozoite cell traversal and transmission. Nat Commun. (2018) 9:2908. doi: 10.1038/s41467-018-05374-3
64. Nussenzweig R, Vanderberg J, Most H. Protective immunity produced by the injection of x-irradiated sporozoites of Plasmodium berghei. IV Dose response, specificity and humoral immunity. Mil Med. (1969) 134:1176–82. doi: 10.1093/milmed/134.9.1176
65. Otto TD, Bohme U, Jackson AP, Hunt M, Franke-Fayard B, Hoeijmakers WA, et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. (2014) 12:86. doi: 10.1186/s12915-014-0086-0
66. Hoo R, Zhu L, Amaladoss A, Mok S, Natalang O, Lapp SA, et al. Integrated analysis of the Plasmodium species transcriptome. EBioMedicine. (2016) 7:255–66. doi: 10.1016/j.ebiom.2016.04.011
67. Longley RJ, Hill AV, Spencer AJ. Malaria vaccines: identifying Plasmodium falciparum liver-stage targets. Front Microbiol. (2015) 6:965. doi: 10.3389/fmicb.2015.00965
68. Lin JW, Annoura T, Sajid M, Chevalley-Maurel S, Ramesar J, Klop O, et al. A novel ‘gene insertion/marker out’ (GIMO) method for transgene expression and gene complementation in rodent malaria parasites. PLoS ONE. (2011) 6:e29289. doi: 10.1371/journal.pone.0029289
69. Zhang C, Xiao B, Jiang Y, Zhao Y, Li Z, Gao H, et al. Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. mBio. (2014) 5:e01414–14. doi: 10.1128/mBio.01414-14
70. Burda PC, Schaffner M, Kaiser G, Roques M, Zuber B, Heussler VT. A Plasmodium plasma membrane reporter reveals membrane dynamics by live-cell microscopy. Sci Rep. (2017) 7:9740. doi: 10.1038/s41598-017-09569-4
71. McCall MBB, Wammes LJ, Langenberg MCC, van Gemert GJ, Walk J, Hermsen CC, et al. Infectivity of Plasmodium falciparum sporozoites determines emerging parasitemia in infected volunteers. Sci. Transl. Med. (2017) 9:eaag2490. doi: 10.1126/scitranslmed.aag2490
72. Gural N, Mancio-Silva L, He J, Bhatia SN. Engineered livers for infectious diseases. Cell Mol Gastroenterol Hepatol. (2018) 5:131–44. doi: 10.1016/j.jcmgh.2017.11.005
73. Juza RM, Pauli EM. Clinical and surgical anatomy of the liver: a review for clinicians. Clin Anat. (2014) 27:764–9. doi: 10.1002/ca.22350
74. Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. (2006) 43:S54–62. doi: 10.1002/hep.21060
75. Sodeman T, Schnitzer B, Durkee T, Jcontacos P. Fine structure of the exoerythrocytic stage of Plasmodium cynomolgi. Science. (1970) 170:340–1. doi: 10.1126/science.170.3955.340
76. Hollingdale MR, Leland P, Schwartz AL. In vitro cultivation of the exoerythrocytic stage of Plasmodium berghei in a hepatoma cell line. Am J Trop Med Hyg. (1983) 32:682–4. doi: 10.4269/ajtmh.1983.32.682
77. Prudencio M, Mota MM, Mendes AM. A toolbox to study liver stage malaria. Trends Parasitol. (2011) 27:565–74. doi: 10.1016/j.pt.2011.09.004
78. Ng S, Schwartz RE, March S, Galstian A, Gural N, Shan J, et al. Human iPSC-derived hepatocyte-like cells support Plasmodium liver-stage infection in vitro. Stem Cell Reports. (2015) 4:348–59. doi: 10.1016/j.stemcr.2015.01.002
79. Castell JV, Jover R, Martinez-Jimenez CP, Gomez-Lechon MJ. Hepatocyte cell lines: their use, scope and limitations in drug metabolism studies. Expert Opin Drug Metab Toxicol. (2006) 2:183–212. doi: 10.1517/17425255.2.2.183
80. Donato MT, Lahoz A, Castell JV, Gomez-Lechon MJ. Cell lines: a tool for in vitro drug metabolism studies. Curr Drug Metab. (2008) 9:1–11. doi: 10.2174/138920008783331086
81. Hu C, Li L. In vitro culture of isolated primary hepatocytes and stem cell-derived hepatocyte-like cells for liver regeneration. Protein Cell. (2015) 6:562–74. doi: 10.1007/s13238-015-0180-2
82. Hollingdale MR, Leland P, Sigler CI. In vitro cultivation of the exoerythrocytic stage of Plasmodium berghei in irradiated hepatoma cells. Am J Trop Med Hyg. (1985) 34:21–3. doi: 10.4269/ajtmh.1985.34.21
83. Hollingdale MR, Nardin EH, Tharavanij S, Schwartz AL, Nussenzweig RS. Inhibition of entry of Plasmodium falciparum and P. vivax sporozoites into cultured cells; an in vitro assay of protective antibodies. J Immunol. (1984) 132:909–13.
84. Schofield L, Ferreira A, Altszuler R, Nussenzweig V, Nussenzweig RS. Interferon-gamma inhibits the intrahepatocytic development of malaria parasites in vitro. J Immunol. (1987) 139:2020–5.
85. Spitalny GL, Verhave JP, Meuwissen JH, Nussenzweig RS. Plasmodium berghei: T cell dependence of sporozoite-induced immunity in rodents. Exp Parasitol. (1977) 42:73–81. doi: 10.1016/0014-4894(77)90063-7
86. Rodriguez-Galan A, Salman AM, Bowyer G, Collins KA, Longley RJ, Brod F, et al. An in vitro assay to measure antibody-mediated inhibition of P. berghei sporozoite invasion against P. falciparum antigens. Sci Rep. (2017) 7:17011. doi: 10.1038/s41598-017-17274-5
87. Chakravarty S, Baldeviano GC, Overstreet MG, Zavala F. Effector CD8+ T lymphocytes against liver stages of Plasmodium yoelii do not require gamma interferon for antiparasite activity. Infect Immun. (2008) 76:3628–31. doi: 10.1128/IAI.00471-08
88. White KL, Snyder HL, Krzych U. MHC class I-dependent presentation of exoerythrocytic antigens to CD8+ T lymphocytes is required for protective immunity against Plasmodium berghei. J Immunol. (1996) 156:3374–81.
89. Sinden RE, Suhrbier A, Davies CS, Fleck SL, Hodivala K, Nicholas JC. The development and routine application of high-density exoerythrocytic-stage cultures of Plasmodium berghei. Bull World Health Organ. (1990) 68:115–25.
90. Sano G, Hafalla JC, Morrot A, Abe R, Lafaille JJ, Zavala F. Swift development of protective effector functions in naive CD8(+) T cells against malaria liver stages. J Exp Med. (2001) 194:173–80. doi: 10.1084/jem.194.2.173
91. Craig AG, Grau GE, Janse C, Kazura JW, Milner D, Barnwell JW, et al. The role of animal models for research on severe malaria. PLoS Pathog. (2012) 8:e1002401. doi: 10.1371/journal.ppat.1002401
92. Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. (2001) 7:927–33. doi: 10.1038/90968
93. Sacci JB Jr, Alam U, Douglas D, Lewis J, Tyrrell DL, Azad AF, et al. Plasmodium falciparum infection and exoerythrocytic development in mice with chimeric human livers. Int. J. Parasitol. (2006) 36, 353–360. doi: 10.1016/j.ijpara.2005.10.014
94. Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, et al. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. (2007) 25:903–10. doi: 10.1038/nbt1326
95. Vaughan AM, Mikolajczak SA, Wilson EM, Grompe M, Kaushansky A, Camargo N, et al. Complete Plasmodium falciparum liver-stage development in liver-chimeric mice. J Clin Invest. (2012) 122:3618–28. doi: 10.1172/JCI62684
96. Soulard V, Bosson-Vanga H, Lorthiois A, Roucher C, Franetich J-F, Zanghi G, et al. Plasmodium falciparum full life cycle and Plasmodium ovale liver stages in humanized mice. Nat Commun. (2015) 6:7690. doi: 10.1038/ncomms8690
97. Ng S, March S, Galstian A, Gural N, Stevens KR, Mota MM, et al. Towards a Humanized mouse model of liver stage malaria using ectopic artificial livers. Sci Rep. (2017) 7:45424. doi: 10.1038/srep45424
98. Wijayalath W, Majji S, Villasante EF, Brumeanu TD, Richie TL, Casares S. Humanized HLA-DR4.RagKO.IL2RγcKO.NOD (DRAG) mice sustain the complex vertebrate life cycle of Plasmodium falciparum malaria. Malar. J. (2014) 13:386. doi: 10.1186/1475-2875-13-386
99. Van Braeckel-Budimir N, Harty JT. CD8 T-cell-mediated protection against liver-stage malaria: lessons from a mouse model. Front Microbiol. (2014) 5:272. doi: 10.3389/fmicb.2014.00272
100. Longley RJ, Bauza K, Ewer KJ, Hill AV, Spencer AJ. Development of an in vitro assay and demonstration of Plasmodium berghei liver-stage inhibition by TRAP-specific CD8+ T cells. PLoS ONE. (2015) 10:e0119880. doi: 10.1371/journal.pone.0119880
101. Prudencio M, Rodrigues CD, Ataide R, Mota MM. Dissecting in vitro host cell infection by Plasmodium sporozoites using flow cytometry. Cell Microbiol. (2008) 10:218–24. doi: 10.1111/j.1462-5822.2007.01032.x
102. Sattabongkot J, Yimamnuaychoke N, Leelaudomlipi S, Rasameesoraj M, Jenwithisuk R, Coleman RE, et al. Establishment of a human hepatocyte line that supports in vitro development of the exo-erythrocytic stages of the malaria parasites Plasmodium falciparum and P. vivax. Am J Trop Med Hyg. (2006) 74:708–15. doi: 10.4269/ajtmh.2006.74.708
103. Ng S, March S, Galstian A, Hanson K, Carvalho T, Mota MM, et al. Hypoxia promotes liver-stage malaria infection in primary human hepatocytes in vitro. Dis Model Mech. (2014) 7:215–24. doi: 10.1242/dmm.013490
104. Ma J, Trop S, Baer S, Rakhmanaliev E, Arany Z, Dumoulin P, et al. Dynamics of the major histocompatibility complex class I processing and presentation pathway in the course of malaria parasite development in human hepatocytes: implications for vaccine development. PLoS ONE. (2013) 8:e75321. doi: 10.1371/journal.pone.0075321
105. Depinay N, Franetich JF, Gruner AC, Mauduit M, Chavatte JM, Luty AJ, et al. Inhibitory effect of TNF-alpha on malaria pre-erythrocytic stage development: influence of host hepatocyte/parasite combinations. PLoS ONE. (2011) 6:e17464. doi: 10.1371/journal.pone.0017464
106. Mota MM, Rodriguez A. Plasmodium yoelii: efficient in vitro invasion and complete development of sporozoites in mouse hepatic cell lines. Exp Parasitol. (2000) 96:257–9. doi: 10.1006/expr.2000.4570
107. Dumoulin PC, Trop SA, Ma J, Zhang H, Sherman MA, Levitskaya J. Flow cytometry based detection and isolation of Plasmodium falciparum liver stages in vitro. PLoS ONE. (2015) 10:e0129623. doi: 10.1371/journal.pone.0129623
108. Kaushansky A, Rezakhani N, Mann H, Kappe SH. Development of a quantitative flow cytometry-based assay to assess infection by Plasmodium falciparum sporozoites. Mol Biochem Parasitol. (2012) 183:100–3. doi: 10.1016/j.molbiopara.2012.01.006
109. Silvie O, Rubinstein E, Franetich J-F, Prenant M, Belnoue E, Rénia L, et al. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat Med. (2002) 9:93–6. doi: 10.1038/nm808
110. Kaushansky A, Douglass AN, Arang N, Vigdorovich V, Dambrauskas N, Kain HS, et al. Malaria parasites target the hepatocyte receptor EphA2 for successful host infection. Science. (2015) 350:1089–92. doi: 10.1126/science.aad3318
111. Rodrigues CD, Hannus M, Prudencio M, Martin C, Goncalves LA, Portugal S, et al. Host scavenger receptor SR-BI plays a dual role in the establishment of malaria parasite liver infection. Cell Host Microbe. (2008) 4:271–82. doi: 10.1016/j.chom.2008.07.012
112. Tuju J, Kamuyu G, Murungi LM, Osier FHA. Vaccine candidate discovery for the next generation of malaria vaccines. Immunology. (2017) 152:195–206. doi: 10.1111/imm.12780
113. Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. (2003) 1:E5. doi: 10.1371/journal.pbio.0000005
114. Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. (2003) 301:1503–8. doi: 10.1126/science.1087025
115. Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, Berriman M, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. (2005) 307:82–6. doi: 10.1126/science.1103717
116. Izak D, Klim J, Kaczanowski S. Host-parasite interactions and ecology of the malaria parasite-a bioinformatics approach. Brief Funct Genomics. (2018) 17:451–7. doi: 10.1093/bfgp/ely013
117. Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. (2000) 287:1816–20. doi: 10.1126/science.287.5459.1816
118. Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, Scarselli M, et al. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science. (2005) 309:148–50. doi: 10.1126/science.1109869
119. Thorpe C, Edwards L, Snelgrove R, Finco O, Rae A, Grandi G, et al. Discovery of a vaccine antigen that protects mice from Chlamydia pneumoniae infection. Vaccine. (2007) 25:2252–60. doi: 10.1016/j.vaccine.2006.12.003
120. Giefing C, Meinke AL, Hanner M, Henics T, Bui MD, Gelbmann D, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med. (2008) 205:117–31. doi: 10.1084/jem.20071168
121. Ariel N, Zvi A, Grosfeld H, Gat O, Inbar Y, Velan B, et al. Search for potential vaccine candidate open reading frames in the Bacillus anthracis virulence plasmid pXO1: in silico and in vitro screening. Infect Immun. (2002) 70:6817–27. doi: 10.1128/IAI.70.12.6817-6827.2002
122. Ross BC, Czajkowski L, Hocking D, Margetts M, Webb E, Rothel L, et al. Identification of vaccine candidate antigens from a genomic analysis of Porphyromonas gingivalis. Vaccine. (2001) 19:4135–42. doi: 10.1016/S0264-410X(01)00173-6
123. Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. (2007) 39:126–30. doi: 10.1038/ng1924
124. Polhemus ME, Magill AJ, Cummings JF, Kester KE, Ockenhouse CF, Lanar DE, et al. Phase I dose escalation safety and immunogenicity trial of Plasmodium falciparum apical membrane protein (AMA-1) FMP2.1, adjuvanted with AS02A, in malaria-naïve adults at the Walter Reed Army Institute of Research. Vaccine. (2007) 5, 4203–4212. doi: 10.1016/j.vaccine.2007.03.012
125. Frimpong A, Kusi KA, Ofori MF, Ndifon W. Novel strategies for malaria vaccine design. Front Immunol. (2018) 9:2769. doi: 10.3389/fimmu.2018.02769
126. Caron E, Kowalewski DJ, Chiek Koh C, Sturm T, Schuster H, Aebersold R. Analysis of major histocompatibility complex (MHC) immunopeptidomes using mass spectrometry. Mol Cell Proteomics. (2015) 14:3105–17. doi: 10.1074/mcp.O115.052431
127. Tsiatsiani L, Heck AJ. Proteomics beyond trypsin. FEBS J. (2015) 282:2612–26. doi: 10.1111/febs.13287
128. Mou Z, Li J, Boussoffara T, Kishi H, Hamana H, Ezzati P, et al. Identification of broadly conserved cross-species protective Leishmania antigen and its responding CD4+ T cells. Sci. Transl. Med. (2015) 7:310ra167. doi: 10.1126/scitranslmed.aac5477
129. Draheim M, Wlodarczyk MF, Crozat K, Saliou JM, Alayi TD, Tomavo S, et al. Profiling MHC II immunopeptidome of blood-stage malaria reveals that cDC1 control the functionality of parasite-specific CD4 T cells. EMBO Mol Med. (2017) 9:1605–21. doi: 10.15252/emmm.201708123
130. Bastiaens GJ, van Meer MP, Scholzen A, Obiero JM, Vatanshenassan M, van Grinsven T, et al. Safety, immunogenicity, and protective efficacy of intradermal immunization with aseptic, purified, cryopreserved Plasmodium falciparum sporozoites in volunteers under chloroquine prophylaxis: a randomized controlled trial. Am J Trop Med Hyg. (2016) 94:663–73. doi: 10.4269/ajtmh.15-0621
131. Douglas AD, Williams AR, Illingworth JJ, Kamuyu G, Biswas S, Goodman AL, et al. The blood-stage malaria antigen PfRH5 is susceptible to vaccine-inducible cross-strain neutralizing antibody. Nat Commun. (2011) 2:601. doi: 10.1038/ncomms1615
132. Ewer KJ, Sierra-Davidson K, Salman AM, Illingworth JJ, Draper SJ, Biswas S, et al. Progress with viral vectored malaria vaccines: a multi-stage approach involving “unnatural immunity”. Vaccine. (2015) 33:7444–51. doi: 10.1016/j.vaccine.2015.09.094
133. Gomes AR, Bushell E, Schwach F, Girling G, Anar B, Quail MA, et al. A genome-scale vector resource enables high-throughput reverse genetic screening in a malaria parasite. Cell Host Microbe. (2015) 17:404–13. doi: 10.1016/j.chom.2015.01.014
134. Damfo SA, Reche P, Gatherer D, Flower DR. In silico design of knowledge-based Plasmodium falciparum epitope ensemble vaccines. J Mol Graph Model. (2017) 78:195–205. doi: 10.1016/j.jmgm.2017.10.004
Keywords: Plasmodium falciparum, pre-erythrocytic malaria, liver-stage, infectivity, vaccine design, antigen identification, immunopeptidomics
Citation: Bettencourt P (2020) Current Challenges in the Identification of Pre-Erythrocytic Malaria Vaccine Candidate Antigens. Front. Immunol. 11:190. doi: 10.3389/fimmu.2020.00190
Received: 29 October 2019; Accepted: 24 January 2020;
Published: 21 February 2020.
Edited by:
Ed C. Lavelle, Trinity College Dublin, IrelandReviewed by:
Sean C. Murphy, University of Washington, United StatesIrene Cruz Talavera, University of Washington, United States, in collaboration with reviewer SM
Adrian John Frederick Luty, Institut de recherche pour le développement (IRD), France
Copyright © 2020 Bettencourt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paulo Bettencourt, cGF1bG8uYmV0dGVuY291cnRAbmRtLm94LmFjLnVr
 Paulo Bettencourt
Paulo Bettencourt