- 1Division of Environmental Genetics and Molecular Toxicology, Department of Environmental and Public Health Sciences, University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 2Division of Biostatistics and Bioinformatics, Department of Environmental and Public Health Sciences, University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 3Pulmonary & Critical Care Medicine, Oregon Health & Science University, Portland, OR, United States
- 4Genomics, Epigenomics and Sequencing Core, Department of Environmental and Public Health Sciences, University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 5Immunobiology Graduate Program, Cincinnati Children's Hospital, Cincinnati, OH, United States
Conventional T cells exhibit a delayed response to the initial priming of peptide antigens presented by major histocompatibility complex (MHC) proteins. Unlike conventional T cells, mucosal-associated invariant T (MAIT) cells quickly respond to non-peptidic metabolite antigens presented by MHC-related protein 1 (MR1). To elucidate the MR1-dependent activation program of MAIT cells in response to mycobacterial infections, we determined the surface markers, transcriptomic profiles, and effector responses of activated human MAIT cells. Results revealed that mycobacterial-incubated antigen-presenting cells stimulated abundant human CD8+ MAIT cells to upregulate the co-expression of CD69 and CD26, as a combinatorial activation marker. Further transcriptomic analyses demonstrated that CD69+CD26++ CD8+MAIT cells highly expressed numerous genes for mediating anti-mycobacterial immune responses, including pro-inflammatory cytokines, cytolytic molecules, NK cell receptors, and transcription factors, in contrast to inactivated counterparts CD69+/−CD26+/− CD8+MAIT cells. Gene co-expression, enrichment, and pathway analyses yielded high statistical significance to strongly support that activated CD8+ MAIT cells shared gene expression and numerous pathways with NK and CD8+ T cells in activation, cytokine production, cytokine signaling, and effector functions. Flow cytometry detected that activated CD8+MAIT cells produced TNFα, IFNγ, and granulysin to inhibit mycobacterial growth and fight mycobacterial infection. Together, results strongly support that the combinatorial activation marker CD69+CD26++ labels the activated CD8+MAIT cells that develop an innate-like activation program in anti-mycobacterial immune responses. We speculate that the rapid production of anti-mycobacterial effector molecules facilitates MAIT cells to fight early mycobacterial infection in humans.
Introduction
Innate and adaptive immune systems have framed a classical dichotomy of immune responses in health and diseases. Innate immune cells rapidly recognize and respond to microbial and endogenous ligands with germline-encoded innate receptors (1). In contrast, conventional naïve T cells undergo a prolonged clonal expansion in responses to major histocompatibility complex (MHC)-presented peptide antigens (1, 2). It has been demonstrated that CD4+ and CD8+ T cells are crucial in combating mycobacterial infections in T cell-deficiency and adoptive transfer mouse models (3, 4). In humans, CD4+ T cells are essential in fighting mycobacterial infections evidenced by a much higher risk of active tuberculosis in HIV coinfection with defective CD4+ T cell responses (5) and conventional CD8+ T cells suppress mycobacterial growth (4). However, a large number of mycobacterial-reactive CD8+ T cells in humans are activated by non-peptidic antigens presented by MHC class I-related protein I (MR1) and are identified as mucosal-associated invariant T (MAIT) cells (6–12). Therefore, labeling the mycobacterial-reactive MAIT cells and defining their activation program regarding its similarities to conventional T cell or innate cells will facilitate the understanding of their unique activation pathways and effector functions in anti-mycobacterial responses.
Previous observations support that MAIT cell response is “innate-like” upon the stimulation with mouse (8–12) and bacterial antigens (6–8), as different from conventional T cells. The fundamental explanation of this “innate-like” MAIT cell activation initially bases on the evidence that MAIT cells express an invariant TCRα chain to interact with a conserved metabolite antigen presented by a monomorphic MR1 protein (12–17). This conservation interestingly contrasts to the tri-molecular interaction for conventional T cell activation using heterogeneous TCRs to recognize variable peptide antigens presented by heterogeneous HLA molecules (1, 2). In a new bacterial infection, MAIT cells can be instantly activated by MR1-mediated antigen presentation bypassing a long priming stage for conventional T cells (8–12). Recently, MAIT cells were demonstrated to protect mice against mycobacterial infections (8, 18) and respond to mycobacterial-infected human cells (7, 8). Surprisingly, abundant MAIT cells in uninfected and latently infected individuals exhibit strong reactivity to mycobacteria and potentially complement conventional T cells in anti-mycobacterial defenses (7, 8). Moreover, reduced frequency of MAIT cells in active tuberculosis (7, 8, 19, 20) seriously demands the elucidation of MAIT cell activation programs and effector responses in mycobacterial infections. Although riboflavin precursor metabolites have been identified from multiple non-tuberculous bacteria (13, 16, 17) and the general activation program of MAIT cells has been defined using E.coli, cytokine, or anti-CD3 stimulation (21–26), it remains uncertain whether mycobacteria efficiently activate human MAIT cells and which surface markers specifically label activated MAIT cells in mycobacterial infection. Moreover, whether activated human MAIT cells show an innate-like transcriptomic program to elicit strong anti-mycobacterial immune responses is unknown. Therefore, applying MAIT cell-specific stimulants, defining the activation program, and determining the effector responses of human MAIT cells in mycobacterial infections will be highly translational to clinically label MAIT cells and mechanistically understand the anti-mycobacterial immune responses of MAIT cells in diseases.
In this study, we elucidated the activation programs of MAIT cells in mycobacterial infections to determine their similarity to CD8+ T and natural killer (NK) cells. Specifically, we activated the dominant CD8+MAIT cell population with mycobacterial stimulation and used the combinatory marker CD69+CD26++ to separate activated MAIT cells for transcriptomic analyses. Results revealed that CD69+CD26++ CD8+MAIT cells shared key gene expression and activation pathways with conventional CD8+ T cells and NK cells, supporting a unique “innate-like” activation program to induce early anti-mycobacterial responses.
Materials and Methods
Preparation of Human Monocyte-Derived Dendritic Cells and MR1-Overexpressing Cells
Blood samples free of detectable infectious or non-infectious diseases were obtained from healthy donors with written informed consent at the Hoxworth Blood Center in the University of Cincinnati. De-identified blood samples were processed according to the protocols approved by the Institutional Review Board of the University of Cincinnati. We isolated human peripheral blood mononuclear cells (PBMCs) and prepared human monocyte-derived dendritic cells (MoDCs) as performed (27, 28). Briefly, upon the isolation of PBMCs using Ficoll-paque gradient (GE Healthcare), we enriched monocytes by plastic adherence and differentiated monocytes using human recombinant granulocyte-macrophage colony-stimulating factor (300 U/ml) and interleukin 4 (200 U/ml) for 5 days (27, 28). The matured MoDCs were further activated with lipopolysaccharide (LPS) at 50 ng/ml for 24 h or incubated with bacteria. We also overexpressed the human MR1 protein by transducing the wild type human MR1 gene with the retroviral vector pMXIP (12) into human HLA-defective myeloid cell line K562 (K562.hMR1) (29) as we reported (11). The expression of the recombinant human MR1 protein was confirmed with flow cytometry, and its function was tested by MAIT cell line activation in bacterial incubation.
Bacterial Incubation of Antigen-Presenting Cells
MoDCs and K562.hMR1 cells were incubated with Listeria monocytogenes (L. monocytogenes strain J0161, Bei resources), Escherichia coli (E. coli strain BL21, New England BioLabs), Mycobacterium bovis- (M. bovis-) derived Bacille Calmette-Guerin (BCG) vaccine (Pasteur strain) (30), and avirulent Mycobacterium tuberculosis (M. tuberculosis, strain H37Ra, Colorado State University, Fort Collins, CO) (31). Both mycobacterial strains BCG and H37Ra were cultured for 5–6 days in middlebrook 7H9 complete medium at 37°C using an orbital shaker with a speed setting at 270 rpm. E.coli and L. monocytogenes were cultured overnight at 37°C in the Luria-Bertani broth using an orbital shaker at 100 rpm. Bacteria were harvested at a log-growing phase, washed with phosphate buffer saline (PBS), and measured for their absorbance (optical density at wavelength 600 nanometres, OD600) according to the report (32). OD600 provides a semi-quantitative method to estimate bacterial cell numbers sufficient for MAIT cell activation (32). Human MoDCs or K562.hMR1 cells were incubated with E.coli in an estimated cell to bacteria ratio of 1:5 and 1:40 and with BCG in a ratio of 1:0 and 1:100. The blockage of activation was performed with an anti-MR1 antibody (clone 26.5, mouse IgG2a, at 2 μg/ml) that blocks MR1-dependent MAIT cell activation (10–12). Anti-HLAI antibody (clone W6/32, mouse IgG2a, Biolegend, at 2 μg/ml) was used as an isotype control for the anti-MR1 antibody and was also used to block the irrelevant effect of MHC class I proteins with similar structures as MR1 (33). Moreover, the chemical inhibitor cyclosporine A (CsA), mainly blocking TCR-mediated calcium signaling pathway for T cell activation (34, 35), was applied at 0.5 μg/ml.
Enzyme-Linked Immunospot
Upon incubation with bacteria overnight, MoDCs and K562.hMR1 cells were washed and incubated with the MAIT cell line (D466F5) (7) in a ratio of 5:1 and 1:4, respectively, by considering the estimated sizes of these cell types for optimal cell contact. The enzyme-linked immunospot (ELISPOT) assay was performed, as we reported (27). Briefly, both bacterial-incubated antigen-presenting cells and MAIT cells were co-cultured for 5 or 15 h on the multiscreen filter plate (Millipore) coated with anti-human IFNγ antibody (Mabtech). IFNγ+ MAIT cell spots were then developed with an indirect immunostain approach using a biotinylated anti-human IFNγ antibody (Mabtech), ExtraAvidin conjugated by alkaline phosphatase (Sigma), and substrates BCIP/NBT (Sigma). We used CTL-ImmunoSpot S6 Micro Analyzer to visualize and quantify IFNγ+ MAIT cell spots. Directional differences between bacterial-incubated and non-incubated conditions and between without and with anti-MR1 blockage were statistically analyzed using a paired t-test.
Isolation and Activation of Primary Human MAIT Cells
Upon removing adhered monocytes, human PBMCs were incubated with anti-Vα7.2 antibody (3C10) conjugated with PE (Biolegend) and followed by a positive selection with anti-PE antibody-conjugated magnetic beads (MACS, Miltenyi Biotec), according to the manufacturer's instructions. Bacterial-incubated K562.hMR1 cells were washed and further incubated with anti-Vα7.2-enriched PBMCs with abundant primary human MAIT cells in a ratio of 1:4 for 15 h with the presence of an anti-CD28 (clone CD28.2) antibody at 2 μg/ml. In addition to the stimulation with bacterial-incubated MR1-expressing K562 cells, anti-Vα7.2-enriched (MAIT cell-enriched) and anti-Vα7.2-depleted (conventional T cell-enriched) PBMCs were also activated with pre-coated anti-CD3 (clone UCHT1) at 1 μg/ml and anti-CD28 (clone CD28.2) at 2 μg/ml as positive controls. Hence, anti-CD28 (clone CD28.2) at 2 μg/ml was used for all K562.hMR1- and anti-CD3-mediated stimulation of T cells. For intracellular cytokine staining, Brefeldin A (10 μg/ml), which inhibits protein transport from the endoplasmic reticulum to the Golgi complexes, was added 2 h prior to cell harvesting for intracellular staining.
Antibodies and Flow Cytometry for Human MAIT Cells
Upon the co-culture for 15 h, surface markers, intracellular cytokines, and transcription factors of MAIT cells were stained following the manufacturer's instructions (Biolegend unless noted). In brief, cells were washed twice with staining buffer (PBS with 2% FBS) then blocked with anti-human Fc receptor antibodies, including anti-CD64 (mouse IgG1, clone 10.1), CD32 (mouse IgG2b, clone FUN-2), CD16 (mouse IgG1, clone 3G8), and additional Fc receptor blocking solution human TruStain FcX. For surface staining, cells were incubated with the combination of monoclonal antibodies, including phycoerythrin (PE)-Vα7.2, biotin-CD4 (OKT4) and streptavidin-conjugated quantum dot 525 or brilliant violet 510-CD4, brilliant violet 711-CD8α (RPA-T8), Allophycocyanin/Cyanine7 (APC/Cy7)-CD161 (HP-3G10), brilliant violet 605 or 421-CD69 (FN50), and PE/Cy5-CD26 (BA5b), for 30 min at 4°C in dark. PE-CD3 (OKT3) was used to replace PE-Vα7.2 for Vα7.2-depleted T cells. The MR1 tetramer loaded with the E.coli metabolite 5-amino-6-D-ribitylaminouracil (5-A-RU) (16, 36) and labeled with brilliant violet 421 was obtained from the NIH tetramer facility. For the staining of intracellular cytokines and transcription factors, cells were first incubated with antibodies against surface markers. Then, cells were fixed and permeabilized using the Fix/Perm Kit (Biolegend) and further stained in the 1 x Perm buffer for 30 min on ice with anti-cytokine and anti-transcription factor antibodies, including PE/Cy7-TNF-α (MAb11), APC-IFNγ (4S.B3), Alexa fluor 647-granulysin (DH2), PE/Cy7-Tbet (4B10), and Alexa fluor 488-Eomes (644730, R&D systems). Flow cytometry used BD Fortessa and Millipore Guava EasyCyte 12 channel high throughput flow cytometer according to the manufacturer's instructions. Flow cytometry data were further compensated and analyzed using Millipore Guava incyte and FlowJo software programs. Directional differences between mycobacterial stimulation and Listeria control was statistically tested using a paired t-test.
RNA-Seq of Human MAIT Cells
Anti-Vα7.2-enriched cells were co-cultured with BCG-incubated K562.hMR1 cells or stimulated with anti-CD3 antibody plus an anti-CD28 antibody for inducing co-stimulation in all conditions. Upon co-culture for 15 h, cells were first gated on Vα7.2+CD161+CD4−CD8+ to enrich this major MAIT cell subset and further sorted based on CD69+CD26++ and CD69+/−CD26+/− to represent stimulated MAIT cells and non-stimulated MAIT-enriched cells. Similar to our report (27, 28), around one thousand cells were collected and lysed in the Lysis Buffer for total RNA extraction using mirVana kit (ThermoFisher, Grand Island, NY). RNA integrity was measured by a Bioanalyzer using Agilent RNA 6000 Pico Kit (Agilent, Santa Clara, CA) and showed a high quality of samples. Next, NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs, Ipswich, MA) was used for polyA RNA purification. The library for RNA-seq was prepared by using NEBNext Ultra Directional RNA Library Prep kit (New England BioLabs, Ipswich, MA). During the second cDNA synthesis, dUTP was incorporated to maintain strand specificity. The library was enriched and indexed via 15 cycles of PCR. The amplified libraries, together with the negative control, were cleaned up for Bioanalyzer QC analysis. To study differential gene expression, individually indexed and compatible libraries at 15 pM total were proportionally pooled for clustering on single-read flow cells in cBot system (Illumina, San Diego, CA). The clustered libraries were sequenced to generate 51 bp reads at ~25 million per sample. RNA-seq data can be accessed at the GEO database (accession number GSE124381).
Bioinformatic Analysis of Differentially Expressed Genes (DEGs)
Similar to our previous report (27), transcriptomic data were analyzed to identify DEGs between CD69+CD26++ CD8+MAIT cells and CD69+/−CD26+/− CD8+MAIT-enriched cells representing activated MAIT cells and inactivated MAIT-enriched cells. The list of DEGs of MAIT cells were analyzed for the gene co-expression, enrichment, and activation pathways in comparison to innate immune cells and conventional T cells. Sequence reads were first aligned to the genome and converted to intensity counts. Resulted intensity counts were compared between CD69+CD26++ and CD69+/−CD26+/− cells from three donors using the edgeR program on the Bioconductor R platform to identify DEGs based on the absolute fold change (>2 folds) and p-values (<0.05). DEGs were shown with volcano plots, and representative altered genes were shown with a heatmap generated with edgeR and ggplot2 programs on the R platform. To predict the co-expression and shared functional clusters of DEGs with innate immune cells and conventional T cells, we used a ToppCluster software package to search DEGs against several databases, including KEGG and REACTOME, and obtained multiple clusters of genes overlapped with other activated immune cells at a statistically significant level. ToppCluster uses a hypergeometric test to obtain functional enrichment (https://toppcluster.cchmc.org/) (37). Enrichment analyses were performed using the Gene Set Enrichment Analysis (GESA) software by searching multiple available expression databases according to the instruction (38). The shared gene clusters between MAIT cells and other immune cells were further input to software Cytoscape Version 3.3.0 (www.cytoscape.org/), a broadly used open-source software platform for visualizing complex networks. We also applied Cytoscape to search various databases, including PANTHER, MSigDB, KEGG, NCI Pathway, and Reactome databases, to identify comprehensive pathways for the activation of conventional T cells and NK cells. The DEGs were annotated using the representative pathways of T cell and NK cell activation.
Mycobacterial Growth Inhibition in Antigen-Presenting Cells
The mCherry-labeled BCG was obtained from Dr. Russell's lab (39) and used to incubate MoDCs and K562.hMR1 cells at a ratio of 50:1 for 2 h. BCG-incubated cells were washed for 4 times and further co-cultured with rested or anti-CD3/CD28-activated primary MAIT cells upon magnetic enrichment with anti-Vα7.2 antibody. After overnight co-culture, we measure the remained mCherry-labeled BCG with antigen-presenting cells (% mCherry+ cells).
Results
Rapid Activation of a Human MAIT Cell Line by Dendritic Cells and MR1-Expressing Cells
Different from published monocytes (8, 24) and a lung epithelial cell line (7), antigen-presenting cells in this study used a human MR1-overexpressed human hematopoietic cell line K562 (K562.hMR1) with defective MHC expression (12, 40) and monocyte-derived dendritic cells (MoDCs) with strong co-stimulation. K562.hMR1 cells allow us to specifically detect bacterial-activated human MAIT cells and delineate MR1-dependent MAIT responses to bacterial infection, as different from bacterial-irrelevant cell activation by anti-CD3 antibody, cytokines, or autologous monocytes (22–24). MoDCs were differentiated using granulocyte-macrophage colony-stimulating factor and interleukin 4 as we reported (27, 28). Prior to the co-culture with T cells, MoDCs were overnight incubated with live E.coli or M. bovis (BCG) at indicated ratios of cell:bacteria. The bacterial-incubated MoDCs were then washed and co-cultured with a human MAIT cell line (D466F5), which was derived from MAIT cells of an active tuberculosis patient (7). As measured in an Enzyme-linked Immunospot (ELISPOT) assay, the number of IFNγ+MAIT cell spots significantly enhanced upon the overnight stimulation (15 h of co-culture) of BCG- and E.coli-incubated MoDCs, to an extent much higher than the non-infected condition or the background response of MoDCs (Figures 1A,B). An anti-MR1 antibody (11) significantly blocked MR1-mediated MAIT cell activation, to an extent comparable to the blockage of cyclosporine that inhibits calcium-mediated signaling cascades in T cell receptor-mediated pathways (35). Statistical analyses support MAIT cell activation upon overnight co-culture and its dependence on MR1-mediated antigen presentation by MoDCs (Figure 1B). Results from three independent assays with a short (5 h) and overnight (15 h) co-culture and anti-MR1 antibody blockage confirmed the activation of MAIT cells and MR1 dependence (Figure 1C). These data demonstrated that MAIT cells were rapidly activated by bacterial-incubated MoDCs within hours in a manner partially dependent on MR1-mediated antigen presentation. However, T cell activation using autologous MoDCs or monocytes is potentially confounded by autoreactivity, as shown with a high CD69 expression of MAIT cells irrelevant to bacterial infection (24). Using bacterial-incubated K562.hMR1 cells, we showed that the human MAIT cell line was also activated in a manner largely blocked by an anti-MR1 antibody and the chemical inhibitor cyclosporine (Figures 1D,E). Three independent assays similarly demonstrated that bacteria-incubated K562.hMR1 rapidly activated MAIT cells with 5 h of co-culture (Figure 1F). Although the MAIT cell line D466F5 was derived from an active tuberculosis patient, its TCR (TRAV1-2/TRBV6) was not evidenced being biased to M. tuberculosis antigens in this assay (Figure 1) or in other reports (7, 41). Together, these results established a rapid assay of MR1-dependent human MAIT cell activation using K562.hMR1 cells, allowing us to identify surface markers and activation pathways of primary human MAIT cells in the following assays.
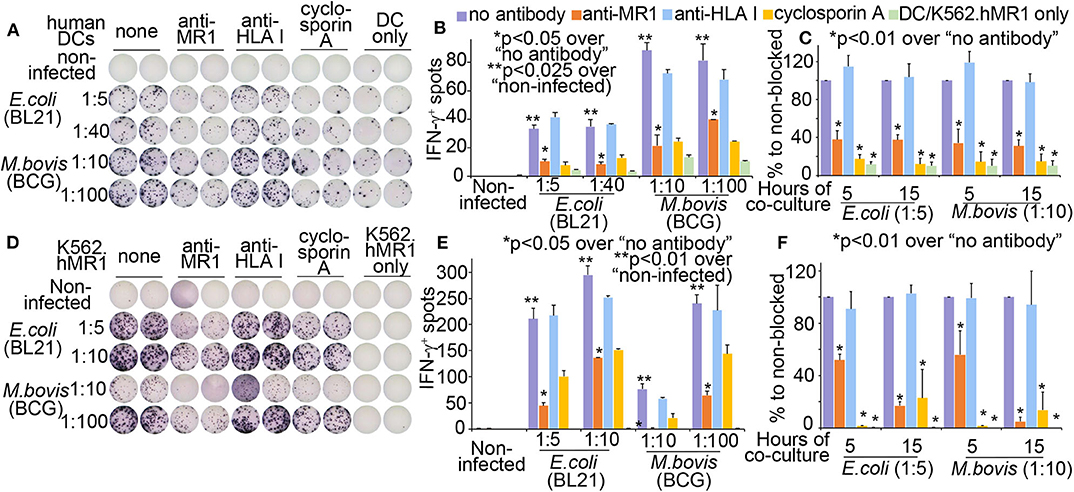
Figure 1. Monocyte-derived dendritic cells (MoDCs) and human MR1-overexpressed K562 cells (K562.hMR1) to activate a human MAIT cell line. Differentiated MoDCs were incubated with E.coli and M. bovis strains overnight. Bacterial cell number was estimated by OD600 absorbance and ratios of bacteria to DCs were shown. Bacterial-incubated MoDCs were washed and further co-cultured with the MAIT cell line (D466F5) (7) for 15 h. Anti-MR1 (clone 26.5, 2 μg/ml), Anti-HLAI antibody (clone W6/32 at 2 μg/ml), cyclosporine A (0.5 μg/ml), and MoDC only were used as controls. IFNγ+ MAIT cells measured by an ELISPOT assay were shown from one independent assay with MoDCs from one donor (A). Means from technical duplicates in panel A are shown with standard errors and statistically analyzed using a paired t-test (B). Three independent assays of DC-MAIT cell co-culture were performed with antibody blockage for 5 or 15 h. Data were normalized to non-blocked condition, shown with standard errors, and statistically analyzed using a paired t-test (C). At the same setting to MoDCs, bacterial-incubated K562.hMR1 cells were co-cultured with the MAIT cell line (D466F5) to stimulate IFNγ+ spots (D). IFNγ+ MAIT spots in technical duplicates were similarly analyzed (E). Three independent assays were similarly performed and analyzed (F).
CD69+CD26++ Labels Activated Human CD8+MAIT Cells
To date, it remains unclear which surface markers specifically detect and separate activated MAIT cells. The discovery of these markers is crucial for the translational detection of MAIT cells and allows further functional or mechanistic characterization of MAIT cells in bacterial infections and various diseases. Previous studies show that the early T cell activation marker CD69 expresses on stimulated MAIT cells upon bacterial infection; however, CD69 also upregulates in uninfected conditions (8, 12, 24). Moreover, dipeptidyl peptidase (DPP4) named CD26 upregulates on MAIT cells in mycobacterial infection (42). However, CD26 also expresses on MAIT cells in the absence of stimulation (42) and on NK cells (43). Therefore, CD69 or CD26 alone appears unable to separate stimulated MAIT cells from the non-stimulated cells completely. To quantify and define activated MAIT cells, we showed that the co-expression of CD69 and CD26 as a combinatorial marker CD69+CD26++ separated the activated CD8+MAIT cells from the inactivated CD69+/−CD26+/− CD8+MAIT-enriched cells (Figure 2). The latter population was defined based on the remaining subset in the conditions without bacterium or with the negative control Listeria (Figure 2A). In this assay, we co-cultured Vα7.2-enriched human PBMCs with bacterial-incubated K562.hMR1 cells and an anti-CD28 antibody. Upon overnight co-culture, cells were gated at Vα7.2+CD161+CD4−CD8+ or Vα7.2+CD161+CD4−CD8− to separate CD69+CD26++ cells from CD69+/−CD26+/− cells (Figure S1), as Vα7.2+CD161+ gating has been used in multiple studies to detect MAIT cells (8, 42, 44–46), especially bacterial-activated MAIT cells (7, 8, 23). Notably, the percentage of CD69+CD26++ MAIT cells was dramatically upregulated through co-culture with MR1-expressing cells, which were pre-incubated with E.coli, M. bovis (BCG), and avirulent M. tuberculosis (H37Ra) (Figure 2). Moreover, CD69+CD26++ MAIT cells were clearly separated from CD69+/−CD26+/− CD8+MAIT-enriched cells. The latter subset remained in non-optimal stimulatory conditions or non-stimulated conditions, such as conditions without bacteria or with non-antigenic Listeria monocytogenes (13, 16). Direct stimulation of Vα7.2-enriched cells with anti-CD3/CD28 antibodies as a positive control also upregulated CD69+CD26++ CD8+MAIT cells (Figure 2). Similarly, in the Vα7.2+CD161+CD4−CD8− population, CD69+CD26++ cells were also upregulated upon various bacterial-stimulations (Figure 2). Moreover, these CD69+CD26++ MAIT cells nearly disappeared upon anti-MR1 blockage in contrast to the isotype control using an anti-HLA class I antibody (Figure S2), since stimulation using K562.hMR1 cells likely skewed to the induction of MR1-dependent MAIT cell activation. Thus, mycobacterial stimulation consistently upregulates the CD69+CD26++ CD8+MAIT cells dependent on the MR1 molecule, while reversely reduce the CD69+/−CD26+/− CD8+MAIT-enriched cells (Figure 2B), strongly supporting that CD69+CD26++ is a specific and sensitive marker to label activated MAITs. These results support that the MR1-dependent MAIT cell activation labeled with a combinatorial marker CD69+CD26++ can be broadly applied as a promising translational measurement to detect the activated MAIT cells in human.
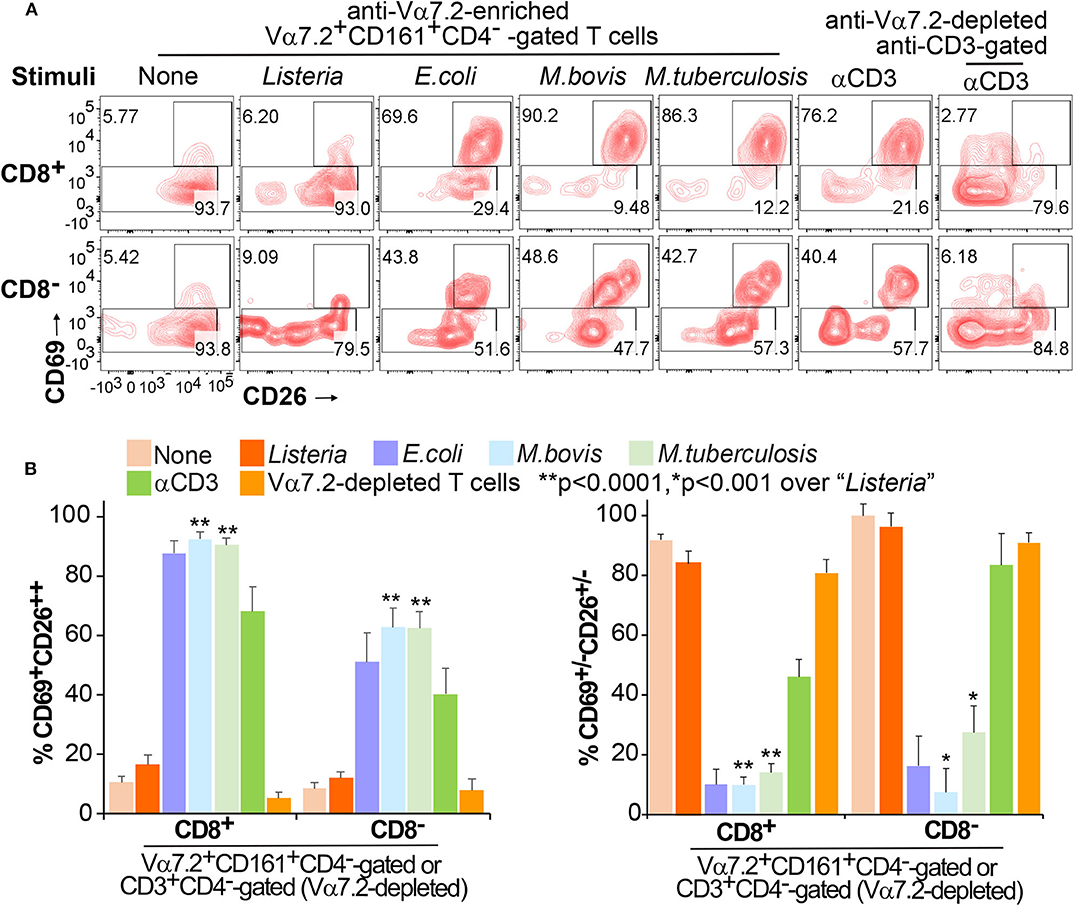
Figure 2. Differentiation of Vα7.2+CD161+CD4−CD8+CD69+CD26++ MAIT cells. Anti-Vα7.2-enriched PBMCs were co-cultured with bacterial-incubated K562.hMR1 cells and anti-Vα7.2-depleted PBMCs were stimulated with anti-CD3 for 15 h. All groups contained an anti-CD28 antibody for co-stimulation. Flow cytometry gated Vα7.2+CD161+CD4−CD8+ (or CD8−) in anti-Vα7.2-enriched PBMCs and CD3+CD4−CD8+ (or CD8−) in anti-Vα7.2-depleted PBMCs. % CD69+CD26++ MAIT cells (upper) and CD69+/−CD26+/− cells (bottom) are annotated. Contour plots show data from one independent assay using PBMCs from one healthy donor (A). Data from four independent assays were plotted with standard errors and analyzed with paired t-tests (B).
Recently, the MR1 tetramer loaded with the E.coli metabolite 5-amino-6-D-ribitylaminouracil (5-A-RU) (16, 36) was used to stain non-stimulated or metabolite antigen-stimulated MAIT cells from healthy donors or patients (45, 47–50). However, whether the MR1-5-A-RU tetramer also serves as a reliable tool to stain activated MAIT cells in bacterial infection is unknown. To explore this possibility, we co-cultured fresh peripheral blood mononuclear cells (PBMCs) without anti-Vα7.2 and magnetic enrichment with bacterial-incubated K562.hMR1 cells for 15 h. Results showed that the MR1-5-A-RU tetramer was able to stain non-stimulated MAIT cells in PBMCs without bacterial incubation or with Listeria incubation. We also showed Vα7.2+MR1-5-A-RU+ PBMCs were mostly Vα7.2+CD161++ (Figure S3A), as consistent with previous reports (24, 45, 47, 48). Minimal background staining of the MR1-5-A-RU tetramer in Vα7.2− PBMCs was also similar to that in reports (24, 48). However, MR1-5-A-RU tetramer staining was surprisingly weak or negative for bacterial-stimulated MAIT cells (Figure S3A). Expended gating showed that CD69+CD26++ cells re-emerged from Vα7.2+/−MR1-5-A-RU+/− population (Figure S3B), indicating that mycobacterial-activated MAIT cells downregulated Vα7.2 expression and MR1-5-A-RU staining signals consistent to the report (51). Regardless, activated MAIT cells in the same PBMC samples were still feasibly recognized by Vα7.2 and CD161 antibodies, as eventually labeled by the combinatorial marker of CD69+CD26++ (Figure S3C) at a similarly high percentage to the anti-Vα7.2-enriched and rested MAIT cells (Figure 2). Therefore, the combinatorial marker CD69+CD26++ is able to detect activated MAIT cells dependent on MR1-mediated antigen-presentation from Vα7.2+CD161+ population in bacterial infections. Importantly, CD69+CD26++ can define human MAIT cells based on MR1 dependence, similar to the definition of CD1-restricted T cells (52, 53) and previous definition of MAIT cells by MR1-dependent activation (7, 8, 10–12).
CD69+CD26++ Human CD8+MAIT Cells Display a Transcriptome With Multiple Upregulated Activation Markers
Our finding of the activation marker CD69+CD26++ allows us to separate activated MAIT cells (CD69+CD26++) unprecedentedly from inactivated MAIT-enriched cells (CD69+/−CD26+/−) in mycobacterial infection or in disease conditions to elucidate the activation program of Vα7.2+CD161+CD4−CD8+ MAIT cells. Recent publications used a MAIT cell antigen (21), anti-CD3 antibody (22), E.coli infection (23), and MR1-independent stimulants (22, 23) to determine the transcriptomes of activated MAIT cells. Different from these activation conditions, we would determine the transcriptomic program of abundant human CD8+ MAIT cells in response to the initial priming of mycobacterial infection in an MR1-dependent manner. MR1-dependent activation also allow us to specifically define MAIT cells, as the similar definition used for CD1-restricted T cells (52, 53) and in early studies of MAIT cells (7, 8, 10–12). Anti-Vα7.2-enriched human PBMCs (n = 3 donors) were co-cultured with BCG-incubated K562.hMR1 cells or stimulated with anti-CD3 antibody for 15 h similarly using anti-CD28 for co-stimulation in all conditions. BCG is the only licensed vaccine against tuberculosis in children and partially protects adults (54). BCG vaccination and M. tuberculosis infection enhance MAIT cell frequency in primates, similar to BCG vaccination in humans (55, 56). BCG stimulation also allows us to sort activated primary human MAIT cells through the core facility at an allowed biosafety level for transcriptomic analyses, which inform the MAIT cell activation program in mycobacterial infection. As in Figure S1, we sorted CD69+CD26++ and CD69+/−CD26+/− cells for transcriptomic analyses. We further isolated polyA RNA from sorted cells and performed quality control (27, 28). Then polyA RNA was reversed transcribed, barcoded, and sequenced using the Illumina Hiseq approach as we reported (27, 28). After the determination of gene identities and quantification of intensity counts, we deposited the transcriptomic data to the GEO database (accession number GSE124381) and normalized transcriptomes using the edgeR program. As a result, CD8+ MAIT cell transcriptomes detected a total of 14,077 transcripts with minimally 1 count per million (cpm) from at least one out of three donors. Differentially expressed genes (DEGs) were defined with more than 2-fold difference of intensity counts between CD69+CD26++ activated CD8+ MAIT cells and CD69+/−CD26+/− inactivated CD8+ MAIT-enriched cells. We used a p-value (<0.05) to test different gene expression between activated and inactivated Vα7.2+CD161+CD4−CD8+ cells from three donors by considering the variability in different donors, in vitro bacterial stimulation, T cell activation, cell sorting, and RNA preparation in different assays. To validate the results, we used a heatmap to list multiple clusters of representative genes relevant to the activation of MAIT cells, conventional T cells, and NK cells, as referred below, validating our transcriptomic datasets of the dominantly activated CD69+CD26++ CD8+MAIT cells (Figure 3A). For example, these signature genes encode surface markers CD69, CD8A, DPP4 (CD26), CD3Z, and KLRB1 (CD161); cytokine and receptors TNF (TNFα), IFNGR1, IL18R1 (IL-18Rα) and IL21R; cytolytic effector molecules GNLY (granulysin), PRF1 (perforin) (44), and FAS (CD95) (57), signaling molecules SLA, SLAMF1, and NFKB1; transcription factors TBX21 (Tbet), RORC (RORγt) (46), IKZF2 (Helios), RUNX2, and ZBTB16 (PLZF) (24, 58) (Figure 3A).
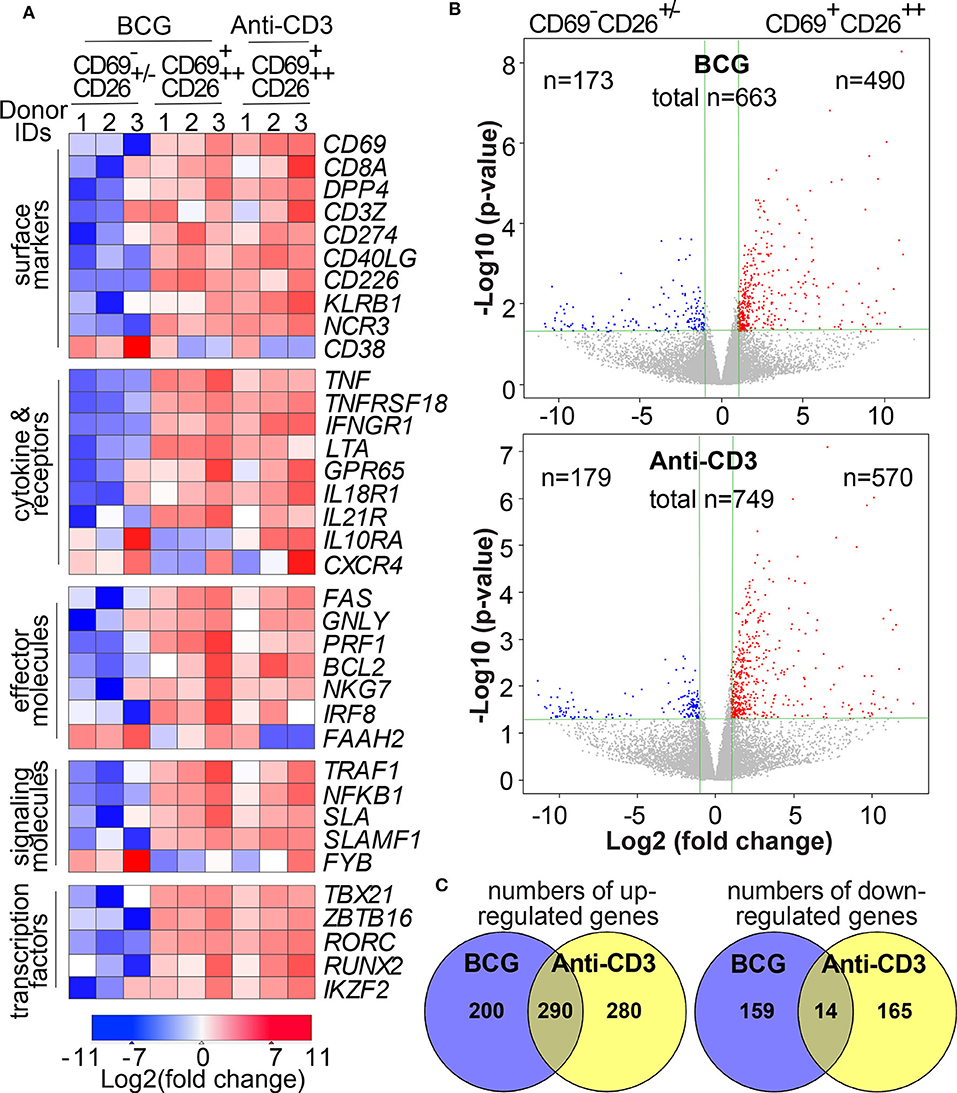
Figure 3. Transcriptomes of Vα7.2+CD161+CD4−CD8+CD69+CD26++ MAIT cells depict multiple activation markers. Upon overnight stimulation using BCG-incubated K562.hMR1 cells or anti-CD3 antibody with anti-CD28 co-stimulation in both conditions, anti-Vα7.2-enriched cells were gated on Vα7.2+CD161+CD4−CD8+ cells and further sorted into two subsets, CD69+CD26++ activated MAITs and CD69+/−CD26+/− inactivated MAIT-enriched cells as in Figure 2 and Figure S1. Representative gene expression relevant to MAIT cell activation was compared between BCG-activated MAIT cells and inactivated cells from three healthy donors using anti-CD3 as an activation control. Relative expression levels of these genes are shown in a heatmap with the color scheme representing Log2(fold change) for each gene (A). Volcano plots show up- and down-regulated differentially expressed genes (DEGs) with fold changes (vertical green lines for 2 folds) and p-values (horizontal green lines for p = 0.05) (B). Venn diagrams show overlapped numbers of DEGs between BCG and anti-CD3 stimulation conditions (C).
CD69+CD26++ Human CD8+MAIT Cells Co-express Genes and Share Functional Pathways With Activated T Cells and Innate Immune Cells
Although MAIT cell transcriptomes had been shown different from other T cells or between MAIT cell subsets (21, 59), we would address whether mycobacterial infection stimulated an activated transcriptomic program in MAIT cells. Specifically, we determined whether genes differentially expressed between activated MAIT cells vs. inactivated cells. Further, we characterized whether these DEGs in MAIT cells were similar to other adaptive or innate immune cells to support an innate-like activation program associated with anti-mycobacterial functions. To perform these analyses, we started with volcano plots that revealed hundreds of DEGs between activated CD69+CD26++ CD8+MAIT cells and inactivated CD69+/−CD26+/− CD8+MAIT-enriched cells in BCG and anti-CD3 stimulations, respectively (Figure 3B). The completed lists of up- and down-regulated genes in BCG (n = 663 in Table S1) and anti-CD3 (n = 749 in Table S2) stimulations were further used for downstream Venn diagram, enrichment, and pathway analyses. Venn diagrams showed large numbers of different and shared genes in BCG and anti-CD3/CD28 stimulation (Figure 3C). In this comparison, 200 upregulated genes (41%) were unique in BCG stimulation and 280 genes (49%) in anti-CD3/CD28 activation. In parallel, 159 downregulated genes (92%) were unique in BCG stimulation and 165 genes (92%) in anti-CD3/CD28 activation. Results supported that BCG stimulation induces a transcriptomic program partially different from that induced by anti-CD3/CD28 antibodies (58), despite that the activated cells were sorted with the same surface markers. To further statistically determine the gene clusters co-expressed with other cell types, we searched the identified BCG-induced DEGs against the gene expression databases in MSigDB (http://software.broadinstitute.org/gsea/msigdb) using the Toppcluster program (37). We identified more than 3,000 of reported gene expression datasets, including conventional CD8+ T cells, NKT, NK cells, macrophages, dendritic cells, and other cell types. These T and innate cells co-expressed a large number of genes with BCG-stimulated MAIT cells at a statistically significant level (with a false discovery rate FDR-corrected p < 0.01), as shown with multiple representative gene sets (Figure 4A). We also use enrichment analyses to show multiple significantly co-expressed datasets, including NK cells, cytotoxic T cells, NKT cells, and monocytes (Figure 4B). To further annotate functional pathways of activated MAIT cells overlapped with other immune cells, we searched BCG-induced DEGs against various cellular pathways using Toppcluster and constructed a pathway network using the Cytoscape program (http://www.cytoscape.org), based on the cutoff of an FDR-corrected p-value at 0.01 (Figure 4C). This network shows that MAIT cells share gene expression in numerous pathways with multiple cell types, including T and NK cell activation, NFκB signaling, MAPK signaling, cytokine productions, and effector functions, etc. Hence, our data reveal that MAIT cell activation shares gene clusters and functional pathways with conventional T cells and innate immune cells.
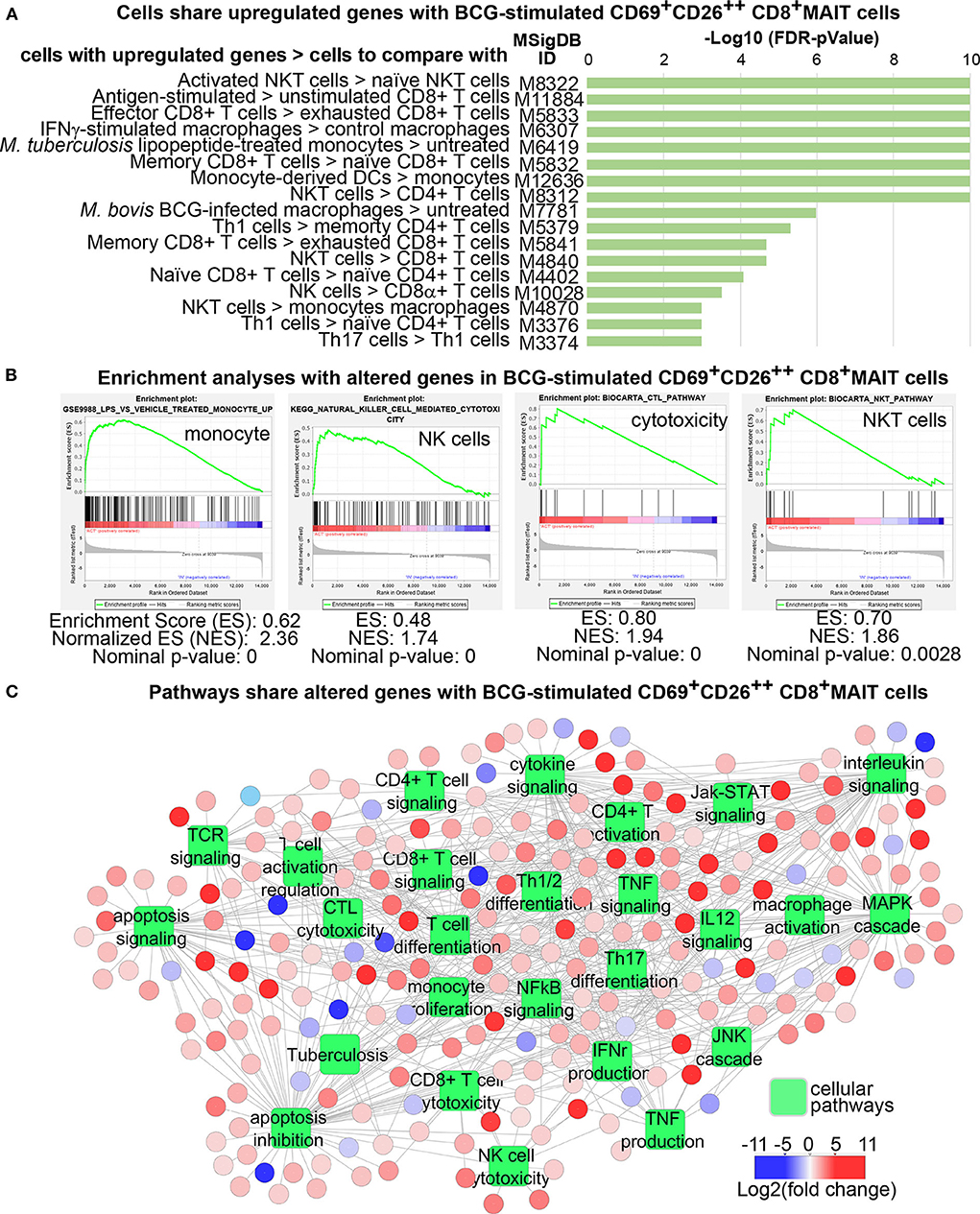
Figure 4. Transcriptomes of Vα7.2+CD161+CD4−CD8+CD69+CD26++ MAIT cells display gene clusters and functional pathways shared with T cells and innate immune cells. Upregulated genes from BCG-stimulated MAIT cells were used to search against the Molecular Signatures Database (MSigDB) and identified significant gene co-expression with more than 3,000 gene datasets (p < 0.01). Some representative datasets with various p-values are shown (A). Enrichment analyses identified multiple example datasets (B). DEGs from BCG stimulation of MAIT cells significantly overlap with genes in various cellular pathways of activated immune cells (p < 0.01) (C).
CD69+CD26++ Human CD8+MAIT Cells Display a Gene Profile Co-expressed in the T Cell Activation Pathway
From the comprehensive gene profiles shared between MAIT and other immune cells (Figure 4B), it is critical to annotate these shared genes in T cell activation pathways. Therefore, we used Cytoscape to obtain a map of canonical T cell activation pathways (60) to annotate BCG-altered genes. The upregulated genes included CD3Z (CD247), CD8A, and LCP2, downstream transcription factors NFKB1A, NFKB1, and REL, which together supported a TCR-mediated NFκB pathway (60) (Figure 5A). Other genes include PTPN6 (SHP1) important for TCR signaling (61), TNF, BCL2, TRAF1, IL15Ra, TNFRSF9 (4-1BB), and FAS for regulating effector function. Moreover, MAIT cells stimulated with anti-CD3/CD28 antibodies induced additional signaling molecules, including ITK, LCK, and VAV1 (Figure 5A). Anti-CD3/CD28 antibodies appear capable of stimulating both NFκB- and MAP3 Kinase-mediated signaling pathways (62, 63), likely promoting more comprehensive singling transduction in comparison to BCG stimulation. These data demonstrate that activated MAIT cells share common signature genes in canonical activation pathways with conventional T cells.
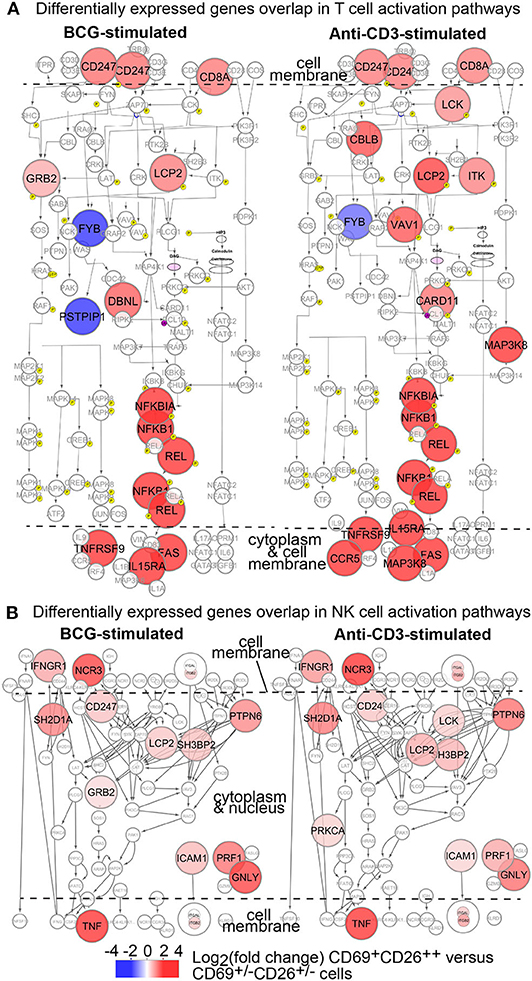
Figure 5. Transcriptomes of Vα7.2+CD161+CD4−CD8+CD69+CD26++ MAIT cells depict overlapped genes in conventional T cell and NK cell activation. DEGs from BCG or anti-CD3 stimulation of MAIT cells are annotated in canonical T cell (A) and NK cell (B) activation pathways as colored.
CD69+CD26++ Human CD8+MAIT Cells Display a Gene Profile Co-expressed in the NK Cell Activation Pathway
It is known that MAIT cells express NK cell receptors CD161 and NCR3, as in our findings (Figures 2, 3). To understand whether activated MAIT cells also share gene signatures in activated NK cells, we similarly obtained canonical NK cell activation pathways through database search and comprehensively annotated BCG-altered MAIT cell genes (Figure 5B). Co-expressed genes included those encoding cell surface molecules, an IFNγ receptor (IFNGR1) for cytokine stimulation and NK cell chemotaxis (64), and a natural cytotoxicity triggering receptor NCR3 (NKp30) for CD3ξ molecule (CD247) interaction and NK cell differentiation (65). Co-expressed genes also encoded multiple signaling molecules, such as SH2D1A (SAP) and PTPN6 (SHP1), which are critical in both NK and T cells (61, 66). Ultimately, the upregulation of effector molecules, such as cytokine TNF (67, 68), cytolytic molecule perforin (PRF1) (57), and adhesion molecule ICAM1, enhance the protection against infection, killing of bacterial-infected cells, and cell migration to infected tissues. Therefore, activated CD8+MAIT cells also share signature gene expression in the NK cell activation pathway (Figure 5B).
CD69 and CD26 Label Pro-inflammatory Responses of Human CD8+MAIT Cells in M. tuberculosis Infection
Pro-inflammatory responses have been demonstrated as critical effector functions against mycobacterial infection (3, 69, 70). Our transcriptomic analyses suggest an enhanced pro-inflammatory response of MAIT cells upon BCG stimulation, including an enhanced expression of TNF, TBX21 (encoding the transcription factor Tbet), and IFNGR1. TNFα bears a prominent early anti-tuberculosis function, which was shown in mouse studies (68) and also supported by severe tuberculosis in human autoimmune diseases with anti-TNFα therapies (67). IFNγ is another critical cytokine in controlling the infection of M. tuberculosis (71). To determine whether primary human MAIT cells are quickly activated to produce anti-mycobacterial pro-inflammatory cytokines, we similarly stimulated MAIT cells with bacterial-incubated K562.hMR1 cells (Figure 2) and validated activated MAIT cells by gating on Vα7.2+CD161+CD4−CD8+CD69+CD26++ cells (Figure S1). Results showed the upregulated frequency of CD26++TNFα+ (Figures 6A,B) and CD69+TNFα+ (Figures 6C,D) MAIT cells in Vα7.2+CD161+CD4−CD8+ upon bacterial and anti-CD3/CD28 stimulation. Further gating on CD69+CD26++ from bacterial-stimulated conditions annotated high TNFα expression subset in comparing to un-stimulated CD69+/−CD26+/− subset in Listeria-incubated condition (Figures 6E,F). Consistently, we demonstrated similar results with IFNγ (Figure 6), which display a high degree of enhancement. The expression of transcription factor Tbet is also upregulated (Figure 6), similar to the enhanced TBX21 expression in the transcriptome of BCG-stimulated MAIT cells. Together, anti-mycobacterial pro-inflammatory responses are enhanced in response to E.coli and mycobacterial infections.
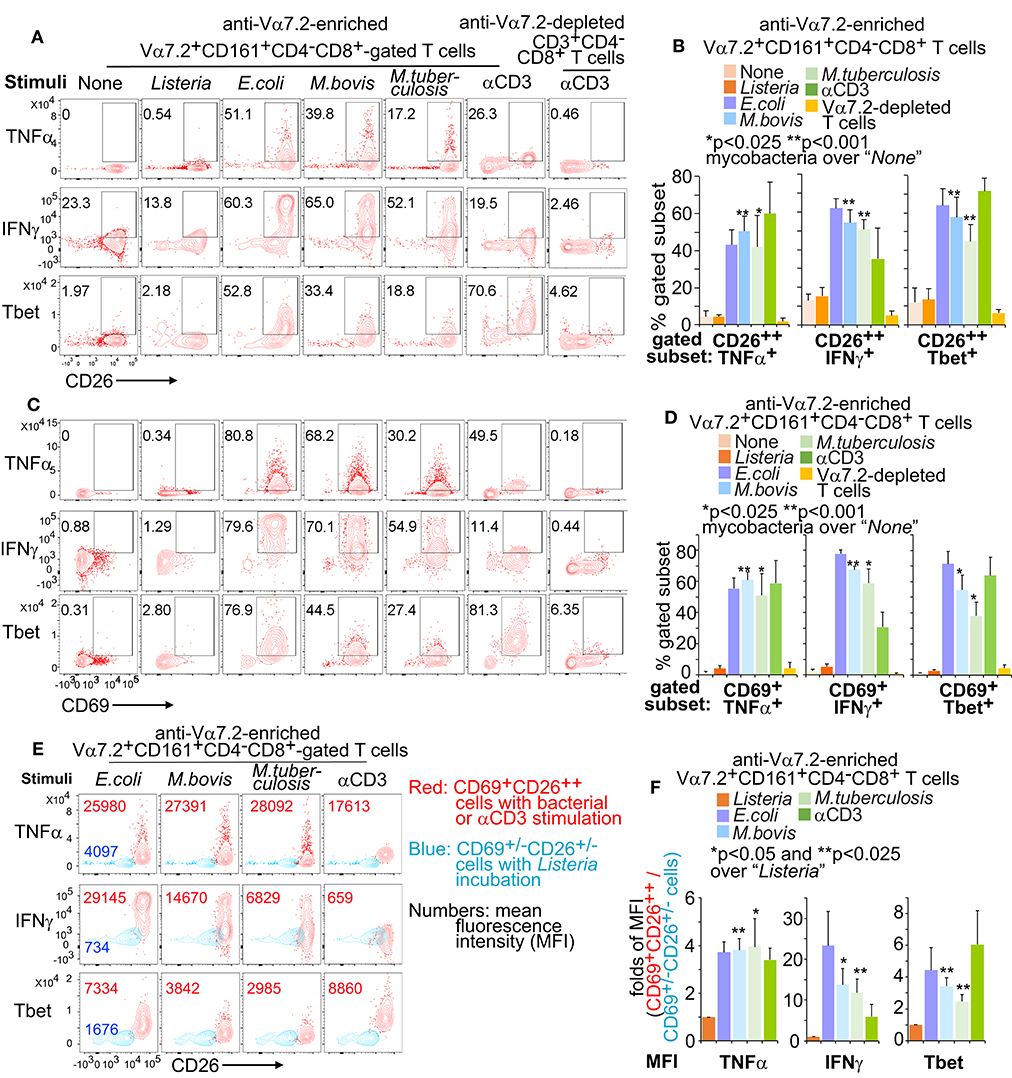
Figure 6. Pro-inflammatory cytokines from activated Vα7.2+CD161+CD4−CD8+ cells. Anti-Vα7.2-enriched PBMCs were similarly activated with bacterial-incubated K562.hMR1 cells or anti-CD3 with anti-CD28 presented in all conditions as in Figure 2. Flow cytometry gated Vα7.2+CD161+CD4−CD8+ cells from anti-Vα7.2-enriched PBMCs and gated CD3+CD4−CD8+ cells from anti-Vα7.2-depleted PBMCs. Contour plots show % positive cells on gated Vα7.2+CD161+CD4−CD8+CD26++ in one independent assay with PBMCs from one healthy donor (A). Data from four independent assays were plotted with standard errors and analyzed with the paired t-test (B). Vα7.2+CD161+CD4−CD8+CD69+ MAIT cells are also plotted (C) and statistically tested (D). Vα7.2+CD161+CD4−CD8+CD69+CD26++-gated activated MAIT cells in bacterial or anti-CD3 stimulation (Red) were further compared with Vα7.2+CD161+CD4−CD8+CD69+/−CD26+/− inactivated cells in Listeria incubation (Blue) by displaying the contour plots and mean fluorescence intensity (MFI) for noted molecules. Data from one independent assay using a blood sample from a healthy donor were shown (E). Fold difference of MFI between gated activated and inactivated cells were plotted with standard errors for data from four independent assays. Paired t-tests were performed to test the difference between mycobacterial stimulation and listeria incubation (F).
Stimulated Human MAIT Cells Enhanced Cytolytic Functions and Inhibited Mycobacterial Growth
Human MAIT cells express cytolytic molecules at an un-stimulated condition or upon the activation of fixed E.coli (24, 44, 72). However, it is unclear whether mycobacterial infection also stimulates MAIT cells to express cytolytic molecules and display a protective function. Overnight incubation of MAIT cells with BCG- or H37Ra-stimulated K562.hMR1 cells enhanced the % CD26++ and CD69+ MAIT cells expressing granulysin and Eomesodermin (Eomes) (Figures 7A–D). The gated CD69+CD26++ MAIT cells also upregulated the fluorescent staining intensity of granulysin and Eomes (Figures 7E,F). Results support that bacterial stimulation further upregulates cytolytic molecules over the basal level of expression. To determine whether both pro-inflammatory cytokines and cytolytic function together play a role to inhibit mycobacterial growth, we co-cultured the unstimulated or anti-CD3-stimulated Vα7.2-enriched cells with K562.hMR1 cells or human MoDCs pre-infected with mCherry-labeled BCG (39). Results demonstrated that either rested or pre-stimulated MAIT-enriched cells inhibited the growth of the mCherry-labeled BCG detected with flow cytometry (Figure 7G). Since pro-inflammatory cytokines and cytolytic functions of conventional T cells have been shown to fight mycobacterial infections (67, 68), our data support human MAIT cells similarly inhibit mycobacterial growth as in a previous report with mouse MAIT cells (18).
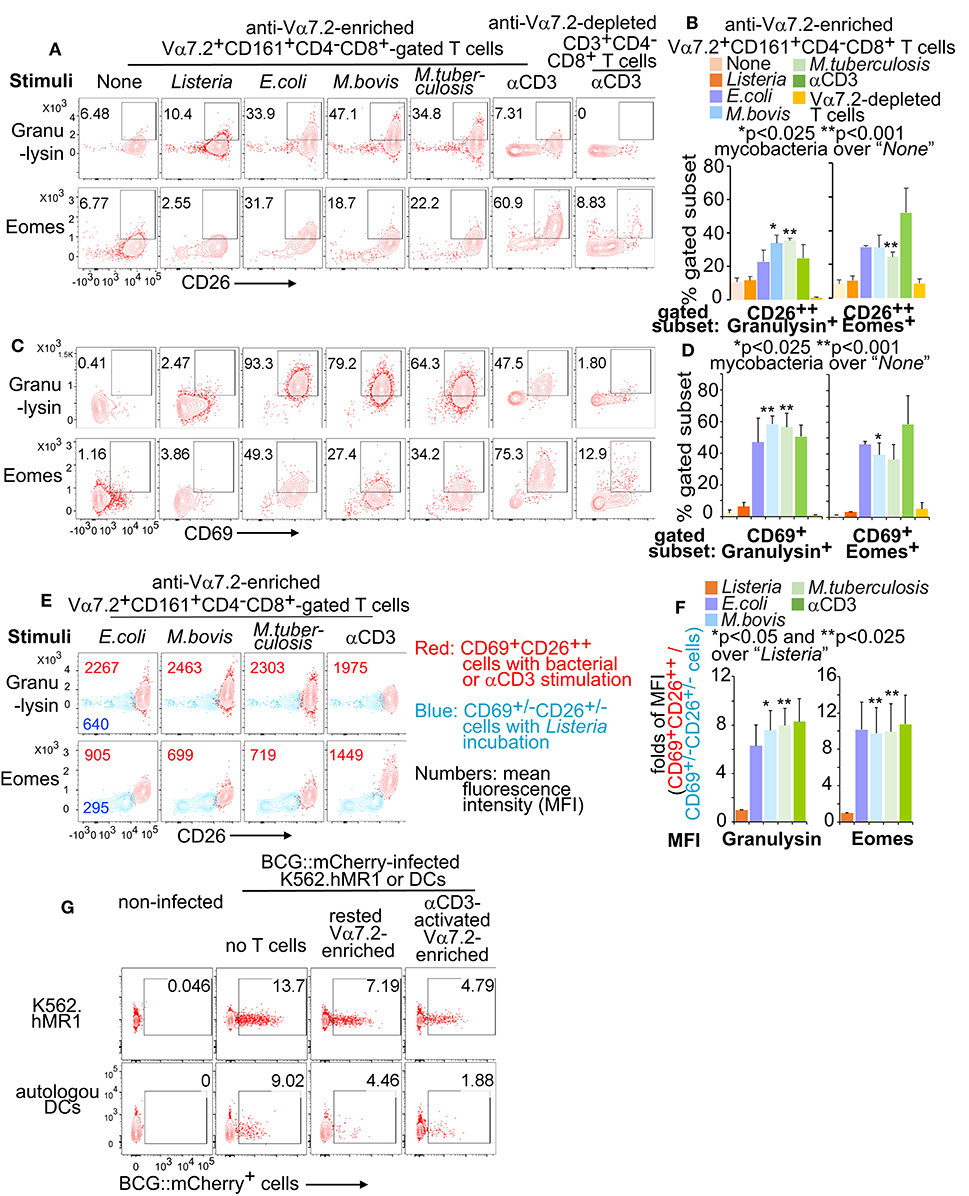
Figure 7. Cytolytic molecules from activated Vα7.2+CD161+CD4−CD8+ cells. Anti-Vα7.2-enriched PBMCs were activated and gated as described in Figure 6. Contour plots show % positive cells of cytolytic molecules on gated Vα7.2+CD161+CD4−CD8+CD26++ in one independent assay with PBMCs from one healthy donor (A). Data from three independent assays were plotted with standard errors and analyzed with paired t-tests (B). Vα7.2+CD161+CD4−CD8+CD69+ MAIT cells are also plotted (C) and statistically tested (D). Vα7.2+CD161+CD4−CD8+CD69+CD26++ activated MAIT cells (Red) were compared with Vα7.2+CD161+CD4−CD8+CD69+/−CD26+/− inactivated cells (Blue) with contour plots and mean fluorescence intensity (MFI) for noted molecules as in Figure 6. Data from one independent assay were shown (E). Fold difference of MFI between activated and inactivated cells from four independent assays were plotted and statistically tested with paired t-tests (F). The BCG.mCherry-incubated MoDCs and K562.hMR1 cells were washed for 4 times and co-cultured with rested or anti-CD3/CD28-activated MAIT-enriched cells for 18 h. Data show remained % mCherry+ cells from an assay using a blood sample of a healthy donor and three independent assays show similar results (G).
Discussion
The hypothesis of “innate-like T cells” was perhaps raised from the early observation of MAIT cell activation that depends on the interaction of a monomorphic MR1 protein, an antigen, and a semi-invariant TCR (9–14, 73–75). This conserved genetics and physical interaction in the MR1-antigen-TCR complex had likely provided a primordial mechanism that preserved innate features in MAIT cell activation and preceded the peptide antigen presentation (9–14), as similarly exemplified in CD1-restricted T cell activation (53, 75–77). In this study, we defined a combinatorial marker to label rapidly activated MAIT cells for the characterization of transcriptomic programs and effector functions against mycobacterial infections. Results reveal gene expression profiles of mycobacterial-activated MAIT cells share with activated CD8+ T cells and NK cells (Figures 3–5). Effector responses, including the enhanced production of pro-inflammatory cytokines (TNFα and IFNγ) and the cytolytic molecule (granulysin) (Figures 3, 4, 6, 7), and the inhibition of mycobacterial growth (Figure 7), are also similar to activated CD8+ T cells and NK cells (78). This unique innate-like activation program of MAIT cells is expected to provide a niche of early protection against M. tuberculosis infection, by potentially bridging innate (79) and adaptive immune responses (4, 5).
An activation marker is crucial for labeling and defining MAIT cells to further estimate MAIT effector functions in mycobacterial infections and other diseases. Previous reports mainly focused on detecting MAIT cells from blood samples using Vα7.2, CD161, or MR1 tetramer in humans and mice (7, 8, 24, 45, 47, 48). Nonetheless, it is challenging to isolate activated MAIT cells from the inactivated in bacterially infected conditions, due to the lack of activation-related surface markers for a complete separation and the potential downregulation of TCR expression that minimizing the signals of anti-Vα7.2 and tetramer staining on activated MAIT cells (Figure S3). Our results showed the TCR downregulation on mycobacterial-stimulated MAIT cells similar to that on activated conventional T cells (80), NKT cells (81), and MAIT cells (51), potentially contributing to a negative feedback regulation of TCR-mediated T cell responses (82). To overcome the difficulty of detecting activated MAIT cells, we used the combinatorial marker CD69+CD26++ to label a high percentage of Vα7.2+CD161++CD4−CD8+ cells at an MR1-dependent activation condition (Figure 2) as blocked by the anti-MR1 antibody (Figure S2). This combinatorial marker is highly translational to specifically detect activated vs. inactivated MAIT subsets from individual tuberculosis patients without using controls of other bacterial-infected patients. Likewise, CD69+CD26++ can be determined whether it is a candidate marker to label activated MAIT cells in M. tuberculosis-infected animal models. Particularly, the animal models are highly demanded to replicate abundant MAIT cell frequency, CD8+ MAIT cell responses, and M. tuberculosis-infected lung pathology in humans. As the major MAIT cell subset in humans (7, 8, 55), CD8+ MAIT cells can be stimulated to generate a higher percentage of CD69+CD26++ cells than CD4−CD8− MAIT cells in mycobacterial stimulation (p < 0.01). Interestingly, CD26 expression on CD8+ T cells has been associated with pro-inflammatory responses as a potential co-stimulatory molecule (83) and cytolytic responses as CD26 co-localizes with granulysin, perforin, and granzymes intracellularly (84). Thus, the high expression of CD26 and the differentiation of a CD69+CD26++ subset can perhaps label the activated CD8+ MAIT cells that produce pro-inflammatory and cytolytic molecules upon M. tuberculosis stimulation as in this study. Indeed, rapidly activated MAIT cells in M. tuberculosis stimulation consist of a higher percentage of CD8+ MAIT cells but a lower percentage of the CD4−CD8− subset than inactivated MAIT cells (Figure 2), consistent to the responses of CD8+ and CD4−CD8− MAIT cells in E.coli stimulation (49). Moreover, the differentiation kinetics and effector functions of CD8+ and CD4−CD8− MAIT cells in chronic M. tuberculosis infection require comprehensive investigations, as extended E.coli stimulation can decrease CD8+ MAIT cell frequency (49). To understand the early activation program of CD8+MAIT cells in mycobacterial stimulation, we used a heatmap to show the expression of multiple signature genes from these CD69+CD26++ CD8+MAIT cells, including surface markers, cytokines, cytolytic molecules, and transcription factors. These molecules have been mostly reported in activated MAIT, other T cells, and NK cells (8, 24, 42–46, 57, 58), validating the activation of CD69+CD26++ MAIT cells in mycobacterial infection (Figure 3A). Our results are also complementary to the findings that MAIT cells can be activated in a manner independent of MR1-mediated antigen presentation. For example, the stimulation with cytokine IL-18 also upregulates the expression of CD69 and IFNγ, independent on anti-MR1 blockage at viral-infected conditions (85, 86). The cellular context and molecular stimulants for MR1-dependent and independent MAIT cell activation are expected to be complementary and similarly important for inducing anti-microbial responses.
Recently, MAIT cell transcriptomes were compared with other T cells (59), associated with tissue repair function (21, 22), and coordination of immune responses (23). However, it remains unclear whether MAIT cells rapidly activated in pathogenic bacterial infections show an innate-like activation program to mediate protective responses. To understand the innateness of MAIT cell activation in mycobacterial infection beyond the conserved molecular interaction of MR1-antigen-TCR complex (12–14, 73, 74), we determined MAIT cell transcriptomes between activated MAIT cells Vα7.2+CD161+CD4−CD8+CD69+CD26++ and inactivated MAIT-enriched cells Vα7.2+CD161+CD4−CD8+CD69+/−CD26+/− in BCG stimulation. Our findings provide multiple lines of evidence to support the innate-like activation program of MAIT cells in mycobacterial stimulation. First, we compared DEGs (n = 663) from BCG-stimulated MAIT cells with various innate or adaptive immune cells and revealed thousands of datasets with co-expressed genes at high statistical significance (Figures 4A,B). Second, DEGs of CD69+CD26++ MAIT cells also significantly overlap with various pathways typically shown in activated CD8+ T cells, NK cells, and NKT cells (Figure 4C). These overlapped pathways broadly involve activation, differentiation, proliferation, cytotoxicity, cytokine production (e.g., IFNγ, TNF), apoptosis, and intracellular signaling, such as NFκB-, MAPK-, Jak-STAT-mediated signaling, in conventional T cells and innate immune cells (Figure 4C). Third, DEGs of activated MAIT cells overlap with both T cells and NK cells in their activation pathways (Figure 5). For example, CD3ζ (CD247) is important in coupling the antigen recognition of T cells and the CD16 surface ligation of NK cells to intracellular signaling pathways (87, 88). CD8α is critical in cytotoxic T cell and NK cell responses (89). NFκB contributes to the pro-inflammatory response and cell survival of T cells (90, 91), and receptor-mediated signaling transduction in innate immune cells (92). Upregulated genes also include multiple TNF superfamily factors, which usually interact with the NFκB-mediated pathway in conventional T cells and innate immune cells (93). Shared with NK cell activation pathways as well, genes encoding cell surface receptors IFNGR1 and NCR3 (NKp30) are crucial in other innate-like T cell populations, such as γδT cells (94, 95). Together, co-expression, enrichment, overlapped functional pathways, and shared genes in activation pathways between activated MAIT cells and other innate or adaptive immune cells strongly support the innateness of MAIT cell activation.
We expect this innate-like activation program and the combinatorial activation marker can be translated to understand early anti-mycobacterial MAIT cell immune responses in humans and animal models. First, our observation supports that rapid MAIT cell responses are potentially important in early protection against M. tuberculosis infection, relevant to previous findings of the conserved tri-molecular interaction in humans (73, 74) and the early inhibition of mycobacterial growth in mice (8, 18). BCG vaccination and M. tuberculosis infections are suggested to transiently enhance MAIT cell frequency in rhesus macaques (56), similar to transient MAIT cell responses to BCG vaccination in humans (55). This early MAIT cell response in vivo is consistent to the innate-like activation program of MAIT cells in this study, although the kinetics, trends, and distributions of MAIT cells or their activation in human M. tuberculosis infections requires further investigations (7, 96–98). Second, we observed that both BCG and avirulent H37Ra similarly activated primary human MAIT cells. However, virulent M. tuberculosis is usually less efficient in stimulating conventional T cells (99, 100), due to its virulence in causing necrosis and reducing antigen presentation capability of infected cells (101, 102), except for T cell responses specific to the unique antigens of virulent M. tuberculosis (103). For MAIT cells, BCG and M. tuberculosis have been suggested to comparably enhance peripheral MAIT cell frequency in the early stage of vaccination or infection of rhesus macaques (56), supporting a rapid MAIT cell activation. However, different from BCG vaccination, chronic lung pathology in M. tuberculosis infection may contribute to the redistribution of MAIT cells from blood to lung tissues and pleural space (42, 96, 97). Third, we showed that CD69+CD26++ CD8+MAIT cells upregulated the gene and protein expression of multiple pro-inflammatory and cytolytic molecules, such as TNFα, IFNγ, granulysin, perforin, and corresponding master transcription factors Tbet and Eomes, upon overnight mycobacterial stimulation. Anti-mycobacterial responses have been usually reflected by the production of pro-inflammatory cytokines from CD4+ and CD8+ T cells, and innate cells (3, 69, 70), together with the cytolytic function of CD8+ T cells (4). Relevant to the enhanced production of various pro-inflammatory cytokines and cytolytic molecules of early activated MAIT cells, co-culture of MAIT cells with mycobacterial-incubated K562.hMR1 cells and MoDCs inhibits the growth of mCherry-labeled BCG. These data support the potential protection of human MAIT cells against early mycobacterial infection, similar to the inhibition of BCG growth by mouse MAIT cells (18). Moreover, rapidly stimulated effector functions of activated CD8+MAIT cells likely facilitate the kicking in of adaptive immune responses to provide a full set of immune protection, representing a unique niche in anti-mycobacterial immune responses.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/ with an accession number GSE124381.
Ethics Statement
Blood samples free of detectable infectious or non-infectious diseases were obtained from healthy donors with written informed consent at the Hoxworth Blood Center in the University of Cincinnati. De-identified blood samples were processed according to the protocols approved by the Institutional Review Board of the University of Cincinnati.
Author Contributions
All authors: reviewed the manuscript. MS and SZ: perform assays, initial data analyses, and interpretation. LN: initial transcriptomic data analyses. DL: generation of MAIT cell lines. XZ: RNA sample preparation and sequencing. SH: study design, data analyses, and manuscript writing.
Funding
This work was supported by grants from National Institute of Allergy and Infectious Diseases (AI115358), American Lung Association (IA-629987), and National Institute of Environmental Health Sciences (ES006096).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Branch Moody for mycobacterial BCG and H37Ra strains, David G. Russell for the mCherry-labeled BCG strain, and Robert Giulitto for human blood samples, human blood donors in the Hoxworth Blood Center. Thank Harinder Singh and Tesfaye Mersha for suggestions on transcriptomic analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01136/full#supplementary-material
Abbreviations
MAIT, mucosal-associated invariant T; MHC, major histocompatibility complex; MR1, MHC-related protein 1; CD, cluster of differentiation; NK, natural killer; NKT, natural killer T cells; TNFα, tumor necrosis factor α; IFNγ, interferon γ; TCR, T cell receptor; HLA, human leukocyte antigens; PBMCs, peripheral blood mononuclear cells; MoDCs, monocyte-derived dendritic cells; LPS, lipopolysaccharide; BCG, Bacille Calmette-Guerin; M. bovis, Mycobacterium bovis; M. tuberculosis, Mycobacterium tuberculosis; E. coli, Escherichia coli; L. monocytogenes, Listeria monocytogenes; ELISPOT, enzyme-linked immunospot; Vα7.2, variable segment 7.2 of the T cell receptor alpha chain; APC/Cy7, Allophycocyanin/Cyanine7; PE, phycoerythrin; DEGs, differentially expressed genes; FDR, false discovery rate; Tbet, T-box transcription factor; Eomes, omesodermin.
References
1. Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. (1989) 54(Pt 1):1–13. doi: 10.1101/SQB.1989.054.01.003
2. Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. (2015) 33:169–200. doi: 10.1146/annurev-immunol-032414-112334
3. Cooper AM. Cell-mediated immune responses in tuberculosis. Annu Rev Immunol. (2009) 27:393–422. doi: 10.1146/annurev.immunol.021908.132703
4. Behar SM. Antigen-specific CD8(+) T cells and protective immunity to tuberculosis. Adv Exp Med Biol. (2013) 783:141–63. doi: 10.1007/978-1-4614-6111-1_8
5. Selwyn PA, Hartel D, Lewis VA, Schoenbaum EE, Vermund SH, Klein RS, et al. A prospective study of the risk of tuberculosis among intravenous drug users with human immunodeficiency virus infection. N Engl J Med. (1989) 320:545–50. doi: 10.1056/NEJM198903023200901
6. Young MH, U'Ren L, Huang S, Mallevaey T, Scott-Browne J, Crawford F, et al. MAIT cell recognition of MR1 on bacterially infected and uninfected cells. PLoS ONE. (2013) 8:e53789. doi: 10.1371/journal.pone.0053789
7. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. (2010) 8:e1000407. doi: 10.1371/journal.pbio.1000407
8. Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. (2010) 11:701–8. doi: 10.1038/ni.1890
9. Hansen TH, Huang S, Arnold PL, Fremont DH. Patterns of nonclassical MHC antigen presentation. Nat Immunol. (2007) 8:563–8. doi: 10.1038/ni1475
10. Huang S, Gilfillan S, Kim S, Thompson B, Wang X, Sant AJ, et al. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J Exp Med. (2008) 205:1201–11. doi: 10.1084/jem.20072579
11. Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, et al. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J Biol Chem. (2005) 280:21183–93. doi: 10.1074/jbc.M501087200
12. Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, et al. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci USA. (2009) 106:8290–5. doi: 10.1073/pnas.0903196106
13. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. (2012) 491:717–23. doi: 10.1038/nature11605
14. Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. (1993) 178:1–16. doi: 10.1084/jem.178.1.1
15. Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. (2003) 422:164–9. doi: 10.1038/nature01433
16. Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. (2014) 509:361–5. doi: 10.1038/nature13160
17. Harriff MJ, McMurtrey C, Froyd CA, Jin H, Cansler M, Null M, et al. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci Immunol. (2018) 3:eaao2556. doi: 10.1126/sciimmunol.aao2556
18. Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. (2012) 80:3256–67. doi: 10.1128/IAI.00279-12
19. Jiang J, Wang X, An H, Yang B, Cao Z, Liu Y, et al. MAIT cell function is modulated by PD-1 signaling in patients with active tuberculosis. Am J Respir Crit Care Med. (2014) 190:329–39. doi: 10.1164/rccm.201401-0106OC
20. Kwon YS, Cho YN, Kim MJ, Jin HM, Jung HJ, Kang JH, et al. Mucosal-associated invariant T cells are numerically and functionally deficient in patients with mycobacterial infection and reflect disease activity. Tuberculosis. (2015) 95:267–74. doi: 10.1016/j.tube.2015.03.004
21. Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. (2019) 28:3249–62.e3245. doi: 10.1016/j.celrep.2019.07.039
22. Leng T, Akther HD, Hackstein CP, Powell K, King T, Friedrich M, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. (2019) 28:3077–91. doi: 10.1016/j.celrep.2019.08.050
23. Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, et al. TCR- or cytokine-activated CD8(+) mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. (2019) 28:3061–76.e5. doi: 10.1016/j.celrep.2019.08.054
24. Dias J, Leeansyah E, Sandberg JK. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc Natl Acad Sci USA. (2017) 114:E5434. doi: 10.1073/pnas.1705759114
25. Dias J, Sobkowiak MJ, Sandberg JK, Leeansyah E. Human MAIT-cell responses to Escherichia coli: activation, cytokine production, proliferation, and cytotoxicity. J Leukoc Biol. (2016) 100:233–40. doi: 10.1189/jlb.4TA0815-391RR
26. Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. (2013) 121:1124–35. doi: 10.1182/blood-2012-07-445429
27. Sharma M, Zhang X, Zhang S, Niu L, Ho SM, Chen A, et al. Inhibition of endocytic lipid antigen presentation by common lipophilic environmental pollutants. Sci. Rep. (2017) 7:2085. doi: 10.1038/s41598-017-02229-7
28. Sharma M, Zhang X, Huang S. Integrate imaging flow cytometry and transcriptomic profiling to evaluate altered endocytic CD1d trafficking. J Vis Exp. (2018) 140:57528. doi: 10.3791/57528
29. Roder JC, Ahrlund-Richter L, Jondal M. Target-effector interaction in the human and murine natural killer system: specificity and xenogeneic reactivity of the solubilized natural killer-target structure complex and its loss in a somatic cell hybrid. J Exp Med. (1979) 150:471–81. doi: 10.1084/jem.150.3.471
30. Layre E, Lee HJ, Young DC, Martinot AJ, Buter J, Minnaard AJ, et al. Molecular profiling of Mycobacterium tuberculosis identifies tuberculosinyl nucleoside products of the virulence-associated enzyme Rv3378c. Proc Natl Acad Sci USA. (2014) 111:2978–83. doi: 10.1073/pnas.1315883111
31. Roura-Mir C, Wang L, Cheng TY, Matsunaga I, Dascher CC, Peng SL, et al. Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through TLR-2. J Immunol. (2005) 175:1758–66. doi: 10.4049/jimmunol.175.3.1758
32. Biesta-Peters EG, Reij MW, Joosten H, Gorris LG, Zwietering MH. Comparison of two optical-density-based methods and a plate count method for estimation of growth parameters of Bacillus cereus. Appl Environ Microbiol. (2010) 76:1399–405. doi: 10.1128/AEM.02336-09
33. Wang M, Tang ST, Stryhn A, Justesen S, Larsen MV, Dziegiel MH, et al. Identification of MHC class II restricted T-cell-mediated reactivity against MHC class I binding Mycobacterium tuberculosis peptides. Immunology. (2011) 132:482–91. doi: 10.1111/j.1365-2567.2010.03383.x
34. Flanagan WM, Corthesy B, Bram RJ, Crabtree GR. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. (1991) 352:803–7. doi: 10.1038/352803a0
35. Matsuda S, Koyasu S. Mechanisms of action of cyclosporine. Immunopharmacology. (2000) 47:119–25. doi: 10.1016/S0162-3109(00)00192-2
36. Eckle SB, Birkinshaw RW, Kostenko L, Corbett AJ, McWilliam HE, Reantragoon R, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med. (2014) 211:1585–600. doi: 10.1084/jem.20140484
37. Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinform. (2007) 8:392. doi: 10.1186/1471-2105-8-392
38. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
39. Sukumar N, Tan SM, Aldridge BB, Russell DG. Exploitation of Mycobacterium tuberculosis reporter strains to probe the impact of vaccination at sites of infection. PLoS Pathog. (2014) 10:e1004394. doi: 10.1371/journal.ppat.1004394
40. Huang S, Cheng TY, Young DC, Layre E, Madigan CA, Shires J, et al. Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proc Natl Acad Sci USA. (2011) 108:19335–40. doi: 10.1073/pnas.1112969108
41. Gold MC, McLaren JE, Reistetter JA, Smyk-Pearson S, Ladell K, Swarbrick GM, et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. (2014) 211:1601–10. doi: 10.1084/jem.20140507
42. Sharma PK, Wong EB, Napier RJ, Bishai WR, Ndung'u T, Kasprowicz VO, et al. High expression of CD26 accurately identifies human bacteria-reactive MR1-restricted MAIT cells. Immunology. (2015) 145:443–53. doi: 10.1111/imm.12461
43. Gorrell MD, Gysbers V, McCaughan GW. CD26: a multifunctional integral membrane and secreted protein of activated lymphocytes. Scand J Immunol. (2001) 54:249–64. doi: 10.1046/j.1365-3083.2001.00984.x
44. Leeansyah E, Svard J, Dias J, Buggert M, Nystrom J, Quigley MF, et al. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PLoS Pathog. (2015) 11:e1005072. doi: 10.1371/journal.ppat.1005072
45. Ben Youssef G, Tourret M, Salou M, Ghazarian L, Houdouin V, Mondot S, et al. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J Exp Med. (2018) 215, 459–479. doi: 10.1084/jem.20171739
46. Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. (2011) 117:1250–9. doi: 10.1182/blood-2010-08-303339
47. Gherardin NA, Loh L, Admojo L, Davenport AJ, Richardson K, Rogers A, et al. Enumeration, functional responses and cytotoxic capacity of MAIT cells in newly diagnosed and relapsed multiple myeloma. Sci Rep. (2018) 8:4159. doi: 10.1038/s41598-018-22130-1
48. Gherardin NA, Keller AN, Woolley RE, Le Nours J, Ritchie DS, Neeson PJ, et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. (2016) 44:32–45. doi: 10.1016/j.immuni.2015.12.005
49. Dias J, Boulouis C, Gorin JB, van den Biggelaar RHGA, Lal KG, Gibbs A, et al. The CD4(-) CD8(-) MAIT cell subpopulation is a functionally distinct subset developmentally related to the main CD8(+) MAIT cell pool. Proc Natl Acad Sci USA. (2018) 115:E11513–22. doi: 10.1073/pnas.1812273115
50. Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. (2015) 212:1095–108. doi: 10.1084/jem.20142110
51. Salio M, Gasser O, Gonzalez-Lopez C, Martens A, Veerapen N, Gileadi U, et al. Activation of human mucosal-associated invariant T cells induces CD40L-dependent maturation of monocyte-derived and primary dendritic cells. J Immunol. (2017) 199:2631–8. doi: 10.4049/jimmunol.1700615
52. Macho-Fernandez E, Brigl M. The extended family of CD1d-restricted NKT cells: sifting through a mixed bag of TCRs, antigens, and functions. Front Immunol. (2015) 6:362. doi: 10.3389/fimmu.2015.00362
53. Van Rhijn I, van Berlo T, Hilmenyuk T, Cheng TY, Wolf BJ, Tatituri RV, et al. Human autoreactive T cells recognize CD1b and phospholipids. Proc Natl Acad Sci USA. (2016) 113:380–5. doi: 10.1073/pnas.1520947112
54. Nemes E, Geldenhuys H, Rozot V, Rutkowski KT, Ratangee F, Bilek N, et al. Prevention of M. tuberculosis infection with H4:IC31 vaccine or BCG revaccination. N Engl J Med. (2018) 379:138–49. doi: 10.1056/NEJMoa1714021
55. Suliman S, Murphy M, Musvosvi M, Gela A, Meermeier EW, Geldenhuys H, et al. MR1-independent activation of human mucosal-associated invariant T cells by mycobacteria. J Immunol. (2019) 203:2917–27. doi: 10.4049/jimmunol.1900674
56. Greene JM, Dash P, Roy S, McMurtrey C, Awad W, Reed JS, et al. MR1-restricted mucosal-associated invariant T (MAIT) cells respond to mycobacterial vaccination and infection in nonhuman primates. Mucosal Immunol. (2016) 10:802–13. doi: 10.1038/mi.2016.91
57. Golstein P, Griffiths GM. An early history of T cell-mediated cytotoxicity. Nat Rev Immunol. (2018) 18:527–35. doi: 10.1038/s41577-018-0009-3
58. Gerart S, Siberil S, Martin E, Lenoir C, Aguilar C, Picard C, et al. Human iNKT and MAIT cells exhibit a PLZF-dependent proapoptotic propensity that is counterbalanced by XIAP. Blood. (2013) 121:614–23. doi: 10.1182/blood-2012-09-456095
59. Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. (2019) 216:133–51. doi: 10.1084/jem.20181483
60. Vallois D, Dobay MP, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. (2016) 128:1490–502. doi: 10.1182/blood-2016-02-698977
61. Paster W, Bruger AM, Katsch K, Gregoire C, Roncagalli R, Fu G, et al. A THEMIS:SHP1 complex promotes T-cell survival. EMBO J. (2015) 34:393–409. doi: 10.15252/embj.201387725
62. Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. (2013) 13:257–69. doi: 10.1038/nri3403
63. Martelli MP, Lin HM, Zhang WQ, Samelson LE, Bierer BE. Signaling via LAT (linker for T-cell activation) and Syk/ZAP70 is required for ERK activation and NFAT transcriptional activation following CD2 stimulation. Blood. (2000) 96:2181–90. doi: 10.1182/blood.V96.6.2181.h8002181_2181_2190
64. Pak-Wittel MA, Yang L, Sojka DK, Rivenbark JG, Yokoyama WM. Interferon-gamma mediates chemokine-dependent recruitment of natural killer cells during viral infection. Proc Natl Acad Sci USA. (2013) 110:E50–9. doi: 10.1073/pnas.1220456110
65. Siewiera J, Gouilly J, Hocine HR, Cartron G, Levy C, Al-Daccak R, et al. Natural cytotoxicity receptor splice variants orchestrate the distinct functions of human natural killer cell subtypes. Nat Commun. (2015) 6:10183. doi: 10.1038/ncomms10183
66. Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C. Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244). Blood. (2005) 105:4722–9. doi: 10.1182/blood-2004-09-3796
67. Keane J, Gershon S, Wise RP, Mirabile-Levens E, Kasznica J, Schwieterman WD, et al. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N Engl J Med. (2001) 345:1098–104. doi: 10.1056/NEJMoa011110
68. Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, Lowenstein CJ, et al. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. (1995) 2:561–72. doi: 10.1016/1074-7613(95)90001-2
69. Prezzemolo T, Guggino G, La Manna MP, Di Liberto D, Dieli F, Caccamo N. Functional signatures of human CD4 and CD8 T cell responses to Mycobacterium tuberculosis. Front Immunol. (2014) 5:180. doi: 10.3389/fimmu.2014.00180
70. Rook GA, Dheda K, Zumla A. Immune responses to tuberculosis in developing countries: implications for new vaccines. Nat Rev Immunol. (2005) 5:661–7. doi: 10.1038/nri1666
71. O'Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. The immune response in tuberculosis. Annu Rev Immunol. (2013) 31:475–527. doi: 10.1146/annurev-immunol-032712-095939
72. Kurioka A, Ussher JE, Cosgrove C, Clough C, Fergusson JR, Smith K, et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. (2015) 8:429–40. doi: 10.1038/mi.2014.81
73. Huang S, Moody DB. Donor-unrestricted T cells in the human CD1 system. Immunogenetics. (2016) 68:577–96. doi: 10.1007/s00251-016-0942-x
74. Huang S. Targeting innate-like T cells in tuberculosis. Front Immunol. (2016) 7:594. doi: 10.3389/fimmu.2016.00594
75. Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. (2013) 13:101–17. doi: 10.1038/nri3369
76. Brutkiewicz RR, Lin Y, Cho S, Hwang YK, Sriram V, Roberts TJ. CD1d-mediated antigen presentation to natural killer T (NKT) cells. Crit Rev Immunol. (2003) 23:403–19. doi: 10.1615/CritRevImmunol.v23.i56.30
77. Kumar A, Suryadevara N, Hill TM, Bezbradica JS, Van Kaer L, Joyce S. Natural killer T cells: an ecological evolutionary developmental biology perspective. Front Immunol. (2017) 8:1858. doi: 10.3389/fimmu.2017.01858
78. Lazarevic V, Glimcher LH, Lord GM. T-bet: a bridge between innate and adaptive immunity. Nat Rev Immunol. (2013) 13:777–89. doi: 10.1038/nri3536
79. Verrall AJ, Schneider M, Alisjahbana B, Apriani L, van Laarhoven A, Koeken V, et al. Early clearance of Mycobacterium tuberculosis is associated with increased innate immune responses. J Infect Dis. (2019) 221:1342–50. doi: 10.1093/infdis/jiz147
80. Cemerski S, Das J, Locasale J, Arnold P, Giurisato E, Markiewicz MA, et al. The stimulatory potency of T cell antigens is influenced by the formation of the immunological synapse. Immunity. (2007) 26:345–55. doi: 10.1016/j.immuni.2007.01.013
81. Wilson MT, Johansson C, Olivares-Villagomez D, Singh AK, Stanic AK, Wang CR, et al. The response of natural killer T cells to glycolipid antigens is characterized by surface receptor down-modulation and expansion. Proc Natl Acad Sci USA. (2003) 100:10913–8. doi: 10.1073/pnas.1833166100
82. Gallegos AM, Xiong H, Leiner IM, Susac B, Glickman MS, Pamer EG, et al. Control of T cell antigen reactivity via programmed TCR downregulation. Nat Immunol. (2016) 17:379–86. doi: 10.1038/ni.3386
83. Hatano R, Ohnuma K, Yamamoto J, Dang NH, Morimoto C. CD26-mediated co-stimulation in human CD8(+) T cells provokes effector function via pro-inflammatory cytokine production. Immunology. (2013) 138:165–72. doi: 10.1111/imm.12028
84. Lettau M, Dietz M, Vollmers S, Armbrust F, Peters C, Dang TM, et al. Degranulation of human cytotoxic lymphocytes is a major source of proteolytically active soluble CD26/DPP4. Cell Mol Life Sci. (2020) 77:751–64. doi: 10.1007/s00018-019-03207-0
85. Loh L, Wang Z, Sant S, Koutsakos M, Jegaskanda S, Corbett AJ, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc Natl Acad Sci USA. (2016) 113:10133–8. doi: 10.1073/pnas.1610750113
86. van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. MAIT cells are activated during human viral infections. Nat Commun. (2016) 7:11653. doi: 10.1038/ncomms11653
87. Baniyash M. TCR zeta-chain downregulation: curtailing an excessive inflammatory immune response. Nat Rev Immunol. (2004) 4:675–87. doi: 10.1038/nri1434
88. Galandrini R, Palmieri G, Paolini R, Piccoli M, Frati L, Santoni A. Selective binding of shc-SH2 domain to tyrosine-phosphorylated zeta- but not gamma-chain upon CD16 ligation on human NK cells. J Immunol. (1997) 159:3767–73.
89. Ahmad F, Hong HS, Jackel M, Jablonka A, Lu IN, Bhatnagar N, et al. High frequencies of polyfunctional CD8+ NK cells in chronic HIV-1 infection are associated with slower disease progression. J Virol. (2014) 88:12397–408. doi: 10.1128/JVI.01420-14
90. Paul S, Schaefer BC. A new look at T cell receptor signaling to nuclear factor-kappa B. Trends Immunol. (2013) 34:269–81. doi: 10.1016/j.it.2013.02.002
91. Oh H, Ghosh S. NF-kappaB: roles and regulation in different CD4(+) T-cell subsets. Immunol Rev. (2013) 252:41–51. doi: 10.1111/imr.12033
92. Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. (2012) 4:a006049. doi: 10.1101/cshperspect.a006049
93. Brenner D, Blaser H, Mak TW. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol. (2015) 15:362–74. doi: 10.1038/nri3834
94. Silva-Santos B, Strid J. Working in “NK mode”: natural killer group 2 member D and natural cytotoxicity receptors in stress-surveillance by gammadelta T cells. Front Immunol. (2018) 9:851. doi: 10.3389/fimmu.2018.00851
95. Campbell KS, Hasegawa J. Natural killer cell biology: an update and future directions. J Allergy Clin Immunol. (2013) 132:536–44. doi: 10.1016/j.jaci.2013.07.006
96. Wong EB, Gold MC, Meermeier EW, Xulu BZ, Khuzwayo S, Sullivan ZA, et al. TRAV1-2(+) CD8(+) T-cells including oligoconal expansions of MAIT cells are enriched in the airways in human tuberculosis. Commun Biol. (2019) 2:203. doi: 10.1038/s42003-019-0442-2
97. Jiang J, Chen X, An H, Yang B, Zhang F, Cheng X. Enhanced immune response of MAIT cells in tuberculous pleural effusions depends on cytokine signaling. Sci Rep. (2016) 6:32320. doi: 10.1038/srep32320
98. Estevez O, Anibarro L, Garet E, Martinez A, Pena A, Barcia L, et al. Multi-parameter flow cytometry immunophenotyping distinguishes different stages of tuberculosis infection. J Infect. (2020) 20:S0163–4453. doi: 10.1016/j.jinf.2020.03.064
99. Kim WS, Kim JS, Cha SB, Han SJ, Kim H, Kwon KW, et al. Virulence-dependent alterations in the kinetics of immune cells during pulmonary infection by Mycobacterium tuberculosis. PLoS ONE. (2015) 10:e0145234. doi: 10.1371/journal.pone.0145234
100. Heuer M, Behlich AS, Lee JS, Ribechini E, Jo EK, Lutz MB. The 30-kDa and 38-kDa antigens from Mycobacterium tuberculosis induce partial maturation of human dendritic cells shifting CD4(+) T cell responses towards IL-4 production. BMC Immunol. (2013) 14:48. doi: 10.1186/1471-2172-14-48
101. Behar SM, Divangahi M, Remold HG. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol. (2010) 8:668–74. doi: 10.1038/nrmicro2387
102. Gan H, Lee J, Ren F, Chen M, Kornfeld H, Remold HG. Mycobacterium tuberculosis blocks crosslinking of annexin-1 and apoptotic envelope formation on infected macrophages to maintain virulence. Nat Immunol. (2008) 9:1189–97. doi: 10.1038/ni.1654
103. Kohli S, Singh Y, Sharma K, Mittal A, Ehtesham NZ, Hasnain SE. Comparative genomic and proteomic analyses of PE/PPE multigene family of Mycobacterium tuberculosis H(3)(7)Rv and H(3)(7)Ra reveal novel and interesting differences with implications in virulence. Nucleic Acids Res. (2012) 40:7113–22. doi: 10.1093/nar/gks465
Keywords: mucosal-associated invariant T (MAIT) cells, transcriptome, MHC-related protein 1 (MR1), Mycobacterium tuberculosis, innate-like activation
Citation: Sharma M, Zhang S, Niu L, Lewinsohn DM, Zhang X and Huang S (2020) Mucosal-Associated Invariant T Cells Develop an Innate-Like Transcriptomic Program in Anti-mycobacterial Responses. Front. Immunol. 11:1136. doi: 10.3389/fimmu.2020.01136
Received: 18 March 2020; Accepted: 11 May 2020;
Published: 09 June 2020.
Edited by:
Edwin Leeansyah, Karolinska Institutet (KI), SwedenReviewed by:
Joana Dias, Vaccine Research Center (NIAID), United StatesWeiming Yuan, University of Southern California, Los Angeles, United States
Copyright © 2020 Sharma, Zhang, Niu, Lewinsohn, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shouxiong Huang, c2hvdXhpb25nLmh1YW5nQHVjLmVkdQ==
 Manju Sharma
Manju Sharma Shuangmin Zhang1
Shuangmin Zhang1 Liang Niu
Liang Niu David M. Lewinsohn
David M. Lewinsohn Shouxiong Huang
Shouxiong Huang