- 1Department of Microbiology and Immunology, Center for Infectious Diseases, Stony Brook University Renaissance School of Medicine, Stony Brook, NY, United States
- 2Department of Immunology, UConn Health, Farmington, CT, United States
- 3Centro de Investigacion en Medicina Traslacional Severo Amuchastegui, Instituto Universitario de Ciencias Biomédicas de Córdoba, Córdoba, Argentina
- 4Centre for Immunobiology, Institute of Infection, Immunity, and Inflammation, University of Glasgow, Glasgow, United Kingdom
- 5The Beatson Institute for Cancer Research, Glasgow, United Kingdom
- 6Department of Microbiology, New York University, New York City, NY, United States
While immune responses have been rigorously examined after intravenous Listeria monocytogenes (Lm) infection, less is understood about its dissemination from the intestines or the induction of adaptive immunity after more physiologic models of foodborne infection. Consequently, this study focused on early events in the intestinal mucosa and draining mesenteric lymph nodes (MLN) using foodborne infection of mice with Lm modified to invade murine intestinal epithelium (InlAM Lm). InlAM Lm trafficked intracellularly from the intestines to the MLN and were associated with Batf3-independent dendritic cells (DC) in the lymphatics. Consistent with this, InlAM Lm initially disseminated from the gut to the MLN normally in Batf3–/– mice. Activated migratory DC accumulated in the MLN by 3 days post-infection and surrounded foci of InlAM Lm. At this time Batf3–/– mice displayed reduced InlAM Lm burdens, implicating cDC1 in maximal bacterial accumulation in the MLN. Batf3–/– mice also exhibited profound defects in the induction and gut-homing of InlAM Lm-specific effector CD8 T cells. Restoration of pathogen burden did not rescue antigen-specific CD8 T cell responses in Batf3–/– mice, indicating a critical role for Batf3 in generating anti-InlAM Lm immunity following foodborne infection. Collectively, these data suggest that DC play diverse, dynamic roles in the early events following foodborne InlAM Lm infection and in driving the establishment of intestinal Lm-specific effector T cells.
Introduction
Listeria monocytogenes (Lm) is a Gram-positive, intracellular pathogen found ubiquitously throughout the environment. As an enteric pathogen, Lm may lead to a mild gastrointestinal distress in healthy human hosts; however, those who are elderly, pregnant, or otherwise immunocompromised, and contract listeriosis risk the development of meningitis, sepsis, or stillbirths. In the United States, listeriosis is typically associated with mortality rates ranging 20–50% among hospitalized individuals (1). Lm was recently responsible for the largest foodborne outbreak in the world, in which over 1,000 people became ill and over 200 deaths occurred in South Africa as a result of contaminated meat (2). Consequently, Lm remains a major public health risk. Lm also has the ability to induce potent CD8 T cell responses and break tolerance to tumor antigens, making it a promising candidate as a vector for anti-cancer vaccines (3). Therefore, the potential use of Lm for the development and generation of mucosal vaccines against enteric pathogens or gastrointestinal cancers has engendered the evaluation of intestinal immune responses in the context of foodborne Lm infection or oral immunization.
The generation of robust adaptive immunity at intestinal inductive sites is dependent on interactions between professional antigen-presenting cells (APC) and T cells. Dendritic cells (DC) in particular survey tissues and phagocytose pathogens before transiting through afferent lymphatic vessels to draining lymph nodes, where antigen is presented to naïve T cells to elicit an effector T cell response (4). DC are a heterogenous population with discrete functions, ontogeny and anatomic compartmentalization (5). Lymphoid tissue-residing CD8α+ DC have been linked both developmentally and functionally to migratory CD103+ DC through their dependence on IRF8 and BATF3 for their development and the ability to cross-present antigens to CD8 T cells (5–7). Together, these Batf3-dependent DC are classified as type 1 conventional DC (cDC1) (8). Migratory small intestinal lamina propria (SI-LP) and MLN cDC1 retain the ability to induce the expression of the gut-homing markers CCR9 and α4β7 upon T cell activation, resulting in gut tropism and localization to the small intestines (9, 10). Meanwhile gut migratory, Irf4-dependent CD11b+ CD103+ DC (cDC2), have been implicated in TH17 cell differentiation (11, 12). Finally, a subset of migratory CD11b+ CD103– DC are important in the differentiation of effector T cells that produce IFNγ and IL-17A (13, 14).
DC are necessary for the formation of cytotoxic responses to eliminate Lm after intravenous (i.v.) infection (15). After i.v Lm infection, splenic marginal zone B cells produce IL-10, which acts on CD169+ macrophages to promote bacterial uptake and survival (16). Lm-containing CD169+ macrophages trans-infect cDC1 to mediate Lm transport to the splenic T cell zones. Thus, CD169+ macrophages and cDC1 provide critical non-redundant roles to initiate infection and promote resolution (16–18). Mice lacking cDC1 are resistant to Lm infection with reduced pathogen burden but display diminished Lm-specific CD8 T cell responses (6, 19). However, restoration of pathogen burden by increasing the infection dose in these mice rescues the CD8 T cell response, revealing that cDC1 are not required for T cell responses to Lm after i.v. infection (17). Despite these findings, cDC2 appear to play minimal roles in the generation of protective immunity to Lm, as mice lacking cDC2 do not exhibit defective CD8 T cell responses (16). Although the roles of resident cDC1 and cDC2 after i.v Lm infection have been assessed, the functions of migratory cDC in the early establishment of Lm infection and the induction of T cell responses after foodborne infection have not been extensively studied (20). Some evidence has emerged from foodborne infection of a susceptible Balb/cBy model that suggests that most Lm replicates extracellularly (21). While these studies noted that only a minor amount of Lm is intracellular following foodborne infection, the intracellular fraction was necessary for invasion into the small intestines, as well as extraintestinal dissemination. Monocytes, which are critical for Lm containment, were identified as a major cell type associated with Lm (22). Intracellular bacteria could also be found within intestinal cDC (23). However, monocytes and ex vivo DC did not support intracellular growth suggesting these cells do not serve as a major niche needed to propagate Lm infection (21, 22).
In this study, foodborne infection with murinized Lm containing a mutation in its internalin A gene (further referred to as InlAM Lm) that recapitulates physiologic infection in humans was used to investigate intestinal immune responses in Balb/c and B6 mice (24–26). After foodborne infection, InlAM Lm transited intracellularly from the intestines to the MLN. However, maximal accumulation of bacteria in the MLN appeared dependent on Batf3, suggesting a role for cDC1 in this process. Finally, InlAM Lm-specific CD8 T cell responses were diminished in Batf3–/– mice and restoration of pathogen burden was unable to restore InlAM Lm-specific CD8 T cell responses, suggesting a dynamic role for DC subsets in driving effector CD8 T cell responses after foodborne InlAM Lm infection.
Materials and Methods
Mice
Balb/c and Batf3–/– Balb/c mice were utilized for all experiments, unless otherwise noted. For experiments involving OT-I cell transfers and thoracic duct cannulations, C57BL/6 (B6) were used. Batf3–/– and OT-I TCR transgenic Rag1–/– CD45 congenic mice were bred in-house. For experiments assessing the role of IRF4-dependent DC after foodborne InlAM Lm infection, Itgax-cre Irf4fl/fl and littermate control Irf4fl/fl mice (B6 background) were used. Both male and female mice 8–19 weeks of age were used. Mice were age- and sex-matched to controls from The Jackson Laboratory (Balb/c) or Charles River/NCI (B6). All mice were housed and treated in accordance with the Institutional Animal Care and Use Committee guidelines and approved by Stony Brook University and UConn Health. All survival surgeries were performed at the University of Glasgow, in accordance with the Animal Welfare Ethical Review Board. The Review Board and UK Home Office approved both the individual license (IBCA6BF4E) and project license (P64BCA712), which are compliant with the Animals (Scientific Procedures) Act 1986. This Act amended European Directive 2010/63/EU on the protection of animals used for scientific purposes.
Foodborne InlAM Lm Infection
For infections in Balb/c mice, InlAM Lm (strain 10403s or EGDe as indicated) was utilized. Mice were deprived of food and water 4–6 h prior to infection, then housed individually and given ∼0.5 cm3 pieces of white bread inoculated with 2 × 109 CFU InlAM Lm in PBS (unless otherwise noted). For infections involving B6 mice, InlAM Lm expressing a truncated ovalbumin (InlAM Lm-OVA; strain 10403s) was used.
Treatments
Mice received gentamicin (1 mg, subcutaneous) or an equal volume of PBS at 4 h post-infection (hpi), 20 mg streptomycin (oral gavage) at day -1, or 250 μg anti-Ly6G mAb (IA8) or control IgG2 at days -1, 0, 1, and 2 (intraperitoneal injection).
Microscopy
At 3 days post-infection (dpi), MLN were harvested, fixed in paraformaldehyde-lysine-periodate (PLP) solution, washed with PBS, and dehydrated in 30% sucrose. MLN were snap-frozen in OCT compound (Tissue-Tek), and 30 μm sections were cut using a cryostat (Leica CM1850). Sections were fixed in acetone for 10 min and stained with primary antibodies diluted in PBS with 2% GS and 2% FCS for 1.5 h at room temperature, washed, and stained with secondary or fluorophore-conjugated antibodies diluted in PBS with 2% GS and 2% FCS for 1 h at room temperature. Images were acquired with an LSM780 (Carl Zeiss, Oberkochen, Germany) and processed with Imaris software (Bitplane, Belfast, United Kingdom). All antibodies used are listed in Supplementary Table 1.
Enumerating Listeria Burden
Liver, spleen, and MLN were processed in 1% saponin. Small intestinal contents were flushed using RPMI containing 5% heat-inactivated bovine serum, homogenized using a GentleMACS (Miltenyi Biotec, Auburn, CA, United States), and lysed with 1% saponin. All suspensions were incubated at 4°C for at least 1 h. All bacterial burdens were plated onto Brain Heart Infusion (BHI) agar containing 200 μg/mL streptomycin, incubated at 37°C, and enumerated 24–48 h later.
Dendritic Cell Isolation, Sorting, and Burden
At 3 dpi, MLN were harvested, pooled together, and digested with 100 U/mL collagenase 37°C for 30 min. MLN were crushed through a 70 μm filter, counted, and stained in PBS containing heat-inactivated bovine serum with fluorochrome-conjugated antibodies. DC subsets were sorted on a FACSAria (Becton, Dickinson and Company, Franklin Lakes, NJ, United States). Sorted subsets were directly plated onto BHI agar containing 200 μg/mL streptomycin to enumerate bacterial burden. All antibodies used are listed in Supplementary Table 1.
Thoracic Duct Cannulation
Mesenteric lymphadenectomy (MLNx) was performed on 6-week old B6 males as outlined previously (27, 28). 6 weeks after MLNx, mice were infected with 2 × 109 CFU InlAM Lm (strain 10403s). At 1 dpi, mice received 200–300 μL of olive oil via oral gavage, to visualize the lymphatics. A polyurethane cannula (2Fr; Linton Instrumentation, Diss, United Kingdom) was surgically inserted into the thoracic duct. Lymph was collected into 1mL PBS containing 5,000 U/mL heparin sodium (Wockhardt United Kingdom, Wrexham, United Kingdom) on ice overnight. Collected lymph was stained using fluorochrome-conjugated antibodies and sorted on a FACSAria (Becton, Dickinson and Company, Wokingham, United Kingdom). Sorted DC were lysed with Triton X, plated directly onto BHI agar containing 200 μg/mL streptomycin to enumerate bacterial burden. For all surgeries, mice were anesthetized with isoflurane (Abbott Labs, Abbott Park, IL, United States).
Lymphocyte Isolation and Flow Cytometry
Single-cell suspensions were prepared from the MLN and spleen by mechanical dissociation. Isolation of DC also utilized enzymatic processing with 100 U/mL collagenase at ambient temperature for 25 min and the addition of 0.1M EDTA prior to mechanical dissociation. For the spleen, red blood cells were lysed in RBC Lysis Buffer (BioLegend, San Diego, CA, United States). Intraepithelial lymphocytes (IEL) were isolated with dithioerythritol as described previously (29, 30). Cells were stained with fluorochrome-conjugated antibodies for 30 min at 4°C in the dark. Panels containing MHC class I tetramers were stained for 1 h in the dark at ambient temperature. Cells were fixed with 2% paraformaldehyde (PFA) for 20 min at 4°C in the dark. Samples were acquired on a LSRFortessa (Becton, Dickinson and Company, Franklin Lakes, NJ, United States), and analyzed with FlowJo (software version v10, Becton, Dickinson, and Company, Ashland, OR, United States). All antibodies used are listed in Supplementary Table 1.
OT-I Transfer Experiments
For location of antigen presentation, 1 × 106 CFSE-labeled CD45.1 OT-I cells were transferred into B6 mice at 1 dpi. 16 h later, MLN and spleen were harvested, processed, and cultured in complete RPMI containing 10 IU human IL-2 for 3 days at 37°C, 5% CO2, and CFSE dilution was analyzed as previously described (31, 32). For duration of antigen presentation, 1 × 106 CFSE-labeled CD45.1 OT-I cells were transferred into infected B6 mice at the indicated times. 3 days after transfer, the MLN and spleen were harvested and processed to assess CFSE dilutions. All antibodies used are listed in Supplementary Table 1.
Statistics
All statistical analyses were performed using Prism 8.3.1 (GraphPad).
Results
InlAM Lm Disseminates Intracellularly From the Intestines to the MLN After Foodborne Infection
Infection route plays an important role in bacterial invasion and dissemination. As such, InlAM Lm dissemination and propagation were analyzed in different tissues after foodborne infection of Balb/c or C57BL/6 (B6) mice with InlAM Lm-inoculated bread. One day after infection, bacteria were present in the intestine and MLN but were absent from the spleen or liver (Figure 1A and Supplementary Figure S1A). However, by 2 dpi, InlAM Lm had disseminated to the liver and spleen. The bacterial burden peaked at 2–3 dpi in all tissues examined and was largely cleared between 5 and 8 dpi. The kinetic analysis of InlAM Lm dissemination after foodborne infection of Balb/c and B6 mice suggests a stepwise pattern of dissemination in which InlAM Lm initially mobilizes from the intestines to the draining MLN before dissemination to extraintestinal sites like the spleen and liver. No evidence emerged of a rapid and direct mobilization of InlAM Lm to the liver. Thus, foodborne InlAM Lm infection recapitulates the dissemination pattern expected for an intracellular pathogen acquired through the gastrointestinal tract.
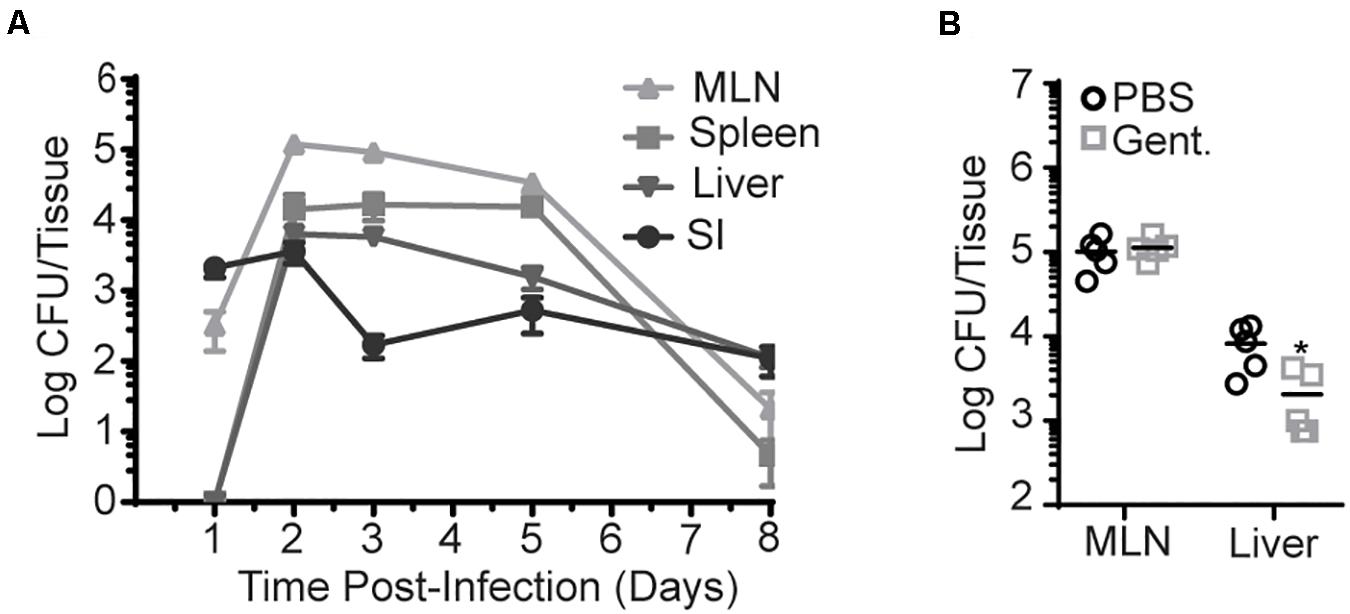
Figure 1. InlAM Lm accumulation in the MLN is intracellular after foodborne infection. Bacterial burden was enumerated from the organs of Balb/c mice after oral infection with InlAM Lm 10403s. (A) Bacterial burden was enumerated from the MLN, spleen, liver, and small intestines from 1-8 dpi after infection with 2 × 109 CFU InlAM Lm. 1 and 3 dpi burdens have been repeated at least 3 times. The complete time course has been performed 1 time in Balb/c mice and 1 time in B6 mice (Supplementary Figure S1A), and 1, 3, and 5 dpi have been depicted in multiple independent experiments reported herein. (B) Mice were foodborne infected with 2 × 109 CFU InlAM Lm and were treated with either gentamicin or PBS at 4 hpi. At 3 dpi, MLN and livers were harvested, and bacterial burdens were quantified. (A) Data depicts 5 mice per timepoint. (B) Data are representative of 2 independent experiments containing 5 mice per group. Statistics were determined by Mann-Whitney test: *p < 0.05.
To assess this more directly, gentamicin was administered to foodborne infected mice to kill extracellular InlAM Lm. As gentamicin is unable to effectively enter cells at the concentration used, intracellular InlAM Lm remains viable in its presence. Gentamicin was administered via subcutaneous injection to Balb/c mice 4 h post-foodborne InlAM Lm infection to allow initial invasion, and bacterial burdens were enumerated at 3 dpi. Subcutaneous injection was chosen as gentamicin is not efficiently absorbed by the intestines (33). At an infection dose of 2 × 109 CFU, the InlAM Lm burden was unaffected in the MLN but decreased approximately fourfold in the liver after antibiotic treatment (Figure 1B). Thus, InlAM Lm is protected from the effects of gentamicin by containment inside cells within the MLN, while a substantial proportion of Lm in the liver has an extracellular component to their lifecycle that promotes gentamicin susceptibility. Increasing the infection dose leads to more extracellular InlAM Lm in the spleen and liver after i.v. Lm infection (34). To confirm gentamicin is effective in the MLN and determine whether increased infectious dose drives an extracellular phase after foodborne InlAM Lm infection of resistant mice, mice were infected with 2 × 1010 CFU of InlAM Lm prior to gentamicin treatment. In this context, gentamicin reduced the recoverable InlAM Lm burden from the MLN and liver at 3 dpi (Supplementary Figure S1B). As neutrophils have been shown to be critical for clearance of extracellular InlAM Lm but dispensable for control of intracellular Lm, the capacity of neutrophil depletion to impact bacterial burdens after foodborne InlAM Lm was tested (34, 35). As such, mice were treated with neutrophil depleting anti-Ly6G mAb (IA8) after foodborne infection. Consistent with the results after gentamicin treatment, neutrophils were dispensable in the MLN but required in the liver for InlAM Lm control (Supplementary Figure S1C). This pattern of predominately intracellular compartmentalization in lymphoid tissues and extracellular compartmentalization in the liver is consistent with Lm replication after i.v. infection. Collectively, these data support the notion that bacteria shuttles intracellularly to the MLN after foodborne infection of Balb/c mice (21). It should be noted that increasing the infection dose does not lead to more extracellular InlAM Lm after foodborne infection of susceptible Balb/cBy mice (21). This model of foodborne infection also showed that a large proportion of gastrointestinal InlAM Lm is extracellular (21). At similar infectious doses of 2 × 109 CFU, Balb/cBy mice had more replicative InlAM Lm in the MLN and liver than either Balb/c or B6 mice (Supplementary Figure S1D). Balb/cBy were also more susceptible to foodborne InlAM Lm infection as only 20% of mice survived until 7 dpi, while B6 and Balb/c mice fully recover at this dose (Supplementary Figure S1E). Taken in context together, these data suggest that the extracellular nature of InlAM Lm infection of Balb/cBy mice may be associated with increased bacterial burden. Therefore, differences in bacterial burden and cellular compartmentalization among Balb/cBy and Balb/c mice could explain the differences noted between foodborne InlAM Lm infection models.
Migratory DC Are Activated and Accumulate Around Regions of InlAM Lm Foci in the MLN After Foodborne Infection
Intracellular bacterial accumulation in the MLN suggested that InlAM Lm may transit to the MLN from the intestines via migratory innate immune cells after foodborne infection. While intestinal macrophages are thought to largely remain tissue-resident within the intestines, DC have been shown to readily migrate from the intestines to the MLN during normal homeostasis or after perturbations of homeostasis such as infection or inflammation (7, 36). As such, resident and migrant DC subsets were evaluated in the MLN of Balb/c mice at steady state and 3 days after foodborne InlAM Lm infection. By 3 dpi, migratory (identified by MHCIIhi CD11c+) but not resident (identified by MHCII+ CD11c+) DC accumulated in the MLN of foodborne infected mice (Figure 2A). Migratory DC from the intestines are typically separated into 3 major subsets based on CD103 and CD11b expression (CD103+ CD11b–, CD103+ CD11b+, and CD103– CD11b+) and each of these were increased in the MLN at 3 dpi (Figure 2B). While all 3 migratory subsets expressed high levels of MHCII, migratory CD103+ CD11b– cDC1 expressed the highest levels of the costimulatory molecule CD80, suggesting that the migrant cDC1 were mature and activated for antigen presentation (Figure 2C).
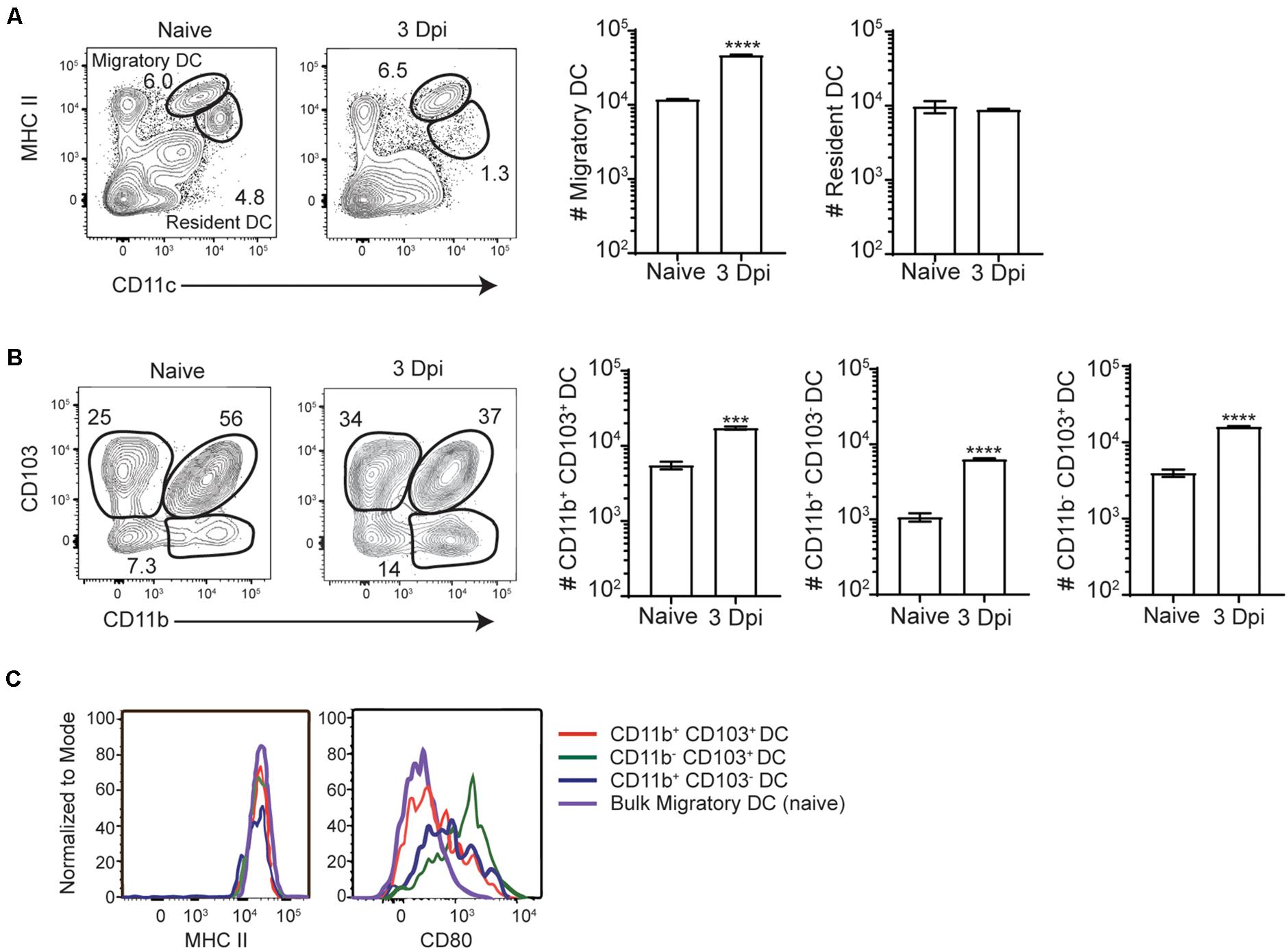
Figure 2. Migratory DC are activated and accumulate in the MLN after foodborne InlAM Lm infection. MLN were harvested from Balb/c mice after foodborne infection with 2 × 109 CFU InlAM Lm 10403s and dendritic cells subsets were analyzed via flow cytometry. (A) Representative flow plots from the MLN of naïve and 3 dpi mice depicting the gating scheme used to identify migratory and resident DC subsets. The populations represented are gated on B220– TCRβ– single, live leukocytes. (B) Cell counts were calculated for migratory dendritic cell subsets at naïve and 3 dpi time points after infection. DC subsets were identified as noted previously, and cell numbers were calculated using cell frequencies and ViCell counts of total viable cells. (C) Representative histograms depicting CD80 and MHCII expression levels on migratory DC subsets. Cell subsets were gated as demonstrated in panels (A,B). Data are representative of two independent experiments with 3–4 mice per group. Unpaired T-tests are reported: ***p < 0.001; ****p < 0.0001.
To further evaluate the anatomical events surrounding intracellular trafficking of InlAM Lm to the MLN, confocal microscopy provided additional context for the localization of InlAM Lm with the early immune response in the MLN after foodborne InlAM Lm infection. InlAM Lm was localized to the deeper cortex in the T cell areas, where it was invariably associated with foci of CD11b+ CD11c– cells (Figures 3a–h). Further assessment identified CD11c+ cells were associated with regions of InlAM Lm foci (Figures 3a–h). InlAM Lm visualized closer to the capsule and subcapsular sinus were surrounded by CD103+ and CD11c+ cells (Figures 3i–k). Thus, DC appear to be intricately involved in the early immune response to foodborne InlAM Lm by surrounding foci of InlAM Lm replication in tight association with T cells suggesting a potential site of antigen presentation.
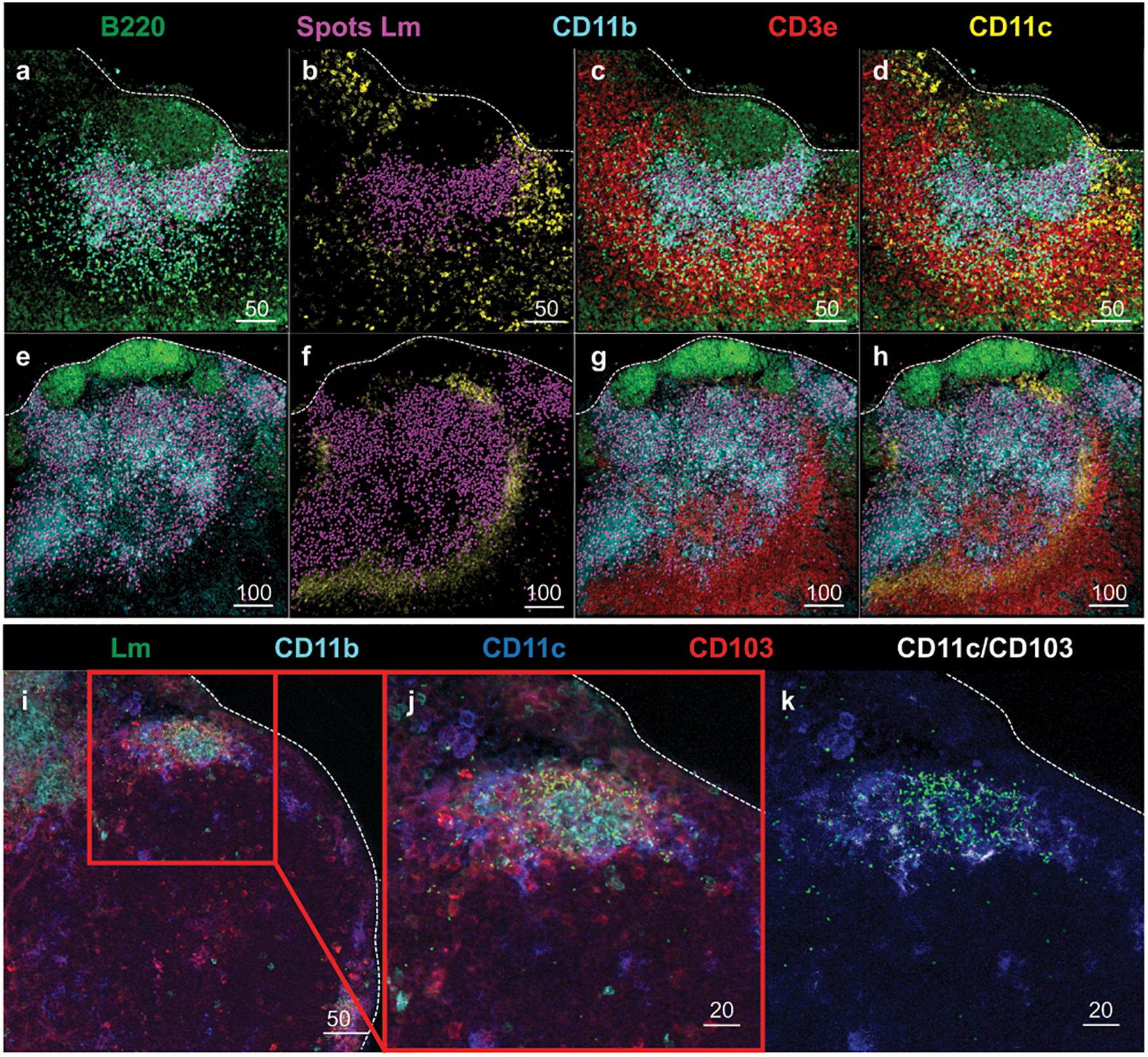
Figure 3. DC subsets surround regions of InlAM Lm foci in the MLN after foodborne infection. (a–h) MLN from 2 Balb/c mice were harvested 2 days after foodborne InlAM Lm infection. MLN cryosections were stained to identify B220 (green), InlAM Lm (magenta), CD11b (cyan), CD3e (red) and CD11c (yellow). Lm staining was highlighted with magenta spheres using Imaris software. Scale bar represents 50 μm (a–d) and 100 μm (e–h). (i–k) MLN from 2 Balb/c mice were harvested 3 days after foodborne InlAM Lm infection. MLN cryosections were stained to identify InlAM Lm (green), CD11b (cyan), CD11c (blue) and CD103 (red). Colocalization of CD11c and CD103 (CD11c/CD103) was highlighted in white using Imaris software. Scale bar represents 50 μm (i) and 20 μm (j,k). All images are representative of at least three independent experiments, with 7–8 images taken for each timepoint.
DC Play Diverse Roles in InlAM Lm Dynamics After Foodborne Infection
After i.v. Lm infection, CD169+ macrophages in the spleen rapidly capture Lm and trans-infect splenic CD8α+ cDC1 (18). Splenic cDC1 have been implicated in the transport of Lm to their replicative niche in T cell zones and are critical for the establishment of productive infection (17, 18). As such, the role of cDC1 in the transit of InlAM Lm and establishment of productive infection after foodborne infection was evaluated in Batf3–/– mice, which lack resident CD8α+ and migrant CD103+ cDC1 (Figure 4A) (6, 17). The small intestine and MLN were assessed for replicating InlAM Lm 1 day after foodborne infection of wild-type and Batf3–/– Balb/c mice to assess colonization in the intestines and early dissemination to the MLN. While Lm burden is low in the spleen and liver of Batf3–/– mice after i.v. infection (17), Batf3 deficiency did not reduce InlAM Lm burden in the small intestine or MLN 1 day after infection (Figure 4B). This data indicates that early intestinal colonization and bacterial dissemination to the MLN after foodborne InlAM Lm infection occurred in a Batf3-independent manner suggesting that cDC1 are not involved in these processes. However, by 3 dpi, Batf3–/– MLN and spleens contained significantly less InlAM Lm than WT MLN and spleens, suggesting that maximal bacterial accumulation in lymphoid tissues is Batf3-dependent (Figure 4C). Of note, the reduction observed in the MLN and spleen of Batf3–/– mice after foodborne InlAM Lm was much less substantial than that reported after i.v. infection (17). Moreover, lack of cDC1 did not diminish the InlAM Lm burden from the liver. Collectively, these findings suggest that while cDC1 are dispensable for early intestinal colonization and dissemination to the MLN, they contribute to the maximal InlAM Lm accumulation in lymphoid tissues.
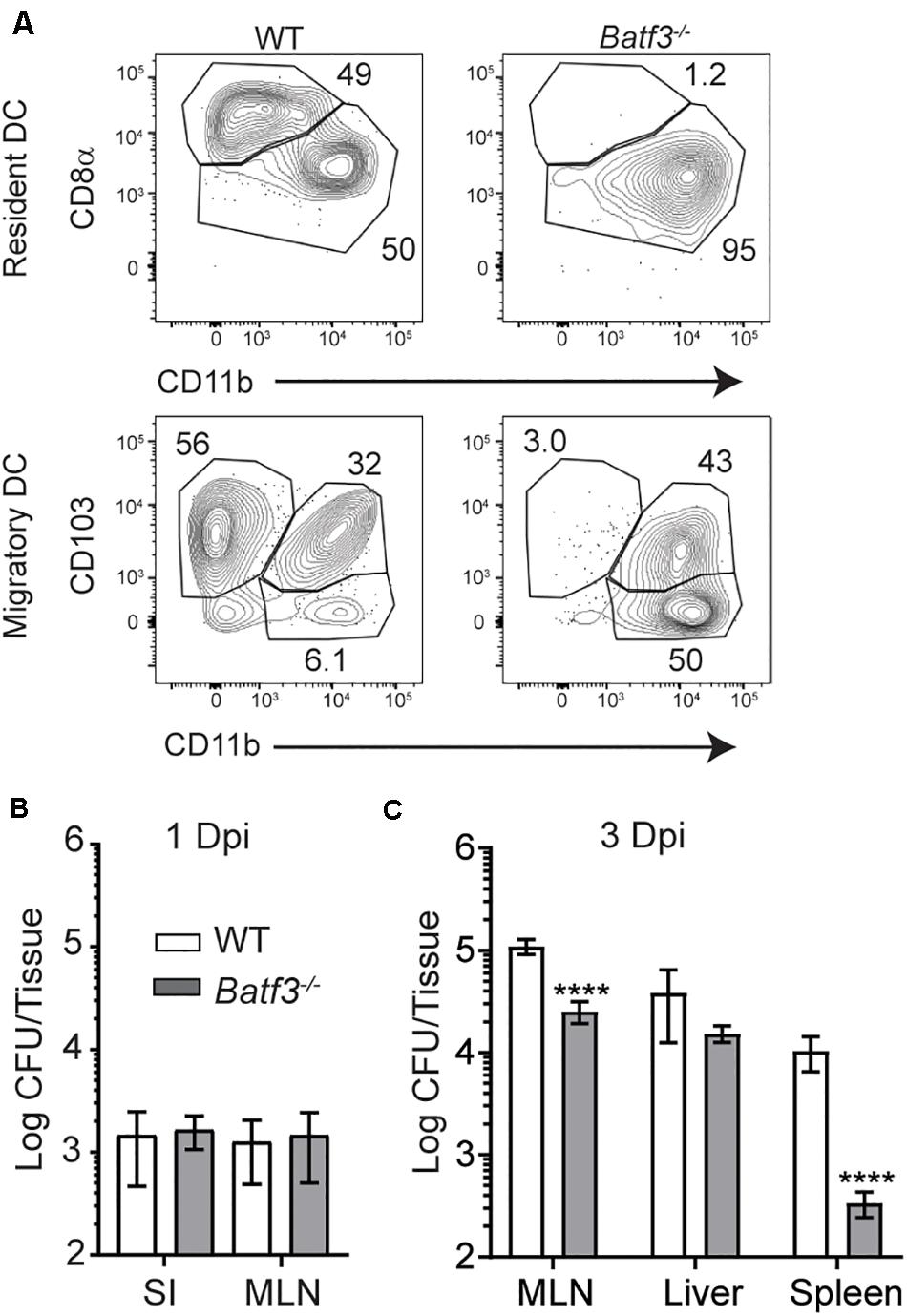
Figure 4. Defects in InlAM Lm accumulation in the MLN after foodborne infection of Batf3–/– mice. (A) WT and Batf3–/– mice were infected with 1 × 1010 CFU InlAM Lm 10403s. At 2 dpi, MLN cell suspensions were assessed by flow cytometry for DC. DC were identified as CD45+ CD19– TCRβ– F4/80– Ly6C– MHCIIhi CD11cint single, live leukocytes (migratory DC) or CD45+ CD19– TCRβ– F4/80– Ly6C– MHCII+ CD11chi single, live leukocytes (resident DC). Resident DC subsets were identified based on their expression of CD8α and CD11b, while migratory DC were identified based on their expression of CD103 and CD11b. (B,C) WT and Batf3–/– mice were infected with 2 × 109 CFU InlAM Lm 10403s. (B,C) The indicted tissues at 1 (B) or 3 (C) dpi were harvested, processed in saponin, and plated on BHI agar plates. Bacterial burdens were enumerated 24–48 h after plating. Data are cumulative of 2 experiments with n = 14–18 mice per group. Mann-Whitney tests were used to analyze the data: ****p < 0.0001.
Based on the role of Batf3 in bacterial accumulation in the MLN, the DC subsets carrying InlAM Lm in the MLN were assessed at 3 dpi. CD8α+, CD11b+, and CD8α– CD11b– DC were sorted from pooled MLN of foodborne-infected WT mice, and bacterial burden was enumerated for each subset. A majority of the InlAM Lm associated with sorted DC was found associated with the CD11b+ DC subset, both after foodborne infection with 2 × 109 CFU and 1 × 1010 CFU (Figures 5A,B). Because CD11b+ DC encompass the largest proportion of the sorted subsets (Figure 5C), these findings imply that most of the bacteria associated with DC in the MLN are found within these cells. MLN-derived neutrophils and monocytes were also included in some experiments to serve as a positive control for the assay (Figure 5 and data not shown). Consistent with previous studies (22) these cells are associated with high numbers of InlAM Lm. However, these cells are thought to be primarily recruited from the blood circulation (37–39). When bacterial burdens were assessed in mice lacking CD103+ CD11b+ DC, no differences were noted in the MLN 3 days after infection when compared to littermate controls (Figure 5D) (11, 12). This suggests that cDC2 are not required for bacterial accumulation in the MLN by 3 dpi. To assess whether CD11b+ cells were responsible for trafficking InlAM Lm from the small intestines to the MLN, thoracic duct cannulation was used to determine whether DC subsets actively migrating in lymph fluid contained replicative InlAM Lm. MLN were surgically removed from mice which were allowed to recover to redirect the flow of lymph fluid from the intestines to the thoracic duct. Lymphadenectomized mice received foodborne InlAM Lm, and “pseudo-afferent” lymph fluid was collected from the thoracic duct over a period of ∼12 h 1 day after infection. Migratory DC subsets were sorted from the lymph, and bacterial burden was assessed for each DC subset (40). InlAM Lm was detectable in both CD103+ CD11b+ and CD103– CD11b– DC early after foodborne infection (Figure 5E). Taken together, these data indicate that intestinal colonization and early accumulation in the MLN can occur in a Batf3-independent manner. Moreover, these findings suggest that multiple DC subsets may be able to transit Lm to the MLN.
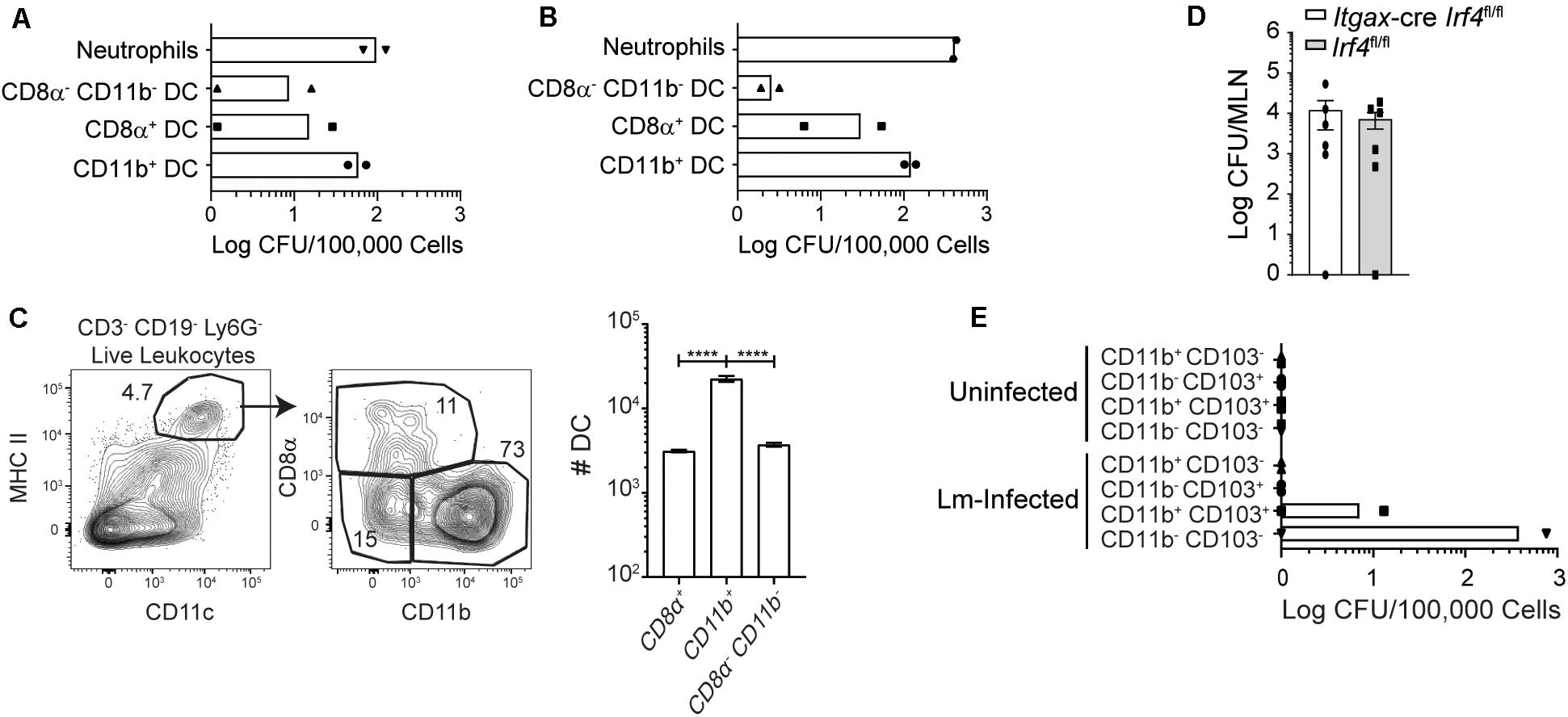
Figure 5. Most InlAM Lm within DC subsets is associated with Batf3-independent DC. (A,C) Mice were infected with 2 × 109 CFU InlAM Lm 10403s. (A) At 3 dpi, MLN from 10 Balb/c mice were pooled together for cell sorting. Sorted subsets were plated on BHI agar containing streptomycin, and bacterial burdens were enumerated 24–48 h later. DC subsets were distinguished based on their expression of CD8α and CD11b. The graph depicts the mean and is cumulative of 2 independent experiments. Neutrophils were included as positive controls for both sets of experiments. (B) Balb/c mice were infected with 1 × 1010 CFU InlAM Lm 10403s. At 3 dpi, 10 MLN were pooled together for cell sorting. Sorted DC subsets were plated onto BHI agar containing streptomycin, and burdens were enumerated 24–48 h after plating. The graph depicts the mean and is cumulative of 2 independent experiments. (C) MLN were harvested from Balb/c mice at 3 dpi, and dendritic cells subsets were analyzed via flow cytometry. Representative flow plots gated on B220– TCRβ– single, live leukocytes are shown. Graph depicts the quantification of dendritic cell subsets. (D) Itgax-cre Irf4fl/fl and littermate (Irf4fl/fl) controls were infected with 2 × 109 CFU InlAM Lm-OVA 10403s. At 3 dpi, MLN were harvested, processed in saponin, and plated on BHI agar containing streptomycin. Bacterial burdens were enumerated 24–48 h after plating. Data are cumulative of 2 experiments. (E) Mesenteric lymphadenectomy was performed on B6 mice. After 4–6 weeks of recovery, mice were infected with 2 × 109 CFU InlAM Lm 10403s, and thoracic duct cannulation was performed at 1 dpi. Migratory DC subsets were sorted from the recovered lymph fluid, plated onto BHI agar plates containing streptomycin, and bacterial burdens were enumerated 24–48 h later. One-way ANOVA with Bonferroni’s multiple comparisons test was used to analyze data. ****p < 0.0001.
Lm-Specific CD8 T Cell Responses Are Greatly Reduced in Batf3–/– Mice After Foodborne Infection
As InlAM Lm infection was limited to gut tissues 1 day after foodborne infection, the location and duration of antigen presentation was analyzed by evaluating the proliferation of CFSE-labeled OT-I cells in different tissues after foodborne infection with InlAM Lm-OVA. To assess major sites of antigen presentation after foodborne infection, CFSE-labeled CD45.1 OT-I T cells were transferred into CD45.2 B6 mice that had been infected with InlAM Lm-OVA 1 day prior. MLN and spleens were harvested 16 h after adoptive transfer and cultured in vitro for 72 h. CFSE was diluted extensively in OT-I cells in the MLN but not in the spleen (Supplementary Figure S2A). This suggests that after foodborne InlAM Lm infection, antigen presentation occurs early in the MLN. The duration of antigen presentation after foodborne infection was also assessed by transferring CFSE-labeled CD45.1 OT-I cells into foodborne-infected CD45.2 B6 mice at various times after infection (Supplementary Figure S2B). Donor cells were isolated 3 days after transfer and evaluated for CFSE dilution. Very few CFSE+ OT-I cells remained in the MLN or spleen after being transferred 3 days after foodborne infection, indicating that the cells underwent extensive proliferation. Transferring OT-I cells from day 6 after infection through day 14 resulted in a dramatic increase in the percentage of undiluted CFSE+ OT-I cells (Supplementary Figure S2B). These findings indicate that T cell proliferation declined over time and was undetectable by day 14 after infection. Thus, antigen was initially available to naïve T cells in mucosal tissues and was presented to T cells for 6–10 days after foodborne infection.
Batf3-dependent cDC1 drive the induction of cell-mediated immunity through their ability to cross-present cell-associated antigen to CD8 T cells (9, 10, 17). However, their role in the formation of adaptive immune responses after foodborne infection is not well understood and their requirement in the induction of CD8 T cell responses to Lm is not absolute after i.v. infection (17). Flow cytometric analyses of blood at 8 dpi yielded severely diminished yet detectable InlAM Lm-specific CD8 T cell responses in Batf3–/– mice compared to WT mice (Figure 6A). InlAM Lm-specific CD8 T cells from Batf3–/– mice also expressed lower levels of the integrin α4β7, suggesting that Batf3-dependent cDC1 also impart gut-homing capability (Figure 6A). This mirrors findings in in vitro studies which noted that CD103+ DC were required to drive effector CD8 T cell expression of the gut homing chemokine receptor CCR9 and integrin α4β7 (9). Further analysis of peak T cell responses at 9 dpi revealed significantly lower proportions of LLO91-Kd+ CD8 T cells in both the MLN and spleens of Batf3–/– mice (Figures 6B,C). Additionally, the differentiation of CD8 effector T cell populations was skewed toward memory potential in Batf3–/– mice, which contained higher proportions of CD127+ KLRG-1– memory precursor effector cells (MPEC) in both their MLN and spleens (Figure 6D). Meanwhile, an evaluation of InlAM Lm-specific CD8 T cells responses at 9 dpi in the MLN of mice lacking migratory cDC2 in their intestinal LP and MLN (Supplementary Figure S3A) yielded no distinct differences when compared to littermate controls (Supplementary Figure S3B) (11, 12). Together, these findings suggest that cDC1, but not cDC2, are necessary to exert full, robust anti-InlAM Lm adaptive immune responses after foodborne InlAM Lm infection.
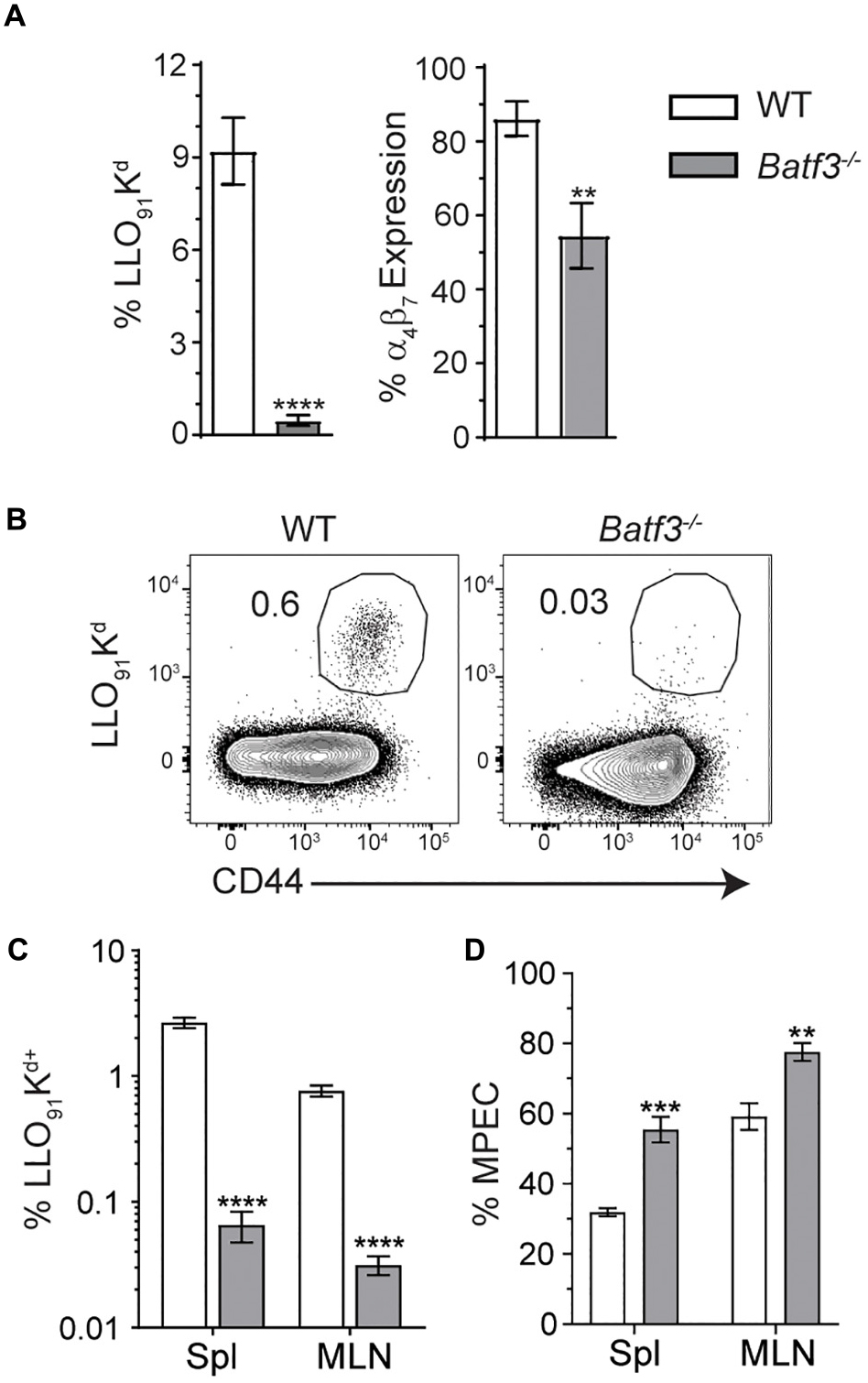
Figure 6. Batf3–/– mice exhibit defects in InlAM Lm-specific adaptive immune responses after foodborne infection. WT and Batf3–/– mice were infected with 2 × 109 CFU InlAM Lm 10403s. (A) At 8 dpi, blood was collected from the tail veins of both groups of mice, and the samples were processed and stained for flow cytometry. LLO91-Kd tetramer staining was utilized to analyze InlAM Lm-specific CD8 T cell responses, while α4β7 indicated gut homing potential. (B–D) At 9 dpi, spleens and MLN were analyzed by flow. LLO91-Kd tetramer staining indicated InlAM Lm-specific CD8 T cells. MPEC were identified as KLRG-1– CD127+ LLO91-Kd+ CD8 T cells. (B) Representative tetramer staining is shown. Data are representative of at least 2 independent experiments with 7–8 mice per group. Unpaired T-tests were used to compare data sets: **p < 0.01; ***p < 0.001; ****p < 0.0001.
After i.v. Lm infection, defects in Lm-specific CD8 T cell responses in Batf3–/– mice were observed. However, when the infection dose administered to Batf3–/– mice was increased, bacterial burden and Lm-specific CD8 T cell responses were restored suggesting that the defect in the CD8 T cell response was due to diminished bacterial burdens in Batf3–/– mice (17). Therefore, whether restoration of pathogen burden could be achieved in Batf3–/– mice after foodborne infection was determined with two distinct approaches. One cohort of Batf3–/– mice received an infection dose of 1 × 1011 CFU while another cohort of Batf3–/– were treated with 20 mg streptomycin by gavage prior to infection with 2 × 105 CFU InlAM Lm. Treatment with streptomycin prior to infection eliminates colonization resistance provided by the normal intestinal microbiota and promotes intestinal invasion and dissemination after low dose foodborne Lm infection (41). Both approaches to restore pathogen burden in the MLN and spleen of Batf3–/– mice after foodborne infection yielded similar levels of InlAM Lm in the MLN and spleen as control mice (Figure 7A). Thus, CD8 T cell responses could be evaluated in WT and Batf3–/– mice with similar pathogen loads. Despite their ability to restore pathogen burden, neither high-dose infection nor pretreatment with streptomycin were capable of rescuing InlAM Lm-specific CD8 T cell responses in the MLN or spleen of Batf3–/– mice (Figures 7B–E). This inability to rescue InlAM Lm-specific CD8 T cell responses was not restricted to lymphoid tissues, as intestinal intraepithelial lymphocyte (IEL) responses were also diminished using both methods of pathogen burden restoration (Supplementary Figure S4). These findings demonstrate that Batf3 is critical for the induction of InlAM Lm-specific CD8 T cell responses after foodborne infection, and that restoration of pathogen burden in Batf3–/– mice is unable to rescue the CD8 T cell defect. This is in contrast to the restoration of pathogen burden during i.v. Lm infection, where Lm-specific CD8 T cell responses were rescued in Batf3–/– mice (16, 17). Altogether, these data suggest that cDC1 may play an important role in driving effector CD8 T cell responses after foodborne InlAM Lm infection.
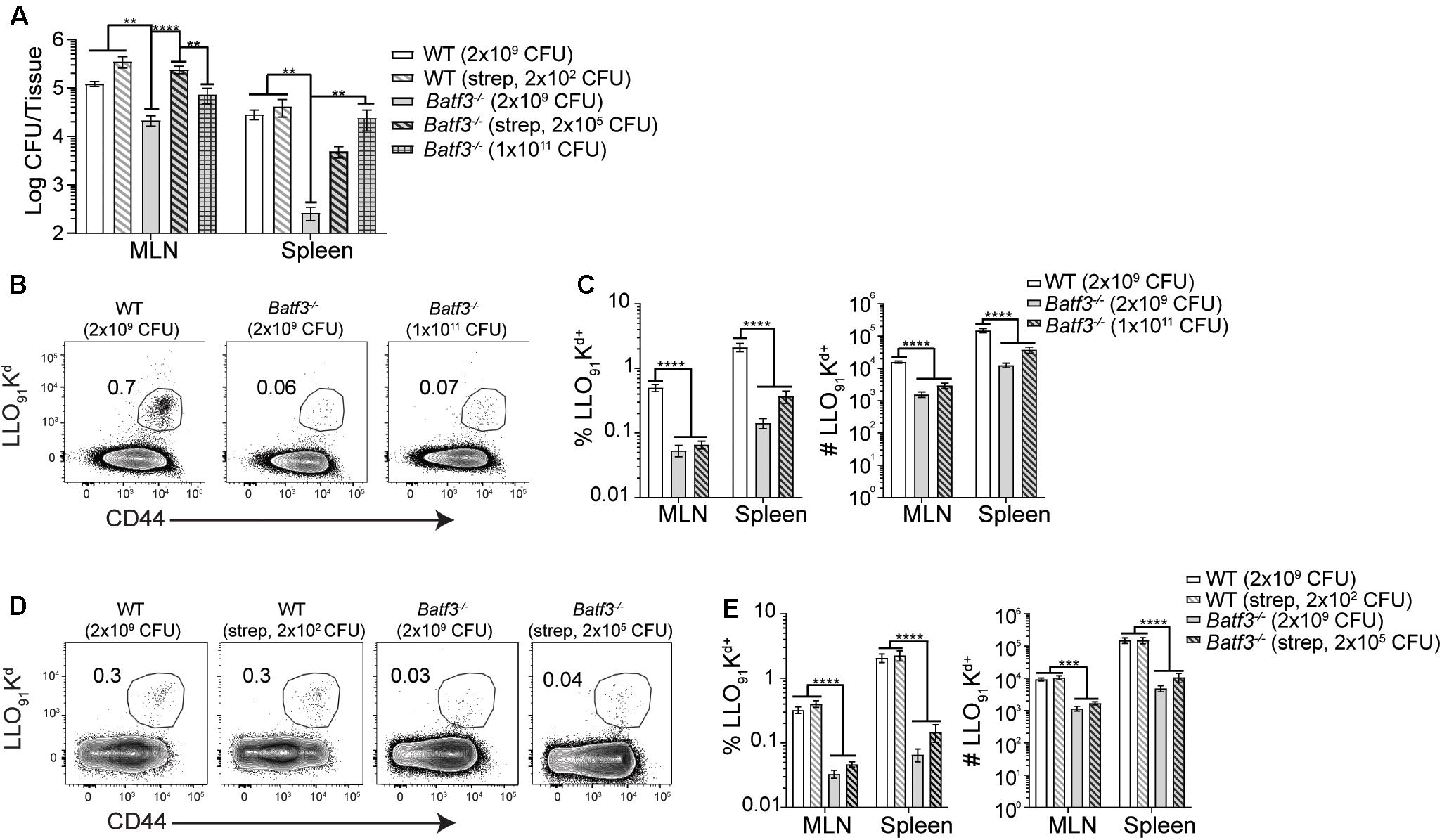
Figure 7. Induction of CD8 T cell responses are not rescued by restoring pathogen load. 20 mg of streptomycin was administered to the indicated groups of WT and Batf3–/– Balb/c mice 1 day prior to infection. Mice were infected with the indicated doses of InlAM Lm 10403s. (A) At 3 dpi, MLN and spleens were harvested, processed, and plated on BHI agar to enumerate InlAM Lm burden. Data are pooled from 5 to 6 individual experiments and included both male and female mice (n = 7–41). (B–E) At 9 dpi, MLN and spleens were harvested and processed for flow cytometry. LLO91-Kd tetramer staining was utilized to analyze InlAM Lm-specific CD8 T cell responses. Data are pooled from 4 to 6 individual experiments and included both male and female mice (n = 12–20 mice per group). (B,D) Representative tetramer staining is shown. Bacterial burdens were compared using one-way ANOVA with Kruskal-Wallis non-parametric test. Cell populations were compared using one-way ANOVA with Bonferroni’s multiple comparisons test: **p < 0.01; ***p < 0.001; ****p < 0.0001.
Discussion
Route of infection and pathogen dissemination from the site of infection play an important role in the generation of anti-pathogen immune responses. A physiologic infection method that more closely resembles human exposure to foodborne pathogens and oral vaccines to assess early immune events that impact dissemination of the bacterial pathogen InlAM Lm and induction of T cell immunity was utilized. After foodborne infection, InlAM Lm appears restricted to the gut tissues before disseminating to extraintestinal sites like the liver and spleen. Balb/c and B6 mice infected with 2 × 109 CFU of InlAM Lm did not contain detectable replicating bacteria in the liver or spleen until 2 days after foodborne infection. While foodborne infection of Balb/cBy (a substrain of Balb/c) mice results in an extracellular lifecycle phase in dissemination to or replication within the MLN (21), foodborne infection of Balb/c mice yielded predominately intracellular InlAM Lm in the MLN as gentamicin treatment or depletion of neutrophils did not reduce the bacterial burden. Balb/cBy mice are known to be highly susceptible to InlAM Lm infection (42, 43), and foodborne infection with 2 × 109 CFU of InlAM Lm resulted in substantially higher bacterial burdens in Balb/cBy mice that was associated with reduced survival. The presence of extracellular InlAM Lm replication may contribute to the enhanced susceptibility of Balb/cBy mice or be a consequence of higher bacterial burdens associated with enhanced susceptibility. Regardless, both models of foodborne infection found that InlAM Lm was largely extracellular in the liver, which is also consistent with what has been reported after i.v. Lm infection (44).
The implication that InlAM Lm is primarily intracellular within the MLN led to the hypothesis that InlAM Lm is trafficked via immune cells migrating from the gut lamina propria. As DC represent the major subset of lymph migrant immune cells from the gut, DC subsets responding to foodborne InlAM Lm infection were assessed. Flow cytometric analysis after foodborne InlAM Lm infection showed that migratory DC accumulated in the MLN by 3 dpi, while confocal microscopy revealed that InlAM Lm in the MLN was mostly contained within foci in close proximity to CD11b+ and CD11c+ cells (some of which also expressed CD103). Additionally, splenic cDC1 are known to shuttle Lm from CD169+ macrophages to the T cell zones after i.v. infection (17, 18). Assessment of bacterial burden in Batf3–/– and wild-type mice demonstrated comparable InlAM Lm burden in the small intestine and MLN at 1 dpi suggesting that cDC1 are dispensable for initial intestinal colonization and early accumulation in the MLN. However, reductions in the MLN and spleen were observable by 3 dpi, suggesting that cDC1 are needed for maximal accumulation of foodborne InlAM Lm. Interestingly, there were no significant differences in bacterial burden in the livers of Batf3–/– and WT mice, suggesting that cDC1 are dispensable for hepatic bacterial accumulation after foodborne InlAM Lm infection and this is consistent with the large reservoir of extracellular replication observed in that tissue. Of note, this is in contrast with i.v. Lm infection of Batf3–/– mice, in which the inability to establish infection is observed in the spleen and liver suggesting that route of infection impacts the role of cDC1 in potentiating InlAM Lm infection (17).
Due to the finding that Batf3–/– mice display defects in maximal bacterial accumulation in the MLN after foodborne InlAM Lm infection, the DC subsets which contain InlAM Lm in the MLN after foodborne infection were determined. CD11b+ DC contained the majority of the DC-associated InlAM Lm found in the MLN at 3 dpi and represented the largest DC subset at this time. InlAM Lm was also found within pseudo-afferent lymph CD11b+ CD103+ and CD103– CD11b– DC 1 day after foodborne InlAM Lm infection. CD103– CD11b– DC migrate in intestinal lymph and are capable of presenting antigen to both CD4 and CD8 T cells (14). Thus, replicative InlAM Lm can transit from the gut to the MLN in association with CD11b+ CD103+ and CD103– CD11b– DC subsets consistent with the findings that suggest early intestinal colonization and accumulation in the MLN is able to occur independently of cDC1. As bacterial burdens in Itgax-cre Irf4fl/fl mice, which lack CD103+ CD11b+ DC, were comparable to those of their littermate controls at 3 dpi, this subset does not appear to be required for Lm accumulation or control within the MLN. Gentamicin treatment did not reduce bacterial burden in the MLN when administered at 4 hpi, suggesting that DC from pseudo-afferent lymph harbored intracellular replicative InlAM Lm during transit to the MLN. Collectively, these data suggest that while cDC2 may be capable of carrying Lm to the MLN, neither cDC1 nor cDC2 are required for the initial dissemination of InlAM Lm from the gut to the MLN. Whether redundancies allow cDC2 to perform this function in the absence of cDC1 and vice versa is unclear and limitations with the thoracic duct cannulation procedure and the sensitivity of the assay preclude eliminating other cells as a source of initial transit. However, collectively, these findings reinforce the idea that diverse DC subsets may be involved in bacterial transit following foodborne infection. Despite the finding that InlAM Lm is most prevalent among cDC2 in the MLN, it is well-established that cDC1 play a critical role in the generation of CD8 T cell responses due to their ability to cross-present cell-associated antigens (19, 45–47). Therefore, these findings suggest that diverse DC subsets may traffic InlAM Lm to the MLN, where cDC1 process and present antigens to naïve CD8 T cells, resulting in robust adaptive immune responses. Thus, these findings are consistent with the notion of a tight bottleneck during invasion of the gastrointestinal tract (48) and suggest that diverse DC subsets dynamically associate with InlAM Lm early after foodborne infection in order to disseminate the pathogen and induce CD8 T cell responses.
Foodborne infection of Balb/cBy mice demonstrated that a large proportion of InlAM Lm adhered to Ly6Chi monocytes (22). However, most InlAM Lm was contained intracellularly after foodborne infection of Balb/c mice in this study and most monocytes in the MLN are likely derived from the blood, the relevance of this population in vivo was not assessed. It is well documented that monocytes are critical for control of Lm (49) and further studies might address differences in this important cell type to pathogen resistance in Balb/c or B6 mice and susceptibility in Balb/cBy mice. Furthermore, intestinal CX3CR1+ mononuclear phagocytes act as vessels to traffic Salmonella enterica from the intestines to the MLN (50). However, whether these cells more closely align with macrophages or dendritic cells is open to interpretation (51). Finally, loss of CD169+ macrophages did not impact bacterial burden in the MLN or small intestines after foodborne InlAM Lm infection (18). This suggests that while these marginal zone macrophages play an important role filtering the blood after i.v. infection, they do not play a similar role in filtering intestinal lymph containing cell-autonomous InlAM Lm after foodborne infection.
These findings suggest that cDC1 play an important role in the generation of robust, protective InlAM Lm-specific CD8 T cell responses after foodborne infection. Numerous publications have outlined the importance of cDC1 in generating CD8 T cell responses to intracellular pathogens or apoptotic cell-associated antigens (19, 45–47, 52). A recent study has also demonstrated that Batf3 is dispensable for InlAM Lm-specific effector CD8 T cells after foodborne infection (53), suggesting the validity of our approach to assess cDC1 in effector CD8 T cell responses. This study revealed that Batf3–/– mice displayed distinct defects in both effector T cell accumulation and α4β7 expression. Expression of α4β7 is imprinted on T cells by DC, and allows for the migration of lymphocytes to the intestines via interactions with its ligand, MAdCAM-1 (54). Furthermore, the few antigen-specific CD8 T cells present in Batf3–/– mice were skewed toward MPEC differentiation. Altogether, these findings indicate that Batf3 is needed to generate these responses and suggest cDC1 are needed for the induction of effector CD8 T cells after foodborne InlAM Lm infection. Previous studies have noted that the defects in T cell responses in Batf3–/– mice after i.v. Lm infection were attributable to low pathogen burden. Subsequent restoration of pathogen burden through increased i.v. infection dose was able to rescue Lm-specific CD8 T cell responses in Batf3–/– mice, suggesting that other antigen presenting cells are capable of eliciting a CD8 T cell response to Lm infection (17). As methods used to normalize bacterial burden were unable to restore the effector CD8 T cell response after foodborne infection, Batf3 is playing a critical role in the development of effector CD8 T cells after foodborne infection. Additionally, the heterogeneity of splenic and MLN DC subsets could impact the differences noted between foodborne and i.v. infection models after pathogen burden restoration. Splenic DC are comprised of only lymphoid-resident DC, while both migratory and lymphoid-resident DC are present in the MLN. While splenic cDC2 in Batf3–/– mice may be more poised to present antigen upon sufficient pathogen burden after i.v. infection, differences in the composition and antigen acquisition, processing and presentation capabilities within the Batf3–/– MLN could limit the ability to drive CD8 T cell responses after foodborne infection. Previous studies of foodborne infection found that once inside cDC, GFP-expressing InlAM Lm was not able to efficiently escape the vacuole and gain access to the cytosol (23). This inability to successfully enter the cytosol could limit the potential of cDC to process and present antigen. In this context, cDC2, which are poor at cross-presenting antigens in vivo (55) might be inefficient at inducing a CD8 T cell response. The inability to cross-present antigen in vivo was further reflected in the finding that mice lacking cDC2 did not display defects in InlAM Lm-specific CD8 T cell responses compared to littermate controls. Additionally, methods of pathogen burden restoration could also result in larger proportions of extracellular bacteria in the Batf3–/– mice, which may be shuttled to the MLN via other cell types or cell-autonomously. Although more studies will need to be performed to examine this, these potential alternate methods of transit could impact antigen uptake and subsequently, presentation to naïve T cells. Finally, while previous studies have demonstrated a profound Batf3 cell-extrinsic effect on the induction of Lm-specific CD8 T cell responses (16, 17), mice used in this study also lack Batf3 expression in CD8 T cells that may influence the T cell responses at higher infectious doses.
Collectively, this study has demonstrated that DC play diverse and dynamic roles in early responses to foodborne InlAM Lm and suggested that cDC1 are needed for the induction of InlAM Lm-specific effector CD8 T cell response. Recent works have demonstrated that vaccines stimulating cDC1 can induce effective anti-tumor CD8 T cell responses and that cDC1 are critical for the formation of robust tissue-resident memory T (TRM) cell responses in the skin after vaccinia virus infection, making these DC potential targets for future vaccine development (56, 57). However, other studies have now implicated IRF4-dependent cDC2 in lung TRM formation after influenza infection (58). Therefore, understanding mucosal DC is critical to learning more about their influence on the formation of effector and memory T cell populations, and how the generation of these T cell populations can be utilized for the development of potent vaccines.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee’s at Stony Brook University and UConn Health and the Animal Welfare Review Board and UK Home Office for cannulations performed at the University of Glasgow.
Author Contributions
JI, DX, PR, ZQ, PP, CK, AA, AB-B, and Q-MP performed the experiments. SM, LL, KK, LP, and BS contributed to the study design. JI and BS designed the experiments, analyzed and interpreted the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by The Society for Mucosal Immunology’s Technique-Sharing Travel Award (JI), NIH grants T32 GM008468 (JI), T32 AI007539 (JI), K12 GM102778 (ZQ), P01 AI056172 (LP), R01 AI076457 (BS), and R21 AI137929 (BS), and grant MR/N023625/1 from the UK Medical Research Council (SM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the NIAID Tetramer Core Facility for the LLO91–99 H-2Kd and OVA257–264 H-2Kb tetramers.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.575967/full#supplementary-material
References
1. Zhu Q, Gooneratne R, Hussain MA. Listeria monocytogenes in fresh produce: outbreaks, prevalence and contamination levels. Foods. (2017) 6:21. doi: 10.3390/foods6030021
2. Salama PJ, Embarek PKB, Bagaria J, Fall IS. Learning from Listeria: safer food for all. Lancet. (2018) 391:2305–6. doi: 10.1016/S0140-6736(18)31206-6
3. Wood LM, Paterson Y. Attenuated Listeria monocytogenes: a powerful and versatile vector for the future of tumor immunotherapy. Front Cell Infect Microbiol. (2014) 4:51. doi: 10.3389/fcimb.2014.00051
4. Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, et al. Immunobiology of dendritic cells. Annu Rev Immunol. (2000) 18:767–811. doi: 10.1146/annurev.immunol.18.1.767
5. Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. (2013) 31:563–604. doi: 10.1146/annurev-immunol-020711-074950
6. Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. (2010) 207:823–36. doi: 10.1084/jem.20091627
7. Persson EK, Scott CL, Mowat AM, Agace WW. Dendritic cell subsets in the intestinal lamina propria: ontogeny and function. Eur J Immunol. (2013) 43:3098–107. doi: 10.1002/eji.201343740
8. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. (2014) 14:571–8. doi: 10.1038/nri3712
9. Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, Forster R, et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. (2005) 202:1063–73. doi: 10.1084/jem.20051100
10. Johansson-Lindbom B, Svensson M, Wurbel MA, Malissen B, Marquez G, Agace W. Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): requirement for GALT dendritic cells and adjuvant. J Exp Med. (2003) 198:963–9. doi: 10.1084/jem.20031244
11. Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, et al. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. (2013) 38:958–69. doi: 10.1016/j.immuni.2013.03.009
12. Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. (2013) 38:970–83. doi: 10.1016/j.immuni.2013.04.011
13. Bar-On L, Birnberg T, Lewis KL, Edelson BT, Bruder D, Hildner K, et al. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc Natl Acad Sci USA. (2010) 107:14745–50. doi: 10.1073/pnas.1001562107
14. Cerovic V, Houston SA, Scott CL, Aumeunier A, Yrlid U, Mowat AM, et al. Intestinal CD103(-) dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. (2013) 6:104–13. doi: 10.1038/mi.2012.53
15. Williams MA, Schmidt RL, Lenz LL. Early events regulating immunity and pathogenesis during Listeria monocytogenes infection. Trends Immunol. (2012) 33:488–95. doi: 10.1016/j.it.2012.04.007
16. Liu D, Yin X, Olyha SJ, Nascimento MSL, Chen P, White T, et al. IL-10-dependent crosstalk between murine marginal zone B cells, macrophages, and CD8alpha(+) dendritic cells promotes Listeria monocytogenes infection. Immunity. (2019) 51:64–76.e7. doi: 10.1016/j.immuni.2019.05.011
17. Edelson BT, Bradstreet TR, Hildner K, Carrero JA, Frederick KE, Kc W, et al. CD8alpha(+) dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity. (2011) 35:236–48. doi: 10.1016/j.immuni.2011.06.012
18. Perez OA, Yeung ST, Vera-Licona P, Romagnoli PA, Samji T, Ural BB, et al. CD169(+) macrophages orchestrate innate immune responses by regulating bacterial localization in the spleen. Sci Immunol. (2017) 2:eaah5520. doi: 10.1126/sciimmunol.aah5520
19. Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. (2008) 322:1097–100. doi: 10.1126/science.1164206
20. Qiu Z, Khairallah C, Sheridan BS. Listeria monocytogenes: a model pathogen continues to refine our knowledge of the CD8 T cell response. Pathogens. (2018) 7:55. doi: 10.3390/pathogens7020055
21. Jones GS, Bussell KM, Myers-Morales T, Fieldhouse AM, Bou Ghanem EN, D’Orazio SE. Intracellular Listeria monocytogenes comprises a minimal but vital fraction of the intestinal burden following foodborne infection. Infect Immun. (2015) 83:3146–56. doi: 10.1128/IAI.00503-15
22. Jones GS, D’Orazio SE. Monocytes are the predominant cell type associated with Listeria monocytogenes in the Gut, but they do not serve as an intracellular growth niche. J Immunol. (2017) 198:2796–804. doi: 10.4049/jimmunol.1602076
23. Jones GS, Smith VC, D’Orazio SEF. Listeria monocytogenes replicate in bone marrow-derived CD11c(+) cells but not in dendritic cells isolated from the murine gastrointestinal tract. J Immunol. (2017) 199:3789–97. doi: 10.4049/jimmunol.1700970
24. Bonazzi M, Lecuit M, Cossart P. Listeria monocytogenes internalin and E-cadherin: from bench to bedside. Cold Spring Harb Perspect Biol. (2009) 1:a003087. doi: 10.1101/cshperspect.a003087
26. Wollert T, Pasche B, Rochon M, Deppenmeier S, van den Heuvel J, Gruber AD, et al. Extending the host range of Listeria monocytogenes by rational protein design. Cell. (2007) 129:891–902. doi: 10.1016/j.cell.2007.03.049
27. Milling S, Yrlid U, Cerovic V, MacPherson G. Subsets of migrating intestinal dendritic cells. Immunol Rev. (2010) 234:259–67. doi: 10.1111/j.0105-2896.2009.00866.x
28. Milling SW, Jenkins C, MacPherson G. Collection of lymph-borne dendritic cells in the rat. Nat Protoc. (2006) 1:2263–70. doi: 10.1038/nprot.2006.315
29. Qiu Z, Sheridan BS. Isolating lymphocytes from the mouse small intestinal immune system. J Vis Exp. (2018) 132:57281. doi: 10.3791/57281
30. Sheridan BS, Lefrancois L. Isolation of mouse lymphocytes from small intestine tissues. Curr Protoc Immunol. (2012) Chapter 3:Unit3.19. doi: 10.1002/0471142735.im0319s99
31. Turner DL, Cauley LS, Khanna KM, Lefrancois L. Persistent antigen presentation after acute vesicular stomatitis virus infection. J Virol. (2007) 81:2039–46. doi: 10.1128/JVI.02167-06
32. Zammit DJ, Turner DL, Klonowski KD, Lefrancois L, Cauley LS. Residual antigen presentation after influenza virus infection affects CD8 T cell activation and migration. Immunity. (2006) 24:439–49. doi: 10.1016/j.immuni.2006.01.015
33. Edson RS, Terrell CL. The aminoglycosides: streptomycin, kanamycin, gentamicin, tobramycin, amikacin, netilmicin, and sisomicin. Mayo Clin Proc. (1987) 62:916–20. doi: 10.1016/s0025-6196(12)65048-4
34. Witter AR, Okunnu BM, Berg RE. The essential role of neutrophils during infection with the intracellular bacterial pathogen Listeria monocytogenes. J Immunol. (2016) 197:1557–65. doi: 10.4049/jimmunol.1600599
35. Aoshi T, Carrero JA, Konjufca V, Koide Y, Unanue ER, Miller MJ. The cellular niche of Listeria monocytogenes infection changes rapidly in the spleen. Eur J Immunol. (2009) 39:417–25. doi: 10.1002/eji.200838718
36. Cerovic V, Bain CC, Mowat AM, Milling SW. Intestinal macrophages and dendritic cells: what’s the difference? Trends Immunol. (2014) 35:270–7. doi: 10.1016/j.it.2014.04.003
37. Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. (2003) 19:71–82. doi: 10.1016/s1074-7613(03)00174-2
38. Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. (2008) 26:421–52. doi: 10.1146/annurev.immunol.26.021607.090326
39. Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. (2011) 11:762–74. doi: 10.1038/nri3070
40. Scott CL, Wright PB, Milling SW, Mowat AM. Isolation and identification of conventional dendritic cell subsets from the intestine of mice and men. Methods Mol Biol. (2016) 1423:101–18. doi: 10.1007/978-1-4939-3606-9_7
41. Becattini S, Littmann ER, Carter RA, Kim SG, Morjaria SM, Ling L, et al. Commensal microbes provide first line defense against Listeria monocytogenes infection. J Exp Med. (2017) 214:1973–89. doi: 10.1084/jem.20170495
42. Cheers C, McKenzie IF. Resistance and susceptibility of mice to bacterial infection: genetics of listeriosis. Infect Immun. (1978) 19:755–62.
43. Cheers C, McKenzie IF, Pavlov H, Waid C, York J. Resistance and susceptibility of mice to bacterial infection: course of listeriosis in resistant or susceptible mice. Infect Immun. (1978) 19: 763–70.
44. Conlan JW. Early pathogenesis of Listeria monocytogenes infection in the mouse spleen. J Med Microbiol. (1996) 44:295–302. doi: 10.1099/00222615-44-4-295
45. Desai P, Tahiliani V, Abboud G, Stanfield J, Salek-Ardakani S. Batf3-dependent dendritic cells promote optimal CD8 T cell responses against respiratory poxvirus infection. J Virol. (2018) 92:JVI.00495-18. doi: 10.1128/JVI.00495-18
46. Desch AN, Randolph GJ, Murphy K, Gautier EL, Kedl RM, Lahoud MH, et al. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J Exp Med. (2011) 208:1789–97. doi: 10.1084/jem.20110538
47. Torti N, Walton SM, Murphy KM, Oxenius A. Batf3 transcription factor-dependent DC subsets in murine CMV infection: differential impact on T-cell priming and memory inflation. Eur J Immunol. (2011) 41:2612–8. doi: 10.1002/eji.201041075
48. Zhang T, Abel S, Abel Zur Wiesch P, Sasabe J, Davis BM, Higgins DE, et al. Deciphering the landscape of host barriers to Listeria monocytogenes infection. Proc Natl Acad Sci USA. (2017) 114:6334–9. doi: 10.1073/pnas.1702077114
49. Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. (2003) 19:59–70. doi: 10.1016/s1074-7613(03)00171-7
50. Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, Cuesta A, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. (2013) 494:116–20. doi: 10.1038/nature11809
51. Bravo-Blas A, Utriainen L, Clay SL, Kastele V, Cerovic V, Cunningham AF, et al. Salmonella enterica serovar typhimurium travels to mesenteric lymph nodes both with host cells and autonomously. J Immunol. (2019) 202:260–7. doi: 10.4049/jimmunol.1701254
52. Mashayekhi M, Sandau MM, Dunay IR, Frickel EM, Khan A, Goldszmid RS, et al. CD8alpha(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity. (2011) 35:249–59. doi: 10.1016/j.immuni.2011.08.008
53. Qiu Z, Khairallah C, Romanov G, Sheridan BS. Cutting edge: Batf3 expression by CD8 T cells critically regulates the development of memory populations. J Immunol. (2020) 205:901–6. doi: 10.4049/jimmunol.2000228
54. Mora JR, Bono MR, Manjunath N, Weninger W, Cavanagh LL, Rosemblatt M, et al. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. (2003) 424:88–93. doi: 10.1038/nature01726
55. den Haan JM, Lehar SM, Bevan MJ. CD8(+) but not CD8(-) dendritic cells cross-prime cytotoxic T cells in vivo. J Exp Med. (2000) 192:1685–96.
56. Hammerich L, Marron TU, Upadhyay R, Svensson-Arvelund J, Dhainaut M, Hussein S, et al. Systemic clinical tumor regressions and potentiation of PD1 blockade with in situ vaccination. Nat Med. (2019) 25:814–24. doi: 10.1038/s41591-019-0410-x
57. Iborra S, Martinez-Lopez M, Khouili SC, Enamorado M, Cueto FJ, Conde-Garrosa R, et al. Optimal generation of tissue-resident but not circulating memory T cells during viral infection requires crosspriming by DNGR-1(+) dendritic cells. Immunity. (2016) 45:847–60. doi: 10.1016/j.immuni.2016.08.019
Keywords: dendritic cells, CD8 T cells, intestine (small), Listeria (L.) monocytogenes, Batf3
Citation: Imperato JN, Xu D, Romagnoli PA, Qiu Z, Perez P, Khairallah C, Pham Q-M, Andrusaite A, Bravo-Blas A, Milling SWF, Lefrancois L, Khanna KM, Puddington L and Sheridan BS (2020) Mucosal CD8 T Cell Responses Are Shaped by Batf3-DC After Foodborne Listeria monocytogenes Infection. Front. Immunol. 11:575967. doi: 10.3389/fimmu.2020.575967
Received: 24 June 2020; Accepted: 24 August 2020;
Published: 11 September 2020.
Edited by:
Cecilia Johansson, Imperial College London, United KingdomReviewed by:
Katharina Lahl, Lund University, SwedenKonjit Getachew, Lund University and Hongkui Xiao, Technical University of Denmark in
collaboration with reviewer KL
Timothy L. Denning, Georgia State University, United States
Copyright © 2020 Imperato, Xu, Romagnoli, Qiu, Perez, Khairallah, Pham, Andrusaite, Bravo-Blas, Milling, Lefrancois, Khanna, Puddington and Sheridan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian S. Sheridanm, YnJpYW4uc2hlcmlkYW5Ac3Rvbnlicm9vay5lZHU=
 Jessica Nancy Imperato
Jessica Nancy Imperato Daqi Xu
Daqi Xu Pablo A. Romagnoli
Pablo A. Romagnoli Zhijuan Qiu1
Zhijuan Qiu1 Pedro Perez
Pedro Perez Camille Khairallah
Camille Khairallah Alberto Bravo-Blas
Alberto Bravo-Blas Simon W. F. Milling
Simon W. F. Milling Kamal M. Khanna
Kamal M. Khanna Lynn Puddington
Lynn Puddington Brian S. Sheridan
Brian S. Sheridan