- 1Department of Clinical Laboratory, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
- 2Department of Clinical Laboratory, The First Hospital of Jilin University, Jilin, China
- 3Department of Rheumatology and Clinical Immunology, Peking Union Medical College Hospital, Key Laboratory of Rheumatology and Clinical Immunology, Ministry of Education, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
Objectives: Systemic sclerosis (SSc) is an uncommon autoimmune disease that varies with ethnicity. Single nucleotide polymorphisms (SNPs) in the GTFSI, NFKB1, and TYK2 genes have been reported to be associated with SSc in other populations and in individuals with various autoimmune diseases. This study aimed to investigate the association between these SNPs and susceptibility to SSc in a Chinese Han population.
Method: A case-control study was performed in 343 patients with SSc and 694 ethnically matched healthy controls. SNPs in GTF2I, NFKB1, and TYK2 were genotyped using a Sequenom MassArray iPLEX system. Association analyses were performed using PLINK v1.90 software.
Result: Our study demonstrated that the GTF2I rs117026326 T allele and the GTF2I rs73366469 C allele were strongly associated with patients with SSc (P = 6.97E-10 and P = 1.33E-08, respectively). Patients carrying the GTF2I rs117026326 TT genotype and the GTF2I rs73366469 CC genotype had a strongly increased risk of SSc (P = 6.25E-09 and P = 1.67E-08, respectively), and those carrying the NFKB1 rs1599961 AA genotype had a suggestively significantly increased risk of SSc (P = 0.014). Moreover, rs117026326 and rs73366469 were associated with SSc in different genetic models (additive model, dominant model, and recessive model) (P < 0.05) whereas rs1599961 was associated with SSc in the dominant genetic model but not in the addictive and recessive models (P = 0.0026). TYK2 rs2304256 was not significantly associated with SSc in this study.
Conclusion: GTF2I rs117026326 and rs73366469 SNPs were strongly associated with SSc in this Chinese Han population. NFKB1 rs1599961 showed a suggestive association with SSc, and no significant association was found between TYK2 rs2304256 and SSc in this Chinese Han population.
Introduction
Systemic sclerosis (SSc) is an autoimmune disease characterized by limited or diffuse skin thickening, tissue fibrosis, and the immune response. Most patients with SSc develop skin thickening with variable organ involvement, including interstitial lung disease (ILD), pulmonary arterial hypertension (PAH), and renal crisis (1). The etiology and pathogenesis of SSc involves genetic, epigenetic, and environmental factors. Genetic factors in particular play an important role in susceptibility to the disease (2).
At present, there have been many genetic studies on SSc, including genome-wide association studies (GWAS) and Immunochip analyses (3–5). Several immune loci related to SSc susceptibility have been identified. Although great progress has been made in the past few years, our understanding of the genetic background of SSc remains limited, and the number of convincing SSc genetic markers only accounts for a small part of the total genetic variance. Therefore, further genetic research is of the utmost importance to better understand the pathogenic process of SSc, such as performing an association analysis of candidate single nucleotide polymorphisms (SNPs).
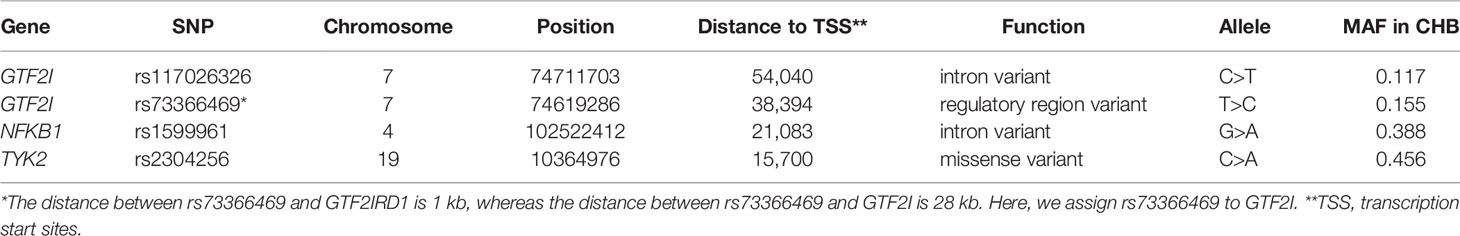
GTF2I is a gene located on the long arm of chromosome 7 (7q11.23) and encodes for a signal-induced transcription factor that functions as a universal regulator of numerous cellular processes. Many genetic studies have found that GTF2I is associated with multiple autoimmune rheumatic diseases. A study from six East Asian cohorts identified SNPs located at the GTF2I region that were associated with systemic lupus erythematosus (SLE) susceptibility (6). A study on rheumatoid arthritis (RA) in Asia confirmed that the GTF2I region may be a pathogenic variant and identified the largest-ever effect by heterogeneity mapping (7). A recent study has reported that this region was associated with susceptibility to SLE and SSc in a Japanese population (7).
Conversely, the deregulation of nuclear factor κB (NF-κB) can lead to multiple autoimmune disorders, including type 1 diabetes (T1D), SLE, and RA (8). NF-κB is a ubiquitous transcription factor of the Rel proto-oncogene family. It regulates the expression of several genes involved in inflammation and immune response. The NF-κB family is composed of five related proteins, including p50 or NF-κB1. The NF-κB signaling pathway plays an important role in the development and progress of RA both in vitro and in vivo (9). A study found that NF-κB1-deficient mice developed more fibrosis compared with wild-type mice (10). Moreover, NFKB1 loci have shown a suggestive association with SSc in a Middle Eastern population (11).
The Janus kinase family (JAK1, 2, and 3 and TYK2) has been recognized as important regulators of inflammation and immune processes. The kinase encoded by TYK2 can regulate the signal transduction of a variety of pro-inflammatory cytokines, including IL12, IL23, and type 1 interferon (IFNα). A recent fine mapping genetic study of RA found that three TYK2 protein coding variants are the most likely pathogenic variants that lead to the associated signals in this region (12). Simultaneously, functional prediction tools confirmed that the TYK2 variant can also cause damage such as SLE and inflammatory bowel disease. Furthermore, the TYK2 variant was reported to be associated with SSc in the European population (4).
Considering the genetic overlap of autoimmune diseases and the association of these genes with SSc in other populations, we hypothesized that some of the related tag SNPs of GTF2I, NFKB1, and TYK2 may also contribute to susceptibility to SSc in the Chinese Han population. Therefore, this study aimed to determine whether SNPs in GTF2I, NFKB1, and TYK2 were associated with SSc in a Chinese Han population and to explore the correlation of these loci and SSc clinical characteristics.
Methods
Study Populations
The SSc cohort employed in this study included 342 clinically diagnosed patients with SSc who fulfilled the 2013 ACR/EULAR classification criteria for SSc (13). Patients were classified with diffuse cutaneous systemic sclerosis (dcSSc) and limited cutaneous systemic sclerosis (lcSSc), according to the LeRoy criteria (14). Tests for the two SSc-related specific autoantibodies anti-topoisomerase I (ATA) and anti-centromere (ACA), which are most commonly tested in clinical applications, were performed in accordance with standard clinical laboratory methods. In total, 694 healthy subjects admitted to the Peking Union Medical College Hospital Health Examination Center for physical examination were recruited as negative controls for this study. ILD was identified with high-resolution computed tomography, and PAH was identified with right heart catheterization (PAH was diagnosed as a mean pulmonary pressure of ≥25 mmHg). All serum samples were stored at −80°C until use. All samples were obtained from patients recruited from the Peking Union Medical College Hospital (PUMCH). This study was approved by the Ethics Committee of the PUMCH, and all recruited participants provided informed consent.
DNA Extraction and Genotyping
The genomic DNA of each patient was extracted from peripheral blood samples using Tiangen DNA kits (Tiangen, Beijing, China). MassArray Assay Design 4.0 software (Sequenom, San Diego, CA, USA) was used to design the primers for the multiplex polymerase chain reaction (PCR) and locus-specific extension. Approximately 10–20 ng of DNA was amplified by multiplex PCR, and the DNA was genotyped using a Sequenom MassArray system (San Diego, CA) according to the manufacturer’s instructions. The final product was desalted and added to a 384-element Spectro CHIP array (Sequenom). Allele detection was performed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Mass spectrometry data and genotype were analyzed using MassArray Typer 4.0 software.
Statistical Analysis
PLINK v1.90, SPSS Statistics v.25 (IBM), and Prism v.8 (GraphPad) were used to analyze the data. The Chi-square test was applied to analyze Hardy–Weinberg equilibrium (HWE). If the P value of the SNP in the healthy control group was less than 0.05, the control group deviated from HWE and would be excluded. The associations between alleles and SSc were assessed with a logistic regression analysis, and the associations between genotypes and SSc were assessed with a Chi-square test. Three logistic regression models (additive, dominant, and recessive) and subgroup stratification analyses were assessed with a logistic regression analysis. All regressions were subjected to age adjustment. The odds ratio (OR) and confidence interval (95% CI) were calculated, and P values were statistically significant when <0.05 and were bolded in Tables 3–5. Finally, haplotype analysis and linkage disequilibrium were conducted using Haploview software v4.2.
Results
Clinical Characteristics of Patients With SSc and Healthy Controls
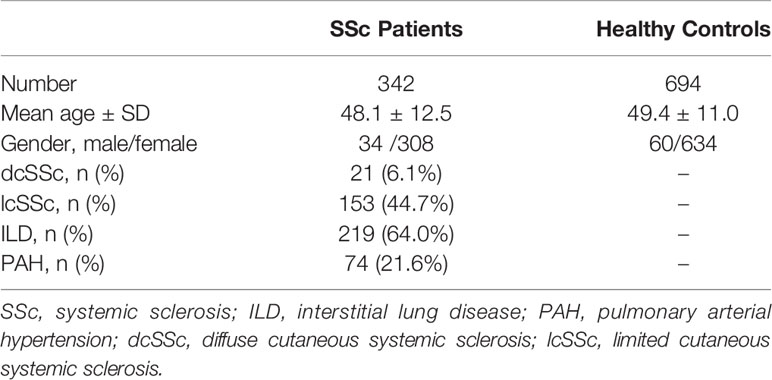
Our study included 342 patients with SSc [mean age ± standard deviation (SD): 48.1 ± 12.5 years; 34 males and 308 females] and 694 healthy controls (mean age ± SD: 49.4 ± 11.0 years; 60 males and 634 females) (Table 1). Among the 342 patients with SSc, 21 had dcSSc and 153 had lcSSc (168 patients with SSc had no classification result). In total, 219 (64.0%) patients with SSc had ILD and 74 (21.6%) had PAH.
Allele and Genotype Frequencies in SSc Patients and Healthy Controls
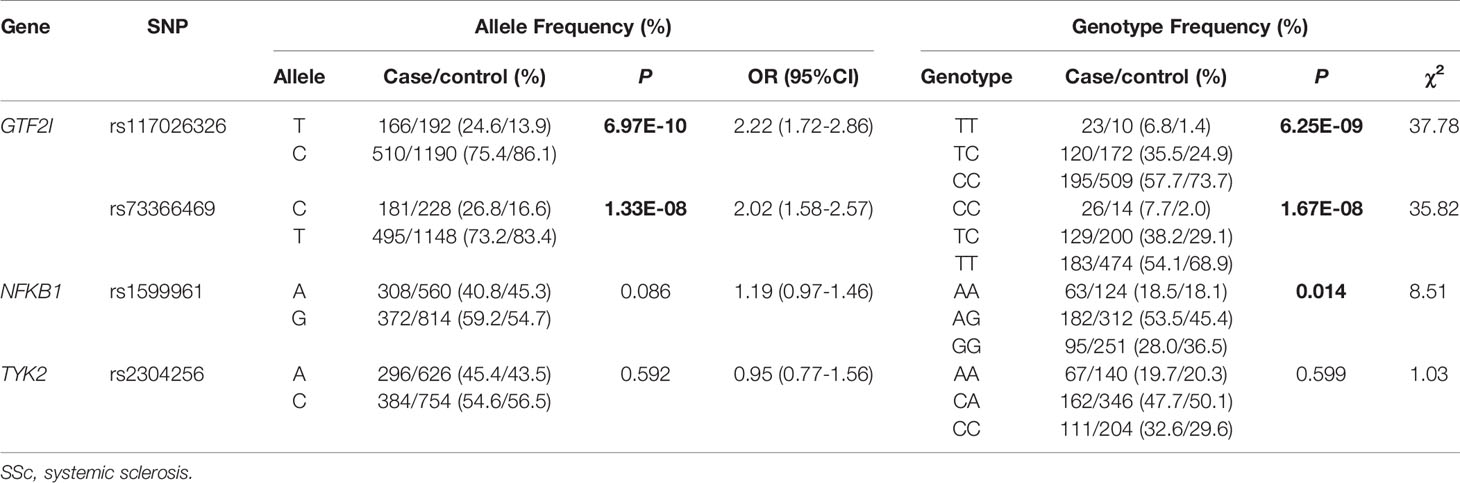
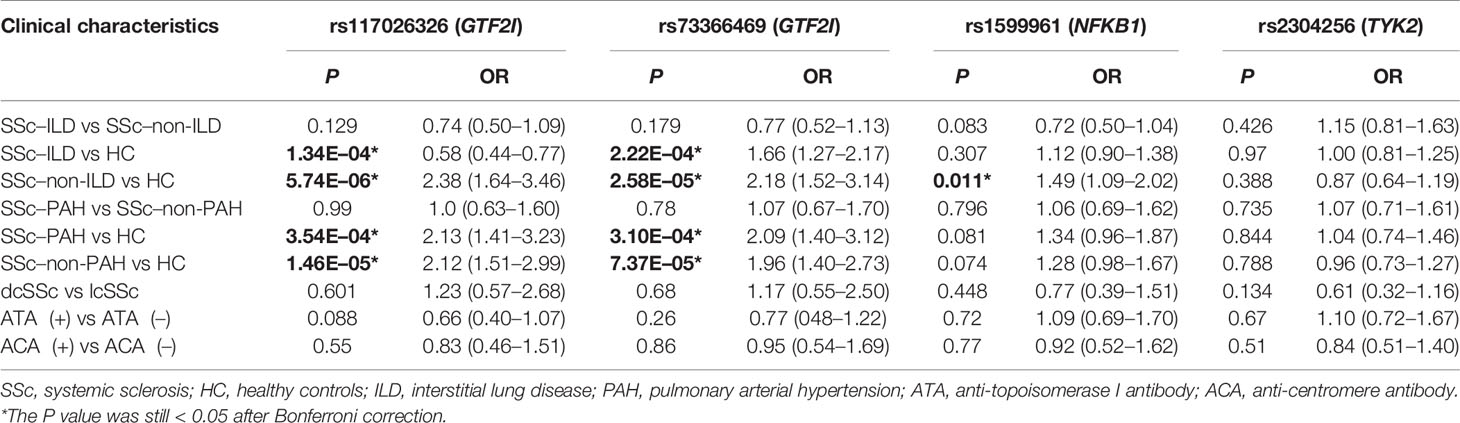
DNA from all subjects in the study was genotyped for the four selected SNPs, and the findings are summarized in Table 2. Four SNPs in the control group fulfilled the HWE criteria (P > 0.05). The allele and genotype frequencies in the patients with SSc and healthy controls are presented in Table 3. The rs117026326 and rs73366469 loci of the GTF2I gene were strongly associated with SSc. The T allele frequency of rs117026326 was higher in patients with SSc than in controls (24.6% vs. 13.9%, OR: 2.22, 95% CI: 1.72–2.86, P = 6.97E-10). The C allele frequency of rs73366469 was higher in patients with SSc than in controls (26.8% vs. 16.6%, OR: 2.02, 95% CI: 1.58–2.57, P = 1.33E-08). Furthermore, the genotype frequencies differed between patients with SSc and healthy controls. The TT genotype of rs117026326 had a significantly increased risk of SSc compared with the TC and CC genotypes (P = 6.25E-09). The CC genotype of rs73366469 had a significantly increased risk of SSc compared with the TC and TT genotypes between patients with SSc and healthy controls (P = 1.67E-08). Additionally, despite the lack of significant differences in the allele frequency in rs1599961 of the NFKB1 gene, the AA genotype had a suggestively significantly increased risk of SSc compared with the AG and GG genotypes between patients with SSc and healthy controls (P = 0.014). The genotype distributions of the three significant SNPs ((A) rs117026326, (B) rs73366469, and (C) rs1599961) in the patients with SSc and healthy controls are summarized in Figure 1. However, we did not detect any significant differences in the allele and genotype frequency distributions of rs2304256 of the TYK2 gene between patients with SSc and healthy controls (P > 0.05).
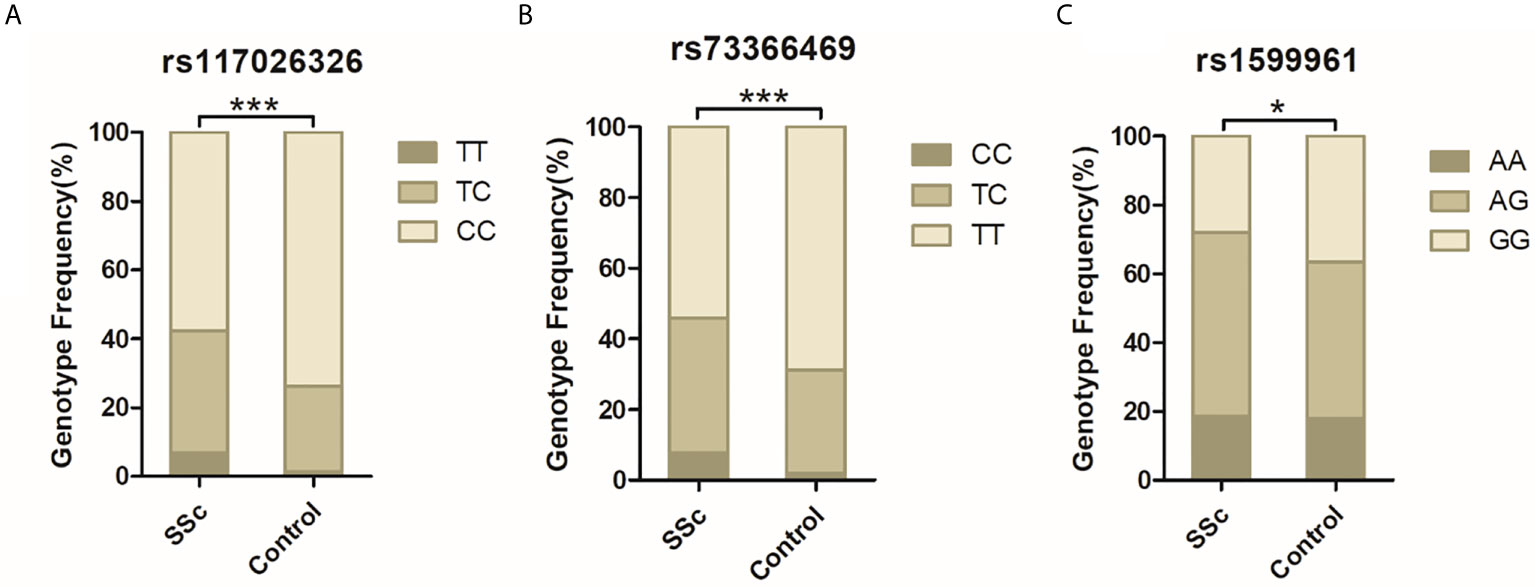
Figure 1 The genotype distributions of the three significant SNPs [(A) rs117026326, (B) rs73366469, and (C) rs1599961] in the patients with SSc and healthy controls are summarized. ***P < 0.001. *P < 0.05.
Genetic Models Determined by Logistic Regression Analysis
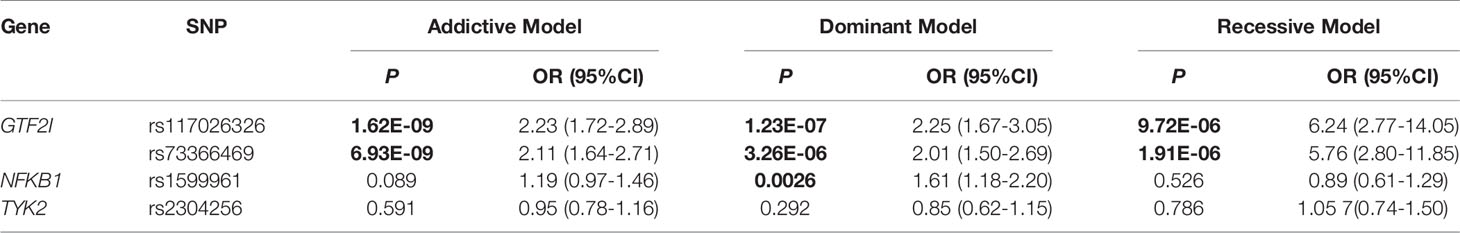
Three genetic models were analyzed to explore the effect of different genotypes on SSc (Table 4). rs117026326 was significantly associated with SSc in the genetic addictive, dominant, and recessive models (P = 1.62E-09, 1.23E-07, and 9.72E-06, respectively). rs73366469 was significantly associated with SSc in the genetic addictive, dominant, and recessive models (P = 6.93E-09, 3.26E-06, and 1.91E-06, respectively). rs1599961 was significantly associated with SSc in the genetic dominant model (P = 0.0026) but not in the genetic addictive or recessive models (P > 0.05), which indicates that the AA and AG genotypes had a significantly increased risk of SSc compared with the GG genotype between patients with SSc and healthy controls. However, rs2304256 was not associated with SSc in any of the three genetic models (P > 0.05).
The above data revealed that rs117026326 might be consistent with rs73366469. A linkage disequilibrium analysis was performed to evaluate this potential, and the results indicated that the two SNPs were in median linkage disequilibrium (D’ = 0.942, r2 = 0.756).
Associations Between SNPs and Patients With SSc With Different Clinical Characteristics
To further investigate the association between these SNPs and SSc susceptibility, we performed subgroup analyses (Table 5). We compared the frequencies of SNPs in patients with SSc with ILD with those of patients with SSc without ILD and with those of healthy controls. We performed the same subgroup analyses for PAH. We also compared the frequencies of SNPs in patients with dcSSc patients with those in patients with lcSSc as well as those in autoantibody-positive patients with those in autoantibody-negative patients. According to our results, SSc–ILD was associated with GTF2I rs117026326 (P = 1.34E-04) and GTF2I rs73366469 (P = 2.22E-04). SSc-non-ILD was associated with GTF2I rs117026326 (P = 5.74E-06), GTF2I rs73366469 (P = 2.58E-05), and NFKB1 rs1599961 (P = 0.011). SSc–PAH was associated with GTF2I rs117026326 (P = 3.54E-04) and GTF2I rs73366469 (P = 3.10E-04). SSc–non-PAH was associated with GTF2I rs117026326 (P = 1.46E-05) and GTF2I rs73366469 (P = 7.37E-05). No statistically significant association was found between TYK2 rs2304256 and any SSc clinical characteristics (P > 0.05). No statistically significant association was found between the four SNPs and the two autoantibodies (P > 0.05).
Functional Annotations of SNPs
We annotated the epigenetic regulatory features for the fours SNPs using Haploreg (15) and GWAS4D (16) online tools. Supplementary Table 1 shows the results of the functional annotations for binding motifs and epigenetic marks by Haploreg. GWAS4D incorporates 127 tissue/cell type-specific epigenomic data, integrates and refines transcription factor (TF) motifs from eight public resources, uniformly processes Hi-C data, etc. The results of annotation with GWAS4D are shown in Supplementary Tables 2–7.
Discussion
To our knowledge, this is a candidate gene association analysis that describes the association between functional GTF2I, NFKB1, and TYK2 polymorphisms and SSc in a Chinese Han population. This study demonstrated that the GTF2I polymorphisms were strongly associated with SSc in the Chinese Han population and that the NFKB1 polymorphism was suggestively associated with SSc in the Chinese Han population.
GTF2I encodes general transcription factor Iii (TFII-I). It binds to the initiator element (Inr) and E-box element in promoters and functions as a regulator of transcription (17). TFII-I is involved in the regulation of transcription, signal transduction, and immune cell signaling (18). Among its related pathways are RNA polymerase II transcription initiation and promoter clearance and the B cell receptor signaling pathway (KEGG). The GTF2I gene can regulate immunoglobulin heavy chain transcription in B cells (19). Makeyev et al. confirmed that the impaired expression of GTF2I could contribute to the etiology of Williams syndrome (20). Novel studies have shown that GTF2I is involved in an array of human diseases including neurocognitive disorders and cancer (21, 22). Our previous GWAS showed that GTF2I was the most strongly associated gene in Chinese Han patients with primary Sjögren’s syndrome (pSS) (23). Several association studies of candidate genes have revealed that the GTF2I gene is an important genetic susceptibility factor in SLE and RA (7, 24). Furthermore, SSc, pSS, SLE, and RA are all connective tissue diseases and share many clinical features, particularly in aspects of immune activation (25). These diseases usually coexist in patients in the form of overlap syndrome. Therefore, the GTF2I gene may also play an essential role in SSc. Our results indeed confirmed that GTF2I SNPs rs117026326 and rs73366469 were strongly associated with SSc in the Chinese Han population. Interestingly, rs73366469 is located within conserved enhancers and overlaps with transcription start sites (TSS) for GTF2I, thereby demonstrating the meaningful regulation of GTF2I (24). Additionally, rs117026326 and rs73366469 displayed consistent allele, genotype, genetic model, and clinical association results because of their medium linkage disequilibrium. Therefore, the two SNPs were most likely to have regulatory functions, which can also be verified by functional annotation using the online tools.
We also compared SSc accompanied with ILD and SSc accompanied without ILD and found no association between the two factors. And there was no previous study assessing the association between GTF2I and ILD. The significant result identified when the two groups were compared with healthy controls was possibly a result of the role of SSc disease itself instead of the role of ILD or non-ILD. The same principle is also applicable to the interpretation of the PAH results.
NFKB1 encodes a 105 kD protein that can undergo cotranslational processing by the 26S proteasome to produce a 50 kD protein. The 50-kD protein is a DNA binding subunit of the NF-kappa-B (NF-κB) protein complex. NF-κB is a transcription regulator that is activated by various intra- and extracellular stimuli such as cytokines, oxidant-free radicals, and bacterial or viral products. The inappropriate activation of NF-κB has been associated with a number of inflammatory diseases whereas persistent NF-κB inhibition leads to inappropriate immune cell development. Increased CD8+ T cell apoptosis in SSc was reported to be associated with low levels of NF-κB (26). A meta-analysis suggested a possible association between NFKB1 polymorphisms and certain autoimmune and inflammatory diseases in the Asian population but not in the Caucasian population (27). A GWAS of SSc in Iranian and Turkish populations identified a suggestive association between NFKB1 loci and SSc (11). However, NFKB1 polymorphisms showed no association with RA and ankylosing spondylitis (AS) (28, 29) or with SSc in Brazil (30), which may be due to the genetic differences of the populations. Then, we used the Haploreg online tool to annotated NFKB1 rs1599961. We found that high-LD SNPs rs1585213(r2 ≥ 1), rs3774933(r2 = 0.99), and rs4647980(r2 = 0.84) have multiple protein binding confirmations. It is speculated that these SNPs may affect the transcription factors such as ZNF263, CEBPB, FOXA1 and POL24H8, and then affect the transcription of downstream genes.
In the present study, people carrying the NFKB1 rs1599961 AA genotype had a suggestively significantly increased risk of SSc in the Chinese Han population; moreover, rs1599961 was related to SSc in the dominant genetic model but not in the addictive or recessive models. This result may have occurred because the frequency of the GG genotype was lower in patients with SSc than in healthy controls. The distribution of rs1599961 significantly differed when comparing patients with SSc without ILD with healthy controls. Thus, NFKB1 rs1599961 could be a candidate locus involved in SSc, particularly in Chinese Han patients without ILD.
TYK2 encodes a tyrosine kinase from the Janus kinase family. This protein can bind to the intracellular domain of Th1 and Th2 cytokine receptors and conduct signal transduction through the phosphorylation of receptor subunits. Previous studies have found that TYK2 polymorphisms are associated with a variety of autoimmune diseases such as RA, SLE, T1D, and multiple sclerosis (31–34). An Immunochip study of SSc found a suggestive association in the TYK2 region (35). Furthermore, rs2304256 was revealed as a nonsynonymous variant (C>A) located in TYK2 exon 8, causing the substitution of valine with phenylalanine (c.1084 G>T, Val362Phe) (36). This substitution thus affected the processing of pre-mRNA at exon 8. In addition, the eQTL results showed that rs2304256 enhanced TYK2 expression in blood. In summary, TYK2 rs2304256 is not neutral but has a potential impact on autoimmune diseases. TYK2 rs2304256 was reported to be strongly associated with SSc in a European population (4). However, this significant result was not replicated in the Chinese Han SSc population. No significant association was found between TYK2 rs2304256 and SSc in the current Chinese Han population in any of the analyses, which may be the result of racial differences.
In conclusion, GTF2I rs117026326 and rs73366469 were strongly associated with SSc in this Chinese Han population. NFKB1 rs1599961 showed a suggestive association with SSc whereas no significant association was found between TYK2 rs2304256 and SSc in this Chinese Han population.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the PUMCH. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
CL wrote this article. SY and HC conducted laboratory tests. ZW provided assistance with clinical information. LL and LC conducted data collection and data analysis. HL revised this article. YL designed experiments and guidance. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the grants from The National Key Research and Development Program of China (2018YFE0207300), the National Natural Science Foundation of China (Grants 81671618 and 81871302) and the CAMS Innovation Fund for Medical Sciences (CIFMS 2017-I2M-3-001and CIFMS 017-I2M-B&R-01). This work was supported by Beijing Key Clinical Specialty for Laboratory Medicine - Excellent Project (No. ZK201000).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.640083/full#supplementary-material
References
1. Denton CP, Khanna D. Systemic Sclerosis. Lancet (2017) 390(10103):1685–99. doi: 10.1016/s0140-6736(17)30933-9
2. Gabrielli A, Avvedimento EV, Krieg T. Scleroderma. N Engl J Med (2009) 360(19):1989–2003. doi: 10.1056/NEJMra0806188
3. Lopez-Isac E, Acosta–Herrera M, Kerick M, Assassi S, Satpathy AT, Granja J, et al. GWAS for Systemic Sclerosis Identifies Multiple Risk Loci and Highlights Fibrotic and Vasculopathy Pathways. Nat Commun (2019) 10(1):4955. doi: 10.1038/s41467-019-12760-y
4. Lopez-Isac E, Campillo–Davo D, Bossini–Castillo L, Guerra SG, Assassi S, Simeon CP, et al. Influence of TYK2 in Systemic Sclerosis Susceptibility: A New Locus in the IL-12 Pathway. Ann Rheum Dis (2016) 75(8):1521–6. doi: 10.1136/annrheumdis-2015-208154
5. Radstake TR, Gorlova O, Rueda B, Martin JE, Alizadeh BZ, Palomino-Morales R, et al. Genome-Wide Association Study of Systemic Sclerosis Identifies CD247 as a New Susceptibility Locus. Nat Genet (2010) 42(5):426–9. doi: 10.1038/ng.565
6. Sun C, Molineros JE, Looger LL, Zhou XJ, Kim K, Okada Y, et al. High-Density Genotyping of Immune-Related Loci Identifies New SLE Risk Variants in Individuals With Asian Ancestry. Nat Genet (2016) 48(3):323–30. doi: 10.1038/ng.3496
7. Kim K, Bang SY, Ikari K, Yoo DH, Cho SK, Choi CB, et al. Association-Heterogeneity Mapping Identifies an Asian-Specific Association of the GTF2I Locus With Rheumatoid Arthritis. Sci Rep (2016) 6:27563. doi: 10.1038/srep27563
8. Brown KD, Claudio E, Siebenlist U. The Roles of the Classical and Alternative Nuclear Factor-Kappab Pathways: Potential Implications for Autoimmunity and Rheumatoid Arthritis. Arthritis Res Ther (2008) 10(4):212. doi: 10.1186/ar2457
9. Handel ML, McMorrow LB, Gravallese EM. Nuclear Factor-Kappa B in Rheumatoid Synovium. Localization of p50 and P65. Arthritis Rheum (1995) 38(12):1762–70. doi: 10.1002/art.1780381209
10. Fullard N, Moles A, O'Reilly S, van Laar JM, Faini D, Diboll J, et al. The c-Rel Subunit of NF-κB Regulates Epidermal Homeostasis and Promotes Skin Fibrosis in Mice. Am J Pathol (2013) 182(6):2109–20. doi: 10.1016/j.ajpath.2013.02.016
11. González-Serna D, López–Isac E, Yilmaz N, Gharibdoost F, Jamshidi A, Kavosi H, et al. Analysis of the Genetic Component of Systemic Sclerosis in Iranian and Turkish Populations Through a Genome-Wide Association Study. Rheumatology (Oxford) (2019) 58(2):289–98. doi: 10.1093/rheumatology/key281
12. Diogo D, Bastarache L, Liao KP, Graham RR, Fulton RS, Greenberg JD, et al. TYK2 Protein-Coding Variants Protect Against Rheumatoid Arthritis and Autoimmunity, With No Evidence of Major Pleiotropic Effects on non-Autoimmune Complex Traits. PLoS One (2015) 10(4):e0122271. doi: 10.1371/journal.pone.0122271
13. van den Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A, et al. 2013 Classification Criteria for Systemic Sclerosis: An American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Ann Rheum Dis (2013) 72(11):1747–55. doi: 10.1136/annrheumdis-2013-204424
14. LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA Jr, et al. Scleroderma (Systemic Sclerosis): Classification, Subsets and Pathogenesis. J Rheumatol (1988) 15(2):202–5.
15. Ward LD, Kellis M. HaploReg: A Resource for Exploring Chromatin States, Conservation, and Regulatory Motif Alterations Within Sets of Genetically Linked Variants. Nucleic Acids Res (2012) 40(Database issue):D930–4. doi: 10.1093/nar/gkr917
16. Huang D, Yi X, Zhang S, Zheng Z, Wang P, Xuan C, et al. GWAS4D: Multidimensional Analysis of Context-Specific Regulatory Variant for Human Complex Diseases and Traits. Nucleic Acids Res (2018) 46(W1):W114–w120. doi: 10.1093/nar/gky407
17. Roy AL. Pathophysiology of TFII-I: Old Guard Wearing New Hats. Trends Mol Med (2017) 23(6):501–11. doi: 10.1016/j.molmed.2017.04.002
18. Cetkovská K, Šustová H, Kosztyu P, Uldrijan S, et al. A Novel Interaction Between TFII-I and Mdm2 With a Negative Effect on TFII-I Transcriptional Activity. PLoS One (2015) 10(12):e0144753. doi: 10.1371/journal.pone.0144753
19. Rajaiya J, Nixon JC, Ayers N, Desgranges ZP, Roy AL, Webb CF. Induction of Immunoglobulin Heavy-Chain Transcription Through the Transcription Factor Bright Requires TFII-I. Mol Cell Biol (2006) 26(12):4758–68. doi: 10.1128/MCB.02009-05
20. Makeyev AV, Bayarsaihan D. New TFII-I Family Target Genes Involved in Embryonic Development. Biochem Biophys Res Commun (2009) 386(4):554–8. doi: 10.1016/j.bbrc.2009.06.045
21. Liu DK, Wang J, Guo Y, Sun ZX, Wang GH. Identification of Differentially Expressed Genes and Fusion Genes Associated With Malignant Progression of Spinal Cord Gliomas by Transcriptome Analysis. Sci Rep (2019) 9(1):13583. doi: 10.1038/s41598-019-50072-9
22. Radovich M, Pickering CR, Felau I, Ha G, Zhang H, Jo H, et al. The Integrated Genomic Landscape of Thymic Epithelial Tumors. Cancer Cell (2018) 33(2):244–58.e10. doi: 10.1016/j.ccell.2018.01.003
23. Li Y, Zhang K, Chen H, Sun F, Xu J, Wu Z, et al. A Genome-Wide Association Study in Han Chinese Identifies a Susceptibility Locus for Primary Sjögren’s Syndrome at 7q11.23. Nat Genet (2013) 45(11):1361–5. doi: 10.1038/ng.2779
24. Meng Y, He Y, Zhang J, Xie Q, Yang M, Chen Y, et al. Association of GTF2I Gene Polymorphisms With Renal Involvement of Systemic Lupus Erythematosus in a Chinese Population. Medicine (Baltimore) (2019) 98(31):e16716. doi: 10.1097/md.0000000000016716
25. Wahren-Herlenius M, Dörner T. Immunopathogenic Mechanisms of Systemic Autoimmune Disease. Lancet (2013) 382(9894):819–31. doi: 10.1016/s0140-6736(13)60954-x
26. Kessel A, Rosner I, Rozenbaum M, Zisman D, Sagiv A, Shmuel Z, et al. Increased CD8+ T Cell Apoptosis in Scleroderma Is Associated With Low Levels of NF-kappa B. J Clin Immunol (2004) 24(1):30–6. doi: 10.1023/B:JOCI.0000018060.36183.bb
27. Zou YF, Wang F, Feng XL, Tao JH, Zhu JM, Pan FM, et al. Association of NFKB1 -94ins/delATTG Promoter Polymorphism With Susceptibility to Autoimmune and Inflammatory Diseases: A Meta-Analysis. Tissue Antigens (2011) 77(1):9–17. doi: 10.1111/j.1399-0039.2010.01559.x
28. Orozco G, Sánchez E, Collado MD, López-Nevot MA, Paco L, García A, et al. Analysis of the Functional NFKB1 Promoter Polymorphism in Rheumatoid Arthritis and Systemic Lupus Erythematosus. Tissue Antigens (2005) 65(2):183–6. doi: 10.1111/j.1399-0039.2005.00341.x
29. Kim TH, Stone MA, Rahman P, Yoo DH, Park YW, Payne U, et al. Interleukin 1 and Nuclear Factor-KappaB Polymorphisms in Ankylosing Spondylitis in Canada and Korea. J Rheumatol (2005) 32(10):1907–10.
30. Salim PH, Jobim M, Bredemeier M, Chies JA, Brenol JC, Jobim LF, et al. Interleukin-10 Gene Promoter and NFKB1 Promoter Insertion/Deletion Polymorphisms in Systemic Sclerosis. Scand J Immunol (2013) 77(2):162–8. doi: 10.1111/sji.12020
31. Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, et al. High-Density Genetic Mapping Identifies New Susceptibility Loci for Rheumatoid Arthritis. Nat Genet (2012) 44(12):1336–40. doi: 10.1038/ng.2462
32. Suarez-Gestal M, Calaza M, Endreffy E, Pullmann R, Ordi–Ros J, Sebastiani GD, et al. Replication of Recently Identified Systemic Lupus Erythematosus Genetic Associations: A Case-Control Study. Arthritis Res Ther (2009) 11(3):R69. doi: 10.1186/ar2698
33. Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG. The Imprinted DLK1-MEG3 Gene Region on Chromosome 14q32.2 Alters Susceptibility to Type 1 Diabetes. Nat Genet (2010) 42(1):68–71. doi: 10.1038/ng.493
34. Mero IL, Lorentzen AR, Ban M, Smestad C, Celius EG, Aarseth JH, et al. A Rare Variant of the TYK2 Gene is Confirmed to be Associated With Multiple Sclerosis. Eur J Hum Genet (2010) 18(4):502–4. doi: 10.1038/ejhg.2009.195
35. Mayes MD, Bossini–Castillo L, Gorlova O, Martin JE, Zhou X, Chen WV, et al. Immunochip Analysis Identifies Multiple Susceptibility Loci for Systemic Sclerosis. Am J Hum Genet (2014) 94(1):47–61. doi: 10.1016/j.ajhg.2013.12.002
Keywords: systemic sclerosis, single nucleotide polymorphism, genetic susceptibility, GTF2I, NFKB1
Citation: Liu C, Yan S, Chen H, Wu Z, Li L, Cheng L, Li H and Li Y (2021) Association of GTF2I, NFKB1, and TYK2 Regional Polymorphisms With Systemic Sclerosis in a Chinese Han Population. Front. Immunol. 12:640083. doi: 10.3389/fimmu.2021.640083
Received: 10 December 2020; Accepted: 07 June 2021;
Published: 23 June 2021.
Edited by:
Dimitrios Petrou Bogdanos, University of Thessaly, GreeceReviewed by:
Fusheng Zhou, Anhui Medical University, ChinaGeorge Bertsias, University of Crete, Greece
Copyright © 2021 Liu, Yan, Chen, Wu, Li, Cheng, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongzhe Li, eW9uZ3poZWxpcHVtY2hAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Chenxi Liu
Chenxi Liu Songxin Yan1†
Songxin Yan1† Haolong Li
Haolong Li