- 1Blood Center of Zhejiang Province, Key Laboratory of Blood Safety Research of Zhejiang Province, Hangzhou, China
- 2Division of Biomedical Informatics and Personalized Medicine, and Department of Immunology and Microbiology, University of Colorado Anschutz Medical Campus, Aurora, CO, United States
Killer cell immunoglobulin-like receptors (KIR) interact with human leukocyte antigen (HLA) class I molecules, modulating critical NK cell functions in the maintenance of human health. Characterizing the distribution and characteristics of KIR and HLA allotype diversity across defined human populations is thus essential for understanding the multiple associations with disease, and for directing therapies. In this study of 176 Zhejiang Han individuals from Southeastern China, we describe diversity of the highly polymorphic KIR and HLA class I genes at high resolution. KIR-A haplotypes, which carry four inhibitory receptors specific for HLA-A, B or C, are known to associate with protection from infection and some cancers. We show the Chinese Southern Han from Zhejiang are characterized by a high frequency of KIR-A haplotypes and a high frequency of C1 KIR ligands. Accordingly, interactions of inhibitory KIR2DL3 with C1+HLA are more frequent in Zhejiang Han than populations outside East Asia. Zhejiang Han exhibit greater diversity of inhibitory than activating KIR, with three-domain inhibitory KIR exhibiting the greatest degree of polymorphism. As distinguished by gene copy number and allele content, 54 centromeric and 37 telomeric haplotypes were observed. We observed 6% of the population to have KIR haplotypes containing large-scale duplications or deletions that include complete genes. A unique truncated haplotype containing only KIR2DL4 in the telomeric region was also identified. An additional feature is the high frequency of HLA-B*46:01, which may have arisen due to selection pressure from infectious disease. This study will provide further insight into the role of KIR and HLA polymorphism in disease susceptibility of Zhejiang Chinese.
Introduction
Natural killer (NK) cells are innate immune effectors having multiple important roles in human health. NK cells are critical in controlling the outcome of infections, tumors and fetal implantation, as well as organ transplantation and immunotherapies (1–5). In these roles, NK cell functions are modulated by interactions of their cell surface receptors with ligands expressed by tissue cells (6, 7). In humans, the interactions include polymorphic combinations of killer cell immunoglobulin-like receptors (KIR) with human leukocyte antigen (HLA) class I ligands (8, 9). The extreme genetic variation of KIR and HLA class I directly impacts NK cell functions, and associates with multiple immune-mediated diseases (10, 11). Also directly affected by combinatorial diversity, and influencing effector functions in health and disease, maturation of NK cells is guided through appropriate interactions of KIR with HLA class I (12–14).
The KIR locus of chromosome 19 (19q13.4) comprises up to 13 genes encoding receptors having distinct ligand specificities and inhibitory or activating functions (15, 16). Encoded by each KIR haplotype are from one to four inhibitory receptors and up to five activating receptors specific for HLA-A, B or C. Four framework genes, present in most individuals, form the basic structure of the locus. Their arrangement divides the KIR locus into centromeric (KIR3DL3 to KIR3DP1) and telomeric (KIR2DL4 to KIR3DL2) oriented regions. Structural diversity of the KIR locus is formed through recombination events that duplicate or delete entire genes, or deletions that create in-frame gene fusions (17, 18). This genetic variation thus has direct consequences for NK cell functions, which are further diversified by the alleles of KIR and their HLA class I ligands (19, 20). Within HLA-A, B and C, only those allotypes that contain one of four amino acid motifs, termed A3/11, Bw4, C1 or C2, in the external-facing α-helices can be ligands for KIR. Polymorphism both within and outside these motifs diversifies the interactions of KIR with HLA and individualize NK cell functions (21, 22). There are two broad forms of KIR haplotype, termed KIR-A and KIR-B (23). KIR-A haplotypes have fixed gene content, encoding four inhibitory KIR specific for HLA-A, B or C, as well as KIR2DS4, which is an activating receptor specific for some HLA-A and C allotypes (24). The majority of KIR2DS4 alleles are not expressed due to a deletion in exon 5 (25). KIR-B haplotypes contain a variable number of genes for activating KIR and fewer for inhibitory KIR. The two haplotype groups display distinct functional characteristics. For example, although KIR2DL2 and KIR2DL3 are encoded by the same locus, they differ in strength and specificity and characterize KIR-B and KIR-A haplotypes, respectively (26). Accordingly, KIR-A and KIR-B haplotypes also have distinct disease association profiles (27). KIR-A haplotypes associate with effective immunity against acute viral infections, but with increased risk of preeclampsia during pregnancy (28, 29). KIR-B haplotypes protect women from this pregnancy disorder and improve the outcome of hematopoietic stem cell transplantation (30, 31). Additional to the KIR-A and KIR-B haplotype designations, specific alleles that can be characteristic to the haplotypes associate with disease susceptibility or protection. A total of 1,110 alleles encoding 543 allotypes have been reported according to the IPD-KIR database release version 2.10.0 (32).
The development of next-generation sequencing (NGS) technologies for genotyping KIR and HLA at high-resolution has helped identify the substantial variation of KIR allotypes and their interactions with HLA ligands across human populations (33–35). It is therefore of great significance to characterize this combinatorial diversity across all human populations. Although China, as one of the four ancient civilizations of the world, accounts for approximately 20% of the human population, analysis of the KIR locus at high-resolution from East Asia is limited to two studies (36, 37). In this study, we fully characterize the allele, haplotype and combinatorial diversity of KIR and HLA class I in the Zhejiang Han population from the Southeast coast of China (27°02’ to 31°11’ North, 118°01’ to 123°10’ East).
Materials and Methods
Samples and DNA Extraction
176 blood samples were collected from healthy blood donors in the Blood Center of Zhejiang province, China. The ethnic background of all individuals is Zhejiang Han. Informed consent was obtained from all participants. The Zhejiang Han are a population of approximately 60 million individuals living on the Southeast coast of China and are a subgroup of the Chinese Southern Han ethnic group. This study was approved by the regional ethics committee, Blood Center of Zhejiang Province. Genomic DNA was extracted using MagNA Pure LC DNA Isolation Kits (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer’s instructions. The final DNA concentration was adjusted to 60 ng/μl and optical density at 260/280 was approximately 1.8.
High-Resolution Sequencing for KIR and HLA
A biotinylated DNA probe-based capture method was applied to sequence KIR and HLA genes to high-resolution, as described previously (33). Briefly, 500 ng genomic DNA for each sample was randomly fragmented using enzyme digestion (New England Biolabs, Boston, MA) and then labeled with TruSeq DNA CD indexes (Illumina Inc, San Diego, CA). Fragments of 800–1,200 bp length were obtained by size selection and the individual samples pooled at equal quantity into one tube. Fragments containing target HLA and KIR genes were captured using specific probes, prepared to a final concentration of 12 pmol/L, as determined by Qubit instrument, then sequenced using a MiSeq instrument and a v3 Reagent Kit (600-cycle; Illumina Inc, San Diego, CA, USA).
Sequence Data Analysis
KIR gene content, copy numbers, and allele genotypes were determined using the Pushing Immunogenetics to the Next Generation pipeline (PING) as previously reported (33). The copy number was determined from the ratio of reads mapping to each KIR gene to those mapping to KIR3DL3, a reference gene present only one copy on each haplotype. Alleles for 13 KIR genes were analyzed, of which KIR2DL2 and KIR2DL3 are alleles at the same locus (KIR2DL2/3), as are KIR2DS3 and KIR2DS5 (KIR2DS3/5), and KIR3DL1 and KIR3DS1 (KIR3DL1/S1). KIR3DL3 alleles were determined using PHASE 2.1 (38). The HLA alleles were determined using TypeStream Visual NGS Analysis Software (TSV2.0, One Lambda Inc, Canoga Park, CA). The presence of KIR ligands, A3/11, Bw4, and C1/C2 was determined according to the HLA alleles.
Frequencies of Alleles, Genotypes, and Haplotypes
KIR genotypes were named based on the presence or absence of KIR genes according to the allelefrequencies.net database (39). Carrier frequencies of KIR genes and genotypes were calculated as their percentage of the total number of individuals. The frequencies of alleles or haplotypes were calculated by direct counting, and the number observed divided by 2N (alleles duplicated on a single haplotype were not included, and absence was counted as a distinct allele). The composition and frequencies of HLA haplotypes were determined using Arlequin 3.5.2.2 (40).
KIR/HLA Interaction Analysis
Interactions between inhibitory KIR and HLA ligands were analyzed. The number of distinct allotype interactions was counted for each KIR for every individual and the mean number was calculated per population. Broadly, KIR2DL1 recognizes C2+HLA-C, and KIR2DL2/3 recognizes C1+HLA-C, and some C2+HLA-C. KIR3DL1 recognizes the Bw4 motif at residues 77–80 of some HLA-A and HLA-B allotypes. KIR3DL2 recognizes the A3/11 motif carried by HLA-A*03 and HLA-A*11 allotypes (41, 42). Major exceptions are KIR2DL1*022, which interacts with C1+HLA, and HLA-B*46 and B*73, which carry C1 motifs (20, 26).
Statistical Analysis
Hardy–Weinberg equilibrium (HWE) was determined for each gene using Fisher’s exact test implemented in the Arlequin software 3.5.2.2. The distributions of genes, alleles, haplotypes, and KIR/HLA interactions in the Zhejiang Han population were compared to other populations as described. Differences between populations were assessed using GraphPad software, by means of the χ2 test for categorical variables. The p values were calculated using Fisher’s exact test (pf), applied when appropriate. Bonferroni correction for multiple comparisons was applied. p <0.05 was regarded as significant.
Results
High Frequency of KIR-A Haplotypes in Zhejiang Han
To analyze the KIR locus of the Zhejiang Han at high resolution, we first established which KIR genes are present, and in how many copies, for every individual. The four framework genes [KIR3DL3, KIR2DL4, KIR3DP1 and KIR3DL2 (15)] were detected in all 176 individuals of the cohort (Figure 1A). KIR2DL4 and KIR3DP1 were each observed with one to three copies and one individual in the cohort has only one copy of KIR3DL2 (Figure 1A). Only four individuals have less than two copies of any of these framework genes, six individuals harbor three copies, and none were observed with four copies. These observations indicate that, although structural diversity of the KIR locus is present, it is rare in the Zhejiang Han population. KIR2DL1, KIR2DL3, KIR3DL1, KIR2DS4, and KIR2DP1 were all observed at a frequency greater than 90% and at two copies in at least 64.8% of the population (Figures 1A, B). These five genes are not present on every KIR haplotype, and when they occur together, they characterize the KIR A haplotype (15, 51). KIR2DL3, which occurs in the centromeric KIR region, showed higher frequency than KIR3DL1 and KIR2DS4, which are located in the telomeric KIR region (81.8% vs 64.8%), indicating a higher frequency of centromeric KIR-A haplotype motifs (Cen-A) than telomeric KIR-A motifs (Tel-A). Accordingly, the genes characteristic of KIR-B haplotypes have frequencies less than 25% in centromeric and approximately 35% in telomeric KIR region (Figure 1B). We compared the distribution of KIR genes of Zhejiang Han with populations representing Asia (37, 43, 46, 52–54), Europe (47, 48, 55, 56), Africa (49, 57), America (50) and Oceania (58, 59) (Supplementary Table 1). As a control, this analysis showed the KIR gene frequencies to be similar to our previous study of an independent Zhejiang Han cohort that was performed at lower resolution (60). The KIR gene frequencies are also similar to those of other populations from East Asia including Japan (37), South Korea (43) and Singapore (53), but distinct from Southeast Asians (46) and populations from other continents (Supplementary Table 1). Accordingly, the frequencies of Cen-B specific KIR genes (KIR2DS2, KIR2DL2, KIR2DL5B) in the Zhejiang Han are significantly lower than all populations outside East Asia (Supplementary Table 1).
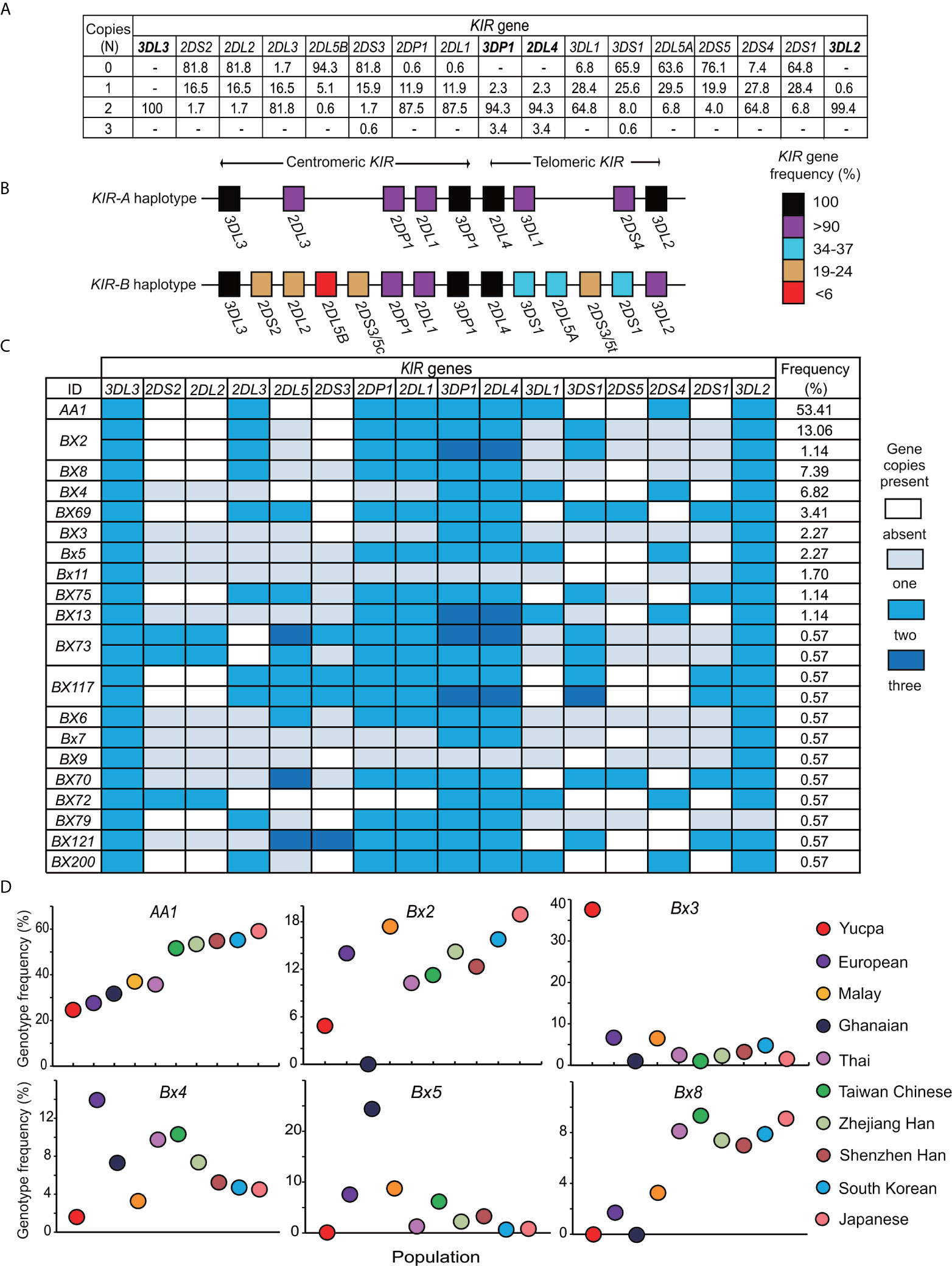
Figure 1 Distribution of genotypes derived from 176 Zhejiang Han individuals. (A) Shown for each KIR gene is the frequency (%) of individuals carrying the number of copies indicated at the left (0–3). Dash “–” represents not observed. The four framework genes are indicated in bold text. (B) Shown is the percentage of KIR-A and KIR-B haplotypes that carry each of the genes indicated. Shades of red to black represent the percentages, and the key is shown at the right. (C) Shown are KIR gene content genotypes observed in the Zhejiang Han individuals. The colors of the boxes indicate the gene copy numbers, as given in the key at the right. Genotype IDs based on the presence or absence of KIR genes (39) are shown on the left. The frequencies (%) of the corresponding genotypes are listed in the right. (D) Shown are the carrier frequencies of KIR gene content genotypes -AA1, Bx2, Bx3, Bx4, Bx5, Bx8 in ten different populations. They are Chinese Southern Han from Shenzhen (36), Japanese (37), South Korean (43), Taiwan Chinese (44), Malay (45), Thai (46), Europeans (47, 48), Sub-Saharan Africans Ghanaians (49) and South American Yucpa (50). The colored circles represent the populations, as shown at the right.
As determined by the copy number of each of the 13 KIR genes, twenty-three distinct genotypes were observed (Figure 1C). Of these genotypes, KIR-AA is the most frequent (53.4%) in Zhejiang Han (Figure 1C). The KIR-AA genotype is common across East Asia, also being observed frequently in other Chinese Southern Han (36), Taiwan Chinese (44), South Korean (43), and Japanese (37). The KIR-AA genotype is less frequent outside East Asia, as illustrated by populations representing Southeast Asia, Europe, South America, and sub-Saharan Africa (Figure 1D). The other 22 KIR genotypes observed in Zhejiang Han are designated KIR-Bx, the distributions of which are also similar among East Asians and distinct from populations of other continents (Figure 1D). For example, Bx2, which is the most frequent KIR-B genotype in Zhejiang Han, was not observed in sub-Saharan Africans, whereas Bx5 is frequent in sub-Saharan Africans, but very rare in Zhejiang Han (Figure 1D). Another example is Bx3, which was observed with high frequency in Amerindians, but almost absent from all other populations (Figure 1D). These examples of distinct genotype profiles characterizing major human population groups illustrate the rapid evolution of the KIR locus (27, 61).
Genes for Inhibitory KIR Exhibit More Allelic Diversity Than Those for Activating KIR
A total of 107 KIR alleles were observed in the Zhejiang Han cohort (Figure 2A and Supplementary Table 2) and no novel alleles were identified. The allele frequency distributions for all the KIR genes were in accordance with Hardy–Weinberg equilibrium. The inhibitory KIR are more diverse and polymorphic (88 alleles in total) than the activating KIR (14 alleles). The most diverse gene encoding an activating KIR is KIR2DS4, which is the only activating KIR carried by the KIR-A haplotype, with five alleles observed (Figure 2B). KIR2DS4*00101 is the most frequent allele and is the only one that can encode a functional allotype on the cell surface, whereas the remainder are non-expressed alleles because of a 22bp deletion in exon 5 (25). Other activating KIR are encoded by KIR-B haplotypes and show little diversity. KIR2DS3/5 can be present on either a Cen-B or Tel-B haplotype; two KIR2DS3 alleles were observed, one is KIR2DS3*00103 observed in Cen-B, and the other is KIR2DS3*00201 observed in Tel-B. Although the activating KIR are less polymorphic, their allele frequency distributions do vary across populations. For example in Zhejiang Han, functional KIR2DS4 allotypes are the most frequent, whereas in some populations, such as in Oceania (62), the non-expressed KIR2DS4 alleles predominate.
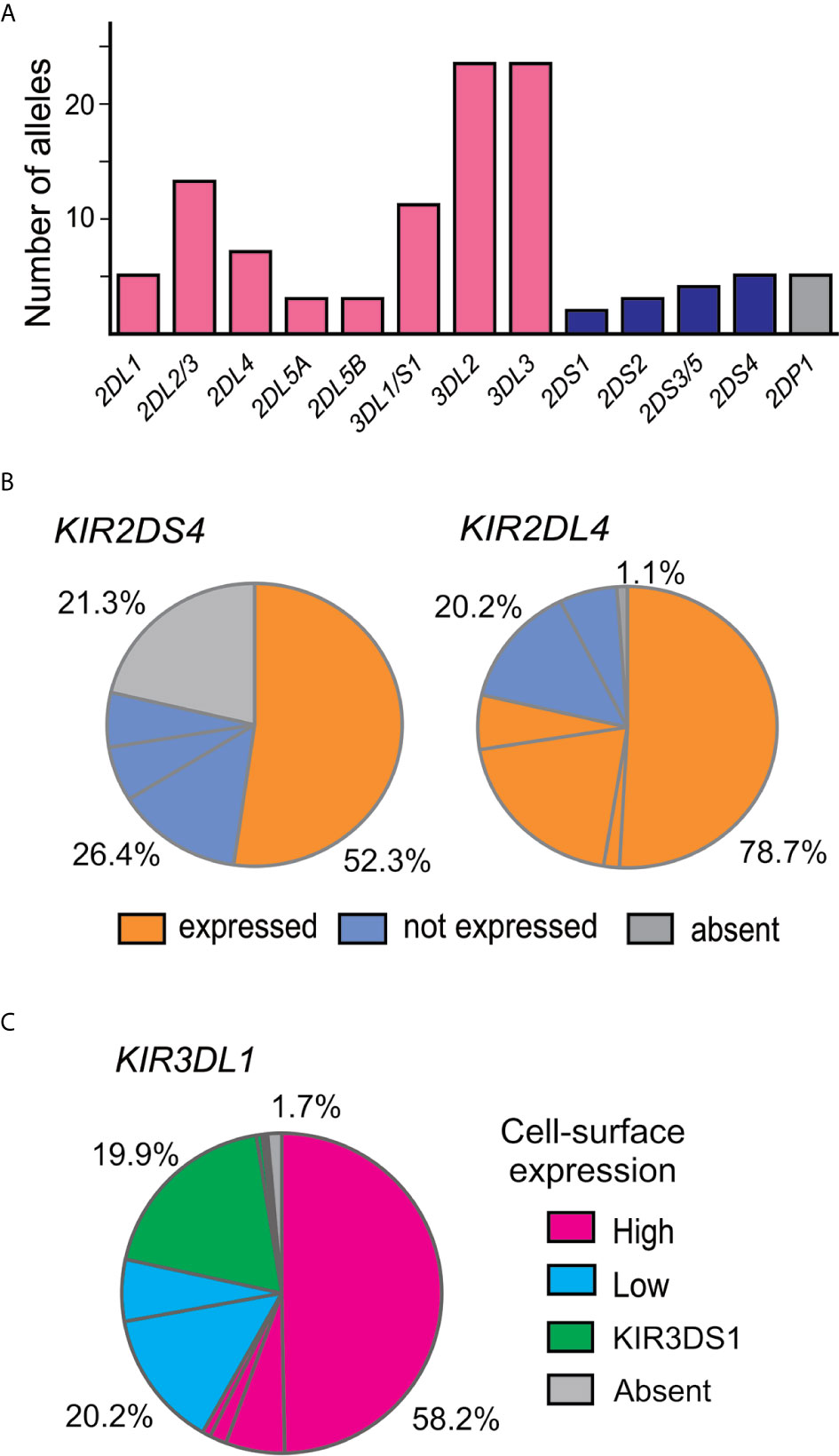
Figure 2 KIR alleles present in the Zhejiang Han population. (A) Shown is the total number of KIR alleles observed in 176 Zhejiang Han. Genes encoding inhibitory KIR are indicated by red bars, activating by blue bars, and pseudogenes by gray bars. (B) Shown are the ratios of expressible and non-expressed alleles of KIR2DS4 and KIR2DL4 in Zhejiang Han. Each pie segment represents a distinct allele, orange indicates allele can be expressed at the cell surface, blue indicates not expressed, grey indicates gene absent. (C) Shown is the KIR3DL1 allele expression frequency spectrum in Zhejiang Han. Magenta indicates high expression alleles, cyan—low expression, green—KIR3DS1 (activating receptor) and gray indicates gene absence or unknown expression pattern.
Among genes encoding inhibitory KIR, those with three domains show greatest variability (25, 32). We observed 23 alleles each of KIR3DL3 and KIR3DL2 and 11 alleles of KIR3DL1/S1, which encode 16, 17, and 11 allotypes, respectively (Supplementary Table 2). Thus, each allele of KIR3DL1 we observed here (Figure 2C) encodes a distinct allotype and the most abundant have high-expressing, high-binding, phenotypes (37, 63, 64). KIR3DL1*015 is the most frequent KIR3DL1 allotype in Zhejiang Han. Comparison with other representative populations shows that KIR3DL1*015 is present worldwide, reaching highest frequency in East Asians (Figure 3). By contrast, KIR3DL1*004, which is a poorly expressed phenotype found in Europeans and Africans, was not observed in Zhejiang Han (Figure 3) and neither was it observed in another Chinese Southern Han group (36). We previously reported KIR3DL1 allele frequencies from an independent cohort of Zhejiang Han (66). Because KIR3DS1 was not analyzed in that cohort, the reported KIR3DL1 allele frequencies are higher than in the present cohort, however, the frequencies are similar when KIR3DS1 is considered. The present analysis also showed that KIR3DL3 has the most extensive cross-population diversity. Here, KIR3DL3*010 clearly characterizes East Asian populations, whereas 3DL3*003 for example characterizes Amerindians, but is rare in East Asians (Figure 3).
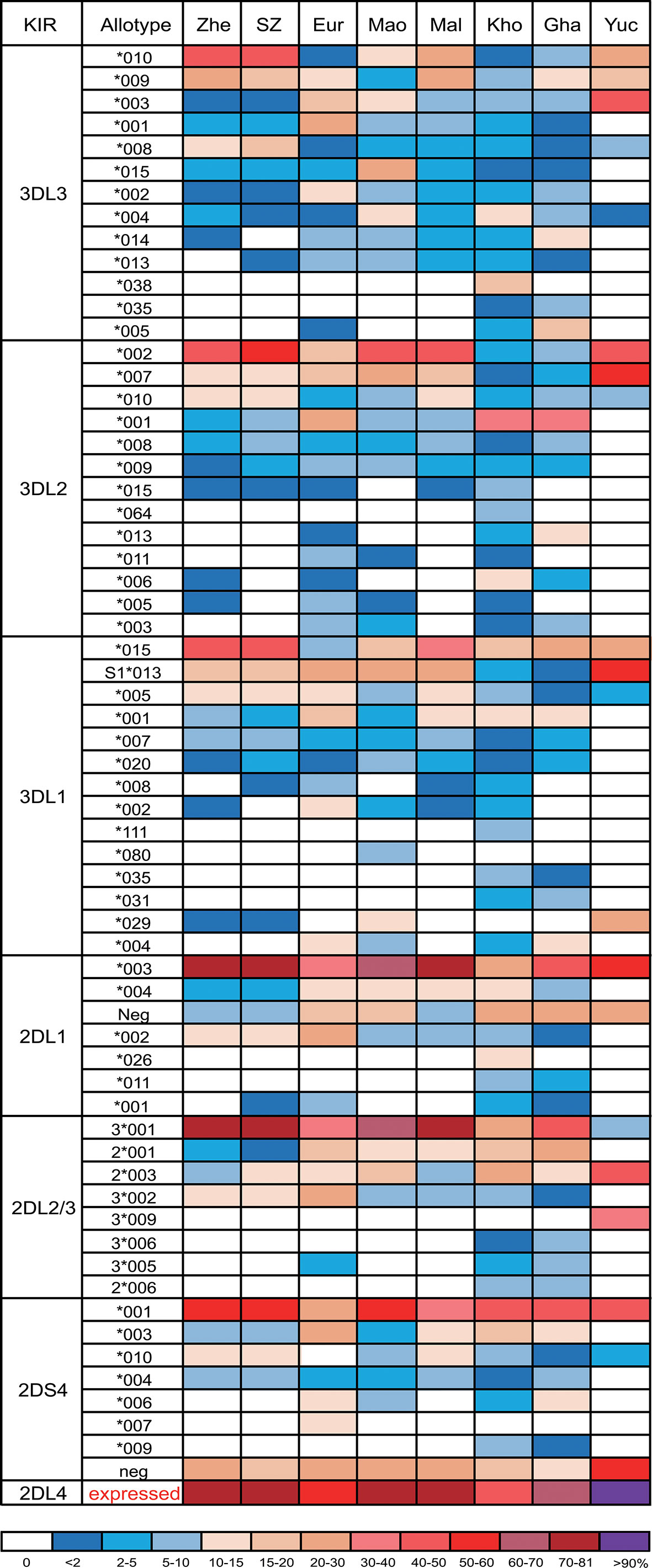
Figure 3 KIR allotype diversity across human populations. Shown are the frequencies of KIR3DL1-3, KIR2DL1-4, and KIR2DS4 allotypes from seven human populations. Shades of dark blue to dark red represent the allotype frequencies from low to high as indicated in the key at the bottom. Zhe, Zhejiang Han; SZ, Shenzhen Han (36); Eur, European (65); Mao, Māori (62); Mal, Malay (45); Kho, Southern Africa Khomani (34); Gha, sub-Saharan Africa Ghanaian (49); Yuc, Amerindian Yucpa (50). For KIR2DL4, the sum of expressible allotypes is shown. * standard WHO nomenclature for KIR alleles.
Although KIR2DL1 is the most polymorphic two-domain KIR, having 173 known alleles [IPD-IMGT/KIR Database version 2.10.0, December 2020 (32)], it has low diversity in Zhejiang Han, with five alleles observed (Figure 2A). The most prominent of these alleles is KIR2DL1*00302 at an allele frequency of 77.8% (Supplementary Table 2), corresponding with a similar frequency of KIR2DL3*00101. This strong linkage disequilibrium between the two alleles is shared across multiple populations worldwide (34, 37, 45, 50, 62, 65). KIR2DL4, which can be an activating receptor specific for HLA-G (67, 68), has six alleles in Zhejiang Han (Supplementary Table 2). Of 107 KIR2DL4 alleles, encoding 54 distinct polypeptide sequences, 18 do not express a receptor on the cell surface due to a single nucleotide deletion at position 811 in Exon 7 (69). The non-expressed alleles include KIR2DL4*008 and KIR2DL4*011, which were observed at a total frequency of 20.2% in Zhejiang Han (Figure 2B). The four expressible KIR2DL4 alleles observed thus constitute a majority. This majority varies little across human populations, the most extreme being Amerindians, where >95% KIR2DL4 alleles are expressible, and the exception being KhoeSan, where there is an approximately equal ratio between expressible and non-expressible forms of KIR2DL4 (Figure 3).
KIR-A Haplotypes Are More Frequent Than KIR-B Haplotypes Both in Centromeric and Telomeric Regions
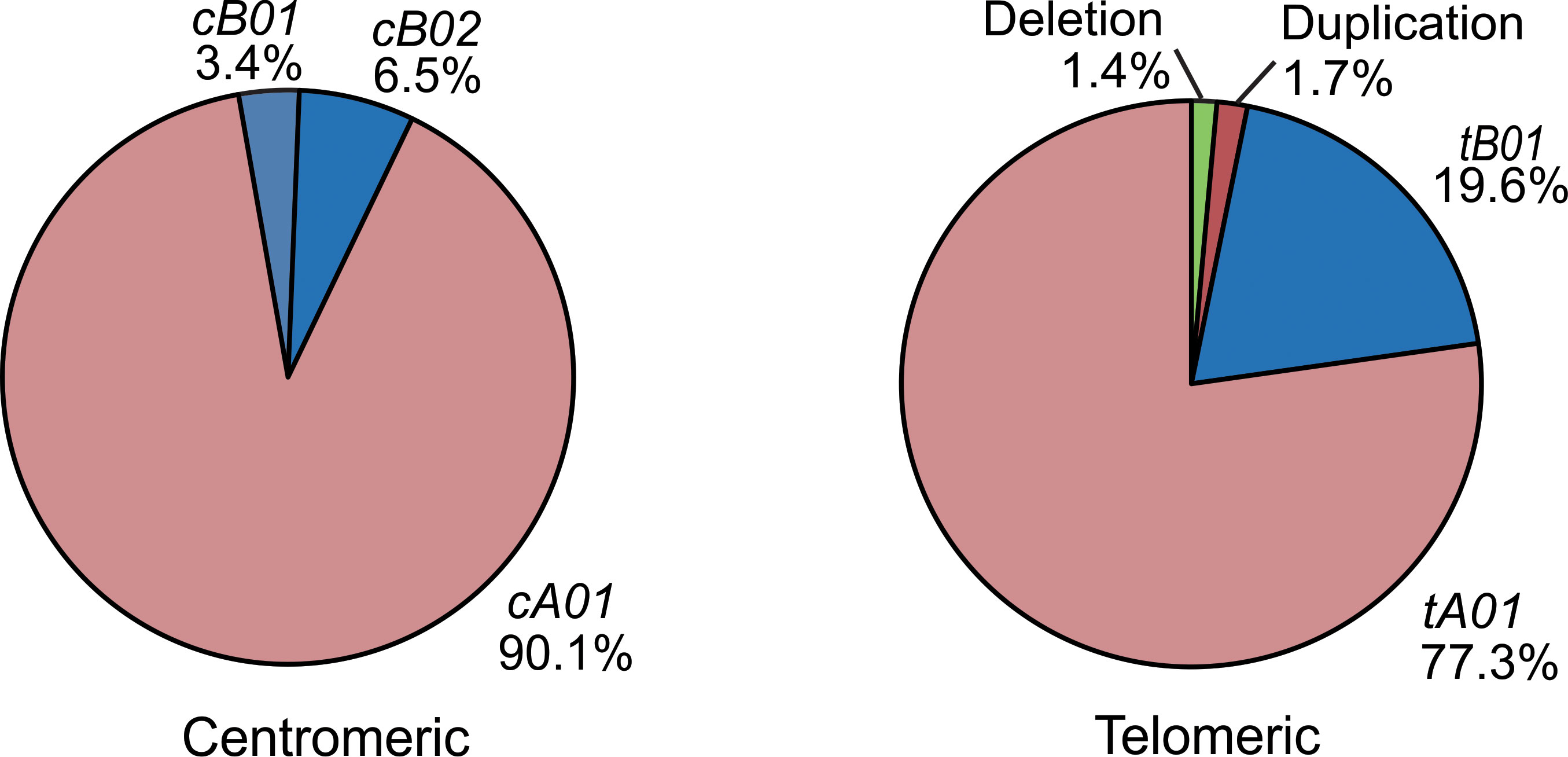
Based on the copy number and allele information obtained from high resolution analysis, 54 centromeric haplotypes and 37 telomeric haplotypes were observed in Zhejiang Han (Supplementary Table 3). As defined by gene content, three common centromeric haplotype groups, cA01, cB01, and cB02, and two telomeric groups, tA01 and tB01, were observed, as well as rarer forms having duplicated or deleted segments (Figure 4). When accounting for allele diversity, there are 41 haplotypes in the cA01 group (Supplementary Table 3A), and 23 in the tA01 group (Supplementary Table 3B). The most frequent cA01 haplotype at the allele level in Zhejiang Han is KIR3DL3*01002-2DL3*00101-2DP1*00201-2DL1*00302, with a frequency of 19.03% (Figure 5A). The five most common cA01 haplotypes (64.5% in total) differ only in their KIR3DL3 alleles (Figure 5A). The high polymorphism of KIR3DL3 alleles diversifies the centromeric region, and the number of the centromeric haplotypes would decrease from 54 to 19 if KIR3DL3 alleles were excluded (Supplementary Table 3A). The most frequent tA01 haplotype at the allele level in Zhejiang Han is KIR2DL4*00102-3DL1*01502-2DS4*00101-3DL2*00201 (Figure 5B), with a frequency of 45.5%. Within the telomeric haplotypes, KIR2DS3/5t and KIR2DL5A alleles show strong linkage disequilibrium, in which KIR2DS5*00201 and KIR2DL5A*00101 are always found together, as are KIR2DS3*00201 and KIR2DL5A*00501. These patterns of linkage disequilibrium are the same as those found in other populations outside of Africa.
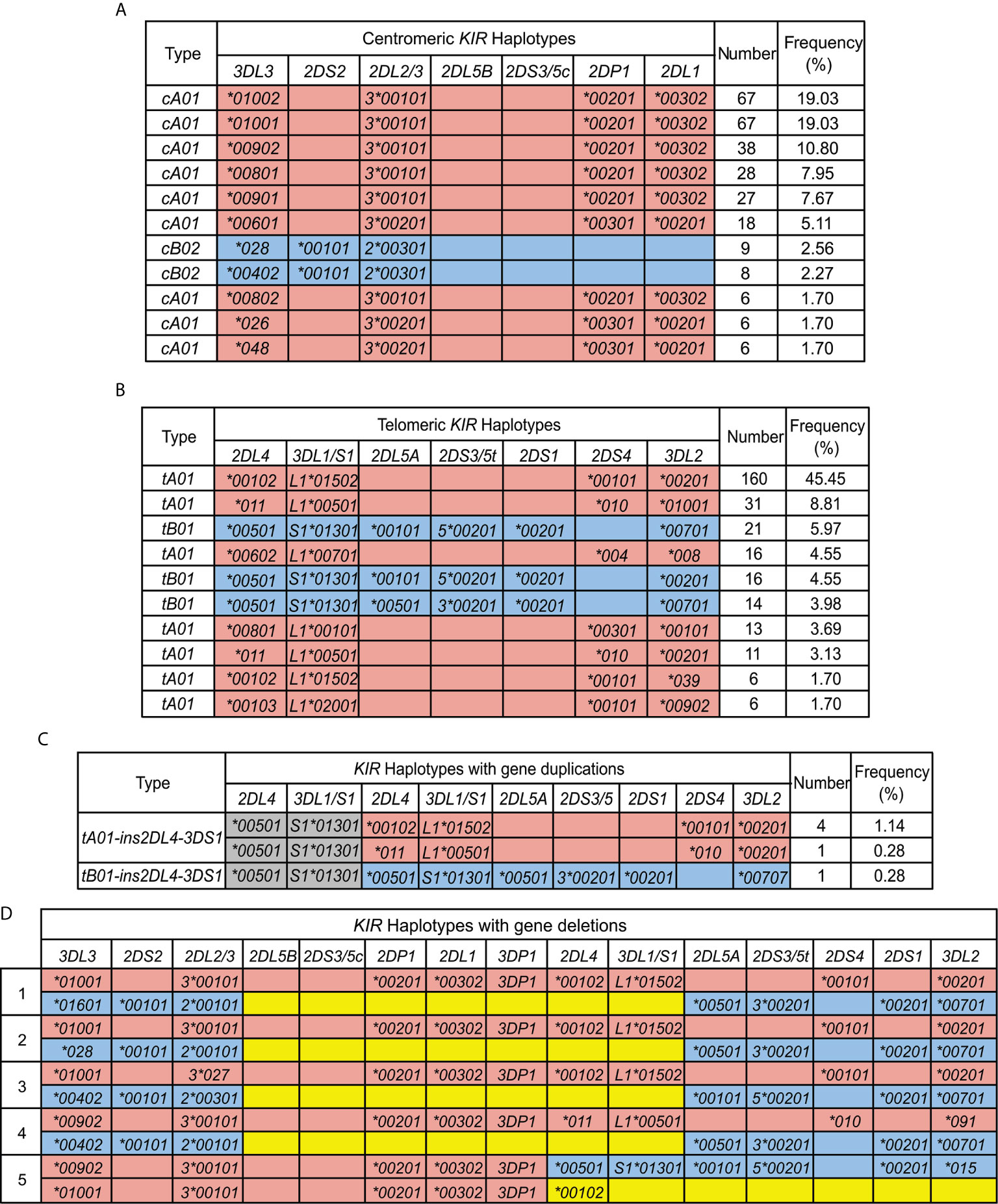
Figure 5 KIR haplotypes identified in 176 Zhejiang Han. (A, B). Shown are the ten most frequent centromeric and telomeric haplotypes and their frequencies observed in Zhejiang Han. (C) Shown are the extended telomeric haplotypes observed in Zhejiang Han. (D) Shown are the two complete haplotypes for each of the five individuals who exhibit contracted KIR haplotypes. KIR-A haplotype are shaded red and KIR-B haplotypes are blue. Gray indicates insertion. Yellow indicates deletion. Blank boxes indicate gene absence. The frequencies of haplotypes are shown at the right. * standard WHO nomenclature for KIR alleles.
Six individuals having an insertion/duplication of KIR3DP1-2DL4-3DS1 were observed in Zhejiang Han (Figure 5C). The corresponding haplotypes comprised two distinguished at the gene content level, and three as determined by allele content. We also identified four individuals with haplotypes lacking KIR3DP1-2DL4-3DL1/S1 (Figure 5D), with three of these haplotypes (individuals 1, 2 and 4: Figure 5D) distinguished only by their alleles of KIR3DL3. These haplotypes likely formed by uneven exchange during meiotic recombination, and have also been observed in other populations (17, 45, 70, 71). Although it remains to be confirmed through full haplotype sequence analysis, the insertions/duplications and the deletions occur in approximately the same location 5’ from KIR3DP1, suggesting the crossovers can occur frequently through shared mechanisms (72). In addition, we identified a unique haplotype that lacks any KIR genes in the telomeric region 3’ from KIR2DL4, leaving only six genes on the entire KIR haplotype (individual 5; Figure 5D). That the allele is KIR2DL4*00102 suggests the parental haplotype that lost these genes was similar to the most frequent KIR-A in Zhejiang Han (Figure 5B).
High Frequency of Strong Inhibitory KIR/HLA Interactions Identified in Zhejiang Han
Alleles for highly polymorphic HLA-class I (HLA-A, -B, -C) were obtained from high resolution sequencing. A total of 25 HLA-A alleles, 43 HLA-B alleles, and 27 HLA-C alleles were observed (Supplementary Table 4), and the distributions for all the HLA genes complied with Hardy–Weinberg equilibrium. Only some HLA-class I allotypes act as ligands for KIR (9, 27). The two most frequent alleles of HLA-A, -B, -C respectively in Zhejiang Han are A*11:01 (25.3%) and A*24:02 (13.1%), B*40:01 (11.6%) and B*46:01 (10.5%), C*07:02 (18.2%) and C*01:02 (16.8%). All the allotypes they encode can interact with KIR, except for B*40:01. Every HLA-C allotype is a ligand for KIR, and in total, 44.1% of HLA-A and 48.6% of HLA-B allotypes by frequency in the Zhejiang Han can act as KIR ligands (Figures 6A, B). Most of the HLA-C allotypes (82.4%) contain a C1 motif (C1+HLA-C) in Zhejiang Han and HLA-B*46:01, which also carries a C1 motif was also observed at high frequency (10.5%). HLA-B*46:01 is one of only two HLA-B allotypes that carry a C1 motif (C1+HLA-B), the other is HLA-B*73:01, and both have geographical distributions focused in Asia, B*46:01 in Southeast Asia and B*73:01 in West Asia (39). C1+HLA-C and C1+HLA-B were both observed at higher frequencies than non-Asian populations (Figure 6C). Accordingly, HLA-C2 shows extremely low frequency in Zhejiang Han, and significantly lower than other populations (Figure 6C, Supplementary Table 5). Moreover, A3/11, specifically encoded by A*11:01, was observed at higher frequency in Zhejiang Han than any other populations. We identified 166 HLA haplotypes (Supplementary Table 4). Most of the observed haplotypes (77.3%) encode more than two KIR ligands, including A*11:01, which was detected in five of the ten most frequent haplotypes (Figure 6D). These findings are similar to those recently reported for Chinese Southern Han from Shenzhen (36).
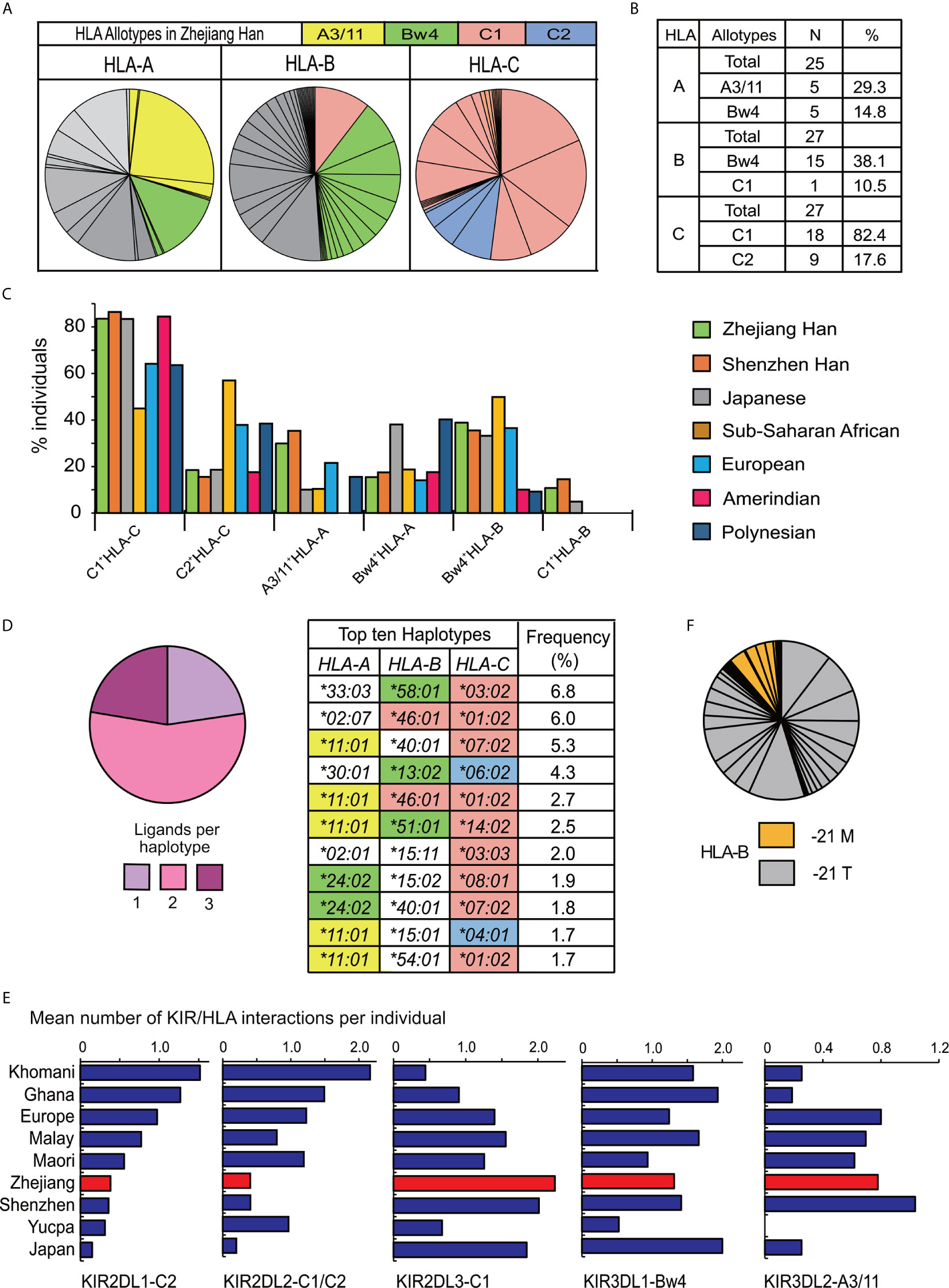
Figure 6 The distribution of HLA in Zhejiang Han. (A) Shown are the frequency spectra of HLA-A, -B, and -C allotypes from 176 Zhejiang Han individuals. Each pie segment represents a distinct allotype (HLA allotype frequencies are given in Supplementary Table 4), colored segments signify the allotypes that are KIR ligands: Yellow—A3/11 (HLA-A3 and -A11); Green—Bw4; Red—C1; Blue—C2; Gray—allotypes that are not KIR ligands. (B) Shown are the numbers of distinct allotypes observed and the combined frequencies of A3/11, Bw4, C1, or C2 allotypes for each HLA class I gene. (C) Shown are the distributions of KIR ligands across seven representative populations. The allotype are shown at the bottom. The populations are listed at the right: Chinese Southern Han from Shenzhen, Japanese, South American Yucpa, European, Sub-Saharan African, and Oceania Polynesian (36, 45, 49, 50, 62, 65). (D) (Left) Shown are the combined frequencies in Zhejiang Han of HLA class I haplotypes encoding one (lilac), two (pink) or three (purple) KIR ligands. (Right) the ten most frequent HLA class I haplotypes observed, colored shading indicates HLA class I alleles that encode KIR ligands, as described in panel (A). (E) Shown is the mean number of distinct interactions per individual observed for each of the inhibitory KIR that interact with polymorphic HLA class I. From left to right: KIR2DL1 interactions with C2+HLA-C [or with C1+HLA-C for KIR2DL1*022 allotype (20)] KIR2DL2 with C1+HLA-B or C, or with C2+HLA-C; KIR2DL3 with C1+HLA-B or C; KIR3DL1 with Bw4+HLA-A or B; KIR3DL2 with HLA-A3/11. The values are shown for nine representative populations: Southern Africa Khomani (34); sub-Saharan Africa Ghanaian (49); European (65); Malay (45); Māori (62); Zhejiang Han; Shenzhen Han (36); Amerindian Yucpa (50); Japanese (37). (F) Shown is the frequency of HLA-B alleles encoding methionine (orange) or threonine (gray) at position -21 in the leader peptide. Each pie segment represents a distinct HLA-B allotype (HLA allotype frequencies are given in Supplementary Table 4).
The high frequency of the KIR-A haplotype, which encodes four inhibitory receptors able to interact with HLA class I ligands (KIR2DL3, KIR2DL1, KIR3DL1 and KIR3DL2), indicates inhibitory KIR-HLA interactions predominate in Zhejiang Han. Here, KIR2DL1*00302, KIR2DL3*00101, KIR3DL1*01502 and KIR3DL2*00201 are the major alleles carried by the KIR-A haplotype, and all the allotypes they encode have strong interactions with their HLA ligands compared to other allotypes (20, 37, 41, 73, 74). Because of the high frequency of KIR-A, Zhejiang Han are rich in KIR-AA genotypes, and each of these individuals therefore has eight strong inhibitory KIR allotypes that can interact with HLA. The mean number of inhibitory KIR-HLA pairs per individual was analyzed, revealing that the mean interaction of KIR2DL3-C1+HLA per individual (2.2) is higher than other populations (Figure 6E). This high frequency is caused by the high frequencies of KIR2DL3 and C1+HLA-C, and the extra C1+HLA-B allotype present (Figure 6A). By contrast, due to the low frequency of the C2+HLA-C allotypes, the frequency of interactions with C2+HLA is low in Zhejiang Han (Figure 6E). In these regards the Zhejiang Han also show very similar repertoires to the Shenzhen Chinese Southern Han (36). Further diversifying NK cell interactions with polymorphic HLA, conserved HLA-E can present peptides derived from HLA-A, C and some B allotypes. The peptides are derived from allotypes possessing methionine at the second residue of the leader sequence (-21M). HLA-E is recognized by inhibitory CD94:NKG2A expressed by NK cell and T cell subsets (75, 76). In Zhejiang Han, the total frequency of HLA-B alleles encoding -21M is 11.6% (Figure 6F). Among them, only HLA-B*38 also encodes a Bw4 ligand for KIR and is the most frequent at 3.7%.
Discussion
Specific genetic variants of KIR, HLA, and their combinations are associated with susceptibility and progression of multiple immune-mediated diseases, including viral infections, autoimmunity, cancer and reproductive disorders (10, 11, 13). Critical to understanding these disease mechanisms are systematic analyses of KIR and HLA allotype diversity across defined human populations. In this study, we used a targeted sequencing method to comprehensively characterize all KIR and HLA class I alleles at high resolution for the Zhejiang Han, who represent the Chinese Southern Han on the Southeastern coast of China. From 176 healthy individuals we identified 23 KIR gene-content genotypes, 107 KIR alleles, 54 centromeric and 37 telomeric KIR haplotypes, and 11 individuals having evidence for large scale gene rearrangements. We also observed 95 HLA class I allotypes, encoded by 166 distinct HLA-A-B-C haplotypes. We show that the Zhejiang Han are characterized by a predominance of interactions of strongly inhibitory KIR with HLA class I, and particularly KIR2DL3 with C1+HLA, supporting the observation that these inhibitory interactions are significantly enhanced in East Asians (36). Because strongly-inhibiting KIR enable maturation of NK cells with strong effector functions (37, 77), one expected consequence is that NK cell responses are enhanced in East Asians relative to other populations.
As observed in other East Asian populations (39), we identified a high frequency of KIR-A haplotypes in Zhejiang Han, where more than 90% of centromeric and 80% telomeric haplotypes have the KIR-A configuration. Previous whole-genome, as well as analyses focused on the KIR genomic region, have shown that the centromeric KIR-A haplotypes are subject to positive natural selection specifically in East Asians (37, 78, 79). By contrast, other studies continue to identify a balance between KIR-A and KIR-B haplotypes across populations worldwide, likely due to competing influences from infection control and reproduction (27, 80). Another outcome of selection favoring KIR-A in East Asia is the low frequency of KIR-B haplotypes, specifically those that contain KIR2DL5B and KIR2DS3. In Amerindians, a complete loss of KIR2DS3 is observed (50), and this may be tolerated because the common 2DS3*00102 allele does not form a functional receptor (81). In addition to gene content, KIR haplotypes characteristic to human populations are distinguished by the alleles they carry (34, 45, 50, 82). Characteristic alleles of Zhejiang Han KIR-A haplotypes include KIR2DL1*003, KIR3DL1*015, and KIR3DL2*002, which all encode strongly inhibiting KIR allotypes, as defined by their ligand binding strength and signal transduction abilities. By contrast the KIR-B haplotypes carry fewer inhibitory receptors and they tend to be allotypes having weak affinity or signal transduction, including KIR2DL1*00401 and KIR3DL2*00701 (41, 74). Thus, even in heterozygous individuals we may expect the functional signature of the KIR-A encoded receptors to dominate. The influence of KIR-B haplotypes being further diminished by the very low frequency of KIR-B homozygous genotypes in this population.
HLA-C evolved to be the dominant KIR ligand in humans (27). Chinese Han have a high frequency of HLA-B*46:01, which is only common in Asia and formed by recombination between HLA-B*15:01 and HLA-C*01:02 (83). HLA-B*46:01 encodes an exceptional HLA-B allotype that can react with KIR2DL2/3, due to possession of a C1 ligand (26), and has risen to high frequency in this geographic region through admixture and natural selection (36, 84). Candidates for this selection include endemic bacterial or viral infections (10, 29, 85–89). An alternative explanation for selection acting on B*46 is that it provides a KIR ligand, whereas B*15:01, from which B*46:01 is derived, does not carry a KIR ligand (36). Co-evolution of KIR with HLA has been driven under natural selection, leading to frequency variation across populations for specific KIR–HLA interactions (49, 50, 90). Our analysis identified many similarities in immunogenetic diversity between the Zhejiang Han and the closely related Shenzhen Han, from Southern China (36). Distinguishing the Zhejiang Han is a high frequency of KIR-A and C1+HLA-C, which is a genetic combination associated with preeclampsia (28). This serious multisystem disorder of pregnancy is most frequent in women homozygous both for KIR-A and C1+HLA-C, if the fetus carries C2+HLA-C (28). However, there is a substantially lower incidence of preeclampsia in China compared with western countries (91–93), which is likely due to the lower frequency C2+ ligands. Moreover, HLA-G expression by extravillous trophoblasts, which promotes vascular remodeling, may help ensure successful placentation thus preventing preeclampsia (94). HLA-G is a ligand for KIR2DL4 (67), and in Zhejiang Han the majority of KIR2DL4 alleles express a functional allotype.
Uneven exchange during recombination events has been a frequent occurrence during KIR gene evolution, creating or amplifying structural diversity of the locus (25, 95). Although our previous analysis of an independent cohort from Zhejiang Han identified similar frequencies of individual KIR genes (60), it was not previously possible to detect any additional structural diversity. In the present analysis of Zhejiang Han, which accounted for gene copy number in addition to gene presence, haplotypes carrying a KIR3DP1-2DL4-3DL1/S1 insertion were observed in six individuals. Acquisition of full sequence data also permitted inference of the allele composition of these haplotypes. Similar haplotypes to those characterized here have been identified in European, Oceanian and other Asian populations, but are very rare in Africans (17). We identified a unique contracted haplotype lacking the telomeric segment, retaining only KIR2DL4 as the 3’ terminal gene. The capture probes we used can tolerate up to 20% sequence divergence (33) and their high density in the target region would imply that we did not miss some highly divergent sequences from this individual. That the allele is KIR2DL4*00102, indicates that this contracted haplotype was formed from the most frequent KIR-A haplotype observed in the Zhejiang Han. As these recombination events most likely occur during meiosis, it remains unlikely that we will observe the reciprocal haplotype formed during that recombination event. That the more frequent structurally divergent haplotypes we observed are all characterized by recombination in approximately similar locations to each other, therefore suggests similar mechanisms or additional recombination hotspots abound in the KIR locus.
In conclusion, characterizing combinatorial diversity of KIR and HLA at high resolution in discrete populations is an important step toward understanding the roles of their polymorphism in disease and in human evolution. The mean number and diversity of KIR and HLA interactions in Zhejiang Han is similar to that of other East Asians, yet highly distinct from African, European and Oceanic populations. These findings will be important for analyzing diseases with susceptibilities that vary across populations and will form a baseline for individualizing immune-based therapies, including transplantation, in Southeastern China.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by Blood Center of Zhejiang Province. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
ST and FZ designed experiments. ST and JW performed lab experiments. YH, KK, PN, and ST analyzed data. PN provided the technology and probes. ST, PN and FZ wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by the Science Research Foundation of Zhejiang Province (LY18H080002) and Science Research Foundation of Zhejiang Healthy Bureau (2018RC003, 2019RC031). PN was supported by National Institutes of Health of the USA (R56 AI151549 and R01 AI128775).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the participants of this study for their generous donation of DNA to facilitate genetic research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.640334/full#supplementary-material
Supplementary Table 3 | Distribution of (A) centromeric and (B) telomeric KIR haplotypes in 176 Zhejiang Han individuals. KIR A haplotypes are shaded red and KIR B haplotypes shaded blue.
Supplementary Table 4 | Distribution of (A) HLA-A, -B, -C alleles and (B) HLA-A-B-C haplotypes in 176 Zhejiang Han individuals. KIR ligands are colored according to Figure 6A, and those expressing -21M are indicated with orange shading.
References
1. Ali A, Gyurova IE, Waggoner SN. Mutually Assured Destruction: The Cold War Between Viruses and Natural Killer Cells. Curr Opin Virol (2019) 34:130–9. doi: 10.1016/j.coviro.2019.02.005
2. Jost S, Altfeld M. Control of Human Viral Infections by Natural Killer Cells. Annu Rev Immunol (2013) 31:163–94. doi: 10.1146/annurev-immunol-032712-100001
3. Lam VC, Lanier LL. NK Cells in Host Responses to Viral Infections. Curr Opin Immunol (2017) 44:43–51. doi: 10.1016/j.coi.2016.11.003
4. Malmberg KJ, Carlsten M, Bjorklund A, Sohlberg E, Bryceson YT, Ljunggren HG. Natural Killer Cell-Mediated Immunosurveillance of Human Cancer. Semin Immunol (2017) 31:20–9. doi: 10.1016/j.smim.2017.08.002
5. Crespo  C, van der Zwan A, Ramalho-Santos J, Strominger JL, Tilburgs T. Cytotoxic Potential of Decidual NK Cells and CD8+ T Cells Awakened by Infections. J Reprod Immunol (2017) 119:85–90. doi: 10.1016/j.jri.2016.08.001
6. Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling Natural Killer Cell Responses: Integration of Signals for Activation and Inhibition. Annu Rev Immunol (2013) 31:227–58. doi: 10.1146/annurev-immunol-020711-075005
7. Trowsdale J. Genetic and Functional Relationships Between MHC and NK Receptor Genes. Immunity (2001) 15:363–74. doi: 10.1016/S1074-7613(01)00197-2
8. Parham P, Moffett A. Variable NK Cell Receptors and Their MHC Class I Ligands in Immunity, Reproduction and Human Evolution. Nat Rev Immunol (2013) 13(2):133–44. doi: 10.1038/nri3370
9. Moretta L, Moretta A. Unravelling Natural Killer Cell Function: Triggering and Inhibitory Human NK Receptors. EMBO J (2004) 233:255–9. doi: 10.1038/sj.emboj.7600019
10. Bashirova AA, Martin MP, McVicar DW, Carrington M. The Killer Immunoglobulin-Like Receptor Gene Cluster: Tuning the Genome for Defense. Annu Rev Genomics Hum Genet (2006) 7:277–300. doi: 10.1146/annurev.genom.7.080505.115726
11. Beziat V, Hilton HG, Norman PJ, Traherne JA. Deciphering the Killer-Cell Immunoglobulin-Like Receptor System At Super-Resolution for Natural Killer and T-cell Biology. Immunology (2017) 150(3):248–64. doi: 10.1111/imm.12684
12. Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK Cell Education by Inhibitory Receptors for MHC Class I. Immunity (2006) 25(2):331–42. doi: 10.1016/j.immuni.2006.06.013
13. Boudreau JE, Hsu KC. Natural Killer Cell Education in Human Health and Disease. Curr Opin Immunol (2018) 50:102–11. doi: 10.1016/j.coi.2017.11.003
14. Goodridge JP, Jacobs B, Saetersmoen ML, Clement D, Hammer Q, Clancy T, et al. Remodeling of Secretory Lysosomes During Education Tunes Functional Potential in NK Cells. Nat Commun (2019) 10(1):514. doi: 10.1038/s41467-019-08384-x
15. Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the Organization and Sequences of Human KIR/ILT Gene Families. Proc Natl Acad Sci USA (2000) 97(9):4778–83. doi: 10.1073/pnas.080588597
16. Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different Patterns of Evolution in the Centromeric and Telomeric Regions of Group A and B Haplotypes of the Human Killer Cell Ig-like Receptor Locus. PloS One (2010) 5:e15115. doi: 10.1371/journal.pone.0015115.g001
17. Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, et al. Meiotic Recombination Generates Rich Diversity in NK Cell Receptor Genes, Alleles, and Haplotypes. Genome Res (2009) 19(5):757–69. doi: 10.1101/gr.085738.108
18. Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, et al. Mechanisms of Copy Number Variation and Hybrid Gene Formation in the KIR Immune Gene Complex. Hum Mol Genet (2010) 19(5):737–51. doi: 10.1093/hmg/ddp538
19. Saunders PM, Vivian JP, O’Connor GM, Sullivan LC, Pymm P, Rossjohn J, et al. A Bird’s Eye View of NK Cell Receptor Interactions With Their MHC Class I Ligands. Immunol Rev (2015) 267(1):148–66. doi: 10.1111/imr.12319
20. Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, et al. Polymorphic HLA-C Receptors Balance the Functional Characteristics of KIR Haplotypes. J Immunol (2015) 195(7):3160–70. doi: 10.4049/jimmunol.1501358
21. Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL, et al. Conserved and Variable Residues Within the Bw4 Motif of HLA-B Make Separable Contributions to Recognition by the NKB1 Killer Cell-Inhibitory Receptor. J Immunol (1997) 158(11):5237–41.
22. Saunders PM, MacLachlan BJ, Widjaja J, Wong SC, Oates CVL, Rossjohn J, et al. The Role of the HLA Class I α2 Helix in Determining Ligand Hierarchy for the Killer Cell Ig-like Receptor 3dl1. J Immunol (2021) 206(4):849–60. doi: 10.4049/jimmunol.2001109
23. Manser AR, Weinhold S, Uhrberg M. Human KIR Repertoires Shaped by Genetic Diversity and Evolution. Immunol Rev (2015) 267:178–96. doi: 10.1111/imr.12316
24. Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, et al. KIR2DS4 is a Product of Gene Conversion With KIR3DL2 That Introduced Specificity for HLA-A*11 While Diminishing Avidity for HLA-C. J Exp Med (2009) 206(11):2557–72. doi: 10.1084/jem.20091010
25. Middleton D, Gonzelez F. The Extensive Polymorphism of KIR Genes. Immunology (2010) 129(1):8–19. doi: 10.1111/j.1365-2567.2009.03208.x
26. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic Polymorphism At Two Positions Distal to the Ligand-Binding Site Makes KIR2DL2 a Stronger Receptor for HLA-C Than KIR2DL3. J Immunol (2008) 180(6):3969–79. doi: 10.4049/jimmunol.180.6.3969
27. Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-Specific Evolution of Killer Cell Immunoglobulin-Like Receptor Recognition of Major Histocompatibility Complex Class I Molecules. Philos Trans R Soc Lond B Biol Sci (2012) 367(1590):800–11. doi: 10.1098/rstb.2011.0266
28. Hiby SE, Walker JJ, O’Shaughnessy K M, Redman CW, Carrington M, Trowsdale J, et al. Combinations of Maternal KIR and Fetal HLA-C Genes Influence the Risk of Preeclampsia and Reproductive Success. J Exp Med (2004) 200(8):957–65. doi: 10.1084/jem.20041214
29. Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK Cell Inhibitory Receptor Genes in Resolving Hepatitis C Virus Infection. Science (2004) 305(5685):872–4. doi: 10.1126/science.1097670
30. Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J, et al. A Kir B Centromeric Region Present in Africans But Not Europeans Protects Pregnant Women From Pre-Eclampsia. Proc Natl Acad Sci (2015) 112(3):845–50. doi: 10.1073/pnas.1413453112
31. Cooley S, Parham P, Miller JS. Strategies to Activate NK Cells to Prevent Relapse and Induce Remission Following Hematopoietic Stem Cell Transplantation. Blood (2018) 131(10):1053–62. doi: 10.1182/blood-2017-08-752170
32. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA Database: Allele Variant Databases. Nucleic Acids Res (2015) 43(Database issue):D423–31. doi: 10.1093/nar/gku1161
33. Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, et al. Defining KIR and HLA Class I Genotypes At Highest Resolution Via High-Throughput Sequencing. Am J Hum Genet (2016) 99(2):375–91. doi: 10.1016/j.ajhg.2016.06.023
34. Nemat-Gorgani N, Hilton HG, Henn BM, Lin M, Gignoux CR, Myrick JW, et al. Different Selected Mechanisms Attenuated the Inhibitory Interaction of KIR2DL1 With C2(+) HLA-C in Two Indigenous Human Populations in Southern Africa. J Immunol (2018) 200(8):2640–55. doi: 10.4049/jimmunol.1701780
35. Wagner I, Schefzyk D, Pruschke J, Schöfl G, Schöne B, Gruber N, et al. Allele-Level KIR Genotyping of More Than a Million Samples: Workflow, Algorithm, and Observations. Front Immunol (2018) 9:2843. doi: 10.3389/fimmu.2018.02843
36. Deng Z, Zhen J, Harrison GF, Zhang G, Chen R, Sun G, et al. Adaptive Admixture of HLA Class I Allotypes Enhanced Genetically Determined Strength of Natural Killer Cells in East Asians. Mol Biol Evol (2021). doi: 10.1093/molbev/msab053
37. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR Polymorphisms in Natural Killer Cell Repertoire Selection and Modulation of Effector Function. J Exp Med (2006) 203(3):633–45. doi: 10.1084/jem.20051884
38. Stephens M, Donnelly P. A Comparison of Bayesian Methods for Haplotype Reconstruction From_ Population Genotype Data. Am J Hum Genet (2003) 73:1162–16. doi: 10.1086/379378
39. Gonzalez-Galarza FF, McCabe A, Santos E, Jones J, Takeshita L, Ortega-Rivera ND, et al. Allele Frequency Net Database (AFND) 2020 Update: Gold-Standard Data Classification, Open Access Genotype Data and New Query Tools. Nucleic Acids Res (2020) 48(D1):D783–8. doi: 10.1093/nar/gkz1029
40. Excoffier L, Lischer HE. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses Under Linux and Windows. Mol Ecol Resour (2010) 10(3):564–7. doi: 10.1111/j.1755-0998.2010.02847.x
41. Augusto DG, O’Connor GM, Lobo-Alves SC, Bass S, Martin MP, Carrington M, et al. Pemphigus is Associated With KIR3DL2 Expression Levels and Provides Evidence That KIR3DL2 may Bind HLA-A3 and A11 In Vivo. Eur J Immunol (2015) 45(7):2052–60. doi: 10.1002/eji.201445324
42. Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is Peptide-Specific. Eur J Immunol (2004) 34(6):1673–9. doi: 10.1002/eji.200425089
43. Whang DH, Park H, Yoon JA, Park MH. Haplotype Analysis of Killer Cell Immunoglobulin-Like Receptor Genes in 77 Korean Families. Hum Immunol (2005) 66(2):146–54. doi: 10.1016/j.humimm.2004.10.013
44. Wu GQ, Zhao YM, Lai XY, Yang KL, Zhu FM, Zhang W, et al. Distribution of Killer-Cell Immunoglobulin-Like Receptor Genes in Eastern Mainland Chinese Han and Taiwanese Han Populations. Tissue Antigens (2009) 74(6):499–507. doi: 10.1111/j.1399-0039.2009.01366.x
45. Tao S, Kichula KM, Harrison GF, Farias TDJ, Palmer WH, Leaton LA, et al. The Combinatorial Diversity of KIR and HLA Class I Allotypes in Peninsular Malaysia. Immunology (2021) 162(4):389–404. doi: 10.1111/imm.13289
46. Chaisri S, Kitcharoen K, Romphruk AV, Romphruk A, Witt CS, Leelayuwat C. Polymorphisms of Killer Immunoglobulin-Like Receptors (Kirs) and HLA Ligands in Northeastern Thais. Immunogenetics (2013) 65(9):645–53. doi: 10.1007/s00251-013-0716-7
47. Hidajat M, Selleslag D, Van Hoof A, Van Droogenbroeck J, Billiet J, Criel A. Killer Immunoglobulin-Like Receptors (Kirs) Genotypes in a Belgian Population. Blood (2004) 104(11):3852. doi: 10.1182/blood.V104.11.3852.3852
48. Arnheim L, Dillner J, Sanjeevi CB. A Population-Based Cohort Study of KIR Genes and Genotypes in Relation to Cervical Intraepithelial Neoplasia. Tissue Antigens (2005) 65(3):252–9. doi: 10.1111/j.1399-0039.2005.00359.x
49. Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, Pando MJ, et al. Co-Evolution of Human Leukocyte Antigen (HLA) Class I Ligands With Killer-Cell Immunoglobulin-Like Receptors (KIR) in a Genetically Diverse Population of sub-Saharan Africans. PloS Genet (2013) 9(10):e1003938. doi: 10.1371/journal.pgen.1003938
50. Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, et al. Co-Evolution of KIR2DL3 With HLA-C in a Human Population Retaining Minimal Essential Diversity of KIR and HLA Class I Ligands. Proc Natl Acad Sci USA (2009) 106(44):18692–7. doi: 10.1073/pnas.0906051106
51. Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human Diversity in Killer Cell Inhibitory Receptor Genes. Immunity (1997) 9:753–63. doi: 10.1016/S1074-7613(00)80394-5
52. Zhu BF, Wang HD, Shen CM, Deng YJ, Yang G, Wu QJ, et al. Killer Cell Immunoglobulin-Like Receptor Gene Diversity in the Tibetan Ethnic Minority Group of China. Hum Immunol (2010) 71(11):1116–23. doi: 10.1016/j.humimm.2010.07.010
53. Lee YC, Chan SH, Ren EC. Asian Population Frequencies and Haplotype Distribution of Killer Cell Immunoglobulin-Like Receptor (KIR) Genes Among Chinese, Malay, and Indian in Singapore. Immunogenetics (2008) 60(11):645–54. doi: 10.1007/s00251-008-0321-3
54. Augusto DG, Amorim LM, Farias TD, Petzl-Erler ML. KIR and HLA Genotyping of Japanese Descendants From Curitiba, a City of Predominantly European Ancestry From Southern Brazil. Hum Immunol (2016) 77(4):336–7. doi: 10.1016/j.humimm.2016.01.011
55. Hiby SE, Ashrafian-Bonab M, Farrell L, Single RM, Balloux F, Carrington M, et al. Distribution of Killer Cell Immunoglobulin-Like Receptors (KIR) and Their HLA-C Ligands in Two Iranian Populations. Immunogenetics (2010) 62(2):65–73. doi: 10.1007/s00251-009-0408-5
56. Seymour LA, Nourse JP, Crooks P, Wockner L, Bird R, Tran H, et al. The Presence of KIR2DS5 Confers Protection Against Adult Immune Thrombocytopenia. Tissue Antigens (2014) 83(3):154–60. doi: 10.1111/tan.12295
57. Nakimuli A, Chazara O, Farrell L, Hiby SE, Tukwasibwe S, Knee O, et al. Killer Cell Immunoglobulin-Like Receptor (KIR) Genes and Their HLA-C Ligands in a Ugandan Population. Immunogenetics (2013) 65(11):765–75. doi: 10.1007/s00251-013-0724-7
58. Velickovic M, Velickovic Z, Dunckley H. Diversity of Killer Cell Immunoglobulin-Like Receptor Genes in Pacific Islands Populations. Immunogenetics (2006) 58(7):523–32. doi: 10.1007/s00251-006-0124-3
59. Velickovic M, Velickovic Z, Panigoro R, Dunckley H. Diversity of Killer Cell Immunoglobulin-Like Receptor Genes in Indonesian Populations of Java, Kalimantan, Timor and Irian Jaya. Tissue Antigens (2009) 73(1):9–16. doi: 10.1111/j.1399-0039.2008.01149.x
60. Jiang K, Zhu FM, Lv QF, Yan LX. Distribution of Killer Cell Immunoglobulin-Like Receptor Genes in the Chinese Han Population. Tissue Antigens (2005) 65(6):556–63. doi: 10.1111/j.1399-0039.2005.00412.x
61. Bruijnesteijn J, de Groot NG, Bontrop RE. The Genetic Mechanisms Driving Diversification of the KIR Gene Cluster in Primates. Front Immunol (2020) 11:582804. doi: 10.3389/fimmu.2020.582804
62. Nemat-Gorgani N, Edinur HA, Hollenbach JA, Traherne JA, Dunn PP, Chambers GK, et al. KIR Diversity in Maori and Polynesians: Populations in Which HLA-B is Not a Significant KIR Ligand. Immunogenetics (2014) 66(11):597–611. doi: 10.1007/s00251-014-0794-1
63. Boudreau JE, Mulrooney TJ, Le Luduec JB, Barker E, Hsu KC. KIR3DL1 and HLA-B Density and Binding Calibrate NK Education and Response to HIV. J Immunol (2016) 196(8):3398–410. doi: 10.4049/jimmunol.1502469
64. Saunders PM, Pymm P, Pietra G, Hughes VA, Hitchen C, O’Connor GM, et al. Killer Cell Immunoglobulin-Like Receptor 3DL1 Polymorphism Defines Distinct Hierarchies of HLA Class I Recognition. J Exp Med (2016) 213(5):791–807. doi: 10.1084/jem.20152023
65. Amorim LM, Augusto DG, Nemat-Gorgani N, Montero-Martin G, Marin W, Shams H, et al. High-Resolution Characterization of KIR Genes in a Large North American Cohort Reveals Novel Details of Structural and Sequence Diversity. Front Immunol (2021) 674778. doi: 10.3389/fimmu.2021.674778
66. Tao SD, He YM, Ying YL, He J, Zhu FM, Lv HJ. KIR3DL1 Genetic Diversity and Phenotypic Variation in the Chinese Han Population. Genes Immun (2014) 15(1):8–15. doi: 10.1038/gene.2013.55
67. Rajagopalan S, Long EO. A Human Histocompatibility Leukocyte Antigen (HLA)-G-specific Receptor Expressed on All Natural Killer Cells. J Exp Med (1999) 189(7):1093–100. doi: 10.1084/jem.189.7.1093
68. Kikuchi-Maki A, Catina TL, Campbell KS. Cutting Edge: KIR2DL4 Transduces Signals Into Human NK Cells Through Association With the Fc Receptor Gamma Protein. J Immunol (2005) 174(7):3859–63. doi: 10.4049/jimmunol.174.7.3859
69. Witt CS, Martin A, Christiansen FT. Detection of KIR2DL4 Alleles by Sequencing and SSCP Reveals a Common Allele With a Shortened Cytoplasmic Tail. Tissue Antigens (2000) 56(3):248–57. doi: 10.1034/j.1399-0039.2000.560307.x
70. Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting Edge: Expansion of the KIR Locus by Unequal Crossing Over. J Immunol (2003) 171(5):2192–5. doi: 10.4049/jimmunol.171.5.2192
71. Gómez-Lozano N, Estefanía E, Williams F, Halfpenny I, Middleton D, Solís R, et al. The Silent KIR3DP1 Gene (CD158c) is Transcribed and Might Encode a Secreted Receptor in a Minority of Humans, in Whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 Genes are Duplicated. Eur J Immunol (2005) 35(1):16–24. doi: 10.1002/eji.200425493
72. Cisneros E, Moraru M, Gómez-Lozano N, Muntasell A, López-Botet M, Vilches C. Haplotype-Based Analysis of KIR-Gene Profiles in a South European Population-Distribution of Standard and Variant Haplotypes, and Identification of Novel Recombinant Structures. Front Immunol (2020) 11:440. doi: 10.3389/fimmu.2020.00440
73. Hilton HG, Vago L, Older Aguilar AM, Moesta AK, Graef T, Abi-Rached L, et al. Mutation At Positively Selected Positions in the Binding Site for HLA-C Shows That KIR2DL1 is a More Refined But Less Adaptable NK Cell Receptor Than KIR2DL3. J Immunol (2012) 189(3):1418–30. doi: 10.4049/jimmunol.1100431
74. Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, et al. Significant Functional Heterogeneity Among KIR2DL1 Alleles and a Pivotal Role of Arginine 245. Blood (2009) 114(25):5182–90. doi: 10.1182/blood-2009-07-231977
75. Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, et al. HLA-E Binds to Natural Killer Cell Receptors CD94/NKG2A, B, and C. Nature (1998) 391(6669):795–9. doi: 10.1038/35869
76. Pereira BI, Devine OP, Vukmanovic-Stejic M, Chambers ES, Subramanian P, Patel N, et al. Senescent Cells Evade Immune Clearance Via HLA-E-mediated NK and CD8(+) T Cell Inhibition. Nat Commun (2019) 10(1):2387. doi: 10.1038/s41467-019-10335-5
77. Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, et al. HLA Alleles Determine Differences in Human Natural Killer Cell Responsiveness and Potency. Proc Natl Acad Sci USA (2008) 105(8):3053–8. doi: 10.1073/pnas.0712229105
78. Augusto DG, Norman PJ, Dandekar R, Hollenbach JA. Fluctuating and Geographically Specific Selection Characterize Rapid Evolution of the Human Kir Region. Front Immunol (2019) 10:989. doi: 10.3389/fimmu.2019.00989
79. Deng Z, Zhao J, Cai S, Qi Y, Yu Q, Martin MP, et al. Natural Killer Cells Offer Differential Protection From Leukemia in Chinese Southern Han. Front Immunol (2019) 10:1646. doi: 10.3389/fimmu.2019.01646
80. Hollenbach JA, Augusto DG, Alaez C, Bubnova L, Fae I, Fischer G, et al. 16(Th) IHIW: Population Global Distribution of Killer Immunoglobulin-Like Receptor (KIR) and Ligands. Int J Immunogenet (2013) 40(1):39–45. doi: 10.1111/iji.12028
81. VandenBussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically Reduced Surface Expression of NK Cell Receptor KIR2DS3 is Attributed to Multiple Residues Throughout the Molecule. Genes Immun (2009) 10(2):162–73. doi: 10.1038/gene.2008.91
82. Alicata C, Ashouri E, Nemat-Gorgani N, Guethlein LA, Marin WM, Tao S, et al. Kir Variation in Iranians Combines High Haplotype and Allotype Diversity With an Abundance of Functional Inhibitory Receptors. Front Immunol (2020) 11:556. doi: 10.3389/fimmu.2020.00556
83. Barber LD, Percival L, Valiante NM, Chen L, Lee C, Gumperz JE, et al. The Inter-Locus Recombinant HLA-B*4601 has High Selectivity in Peptide Binding and Functions Characteristic of HLA-C. J Exp Med (1996) 184(2):735–40. doi: 10.1084/jem.184.2.735
84. Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-Specific Evolution and Adaptation Led to Major Qualitative Differences in the Variable Receptors of Human and Chimpanzee Natural Killer Cells. PloS Genet (2010) 6(11). doi: 10.1371/journal.pgen.1001192
85. Thons C, Senff T, Hydes TJ, Manser AR, Heinemann FM, Heinold A, et al. Hla-Bw4 80(T) and Multiple HLA-Bw4 Copies Combined With KIR3DL1 Associate With Spontaneous Clearance of HCV Infection in People Who Inject Drugs. J Hepatol (2017) 67(3):462–70. doi: 10.1016/j.jhep.2017.03.040
86. Petitdemange C, Becquart P, Wauquier N, Beziat V, Debre P, Leroy EM, et al. Unconventional Repertoire Profile is Imprinted During Acute Chikungunya Infection for Natural Killer Cells Polarization Toward Cytotoxicity. PloS Pathog (2011) 7(9):e1002268. doi: 10.1371/journal.ppat.1002268
87. Townsley E, O’Connor G, Cosgrove C, Woda M, Co M, Thomas SJ, et al. Interaction of a Dengue Virus NS1-derived Peptide With the Inhibitory Receptor KIR3DL1 on Natural Killer Cells. Clin Exp Immunol (2016) 183(3):419–30. doi: 10.1111/cei.12722
88. Naiyer MM, Cassidy SA, Magri A, Cowton V, Chen K, Mansour S, et al. KIR2DS2 Recognizes Conserved Peptides Derived From Viral Helicases in the Context of HLA-C. Sci Immunol (2017) 2(15):eaal5296. doi: 10.1126/sciimmunol.aal5296
89. Hilton HG, McMurtrey CP, Han AS, Djaoud Z, Guethlein LA, Blokhuis JH, et al. The Intergenic Recombinant HLA-B∗46:01 has a Distinctive Peptidome That Includes Kir2dl3 Ligands. Cell Rep (2017) 19(7):1394–405. doi: 10.1016/j.celrep.2017.04.059
90. Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, et al. Global Diversity and Evidence for Coevolution of KIR and HLA. Nat Genet (2007) 39(9):1114–9. doi: 10.1038/ng2077
91. Wang Q, Zhang H, Liang Q, Knibbs LD, Ren M, Li C, et al. Effects of Prenatal Exposure to Air Pollution on Preeclampsia in Shenzhen, China. Environ Pollution (2018) 237:18–27. doi: 10.1016/j.envpol.2018.02.010
92. Roberts CL, Ford JB, Algert CS, Antonsen S, Chalmers J, Cnattingius S, et al. Population-Based Trends in Pregnancy Hypertension and Pre-Eclampsia: An International Comparative Study. BMJ Open (2011) 1(1):e000101. doi: 10.1136/bmjopen-2011-000101
93. Ananth CV, Keyes KM, Wapner RJ. Pre-Eclampsia Rates in the United States, 1980-2010: Age-Period-Cohort Analysis. Bmj (2013) 347(nov07 15):f6564. doi: 10.1136/bmj.f6564
94. Pazmany L, Mandelboim O, Vales-Gomez M, Davis DM, Becker TC, Reyburn HT, et al. Human Leucocyte Antigen-G and its Recognition by Natural Killer Cells. J Reprod Immunol (1999) 43(2):127–37. doi: 10.1016/s0165-0378(99)00028-5
Keywords: KIR, HLA, KIR/HLA interaction, Zhejiang Han population, NK cells
Citation: Tao S, He Y, Kichula KM, Wang J, He J, Norman PJ and Zhu F (2021) High-Resolution Analysis Identifies High Frequency of KIR-A Haplotypes and Inhibitory Interactions of KIR With HLA Class I in Zhejiang Han. Front. Immunol. 12:640334. doi: 10.3389/fimmu.2021.640334
Received: 11 December 2020; Accepted: 07 April 2021;
Published: 30 April 2021.
Edited by:
Eleanor Riley, University of Edinburgh, United KingdomReviewed by:
Kouyuki Hirayasu, Kanazawa University, JapanRoberto Biassoni, Giannina Gaslini Institute (IRCCS), Italy
Copyright © 2021 Tao, He, Kichula, Wang, He, Norman and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Faming Zhu, emZtMDBAaG90bWFpbC5jb20=; Paul J. Norman, cGF1bC5ub3JtYW5AY3VhbnNjaHV0ei5lZHU=
 Sudan Tao
Sudan Tao Yanmin He
Yanmin He Katherine M. Kichula2
Katherine M. Kichula2 Paul J. Norman
Paul J. Norman Faming Zhu
Faming Zhu