- 1Liaoning Key Laboratory of Marine Animal Immunology, Dalian Ocean University, Dalian, China
- 2Functional Laboratory of Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Liaoning Key Laboratory of Marine Animal Immunology and Disease Control, Dalian Ocean University, Dalian, China
- 4Southern Laboratory of Ocean Science and Engineering (Guangdong, Zhuhai), Zhuhai, China
- 5Dalian Key Laboratory of Aquatic Animal Disease Prevention and Control, Dalian Ocean University, Dalian, China
Interferon (IFN) system is considered as the first defense line against viral infection, and it has been extensively studied in vertebrates from fish to mammals. In invertebrates, Vagos from arthropod and IFN-like protein (CgIFNLP) from Crassostrea gigas appeared to function as IFN-like antiviral cytokines. In the present study, the CgIFNLP protein in hemocytes was observed to increase after Poly (I:C) stimulation. After CgIFNLP was knocked down by RNAi, the mRNA expression of IFN-stimulated genes (CgISGs) was significantly inhibited. Both cyclic GMP-AMP synthase (CgcGAS) and stimulator of interferon gene (CgSTING) identified from oyster were able to recognize the double-stranded nucleic acid [Poly (I:C) and dsDNA] and expressed at high level after Poly (I:C) stimulation. The expression of CgIFNLP and interferon regulatory factors (CgIRF1/8) and the nuclear translocation of CgIRF8 were all suppressed in CgcGAS-RNAi or CgSTING-RNAi oysters after Poly (I:C) stimulation. The expression level of CgSTING and TANK binding kinase1 (CgTBK1) did not decrease in CgcGAS-RNAi oysters. After CgSTING was knocked down, the high expression of CgTBK1 induced by Poly (I:C) was prevented significantly. These results indicated that there was a primitive IFN-like antiviral mechanism dependent on the cGAS/STING–TBK1–IRFs regulatory axis in mollusks, which was different from the classic cGAS–STING–TBK1 signal pathway in mammals.
Introduction
Interferon (IFN) system is the vital component of immune response in mammal, and it is recognized as the first line of defense against viral infection. IFNs don’t possess the direct antiviral activity, and they are so named because of the shared property to impede viral replication at numerous stages of the viral lifecycle, which is indeed critical for activating a robust host response against viral infection (1). The released IFNs engage their cognate receptors at the cell surface, then trigger JAK–STAT signaling and ultimately lead to the transcriptional activation of IFN-stimulated genes (ISGs) (2). The proteins encoded by ISGs, such as interferon-induced proteins, myxovirus resistance (Mx), and Viperin, can disrupt the life cycle of virus and reduce the damage caused by virus infections (3).
The signal pathways to mediate the expression of IFNs are relatively conservative, even though the IFNs are principally expressed by different kinds of innate immune cells in mammal (4). The reaction cascade to regulate the production of IFNs can be initiated by the specific binding of pathogen-associated molecular patterns (PAMPs) to pattern recognition receptors (PRRs), such as Toll like receptors (TLRs), retinoic acid-inducible gene I (RIG-I), melanoma differentiation associated protein 5 (MDA5), cyclic GMP-AMP (cGAMP) synthase (cGAS), and nucleotide binding oligomerization domain-like receptors (NLRs) (5–9). For instance, RIG-I/MDA5 specializes in discriminating pathogen-derived RNA (e.g., dsRNA or 5′ppp-RNA) in the cytoplasm, while cGAS recognizes cytosolic DNA in a sequence-independent manner (10, 11). The activated cGAS catalyzes the reaction to generate 2′3′-cGAMP which then activates the stimulator of interferon genes (STING) (12). The STING recruits the kinases TANK binding kinase 1 (TBK1) and Ikappa B Kinase Ɛ (IKKƐ) to phosphorylate IFN regulatory factor IRF-3/7, which translocates into the nucleus to initiate the expression of IFNs (13). The IRF family members are renowned for their involvement in the regulation of IFN system (14). For instance, the IRF-3 and IRF-7 are essential for the regulated expression of IFNs, and IRF-9 is indispensable for gene transcription of ISGs by combining the STAT1 and STAT2 (15).
The IFN mediated antiviral response displays conserved property in fishes and all tetrapods, but not in tunicates or amphioxus (16). It has been suspected that the IFN system derives from earliest jawed vertebrates and evolves from a class II helical cytokine ancestor, along with the IL-10 cytokine family (17–19). Recently, the accumulating pieces of evidence have demonstrated that the nucleic acid-induced antiviral immunity also exists in invertebrates with the similar characteristic of IFN responses in mammals, even there is no identified IFN homologous (20). It is worth noting that Vago appears to function as an IFN-like antiviral cytokine in arthropods, such as DmVago in Drosophila (21), CxVago in Culex (22), and LvVago in Litopenaeus vannamei (23). These Vagos restrict virus infection by the activation of JAK–STAT pathway, indicating the existence of IFN-like system in arthropods. Meanwhile, many elements of IFN signaling pathway were recently discovered in abalone, oyster, and clam, which provided the evidence of functional homolog of IFN in mollusks. For instance, Mx and Viperin were identified in Haliotis discus discus and C. gigas respectively (24, 25), and several IRFs were found to participate in antiviral immune response in Pinctada fucata and C. gigas (26, 27).
The Pacific oyster C. gigas is the dominant farmed oyster species worldwide. In the past decades, the oyster aquaculture suffered from mass mortalities, and the Ostreid herpesvirus (OsHV-1) was considered to be one of the major virus pathogen (28, 29). In our previous works, an IFN-like protein (CgIFNLP) and an IFN receptor-like 3 (CgIFNR-3) were identified from C. gigas (30, 31), and their mRNA expression levels in hemocytes increased significantly after Poly (I:C) stimulation. CgIFNR-3 interacted with CgIFNLP in vitro, and it could activate the expression of human interferon-stimulated response element (ISRE), STAT3, and GAS in the reporter luciferase activity assay (31). In the present study, the cGAS and STING homologs (designated as CgcGAS and CgSTING) were identified from C. gigas with the main objectives (1) to determine their recognition towards double-stranded nucleic acid and the effect on nuclear translocation of CgIRFs, (2) to illuminate their regulatory mechanism for CgIFNLP expression, (3) to examine the expression changes of CgISGs after CgIFNLP was knocked down, and clarify the CgcGAS/CgSTING mediated primitive IFN-like antiviral mechanism in oyster.
Material and Methods
Experimental Animals and Sample Collection
All experiments were performed in accordance with the approval and guidelines of the Ethics Review Committee of Dalian Ocean University. Adult Pacific oysters C. gigas (about 13.0 cm in shell length, average weight of 100 g) collected from a local farm in Dalian, Liaoning Province, China, were cultured in laboratory aquarium tanks with aerated seawater at 15–20°C for one week before processing. Six-week old Kunming mice were provided by the Experimental Animal Center at Dalian Medical University, Dalian, China and raised and handled under pathogen-free conditions.
One hundred and eight oysters were randomly divided into two groups. The oysters in Poly (I:C) stimulation group and control group received individually an injection with 0.1 ml of Poly (I:C) (1 mg/ml, dissolved in seawater) (Sigma-Aldrich, USA) and seawater, respectively. The hemocytes were collected from nine oysters in each group at 0, 3, 6, 12, 24, and 48 h after injection. The hemocytes from three oysters were pooled together as one sample, and there were three replicates for each time point. The hemocytes were harvested by centrifugation at 800 × g, 4°C for 8 min. Meanwhile, the tissues including gonad, adductor muscle, mantle, gills, hemolymphs, labial palps, and hepatopancreas were collected from untreated oysters.
cDNA Cloning and Sequence Analysis
The full-length cDNA fragments of CgcGAS, CgSTING, and CgIRF-8 were obtained by PCR according to the previous report (31). The primers (Table S1, P1–6) were designed according to sequence information (XM_034454330.1, XM_011452302.3, XM_011414344.3) from the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/). BLASTx (http://www.ncbi.nlm.nih.gov/) was used for homology analysis, and ExPASy (http://www.expasy.org/) was employed to predict the amino acid sequences. SMART (http://smart.embl-heidelberg.de/) was used to predict the protein domains. MEGA 6.0 program and Clustal X were used for phylogenetic analysis and multiple sequence alignment, respectively.
RNA Extraction,cDNA Synthesis and Quantitative Real-Time PCR Analysis
Total RNA was isolated from all the tissue and hemocyte samples using Trizol reagent (Thermo Fisher Scientific, USA) and synthesized into cDNA with PrimeScript Reverse Transcriptase following the manufacturer’s introduction (Takara, China). The qRT-PCR was carried out according to the method described previously with the designed primers (Table S1, P7–P26) (32). Briefly, qRT-PCR was performed with the SYBR premix ExTap (RR420, Takara, Japan) on ABI PRISM 7500 Sequence Detection System (Thermo Fisher, USA). The relative expression level was calculated by 2-ΔΔCt method with CgEF (NM_001305313.2) as internal reference. Dissociation curve analysis of amplification products was performed to confirm the specificity of amplification.
Preparation of Recombinant Proteins and Their Polyclonal Antibodies
The ORF of CgcGAS, CgIRF-8, and the CBD (c-di-GMP-binding domain) sequences of CgSTING was amplified using their respective primers (Table S1, P27–P32), and the PCR fragments were cloned into pET-30a, pMAL-c5x and pET-30a expression vector, respectively. The recombinant plasmids were transformed and expressed in Escherichia coli Transetta DE3. The recombinant proteins of CgcGAS (rCgcGAS) and CBD in CgSTING (rCgSTING-CBD) and CgIFNLP (rCgIFNLP) with two His tag and CgIRF-8 (rCgIRF-8) with MBP tag were purified by Ni+ or Maltose affinity chromatography and desalted by extensive dialysis.
The purified proteins of rCgcGAS, rCgIRF-8, and rCgIFNLP were injected into six-weeks female mice to acquire polyclonal antibody as in previous description, respectively (33). Rabbit polyclonal antibody of CgSTING was obtained and purified by Gene Universal (34). The specificity of polyclonal antibodies was examined by Western blot. The hemocyte proteins were extracted from oysters after Poly (I:C) stimulation, separated by SDS-PAGE, and observed by a standard Western blot procedure as described previously (33).
Immunocytochemical Assay
The hemocytes collected from oysters were resuspended in L15 cell culture media (extra addition of 20.2 g/L NaCl, 0.54 g/L KCl, 0.6 g/L CaCl, 3.9 g/L MgCl2, and 1 g/L MgSO4) and deposited on dishes pre-coated with poly-L-lysine, and then incubated at 37°C for 1 h to adhere to the glass slides. The hemocytes on the object slides were fixed with 4% paraformaldehyde, washed with PBS, and permeabilized with 1% Triton X-100 for 5 min. After three times of PBS wash, the hemocytes were blocked with 3% (w/v) fetal bovine serum albumin (BSA) at 37°C for 30 min and incubated with the antibody [anti-CgIRF-8, anti-CgcGAS, 1:500 (v/v) in 3% BSA] at 37°C for 1 h. Then the samples were washed with PBS and incubated with Alexa Fluer 488-labeled goat-anti-mouse antibody (Solarbio life sciences, China) diluted at 1:1,000 (v/v) with 3% BSA at 37°C in the dark for 1 h. The DAPI dihydrochloride (1 mg/ml in PBS; Solarbio Life Sciences, China) was added to stain the nucleus for 5 min followed by three times wash with PBS. Fluorescence was observed using Carl Zeiss Axio Imager A2 microscope (Carl Zeiss, Germany).
PAMP Binding Assay
The PAMP binding activity of rCgcGAS and rCgSTING-CBD was examined by enzyme-linked immunosorbent assay (35) as described previously (36). Briefly, the 96-well microliter plates were coated with dsEGFP (dsDNA) and Poly (I:C) at 4°C for 12 h, respectively. The plate was blocked with 3% BSA and incubated with 100 μl of rCgcGAS or rCgSTING-CBD at 18°C for 3 h. Blank plasmid coded recombination protein rTrx was added in separated wells as a negative control at the same time. After three times of washing with TBST, anti-His tag antibody (ABclonal, USA) and diluted HRP Goat-anti-mouse Ig-alkaline phosphatase conjugate (ABclonal, USA) were added successively and incubated at 37°C for 1 h. After the final wash with TBST, 100 μl of TMB (dihydrochloride) (Solarbio, USA) was added and incubated at room temperature in dark for 15 min. The reaction was stopped by adding 1 M HCl (100 μl/well), and the absorbance at OD450 was measured by Infinite M1000 PRO (Tecan, Switzerland). The dissociation constant (Kd) was calculated using GraphPad Prism 5 with non-linear regression curve fit and a one-site binding model analysis.
RNA Interference
The 3′-terminal cDNA sequences of CgcGAS, CgSTING, CgTBK1, CgIFNLP, and EGFP were amplified by the primers Fi and Ri linked to the T7 promoter (Table S1, P33–42) as templates for the synthesis of dsRNA. The dsRNA was synthesized using T7 polymerase according to the instruction of manufacturer (Takara, China). The dsRNA (100 μg) was injected into adductor muscle of each oyster (nine oysters/group). To strengthen the effect of RNA interference, the second injection was carried out at 12 h after the first injection. After the second injection, the oysters were stimulated with Poly (I:C) (1 mg/ml, 0.1 ml). The hemocytes from three individuals were pooled together as one sample and centrifuged to harvest the hemocytes at 24 h after the second injection. The total RNA was extracted from the hemocytes, and the expression level of target genes was assessed by qRT-PCR.
Statistical Analysis
All the data were expressed as mean ± SEM unless otherwise specified. Statistical analysis was conducted with an ANOVA followed by the Tukey t-test using GraphPad Prism 5.0 software and SPSS software.
Results
The Expression of ISGs After Poly (I:C) Stimulation and CgIFNLP RNAi
The specificity of polyclonal antibody against CgIFNLP and the abundance of CgIFNLP in hemolymph were examined by Western blot. A single band about 15 kDa with the high specificity was observed, which was consistent with the predicted molecular mass of CgIFNLP (Figure 1A). The intensity of CgIFNLP band in hemocyte proteins increased after Poly (I:C) stimulation (Figures 1B, C). The dsRNA targeted CgIFNLP was applied to suppress the CgIFNLP expression, and its RNA expression level decreased to 0.39-fold of that in the EGFP-RNAi group (p < 0.05) (Figure 1D). The mRNA expression levels of CgISG-like molecules, including CgMx, CgViperin, and CgIFNIP-44 (interferon-induced protein 44) in EGFP-RNAi group were up-regulated after Poly (I:C) stimulation, but they were suppressed in CgIFNLP–RNAi oysters, which was 0.30-fold (p < 0.01), 0.40-fold (p < 0.05), and 0.33-fold (p < 0.05) of that in the EGFP-RNAi group, respectively (Figure 1E).
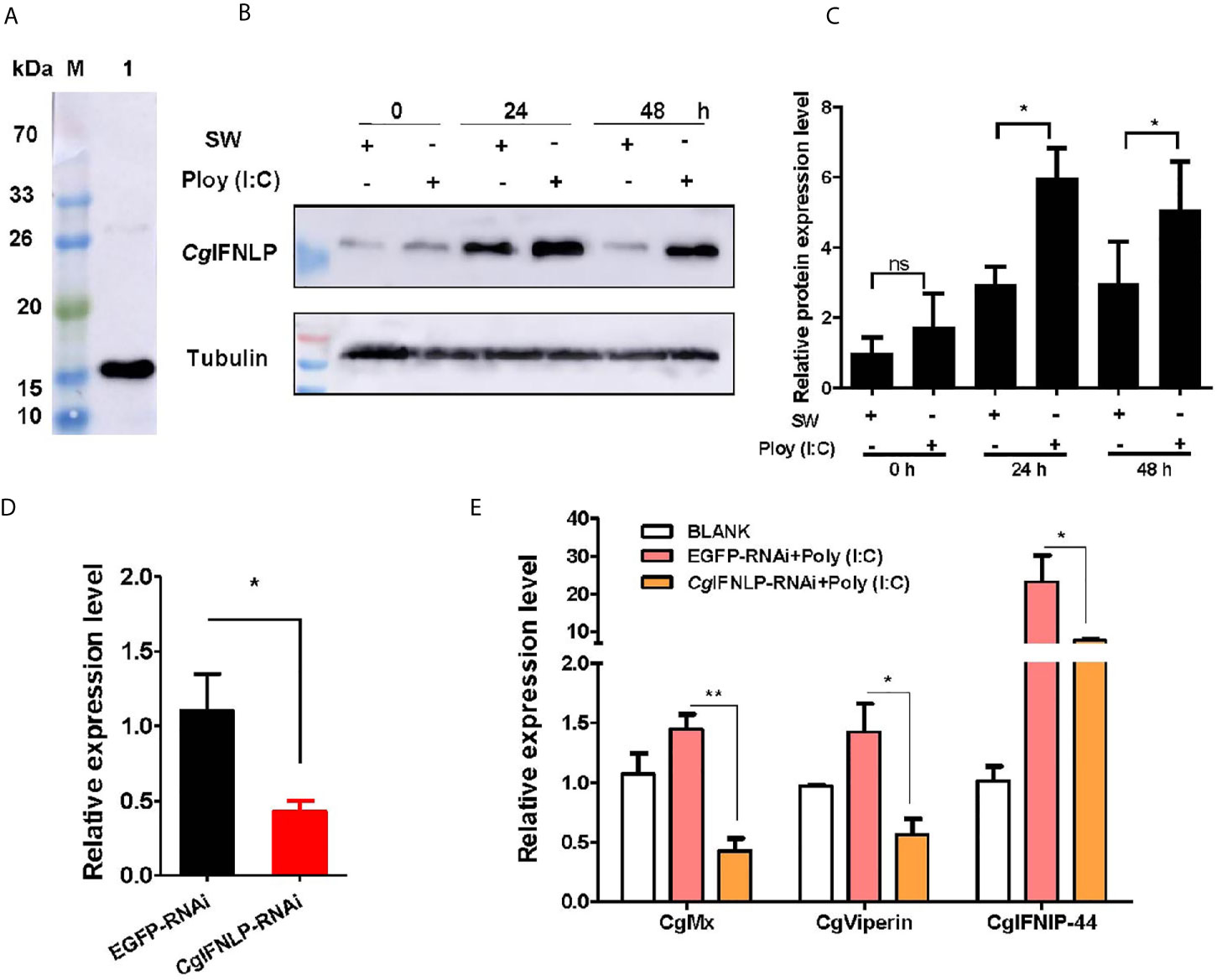
Figure 1 The expression of CgISGs in CgIFNLP–RNAi oysters. (A) The specificity of polyclonal antibody for CgIFNLP examined by Western blot. (B) The protein level of CgIFNLP in hemocytes after Poly (I:C) stimulation. (C) The grayscale analysis of CgIFNLP protein after Poly (I:C) stimulation using ImageJ software. (D) The efficiency of CgIFNLP–RNAi in hemocytes (EGFP–RNAi was used as control). (E) The mRNA expressions of CgMx, CgViperin, and CgIFNIP-44 in CgIFNLP–RNAi oysters after Poly (I:C) stimulation. Vertical bars represent the mean ± SEM (N = 3). *p < 0.05; **p < 0.01; ns, not significant.
CgcGAS and Its Activity to Recognize Viral Nucleic Acid
A cGAS homolog (designated as CgcGAS) was identified from C. gigas. The ORF of CgcGAS was of 1,773 bp encoding a putative polypeptide of 590 amino acids (Figure S1A). There were a Mab-21 and three consecutive Zinc finger domains in the predicted CgcGAS protein (Figure S1A). CgcGAS shared the conserved key amino acids with cGAS sequences from Mus musculu, Homo sapiens, Danio rerio, and Drosophila melanogaster (Figure S1B). In the phylogenetic tree, CgcGAS was firstly clustered with cGAS from C. virginica as a sister branch of the cGAS from Drosophila (Figure S1C). The mRNA transcripts of CgcGAS were detected in all the tested tissues with the highest level in gill, which was nearly 20-fold higher than that in mantle (p < 0.05) (Figure 2A). The expression level of CgcGAS mRNA in hemocytes increased significantly at 12 h with highest expression at 48 h (13.08-fold of that at 0 h, p < 0.05) after Poly (I:C) stimulation (Figure 2B). The recombinant protein of CgcGAS (rCgcGAS) was purified using the Ni-NATA affinity chromatography and examined by 15% SDS-PAGE. An evident band with a molecular weight about 58 kDa was observed, corresponding to CgcGAS with His-tag (6 kDa) (lane 3 in Figure 2C). The purified rCgcGAS was used to prepare polyclonal antibody, and a distinct band about 60 kDa was revealed in hemocyte protein by Western blot (Figure 2D). The abundance of CgcGAS protein in hemocytes increased significantly at 24 and 48 h after Poly (I:C) stimulation (Figures 2E, F). The binding activity of rCgcGAS toward dsRNA and dsDNA was examined by ELISA. rCgcGAS could directly bind dsRNA and dsDNA in a concentration dependent manner with a saturable process from 0 to 3.5 μM (Figures 2G, H). The apparent Kd values of rCgcGAS toward dsRNA and dsDNA, calculated from the saturation curve, were 2.01 × 10−7 and 1.6 × 10−8, respectively (Figures 2G, H).
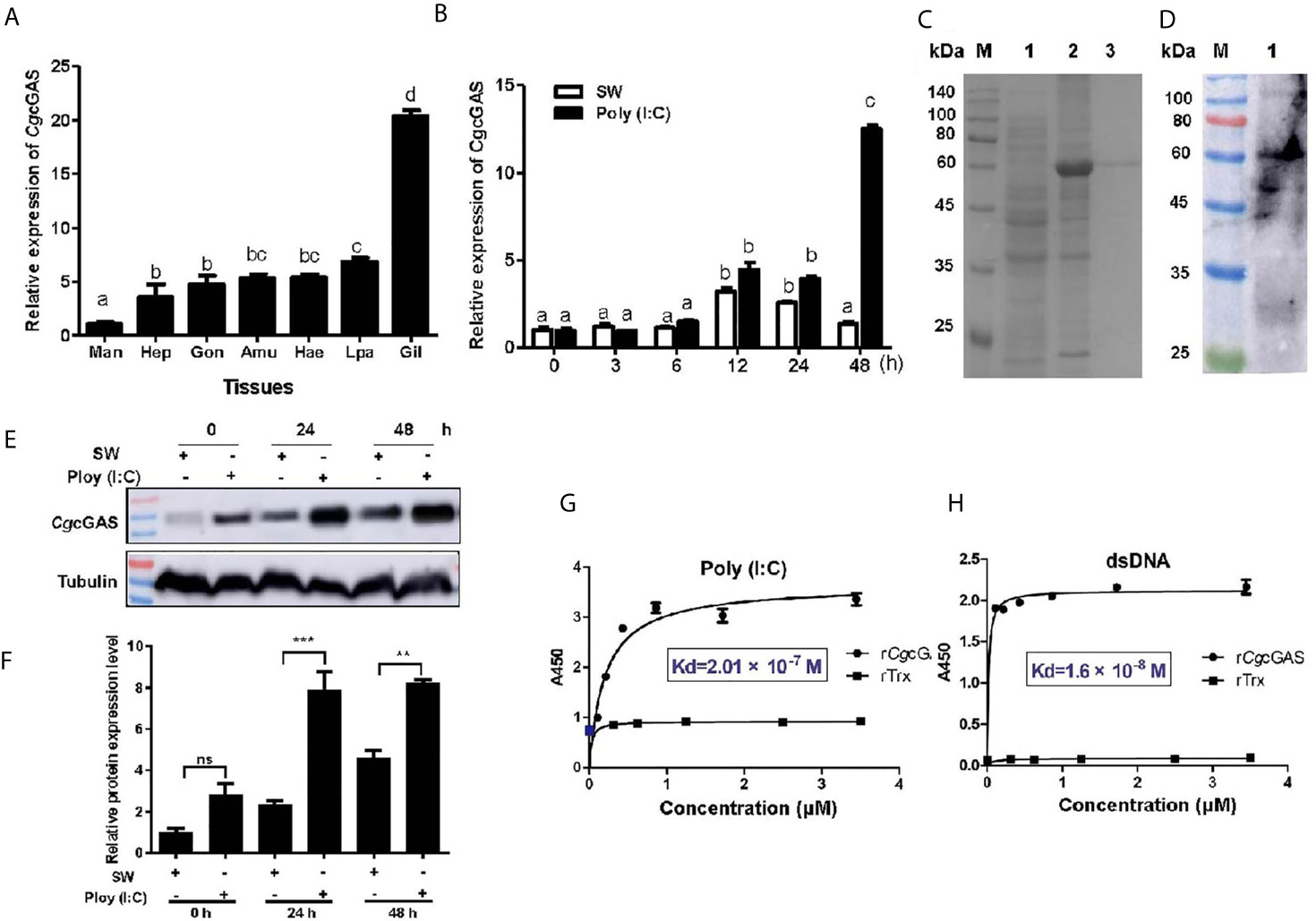
Figure 2 The binding activity of CgcGAS to viral nucleic acid. (A) The tissue distribution of CgcGAS mRNA. Man, mantle; Hep, hepatopancrease; Gon, gonad; Amu, adductor muscle; He, hemolymph; Lpa, labial palp; Gil, gill. (B) The temporal mRNA expression of the CgcGAS after Poly (I:C) stimulation in hemocytes. (C) The recombination protein of CgcGAS. Lane M: standard protein molecular weight marker; Lane 1: negative control (without induction); Lane 2: induced rCgcGAS; Lane 3: purified rCgcGAS. (D) Lane 1: the specificity of polyclonal antibody for CgcGAS detected by Western blot. (E) The protein level of CgcGAS after Poly (I:C) stimulation. (F) The statistical analysis of CgcGAS protein level using ImageJ. (G) The binding activity of rCgcGAS towards Poly (I:C) and (H) dsDNA. A450 was the absorbance value at 450 nm, and the Kd values were calculated by GraphPad Prism 5. **p < 0.01; ***p < 0.001; ns, not significant. The different letters show the existence significant differences comparing with other groups (p < 0.05).
The Regulation of CgcGAS on CgIFNLP Activation and the Nuclear Translocation of CgIRF8
In order to identify the potential immune function of CgcGAS in the regulation of CgIFNLP, the dsRNA of CgcGAS was employed to interfere CgcGAS expression. The mRNA transcript of CgcGAS was down-regulated significantly after dsRNA injection, which was 0.27-fold of that in the EGFP group (p < 0.01, Figure 3A). All the mRNA expression levels of CgIFNLP, CgIRF1, and CgIRF8 decreased obviously in the CgcGAS-RNAi group after Poly (I:C) treatment, which was 0.39-fold (p < 0.001), 0.40-fold (p < 0.001), and 0.41-fold (p < 0.001) of that in the EGFP-RNAi group, respectively (Figure 3B). Unexpectedly, the mRNA expression level of CgSTING and CgTBK1 increased to some extent after CgcGAS was knocked down, which was 1.88-fold (p < 0.05) and 3.1-fold (ns) of that in EGFP-RNAi oysters after Poly (I:C) treatment, respectively (Figure 3B). The recombinant proteins of CgIRF8 (rCgIRF8) were obtained by prokaryotic expression with a molecular weight about 97 kDa (containing MBP-tag of 42 kDa) (Figure 3C). The polyclonal antibody of rCgIRF8 was further prepared, and its specificity was verified by Western blot (Figure 3D). The subcellular localization of CgIRF8 in oyster hemocytes was determined by immunocytochemical assay. The positive signals of CgIRF8 were observed in green, which were mainly distributed in the cytoplasm (Figures 3E, F). After Poly (I:C) stimulation, the green signals of CgIRF8 in hemocyte nucleus increased significantly compared to those in the blank group, which were overlapped with blue signals of nuclei stained by DAPI (Figures 3G, H). However, the green signals of CgIRF8 were still mainly distributed in the hemocyte cytoplasm of CgcGAS-RNAi oysters after Poly (I:C) stimulation (Figures 3G, H).
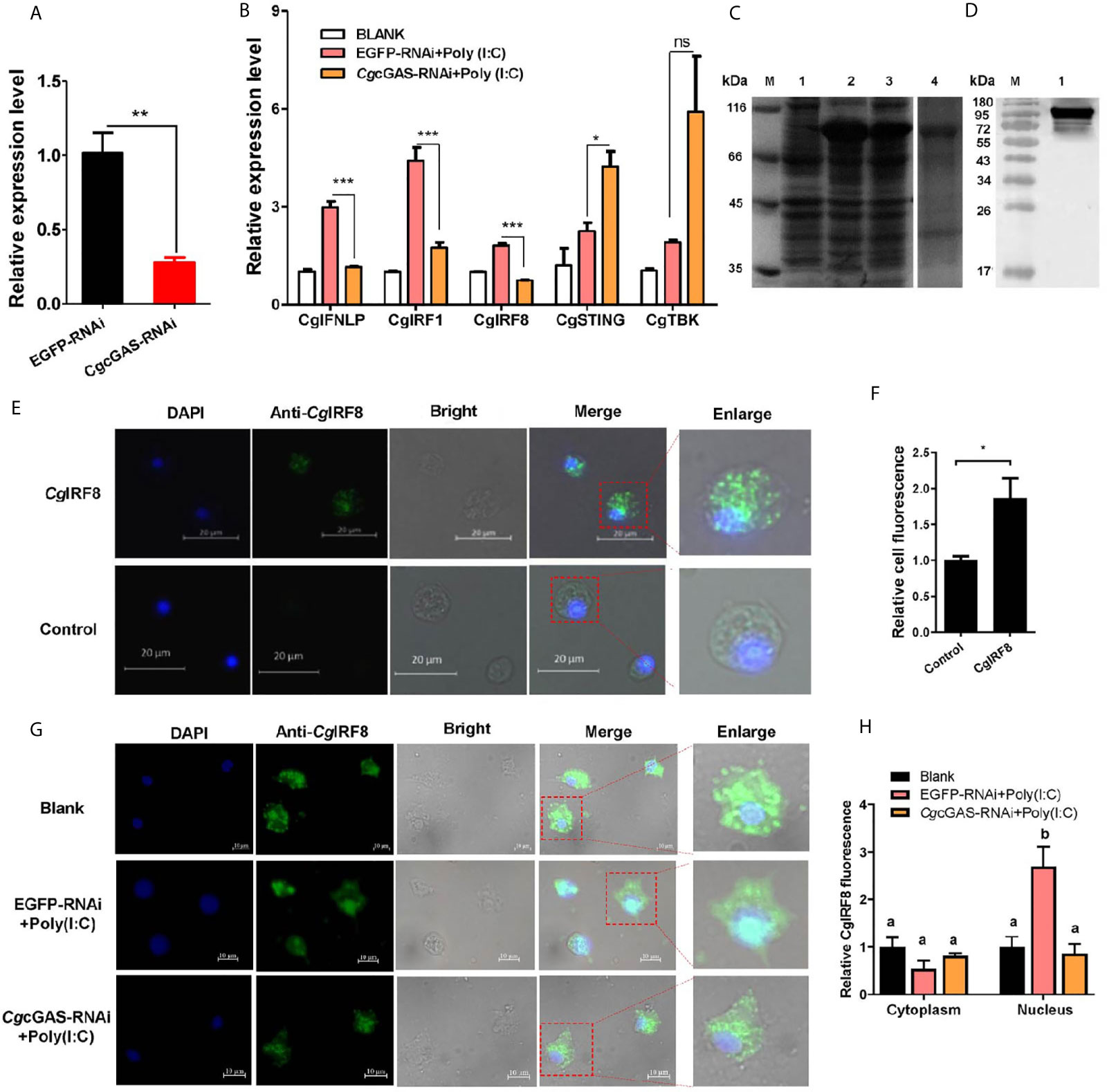
Figure 3 Regulation of CgcGAS on the expression of CgIFNLP and nuclear translocation of CgIRF-8. (A) The efficiency of CgcGAS–RNAi in hemocytes. EGFP–RNAi was used as control. (B) The mRNA expressions of CgIFNLP, CgIRF-1, CgIRF-8, CgSTING, and CgTBK1 in CgcGAS–RNAi oysters after Poly (I:C) stimulation. (C) The recombination protein of CgIRF-8. Lane M: standard protein molecular weight marker; Lane 1: negative control (without induction); Lane 2: induced rCgIRF-8; Lane 3: Supernatant of lysate; Lane 4: purified rCgcGAS. (D) Lane 1: specificity of polyclonal antibody for CgIRF-8 examined by Western blot. (E) The subcellular localization of CgIRF-8 (negative serum as control) in hemocytes and (F) the statistical analysis of relative fluorescence intensity of CgIRF-8 using ImageJ. (G) The subcellular localization of CgIRF-8 in CgcGAS–RNAi hemocytes after Poly (I:C) stimulation and (H) the statistical analysis of fluorescence intensity using ImageJ. Nucleus staining with DAPI is shown in blue signal; anti-CgIRF-8 conjugated to Alexa-fluor 488 is shown in green signal. Vertical bars represent the mean ± SEM. (N = 3). *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant. The different letters show the existence significant differences comparing with other groups (p < 0.05).
CgSTING and Its Recognition to Viral Nucleic Acid
A STING homolog (designated as CgSTING) was identified in C. gigas with an ORF of 1,191 bp encoding a peptide of 396 amino acids (Figure S2A). There was a Toll/IL-1R homologous and a C-di-GMP-bing domain (CBD) in the predicated CgSTING protein, which was consistent with the STINGs from other animals (Figure S2B). The mRNA transcripts of CgSTING were highly expressed in hemocytes, which were 10.80-fold (p < 0.01) of those in hepatopancrease (Figure 4A). No significant difference of expression level was observed in other tissues (Figure 4A). After Poly (I:C) stimulation, the mRNA expression of CgSTING in hemocytes increased significantly at 24 and 48 h, which was 3.81 and 7.25-fold (p < 0.001) of that at 0 h (Figure 4B), respectively. The recombinant protein of CgSTING-CBD with His-tag was obtained by prokaryotic expression and purified using the Ni-NATA affinity chromatography (Figure 4C). The purified rCgSTING-CBD was further used to prepare polyclonal antibody, whose specificity was verified with hemocyte protein by Western blot, and a single distinct band about 43 kDa was observed (Figure 4D). The CgSTING protein in hemocytes also increased after Poly (I:C) stimulation at 24 and 48 h (Figures 4E, F). The PAMP binding assay showed that rCgSTING-CBD could directly bind dsRNA and dsDNA in a concentration dependent manner with a saturable process from 0 to 5 μM (Figures 4G, H). The Kd values of rCgSTING-CBD toward dsRNA and dsDNA were 1.32 × 10−7 and 2.19 × 10−7, respectively (Figures 4G, H).
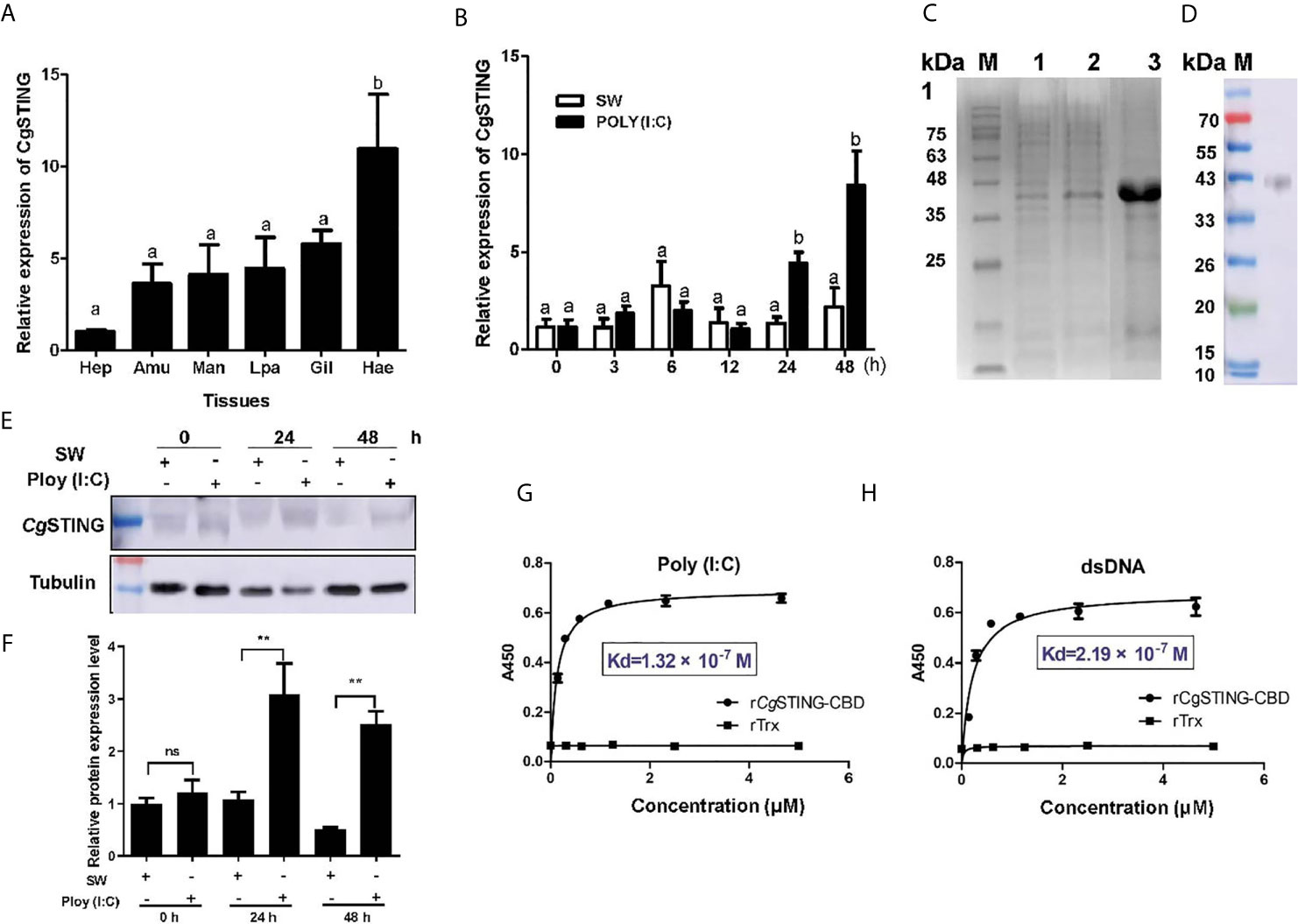
Figure 4 The binding activity of CgSTING to viral nucleic acid. (A) The tissues distribution of the CgSTING mRNA. Man, mantle; Hep, hepatopancrease; Amu, adductor muscle; He, hemolymph; Lpa, labial palp; Gil, gill. (B) The temporal expression profile of CgSTING mRNA after Poly (I:C) stimulation. (C) The recombination protein of CgSTING-CBD. Lane M: standard protein molecular weight marker; Lane 1: negative control (without induction); Lane 2: induced rCgSTING; Lane 3: purified rCgSTING. (D) Lane 1: Specificity detection for polyclonal antibody of CgSTING-CBD by Western blot. (E) The protein level of CgSTING after Poly (I:C) stimulation. (F) The statistical analysis of CgSTING protein level using ImageJ. (G) The binding activity of rCgSTING-CBD to Poly (I:C) and (H) dsDNA. A450 was the absorbance value at 450 nm, and the Kd values were calculated by GraphPad Prism 5. The different letters show the existence significant differences comparing with other groups (p < 0.05). **p < 0.01; ns, not significant.
The Regulation of CgSTING on CgIFNLP Activation and Nuclear Translocation of CgIRF8
The mRNA expression of CgIFNLP, CgTBK1, CgIRF1, and CgIRF8 was examined after CgSTING was knocked down by RNAi with CgSTING-dsRNA. The expression level of CgSTING mRNA in hemocytes decreased at 24 h after the injection with CgSTING-dsRNA, which was 0.34-fold of that in EGFP-RNAi oysters (Figures 5A). The mRNA expressions of CgIFNLP, CgTBK1, CgIRF1, and CgIRF8 in hemocytes were all suppressed in CgSTING-RNAi oysters after Poly (I:C) stimulation, which was 0.33-fold (p < 0.05), 0.62-fold (p < 0.05), 0.66-fold (p < 0.05), and 0.67-fold (p < 0.05) of those in EGFP-RNAi oysters, respectively (Figure 5B). No significant change of CgcGAS expression was observed in CgSTING-RNAi oysters after Poly (I:C) stimulation (Figure 5B). The subcellular localization of CgIRF8 in hemocytes of CgSTING-RNAi oysters was detected by immunocytochemical assay with anti-CgIRF8 polyclonal antibody. The positive signals of CgIRF8 labeled with Alexa Fluer 488 were observed in green, which were mainly distributed in the cytoplasm of oyster hemocytes in blank group. After Poly (I:C) stimulation, the green signals of CgIRF8 were mainly located in hemocyte nucleus of EGFP-RNAi oysters. In CgSTING-RNAi oysters, the green signals were still mainly observed in hemocyte cytoplasm with few distributions in the nucleus (Figures 5C, D). The expression of CgTBK1 in hemocytes increased after Poly (I:C) stimulation at 24 and 48h (Figure 5E). After the injection with dsRNA of CgTBK1, its mRNA expression level decreased to 0.41-fold of that in EGFP-RNAi group (p < 0.01) (Figure 5F). In the EGFP-RNAi group, the expression levels of CgIRF8 and CgIFNLP mRNA in hemocytes both increased after Poly (I:C) stimulation. In CgTBK1–RNAi group, the increase of CgIRF8 and CgIFNLP expressions induced by Poly (I:C) injection was inhibited, which was 0.52-fold (p < 0.001) and 0.61-fold (p < 0.01) of that in the EGFP-RNAi group, respectively (Figure 5G).
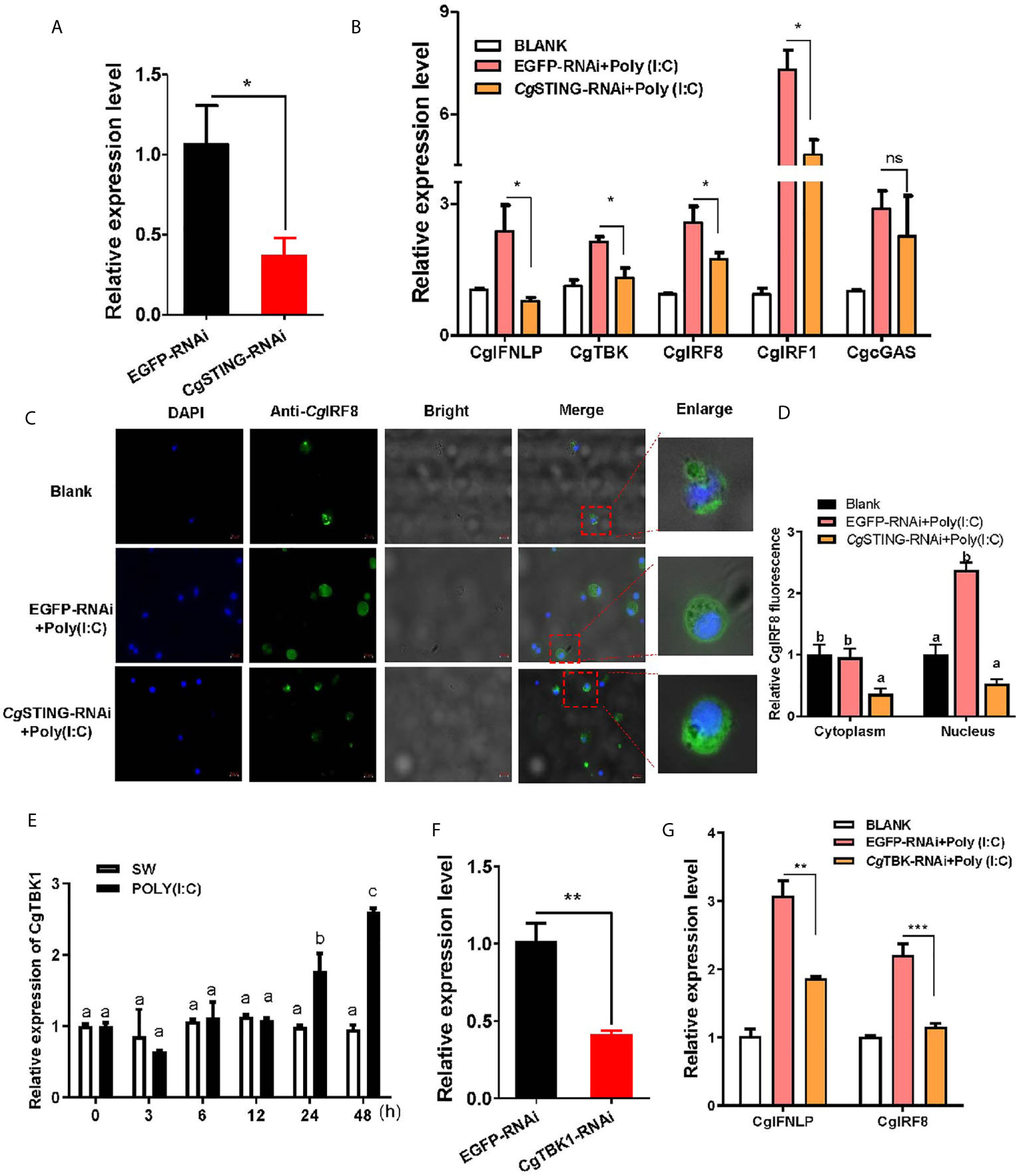
Figure 5 The regulation of CgSTING and CgTBK1 on the expression of CgIFNLP and IRFs. (A) The efficiency of CgSTING-RNAi in hemocytes (EGFP–RNAi was used as control). (B) The mRNA expressions of CgIFNLP, CgTBK1, CgIRF-1, CgIRF-8, and CgcGAS in CgSTING–RNAi oysters after Poly (I:C) stimulation were detected by qRT-PCR. (C) The subcellular localization of CgIRF-8 protein in hemocytes of CgSTING–RNAi oyster after Poly (I:C) stimulation. (D) The statistical analysis of relative fluorescence intensity of CgIRF-8 using ImageJ. Nucleus staining with DAPI was shown in blue signal; anti-CgIRF-8 conjugated to Alexa-fluor 488 was shown in green signal. (E) Temporal expression of CgTBK1 mRNA after Poly (I:C) stimulation. (F) The efficiency of CgTBK1–RNAi in hemocytes. (G) The mRNA expressions of CgIFNLP and CgIRF-8 in CgTBK1–RNAi oysters after Poly (I:C) stimulation. Vertical bars represent the mean ± SEM (N = 3). *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant. The different letters show the existence significant differences comparing with other groups (p < 0.05).
Discussion
IFNs are proteins or glycoproteins that spontaneously orchestrate complex cell-intrinsic and cell-extrinsic antiviral defenses through their effects on the intracellular events of viral cycle. Since IFNs were first defined in 1957, a large number of studies have been carried out on their molecular structural features, expression patterns, signaling pathways, and myriad roles in innate and adaptive immunity. So far, IFNs have been widely discovered in almost all kinds of vertebrates from fish to mammals (16, 37). Therefore, IFNs with conserved sequence are deemed to evolutionarily derive from teleosts (17). In addition to the classic antiviral function, the recombinant fish IFNφ1 were identified to possess broad-spectrum direct antibacterial activities, indicating IFNs’ multiple functions and importance in host defense (38). Recently, there have been increasing pieces of evidence on IFN-like molecule mediated antiviral mechanism in invertebrate (20). In arthropod, a novel viral infection-inducible peptide, Vago, has been characterized in Drosophila, Culex, and L. vannamei (21, 22, 24). Although these Vagos share low sequence similarity with IFNs from vertebrates, they are identified with the similar functions as Type I IFNs in vertebrates. They activate the semblable JAK–STAT pathway to block viral reproduction and protect arthropod from virus infection (21). Even an IFN-like protein and a cytokine receptor-like 3 have been identified from mollusk C. gigas (30, 31), the existence of IFN-like system and the detailed molecular mechanism underlying IFN-like mediated antiviral responses remains bewildered in primitive invertebrates. In this study, the signal pathways to mediate the expression of CgIFNLP were investigated to comprehensively understand its involvement in antiviral immune responses in C. gigas.
In response to viral infections, vertebrate IFNs up-regulate the expression of hundreds of ISGs, whose cumulative action can potently inhibit the replication of virus (39). In the present study, the protein level of CgIFNLP in hemocytes was observed to increase after Poly (I:C) stimulation. In order to confirm the activation mechanism of IFN signaling pathways in oysters, RNAi was applied to inhibit the expression of CgIFNLP. The expression of CgISGs including CgMx, CgViperin, and CgIFNIP-44 was suppressed after CgIFNLP was knocked down. As a classic ISG, MX1 acts prior to genome replication at an early post entry step of the virus life cycle, while viperin inhibits both viral egress and the replication of multiple viruses, and IFNIP-44 has been shown to restrict infection involving the reduction of viral genome transcription or replication (39–41). The results of CgIFNLP–RNAi indicated that CgIFNLP displayed similar characteristics with vertebrate IFNs to resist viral infection dependent on ISG release (2).
In vertebrates, virus-derived DNA is sensed by cGAS in the cytosol, which synthesizes the second messenger 2′3′-cGAMP to bind and activate the signaling adaptor STING (42). The activated STING initiates a downstream signaling event by recruiting and activating TBK1 or IKK, which leads to the phosphorylation of IRF3 to initiate the expression of IFNs and IκB family of inhibitors of transcription factor NF-κB, respectively (13). The cGAS homologs have been reported to be present in lots of vertebrate species and show similar structures with human or murine cGAS (11). In the present study, both cGAS and STING homologs were identified in C. gigas, and they shared sequence conservation with their homologs from vertebrates. The origins of cGAS and STING were considered to be traced back to a choanoflagellate Monosiga brevicollis, the closest relative of metazoans (43). However, it was found that the cGAS ortholog in Drosophila did not play a role during Listeria or IIV6 infection, which was speculated to be ascribed to the lack of the zinc-ribbon domain and a positively charged N terminus functionally important for DNA binding (44). Interestingly, a functional cGAS–STING pathway has been reported in bacteria Vibrio cholerae, which suggested that the eukaryotic cGAS–STING antiviral pathway has ancient evolutionary roots stemming from microbial defenses against phages (45). The cGAS–STING pathway was also confirmed to exist in the sea anemone Nematostella vectensis (46). Unlike the cGAS in mammal and arthropod, CgcGAS in C. gigas was able to bind both dsRNA and dsDNA with higher combining capacity with dsRNA and sensitivity with dsDNA. However, the cGAS homologs in invertebrates have been speculated to lose the function as DNA sensors, and act a nucleotidyl transferase to produce cGAMP and other cyclic dinucleotides (CDNs) (43). The recognition capability of CgcGAS with double-stranded nucleic acid might benefit from its conserved Zinc finger domains and critical amino acid residues. The expression of CgcGAS was highly induced after Poly (I:C) stimulation, indicating its involvement in antiviral immunity of C. gigas. After CgcGAS was knocked down by RNAi, the expression of CgIFNLP, CgIRF-1 and -8 in hemocytes was reduced, which demonstrated the important roles of CgcGAS in CgIFNLP mediating antiviral immune response. Consistently, induction of antiviral IFNs is the major outcome of cGAS–STING activation in vertebrates (42). Moreover, the inhibition of CgcGAS expression suppressed the nuclear translocation of CgIRF-8 after Poly (I:C) stimulation, revealing that the CgcGAS might induce the CgIFNLP expression through mediating the nuclear translocation of CgIRF-8. IRFs are the key transcriptional factors of IFN signaling, and a lot of IRFs have been discovered in invertebrates, such as Branchiostoma belcheri tsingtauense (47), Litopenaeus vannamei (23), Pinctada fucata (27), and C. gigas (48). Three IRFs (CgIRF-1a, -1b and -8) have been identified in C. gigas (48). CgIRF-1a and CgIRF-1b with typical IRF domain (also known as DNA-binding domain) have been certified to regulate the expression of CgIFNLP as a transcriptional regulatory factor (26). In the present study, CgIRF-8 was identified from C. gigas as a crucial factor for CgcGAS, and its mRNA expression was induced after Poly (I:C) stimulation, indicating that both CgIRF-1 and CgIRF-8 are necessary for the regulation of CgIFNLP expression. Unexpectedly, the expression of CgSTING and CgTBK1 did not change significantly after CgcGAS was knocked down, but increased after Poly (I:C) stimulation in CgcGAS-RNAi oysters, which suggested that CgSTING and CgTBK1 were not the downstream molecules of CgcGAS in C. gigas.
Recently, the functional STING homologs were identified in bacteria. They were functional cyclic dinucleotide receptors and showed TIR–STING fusion structure, which confirmed the STING cyclic dinucleotide sensing originated in bacteria (49). Together with the cGAS homologs in prokaryotic antiviral immunity, the functional cGAS–STING signaling might arise from an ancient mechanism of defense against bacteriophages. Moreover, the crystal structure of C. gigas STINGs was also analyzed, which showed conserved TIR–STING fusions and cyclic dinucleotide binding activity (49). Consistently, in the present study, the CgSTING from C. gigas displayed a similar TIR–STING fusion structure shown as TIR and CBD domains. The CBD of CgSTING (rCgSTING-CBD) could bind the Poly (I:C) and dsDNA in vitro. In mammals, the central CBD of STING was determined to mediate the binding to CDNs from cGAS (50). However, the relatively conservative domain in CgSTING suggested that it could act as a nucleic acid identifier, implying the largely elusive and diversiform functions of STINGs from invertebrates. Furthermore, the mRNA expression of CgSTING was found to increase after Poly (I:C) stimulation, indicating its involvement in oyster antiviral immunity. Increasing evidence suggested that the ancestral functions of STING might be related to the activation of antibacterial immunity (13), such as the STING in Litopenaeus vannamei (LvSTING) and Drosophila (dmSTING), which were reported to be involved in the innate immune response against bacterial infection (44, 51). In the present study, after CgSTING was knocked down by RNAi, the expression level of CgIFNLP, CgTBK1, CgIRF-1, and CgIRF-8 mRNA, as well as the nuclear translocation CgIRF-8 induced by Poly (I:C) was significantly suppressed. These results suggested that the CgSTING also mediated the expression of CgIFNLP and CgIRFs, which was consistent with CgcGAS. However, different from the observation in mammals, the CgSTING and CgTBK1 were not the downstream molecules of CgcGAS. The homologs of STING in invertebrate are postulated to loss the function to induce innate immune response against infection for the lack of a carboxy-terminal tail (CTT) domain which is the essential domain for mammalian STING to recruit the critical downstream TBK1 and IRF3 signaling components (43, 50). However, CgTBK1 from C. gigas was reported to interact with CgSTING in HEK293T cells, providing the evidence that CgTBK1 could be activated by direct binding to CgSTING (52). Furthermore, the expression of CgIFNLP and CgIRF-8 in CgTBK1–RNAi oysters also decreased after Poly (I:C) stimulation. The results collectively illustrated that CgSTING might activate CgIFNLP to cope with viral nucleic acid by binding CgTBK1 and further inducing the expression and nuclear migration of CgIRFs. Therefore, it was proposed that the CgcGAS and CgSTING recognized the viral nucleic acid simultaneously and further activated the CgIFNLP expression synergistically.
In summary, CgcGAS and CgSTING were identified to recognize and bind dsRNA and dsDNA. They were involved in the response against Poly (I:C) stimulation and synergistically facilitated the CgIRF-mediated CgIFNLP production, which further induced the expression of CgISGs (Figure S3). The results defined a CgcGAS/STING–TBK1–IRFs regulatory axis to mediate the primitive IFN-like antiviral mechanism in C. gigas.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
XQ, YZ, and ZW designed, performed, and analyzed the experiments, participated in the design of the study, and drafted the manuscript. ZL and YL participated in the design of the study and discussed the results. LW and LS conceived of the study, coordinated the experiment, and helped draft the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by National Key R&D Program (2018YFD0900502), grants (Nos. U1706204, 41961124009, 32002418) from National Science Foundation of China, earmarked fund (CARS-49) from Modern Agro-industry Technology Research System, the Fund for Outstanding Talents and Innovative Team of Agricultural Scientific Research, Key R&D Program of Liaoning Province (2017203004, 2017203001), Liaoning Climbing Scholar, the Distinguished Professor of Liaoning (XLYC1902012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to all the laboratory members for technical advice and helpful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.689783/full#supplementary-material
Abbreviations
IFN, interferon; ISGs, IFN-stimulated genes; cGAS, cyclic GMP-AMP synthase; STING, stimulator of interferon gene; TBK1, TANK binding kinase1; IRF, interferon regulatory factors; Mx, myxovirus resistance; PAMPs, pathogen-associated molecular patterns; PRRs, pattern recognition receptors; TLRs, Toll like receptors; RIG-I, retinoic acid-inducible gene I; MDA5, melanoma differentiation associated protein 5; IKKƐ, Ikappa B Kinase Ɛ; IFNR-3, IFN receptor-like 3; CBD, c-di-GMP-binding domain.
References
1. Decker T, Stockinger S, Karaghiosoff M, Muller M, Kovarik P. Ifns and STATs in Innate Immunity to Microorganisms. J Clin Invest (2002) 109:1271–7. doi: 10.1172/JCI0215770
2. Villarino AV, Kanno Y, O’Shea JJ. Mechanisms and Consequences of Jak-STAT Signaling in the Immune System. Nat Immunol (2017) 18:374–84. doi: 10.1038/ni.3691
3. Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-Lambdas Mediate Antiviral Protection Through a Distinct Class II Cytokine Receptor Complex. Nat Immunol (2003) 4:69–77. doi: 10.1038/ni875
4. Gray PW, Goeddel DV. Structure of the Human Immune Interferon Gene. Nature (1982) 298:859–63. doi: 10.1038/298859a0
5. Leu SW, Shi L, Xu C, Zhao Y, Liu B, Li B, et al. TLR4 Through IFN-β Promotes Low Molecular Mass Hyaluronan-Induced Neutrophil Apoptosis. J Immunol (Baltimore Md. 1950) (2011) 186:556–62. doi: 10.4049/jimmunol.1001630
6. Elion DL, Cook RS. Harnessing RIG-I and Intrinsic Immunity in the Tumor Microenvironment for Therapeutic Cancer Treatment. Oncotarget (2018) 9:29007–17. doi: 10.18632/oncotarget.25626
7. Iurescia S, Fioretti D, Rinaldi M. Targeting Cytosolic Nucleic Acid-Sensing Pathways for Cancer Immunotherapies. Front Immunol (2018) 9:711. doi: 10.3389/fimmu.2018.00711
8. Tan X, Sun L, Chen J, Chen ZJ. Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu Rev Microbiol (2018) 72:447–78. doi: 10.1146/annurev-micro-102215-095605
9. Vanpouille-Box C, Demaria S, Formenti SC, Galluzzi L. Cytosolic DNA Sensing in Organismal Tumor Control. Cancer Cell (2018) 34:361–78. doi: 10.1016/j.ccell.2018.05.013
10. Wu J, Chen ZJ. Innate Immune Sensing and Signaling of Cytosolic Nucleic Acids. Annu Rev Immunol (2014) 32:461–88. doi: 10.1146/annurev-immunol-032713-120156
11. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Sci (New York N.Y.) (2013) 339:786–91. doi: 10.1126/science.1232458
12. Ishikawa H, Ma Z, Barber GN. STING Regulates Intracellular DNA-mediated, Type I Interferon-Dependent Innate Immunity. Nature (2009) 461:788–92. doi: 10.1038/nature08476
13. Margolis SR, Wilson SC, Vance RE. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol (2017) 38:733–43. doi: 10.1016/j.it.2017.03.004
14. Mancino A, Natoli G. Specificity and Function of IRF Family Transcription Factors: Insights From Genomics. J Interferon Cytokine Res Off J Int Soc Interferon Cytokine Res (2016) 36:462–9. doi: 10.1089/jir.2016.0004
15. Negishi H, Taniguchi T, Yanai H. The Interferon (Ifn) Class of Cytokines and the IFN Regulatory Factor (Irf) Transcription Factor Family. Cold Spring Harbor Perspect Biol (2018) 10:a028423. doi: 10.1101/cshperspect.a028423
16. Langevin C, Aleksejeva E, Passoni G, Palha N, Levraud JP, Boudinot P. The Antiviral Innate Immune Response in Fish: Evolution and Conservation of the IFN System. J Mol Biol (2013) 425:4904–20. doi: 10.1016/j.jmb.2013.09.033
17. Vilcek J. Fifty Years of Interferon Research: Aiming at a Moving Target. Immunity (2006) 25:343–8. doi: 10.1016/j.immuni.2006.08.008
18. Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the Vertebrate Ancestral Genome Reveals Dynamic Genome Reorganization in Early Vertebrates. Genome Res (2007) 17:1254–65. doi: 10.1101/gr.6316407
19. Secombes C, Zou J. Evolution of Interferons and Interferon Receptors. Front Immunol (2017) 8:209. doi: 10.3389/fimmu.2017.00209
20. Qiao X, Wang L, Song L. The Primitive Interferon-Like System and its Antiviral Function in Molluscs. Dev Comp Immunol (2021) 118:103997. doi: 10.1016/j.dci.2021.103997
21. Deddouche S, Matt N, Budd A, Mueller S, Kemp C, Galiana-Arnoux D, et al. The DExD/H-box Helicase Dicer-2 Mediates the Induction of Antiviral Activity in Drosophila. Nat Immunol (2008) 9:1425–32. doi: 10.1038/ni.1664
22. Paradkar PN, Trinidad L, Voysey R, Duchemin JB, Walker PJ. Secreted Vago Restricts West Nile Virus Infection in Culex Mosquito Cells by Activating the Jak-STAT Pathway. Proc Natl Acad Sci USA (2012) 109:18915–20. doi: 10.1073/pnas.1205231109
23. Li C, Li H, Chen Y, Chen Y, Wang S, Weng SP, et al. Activation of Vago by Interferon Regulatory Factor (IRF) Suggests an Interferon System-Like Antiviral Mechanism in Shrimp. Sci Rep (2015) 5:15078. doi: 10.1038/srep15078
24. De Zoysa M, Kang HS, Song YB, Jee Y, Lee YD, Lee J. First Report of Invertebrate Mx: Cloning, Characterization and Expression Analysis of Mx cDNA in Disk Abalone (Haliotis Discus Discus). Fish Shellfish Immunol (2007) 23:86–96. doi: 10.1016/j.fsi.2006.09.007
25. Green TJ, Speck P, Geng L, Raftos D, Beard MR, Helbig KJ. Oyster Viperin Retains Direct Antiviral Activity and its Transcription Occurs Via a Signalling Pathway Involving a Heat-Stable Haemolymph Protein. J Gen Virol (2015) 96:3587–97. doi: 10.1099/jgv.0.000300
26. Lu M, Yang C, Li M, Yi Q, Lu G, Wu Y, et al. A Conserved Interferon Regulation Factor 1 (IRF-1) From Pacific Oyster Crassostrea Gigas Functioned as an Activator of IFN Pathway. Fish Shellfish Immunol (2018) 76:68–77. doi: 10.1016/j.fsi.2018.02.024
27. Huang XD, Liu WG, Wang Q, Zhao M, Wu SZ, Guan YY, et al. Molecular Characterization of Interferon Regulatory Factor 2 (IRF-2) Homolog in Pearl Oyster Pinctada Fucata. Fish Shellfish Immunol (2013) 34:1279–86. doi: 10.1016/j.fsi.2013.02.003
28. Le Deuff RM, Renault T. Purification and Partial Genome Characterization of a Herpes-Like Virus Infecting the Japanese Oyster, Crassostrea Gigas. J Gen Virol (1999) 80(Pt 5):1317–22. doi: 10.1099/0022-1317-80-5-1317
29. de Lorgeril J, Lucasson A, Petton B, Toulza E, Montagnani C, Clerissi C, et al. Immune-Suppression by OsHV-1 Viral Infection Causes Fatal Bacteraemia in Pacific Oysters. Nat Commun (2018) 9:4215. doi: 10.1038/s41467-018-06659-3
30. Zhang R, Liu R, Wang W, Xin L, Wang L, Li C, et al. Identification and Functional Analysis of a Novel IFN-Like Protein (CgIFNLP) in Crassostrea Gigas. Fish Shellfish Immunol (2015) 44:547–54. doi: 10.1016/j.fsi.2015.03.015
31. Zhang R, Liu R, Xin L, Chen H, Li C, Wang L, et al. A CgIFNLP Receptor From Crassostrea Gigas and its Activation of the Related Genes in Human JAK/STAT Signaling Pathway. Dev Comp Immunol (2016) 65:98–106. doi: 10.1016/j.dci.2016.06.010
32. Wu Z, Sun J, Wang L, Zong Y, Han Z, Yang W, et al. CgSOCS6 Negatively Regulates the Expression of CgIL17s and CgDefh1 in the Pacific Oyster Crassostrea Gigas. Fish Shellfish Immunol (2019) 93:1084–92. doi: 10.1016/j.fsi.2019.08.055
33. Zong Y, Liu Z, Wu Z, Han Z, Wang L, Song L. A Novel Globular C1q Domain Containing Protein (C1qDC-7) From Crassostrea Gigas Acts as Pattern Recognition Receptor With Broad Recognition Spectrum. Fish Shellfish Immunol (2019) 84:920–6. doi: 10.1016/j.fsi.2018.10.079
34. Gushchina LV, Kwiatkowski TA, Bhattacharya S, Weisleder NL. Conserved Structural and Functional Aspects of the Tripartite Motif Gene Family Point Towards Therapeutic Applications in Multiple Diseases. Pharmacol Ther (2018) 185:12–25. doi: 10.1016/j.pharmthera.2017.10.020
35. Wynne C, Lazzari E, Smith S, McCarthy EM, Gabhann JN, Kallal LE, et al. TRIM68 Negatively Regulates IFN-β Production by Degrading TRK Fused Gene, a Novel Driver of IFN-β Downstream of Anti-Viral Detection Systems. PLoS ONE (2014) 9:e101503. doi: 10.1371/journal.pone.0101503
36. Liu Y, Zhang P, Wang W, Dong M, Wang M, Gong C, et al. A DM9-containing Protein From Oyster Crassostrea Gigas (CgDM9CP-2) Serves as a Multipotent Pattern Recognition Receptor. Dev Comp Immunol (2018) 84:315–26. doi: 10.1016/j.dci.2018.03.003
37. Gan Z, Chen SN, Huang B, Hou J, Nie P. Intronless and Intron-Containing Type I IFN Genes Coexist in Amphibian Xenopus Tropicalis: Insights Into the Origin and Evolution of Type I Ifns in Vertebrates. Dev Comp Immunol (2017) 67:166–76. doi: 10.1016/j.dci.2016.10.007
38. Xiao X, Zhu W, Zhang Y, Liao Z, Wu C, Yang C, et al. Broad-Spectrum Robust Direct Bactericidal Activity of Fish Ifnφ1 Reveals an Antimicrobial Peptide-Like Function for Type I Ifns in Vertebrates. J Immunol (2021) 206:1337–47. doi: 10.4049/jimmunol.2000680
39. Feng J, Wickenhagen A, Turnbull M, Rezelj V, Kreher F, Tilston-Lunel N, et al. Interferon-Stimulated Gene (Isg)-Expression Screening Reveals the Specific Antibunyaviral Activity of ISG20. J Virol (2018) 92:02140–17. doi: 10.1128/JVI.02140-17
40. Helbig KJ, Beard MR. The Role of Viperin in the Innate Antiviral Response. J Mol Biol (2014) 426:1210–9. doi: 10.1016/j.jmb.2013.10.019
41. Busse D, Habgood-Coote D, Clare S, Brandt C, Bassano I, Kaforou M, et al. Interferon-Induced Protein 44 and Interferon-Induced Protein 44-Like Restrict Replication of Respiratory Syncytial Virus. J Virol (2020) 94:00297–20. doi: 10.1128/JVI.00297-20
42. Gamdzyk M, Doycheva DM, Araujo C, Ocak U, Luo Y, Tang J, et al. Cgas/STING Pathway Activation Contributes to Delayed Neurodegeneration in Neonatal Hypoxia-Ischemia Rat Model: Possible Involvement of LINE-1. Mol Neurobiol (2020) 57:2600–19. doi: 10.1007/s12035-020-01904-7
43. Wu X, Wu FH, Wang X, Wang L, Siedow JN, Zhang W, et al. Molecular Evolutionary and Structural Analysis of the Cytosolic DNA Sensor cGAS and STING. Nucleic Acids Res (2014) 42:8243–57. doi: 10.1093/nar/gku569
44. Martin M, Hiroyasu A, Guzman RM, Roberts SA, Goodman AG. Analysis of Drosophila Sting Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep (2018) 23:3537–3550 e3536. doi: 10.1016/j.celrep.2018.05.029
45. Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer-Shaanan Y, Kacen A, et al. Cyclic GMP-AMP Signalling Protects Bacteria Against Viral Infection. Nature (2019) 574:691–5. doi: 10.1038/s41586-019-1605-5
46. Kranzusch PJ, Wilson SC, Lee AS, Berger JM, Doudna JA, Vance RE. Ancient Origin of Cgas-STING Reveals Mechanism of Universal 2’,3’ cGAMP Signaling. Mol Cell (2015) 59:891–903. doi: 10.1016/j.molcel.2015.07.022
47. Yuan S, Zheng T, Li P, Yang R, Ruan J, Huang S, et al. Characterization of Amphioxus Ifn Regulatory Factor Family Reveals an Archaic Signaling Framework for Innate Immune Response. J Immunol (2015) 195:5657–66. doi: 10.4049/jimmunol.1501927
48. Mao F, Lin Y, Zhou Y, He Z, Li J, Zhang Y, et al. Structural and Functional Analysis of Interferon Regulatory Factors (Irfs) Reveals a Novel Regulatory Model in an Invertebrate, Crassostrea Gigas. Dev Comp Immunol (2018) 89:14–22. doi: 10.1016/j.dci.2018.07.027
49. Morehouse B, Govande A, Millman A, Keszei A, Lowey B, Ofir G, et al. STING Cyclic Dinucleotide Sensing Originated in Bacteria. Nature (2020) 586:429–33. doi: 10.1038/s41586-020-2719-5
50. Tanaka Y, Chen ZJ. STING Specifies IRF3 Phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Sci Signaling (2012) 5:ra20. doi: 10.1126/scisignal.2002521
51. Li H, Wang S, Lu K, Yin B, Xiao B, Li S, et al. An Invertebrate STING From Shrimp Activates an Innate Immune Defense Against Bacterial Infection. FEBS Lett (2017) 591:1010–7. doi: 10.1002/1873-3468.12607
Keywords: Crassostrea gigas, interferon-like system, cyclic GMP-AMP (cGAMP) synthase (cGAS), stimulator of interferon genes (STING), antiviral immunity
Citation: Qiao X, Zong Y, Liu Z, Wu Z, Li Y, Wang L and Song L (2021) The cGAS/STING–TBK1–IRF Regulatory Axis Orchestrates a Primitive Interferon-Like Antiviral Mechanism in Oyster. Front. Immunol. 12:689783. doi: 10.3389/fimmu.2021.689783
Received: 01 April 2021; Accepted: 18 May 2021;
Published: 08 June 2021.
Edited by:
Jun Li, Lake Superior State University, United StatesCopyright © 2021 Qiao, Zong, Liu, Wu, Li, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linsheng Song, bHNoc29uZ0BkbG91LmVkdS5jbg==; Lingling Wang, d2FuZ2xpbmdsaW5nQGRsb3UuZWR1LmNu
 Xue Qiao
Xue Qiao Yanan Zong
Yanan Zong Zhaoqun Liu
Zhaoqun Liu Zhaojun Wu
Zhaojun Wu Yuanmei Li
Yuanmei Li Lingling Wang
Lingling Wang Linsheng Song
Linsheng Song