- 1Department of Cellular & Molecular Medicine, University of California San Diego, La Jolla, CA, United States
- 2Department of Chemistry and Biochemistry, University of California San Diego, La Jolla, CA, United States
The COVID-19 pandemic, caused by the coronavirus SARS-CoV-2, is the most severe public health event of the twenty-first century. While effective vaccines against SARS-CoV-2 have been developed, there remains an urgent need for diagnostics to quickly and accurately detect infections. Antigen tests, particularly those that detect the abundant SARS-CoV-2 Nucleocapsid protein, are a proven method for detecting active SARS-CoV-2 infections. Here we report high-resolution crystal structures of three llama-derived single-domain antibodies that bind the SARS-CoV-2 Nucleocapsid protein with high affinity. Each antibody recognizes a specific folded domain of the protein, with two antibodies recognizing the N-terminal RNA binding domain and one recognizing the C-terminal dimerization domain. The two antibodies that recognize the RNA binding domain affect both RNA binding affinity and RNA-mediated phase separation of the Nucleocapsid protein. All three antibodies recognize highly conserved surfaces on the Nucleocapsid protein, suggesting that they could be used to develop affordable diagnostic tests to detect all circulating SARS-CoV-2 variants.
Introduction
The COVID-19 pandemic, caused by the novel coronavirus SARS-CoV-2, has resulted in over 170 million confirmed cases and caused over 3.5 million deaths as of early June 2021 (John Hopkins Coronavirus Resource Center, https://coronavirus.jhu.edu). Despite the development and widespread use of effective vaccines against SARS-CoV-2 (1–3), the virus and its emerging variants will remain an active public health threat for the foreseeable future. In order to detect and quickly respond to new infections and local outbreaks, robust diagnostic tools are required that can quickly and reliably detect active SARS-CoV-2 infections.
Antigen tests are immunoassays that detect the presence of a viral antigen such as an abundant protein, and constitute an effective means of detecting active infections for respiratory pathogens including SARS and SARS-CoV-2 (4–6). Antigen tests are an important complement to PCR-based tests, which detect the presence of viral nucleic acids (genomic or sub-genomic RNA in the case of SARS-CoV-2), but may give positive results after a patient is no longer infectious. While antigen tests are less sensitive than PCR-based tests, they are generally faster and require less specialized equipment than PCR tests, enabling wider deployment than PCR-based tests.
The major structural proteins of SARS-CoV-2 include S (Spike), M (Membrane), E (Envelope), and N (Nucleocapsid). While the Spike protein is exposed on the surface of virions and is the target of all major vaccines, the Nucleocapsid protein is highly abundant in virions and infected cells, and is therefore a common choice for antigen tests. The N protein plays several roles in the SARS-CoV-2 life cycle, including facilitating viral RNA production, suppressing host cells’ innate immune responses, and packaging viral genomic RNA into developing virions. To accomplish these tasks, the N protein possesses a modular structure with an N-terminal RNA-binding domain (RBD) and a C-terminal dimerization domain (CTD), plus three intrinsically disordered regions (IDRs) at the N- and C-termini and between the RBD and CTD. The protein oligomerizes through its CTD and disordered C-terminal tail (7), and the protein also undergoes liquid-liquid phase separation with RNA mediated by its RBD and central disordered region (8–14). In cells, N protein condensates recruit the stress granule proteins G3BP1 and G3BP2, suppressing stress granule assembly (8, 12, 15). During virion production, the N protein assembles into viral RNA-protein complexes (RNPs) with a characteristic barrel shape to package the viral RNA (16, 17), and interacts with the Membrane protein to recruit the viral genome to developing virions (8).
Two recent studies reported the isolation of a total of four single-domain antibodies (sdAbs) that target SARS-CoV-2 N with high affinity (18, 19). Here, we map the epitopes of these sdAbs and show that each sdAb recognizes a specific domain of SARS-CoV-2 N. We report high-resolution crystal structures of two sdAbs bound to the N RBD, and of one sdAb bound to the N CTD. Comparison of the mapped sdAb epitopes with N protein mutations in a set of nearly 500,000 SARS-CoV-2 genomes shows that the sdAbs recognize surfaces that are highly conserved across SARS-CoV-2 isolates and in major variants of concern, suggesting broad utility in recognizing SARS-CoV-2 infections.
Materials and Methods
Cloning and Protein Purification
We previously reported the cloning, expression, and purification of full-length and truncated SARS-CoV-2 N protein (7, 8). Briefly, constructs were amplified by PCR from the IDT 2019-nCoV N positive control plasmid (IDT cat. # 10006625; NCBI RefSeq YP_009724397) and inserted by ligation-independent cloning into UC Berkeley Macrolab vectors 2B-T (AmpR, N-terminal His6-fusion; Addgene #29666) or 2C-T (AmpR, N-terminal His6-MBP fusion; Addgene #29706). Plasmids were transformed into E. coli strain Rosetta 2(DE3) pLysS (Novagen), and grown in the presence of ampicillin and chloramphenicol to an OD600 of 0.8 at 37°C, induced with 0.25 mM IPTG, then grown for a further 16 hours at 18°C prior to harvesting by centrifugation. Harvested cells were resuspended in buffer A (25 mM Tris-HCl pH 7.5, 5 mM MgCl2 10% glycerol, 5 mM β-mercaptoethanol, 1 mM NaN3) plus 500 mM NaCl (for full-length N, 1 M NaCl) and 5 mM imidazole pH 8.0. For purification, cells were lysed by sonication, then clarified lysates were loaded onto a Ni2+ affinity column (Ni-NTA Superflow; Qiagen), washed in buffer A plus 300 mM NaCl and 20 mM imidazole pH 8.0, and eluted in buffer A plus 300 mM NaCl and 400 mM imidazole. For cleavage of His6-tags, proteins were buffer exchanged in centrifugal concentrators (Amicon Ultra, EMD Millipore) to buffer A plus 300 mM NaCl and 20 mM imidazole, then incubated 16 hours at 4°C with TEV protease (20). Cleavage reactions were passed through a Ni2+ affinity column again to remove uncleaved protein, cleaved His6-tags, and His6-tagged TEV protease. Proteins were concentrated in centrifugal concentrators and purified by size-exclusion chromatography (Superdex 200; GE Life Sciences) in gel filtration buffer (25 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 1 mM DTT). Purified proteins were concentrated and stored at 4°C for analysis.
Vectors encoding sdAbs B6, C2, and E2 in pET22b were generously provided by Ellen Goldman (18). These vectors encode each sdAb with a pelB leader sequence at the N-terminus for periplasmic expression, and a His6-tag at the C-terminus for purification (Supplementary Table 1). For sdAb-N3, a codon-optimized gene was synthesized (Integrated DNA Technologies) and inserted into pET22b. All sdAbs were transformed into Rosetta 2(DE3) pLysS, grown in the presence of ampicillin and chloramphenicol to an OD600 of 0.8 at 37°C, induced with 0.5 mM IPTG, then grown for a further 4 hours at 24°C prior to harvesting by centrifugation. Proteins were purified as above using Ni2+ affinity and size exclusion chromatography (Superdex 75, GE Life Sciences), excluding cleavage of the His6-tags.
Nickel Pulldown Assays
For Ni2+ affinity pulldowns, 10 μg sdAb-His6 fusion protein was mixed with 10 μg N protein (FL or truncated) in a total reaction volume of 50 μl, in binding buffer (20 mM HEPES pH 7.5, 200 mM NaCl, 5 mM MgCl2, 5% glycerol, 10 mM Imidazole, and 0.1% NP-40). After a 60 minute incubation at room temperature, 30 μl Ni2+ beads equilibrated in binding buffer (Qiagen Ni-NTA superflow) was added, and the mixture incubated a further 20 minutes at room temperature with rotation. Beads were then washed 3X with 1 mL binding buffer, and bound proteins were eluted by adding 5 μL of 2 M Imidazole plus 20 μL of 2X Laemmli sample buffer. Input (5 μl of the initial reaction) and bound (15 μl of elution fraction) samples were analyzed by SDS-PAGE and Coomassie blue staining.
Crystallography
For crystallization of the N49-174:sdAb-C2 complex, sdAb-C2 and N49-174 were exchanged into crystallization buffer (20 mM HEPES pH 7.0, 200 mM NaCl, 5 mM MgCl2, 1 mM TCEP) and mixed at a 1:1 molar ratio of sdAb to N, for a final protein concentration of 15 mg/ml. Crystals were obtained in sitting drop format by mixing protein 1:1 with well solution containing 0.1 M Tris-HCl pH 8.5, 0.2 M LiSO4, and 20% PEG 4000. Crystals were transferred to a cryoprotectant containing an additional 10% glycerol, then flash-frozen in liquid nitrogen. Diffraction data were collected at Advanced Photon Source, NE-CAT beamline 24ID-E (support statement below). Data was automatically indexed and reduced by the RAPD data-processing pipeline (https://github.com/RAPD/RAPD), which uses XDS (21) for indexing and integration, and the CCP4 programs AIMLESS and TRUNCATE (22) for scaling and structure-factor calculation. The structure was determined by molecular replacement using PHASER (23) with a structure of the SARS-CoV-2 RNA binding domain [PDB ID 7CDZ (24)] and a structure of an alpaca nanobody selected against the Hepatitis C virus E2 glycoprotein [PDB ID 4JVP (25)]. Models were manually rebuilt in COOT (26) and refined in phenix.refine (27) (Supplementary Table 2). Hydrogen atoms were included in the final models for the purposes of robust geometry refinement, and were refined using a riding hydrogen model.
For crystallization of the N49-174:sdAb-B6 complex, sdAb-B6 and N49-174 were exchanged into crystallization buffer (20 mM HEPES pH 7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM TCEP) and mixed at a 1:1 molar ratio of sdAb to N, for a final protein concentration of 15 mg/mL. Crystals were obtained in hanging drop format by mixing protein 1:1 with well solution containing 0.1 M sodium citrate pH 5.6, 0.1 M sodium-potassium tartrate, and 19% PEG 3350. Crystals were transferred to a cryoprotectant containing an additional 10% glycerol, then flash-frozen in liquid nitrogen. Diffraction data were collected at Advanced Photon Source, NE-CAT beamline 24ID-C (support statement below), and data were processed and the structure determined as above. The structure was determined by molecular replacement using PHASER with separate N49-174 and nanobody scaffold chains as above. The model, comprising three identical copies of a 1:1 N49-174:sdAb-B6 complex, was manually rebuilt in COOT and refined with phenix.refine (Supplementary Table 2). Hydrogen atoms were refined using a riding hydrogen model. To maintain stability in the refinement and improve overall geometry, we also applied NCS restraints against equivalent chains [A/B/C (N49-174), and D/E/F (sdAb-B6)].
For crystallization of the N269-364:sdAb-E2 complex, sdAb-E2 and N269-364 were separately subjected to surface lysine demethylation by treatment of the purified protein with borane (50 mM final concentration) and formaldehyde (100 mM) for two hours at 4°C, followed by addition of glycine (25 mM final concentration) to quench the reaction, then buffer-exchange by centrifugal concentration and dilution. Methylated N269-364 and sdAb-E2 were mixed at a ratio of two N269-364 (one dimer) to one sdAb-E2 in crystallization buffer (20 mM HEPES pH 7.0, 200 mM NaCl, 5 mM MgCl2, 1 mM TCEP), according to the inferred ratio of these proteins in copurified samples. Crystals were obtained in hanging drop format by mixing protein 1:1 with well solution containing 0.1 M imidazole pH 8.5, 0.1 M calcium acetate, 12% isopropanol, and 24% PEG 3350. Crystals were transferred to a cryoprotectant containing 10% glycerol, then flash-frozen in liquid nitrogen. Diffraction data were collected at Advanced Light Source, Berkeley Center for Structural Biology beamline 5.0.2 (support statement below), and data were processed by XDS (21) and the CCP4 programs AIMLESS and TRUNCATE (22, 28). The structure was determined by molecular replacement using PHASER with separate N269-364 dimer and nanobody scaffold chains as above. The model, comprising four identical copies of a complex with a dimer of N269-364 bound to a single sdAb-E2, was manually rebuilt in COOT and refined in phenix.refine. Because of strong diffraction anisotropy and translational non-crystallographic symmetry (tNCS), refinement was performed against a pruned dataset with the lowest-intensity ~17% of reflections removed from the dataset by PHASER in NCS mode, and refined only to 2.2 Å resolution as higher resolution bins showed less than 75% completeness after pruning. Since some protomers of N269-364 showed poor electron density in regions distant from the sdAb-E2 interface, we also applied NCS restraints against equivalent chains (A/C/E/G (N269-364), B/D/F/H (N269-364), and I/J/K/L(sdAb-E2)) to maintain stability in the refinement (Supplementary Table 2).
Sequence and Structure Analysis
For examination of N protein variation, a dataset of N protein mutations in 489,605 SARS-CoV-2 genomes was downloaded from the COVID CG project (http://covidcg.org) in February 2021. Highly mutated positions were graphed in Prism v. 9 (Graphpad Software). Mutations in variants of concern were obtained from https://outbreak.info/. Analysis of sdAb-N interfaces was performed with PDBePISA (https://www.ebi.ac.uk/pdbe/pisa/), and graphed alongside N protein variation data.
RNA Binding Assays
RNA binding was performed with 5’-fluorescein (FAM) labeled 7mer single-stranded RNA (5’-CACUGAC-3’) or dsRNA (5’-FAM-CACUGAC-3’ annealed with 5’- GUCAGUG-3’ by slow cooling) in a reaction buffer with 20 mM Tris-HCl pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, and 5% glycerol. RNA at 100 nM was mixed with protein (5, 2.5, 1.25, 0.625, 0.3125, 0.15625, and 0 μM) in a total reaction buffer of 20 μL and incubated 1 hour at 4°C. 10 μL of each reaction was mixed with 2 μL 6X loading dye, and samples were loaded onto a 10% TBE-PAGE gel (pre-run in 0.5X TBE at 200 volts for 1 hour at 4°C), and run at 150 volts for 40 minutes at 4°C. Gels were imaged on a Bio-Rad ChemiDoc XRS+ imager set to detect Cy-2 with a 0.5 second exposure. Bound/unbound fractions were calculated in ImageJ (29) and binding curves were calculated in Prism v. 9 (Graphpad Software) using a cooperative model with Hill coefficient fixed at 5 to stabilize curve-fitting.
Phase Separation Assays
In vitro phase separation assays were performed at 25°C. Unlabeled N protein was mixed with Cy5-labeled protein (linked to an engineered N-terminal cysteine using maleimide linkage) at 1:10 ratio, then mixed with each sdAb at 1:1 ratio. Phase separation of 40 μM N protein (in 5 mM HEPES pH 7.5, 160 mM KCl) was induced by adding to a pre-mixture of three times volumes of UTR265 RNA (13.3 ng/μl in 5 mM HEPES pH 7.5). Samples were mixed in protein LoBind tubes (Eppendorf, 022431064) and then immediately transferred onto 18-well glass-bottom chamber slides (iBidi, 81817). Condensates were imaged after 40 min when the phase separation particles are stable. Images were taken on a CQ1 confocal quantitative image microscope (Yokogawa) with 20x-PH objective. For quantitation, condensates were identified by particle analysis in CQ1 analysis software, then the total fluorescence inside condensates and the total volume of condensates were calculated and divided by the total fluorescence in the field or the total volume of the field to yield a percentage value.
>UTR265 (nt 1-265 of SARS-CoV-2 genome) AUUAAAGGUUUAUACCUUCCCAGGUAACAAACCAACCAACUUUCGAUCUCUUGUAGAUCUGUUCUCUAAACGAACUUU
AAAAUCUGUGUGGCUGUCACUCGGCUGCAUGCUUAGUGCACUCACGCAGUAUAAUUAAUAACUAAUUACUGUCGUUGA
CAGGACACGAGUAACUCGUCUAUCUUCUGCAGGCUGCUUACGGUUUCGUCCGUGUUGCAGCCGAUCAUCAGCACAUCU
AGGUUUCGUCCGGGUGUGACCGAAAGGUAAG.
Beamline Support Statements
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines at the Advanced Photon Source, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on 24-ID-E is funded by a NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
The Berkeley Center for Structural Biology is supported in part by the Howard Hughes Medical Institute. The Advanced Light Source is a Department of Energy Office of Science User Facility under Contract No. DE-AC02-05CH11231. The ALS-ENABLE beamlines are supported in part by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169.
Results
sdAbs Recognize Distinct Domains of the SARS-CoV-2 N Protein
Two recent studies reported the isolation of single-domain antibodies (sdAbs) that recognize the SARS-CoV-2 N protein (Figures 1A, B) (18, 19). One study used an in vitro selection method to isolate one sdAb (here termed sdAb-N3; Supplementary Table 1) that recognizes the N protein with high affinity (EC50 ~50 nM as measured by luciferase-based ELISA assay) (19). The second study isolated three sdAbs (here termed sdAb-B6, sdAb-C2, and sdAb-E2; Supplementary Table 1) from an immunized llama, which each recognize the N protein with extremely high affinity (Kd of 0.8-1.6 nM as measured by surface plasmon resonance) (18). The four sdAbs show no obvious similarity in their three variable complementarity-determining regions (CDRs; Figures 1B, C), suggesting that they likely each recognize distinct epitopes on the N protein.
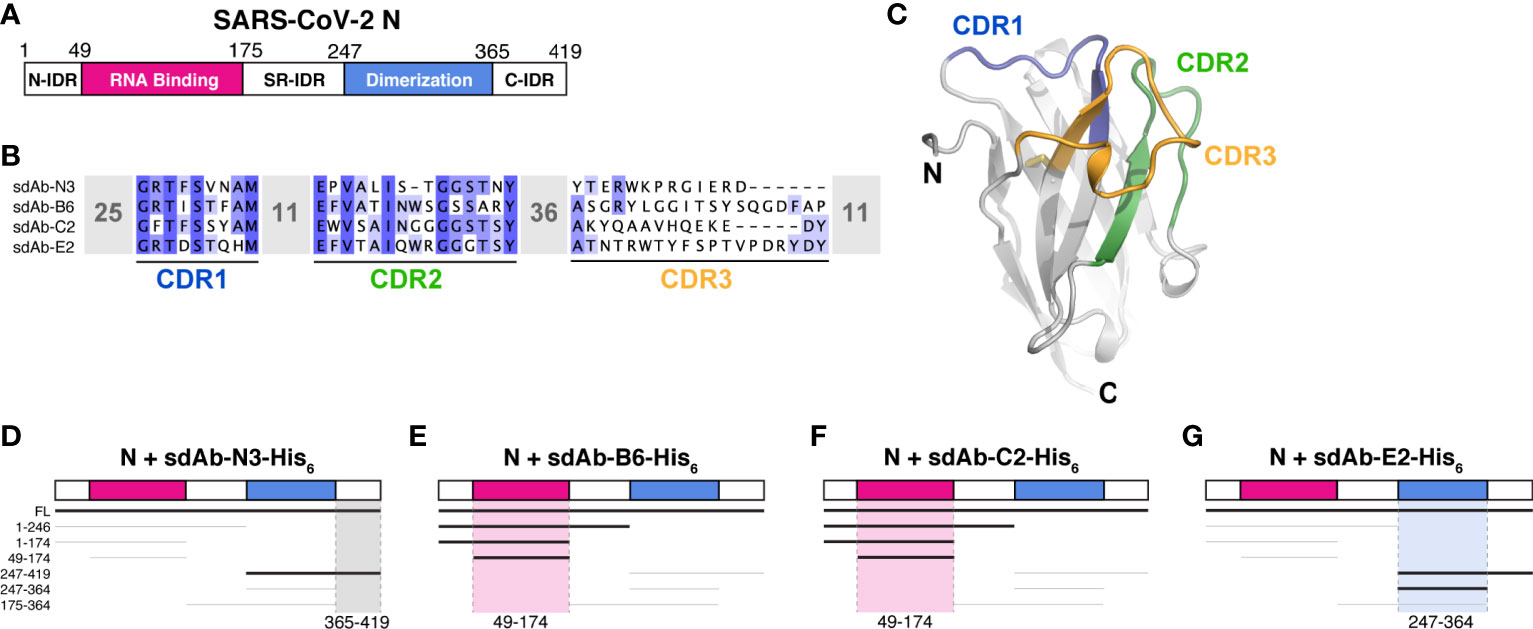
Figure 1 sdAbs target individual domains of the SARS-CoV-2 N protein. (A) Domain schematic of SARS-CoV-2 N, with the RNA-binding domain in magenta, the dimerization domain in blue, and the three intrinsically disordered domains (N-IDR, serine/arginine rich (SR)-IDR, and C-IDR) in white. (B) Sequences of four sdAbs targeting the SARS-CoV-2 N protein (18, 19) showing the variable sequences within the Complementarity-Determining Regions (CDRs). (C) Example sdAb structure (sdAb-B6 shown) with complementarity-determining regions (CDR) 1, 2, and 3 colored blue, green, and orange, respectively. (D–G) Summary of Ni2+ pulldown assays using C-terminally His6-tagged sdAbs and seven truncations of SARS-CoV-2 N (see Supplementary Figure 1). Thick lines indicate a positive interaction; highlighted regions indicate inferred region of SARS-CoV-2 N targeted by each sdAb.
The SARS-CoV-2 N protein possesses a modular structure with an N-terminal RNA-binding domain (RBD) and a C-terminal dimerization domain (CTD), plus three intrinsically disordered regions at the N- and C-termini (N-IDR and C-IDR, respectively) and in the serine/arginine region separating the RBD and CTD (SR-IDR; Figure 1A). We and others have previously shown that the N protein oligomerizes in vitro through its CTD and disordered C-terminal tail (7), and that the protein undergoes liquid-liquid phase separation with RNA mediated by its RBD and central disordered region (8–14). To determine which domain(s) of N are recognized by each sdAb, we performed direct Ni2+ affinity pulldown assays using His6-tagged sdAbs and seven N protein truncations spanning its five distinct domains (Figures 1D–G and Supplementary Figure 1). As expected, we found that all four sdAbs robustly bind full-length N protein. We could further narrow the binding region of each sdAb to a defined subdomain: sdAb-B6 and sdAb-C2 each recognize the N protein RBD (residues 49-174), while sdAb-E2 recognizes the CTD (residues 247-364) and sdAb-N3 recognizes the C-IDR (residues 365-419; Figures 1D–G and Supplementary Figure 1).
Structures of sdAb-N Protein Complexes
We next sought to determine high-resolution structures of N:sdAb complexes. We first co-crystallized sdAbs C2 and B6 with the N protein RNA-binding domain (residues 49-174; referred to hereafter as NRBD), and determined structures of these complexes to 1.42 Å and 2.56 Å resolution, respectively (Supplementary Table 2). The structures reveal that the two sdAbs recognize distinct surfaces on the N protein RBD, away from the RNA binding surface. sdAb-C2 recognizes a concave surface on the N protein RBD, burying 765 Å2 of surface on each partner (Figures 2B, C). The binding interface is made up mostly of the CDR2 (40% of the interface) and CDR3 (53%) loops, with only a minor contribution from CDR1 (7%). The bulk of the interface is hydrophobic, with sdAb-C2 residues A33, W47, A50, Y99, A101, and V103 docking against the NRBD surface. Specific hydrogen bonds are made by sdAb-C2 residues S57 and Q105.
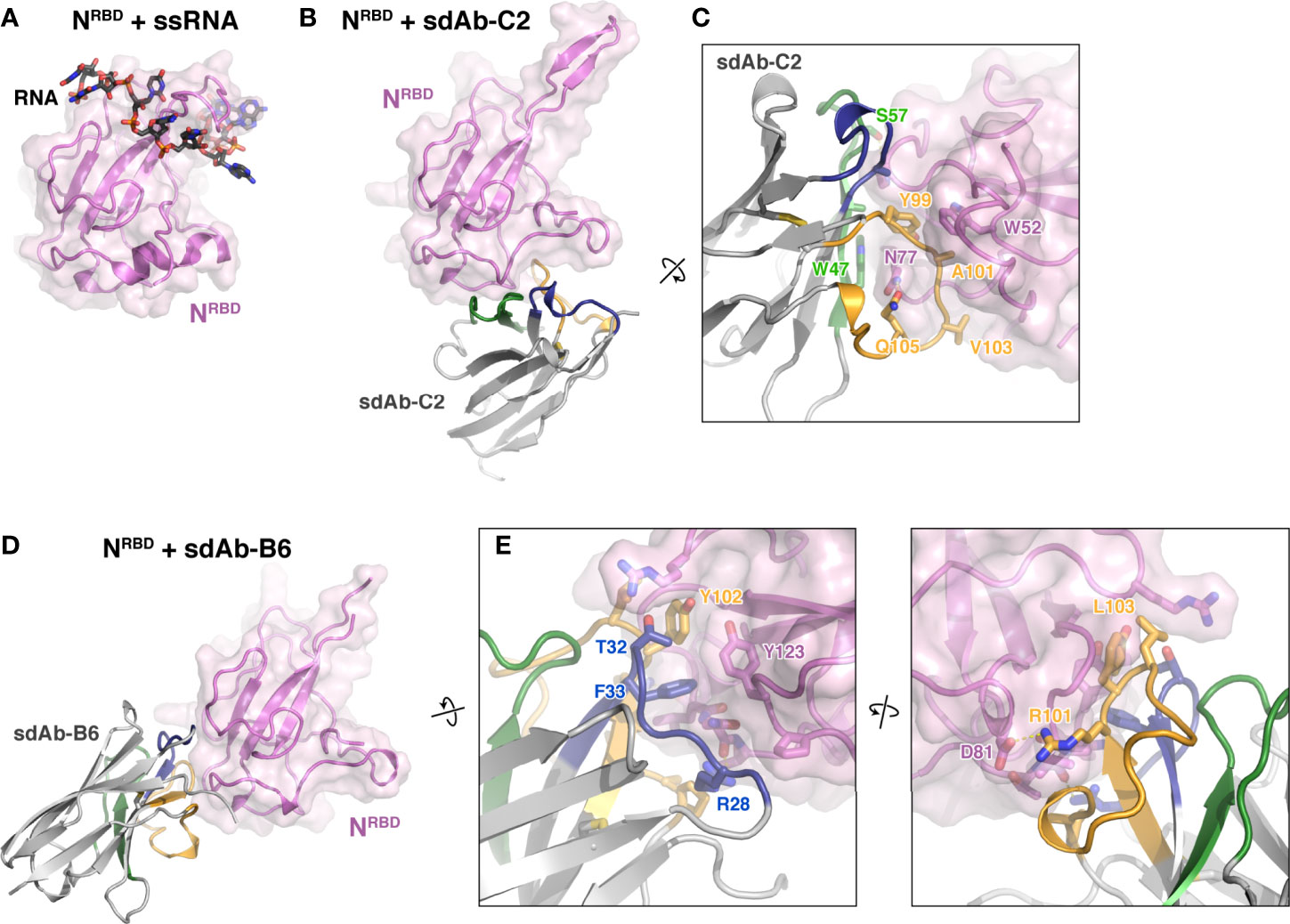
Figure 2 Structures of two sdAbs targeting the SARS-CoV-2 N protein RBD. (A) Structure of the SARS-CoV-2 NRBD (pink) bound to single-stranded RNA (black) [PDB ID 7ACT (30)]. (B, D) show the NRBD in the same orientation. (B) Structure of NRBD (pink) bound to sdAb-C2. CDR1, CDR2, and CDR3 are colored blue, green, and orange as in Figure 1C. (C) Close-up view of the NRBD:sdAb-C2 complex. (D) Structure of NRBD (pink) bound to sdAb-B6. CDR1, CDR2, and CDR3 are colored blue, green, and orange as in Figure 1C. (E) Two close-up views of the NRBD:sdAb-B6 complex.
sdAb-B6 binds a separate surface on NRBD, burying 617 Å2 of surface area on each partner (Figures 2D, E). The interface of sdAb-B6 with N is made up mostly of CDR1 (34% of the interface) and CDR3 (60%), with only a minor contribution from CDR2 (4%). At the core of the interface is a pair of aromatic side-chains from sdAb-B6, F33 and Y102, which together dock into a hydrophobic surface on NRBD. Two arginine residues, R28 and R101, also make specific hydrogen bonds with NRBD.
Next, we co-crystallized sdAb-E2 with the N protein CTD. Initial crystals with the full dimerization domain (residues 247-364) failed to diffract x-rays, so we instead co-crystallized sdAb-E2 with a truncated construct lacking 22 N-terminal residues (residues 269-364; referred to hereafter as NCTD). We determined the structure of the NCTD:sdAb-E2 complex to 2.2 Å resolution in a crystal form with four independent copies of the NCTD dimer, with each dimer bound to a single sdAb-E2 (Figure 3A). All four copies show an identical interface, with sdAb-E2 binding across the flat β-sheet making up the NCTD dimer interface. The interface buries a total of 710 Å2 of surface area on each partner, with CDR2 and CDR3 making up nearly the entire interface (33% and 64% of the interface, respectively). CDR1 structurally stabilizes CDR2 and CDR3, but makes no direct contact with the N protein (Figure 3B). The majority of the interface involves three aromatic residues on CDR3 – W102, Y104, and F105 – with W102 and Y104 laying nearly flat against the NCTD β-sheet. sdAb-E2 also makes specific hydrogen bonds with Q281 of both NCTD protomers, with one Q281 residue making a salt-bridge interaction with both the main chain and side chain of sdAb-E2 S106, and the other Q281 hydrogen bonding sdAb-E2 residue R54 (Figure 3B).
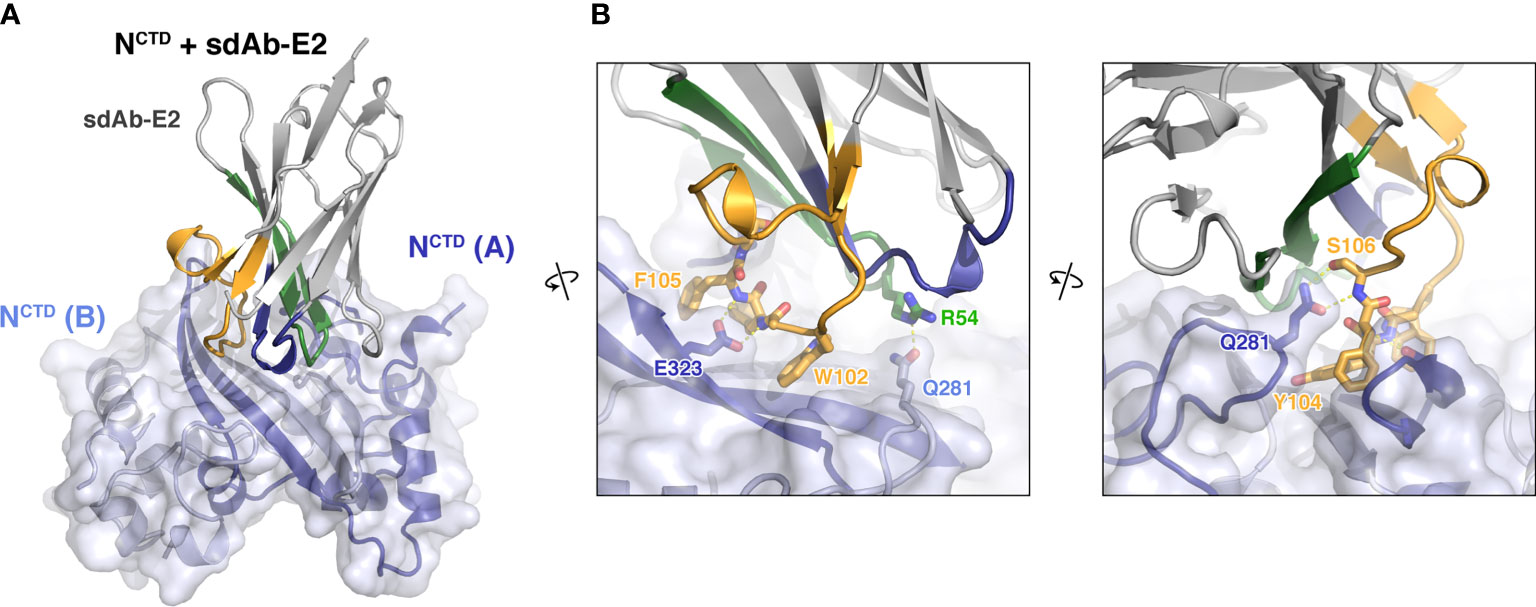
Figure 3 Structure of an sdAb targeting the SARS-CoV-2 N protein CTD. (A) Structure of sdAb-E2 (with CDR1, CDR2, and CDR3 colored blue, green, and orange as in Figure 1C) bound to the NCTD dimer (light blue/dark blue). (B) Two close-up views of the NCTD:sdAb-E2 complex.
Variability of sdAb-Recognition Surfaces on SARS-CoV-2 N
To date, more than 500,000 individual isolates of SARS-CoV-2 have been fully sequenced, enabling a comprehensive examination of variability across the surface of each of its proteins. The sequences of many variants of concern, many of which have higher infectivity than the ancestral SARS-CoV-2 strain, are also known. To determine the likely effectiveness of each sdAb in recognizing variants of concern and the broader population of SARS-CoV-2 variants, we compared the epitopes of each sdAb with a dataset of variants in 489,605 SARS-CoV-2 sequences from the GISAID Initiative (http://www.gisaid.org) and the COVID CG project (http://covidcg.org). These data reveal that the proteins’ three intrinsically disordered regions are mutated more than 20-fold more often than the ordered NRBD or NCTD domains (0.012 mutations/residue/isolate for intrinsically disordered regions vs. 0.0005 mutations/residue/isolate for the ordered domains; Supplementary Figure 2A and Supplementary Table 3), demonstrating strong selective pressure to maintain these domains’ sequence and structure. We also examined N protein mutations in the major SARS-CoV-2 variants of concern worldwide. As with the larger dataset, mutations in these variants strongly cluster in the central intrinsically disordered region, with a few mutations in the NRBD domain (Supplementary Figure 2A).
We compared the epitopes of sdAbs B6, C2, and E2 to the data on N protein variation, and found that the three sdAbs largely bind highly conserved surfaces (Supplementary Figures 2B, C). For sdAb-B6, the most highly mutated residues in its epitope are NRBD residues T135, L139, and T141, which are mutated in only 1372 (0.28%), 976 (0.19%), and 402 (0.08%) of the 489,605 SARS-CoV-2 genome samples, respectively. The sdAb-C2 epitope overlaps two residues on NRBD, D144 and H145, which are mutated in 575 (0.12%) and 2010 (0.41%) SARS-CoV-2 genome samples, respectively. Both sdAb-B6 and sdAb-C2 contact residue P80, which is mutated to arginine in variant P1. This residue is at the edge of both interfaces, however, and its mutation is unlikely to affect NRBD recognition by either sdAb.
On the NCTD, sdAb-E2 contacts three residues – T325, S327, and T334 – that are mutated in 373 (0.08%), 511 (0.10%), and 990 (0.20%) SARS-CoV-2 genome samples, respectively. No major variants of concern show mutations in the NCTD, illustrating the high conservation of this domain and the potential utility of sdAb-E2 for robust N protein recognition. Overall, this analysis suggests that all three sdAbs will recognize the vast majority of SARS-CoV-2 N proteins in active infections, including those encoded by the major variants of concern worldwide. Given the higher variability of the IDR regions compared to the RBD or CTD, sdAb-N3 (which binds the C-IDR) may be less broadly effective at recognizing N protein variants than the three other sdAbs.
sdAb Binding Compromises RNA Binding and Phase Separation of SARS-CoV-2 N
The multiple roles of SARS-CoV-2 N in the viral life cycle rely on its ability to bind RNA and self-assemble through liquid-liquid phase separation. We tested whether addition of sdAbs would affect these biochemical properties, in the context of the full-length N protein. We first tested RNA binding, using electrophoretic mobility shift assays (EMSAs) with short single-stranded or double-stranded RNAs. Structures of the N protein RBD were recently determined by NMR in complex with either single- and double-stranded RNA, revealing distinct binding modes on the same positively-charged surface of the protein (Figure 2A) (30). Using short 7-base single- or double-stranded RNAs also used in this prior study, we performed EMSAs using full-length N protein in the presence or absence of sdAbs B6 and C2. Despite the use of short RNAs that each can only bind a single N protomer, we found that all binding curves are best fit using a cooperative binding model with Hill coefficient ~5, hinting that binding of even short RNAs is intimately linked with self-assembly of the N protein (Figures 4A, B). For full-length N, we measured the binding affinity for ssRNA at 0.56 +/- 0.06 μM, and for dsRNA at 0.48 +/- 0.03 μM, comparable to the affinity measured previously in low-salt buffer (30). Instead of showing a simple shift from an unbound to a bound band as expected for a 1:1 complex, our gels clearly showed a progressive shift toward the well of the gel at higher N protein concentration, consistent with higher-order assembly of the N-RNA complexes (Supplementary Figure 3). We next added equimolar amounts of either sdAb-B6 or sdAb-C2, both of which bind the N protein RBD. Despite neither sdAb’s epitope overlapping the RNA binding site directly, both sdAbs slightly reduced the affinity for ssRNA (to 0.70 +/- 0.04 and 0.63 +/- 0.04 μM, respectively, for sdAb-B6 and sdAb-C2) (Figure 4A). Curiously, while sdAb-C2 had no effect on dsRNA binding (Kd=0.49 +/- 0.06 μM), sdAb-B6 significantly reduced dsRNA binding (Kd=0.82 +/- 0.11 μM) (Figure 4B). Both sdAbs B6 and C2 significantly reduced the progressive band-shift observed with wild-type N protein, especially for the dsRNA binding assays (Supplementary Figure 3). This observation suggests that the two sdAbs affect the N protein-RNA interaction not by interfering with RNA binding of a single N protein protomer, but rather by compromising large-scale self-assembly of N.
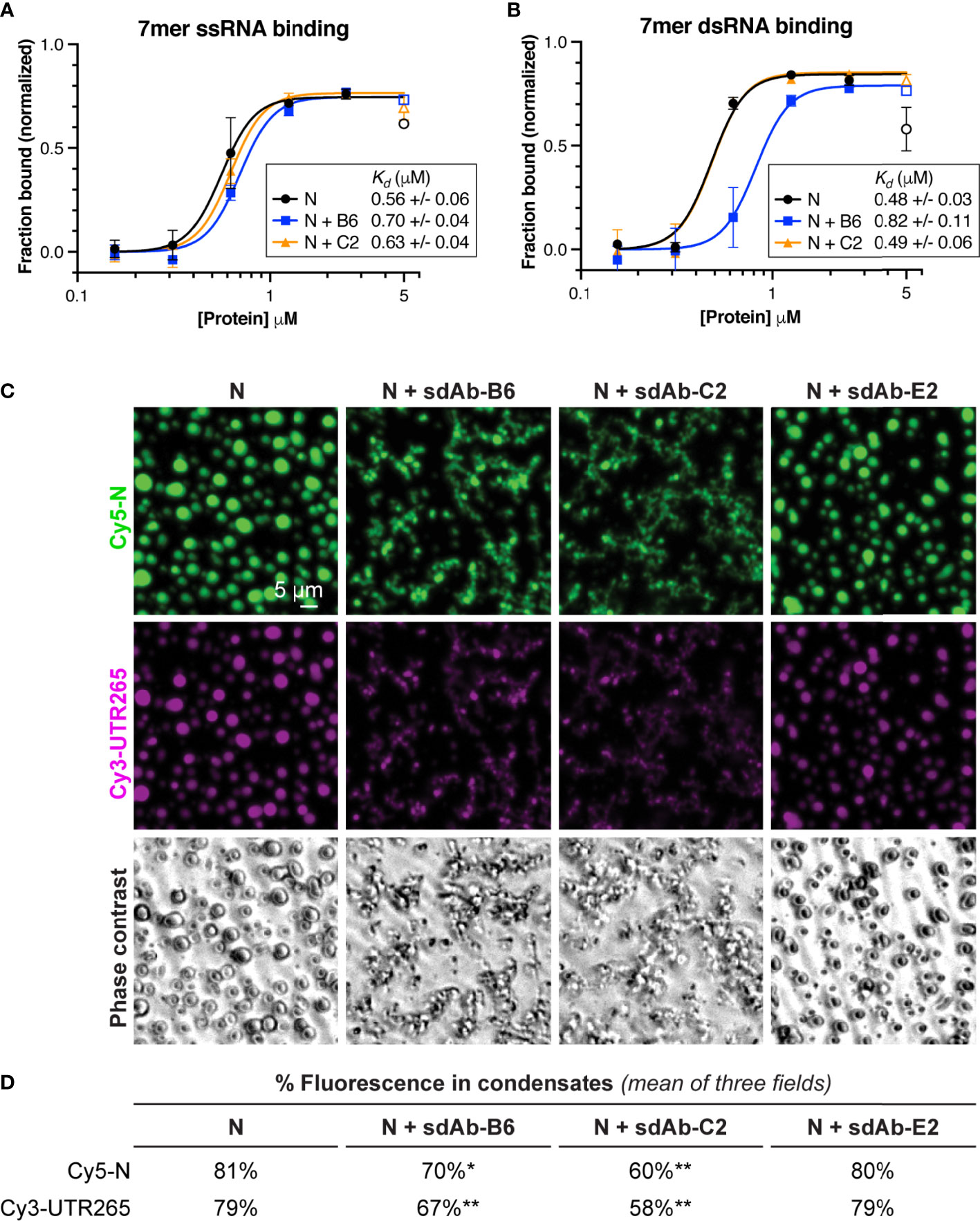
Figure 4 RBD-targeting sdAbs targeting affect RNA binding affinity and RNA-mediated phase separation of SARS-CoV-2 N. (A) Binding curves for SARS-CoV-2 N (full-length protein) binding to a 7-mer single-stranded RNA, in the absence of added sdAbs (black points) or in the presence of equimolar sdAb-B6 (blue points) or sdAb-C2 (orange points). See Supplementary Figure 3A for example electrophoretic mobility shift (EMSA) gels. (B) Binding curves for SARS-CoV-2 N binding to a 7-mer double-stranded RNA, in the absence of added sdAbs (black points) or in the presence of equimolar sdAb-B6 (blue points) or sdAb-C2 (orange points). See Supplementary Figure 3C for example electrophoretic mobility shift (EMSA) gels. (C) In vitro phase separation of Cy5-labeled N (green) and Cy3-labeled UTR265 RNA (magenta), in the absence or presence of equimolar sdAb-B6, sdAb-C2, or sdAb-E2. (D) Quantitation of fluorescence (protein and RNA) in condensates. All measurements represent the mean of three separate microscope fields. Asterisks indicate P-values from unpaired t-tests: *P < 0.05, **P < 0.01.
Our RNA binding results suggested that sdAbs may compromise self-assembly of N into large-scale oligomers, including the condensates that we and others have observed both in vitro and in cells. To directly test whether sdAbs compromise LLPS of the N protein, we mixed Cy5-labeled full-length N with a 265-base RNA derived from the 5′ UTR of SARS-CoV-2 (UTR265), and directly observed the formation of phase-separated condensates using light microscopy (Figures 4C, D). We observed robust assembly of N+RNA condensates that was significantly compromised by addition of equimolar amounts of either sdAb-B6 or sdAb-C2 (Figure 4D). In addition to reducing the overall amount of LLPS, both sdAbs altered the morphology of the observed condensates, from round droplets to more extended, fibrous structures (Figure 4C). Addition of sdAb-E2, which binds the CTD, did not strongly affect formation of N+RNA condensates.
Discussion
Here, we present high-resolution structures of three single-domain antibodies bound to defined domains of the SARS-CoV-2 N protein. Two sdAbs bind the N-terminal RNA binding domain, and one binds the C-terminal dimerization domain. All three sdAbs recognize highly conserved surfaces on the N protein, suggesting that they could be used to recognize SARS-CoV-2 infections in antigen tests. As SARS-CoV-2 transitions from an acute public health threat to an endemic phase, the ability to robustly detect and respond to infections and local outbreaks will remain important, especially in the developing world. Development of antigen tests with the sdAbs characterized here, which can be produced at high levels in E. coli and recognize the highly-abundant N protein with high specificity and affinity, can significantly aid this effort.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: RCSB Protein Data Bank (https://www.rcsb.org), accession numbers 7N0I, 7N0R, and 7R98.
Author Contributions
QY and KC conceived of the study. QY performed protein purification, determined crystal structures, and performed RNA binding assays. SL performed phase separation assays. KC wrote the paper and prepared the figures with advice from QY and SL. All authors contributed to the article and approved the submitted version.
Funding
KC acknowledges support from UC San Diego and the National Institutes of Health (R01GM128464).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Ellen Goldman for the kind gift of vectors encoding sdAb-B6, sdAb-C2, and sdAb-E2; Stacey Ortega and Marc Allaire at Advanced Light Source beamline 5.0.2; the staff of Advanced Light Source NE-CAT beamlines 24ID-C and 24ID-E; Haiyang Yu for helpful experimental suggestions, and members of the Corbett lab for critical feedback.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.719037/full#supplementary-material
References
1. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New Engl J Med (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
2. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. New Engl J Med (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
3. Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, de Groot AM, et al. Interim Results of a Phase 1–2a Trial of Ad26.COV2.S Covid-19 Vaccine. New Engl J Med (2021) 384:1824–35. doi: 10.1056/NEJMoa2034201
4. Sethuraman N, Jeremiah SS, Ryo A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA (2020) 323:2249–51. doi: 10.1001/jama.2020.8259
5. Guglielmi G. Fast Coronavirus Tests: What They Can and Can’t Do. Nature (2020) 585:496–8. doi: 10.1038/d41586-020-02661-2
6. Che XY, Hao W, Wang Y, Di B, Yin K, Xu YC, et al. Nucleocapsid Protein as Early Diagnostic Marker for SARS. Emerg Infect Dis (2004) 10:1947–9. doi: 10.3201/eid1011.040516
7. Ye Q, West AMV, Silletti S, Corbett KD. Architecture and Self-Assembly of the SARS-CoV-2 Nucleocapsid Protein. Protein Sci (2020) 29:1890–901. doi: 10.1002/pro.3909
8. Lu S, Ye Q, Singh D, Villa E, Cleveland DW, Corbett KD. The SARS-CoV-2 Nucleocapsid Phosphoprotein Forms Mutually Exclusive Condensates With RNA and the Membrane-Associated M Protein. Nat Commun (2021) 12:502. doi: 10.1038/s41467-020-20768-y
9. Iserman C, Roden CA, Boerneke MA, Sealfon RSG, McLaughlin GA, Jungreis I, et al. Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. Mol Cell (2020) 80:1078–91.e6. doi: 10.1016/j.molcel.2020.11.041
10. Savastano A, Ibáñez de Opakua A, Rankovic M, Zweckstetter M. Nucleocapsid Protein of SARS-CoV-2 Phase Separates Into RNA-Rich Polymerase-Containing Condensates. Nat Commun (2020) 11:6041. doi: 10.1038/s41467-020-19843-1
11. Carlson CR, Asfaha JB, Ghent CM, Howard CJ, Hartooni N, Safari M, et al. Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for Its Dual Functions. Mol Cell (2020) 80:1092–103.e4. doi: 10.1016/j.molcel.2020.11.025
12. Luo L, Li Z, Zhao T, Ju X, Ma P, Jin B, et al. SARS-CoV-2 Nucleocapsid Protein Phase Separates With G3BPs to Disassemble Stress Granules and Facilitate Viral Production. Sci Bull (2021) 66:1194–204. doi: 10.1016/j.scib.2021.01.013
13. Perdikari TM, Murthy AC, Ryan VH, Watters S, Naik MT, Fawzi NL. SARS-CoV-2 Nucleocapsid Protein Phase-Separates With RNA and With Human hnRNPs. EMBO J (2020) 39:e106478. doi: 10.15252/embj.2020106478
14. Cubuk J, Alston JJ, Jeremías Incicco J, Singh S, Stuchell-Brereton MD, Ward MD, et al. The SARS-CoV-2 Nucleocapsid Protein Is Dynamic, Disordered, and Phase Separates With RNA. Nat Commun (2021) 12:1936. doi: 10.1038/s41467-021-21953-3
15. Nabeel-Shah S, Lee H, Ahmed N, Marcon E, Farhangmehr S, Pu S, et al. SARS-CoV-2 Nucleocapsid Protein Attenuates Stress Granule Formation and Alters Gene Expression via Direct Interaction With Host mRNAs. bioRxiv (2020). 2020.10.23.342113. doi: 10.1101/2020.10.23.342113
16. Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, et al. SARS-CoV-2 Structure and Replication Characterized by In Situ Cryo-Electron Tomography. Nat Commun (2020) 11:5885. doi: 10.1038/s41467-020-19619-7
17. Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell (2020) 183:730–738.e13. doi: 10.1016/j.cell.2020.09.018
18. Anderson GP, Lui JL, Esparza TJ, Voelker BT, Hofmann ER, Goldman ER. Single Domain Antibodies for the Detection of SARS-CoV-2 Nucleocapsid Protein. Anal Chem (2021) 93:7283–91. doi: 10.1021/acs.analchem.1c00677
19. Sherwood LJ, Hayhurst A. Toolkit for Quickly Generating and Characterizing Molecular Probes Specific for SARS-CoV-2 Nucleocapsid as a Primer for Future Coronavirus Pandemic Preparedness. ACS Synthetic Biol (2021) 10:379–90. doi: 10.1021/acssynbio.0c00566
20. Tropea JE, Cherry S, Waugh DS. Expression and Purification of Soluble His(6)-Tagged TEV Protease. Methods Mol Biol (Clifton NJ) (2009) 498:297–307. doi: 10.1007/978-1-59745-196-3_19
21. Kabsch W. XDS. Acta Crystallographica Section D. Biol Crystallogr (2010) 66:125–32. doi: 10.1107/S0907444909047337
22. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr Section D Biol Crystallogr (2011) 67:235–42. doi: 10.1107/S0907444910045749
23. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser Crystallographic Software. J Appl Crystallogr (2007) 40:658–74. doi: 10.1107/S0021889807021206
24. Peng Y, Du N, Lei Y, Dorje S, Qi J, Luo T, et al. Structures of the SARS-CoV-2 Nucleocapsid and Their Perspectives for Drug Design. EMBO J (2020) 39:e105938. doi: 10.15252/embj.2020105938
25. Tarr AW, Lafaye P, Meredith L, Damier-Piolle L, Urbanowicz RA, Meola A, et al. An Alpaca Nanobody Inhibits Hepatitis C Virus Entry and Cell-to-Cell Transmission. Hepatology (2013) 58:932–9. doi: 10.1002/hep.26430
26. Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and Development of Coot. Acta Crystallogr Section D Biol Crystallogr (2010) 66:486–501. doi: 10.1107/S0907444910007493
27. Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards Automated Crystallographic Structure Refinement With Phenix.Refine. Acta Crystallogr Section D Biol Crystallogr (2012) 68:352–67. doi: 10.1107/S0907444912001308
28. Evans PR, Murshudov GN. How Good Are My Data and What Is the Resolution? Acta Crystallogr Section D Biol Crystallogr (2013) 69:1204–14. doi: 10.1107/S0907444913000061
29. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 Years of Image Analysis. Nat Methods (2012) 9:671–5. doi: 10.1038/nmeth.2089
Keywords: SARS-CoV-2, nanobodies, Nucleocapsid (N) protein, X-ray crystallography, RNA binding, phase separation
Citation: Ye Q, Lu S and Corbett KD (2021) Structural Basis for SARS-CoV-2 Nucleocapsid Protein Recognition by Single-Domain Antibodies. Front. Immunol. 12:719037. doi: 10.3389/fimmu.2021.719037
Received: 01 June 2021; Accepted: 29 June 2021;
Published: 26 July 2021.
Edited by:
Goran Bajic, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Patrice Gouet, Université Claude Bernard Lyon 1, FranceMickael Blaise, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2021 Ye, Lu and Corbett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin D. Corbett, a2NvcmJldHRAdWNzZC5lZHU=
 Qiaozhen Ye
Qiaozhen Ye Shan Lu1
Shan Lu1 Kevin D. Corbett
Kevin D. Corbett