- 1Department of Immunology, Leiden University Medical Center, Leiden, Netherlands
- 2Department of Internal Medicine (Nephrology), Leiden University Medical Center, Leiden, Netherlands
- 3Department of Internal Medicine, Erasmus Medical Center Transplantation Institute, University Medical Center Rotterdam, Rotterdam, Netherlands
- 4Eurotransplant Reference Laboratory, Leiden, Netherlands
Matching strategies based on HLA eplets instead of HLA antigens in solid organ transplantation may not only increase the donor pool for highly sensitized patients, but also decrease the incidence of de novo donor-specific antibody formation. However, since not all eplets are equally capable of inducing an immune response, antibody verification is needed to confirm their ability to be bound by antibodies, such that only clinically relevant eplets are considered. The HLA Epitope Registry has documented all theoretically defined HLA eplets along with their antibody verification status and has been the foundation for many clinical studies investigating eplet mismatch in transplantation. The verification methods for eplets in the Registry range from polyclonal sera from multi- and uni-parous women to murine and human monoclonal antibodies (mAbs), and antibodies purified by adsorption and elution from sera of HLA immunized individuals. The classification of antibody verification based on different methods for validation is problematic, since not all approaches represent the same level of evidence. In this study, we introduce a classification system to evaluate the level of evidence for the antibody-verified status of all eplets in the HLA Epitope Registry. We demonstrate that for a considerable number of eplets, the antibody-verified status is solely based on polyclonal serum reactivity of multiparous women or on reactivity of murine mAbs. Furthermore, we noted that a substantial proportion of patient sera analyses and human mAb data presented in the HLA Epitope Registry Database has never been published in a peer-reviewed journal. Therefore, we tested several unpublished human HLA-specific mAbs by luminex single antigen beads assay to analyze their HLA reactivity for eplet antibody verification. Although the majority of analyzed mAbs indeed verified their assigned eplets, this was not the case for a number of eplets. This comprehensive overview of evidence for antibody verification of eplets in the HLA Epitope Registry is instrumental for future investigations towards eplet immunogenicity and clinical studies considering antibody-verified eplet mismatch in transplantation and warrants further standardization of antibody verification using high quality data.
Introduction
Donor-specific antibodies (DSA) are formed against mismatched polymorphic amino acid residues on donor human leukocyte antigens (HLA) and are a major complication in renal transplantation, leading to chronic rejection and graft loss (1, 2). HLA eplets are small configurations of surface-exposed amino acids within a 3-3.5 Ångstrom (Å) radius (3, 4) and resemble the functional epitope, which generally determines the specificity of the antibody through interaction with the complementarity-determining region 3 (CDR3) of the heavy chain of the antibody (5–7). Consideration of HLA eplets instead of HLA antigens may not only refine HLA matching strategies, resulting in decreased DSA formation, but also expand the donor pool for highly sensitized patients and facilitate personalized immunosuppressive treatment based on immunological risk evaluation (8). Indeed, several studies have shown that eplet mismatches are correlated with DSA formation, graft rejection and graft loss (9–14). However, as eplets have been theoretically defined, their clinical relevance needs to be validated by antibody verification (8, 15). Although antibody-verified eplet mismatches have been demonstrated to correlate with DSA formation and graft survival (13, 14), recent reports also indicated that there are still clinically relevant eplets which have not been antibody-verified yet (13, 16).
The HLA Epitope Registry is an online database founded under auspices of the 16th International HLA and Immunogenetics Workshop in 2012, which has documented all theoretically defined eplets, as well as their antibody verification status (17) with the aim to reflect the eplet repertoire incorporated in the widely used HLAMatchmaker software. The Registry has formed a pivotal source of information on eplets that have been defined on HLA as well as on MICA (Human Major Histocompatibility Complex Class I Chain-Related gene A) and has been of great benefit to the field of histocompatibility. Antibody-verified eplets in the Registry have been verified by analyzing reactivity patterns of either polyclonal sera from multi- or uni-parous women, murine monoclonal antibodies (mAb), human mAbs, or antibodies purified by adsorption and elution from sera of HLA immunized individuals (18, 19). The use of different methods for eplet verification is problematic because not all approaches represent the same level of evidence. In most, if not all cases, reactivity of polyclonal sera in luminex single antigen bead (SAB) assays cannot be attributed to a monoclonal response directed against a single eplet, and even adsorption and elution of antibodies from patient sera does not guarantee that the SAB reactivity is caused by antibody reactivity against a single eplet. Also the notion that purification of IgG may reveal ‘‘natural’’ (non-pathogenic) anti-HLA antibodies need to be considered when eluted antibodies are analyzed (20, 21). Additionally, the use of murine mAbs does not provide sufficient evidence for immunogenicity in the human setting, since the immunogenicity of mismatched HLA antigens is affected by the recipient’s HLA type (22). Consequently, murine mAbs may recognize different HLA epitopes than human antibodies. Therefore, we consider the use of human HLA-specific mAbs as the highest level of evidence for eplet antibody verification. In previous versions of the Registry, a subcategory of provisionally antibody-verified eplets was present, which unfortunately was discontinued. Such category is useful for data that hint towards true eplet-antibody interaction, but that are not strong enough for actual antibody verification.
The disparity in the level of evidence of antibody verification hampers the clinical application of evidence-based eplet matching and is not only caused by the different methods of antibody verification, but also by the incorporation of unpublished data in the HLA Epitope Registry, as opposed to experimental evidence from peer-reviewed literature. In this paper, we establish a comprehensive overview of the evidence for the antibody verification status of eplets included in the Registry by evaluating the level of evidence of different experimental methods using a classification system. Furthermore, we show previously unpublished SAB analyses of a number of human HLA-specific mAbs that are included in the Registry. We demonstrate that antibody-verified status of 45% of the eplets is based on analysis of polyclonal sera, murine mAbs or experiments with low resolution HLA typed cells and that several human mAbs have been wrongfully attributed to the verification of certain eplets. Our results illustrate the heterogenous and occasionally nontransparent methods of antibody verification and stress the importance of standardization of experimental procedures for antibody verification of HLA eplets.
Materials and Methods
Review of References in the HLA Epitope Registry Databases
HLA Epitope Registry HLA-ABC, HLA-DRB, HLA-DQ and HLA-DP databases were accessed on http://www.EpRegistry.com.br on 28 January 2021. All literature references for antibody-verified eplets present in these databases were reviewed for their level of evidence according to Table 1. Eplets with one or more references of A1 or A2 level were considered truly antibody-verified. Eplets with level B, C or D were classified as provisionally antibody-verified, a category of verification that was present in first report of the HLA Epitope Registry (18), but has been removed since the second update (23). Human mAb data presented in the database that had not been published in a peer-reviewed journal were considered as not sufficient for antibody verification. In order to provide a thorough overview of the antibody verification status of eplets, recent papers that provide evidence for antibody verification and which were not included in the HLA Epitope Registry at the moment of data extraction were also evaluated for their level of evidence.
HLA-Specific Human Monoclonal Antibodies
For a number of eplets, human mAb data presented in the Registry has not been published in a peer-reviewed journal. Therefore, these human mAbs which were previously produced by cloned B cell hetero-hybridomas derived from pregnancy immunized individuals (24–28), were tested in luminex SAB assay and subsequently analyzed for their HLA-specificity. IgG human mAbs were tested in the Lifecodes HLA class I or HLA class II SAB assay (Immucor, Stamford, CT, USA) according to the manufacturer’s instructions. For IgM mAbs, the PE-conjugated goat anti-human IgG was replaced with a PE-conjugated anti-human IgM detection antibody (One Lambda, Canoga Park, CA, USA) used in 1:100 dilution. All mAbs were tested at a concentration of 10 ug/ml (29), unless the neat sample concentration was below 10 ug/ml. Supplementary Table 1 lists the alleles present in the SAB panel that was used. HLA antibody data were analyzed with Match It! Antibody software version 1.3.0 (Immucor). Results were expressed as background-corrected mean fluorescence intensity (MFI). Bead-specific cut-off based on raw MFI/lowest ranked antigen (LRA) (MFI/LRA) in combination with raw MFI >750 was utilized to assign positive beads. For some mAbs, the reactivity pattern was corroborated by testing with One Lambda SAB assay (LABscreen, One Lambda, Canoga Park, CA, USA).
Lymphocytotoxicity
Lymphocytotoxicity data for mAbs VDK1D12, VN2F1, DMS4G2 and SN66E3 were obtained from the 13th International HLA and Immunogenetics Workshop. In this project, a panel of more than 800 second-field HLA-typed cells were tested in complement dependent cytotoxicity (CDC) assays in twelve laboratories worldwide (30). Only cells with a single SAB reactive allele were included for analysis. The average CDC score for each allele was calculated from the previously determined reactivity grades 1 (negative), 2 (doubtful positive), 4 (weakly positive), 6 (positive) and 8 (strongly positive). Lymphocytotoxicity data for mAbs DK1G8 and VIE6C10 were obtained from earlier performed CDC assays with second-field HLA typed cells, which were carried out as previously described with mAb FK5 (pan HLA class I) as positive control (31). The percentages of target cell lysis were converted to CDC scores (0-10% lysis: 1, 11-20%: 2, 21-50%: 4, 51-80%: 6 and >80%: 8) (32) and average scores for each allele were calculated.
Reactivity Analysis of Human mAbs
HLA Epitope Mismatch Algorithm (HLA-EMMA) version 1.05 (33) was used to determine the solvent accessible amino acid mismatches between the HLA of the antibody producer and the mismatched HLA allele of the immunizer. In case of an ambiguous second field HLA typing, the most likely second field typing was selected based on a high resolution typed panel (n=1305) from Leiden, the Netherlands (http://www.allelefrequencies.net/pop6001c.asp?pop_id=0003257). If the immunizer was unknown, the specificity of the bead with the highest MFI in SAB assay was used to determine amino acid mismatches. Next, we determined whether these solvent accessible amino acid mismatches were uniquely shared by the reactive HLA alleles and absent on the non-reactive HLA alleles. In order to visualize amino acid positions and to establish whether amino acids were within 3-3.5 Å to form an eplet, the following HLA crystal structures were visualized in Swissviewer (34): Protein Data Bank (PBD) 1A6A, 1M6O, 1S9V, 1UVQ, 1X7Q, 1XR9, 3BO8, 3RL1, 3UTQ, 3WL9, 4U1H, 4Z7U, 5IND and 6PCL (downloaded from https://www.rcsb.org/on July 26, 2021). When HLA crystal structures were not available, modelled PBD structures were used; 3PL6, 3WEX, 4I5B, 4NT6 and 4Z7U (downloaded from https://www.phla3d.com.br/ on July 26, 2021). For HLAMatchmaker analysis, ABC Antibody Analysis Program V3.1 and DRDQDP Antibody Analysis Program v3.1 were used (http://www.epitopes.net/).
Review of Eplet Definitions
For every eplet with the antibody-verified status in the HLA Epitope Registry that consisted of more than one polymorphic residue, it was determined whether the involved amino acids were indeed within 3-3.5 Å using Swissviewer (34). If not, antibody reactivity analysis for this eplet was repeated using SAB data from the referenced paper to identify uniquely shared residues. If multiple uniquely shared residues were identified that were not within 3-3.5 Å (eplet definition), the eplet was classified as ‘‘reactivity pattern’’ (see Box 1).
Box 1. Definitions:
Functional epitope: The functional epitope determines the specificity of the antibody through its interaction with the complementarity-determining region 3 (CDR3) of the heavy chain of the antibody.
Eplet: The definition of an eplet resembles the functional epitope and comprises the minimal amino acid configuration on the HLA-molecule that is needed to induce an antibody response. Involved residues must be within 3-3.5 Å.
Structural epitope: The structural epitope comprises all amino acids of the HLA-molecule that are involved in the binding to the antibody paratope and spans a radius of approximately 15 Å.
Reactivity pattern: In some cases, the SAB analysis of a human mAb yields multiple uniquely shared residues or multiple uniquely shared combinations of residues that are not within 3-3.5 Å, indicating that there are multiple possible eplets that could have induced the formation of the antibody. Often, these amino acids are simultaneously present on HLA alleles, which limits the possibilities of determining the actual eplet using SAB or cellular assays. However, the fact that the residues involved always occur together on these HLA alleles, also means that these residues can be regarded as a ‘’reactivity pattern’’ and can be used as a single entity in matching strategies and immunological risk assessment for the vast majority of transplant patients.
Results
Antibody Verification of HLA Class I Eplets by Human mAbs
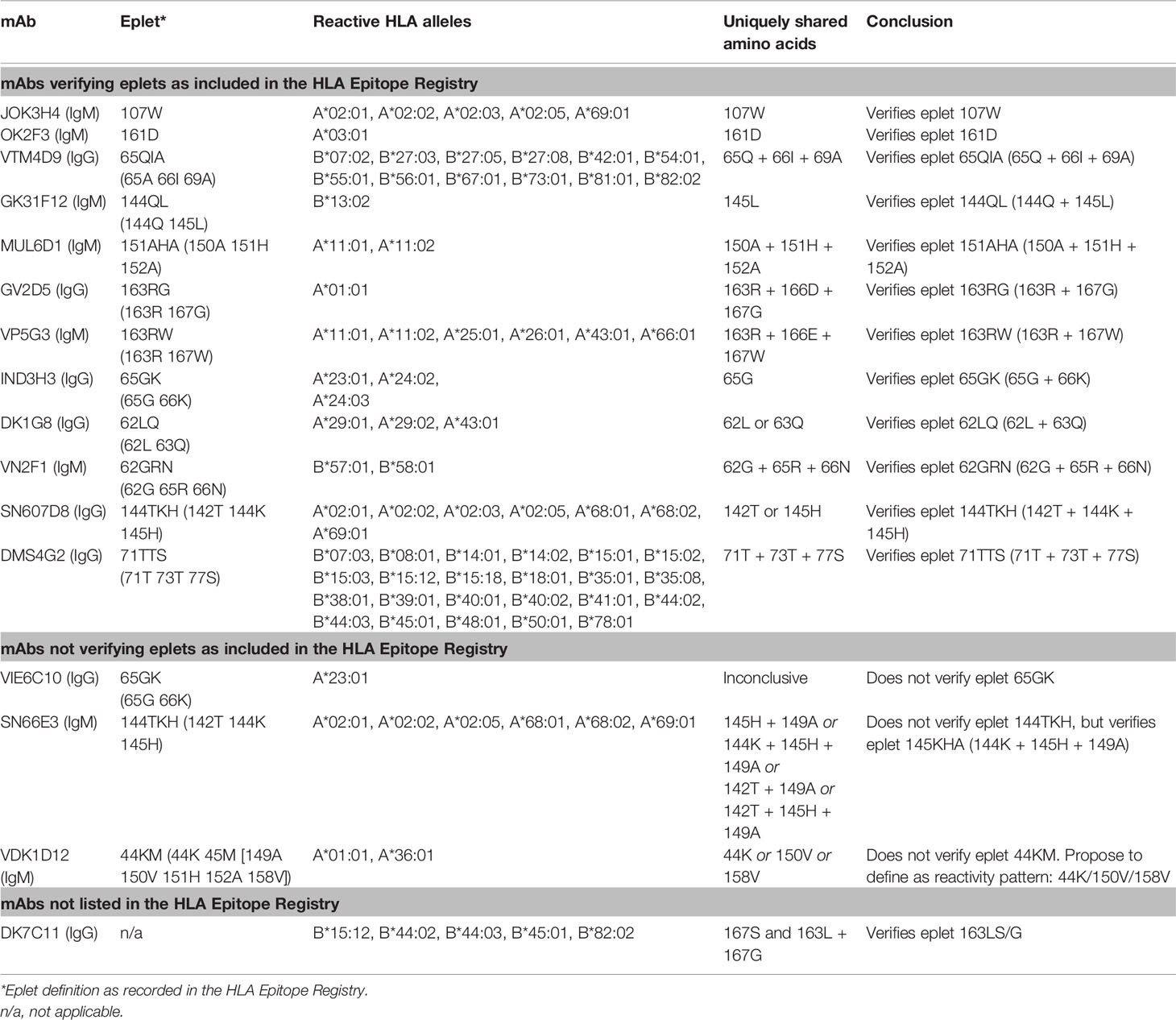
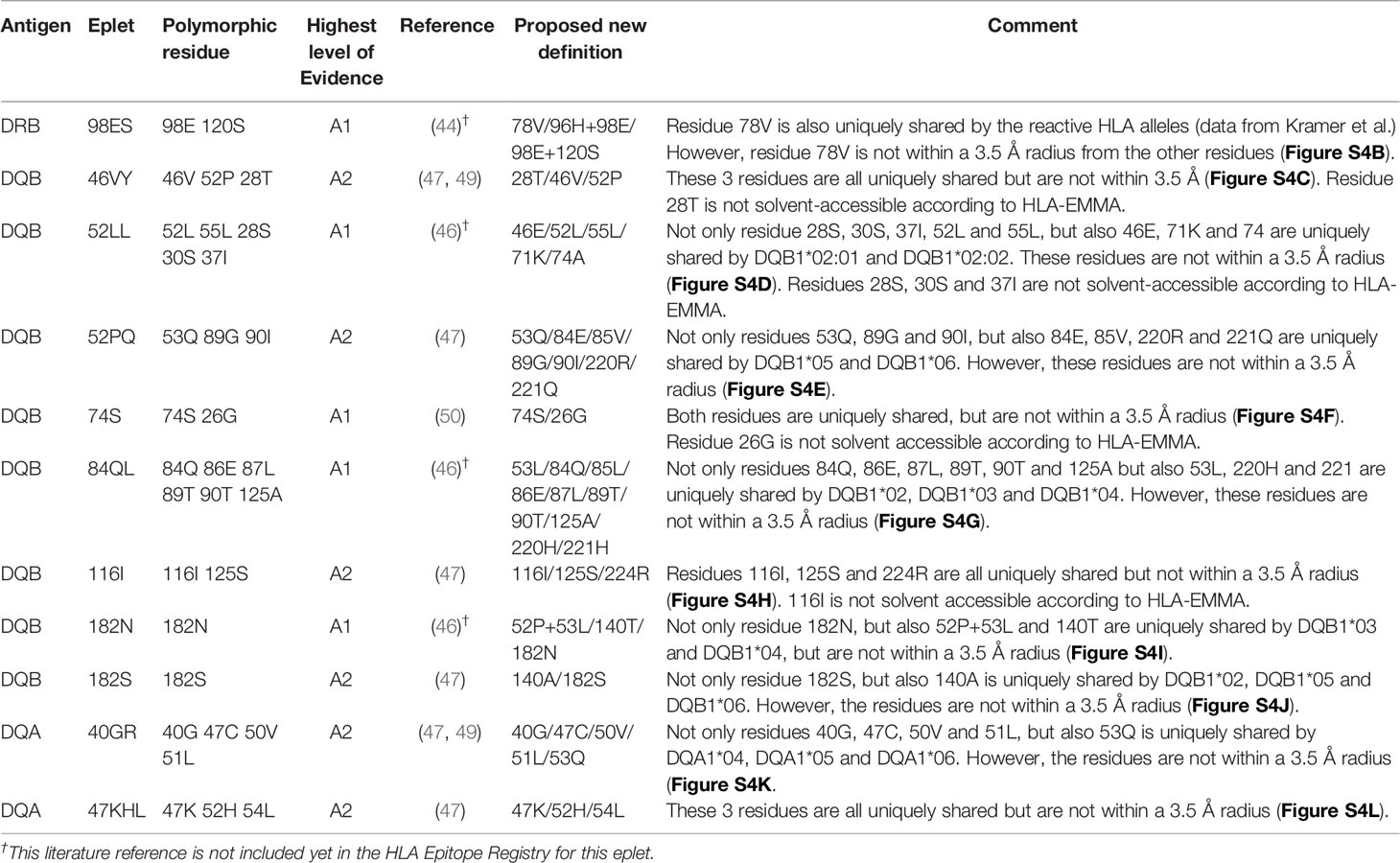
For 13 HLA class I eplets in the HLA Epitope Registry, antibody-verified status was based on data of 15 mAbs that had not been published in a peer-reviewed journal. Therefore, these mAbs were re-tested in luminex SAB assay to determine whether they would indeed provide evidence for antibody verification of these eplets. SAB analysis of the mAbs was performed by comparison of amino acid sequences of the reactive alleles in SAB assay with non-reactive alleles to identify uniquely shared amino acids that could have induced the antibody response. These uniquely shared residues were then mapped to corresponding eplets (Table 2). Overall, 12 human mAbs indeed verified the eplet as listed in the HLA Epitope Registry. mAbs JOK3H4, OK2F3, VTM4D9, GK31F12, MUL6D1, GV2D5, VP5G3 and IND3H3 verified eplets 107W, 161D, 65QIA, 144QL, 151AHA, 163RG, 163RW and 65GK respectively (Figures S1A–H). For mAbs DK1G8 and VN2F1, SAB data did not only show several beads with MFI > 10,000 (uniquely shared by eplets 62LQ and 62GRN respectively), but also included multiple positive reactions with considerably lower MFIs (MFI 814 to 9678). Analysis of previously acquired CDC data demonstrated that cells bearing alleles with these lower MFI values were negative in CDC (Figures 1A, B). Thus, analysis of SAB and CDC data of mAbs DK1G8 and VN2F1 confirmed the verification of eplets 62LQ and 62GRN respectively. Analysis of mAb SN607D8, which is listed as evidence for antibody verification of eplet 144TKH (142T 144K 145H), showed that both 142T and 145H are uniquely shared residues and that 144K is not (Figure 1C). Therefore, it is possible that only one of these two residues, or the combination of 142T and 145H is required for antibody induction. However, since the combination of 142T, 144K and 145H is also uniquely shared and is within 3.5 Å (Figure S2K), it cannot be ruled out that all three residues are crucial. Therefore, we consider the SAB analysis of mAb SN607D8 as evidence for the antibody verification of eplet 144TKH. SAB data of mAb DMS4G2, which is listed in the HLA Epitope Registry for verification of eplet 71TTS, demonstrated a broad spectrum of positive MFI values and showed three alleles that do not bear the eplet but are positive in SAB (MFI 4180-5160) (Figure 1D). However, data from previously performed CDC assays demonstrated that these alleles were negative in CDC. Interestingly, also a number of alleles bearing eplet 71TTS were not reactive in CDC. Therefore, it appears that for this antibody producer, eplet 71TTS has induced the antibody response, but the antibody does not bind equally strong to all eplet-bearing alleles, presumably due to other amino acid residues that play a role in the antibody binding. For instance, although alleles B*14:01 and B*14:02 only have one amino acid mismatch on position 11 (non-exposed), there is a large difference in MFI (14324 vs 3869). We hypothesize that the different amino acid sequence influences the structural and electrostatic properties of the epitope and consequently alters the antibody reactivity, as has been previously reported for the Bw6 epitope (35). Therefore, since the SAB data of mAb DMS4G2 provides evidence that eplet 71TTS can induce antibody formation, we consider this eplet antibody-verified. The positions of the involved amino acid residues on the surface of the respective HLA molecules for all aforementioned eplets are depicted in Figure S2.
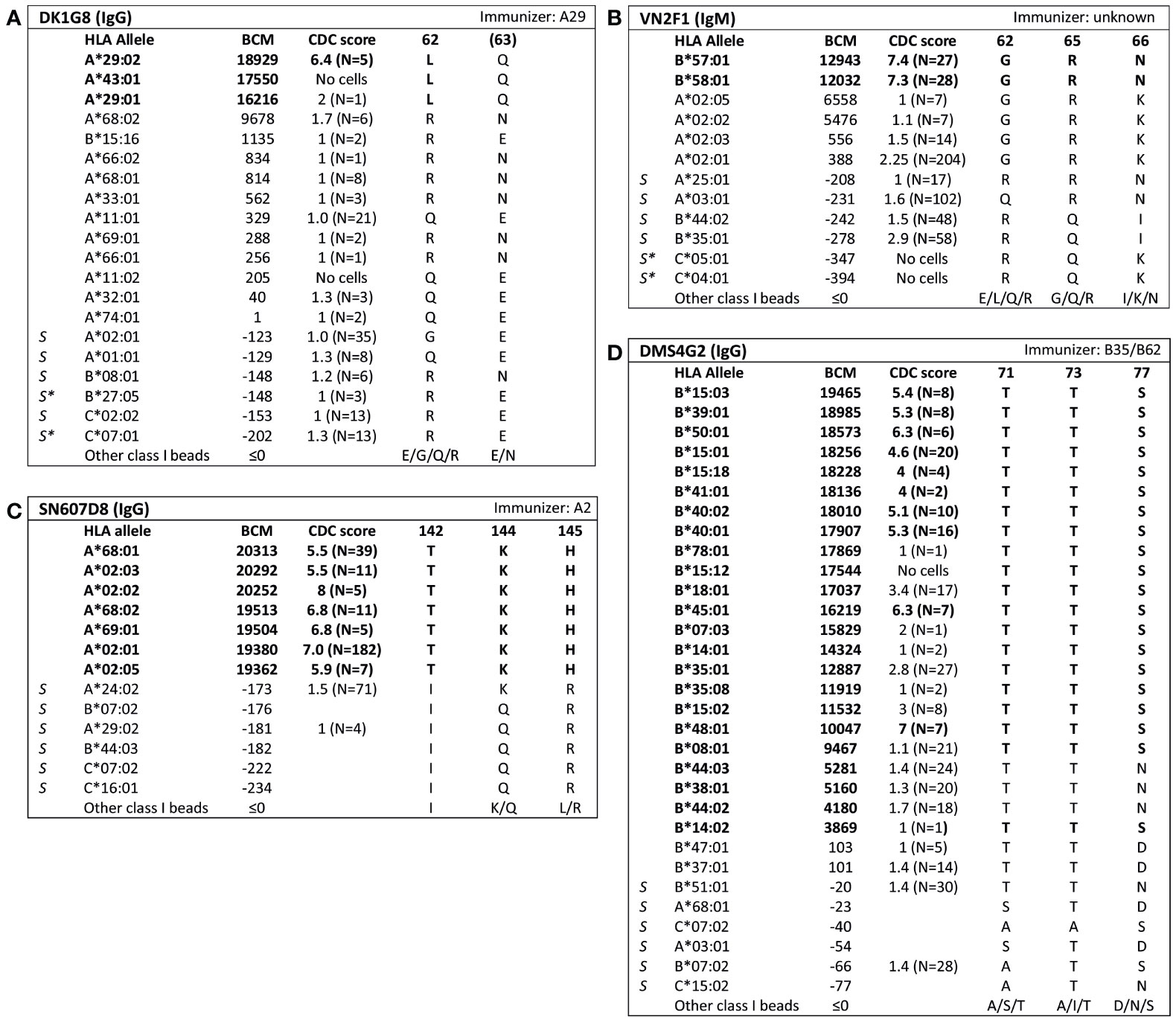
Figure 1 Comparison of the amino acid positions of interest of a selection of HLA class I alleles in the single antigen bead assay and complement dependent cytotoxicity assay for (A) mAb DK1G8, (B) VN2F1, (C) SN607D8 and (D) DMS4G2. mAb concentrations used for testing were 6.3 µg/ml for DMS4G2 and 10 µg/ml for the other mAbs. Self HLA alleles of the antibody producer marked with * are the most likely high resolution HLA typing due to ambiguous second-field typing. Alleles in bold are considered positive. Amino acid residues in bold are uniquely shared by the reactive alleles, or are part of a uniquely shared combination of residues. BCM, background corrected mean fluorescence intensity; CDC, complement dependent cytotoxicity; S, self HLA alleles of antibody producer.
Several Human mAbs Do Not Verify HLA Class I Eplets as Listed in the HLA Epitope Registry
Analysis of SAB data of three HLA class I mAbs did not verify the eplets which they were attributed to by the HLA Epitope Registry. Firstly, according to the Registry, mAb VIE6C10 verifies eplet 65GK. However, SAB and CDC results demonstrated that mAb VIE6C10 is negative for allele A*24:02, which bears eplet 65GK (Figure 2A). These data were confirmed in the One Lambda SAB assay (data not shown). Allele A*24:03, which also bears eplet 65GK, was also negative in SAB, but a previous CDC result showed positivity (N=1). Therefore, mAb VIE6C10 was also tested in a lower concentration of 1 ug/ml in SAB (1:10 dilution), to rule out the prozone effect, which can occur when high-titer antibodies interfere with the detection of IgG in the SAB assay (36). However, this SAB assay yielded similar results (data not shown). Reactivity analysis did not identify any other uniquely shared residue or eplet, and HLAMatchmaker analysis of the SAB data did not identify any eplet either. We therefore conclude that mAb VIE6C10 does not verify eplet 65GK.
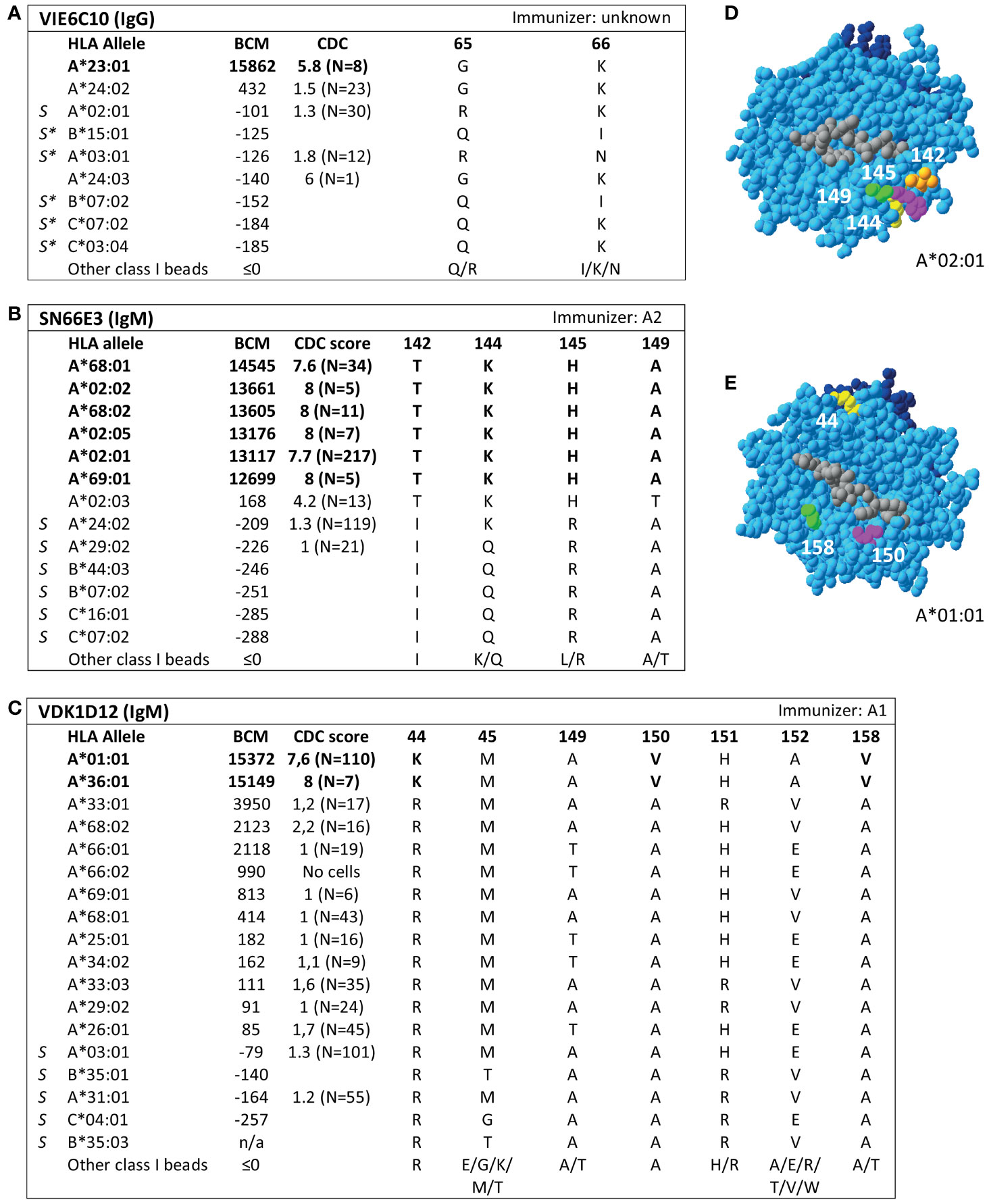
Figure 2 Reactivity analysis of HLA class I specific-monoclonal antibodies (mAb) that do not confirm eplets as defined in the HLA Epitope Registry. Comparison of the amino acid positions of interest of a selection of HLA class I alleles in the single antigen bead assay and complement dependent cytotoxicity assay for (A) mAb VIE6C10, (B) SN66E3 and (C) VDK1D12. Allele B*35:03 is a self-allele that is not present in the single antigen beads assay panel and has only 1 amino acid mismatch on position 116 with the other self-allele B*35:01. (D) Location of amino acids 142T (orange), 144K (yellow), 145H (magenta) and 149A (green) on the crystal structure of A*02:01 (PBD: 3UTQ). (E) Location of amino acids 44K (yellow), 150V (magenta) and 158V (green) on the crystal structure of A*01:01 (PBD: 3BO8). The α chain is depicted in light blue, the β chain in dark blue, and the peptide in grey. mAb concentrations used for testing were 10 µg/ml. Self HLA alleles of the antibody producer marked with * are the most likely high resolution HLA typing due to ambiguous second-field typing. Alleles in bold are considered positive. Amino acid residues in bold are uniquely shared by the reactive alleles, or are part of a uniquely shared combination of residues. BCM, background corrected mean fluorescence intensity; CDC, complement dependent cytotoxicity; PBD, Protein Data Bank; S, self HLA alleles of antibody producer.
Additionally, although mAb SN66E3 is listed in the Registry as one of two mAbs that verifies eplet 144TKH, reactivity analysis did not verify this eplet. Although the 144TKH-bearing allele HLA-A*02:03 is weakly positive in CDC (some cells bearing this allele being completely negative and some being positive), it was negative in SAB analysis. Considering A*02:03 non-reactive, the combinations of 145H + 149A or 142T + 149A are uniquely shared by the reactive alleles (Figure 2B). All three involved residues are within 3.5 Å (Figure 2D) and correspond to eplet 145KHA (144K 145H 149A), which was also identified by HLAMatchmaker analysis. Similarly to the analysis of mAb SN607D8, it is not possible to determine whether the combination of two or three amino acid residues is crucial for antibody induction. Consequently, we consider mAb SN66E3 as evidence for the antibody verification of eplet 145KHA.
Lastly, SAB analysis of mAb VDK1D12 yielded three uniquely shared residues; 44K, 150V and 158V (Figure 2C), which are not within a 3.5 Å or 15 Å distance and therefore cannot form an eplet or structural epitope (Figure 2E). This mAb is listed as evidence for verification of eplet 44KM, which does not fit the eplet definition as the residues (44K 45M [149A 150V 151H 152A] [158V]) exceed the 3.5 Å radius. We therefore conclude that 44KM is not an antibody-verified eplet but propose to consider 44K/150V/158V as an antibody-verified reactivity pattern (see Box 1).
Reactivity Analysis of an Unlisted HLA Class I mAb
Additionally, SAB analysis of mAb DK7C11, which is not included in the Registry, verified eplet 163LS/G (Figure 3). Tested at a concentration of 1 µg/ml, three alleles with 46 ≤ MFI ≤ 1010 became negative (MFI ≤ 0), while the five positive alleles remained positive with MFIs > 4000 (data not shown). The immunizing antigen for this mAb was HLA-B45, which bears the uniquely shared residue 167S. Interestingly, the mAb showed cross-reactivity with HLA-B*15:12, for which the combination of 163L + 167G is uniquely shared. No CDC data on cells bearing HLA-B*15:12 were available. Based on these new mAb data, the level of evidence for antibody verification of eplet 163LS/G is raised from level B (patient sera) to level A1.
Figure 3 Reactivity analysis of mAb DK7C11. (A) Comparison of the amino acid positions of interest of a selection of HLA class I alleles in the single antigen bead assay. Monoclonal antibody concentration used for testing was 10 µg/ml. Amino acid positions in brackets are not solvent-accessible according to HLA-EMMA. Self HLA alleles of the antibody producer marked with * are the most likely high resolution HLA typing due to ambiguous second-field typing. (B) Location of amino acids 163L (yellow) and 1671 (magenta) on the crystal structure of B*44:02 (PBD: 1M6O). Alleles in bold are considered positive. Amino acid residues in bold are part of the combination of residues that is uniquely shared by the reactive alleles. BCM, background corrected mean fluorescence intensity; S, self HLA alleles of antibody producer; PBD, Protein Data Bank.
Reactivity Analysis of HLA Class II-Specific Human mAbs
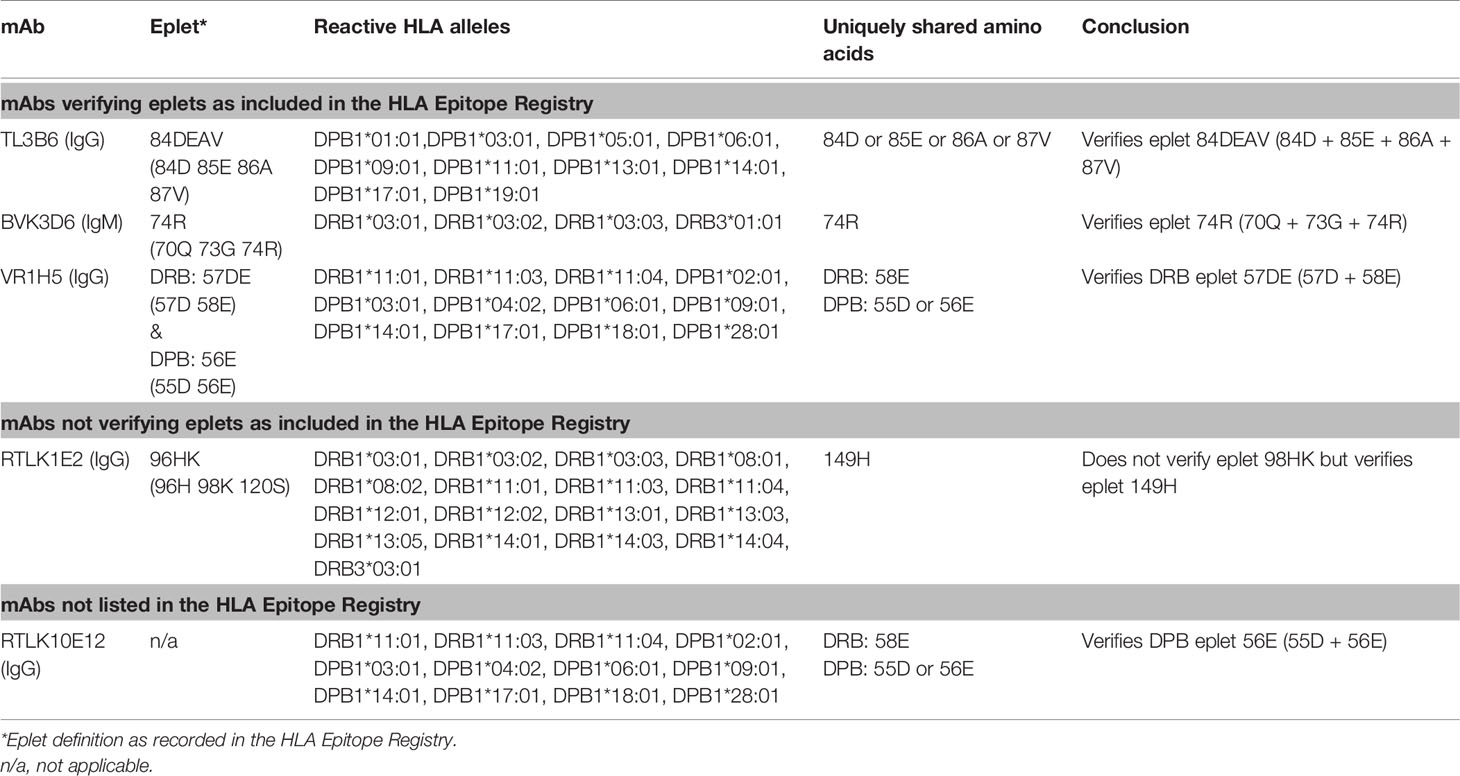
For HLA class II, four mAbs that were included in the HLA Epitope Registry without having been published and one additional mAb were tested in SAB assays (Table 3). HLA-DPB-specific mAb TL3B6 verified eplet 84DEAV (Figures S3A, B) and HLA-DRB-specific mAb BVK3D6 verified eplet 74R (Figures S3C, D). The HLA-DR11 induced mAb VR1H5, which was included in the HLA Epitope Registry as evidence for verification of eplet 57DE on HLA-DR and eplet 56E on HLA-DPB indeed verified eplet 57DE and was cross-reactive with eplet 56E (Figure 4A). Interestingly, mAb RTLK10E12, which is currently not included in the Registry, showed the same reactivity as VR1H5, but was induced by immunizing allele DPB1*09:01 and was cross-reactive with HLA-DR11 (Figure 4B). Hence, analysis of mAbs VR1H5 and RTLK10E12 confirm that both eplets can induce a cross-reactive antibody response and both eplets are therefore considered antibody-verified. SAB analysis of mAb RTLK1E2 was performed with the previously generated recombinant mAb RTLK1E2rec-IgG1 (37). Although RTLK1E2 is listed in the HLA Epitope Registry as evidence for antibody verification of eplet 96HK, SAB analysis showed that this mAb is reactive with allele DRB3*03:01, which does not bear eplet 96HK (Figure 4C). Instead, residue 149H was identified as the uniquely shared residue for mAb RTLK1E2. Since this result was not in line with the data in the HLA Epitope Registry, the mAb was also tested using One Lambda SAB assay. In concordance with our data, this assay demonstrated allele DRB3*03:01 to be reactive as well, and reactivity analysis demonstrated 149H to be uniquely shared (data not shown). Furthermore, the same reactive alleles have previously been described to be positive in a C3d SAB assay (37). Hence, based on analysis of mAb RTLK1E2, eplet 96HK cannot be regarded as antibody-verified. Instead, RTLK1E2 verifies eplet 149H, which was already present in the HLA Epitope Registry, but had not been antibody-verified yet. Localizations of eplet 57DE, 56E and 149H on the surface of HLA molecules are visualized in Figures 4D–F.
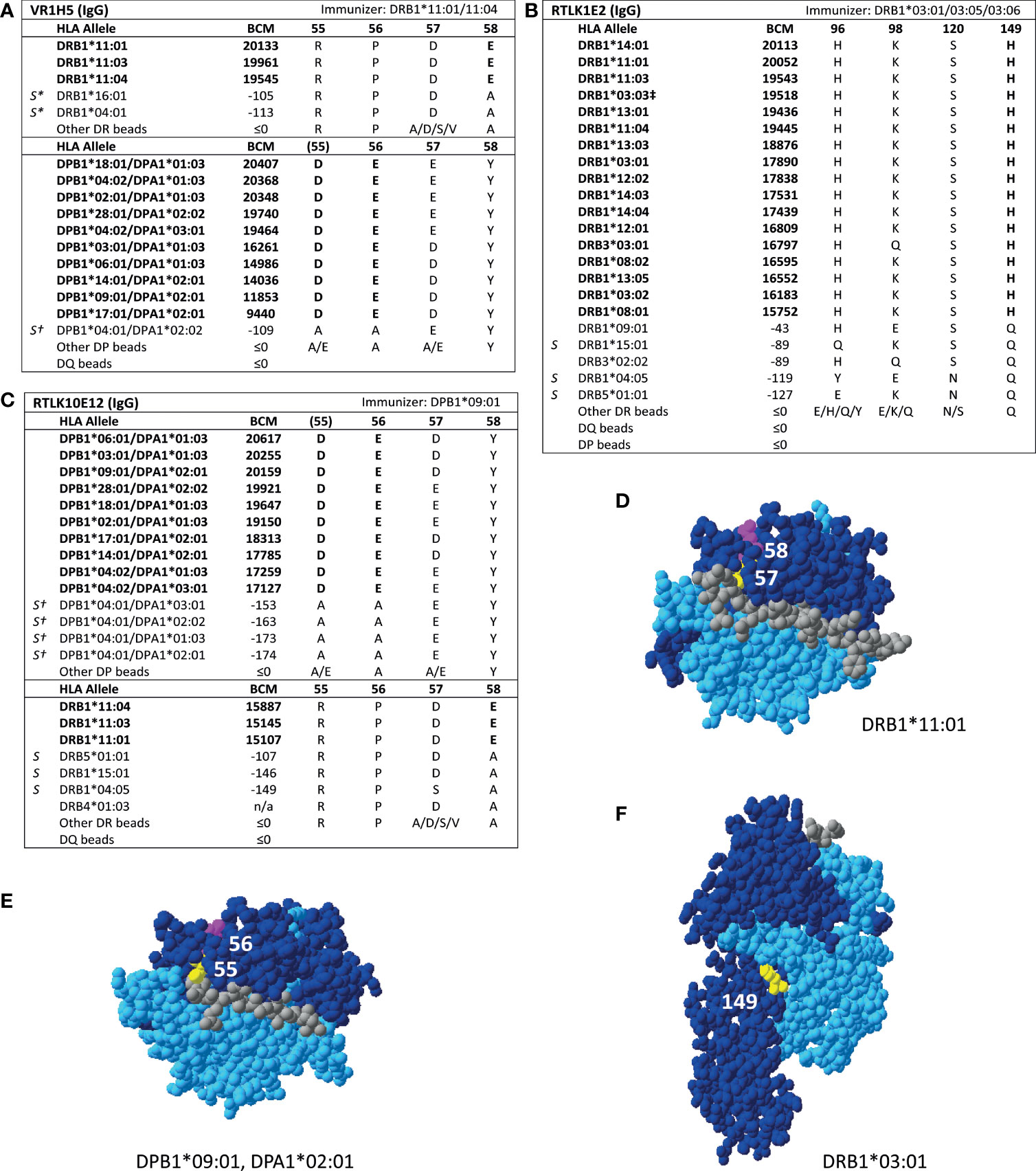
Figure 4 Reactivity analysis of HLA class II specific-monoclonal antibodies. Comparison of the amino acid positions of interest of a selection of DRB1 and DPB1 alleles in the single antigen bead assay for (A) mAb VR1H5, (B) RTLK10E12 and (C) RTLK1E2rec-IgG1 (Kramer et al. HLA. 2019 Nov;94(5):415-424.). Monoclonal antibody concentrations used for testing were 10, 2.5 and 10 µg/ml for mAb VR1H5, RTLK10E12 and RTLK1E2 respectively. Self HLA alleles of the antibody producer marked with * are the most likely high resolution HLA typing due to ambiguous second-field typing. Self-allele DRB4*01:03 for mAb RTLK10E12 is not present in the single antigen beads assay panel. (D) Location of amino acids 57D (yellow) and 58E (magenta) on the crystal structure of DRB1*11:01 (PBD: 6PCL). (E) Location of amino acids 55D (yellow) and 56E (magenta) on the crystal structure of DPA1*02:01/DPB1*09:01 (Modelled PBD: 3WEX). (F) Location of amino acid 149H (yellow) on the crystal structure of DRB1*03:01 (PBD: 1A6A). †DPA1 typing of antibody producer is not known. ‡The sequence of DRB1*03:03 is not fully known. For the unknown sections (residue positions 1-5 and 95-226), the same sequence as DRB1*03:01 is assumed (C. Heylen, Immucor, personal communication, August 4, 2020). Alleles in bold are considered positive. Amino acid residues in bold are uniquely shared by the reactive alleles. BCM, background corrected mean fluorescence intensity; S, self HLA alleles of antibody producer; PBD, Protein Data Bank.
Critical Review of All Evidence for Antibody Verification Status of Eplets in the HLA Epitope Registry
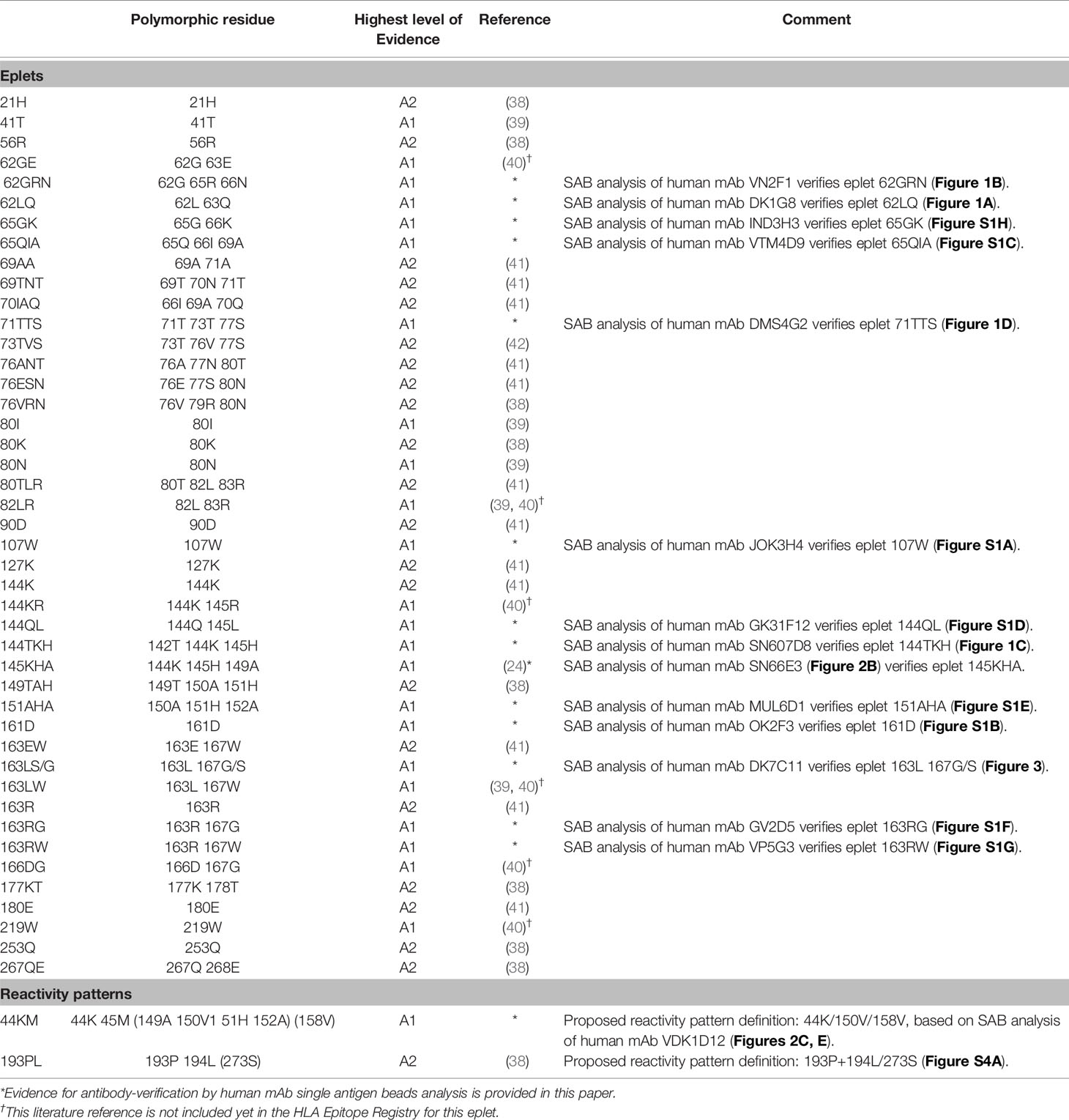
The HLA Epitope Registry databases include a total of 492 eplets of which 72 HLA class I, 36 HLA-DRB, 27 HLA-DQ and 11 HLA-DP have the antibody-verified status. In order to assign a level of evidence for antibody verification status of these eplets, a total of 121 literature references that are incorporated in the Registry were critically reviewed according to the classification in Table 1. Eplets with level A1 or A2 evidence were considered as truly antibody-verified, while level B, C and D were considered as provisionally antibody-verified (Table 4). The complete overview of all reviewed literature and level of evidence classification per eplet can be found in Supplementary Tables 2 and 3 for HLA class I and II respectively.
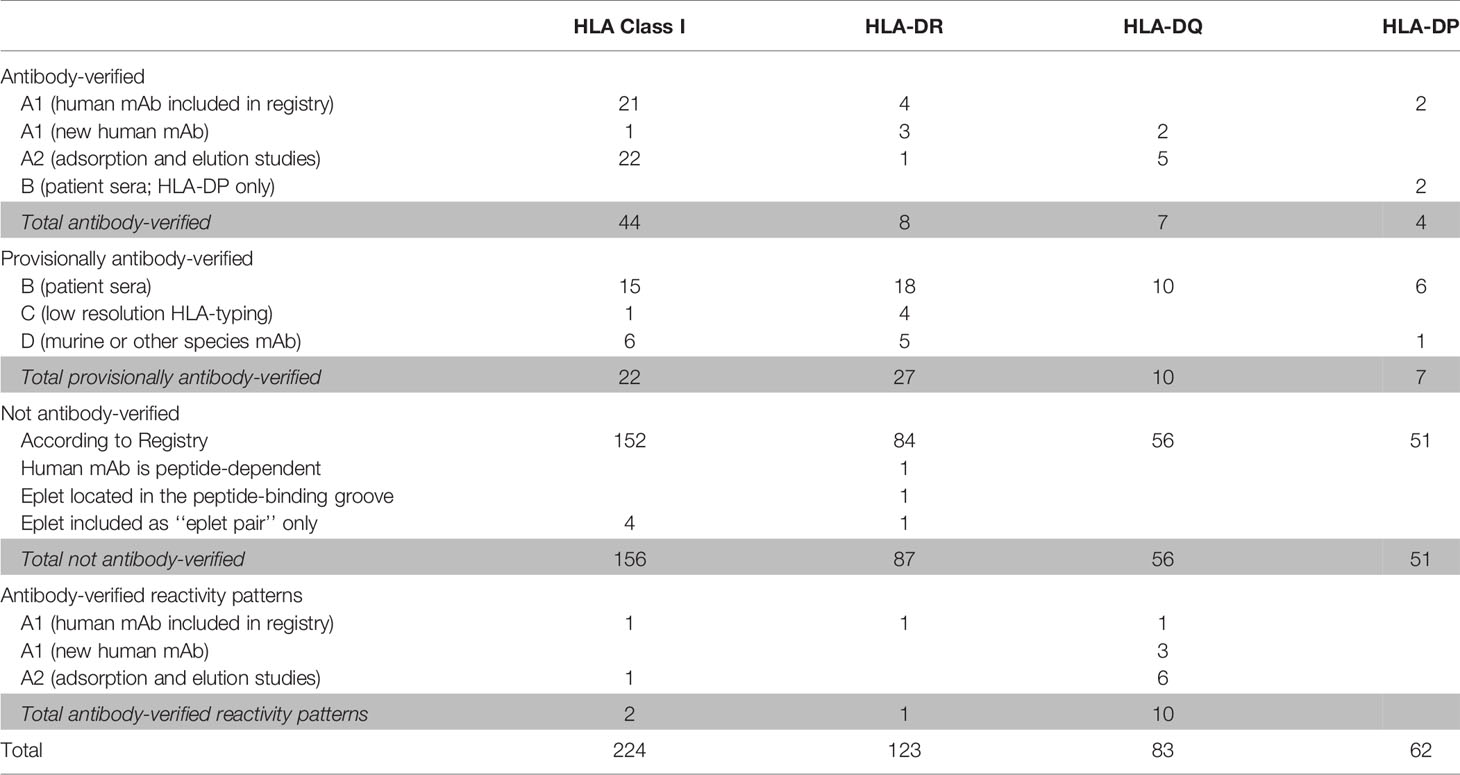
Table 4 Classification of level of evidence for antibody-verification of HLA class I and class II eplets.
For HLA class I, 44 eplets were considered truly antibody-verified based on human mAb data (n=22), including the mAb data presented in this paper, and adsorption and elution studies (n=22). A number of 22 eplets were considered provisionally antibody-verified based on reactivity analysis of patient sera (n=15), CDC with serologically typed cells only (n=1) and murine mAbs (n=6). The HLA Epitope Registry included four eplets that were listed as ‘eplet pairs’. Eplet pairs were considered not antibody-verified since they consist of two separate eplets that are located too far from each other to form a single eplet (> 3.5 Å), and thus are not jointly capable of inducing an antibody response. For two eplets, 44KM and 193PL, the residues that comprise these eplets as defined by the HLA Epitope Registry exceed the 3.5 Å range (Figures 2E and S4A). Since eplet 44KM is verified by human mAb analysis and verification of eplet 193PL is based on adsorption and elution studies, we consider them as antibody-verified reactivity patterns (see Box 1), of which the actual eplets remain unknown. The overall list of HLA class I antibody-verified eplets and reactivity patterns including literature references (24, 38–42) is depicted in Table 5.
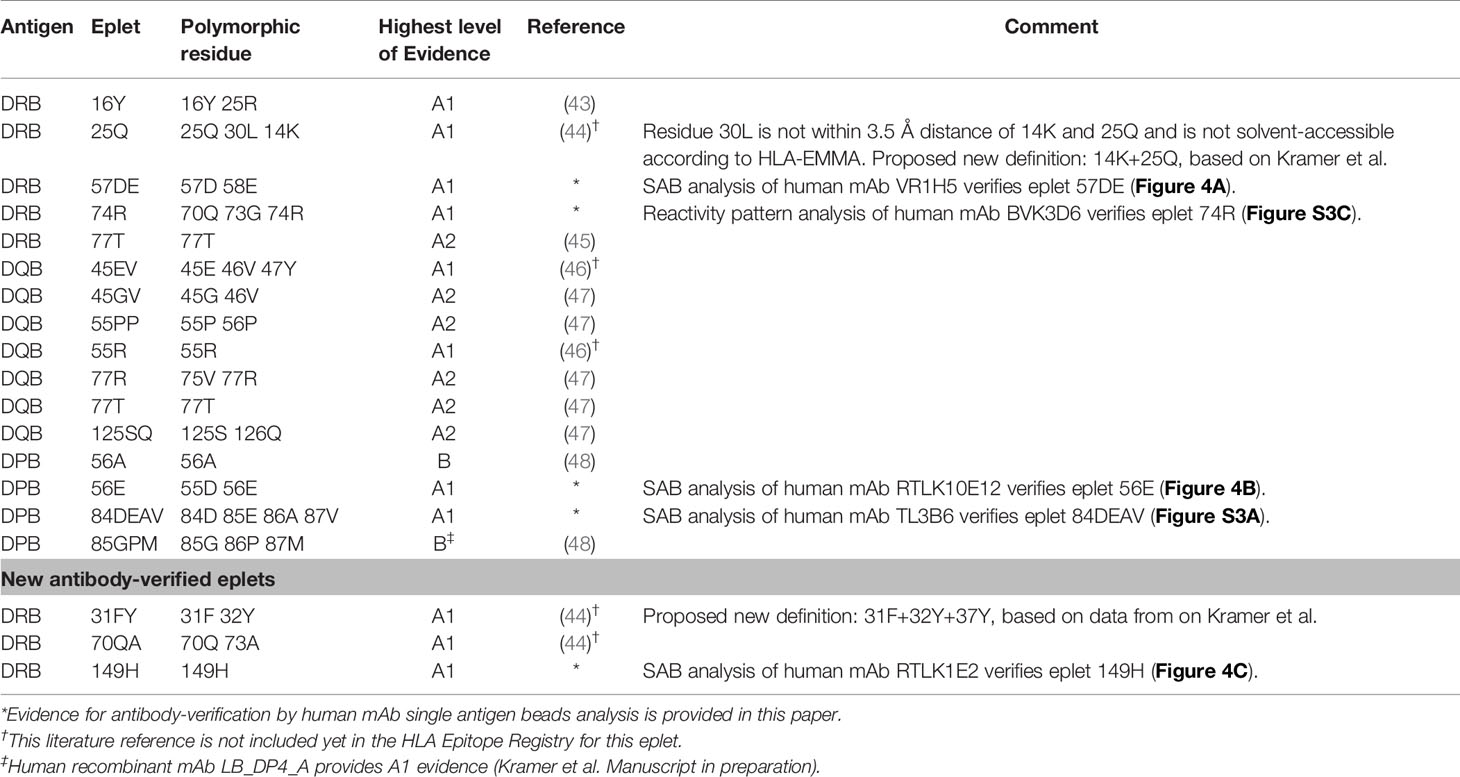
For HLA class II, we observed that for several eplets, especially HLA-DQ, the residues that comprise the eplet as defined by the HLA Epitope Registry exceed the 3.5 Å radius. For these eplets, previously published SAB data were re-analyzed to determine the uniquely shared residues and subsequently the possible eplets, which led to proposed new definitions of these reactivity patterns. The list of HLA class II antibody-verified eplets and antibody-verified reactivity patterns including literature references (43–50) are depicted in Tables 6 and 7 respectively.
For HLA-DR, five eplets were considered antibody-verified based on human mAb data that were included in the Registry (n=4) and based on reactivity analysis of adsorbed and eluted antibodies (n=1). Three previously not-verified eplets could be verified based on recent literature that was not yet included in the Registry (eplets 31FY and 70QA) (44) and based on new human mAb analysis in this current paper (eplet 149H). 27 eplets were considered provisionally antibody-verified based on reactivity analysis of patient sera (n=18), CDC with low resolution HLA typed cells only (n=4) and murine mAbs (n=5). Three eplets listed as antibody-verified by the HLA Epitope Registry were considered not antibody-verified. Eplet 11STS was not considered antibody-verified because the amino acids defining this eplet are located on the bottom of the peptide-binding groove (45), making it very unlikely that it is accessible for the B cell receptor and can induce antibody formation. Eplet 67LQ was not considered antibody-verified as it was solely listed as an eplet pair, and eplet 30C was considered not antibody-verified because binding of the mAb that was used for verification is peptide-dependent (51, 52) and no other evidence was available.
The residues defining eplet 98ES exceed the 3.5 Å distance, which is therefore considered as an antibody-verified reactivity pattern (Figure S4B). Eplet 96HK is provisionally antibody-verified (level B) as human mAb data analysis showed that not eplet 96HK but eplet 149H was uniquely shared (Figure 4C), leaving patient sera tested in SAB assay (published on the HLA Epitope Registry website, not peer-reviewed) as highest level of evidence for eplet 96HK. Furthermore, we propose to redefine eplet 25Q (25Q 30L 14K) to 14K + 25Q, since residue 30L is not solvent accessible and is not a within 3.5 Å radius of residues 14K and 25Q (44).
For HLA-DQ, 10 of the antibody-verified eplets exceed the 3.5 Å radius and are therefore considered as antibody-verified reactivity patterns (Figures S4C–L). We consider seven eplets truly antibody-verified based on new mAb data (n=2) and adsorption and elution experiments (n=5). The remaining 10 eplets are provisionally antibody-verified based on patient sera.
For HLA-DP, two eplets were antibody-verified based on human mAb data and seven eplets were provisionally antibody-verified based on reactivity analysis of patient sera (n=6) and a murine mAb (n=1). An exception regarding antibody verification classification was made for eplets 56A and 85GPM, of which the highest level of evidence is patient sera. These eplets were considered antibody-verified because of the extensive analysis on multiple sera performed by Cano et al. (48), and the fact that these particular HLA-DP epitopes are well established (53). Additionally, unpublished data from our own laboratory provides A1 evidence for eplet 85GPM (Kramer et al. manuscript in preparation).
Overall, we consider 44 HLA class I eplets and 19 HLA class II eplets as being truly antibody-verified and a total of two HLA class I and 11 HLA class II reactivity patterns as being antibody-verified.
Discussion
The HLA Epitope Registry and HLAMatchmaker have formed the foundation for the vast majority of clinical studies investigating the role of HLA eplets in transplantation. In this study, we have critically reviewed the evidence for the antibody verification status of eplets included in the HLA Epitope Registry. The different methodologies that are currently used for antibody-verification do not represent the same level of evidence for the antibody-verified status of eplets. However, while previously a category of ‘provisionally verified’ was present, the current dichotomous yes or no antibody-verified status in the HLA Epitope Registry does not take the heterogeneity in the level of evidence into account. To provide insight on what basis an eplet is considered antibody-verified by the Registry, we have introduced a classification system to score the level of evidence. Our results show that for many eplets, especially for HLA class II, the antibody-verified status is based on sera from multi- or uni-parous women or transplant patients, experiments with only serologically typed cells, or murine mAbs. However, we argue that these methods are not suitable for definitive antibody verification of eplets. Although SAB analysis of sera from immunized individuals can be informative, the reactivity of sera tested in SAB is in most, if not all cases the result of a polyclonal antibody response. These patterns are often broad and do not permit the identification of a single HLA eplet, since the pattern of reactive HLA alleles is caused by multiple antibodies recognizing several HLA epitopes. Even seemingly narrow SAB reactivity may be caused by more than one eplet mismatch. For several other eplets, antibody verification status was based on experiments using serologically typed cells only. These cells are not suitable for state-of-the-art reactivity analysis due to the low resolution of HLA typing, which makes definitive assignment of the inducing eplet very difficult. Furthermore, for 11 eplets only reactivity analysis of murine mAbs was available. Murine mAbs are generated by immunization with HLA but do not necessarily recognize the same epitopes as human mAbs, since immunogenicity of HLA antigens is affected by the recipients’ HLA type (22). Therefore, we argue that if antibody-verified status in the HLA Epitope Registry is solely based on reactivity analysis of patient sera, experiments with serologically typed cells or murine mAbs, this should result in provisional evidence for antibody verification, but not a definitive antibody-verified status. In the first report of the antibody verification of eplets in the HLA Epitope Registry, antibody-verified eplets were classified as ‘confirmed’ or ‘provisional’ depending on the amount and degree of evidence that was available (18). However, this classification was removed in the second update of the Registry (23).
Aside from eplets, the HLA Epitope Registry also includes ‘‘eplet pairs’’, of which a number have been assigned the antibody-verified status. HLA eplets are based on the concept that one or multiple mismatched amino acid residues induce the humoral immune response through interaction with the CDR3 region of the B cell receptor heavy chain. Accordingly, the residues that constitute an eplet should be in a 3.5 Å radius (4). However, eplet pairs consist of two eplets (a combination of a nonself-eplet and a self-eplet) that are located within the 15 Å radius that constitutes the structural epitope, but are not within 3.5 Å from each other (54). Therefore, eplet pairs cannot be regarded as the configuration that induces the antibody response and subsequently, we did not consider eplet pairs for antibody verification.
Our review of the HLA Epitope Registry does not only provide insight in the heterogeneity of the level of evidence of eplet antibody verification, but also demonstrates that a substantial portion of the presented mAb data and patient sera analyses had not been published in peer-reviewed journals. Aiming to substantiate the antibody-verified status of eplets based on human mAbs which reactivity analyses have not been published previously, we tested these mAbs in SAB assays and performed reactivity analysis. For the majority of mAbs tested, the identified uniquely shared amino acids indeed corresponded with the eplet. However, SAB analysis of three mAbs did not confirm the antibody verification of the eplet as assigned by the Registry. The analyses of mAbs SN66E3 and RTLK1E2 supported the verification of two different eplets, while no inducing eplet could be determined for mAb VIE6C10. Furthermore, the reactivity analyses of SN607D8 and SN66E3 identified multiple uniquely shared residues or uniquely shared combinations of two residues, while the corresponding eplets, 144TKH and 145KHA respectively, are defined by three residues. Based on our analyses, it is possible that not all three, but only one or two residues are crucial for the induction of anti-HLA antibodies. For these eplets, this difference in possible eplet definitions is clinically relevant, since there are less common, but intermediately and well-documented alleles that bear only one of the uniquely shared residues (55). Consequently, using the definition that includes all three residues could possibly disregard patients with these less common HLA alleles in respect to HLA eplet matching purposes. Mutation studies of HLA alleles or testing of the mAbs against a panel of cells containing these less common HLA types could provide more insight in the actual configuration of polymorphic residues that comprises these eplets. However currently, experimental possibilities are limited due to the lack of suitable reagents.
Detailed analysis of the localization of antibody-verified eplet configurations on crystalized HLA structures demonstrated that not all antibody-verified eplets in the HLA Epitope Registry comply with the eplet definition. Especially for HLA-DQ, the polymorphic residues that comprise the eplet configuration are often too distant (> 3.5 Å) from each other to form an eplet. Re-analysis of previously published SAB data of human mAbs and eluted antibodies from patient sera demonstrated that for 10 HLA-DQ, one HLA-DR and two HLA class I eplets multiple uniquely shared amino acids could be identified that were not within 3.5 Å. Because these residues are simultaneously present on the Common HLA alleles in the CIWD 3.0.0 (55) with only a few exceptions, we propose to consider these configurations as antibody-verified reactivity patterns instead of eplets. Accordingly, these antibody-verified reactivity patterns can still be considered in HLA matching strategies and molecular mismatch evaluation for the vast majority of transplant patients. For four reactivity patterns there is a small number of Common HLA alleles that can be considered as an exception, which are listed in Supplementary Table 4. For instance, the antibody-verified reactivity pattern 74S/26G is present on all Common DQB1*04 and DQB1*05 alleles. There are also two alleles that bear 26G, but have 74E instead of 74S (DQB1*03:05 and DQB1*03:25). When a patient carrying DQB1*03:01 (which lacks this reactivity pattern) would receive a transplant from a DQB1*03:05 donor, only one of the two residues of this reactivity pattern would be mismatched, namely 26G. At this moment it is not clear whether residue 26G or residue 74S is crucial for antibody induction and therefore it is uncertain whether the 26G mismatch in this case would be clinically relevant. Structural data based on mutation studies and crystallography are required to determine the true binding place of the antibody to be able to determine which of the residues of a reactivity pattern can be considered as the true eplet.
The rationale for this explicit and precise definition of eplets and reactivity patterns also follows from the need to define the most immunogenic eplets in transplantation (56). Multiple studies have tried to identify the most immunogenic eplet mismatches (16, 57–59), which is a crucial step in making eplet-matching in transplantation clinically applicable. Currently, these studies are limited by the use of different versions of the HLA Epitope Registry and/or HLAMatchmaker and consequently the different eplet definitions that are used in the analyses. For example, a recent paper investigating the immunogenicity of HLA-DQ eplets used HLAMatchmaker 2.1 to determine eplet mismatches (59). In this version of HLAMatchmaker, eplets 84QL and 125A are considered as separate eplets. In the current version of the HLA Epitope Registry however, eplet 125A has been removed. In fact, residue 125A has been added to the definition of eplet 84QL, since residues 84Q, 86E, 87L, 89T, 90T and are all uniquely shared by alleles DQB1*02, DQB1*03 and DQB1*04, but are not within 3.5 Å. Hence, according to our proposed classification, eplet 84QL rather is a reactivity pattern. Accordingly, the use of different definitions for the same eplet and the inclusion of eplet pairs in immunogenicity studies distorts the interpretation and comparability of immunogenicity scores. Furthermore, inconsistencies in eplet definition and antibody-verified status between the HLA Epitope Registry and HLAMatchmaker (60) and the lack of documentation of previous versions of the Registry hamper investigations towards eplet mismatch loads and transplant outcomes.
The SAB assays in this study were performed with the Lifecodes SAB assay from Immucor. It has been demonstrated that the beads of the other manufacturer of these assays (One Lambda, Thermofisher) are bound with an admixture of intact and denatured HLA (61, 62), while the Immucor assay predominantly contains intact HLA (63). The presence of denatured HLA on beads can results in detection of antibodies against cryptic epitopes (64). Since these antibodies will not bind to intact HLA, cellular testing of mAbs or eluted antibodies can exclude the possibility of an antibody directed towards a cryptic epitope. The clinical relevance of antibodies against cryptic epitopes in transplantation remains questionable and warrants further investigation.
This critical review of the antibody-verified status of eplets in the HLA Epitope Registry has demonstrated that the level of evidence of antibody-verified eplets is heterogeneous and that not all data have been published in peer-reviewed journals. Analysis of luminex SAB data of human mAbs showed that not all mAbs verified the eplets they were assigned to. Since an increasing number of clinical studies investigate eplet mismatch load as a risk factor for inferior transplant outcomes and seek to identify the most immunogenic eplet mismatches, it is vital to define a set of well-defined antibody-verified eplets in a transparent manner. Our list of antibody-verified eplets and reactivity patterns is the first step towards a uniform and transparent method of eplet definition and antibody verification. However, eplets that are considered provisionally verified, or not antibody-verified could still play a clinically relevant role in transplantation, since antibody verification is limited by the available reagents and patient material. In this respect it is important for our field to collaborate in the yet uncompleted endeavor of eplet antibody verification. For future publications, we propose to set a standard of required data regarding antibody verification that should be published. Preferably, this includes reactivity analysis of human mAb data tested in SAB assay or antibodies eluted from immunized patient sera. The used SAB panel and HLA typing of the antibody producer and immunizer should also be included to make re-analysis and thorough interpretation of the data possible. Finally, we propose to establish an international committee that oversees nomenclature and antibody verification of eplets to facilitate the establishment of a well-documented, transparent list of eplets with antibody verification status classified by the level of evidence, striving for better comparable results in clinical and immunogenicity studies on the road to eplet matching in transplantation.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SB reviewed the literature and analyzed the data. KB performed laboratory experiments. SB and SH wrote the manuscript. CK, JF, MR, AM, and FC critically reviewed the manuscript. All authors contributed to the article and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the HLA typing and screening laboratory from Leiden University Medical Center, Leiden, the Netherlands for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.800946/full#supplementary-material
Abbreviations
Å, Ångstrom; CDC, complement-dependent cytotoxicity; HLA, human leukocyte antigen; HLA-EMMA, HLA Epitope Mismatch Algorithm; mAb, monoclonal antibody; MFI, mean fluorescence intensity; SAB, single antigen beads.
References
1. Zhang R. Donor-Specific Antibodies in Kidney Transplant Recipients. Clin J Am Soc Nephrol (2018) 13(1):182–92. doi: 10.2215/cjn.00700117
2. Loupy A, Lefaucheur C. Antibody-Mediated Rejection of Solid-Organ Allografts. N Engl J Med (2018) 379(12):1150–60. doi: 10.1056/NEJMra1802677
3. Duquesnoy RJ, Askar M. HLAMatchmaker: A Molecularly Based Algorithm for Histocompatibility Determination. V. Eplet Matching for HLA-DR, HLA-DQ, and HLA-DP. Hum Immunol (2007) 68(1):12–25. doi: 10.1016/j.humimm.2006.10.003
4. Duquesnoy RJ. A Structurally Based Approach to Determine HLA Compatibility at the Humoral Immune Level. Hum Immunol (2006) 67(11):847–62. doi: 10.1016/j.humimm.2006.08.001
5. Amit AG, Mariuzza RA, Phillips SE, Poljak RJ. Three-Dimensional Structure of an Antigen-Antibody Complex at 2.8 A Resolution. Science (1986) 233(4765):747–53. doi: 10.1126/science.2426778
6. Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, et al. Forced Usage of Positively Charged Amino Acids in Immunoglobulin CDR-H3 Impairs B Cell Development and Antibody Production. J Exp Med (2006) 203(6):1567–78. doi: 10.1084/jem.20052217
7. Xu JL, Davis MM. Diversity in the CDR3 Region of V(H) Is Sufficient for Most Antibody Specificities. Immunity (2000) 13(1):37–45. doi: 10.1016/s1074-7613(00)00006-6
8. Lemieux W, Mohammadhassanzadeh H, Klement W, Daniel C, Sapir-Pichhadze R. Matchmaker, Matchmaker Make Me a Match: Opportunities and Challenges in Optimizing Compatibility of HLA Eplets in Transplantation. Int J Immunogenet (2021) 48(2):135-44. doi: 10.1111/iji.12525
9. Wiebe C, Pochinco D, Blydt-Hansen TD, Ho J, Birk PE, Karpinski M, et al. Class II HLA Epitope Matching-A Strategy to Minimize De Novo Donor-Specific Antibody Development and Improve Outcomes. Am J Transplant (2013) 13(12):3114–22. doi: 10.1111/ajt.12478
10. Lachmann N, Niemann M, Reinke P, Budde K, Schmidt D, Halleck F, et al. Donor-Recipient Matching Based on Predicted Indirectly Recognizable HLA Epitopes Independently Predicts the Incidence of De Novo Donor-Specific HLA Antibodies Following Renal Transplantation. Am J Transplant (2017) 17(12):3076–86. doi: 10.1111/ajt.14393
11. Wiebe C, Kosmoliaptsis V, Pochinco D, Gibson IW, Ho J, Birk PE, et al. HLA-DR/DQ Molecular Mismatch: A Prognostic Biomarker for Primary Alloimmunity. Am J Transplant (2019) 19(6):1708–19. doi: 10.1111/ajt.15177
12. Snanoudj R, Kamar N, Cassuto E, Caillard S, Metzger M, Merville P, et al. Epitope Load Identifies Kidney Transplant Recipients at Risk of Allosensitization Following Minimization of Immunosuppression. Kidney Int (2019) 95(6):1471–85. doi: 10.1016/j.kint.2018.12.029
13. Sapir-Pichhadze R, Zhang X, Ferradji A, Madbouly A, Tinckam KJ, Gebel HM, et al. Epitopes as Characterized by Antibody-Verified Eplet Mismatches Determine Risk of Kidney Transplant Loss. Kidney Int (2020) 97(4):778–85. doi: 10.1016/j.kint.2019.10.028
14. Senev A, Coemans M, Lerut E, Van Sandt V, Kerkhofs J, Daniëls L, et al. Eplet Mismatch Load and De Novo Occurrence of Donor-Specific Anti-HLA Antibodies, Rejection, and Graft Failure After Kidney Transplantation: An Observational Cohort Study. J Am Soc Nephrol (2020) 31(9):2193–204. doi: 10.1681/asn.2020010019
15. Kramer C, Heidt S, Claas FHJ. Towards the Identification of the Relative Immunogenicity of Individual HLA Antibody Epitopes. Hum Immunol (2019) 80(4):218–20. doi: 10.1016/j.humimm.2019.02.002
16. Mohammadhassanzadeh H, Oualkacha K, Zhang W, Klement W, Bourdiec A, Lamsatfi J, et al. On Path to Informing Hierarchy of Eplet Mismatches as Determinants of Kidney Transplant Loss. Kidney Int Rep (2021) 6(6):1567–79. doi: 10.1016/j.ekir.2021.03.877
17. Duquesnoy RJ, Marrari M, da M, Sousa LCD, de M. Barroso JRP, de S. U. Aita KM, et al. Workshop Report: A Website for the Antibody-Defined HLA Epitope Registry. Int J Immunogenet (2013) 40:54–9. doi: 10.1111/iji.12017
18. Duquesnoy RJ, Marrari M, Mulder A, Sousa LC, da Silva AS, do Monte SJ. First Report on the Antibody Verification of HLA-ABC Epitopes Recorded in the Website-Based HLA Epitope Registry. Tissue Antigens (2014) 83(6):391–400. doi: 10.1111/tan.12341
19. Duquesnoy RJ, Marrari M, Tambur AR, Mulder A, Sousa LC, da Silva AS, et al. First Report on the Antibody Verification of HLA-DR, HLA-DQ and HLA-DP Epitopes Recorded in the HLA Epitope Registry. Hum Immunol (2014) 75(11):1097–103. doi: 10.1016/j.humimm.2014.09.012
20. Ravindranath MH, Terasaki PI, Maehara CY, Jucaud V, Kawakita S, Pham T, et al. Immunoglobulin (Ig)G Purified From Human Sera Mirrors Intravenous Ig Human Leucocyte Antigen (HLA) Reactivity and Recognizes One's Own HLA Types, But May be Masked by Fab Complementarity-Determining Region Peptide in the Native Sera. Clin Exp Immunol (2015) 179(2):309–28. doi: 10.1111/cei.12450
21. Kohler H, Bayry J, Kaveri SV. The Homophilic Domain - An Immunological Archetype. Front Immunol (2016) 7:106. doi: 10.3389/fimmu.2016.00106
22. Doxiadis II, Smits JM, Schreuder GM, Persijn GG, van Houwelingen HC, van Rood JJ, et al. Association Between Specific HLA Combinations and Probability of Kidney Allograft Loss: The Taboo Concept. Lancet (1996) 348(9031):850–3. doi: 10.1016/s0140-6736(96)02296-9
23. Duquesnoy RJ, Marrari M, Marroquim MS, Borges AG, da Mata Sousa LCD, Socorro A, et al. Second Update of the International Registry of HLA Epitopes. I. The HLA-ABC Epitope Database. Hum Immunol (2019) 80(2):103–6. doi: 10.1016/j.humimm.2018.11.007
24. Mulder A, Kardol M, Blom J, Jolley WB, Melief CJ, Bruning H. A Human Monoclonal Antibody, Produced Following In Vitro Immunization, Recognizing an Epitope Shared by HLA-A2 Subtypes and HLA-A28. Tissue Antigens (1993) 42(1):27–34. doi: 10.1111/j.1399-0039.1993.tb02162.x
25. Mulder A, Kardol M, Regan J, Buelow R, Claas F. Reactivity of Twenty-Two Cytotoxic Human Monoclonal HLA Antibodies Towards Soluble HLA Class I in an Enzyme-Linked Immunosorbent Assay (PRA-STAT). Hum Immunol (1997) 56(1-2):106–13. doi: 10.1016/s0198-8859(97)00146-8
26. Mulder A, Kardol MJ, Niterink JGS, Parlevliet JH, Marrari M, Tanke J, et al. Successful Strategy for the Large Scale Production of HLA Human Monoclonal Antibodies. In: Charron D, editor. HLA: Genetic Diversity of HLA: Functional and Medical Implications Volume 2 - Proceedings of the Twelfth International Histocompatibility Conference. Paris, France: EDK (1997). p. 354–6.
27. Mulder A, Eijsink C, Kester MG, Franke ME, Kardol MJ, Heemskerk MH, et al. Impact of Peptides on the Recognition of HLA Class I Molecules by Human HLA Antibodies. J Immunol (2005) 175(9):5950–7. doi: 10.4049/jimmunol.175.9.5950
28. Mulder A, Kardol MJ, Arn JS, Eijsink C, Franke ME, Schreuder GM, et al. Human Monoclonal HLA Antibodies Reveal Interspecies Crossreactive Swine MHC Class I Epitopes Relevant for Xenotransplantation. Mol Immunol (2010) 47(4):809–15. doi: 10.1016/j.molimm.2009.10.004
29. Gu Y, Koh RWK, Lai ML, Pochinco D, Teo RZC, Chan M, et al. Defining the Structural Basis for Human Leukocyte Antigen Reactivity in Clinical Transplantation. Sci Rep (2020) 10(1):18397. doi: 10.1038/s41598-020-75355-4
30. Fernandez-Vina M, Maiers M, Gutierrez M, Mulder A, Lee JH, Stoddard J, et al. 13th IHWS Serology and HLA Phenotypes Joint Report. In: Hansen JA, editor. Immunobiology of the Human MHC - Proceedings of the 13th International Histocompatibility Workshop and Conference - Volume I. Seattle, Washington, USA: Seatttle IHWG Press (2006). p. 890–931.
31. Bruning JW, Claas FH, Kardol MJ, Lansbergen Q, Naipal AM, Tanke HJ. Automated Reading of HLA-A,B,C Typing and Screening. The Propidium Iodide (PI) Method. Hum Immunol (1982) 5(3):225–31. doi: 10.1016/0198-8859(82)90135-5
32. Peña JR, Fitzpatrick D, Saidman SL. Complement-Dependent Cytotoxicity Crossmatch. In: Zachary V, Leffell M, editors. Transplantation Immunology Methods and Protocols. Totowa NJ: Humana Press (2013). p. 257–83.
33. Kramer CSM, Koster J, Haasnoot GW, Roelen DL, Claas FHJ, Heidt S. HLA-EMMA: A User-Friendly Tool to Analyse HLA Class I and Class II Compatibility on the Amino Acid Level. HLA (2020) 96(1):43–51. doi: 10.1111/tan.13883
34. Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An Environment for Comparative Protein Modeling. Electrophoresis (1997) 18(15):2714–23. doi: 10.1002/elps.1150181505
35. Kosmoliaptsis V, Dafforn TR, Chaudhry AN, Halsall DJ, Bradley JA, Taylor CJ. High-Resolution, Three-Dimensional Modeling of Human Leukocyte Antigen Class I Structure and Surface Electrostatic Potential Reveals the Molecular Basis for Alloantibody Binding Epitopes. Hum Immunol (2011) 72(11):1049–59. doi: 10.1016/j.humimm.2011.07.303
36. Weinstock C, Schnaidt M. The Complement-Mediated Prozone Effect in the Luminex Single-Antigen Bead Assay and Its Impact on HLA Antibody Determination in Patient Sera. Int J Immunogenet (2013) 40(3):171–7. doi: 10.1111/j.1744-313X.2012.01147.x
37. Kramer CSM, Franke-van Dijk MEI, Priddey AJ, Pongrácz T, Gnudi E, Car H, et al. Recombinant Human Monoclonal HLA Antibodies of Different IgG Subclasses Recognising the Same Epitope: Excellent Tools to Study Differential Effects of Donor-Specific Antibodies. HLA (2019) 94(5):415–24. doi: 10.1111/tan.13664
38. El-Awar NR, Akaza T, Terasaki PI, Nguyen A. Human Leukocyte Antigen Class I Epitopes: Update to 103 Total Epitopes, Including the C Locus. Transplantation (2007) 84(4):532–40. doi: 10.1097/01.tp.0000278721.97037.1e
39. Duquesnoy RJ, Marrari M, Mulder A, Claas FH, Mostecki J, Balazs I. Structural Aspects of Human Leukocyte Antigen Class I Epitopes Detected by Human Monoclonal Antibodies. Hum Immunol (2012) 73(3):267–77. doi: 10.1016/j.humimm.2011.11.011
40. Duquesnoy RJ, Marrari M, Jelenik L, Zeevi A, Claas FH, Mulder A. Structural Aspects of HLA Class I Epitopes Reacting With Human Monoclonal Antibodies in Ig-Binding, C1q-Binding and Lymphocytotoxicity Assays. Hum Immunol (2013) 74(10):1271–9. doi: 10.1016/j.humimm.2013.05.016
41. El-Awar N, Lee JH, Tarsitani C, Terasaki PI. HLA Class I Epitopes: Recognition of Binding Sites by Mabs or Eluted Alloantibody Confirmed With Single Recombinant Antigens. Hum Immunol (2007) 68(3):170–80. doi: 10.1016/j.humimm.2006.11.006
42. Akaza T, El-Awar N, Nguyen A, Kitawaki J, Terasaki P. HLA Class I Epitopes: C-Locus. Clin Transpl (2006) 95–102.
43. Navarette C, Brown C, de Lange P, Schreuder GMT. 12th International Histocompatibility Workshop HLA Class II Monoclonal Antibodies Study. In: Charron D, editor. HLA: Genetic Diversity of HLA: Functional and Medical Implications Volume 1 - Proceedings of the Twelfth International Histocompatibility Workshop. Paris, France: EDK (1997). p. 11–7.
44. Kramer CSM, Franke-van Dijk MEI, Bakker KH, Uyar-Mercankaya M, Karahan GE, Roelen DL, et al. Generation and Reactivity Analysis of Human Recombinant Monoclonal Antibodies Directed Against Epitopes on HLA-DR. Am J Transplant (2020) 20(12):3341–53. doi: 10.1111/ajt.15950
45. Cai J, Terasaki PI, Mao Q, Pham T, El-Awar N, Lee JH, et al. Development of Nondonor-Specific HLA-DR Antibodies in Allograft Recipients Is Associated With Shared Epitopes With Mismatched Donor DR Antigens. Am J Transplant (2006) 6(12):2947–54. doi: 10.1111/j.1600-6143.2006.01560.x
46. Bezstarosti S, Kramer CSM, Franke-van Dijk MEI, Vergunst M, Bakker KH, Uyar-Mercankaya M, et al. HLA-DQ-Specific Recombinant Human Monoclonal Antibodies Allow for In-Depth Analysis of HLA-DQ Epitopes. Front Immunol (2022) 12:761893. doi: 10.3389/fimmu.2021.761893
47. El-Awar N, Nguyen A, Almeshari K, Alawami M, Alzayer F, Alharbi M, et al. HLA Class II DQA and DQB Epitopes: Recognition of the Likely Binding Sites of HLA-DQ Alloantibodies Eluted From Recombinant HLA-DQ Single Antigen Cell Lines. Hum Immunol (2013) 74(9):1141–52. doi: 10.1016/j.humimm.2013.05.013
48. Cano P, Fernández-Viña M. Two Sequence Dimorphisms of DPB1 Define the Immunodominant Serologic Epitopes of HLA-DP. Hum Immunol (2009) 70(10):836–43. doi: 10.1016/j.humimm.2009.07.011
49. Deng CT, El-Awar N, Ozawa M, Cai J, Lachmann N, Terasaki PI. Human Leukocyte Antigen Class II DQ Alpha and Beta Epitopes Identified From Sera of Kidney Allograft Recipients. Transplantation (2008) 86(3):452–9. doi: 10.1097/TP.0b013e3181804cd2
50. Ge J, Hannestad K. A Cytotoxic Human Hybridoma Monoclonal Antibody (TrJ6) Defining an Epitope Expressed by HLA-DQ4 and -DQ5. Hum Immunol (1994) 39(2):106–12. doi: 10.1016/0198-8859(94)90108-2
51. Wölpl A, Halder T, Kalbacher H, Neumeyer H, Siemoneit K, Goldmann SF, et al. Human Monoclonal Antibody With T-Cell-Like Specificity Recognizes MHC Class I Self-Peptide Presented by HLA-DR1 on Activated Cells. Tissue Antigens (1998) 51(3):258–69. doi: 10.1111/j.1399-0039.1998.tb03100.x
52. Löffler D, Welschof M, Goldmann SF, Wölpl A. Recognition of HLA-DR1/DRB1*0101 Molecules Presenting HLA-A2 Derived Peptides by a Human Recombinant Antibody, Fab-5 A1. Eur J Immunogenet (1998) 25(5):339–47. doi: 10.1046/j.1365-2370.1998.00120.x
53. Simmons DP, Kafetzi ML, Wood I, Macaskill PC, Milford EL, Guleria I. Antibodies Against HLA-DP Recognize Broadly Expressed Epitopes. Hum Immunol (2016) 77(12):1128–39. doi: 10.1016/j.humimm.2016.09.008
54. Marrari M, Mostecki J, Mulder A, Claas F, Balazs I, Duquesnoy RJ. Human Monoclonal Antibody Reactivity With Human Leukocyte Antigen Class I Epitopes Defined by Pairs of Mismatched Eplets and Self-Eplets. Transplantation (2010) 90(12):1468–72. doi: 10.1097/TP.0b013e3182007b74
55. Hurley CK, Kempenich J, Wadsworth K, Sauter J, Hofmann JA, Schefzyk D, et al. Common, Intermediate and Well-Documented HLA Alleles in World Populations: CIWD Version 3.0.0. HLA (2020) 95(6):516–31. doi: 10.1111/tan.13811
56. Heidt S, Claas FHJ. Not All HLA Epitope Mismatches Are Equal. Kidney Int (2020) 97(4):653–5. doi: 10.1016/j.kint.2019.12.017
57. McCaughan JA, Battle RK, Singh SKS, Tikkanen JM, Moayedi Y, Ross HJ, et al. Identification of Risk Epitope Mismatches Associated With De Novo Donor-Specific HLA Antibody Development in Cardiothoracic Transplantation. Am J Transplant (2018) 18(12):2924–33. doi: 10.1111/ajt.14951
58. Hönger G, Niemann M, Schawalder L, Jones J, van Heck MR, van de Pasch LAL, et al. Toward Defining the Immunogenicity of HLA Epitopes: Impact of HLA Class I Eplets on Antibody Formation During Pregnancy. HLA (2020) 96(5):589–600. doi: 10.1111/tan.14054
59. Schawalder L, Hönger G, Kleiser M, van Heck MR, van de Pasch LAL, Vendelbosch S, et al. Development of an Immunogenicity Score for HLA-DQ Eplets: A Conceptual Study. HLA (2021) 97(1):30–43. doi: 10.1111/tan.14110
60. Tassone G, De Santis D, Vukovic I, Downing J, Martinez OP, D'Orsogna LJ. Different Eplet Software Programs Give Discordant and Incorrect Results: An Analysis of HLAMatchmaker vs Fusion Matchmaker Eplet Calling Software. HLA (2020) 96(1):52–63. doi: 10.1111/tan.13897
61. Jucaud V, Ravindranath MH, Terasaki PI. Conformational Variants of the Individual HLA-I Antigens on Luminex Single Antigen Beads Used in Monitoring HLA Antibodies: Problems and Solutions. Transplantation (2017) 101(4):764–77. doi: 10.1097/tp.0000000000001420
62. Grenzi PC, de Marco R, Silva RZ, Campos EF, Gerbase-DeLima M. Antibodies Against Denatured HLA Class II Molecules Detected in Luminex-Single Antigen Assay. Hum Immunol (2013) 74(10):1300–3. doi: 10.1016/j.humimm.2013.06.035
63. Ravindranath MH, Jucaud V, Ferrone S. Monitoring Native HLA-I Trimer Specific Antibodies in Luminex Multiplex Single Antigen Bead Assay: Evaluation of Beadsets From Different Manufacturers. J Immunol Methods (2017) 450:73–80. doi: 10.1016/j.jim.2017.07.016
Keywords: eplet, epitope-matching, transplantation, human leukocyte antigen, antibody verification, reactivity pattern, monoclonal antibody
Citation: Bezstarosti S, Bakker KH, Kramer CSM, de Fijter JW, Reinders MEJ, Mulder A, Claas FHJ and Heidt S (2022) A Comprehensive Evaluation of the Antibody-Verified Status of Eplets Listed in the HLA Epitope Registry. Front. Immunol. 12:800946. doi: 10.3389/fimmu.2021.800946
Received: 24 October 2021; Accepted: 30 December 2021;
Published: 28 January 2022.
Edited by:
Julie Ho, University of Manitoba, CanadaReviewed by:
Rhonda Holdsworth, Australian Red Cross Blood Service, AustraliaMepur Hanumantha-Rao Ravindranath, Children’s Hospital of Los Angeles, United States
Copyright © 2022 Bezstarosti, Bakker, Kramer, de Fijter, Reinders, Mulder, Claas and Heidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastiaan Heidt, cy5oZWlkdEBsdW1jLm5s
†ORCID: Suzanne Bezstarosti, orcid.org/0000-0002-0315-115X
Cynthia S. M. Kramer, orcid.org/0000-0003-1350-2336
Marlies E. J. Reinders, orcid.org/0000-0001-9543-567X
Arend Mulder, orcid.org/0000-0001-7805-7064
Frans H. J. Claas, orcid.org/0000-0003-4157-6201
Sebastiaan Heidt, orcid.org/0000-0002-6700-188X
 Suzanne Bezstarosti
Suzanne Bezstarosti Kim H. Bakker1
Kim H. Bakker1 Cynthia S. M. Kramer
Cynthia S. M. Kramer Johan W. de Fijter
Johan W. de Fijter Arend Mulder
Arend Mulder Frans H. J. Claas
Frans H. J. Claas Sebastiaan Heidt
Sebastiaan Heidt