- 1Department of Molecular Biology, Ariel University, Ariel, Israel
- 2Department of Computer Sciences, Ariel University, Ariel, Israel
- 3Department of Life Sciences, Ben-Gurion University of the Negev, Beer-Sheva, Israel
The closely linked recombination activating genes (RAG1 and RAG2) in vertebrates encode the core of the RAG recombinase that mediates the V(D)J recombination of the immunoglobulin and T-cell receptor genes. RAG1 and RAG2 homologues (RAG1L and RAG2L) are present in multiple invertebrate phyla, including mollusks, nemerteans, cnidarians, and sea urchins. However, the function of the invertebrates’ RAGL proteins is yet unknown. The sea urchins contain multiple RAGL genes that presumably originated in a common ancestral transposon. In this study, we demonstrated that two different RAG1L genes in the sea urchin Paracentrutus lividus (PlRAG1La and PlRAG1Lb) lost their mobility and, along with PlRAG2L, were fully domesticated to carry out different functions. We found that the examined echinoid RAGL homologues have distinct expression profiles in early developmental stages and in adult tissues. Moreover, the predicted structure of the proteins suggests that while PlRAG1La could maintain its endonuclease activity and create a heterotetramer with PlRAG2L, the PlRAG1Lb adopted a different function that does not include an interaction with DNA nor a collaboration with PlRAG2L. By characterizing the different RAG homologues in the echinoid lineage, we hope to increase the knowledge about the evolution of these genes and shed light on their domestication processes.
Introduction
The recombination activating genes, RAG1 and RAG2, are the primary facilitators of the V(D)J recombination of the antibodies and T-cell receptor genes in the vertebrates (1, 2). The vertebrate RAG-1/2 complex recognizes and binds asymmetric recombination signal sequences (RSSs), flanking the V, D, and J gene segments of the Immunoglobulin (Ig) genes according to the 12/23 rule (3). The complex nicks the DNA helix at the junction with the coding gene segments. The intervening DNA is removed, and the randomly chosen coding segment pair is eventually ligated together (4). Just before the ligation of the segments, another diversification process occurs at the junction of the recombined genes segments. Several proteins are involved in the process, including general DNA non-homologous end-joining and repair enzymes and two dedicated V(D)J recombination proteins; the Artemis nuclease (also known as DNA Cross-Link Repair 1C – DCLRE1C) and the Terminal deoxynucleotidyl transferase (TdT). These proteins are involved in the nicking of the hairpin loop at the end of the pre-ligated coding segments and the addition of random N-nucleotides at the junction (5).
RAGs originated from a transposon but are thought to have been evolved predominantly in the vertical way (6–8). According to the transposon/split receptor gene hypothesis, the same transposon gave rise to vertebrate RAG genes and simultaneously caused the split of a primordial Ig gene that later resulted in the formation of the V, D and J segments (7, 9, 10). We recently proposed an alternative hypothesis in which a separate genetic element, not a RAG1/2 transposon, was responsible for the Ig split. Based on the similarity between the RSSs and the Terminal Inverted Repeats (TIRs) of some Transib transposons, we suggested that the invading element which caused the Ig split was a Transib-like transposon (11). This hypothesis allows a more graduate, stepwise evolutionary explanation for the formation of the V(D)J recombination system in which the RAGs were first domesticated to carry out a different function and only later became a recombinase. We proposed that the initial role of the primordial vertebrate RAGs was to guard the vertebrate genome from the invasion of similarly structured harmful transposons.
RAGs-like (RAGL) genes are widely found in invertebrates, both in deuterostomes (echinoderms, cephalochordates and hemichordates) and in protostomes (mollusks, nemerteans and cnidarians). These invertebrate RAGL genes are found as RAG1/2L pairs, residing in transcriptionally convergent tail-to-tail configuration, as unpaired RAG1L genes and more rarely unpaired RAG2L genes, or as RAGL pseudogenes (12, 13). Many of the RAGLs still have some signatures of transposition activity, including Terminal Inverted Repeats (TIRs), the DNA recognition and cleavage sites, and 5 bp-long target site duplications (TSD) that are left on both DNA strands after a transposition (14, 15). A “living fossil” RAG1/2L transposon was discovered in lancelets and was termed protoRAG (16). This finding provided a proof of concept for the assumption the RAG1/2 loci (in both vertebrates and invertebrates) originated from a single transposon and suggested that active RAGL transposons may still exist. In fact, several such candidate transposons were suggested to exist in different taxonomic groups, such as insects (N. geniculatus), oysters and corals (13). On the other hand, some invertebrate RAGL genes lack TIRs and thus are suspected to be domesticated (13, 17).
In sea urchins, both RAG1/2L pairs and unpaired RAG1L genes were identified (10, 12, 13). Furthermore, it was demonstrated that two Strongylocentrotus purpuratus RAG genes, SpRAG1 and SpRAG2L, are co-expressed in early developmental stages and in coelomocytes, and the encoded proteins create a stable complex, similar to the vertebrate RAG proteins (12). The SpRAG1/2L genes encode full-length proteins with all major functional domains as in the vertebrate RAGs, including a typical vertebrate RAG1 core with a DDE catalytic motif (17) and a functional PHD domain in SpRAG2 (18). Furthermore, SpRAG1L binds to the vertebrate RSSs with 12 bp spacers in the presence of vertebrate high mobility group (HMG) proteins (12).
Although we know that RAG1L and RAG2L genes in sea urchins are expressed, little is known about their specific expression patterns and location in early developmental stages and in adult tissues. In fact, the only verified information is from a single pioneer study about RAG1L and RAG2L genes in S. purpuratus (12). It is also unclear whether the transposition of RAGs is still ongoing in sea urchins. Furthermore, attention was so far paid exclusively to the RAG1/2L pair and not to the rest of the unpaired RAGL genes, and the functionality of these genes in sea urchins remains to be uncovered. In this study, we aimed to characterize and compare different RAGL gene sequences in sea urchins and to understand whether and when these genes were domesticated. We specifically chose to focus on two P. lividus RAG1L genes – the first (PlRAG1a) is in the same genetic locus (paired) with PlRAG2L, while the second (PlRAG1b) is unpaired. We found that the paired and unpaired genes are immobile and expressed differently in early developmental stages and in adults. Based on the above, we conclude that the two PlRAGL genes were recently domesticated to perform different, unrelated functions. An analysis of the functional protein structure suggests that PlRAG1a may work together with PlRAG2L as a nuclease while the PlRAG1b act alone and have adopted a different function altogether outside of the nucleus.
Methods
Culturing of sea urchins and obtaining embryos and larvae
Sea urchins (P. lividus) were obtained from the Israeli National Center for Mariculture in Eilat. Urchins were kept in artificial seawater (Red Sea Fish Pharm Ltd., Herzliya, Israel) in a 165-L aquarium in the Molecular Ecology Laboratory at Ariel University, of which 1/10 of the volume was replaced every week. The water salinity was kept around 40 ppt and the temperature between 18 and 22°C. The animals were fed once a week with either fresh or frozen Ulva lactuca algae. For obtaining P. lividus embryos and larvae, adult sea urchins were first injected with 0.5M KCl to induce spawning. Fertilization was made in filtered artificial seawater and developed embryos were maintained up to 72 hours post fertilization (pluteus larva, after which larvae start to feed).
RNA isolation and cDNA creation
Total RNA was isolated with the RNeasy Mini Kit (Qiagen). The purification included a DNase treatment using the RNase-free DNase Set (Qiagen). The yield and purity and integrity of the RNA were measured using NP-80 spectrophotometer (Implen) and by gel electrophoresis. cDNA was created by qPCRBIO cDNA Synthesis Kits (PCR Biosystems, Tamar, Israel) according to manufacturer instructions.
qPCR
Quantitative PCR reactions were executed in duplicates or triplicates in 96-well plates using QuantStudio real-time PCR (Thermo Fisher Scientific) with SYBR Green PCR mix (BioRad). Relative expression quantification was performed using delta-delta-CT with PlTubulin as a reference gene. To normalize the expression level across different tissues/organ, average expression value of the tested gene in each of the tissue/organ samples, was divided by the sum of the total expression of that gene in all tissues. Primers used: for PLRAG1a (Pliv25095.1) - F CCCTGGAAGAACTGGATGATAG R TCTTTGATGGTCGTCTCGATAAC, for PLRAG1b (Pliv07077.1) - F GAGGAGCTGGATGACCAAATTA R ACCATCTGCTCCATCCTTTATG, for PLRAG2L (Pliv25741.1) F GGAGATGCGATGACGGATATT R GATACGCCCTAGCTGTTGTT, for PlArtemisL (Pliv23693.1) - F CAACAGAGGGCAGTCTAACA R TGGTACTGGGTGTGATCATAAG, for PlTdTL (Pliv29804.1) - F CTAAGAACTCTGGGAGACGTAAAG R GGCGTGTTCAGTTCATCAAAG.
In-situ probes preparation
P. Lividus RAG1b template sequence was amplified using the following primers – F AAACCAAAGGACGTCTCGCAAG R TGGAGTTACAGGGAGGCAGTCG. PCR amplicons were purified using QiaQuick PCR purification kit (Qiagen). Amplicons were then cloned into a pDrive cloning vector (Invitrogen) using the Qiagen PCR Cloning Kit (Qiagen) and transformed into E. coli bacteria. Positive colonies were grown in selective liquid LB media overnight, followed by purification using QIAprep Spin Miniprep Kit (Qiagen). The success of the cloning was verified by PCR and sequencing. DIG-labeled RNA probes were generated from the recombinant plasmids using DIG-RNA Labeling Kit (Roche). Sense and antisense probes were generated, using SP6 and T7 promoters flanking the cloning sites. Probes were stored in aliquotes at -80°C until use.
Whole mount In-situ hybridization
The WhMISH was performed according to (19, 20). Briefly, the embryos were fixed with 4% formaldehyde, washed and prehybridized in the 96 well plates for 3 h in a hybridization buffer at 65°C. The hybridization was carried out overnight at 65°C. The embryos were thoroughly post-washed, blocked and incubated with AntiDIG-AP antibody overnight. The washed embryos were stained with NBT/BCIP and observed under a microscope.
Databases and publications screening
The search for RAGL genes was performed in both annotated and not annotated sea urchin genomes by gene name and/or by sequence comparison using the SpRAG1L and SpRAG2L aa sequence. The comparisons were performed with tblastn (aa sequence against translated nucleotide sequences) against the NCBI gene bank and other databases of S. purpuratus (https://www.ncbi.nlm.nih.gov/assembly/GCF_000002235.5/), H. pulcherrimus (https://cell-innovation.nig.ac.jp/cgi-bin/Hpul_public/Hpul_annot_home.cgi), P. lividus (https://www.ncbi.nlm.nih.gov/assembly/GCA_940671915.1/, http://octopus.obs-vlfr.fr/blast/oursin/blast_oursin.php), and L. variegatus (https://www.ncbi.nlm.nih.gov/assembly/GCF_018143015.1/). For RAGL expression data, a comprehensive search was performed in the following TSA databases: GHFM00000000.1, HAMP00000000.1, GHJZ00000000.1, GGVM00000000.1, GAPB00000000.1, GECD00000000.1, GAVR00000000.1, GAZP00000000.1, GAVF00000000.1, GCZS00000000.1, GEDS00000000.1, GFRN00000000.1, GIIR00000000.1, HACU00000000.1 corresponding to S. purpuratus, Echinometra sp., Mesocentrotus franciscanus, Loxechinus albus, Evechinus chloroticus, Arbacia punctulata, Echinarachnius parma, Eucidaris tribuloides, Sphaerechinus granularis sea urchin species. Available literature was screened for RAGLs expression data in the sea urchins S. purpuratus, Strongylocentrotus droebachiensis, L. variegatus, Psammechinus miliariswas, results were organized as a table (Table S4).
RAG expression analysis
To identify expression level we analyzed data from four publicly available RNA-seq datasets (GSE149221 – single cell experiment (21), GSE134350 – single cell experiment (22), GSE97448, PRJNA81157 (23, 24). Those datasets profiled developmental stages or immune challenges of S. purpuratus tissues. When the expression table was available, it was downloaded from the experiment site. In the single-cell experiments, python programs were written to sum up the expression for each gene in all cells. In the case of PRJNA81157, reads were downloaded and mapped to S. purpuratus genome (https://www.ncbi.nlm.nih.gov/assembly/GCF_000002235.5) using hisat2 (25) (version 2.0.5). Bam files were sorted and indexed by SAMtools (26) (version 1.9). The expression level (high, low or no expression) was determined by visually inspection using IGV tool (27). Low expression was regarded as less than 10 reads in the junctions.”
Phylogenetic analysis
For the phylogenetic analysis we used sequences of which transcripts were identified with blast score > 100 and E value < 1e-18. Transcription information was obtained from TSA databases: GEDS00000000.1, GFRN01000000.1, GIIR00000000.1, HACU00000000.1 (P. lividus), GJVT00000000.1, GHFM00000000.1, GAVU00000000.1 (S. purpuratus), GAUR00000000.1 for L. variegatus and transcriptomic databases available in public genomic viewers for H. pulcherrimus https://cell-innovation.nig.ac.jp/cgi-bin/Hpul_public/Hpul_annot_home.cgi, and P. lividus https://www.ncbi.nlm.nih.gov/assembly/GCA_940671915.1/. Phylogenetic relationships of the echinoid RAG1L translated protein sequences were inferred with MUSCLE alignment with default parameters. Tested models included JTT and WAG, resulted in similar tree typologies. We chose to present a maximum likelihood method tree created with Whelan and Goldman (WAG) model (28) with a consensus tree inferred from 500 bootstrap replicates.
Nanopore sequencing of amplified genomes from single sea urchin cells
Single sperm cells and coelomocytes of S. pupuratus were previously isolated using limited dilutions and subjected to whole genome amplification procedure (29). Amplified genome from one sperm and one coelomocyte were subjected to whole genome sequencing using MinION nanopore sequencing platform. The sequencing was performed according to Nanopore SQK-LSK109 protocol for long reads with MinION Nanopore apparatus (Oxford Nanopore Technologies). ~ 200 fmol of purified amplification products were subjected to DNA repair and end-prep using a NEBNext DNA repair mix and NEBNext Ultra II End Repair/dA-Tailing Module (New England Biolabs). DNA purification steps were carried out by the SPRI magnetic beads (Canvax, Spain). Base-calling was done automatically by the MinKnow program. Raw reads were obtained in FAST5 and FASTQ formats from which “pass” quality reads were cleaned from adaptor sequences by porechop (30) and uploaded to SRA database (accession SAMN31216822, SAMN31216823) and subjected to further analysis.
Sequencing data analysis and assembly
Sperm and coelomocyte reads were sorted by quality by samtools (26) and aligned to the last version of S. purpuratus genome (Accession: GCF_000002235.5) by minimap2 (31), aligned reads were sorted and indexed by the samtools (26) and visualized in IGV genome browser (27). Long reads that overlapped with RAGs and/or the surrounding genes were identified by BLAST.
Protein 3-D structure
The structure of the PIRAG1 type A and PIRAG2 complexes with DNA was modeled using th3 or comparative modeling software MODELLER (32). The template used is the structure of BbRAGL-3’TIR synaptic complex with nicked DNA (PDB code 6B40 (33)) from Branchiostoma belcheri. The percentage sequence identity (similarity) between sequence and it template structure is 42.4 (58.9%) % for PIRAG1 Type A and 30.6 (49.8) % for PIRAG2. At this level of sequence similarity, the resulting model is expected to show RMS error rises to about 1.5 Å for about 80% of residues from the correct structure as noted by Fiser A. Template-based protein structure modeling (34). The same nicked DNA of the template structure was used in the model. Inter chain (or subunits) residues with atoms within a cutoff distance of 4.5Å are defined as contacting residues (pyMOL).
Results
Echinoid RAG1L genes evolved through multiple lineages
To characterize the different sea urchin RAG1L genes, we investigated four available sea urchin genomes of S. purpuratus, P. lividus, Hemicentrotus pulcherrimus and Lytechinus variegatus. In contrast to jawed vertebrates, which have a single RAG1 copy, available sea urchin genomes contain multiple copies of RAG1L sequences. The genomes that were screened contained between three sequence matches (in L. variegatus genome) and up to 7 sequences (in P. lividus) with a similarity blast score of more than 100 when compared with the complete SpRAG1L sequence (Table S1). About 60% of the RAG1L sequences were not annotated as genes in the genomic databases and did not appear in any of the expression databases. We, therefore, regarded these sequences as potential pseudogenes, perhaps reminiscence of past transposition activity, and excluded them from our analysis. The sea urchin RAG1L genes can be fitted into one of two main categories according to their genomic organization: the first category consists of RAG1L genes that are found next to RAG2L genes in tail-to-tail transcriptionally convergent orientation (Figure 1A), in similar to the lancelet protoRAG and the vertebrate RAGs loci (from now on will be termed type I). RAG1L type I genes were found in a single copy in each of the sea urchin genomes except for H. pulcherrimus, in which two RAG1L/RAG2L loci were identified on two separate genomic scaffolds (scaffold:2135 BEXV01002133.1 and scaffold:3118 BEXV01003119.1) (Figure 2). The second RAG1L category, consists of solitary RAG1L copies that are spread throughout the genome (Table S2), often in small clusters and sometimes as potential pseudogenes (will be termed type II). Both RAG1L gene types consist of multiple exons, as in the sea urchins P. lividus and S. purpuratus, suggesting they may be transcribed in several alternative isoforms (Figure 1A).
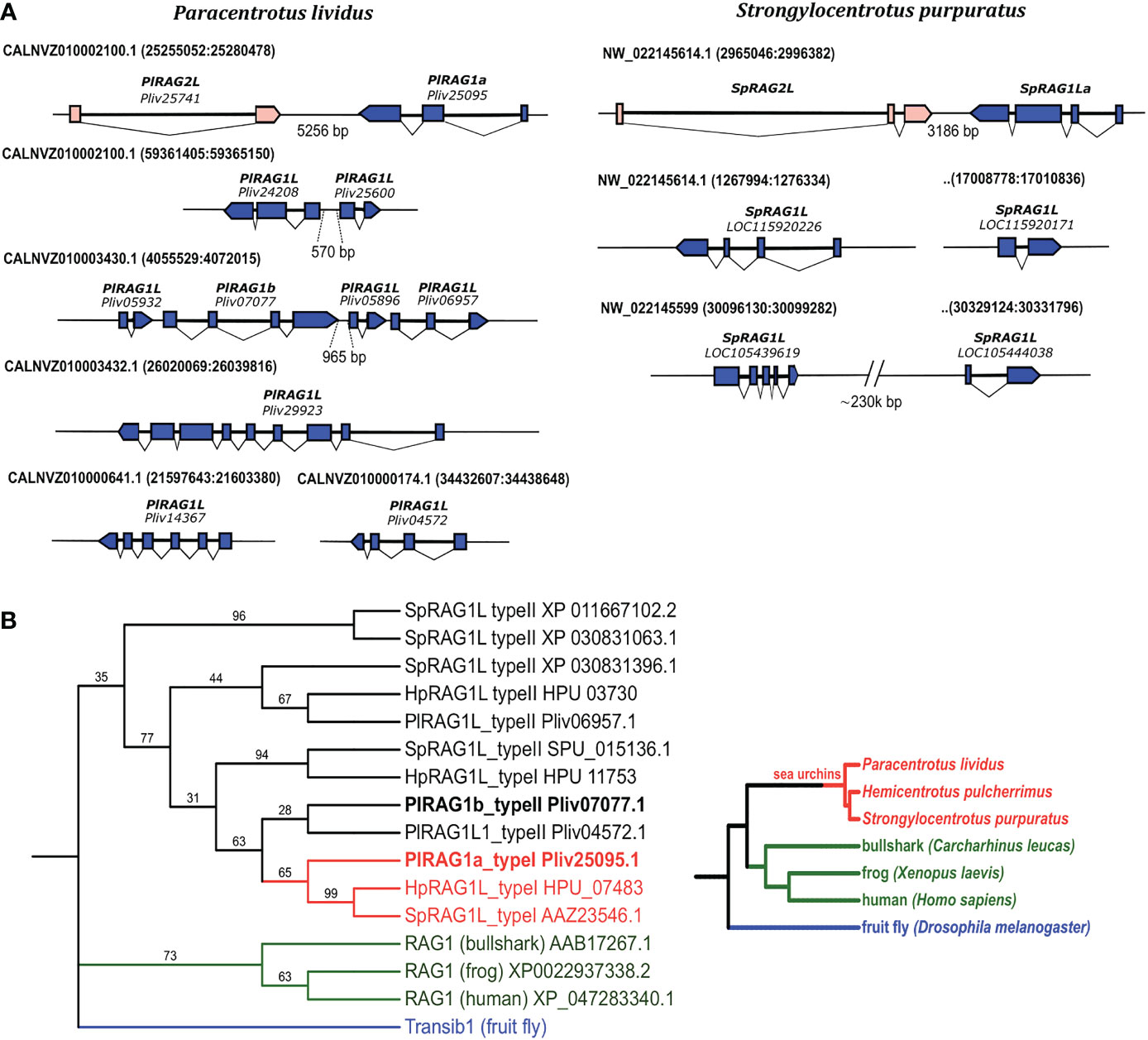
Figure 1 Genomic organization and phylogenetic relationships among echinoid RAG1L sequences. (A). annotated RAGL genes in the genomes of P. lividus and S. purpuratus. (B). phylogenetic relationships among echinoid RAG1L sequences. The phylogeny was made using transcribed sequences, based on maximum likelihood with Whelan and Goldman (WAG) model with consensus tree inferred from 500 bootstrap replicates. Drosophila melanogaster Transib autonomous protein was chosen as the outgroup (repbase https://www.girinst.org/server/RepBase/index.php accessed on July 2022). A corresponding guideline phylogeny of species is presented on the right to the phylogenetic relation of RAG1L translated protein sequences.
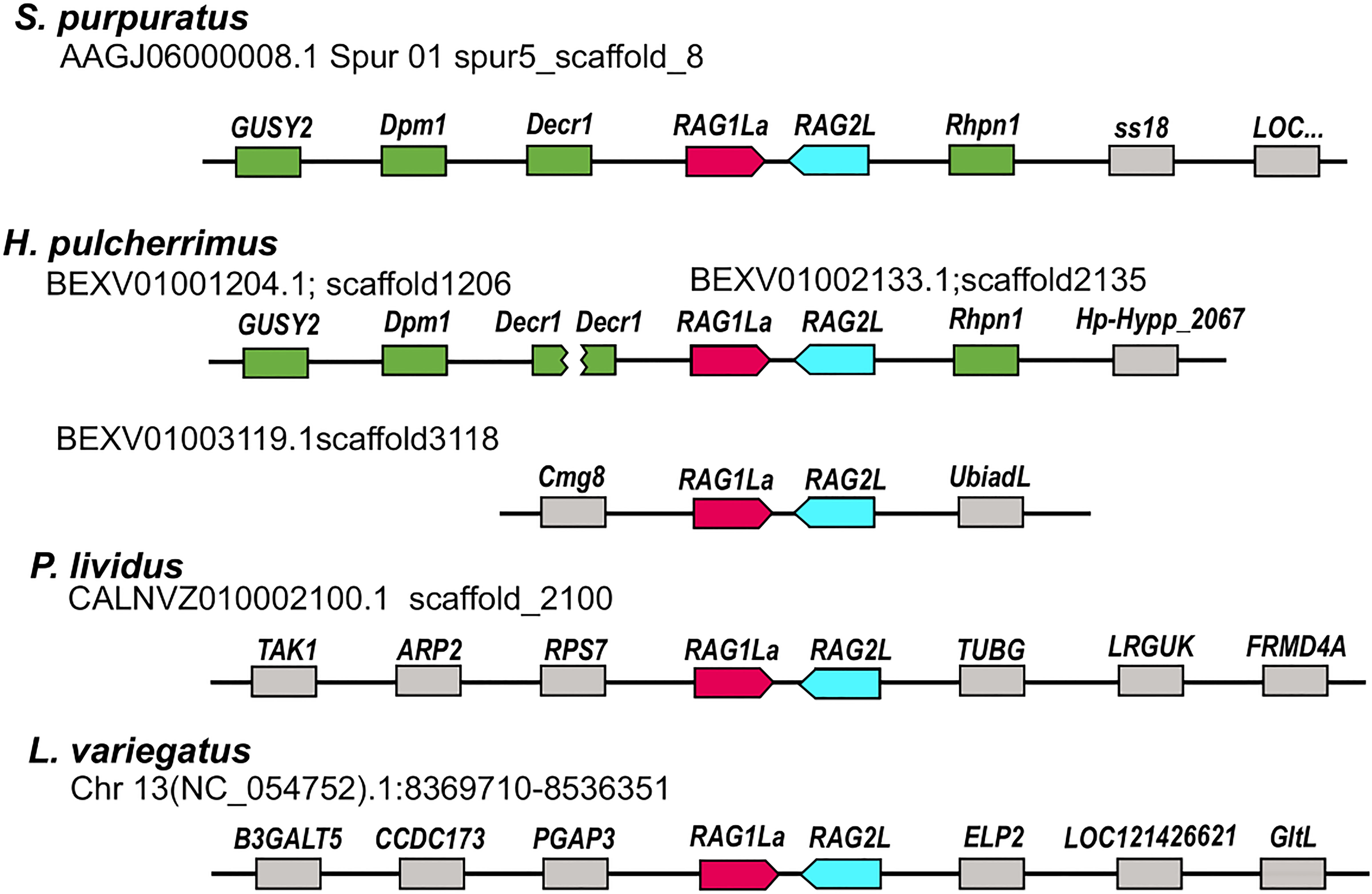
Figure 2 Relative genomic locations of type I RAGL genes in sea urchin genomes. The relative genomic location of RAG1/2 genomic pairs in S. purpuratus, P. lividus, H. pulcherrimus and L. variegatus is presented. Matching neighboring genes are marked in green. The SpRAG1/2L and HpRAG1/2L loci are similarly neighboring rhodophilin-1 (Rhpn1) gene upstream to the RAG2L sequence. On the other side, upstream to the RAGL1 sequence, the RAG loci are adjacent to 2,4-Dienoyl-CoA Reductase-1 (DECR1) gene. The H. pulcherrimus genome scaffold BEXV01002133.1 is ending in the middle of DECR1 gene but another scaffold - BEXV01001204.1 includes the rest of the DECR1 gene sequence as well as two other genes with similarity to Dolichol-phosphate mannosyltransferase subunit 1 (DPM1) and Retinal Guanylate Cyclase 1-like (GUSY1) similarly to the corresponding neighboring RAG1/2L genes of the S. pupuratus genome.
To understand the phylogenetic relationship among echinoid RAG1L genes from the sea urchin species investigated, a phylogenetic tree was constructed based on the RAG1L translated amino acid sequences. Only the sequences previously annotated as genes or expressed (found in corresponding transcriptomic databases) were used for the tree. In the phylogenetic analysis, all echinoid RAG1L type I genes, except for one (HpRAG1L_type I HPU_11753), were clustered in a single clade (Figure 1B in Red), implying their homology to each other. On the other hand, RAG1L type II sequences were clustered in a polyphyletic manner in four clades, of which two were composed of genes from different sea urchin species (Figure 1B in Black). These results suggest that some of the sequences might have originated in separate RAG lineages formed within a common echinoid ancestor/s before the speciation events of the examined species took place.
Mobile RAGL sequences were domesticated in specific sea urchin species
The RAGL genes were previously identified in non-syntenic regions in the genomes of different sea urchin species (35, 36). To expand this analysis based on the newly added and improved genomic data, we compared the relative genomic location of the RAGL genes in all four available sea urchin genomes based on the identification of the neighboring genes. In almost all cases, RAGL genes (both type I and II) were found in non-synonymous positions in the echinoid genomes (Figure 2; Table S2). One exception was the SpRAG1L type I (accession: NW_022145614.1) and the HpRAG1L type I (located on scaffold BEXV01002133.1) that, together with their adjacent echinoid RAG2L, were found to be in corresponding genomic loci (Figure 2 genes marked in green). These findings suggest that RAG1L type I immobilization in these two species already occurred in a common ancestor around 6 to 8 million years ago when the divergence of these species took place (37, 38).
To test whether the sea urchin RAG genes are still mobile today, we performed a nanopore long-read sequencing of amplified genomes of a single sperm cell and a coelomocyte from S. purpuratus that were combined for the alignment of the reads to the S. pupuratus genome (GCF_000002235.5). The alignment suggested that all RAG1L sequences were not mobile and remained in synonymous positions in both S. purpuratus genomes. We note that because of the low coverage (3-5x), we were not able to assemble the genome fully. However, using the nanopore long reads, we were able to identify overlaps between the reads and large genomic areas upstream and downstream of the genes, which confirmed the linkage between the RAG1L genes and the neighboring genomic sequences and genes (Figure 3). We, therefore, considered the data reliable enough to conclude that the RAG1L genes are located within the same loci for this species and not active transposons.

Figure 3 SpRAGL genes are immobile in individual sperm and a coelomocyte. The IGV visualized alignment is shown for SpRAG1L type I and II (LOC115920226, LOC105439619, LOC105444038, LOC115920171) genes and their neighboring genes. Red arrows pointing to the RAG1 genes, purple triangles mark the mismatches. Reads with good mapping quality (over 60) are marked in grey, the reads with lower quality mapping are in white.
PlRAG1a and PlRAG1b: Similar genes with opposing expression patterns
To characterize the expression patterns of P. lividus RAG1L genes, we chose two transcribed genes (P. lividus database accessions: Pliv25095.1 and Pliv07077.1), with complete ORFs, representatives of type I and type II RAG1L and named them PlRAG1a and PlRAG1b accordingly (Figure 1). We tested the expression patterns of these genes using qPCR and whole-mount in-situ hybridization (WMISH) in P. lividus early life stages and adult tissues. We also tested the expression levels of the PlRAG2L gene and two of the vertebrate V(D)J recombination gene homologues; PlArtemis (Pliv23693.1) and Pl Terminal deoxynucleotidyl transferase (TdT) (Pliv29804.1). In the vertebrate lineage, these two genes work together with the RAG1/2 complex and therefore are expressed in various organs and co-expressed in the thymus (39). The translated PlArtemisL protein sequence showed 64% aa identity to the human protein (XP_047281606.1), and the PlTdT had 37,5% aa sequence identity compared to its human counterpart (NP_004079.3).
Our qPCR analysis showed the expression of all five tested genes in adult P. lividus tissue. Among them, the two PlRAGLs showed contrasting expression patterns. While PlRAG1a (type I) was co-expressed with PlRAGL2 mainly in the upper digestive system – in the esophagus and to a lower extent in the gut and in the gonads, the PlRAG1b (type II) was almost exclusively expressed in coelomocytes. Interestingly, similar to PlRAG1a and PlRAGL2, PlArtemis and PlTdT were co-expressed in the esophagus and gut. However, not like PlRAG1a and PlRAG2L, they also showed a similar expression level in the axial organ (Figure 4A). To test the expression patterns of the two P. lividus RAGs in the early P. lividus developmental stages, we performed qPCR and WMISH using embryos and larvae from six post-fertilization time points - 4h, 16h, 25h, 42h, 48h, and 72h post fertilization (PF). The qPCR of the early life stages showed expression of PlRAG1b, peaking at the 48h pluteus larva while PlRAG1a and PlRAG2 were not expressed at all (Figure 4B). The WMISH showed expression as early as the blastula stage where it was identified in the endoderm (16h PF), continuing through the pluteus larvae stages where it was found in the early gut (42h-72h PF) peaking at 42-48h PF (Figure 4C).
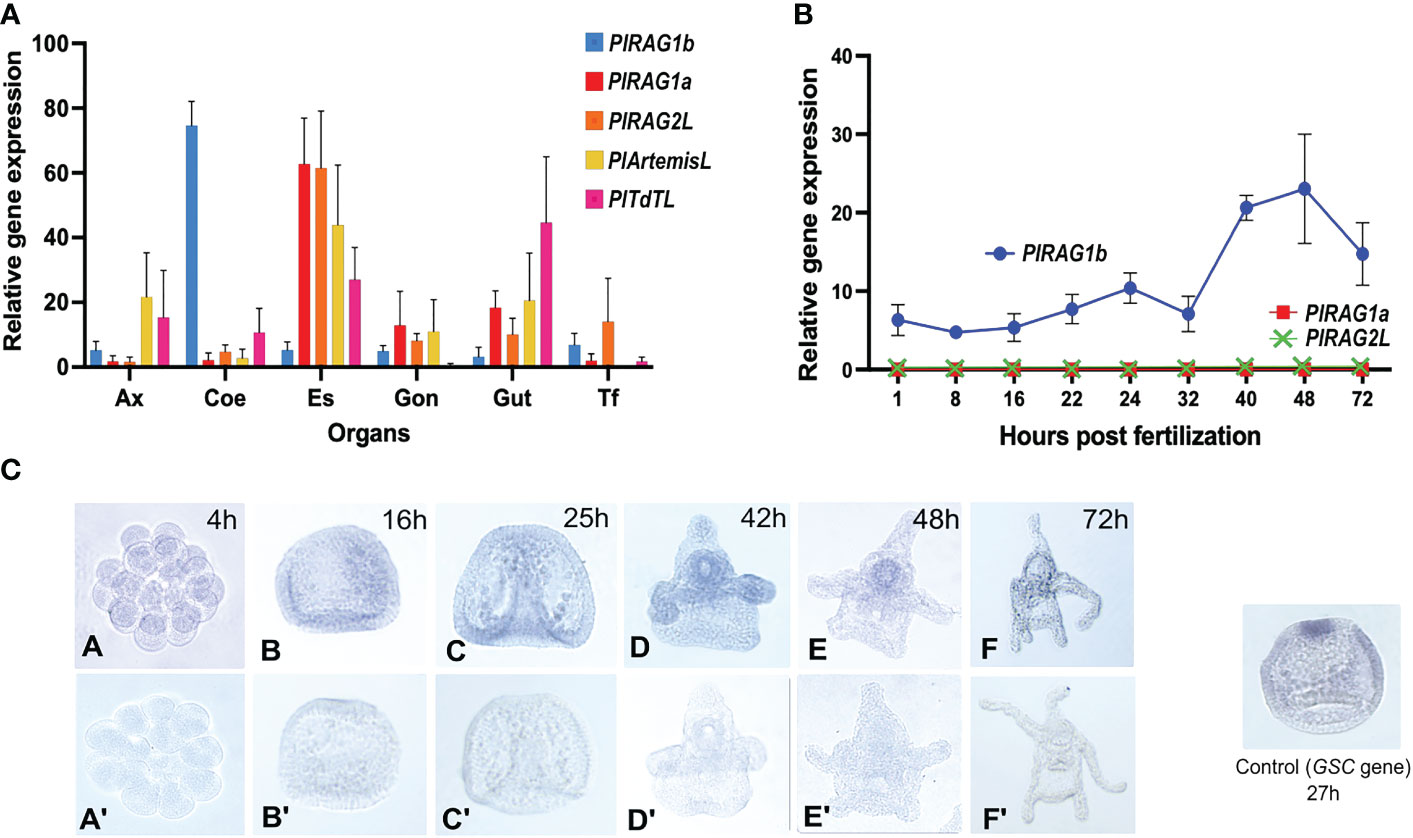
Figure 4 Expression patterns of PlRAGL1a, PlRAGL2, PlRAGL1b, PlArtemisL and PlTdTL genes in adult tissues and early developmental stages of P. lividus (A). Relative gene expression in of the tested genes in adult sea urchin tissues and organs by qPCR. Ax - axial organ, Coe - coelomocytes, Es - esophagus, Gon - gonads, Gut, Tf - tubefeet. (B). Relative gene expression of PlRAG1a and PlRAG1b in the early developmental stages by qPCR. The PlRAG2L expression is showed according to the PRJNA264358, PRJNA376650 and PRJEB10269 RNA-seq databases. (C). PlRAG1b expression in the developmental stages visualized by the in-situ hybridization. Gene accession numbers in octopus database (http://octopus.obs-vlfr.fr/): PlRAG1a - Pliv25095.1 (type I), PlRAG1b- Pliv07077.1 (type II), PlRAG2L- Pliv25741.1, PlArtemisL - Pliv23693.1, PlTdTL - Pliv29804.1.
To sum up the available knowledge on the expression of different RAGs, we searched available sequence read archives (SRAs) of S. purpuratus and transcriptome shotgun assembly (TSA) datasets (Table S3) of in the sea urchins S. purpuratus, Echinometra sp., Mesocentrotus franciscanus, Loxechinus albus, Evechinus chloroticus, Arbacia punctulata, Echinarachnius parma, Eucidaris tribuloides, Sphaerechinus granularis. We also screened the literature for relevant expression (Tables S4). The S. purpuratus SRA datasets analyses (Table S3) concluded in ambiguous results. Some developmental expression datasets, (e.g. GSE149221) clearly showed the presence of SpRAG1L and SpRAG2L, while others (e.g. GSE134350) resulted in non or very few RAGL transcripts. Unexpectedly, adult tissue transcriptomic datasets GSE97448 and PRJNA81157 also showed very little RAGL expression (Table S3). In most datasets in both developmental stages and adult organs the type II RAG1L genes were not present. While separately analyzing the available literature and associated TSA databases, when available, we were able to see the expression of RAGL1 type I and RAG2Ls in early developmental stages in the sea urchins S. purpuratus and Echinometra sp. (Table S4). RAG1L type I was also expressed in adult tissues of the sea urchins Psammechinus miliaris, Strongylocentrotus droebachiensis and L. variegatus. In P. lividus developmental transcriptomic dataset (Table S4), RAG1L type II transcripts were present in all tested developmental stages while transcripts of PlRAG type I and PlRAGL2 were (with the exceptions of low expression in PRJNA264358 and PRJNA376650), not found. Overall, this data agrees with our expression data.
Predicted PlRAG1 protein structures suggest that they differ in their intercellular location and function
The modular core of jawed vertebrate RAG1 protein is made of the domains that help it to bind to the DNA, particularly to recognize a nonamer and heptamer parts of DNA signal sequences and perform its endonuclease activity. Traditionally, both RAG proteins are subdivided into a “functional protein core” (384-1008 of RAG1 and 1-387 of RAG2 in mouse) that was proven to be able to carry out recombination on its own (40) and an N-terminal “non-core” regions. The RAG1 core includes a nonamer-binding domain (NBD), dimerization and DNA-binding domain (DDBD), pre- and RNase H domains (preR and RNH), Zn-binding domains, and the C-terminal domain (CTD) (41–43). The RAG2 core includes a six-bladed beta-propeller (WD40 repeat) domain, which interacts with both RAG1 and the DNA of the coding segment next to the heptamer sequence (44). RAG1 N-terminal “non-core” region contains structurally undefined regions as well as a Zinc Dimerization Domain (ZDD) that plays a role in the dimerization, stability and fidelity of the recombinase, and within it, the RING Zn finger motif that plays a role in polyubiquitination of RAG leading to the enhanced recombination activity (45, 46, 47). Additionally, protein domain prediction tools (e.g. InterPro) also recognize the importin binding domain (Imp-bd) in the N-terminus of RAG1 (48). The non-core region of RAG2 consists of a zinc finger plant homeodomain (PHD), which binds to the N-terminal tail of methylated histone 3 (H3K4me3) and is required for RAG2 correct interaction with the open chromatin during recombination (49).
The protein domain search in the echinoid RAGL-encoding loci was done according to the pre-defined domains listed in (13). As reported earlier, the ZDD was completely missing from all studied echinoid RAGLs (12, 33, 36). For type I RAGs - traces of NBD domain were found (17% and 13% for SpRAG1L and PLRAG1L to the Mus musculus RAG1), while all other domains were present with various (18-67%) levels of similarity (Table 1). The echinoid type II RAG sequences, on the other hand, lacked the non-core region and the NBD in most of the sequences, whereas the “core” domains were either defined or had insufficient similarity or were completely absent (Figure 5).
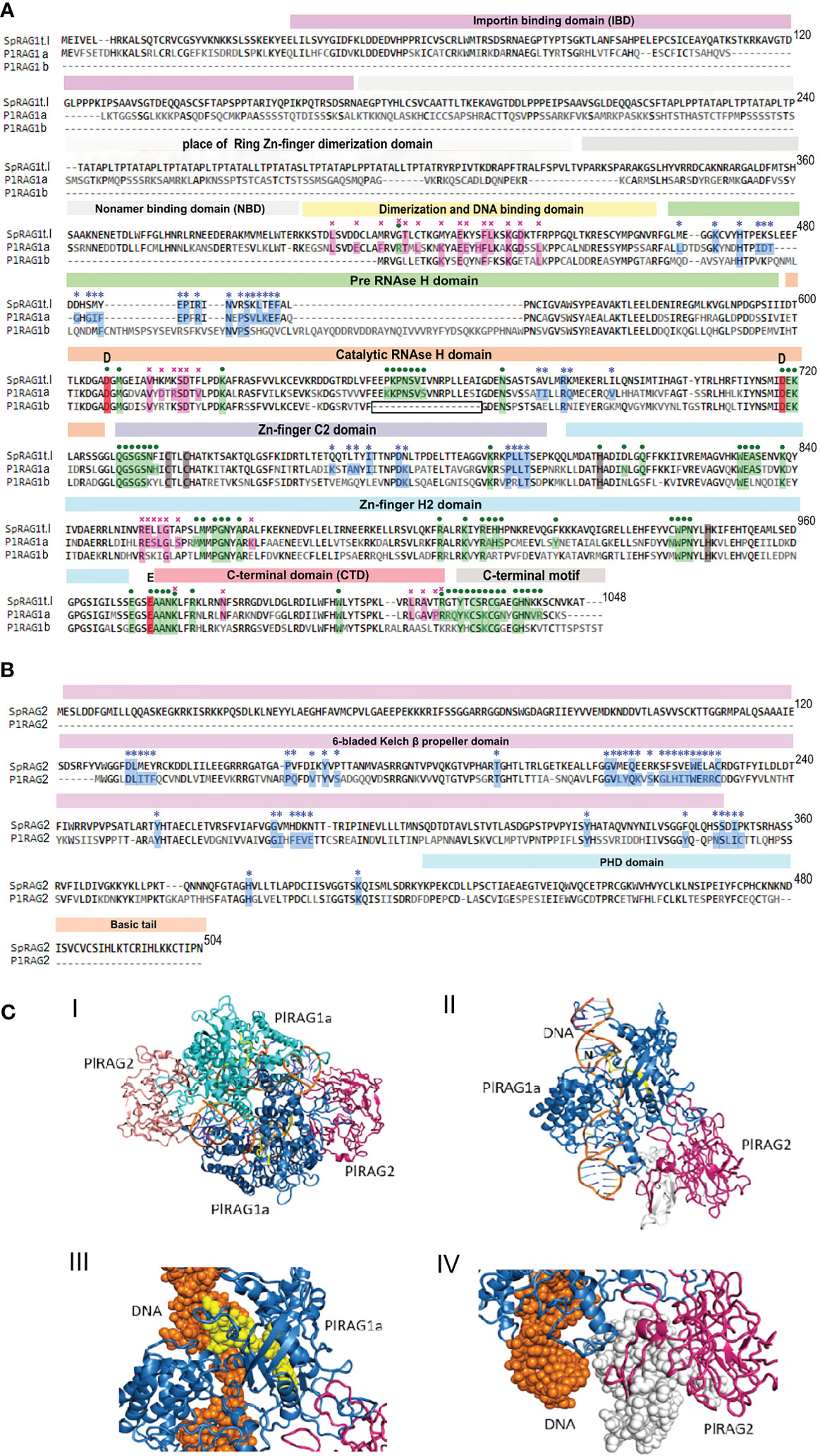
Figure 5 Predicted protein structure of PlRAGLs. (A). sequences alignment of SpRAG1L, PlRAG1a and PlRAG1b with domains (Martin, Vicari et al., 2020). blue stars – contacting residues with RAG2 (RAG1 for RAG2), purple crosses - RAG1 dimerization contacts, green dots - contacting residues of RAG1 with DNA. Grey areas - places that are aligned to the ZDD and NBD domains of Mus musculus RAG1 with low similarity. Boxed section - missing sequence in the catalytic RNH domain in the RAG1Lb protein sequence. (B). SpRAG2L and PlRAG2L sequence alighnment. (C). Model structure of P. lividus Rag1L-Rag2L-nicked DNA complex. (i) The P. lividus Rag1/2L_DNA dimeric structure is shown using cartoon representation. One PlRag1L/PlRag2L complex is colored skyblue/hotpink and the other cyan/salmon, with the DNA colored orange. (ii) Monomeric half of the PlRAGL complex with the exposed DNA nick (marked with ‘N’). The region colored in yellow is missing in the PlRAG1b predicted protein. The region colored in white is missing in the PlRAG2L compared to SpRAG2L (iii) Magnification of the missing DNA binding (in RNAse H domain) region in PlRAG1b (shown in yellow spheres) and its interaction with the nicked DNA helix (shown in orange) (IV) Magnification of the missing region in PlRAG2L (white spheres) as compared with the SpRAG2L. The figure was prepared using the PyMol software 3.
Multiple alignment of the translated protein sequences of the two PlRAGs and the type I S. purpuratus RAG1L (NW_022145614.1) showed high degree of sequence similarity among the proteins (Figure 5A). PlRAGL1a (type I) showed 95% of translated protein sequence coverage and 50% sequence identity compared to only 56% sequence coverage and 50% identity for PlRAGL1b to the SpRAG1l protein sequence. While PlRAGL1a seems to include all mentioned domains as in SpRAG1L, its N-terminal 1-159 aa sequence, which includes the ZDD, showed a lower degree of conservation (29% aa identity) (Figure 5A). On the other hand, the core catalytic domains and the C-terminal region were found to be highly conserved with 67% aa identity. Compared with the S. purpuratus RAG1L type I, the translated PlRAGL1b protein sequence lacks its first 407 aa (1–407) of the N-terminus non-core region, as well as an 18 aa-long section of the catalytic RNH-like domain (Figure 5A, boxed). On the other hand, it includes an additional 20 aa-long and 38 aa-long sequences in the middle of the RNH domain. The translated PlRAG2L protein sequence was found to be shorter than SpRAG2L, with 126 aa-long sequence missing from its N-terminal part. The rest of the protein sequence showed 50% identity to its S. purpuratus counterpart (Figure 5B).
A predicted 3-D model of the dimers complex of RAG1/2L_DNA and monomeric form in P. lividus (Figure 5C panels I and II, respectively) was build using homology modeling technique based on the crystal structure of the Branchiostoma belcheri protoRAG-DNA complex (50). Although missing non-core ZDD, our P. lividus model suggests the dimerization of a PlRAG1a is possible with the predicted 34 contacting residues and the formation of RAG1/2L complex with 37 contacting residues. The predicted RAG1/2L-DNA complex includes 72 contacting residues with the DNA strand (Figure 5A). We further used our P. lividus PlRAG1a- PlRAG2L-DNA model to compare it to the predicted structure of PlRAG1b and to SpRAG2L. In the first comparison, we found that the 18 aa-long missing “catalytic” section in PlRAG1b (Figure 5A, boxed) is a part that in PlRAG1a closely interacts with the area of the nick in the DNA double helix (Figure 5C panel III yellow). This section includes at least seven residues that could directly contact the DNA helix in the predicted RAG1/2L-DNA complex (Figure 5A, boxed section). We, therefore, assume that RAG1Lb, which lacks this section, has a weaker connection with the DNA and probably has no nuclease activity. The PlRAG2L seems to form a complex with PlRAG1a but is missing a 126 aa-long section of its N-terminal side of the core 6-bladed Kelch domain (Figures 5B, C panel IV white). Considering the fact that PlRAG1b has 20 fewer possible contacting residues with PlRAG2L, we suggest that PlRAG1b and PlRAG2L may not at all form a dimer.
Discussion
The interest in the RAGs and their unique evolutionary histories have been growing ever since the discovery of the RAG genes in vertebrates. In recent years additional data has been accumulated about various RAGL genes in different taxonomic groups of invertebrates, portraying a much wider presence of these genes throughout the tree of life. With the absence of the V(D)J recombination targets in the vertebrate lineage and the structural differences, it is yet not clear what is the function of these genes in invertebrate animals. Previous studies revealed that sea urchin RAGLs didn’t contain TIR sequences (12, 13), suggesting that they may not be functional transposons. For over a decade the question was raised whether sea urchin RAGLs genes have been domesticated, but attention was usually paid to the paired RAGL1/2 genes, because of their similar genomic organization to the vertebrate RAG1/2. On the other hand, unpaired RAGL genes were neglected as the potential pseudogenes and past transposon remnants. In this study, we categorized the available RAGL genes of sea urchins based on their genomic organization (paired vs. unpaired). We focused on P. lividus species, for which the RAGL genes were not yet characterized and demonstrate that two different PlRAGL genes, PlRAGLa and PlRAGLb are domesticated and may have different functions.
Although TIR sequences around RAGL genes were not identified in any of the echinoid genomes, the nonsynonymous positions of these genes in different sea urchin species indicate a previous transposition activity. To check whether the RAGL genes are still mobile, we conducted nanopore long-read sequencing of single S. purpuratus cells and mapping of the reads to the existing S. purpuratus genome. Using this method, we confirm that sequences of both types I and II RAGLs are currently immobile in S. purpuratus. Furthermore, the RAG1L type I genes in two closely related sea urchin species- S. purpuratus and H. pulcherrimus were found in corresponding genomic locations, indicating that they have been immobile for at least 6 million years. Based on the assumption that all echinoid RAGL genes were originated in a single ancestor RAG1/2L transposon, which evolved through vertical evolution (8, 13, 51), we assume that the echinoid RAGL gene repertoire was shaped through a combination of intra-species transposition and duplication events in ancestral sea urchin species. This notion is further supported by the tendency for clustering of the genes that includes partial sequences and pseudogenes. Another evolutionary process that probably took place in the echinoid lineage is the acquisition of introns into the different RAGL genes. The sea urchin SpRAG1/2L genes contain multiple exons (12), while most vertebrate RAG genes, except for teleost fishes (52), consist of single exons. Interestingly, the amphioxus ProtoRAG also contains multiple exons but with introns in different sites and different intron phases (16). The presence of introns is a condition for alternative splicing variants (53), as was previously predicted but not proven for SpRAGL genes (12) which could not be shown in this study, due to the low expression rates of the RAGL genes in the sea urchin transcriptomic datasets.
Our finding shows that the PlRAG1a is co-expressed with PlRAG2L as well as with other homologues of vertebrates V(D)J recombination genes (i.e. PlArtemis and PlTdT). We cannot rule out the possibility that the PlRAG1 and PlRAG2 are working together with PlArtemis and PlTdT and maybe other V(D)J homologues, as a nuclease/recombinase complex, which may even be involved in increasing immune receptor diversity as in the vertebrates. However, no direct proof for the existence of such a complex was provided in this work and therefore we can only speculate at this stage. The co-expression of these genes in the adult tissues was identified mainly in the upper digestive system (esophagus and gut), which is not considered a specialized immune system organ. On the other hand, the upper part of the digestive system is perhaps the region, which is the most exposed to the external environment and thus is an area where immune protection is most needed.
Unexpectedly, PlRAG1a showed a different expression pattern from its S. purpuratus type I RAG1L equivalent. The explanation for this may be found in the comparative analysis of the 3-D structure of the RAG1/2L protein-DNA heterotetramer, which included differences in comparison to S. purpuratus. The differences in the complex structure between the two species include minor differences in the RAG1L protein structure (mainly the N-terminal side), and a major difference in the protein structure of the predicted PlRAG2L, which is missing the first 126 aa from its N-terminus when compared with the SpRAG2L sequence.
The predicted protein sequence and structure of PlRAG1a and PlRAG1b genes include two major differences First, the PlRAG1b is missing its N-terminus 1-159 residues, including the Importin binding domain, which functions in the binding of proteins to the importin transporter, which, in turn, carries the relevant proteins from the cytoplasm into the nucleus (54). The absence of the Importin binding domain suggests that the PlRAG1b encoded protein may not be internalized into the nucleus. The complementary evidence for the above notion is the absence of an 18 aa-long segment in the NBD of PlRAG1b. The segment seems to be important for the PlRAG1a interaction and perhaps nicking of the DNA double helix. The absence of these segment increases the chance that the PlRAG1b does not possess any nuclease activity.
While the role of the vertebrate RAGs is strictly immunological, in sea urchins, it may have been domesticated for other purposes. Nevertheless, the evidence for SpRAG1/2 complex formation (17), together with our prediction for the PlRAG1/2L complex, suggests that type I RAG1L proteins in sea urchins may still work as nucleases with or without the help of their corresponding RAG2Ls. One possible immunological target for the echinoid type I RAGL complexes is the Transformers (Trf) gene family (55–57) that was shown to be subjected to somatic gene diversification in individual cells (29) and to be present and functional in several sea urchin species (58). Further studies are needed to reveal the genomic targets of the echinoid RAGL complexes and to elucidate the specific role and mechanism of action.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA888396.
Author contributions
IY and MO contributed to the development of the study IY, MO, DT, HN-G did the experimental part. IY and MO wrote the article. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1066510/full#supplementary-material
References
1. Tonegawa S. Somatic generation of antibody diversity. Nature (1983) 302(5909):575–81. doi: 10.1038/302575a0
2. Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V (D) J recombination. Science (1990) 248(4962):1517–23. doi: 10.1126/science.2360047
3. van Gent DC, Ramsden DA, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V (D) J recombination. Cell (1996) 85(1):107–13. doi: 10.1016/S0092-8674(00)81086-7
4. Yanagi Y, Yoshikai Y, Leggett K, Clark SP, Aleksander I, Mak TW. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature (1984) 308(5955):145–9. doi: 10.1038/308145a0
5. Kallenbach S, Doyen N, d'Andon MF, Rougeon F. Three lymphoid-specific factors account for all junctional diversity characteristic of somatic assembly of T-cell receptor and immunoglobulin genes. Proc Natl Acad Sci (1992) 89(7):2799–803. doi: 10.1073/pnas.89.7.2799
6. Sakano H, Hüppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature (1979) 280(5720):288. doi: 10.1038/280288a0
7. Thompson CB. New insights into V (D) J recombination and its role in the evolution of the immune system. Immunity (1995) 3(5):531–9. doi: 10.1016/1074-7613(95)90124-8
8. Fugmann SD. The origins of the rag genes–from transposition to V (D) J recombination. In: Seminars in immunology. Elsevier (2010) 22(1):10–16. doi: 10.1016/j.smim.2009.11.004
9. Schatz DG. Antigen receptor genes and the evolution of a recombinase. In: Seminars in immunology. Elsevier (2004) 16(4):245–56. doi: 10.1016/j.smim.2004.08.004
10. Carmona LM, Schatz DG. New insights into the evolutionary origins of the recombination-activating gene proteins and V (D) J recombination. FEBS J (2017) 284(11):1590–605. doi: 10.1111/febs.13990
11. Yakovenko I, Agronin J, Smith LC, Oren M. Guardian of the genome: An alternative RAG/Transib Co-evolution hypothesis for the origin of V (D) J recombination. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021.709165
12. Fugmann SD, Messier C, Novack LA, Cameron RA, Rast JP. An ancient evolutionary origin of the Rag1/2 gene locus. Proc Natl Acad Sci (2006) 103(10):3728–33. doi: 10.1073/pnas.0509720103
13. Martin EC, Vicari C, Tsakou-Ngouafo L, Pontarotti P, Petrescu AJ, Schatz DG. Identification of RAG-like transposons in protostomes suggests their ancient bilaterian origin. Mobile DNA (2020) 11:1–20. doi: 10.1186/s13100-020-00214-y
14. Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature (1998) 394(6695):744. doi: 10.1038/29457
15. Hiom K, Melek M, Gellert M. DNA Transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell (1998) 94(4):463–70. doi: 10.1016/S0092-8674(00)81587-1
16. Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, et al. Discovery of an active RAG transposon illuminates the origins of V (D) J recombination. Cell (2016) 166(1):102–14. doi: 10.1016/j.cell.2016.05.032
17. Carmona LM, Fugmann SD, Schatz DG. Collaboration of RAG2 with RAG1-like proteins during the evolution of V (D) J recombination. Genes Dev (2016) 30(8):909–17. doi: 10.1101/gad.278432.116
18. Wilson DR, Norton DD, Fugmann SD. The PHD domain of the sea urchin RAG2 homolog, SpRAG2L, recognizes dimethylated lysine 4 in histone H3 tails. Dev Comp Immunol (2008) 32(10):1221–30. doi: 10.1016/j.dci.2008.03.012
19. Ben-Tabou de-Leon S, Su Y-H, Lin K-T, Li E, Davidson EH. Gene regulatory control in the sea urchin aboral ectoderm: Spatial initiation, signaling inputs, and cell fate lockdown. Dev Biol (2013) 374(1):245–54. doi: 10.1016/j.ydbio.2012.11.013
20. Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expression Patterns (2004) 4(4):449–56. doi: 10.1016/j.modgep.2004.01.009
21. Foster S, Oulhen N, Wessel G. A single cell RNA sequencing resource for early sea urchin development. Development (2020) 147(17). doi: 10.1242/dev.191528
22. Foster S, Teo YV, Neretti N, Oulhen N, Wessel GM. Single cell RNA-seq in the sea urchin embryo show marked cell-type specificity in the Delta/Notch pathway. Mol Reprod Dev (2019) 86(8):931–4. doi: 10.1002/mrd.23181
23. Tu Q, Cameron RA, Davidson EH. Quantitative developmental transcriptomes of the sea urchin strongylocentrotus purpuratus. Dev Biol (2014) 385(2):160–7. doi: 10.1016/j.ydbio.2013.11.019
24. Tu Q, Cameron RA, Worley KC, Gibbs RA, Davidson EH. Gene structure in the sea urchin strongylocentrotus purpuratus based on transcriptome analysis. Genome Res (2012) 22(10):2079–87. doi: 10.1101/gr.139170.112
25. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol (2019) 37(8):907–15. doi: 10.1038/s41587-019-0201-4
26. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. GigaScience (2021) 10(2). doi: 10.1093/gigascience/giab008
27. Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinf (2012) 14(2):178–92. doi: 10.1093/bib/bbs017
28. Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol (2001) 18(5):691–9. doi: 10.1093/oxfordjournals.molbev.a003851
29. Oren M, Rosental B, Hawley TS, Kim G-Y, Agronin J, Reynolds CR, et al. Individual sea urchin coelomocytes undergo somatic immune gene diversification. Front Immunol (2019) 10:1298. doi: 10.3389/fimmu.2019.01298
30. Wick RR, Judd LM, Gorrie CL, Holt KE. Completing bacterial genome assemblies with multiplex MinION sequencing. Microbial Genomics (2017) 3(10). doi: 10.1099/mgen.0.000132
31. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics (2018) 34(18):3094–100. doi: 10.1093/bioinformatics/bty191
32. Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol (1993) 234(3):779–815. doi: 10.1006/jmbi.1993.1626
33. Poole JRM, Huang SF, Xu A, Bayet J, Pontarotti P. The RAG transposon is active through the deuterostome evolution and domesticated in jawed vertebrates. Immunogenetics (2017) 69(6):391–400. doi: 10.1007/s00251-017-0979-5
34. Fiser A. Template-based protein structure modeling. Methods Mol Biol (2010) 673:73–94. doi: 10.1007/978-1-60761-842-3_6
35. Kapitonov VV, Jurka J. RAG1 core and V (D) J recombination signal sequences were derived from transib transposons. PloS Biol (2005) 3(6):e181. doi: 10.1371/journal.pbio.0030181
36. Kapitonov VV, Koonin EV. Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct (2015) 10(1):20. doi: 10.1186/s13062-015-0055-8
37. Lee Y-H. Molecular phylogenies and divergence times of Sea urchin species of strongylocentrotidae, echinoida. Mol Biol Evol (2003) 20(8):1211–21. doi: 10.1093/molbev/msg125
38. Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, et al. TimeTree 5: An expanded resource for species divergence times. Mol Biol Evol (2022) 39(8). doi: 10.1093/molbev/msac174
39. Maezawa S, Nakano S, Kuniya T, Koiwai O, Koiwai K. Double-strand break repair based on short-homology regions is suppressed under terminal deoxynucleotidyltransferase expression, as revealed by a novel vector system for analysing DNA repair by nonhomologous end joining. FEBS Open Bio (2016) 6(1):16–23. doi: 10.1002/2211-5463.12001
40. Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev (1999) 13(23):3070–80. doi: 10.1101/gad.13.23.3070
41. Gwyn LM, Peak MM, De P, Rahman NS, Rodgers KK. A zinc site in the c-terminal domain of RAG1 is essential for DNA cleavage activity. J Mol Biol (2009) 390(5):863–78. doi: 10.1016/j.jmb.2009.05.076
42. Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature (2015) 518(7540):507–11. doi: 10.1038/nature14174
43. Ru H, Chambers MG, Fu TM, Tong AB, Liao M, Wu H. Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell (2015) 163(5):1138–52. doi: 10.1016/j.cell.2015.10.055
44. Callebaut I, Mornon J-P. The V (D) J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci CMLS (1998) 54(8):880–91. doi: 10.1007/s000180050216
45. Rodgers KK, Bu Z, Fleming KG, Schatz DG, Engelman DM, Coleman JE. A zinc-binding domain involved in the dimerization of RAG1. J Mol Biol (1996) 260(1):70–84. doi: 10.1006/jmbi.1996.0382
46. Arbuckle JL, Rahman NS, Zhao S, Rodgers W, Rodgers KK. Elucidating the domain architecture and functions of non-core RAG1: The capacity of a non-core zinc-binding domain to function in nuclear import and nucleic acid binding. BMC Biochem (2011) 12(1):23. doi: 10.1186/1471-2091-12-23
47. Jones JM, Gellert M. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc Natl Acad Sci U S A (2003) 100(26):15446–51. doi: 10.1073/pnas.2637012100
48. Cortes P, Ye ZS, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci U S A (1994) 91(16):7633–7. doi: 10.1073/pnas.91.16.7633
49. Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V (D) J recombination. Nature (2007) 450(7172):1106–10. doi: 10.1038/nature06431
50. Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, et al. Transposon molecular domestication and the evolution of the RAG recombinase. Nature (2019) 569(7754):79. doi: 10.1038/s41586-019-1093-7
51. Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet (2010) 11(1):47–59. doi: 10.1038/nrg2703
52. Wang X, Tan X, Zhang PJ, Zhang Y, Xu P. Recombination-activating gene 1 and 2 (RAG1 and RAG2) in flounder (Paralichthys olivaceus). J Biosci (2014) 39(5):849–58. doi: 10.1007/s12038-014-9469-1
53. Avgan N, Wang JI, Fernandez-Chamorro J, Weatheritt RJ. Multilayered control of exon acquisition permits the emergence of novel forms of regulatory control. Genome Biol (2019) 20(1):1–13. doi: 10.1186/s13059-019-1757-5
54. Lott K, Cingolani G. The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim Biophys Acta (BBA) - Mol Cell Res (2011) 1813(9):1578–92. doi: 10.1016/j.bbamcr.2010.10.012
55. Nair SV, Del Valle H, Gross PS, Terwilliger DP, Smith LC. Macroarray analysis of coelomocyte gene expression in response to LPS in the sea urchin. identification of unexpected immune diversity in an invertebrate. Physiol Genomics (2005) 22(1):33–47. doi: 10.1152/physiolgenomics.00052.2005
56. Terwilliger DP, Buckley KM, Brockton V, Ritter NJ, Smith LC. Distinctive expression patterns of 185/333 genes in the purple sea urchin, strongylocentrotus purpuratus: an unexpectedly diverse family of transcripts in response to LPS, beta-1,3-glucan, and dsRNA. BMC Mol Biol (2007) 8:16. doi: 10.1186/1471-2199-8-16
57. Oren M, Hudgell MAB, D’Allura B, Agronin J, Gross A, Podini D, et al. Short tandem repeats, segmental duplications, gene deletion, and genomic instability in a rapidly diversified immune gene family. BMC Genomics (2016) 17(1):1–19. doi: 10.1186/s12864-016-3241-x
Keywords: RAG1, RAG2, sea urchin immunity, P. lividus, S. purpuratus, gene domestication, RAG transposon, RAG evolution
Citation: Yakovenko I, Tobi D, Ner-Gaon H and Oren M (2023) Different sea urchin RAG-like genes were domesticated to carry out different functions. Front. Immunol. 13:1066510. doi: 10.3389/fimmu.2022.1066510
Received: 10 October 2022; Accepted: 29 December 2022;
Published: 16 January 2023.
Edited by:
Annalisa Pinsino, Institute of Translational Pharmacology (IFT) - National Research Council (CNR), ItalyReviewed by:
Ryohei Furukawa, Keio University, Hiyoshi Campus, JapanMasaki Miyazaki, Kyoto University, Japan
Copyright © 2023 Yakovenko, Tobi, Ner-Gaon and Oren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matan Oren, bWF0YW5vckBhcmllbC5hYy5pbA==; Iryna Yakovenko, aXJlbmlyZW4ueUBnbWFpbC5jb20=
 Iryna Yakovenko
Iryna Yakovenko Dror Tobi
Dror Tobi Hadas Ner-Gaon
Hadas Ner-Gaon Matan Oren
Matan Oren