- 1Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Faculty of Medical Laboratory Science, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China
Background: Endometriosis, classically viewed as a localized disease, is increasingly recognized as a systemic disease with multi-organ effects. This disease is highlighted by systemic inflammation in affected organs and by high comorbidity with immune-mediated diseases.
Results: We provide genomic evidence to support the recognition of endometriosis as an inflammatory systemic disease. This was achieved through our genomics-led target prioritization, called ‘END’, that leverages the value of multi-layered genomic datasets (including genome-wide associations in disease, regulatory genomics, and protein interactome). Our prioritization recovered existing proof-of-concept therapeutic targeting in endometriosis and outperformed competing prioritization approaches (Open Targets and Naïve prioritization). Target genes at the leading prioritization revealed molecular hallmarks (and possibly the cellular basis as well) that are consistent with systemic disease manifestations. Pathway crosstalk-based attack analysis identified the critical gene AKT1. In the context of this gene, we further identified genes that are already targeted by licensed medications in other diseases, such as ESR1. Such analysis was supported by current interests targeting the PI3K/AKT/mTOR pathway in endometriosis and by the fact that therapeutic agents targeting ESR1 are now under active clinical trials in disease. The construction of cross-disease prioritization map enabled the identification of shared and distinct targets between endometriosis and immune-mediated diseases. Shared target genes identified opportunities for repurposing existing immunomodulators, particularly disease-modifying anti-rheumatic drugs (such as TNF, IL6 and IL6R blockades, and JAK inhibitors). Genes highly prioritized only in endometriosis revealed disease-specific therapeutic potentials of targeting neutrophil degranulation – the exocytosis that can facilitate metastasis-like spread to distant organs causing inflammatory-like microenvironments.
Conclusion: Improved target prioritization, along with an atlas of in silico predicted targets and repurposed drugs (available at https://23verse.github.io/end), provides genomic insights into endometriosis, reveals disease-specific therapeutic potentials, and expands the existing theories on the origin of disease.
Introduction
Endometriosis, characterized by the presence of ectopic endometrial-like tissues, is classically viewed as a chronic hormone dependent neuroinflammatory disease locally restricted to the pelvis (1). Endometriosis is estimated to be present in 50–80% of women with pelvic pain, and occurs in up to 50% of women with infertility (2, 3). Clinical manifestations are varied, with high prevalence (affecting 5-10% of reproductive-age women with negative impacts on quality of life), common misdiagnosis (65% of women initially misdiagnosed) or diagnostic delay (delayed by 4-11 years). Currently, endometriosis is increasingly recognized as a systemic disease with multi-organ effects throughout the body [reviewed in (4)]. The full effects are far from clear, but consensus has been reached that patients with endometriosis are likely to have: (i) adipocyte and hepatic metabolic changes, such as low body mass index (5); (ii) neurological alterations that enhance pain sensitivity and increase the risk of developing mood disorders, such as fatigue, depression and anxiety (6); (iii) systemic inflammation that causes widespread inflammatory-like microenvironments in affected tissues or organs (7); and (iv) the tendency to develop immune-mediated diseases (8).
Likely due to the complexity of systemic effects and disease comorbidities described above, there is currently no cure for endometriosis. Available treatments help to reduce symptoms and maintain quality of life. Two treatment options (medical and surgical therapies) are mainly for alleviating endometriosis-associated pain (4). Medical therapy, combining oral contraceptives (or progestins) and non-steroidal anti-inflammatory drugs (known as ‘NSAIDs’), represents the first-line therapy. On average, 25-33% of patients do not respond to first-line therapies. For non-responders with persistent pain, gonadotropin-releasing hormone (GnRH) analogs or aromatase inhibitors represent the second-line therapy. Surgical therapy aims to treat pain and disease-related infertility as well. Medical therapy following surgery represents the third-line therapy that is supposed to reduce or minimize disease recurrence and systemic effects. It should be noted, however, that neither medical therapy nor surgery fully addresses the systemic nature of the disease. Recognizing endometriosis as a systemic disease highlights the importance of new target identification and validation to increase the range of medical therapies, ultimately for better therapeutics.
The etiopathogenesis of endometriosis is complex, involving the interplay between genetic inheritance and environmental influence. Genetic associations arising from genome-wide association studies (GWAS) provide a rich source of genetic targets. For example, GWAS meta-analysis in endometriosis has identified disease risk loci that are likely to affect genes involved in hormone metabolism (9). Such genetic evidence is critical for successful therapeutic development (10, 11). How to harness GWAS findings for use in drug discovery (12) and drug repurposing (13), however, requires a paradigm change in strategies. Recently, we have proposed a strategic framework that generalizes how to establish the link from genetic loci to modulated genes that can be further linked to drug targets (14, 15). Our advocates of genetic target prioritization have driven this field of research (16–22). In this study, we extended this into genomics-led target prioritization (called ‘END’) and demonstrated better performance than the status quo approaches (Figure 1), with the aim of providing genomic evidence for endometriosis as an inflammatory systemic disease. Our genomic prioritization, followed by a range of integrative bioinformatics analyses (including comparisons with immune diseases), enabled us to identify repurposing opportunities for existing immunomodulatory drugs and more importantly, to reveal therapeutic strategies for endometriosis-specific targeting. In a wider context, our study provides a new strategy to advance the use of human genetics and genomics for target identification and validation in inflammatory systemic diseases.
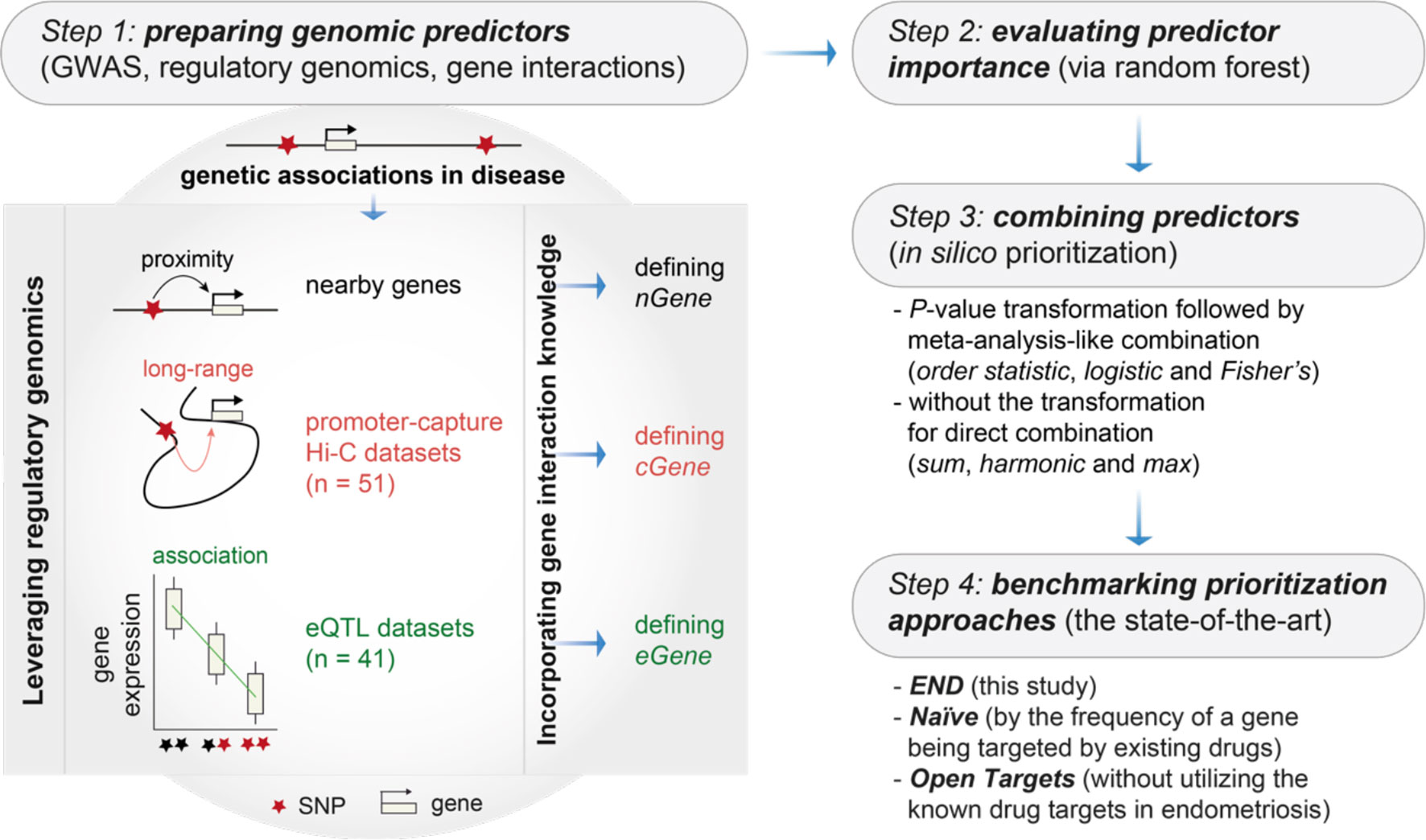
Figure 1 Overview of genomics-led target prioritization in endometriosis (called ‘END’). Steps 1-3 sequentially describe how to prepare, evaluate and combine genomic predictors for in silico target prioritization. The key is the preparation of genomic predictors at Step 1, as illustrated on the right, utilizing genetic associations in disease, regulatory genomics (mainly promoter-capture Hi-C datasets and eQTL datasets) and the knowledge of gene/protein interactions. Step 4 is intended for benchmarking to compare the performance of END with two competing prioritization approaches, including Naïve (a gene prioritized simply according to how often the gene is already targeted by existing drugs) and Open Targets (without utilizing the information of known drug targets). GWAS, genome-wide association studies; nGene, nearby genes; cGene, conformation genes; eGene, expression genes.
Materials and Methods
Genomics-Led Prioritization and Performance Evaluation
The steps designed for genomic prioritization and evaluation were briefly described in Figure 1 and detailed below.
Step 1: Preparing Genomic Predictors
We extended our previous pipeline (23) to prepare genomic predictors utilizing multi-layered genomic datasets and the knowledge of gene/protein interactions. Genomic datasets included: (i) GWAS summary statistics in endometriosis (9, 24–27), considering a typical threshold (P-values < 5×10−8) and SNPs in linkage disequilibrium (R2<0.8) according to the European population (28) to define nearby genes (nGene); (ii) promoter capture Hi-C (29–34), defining conformation genes (cGene); and (iii) eQTL (35–41), defining expression genes (eGene). The knowledge of gene/protein interactions was obtained from the STRING database (high-quality, only with evidence codes ‘experiments’ or ‘databases’) (42), corresponding to a total of 14,325 target genes considered for prioritization (Table S1).
Step 2: Evaluating Predictor Importance
We applied random forest (43) to evaluate predictor importance. Considering the fact that the nGene predictor is conventionally used (thus treated as the baseline), only cGene and eGene predictors that were no less important/informative than nGene remained for the subsequent combination.
Step 3: Combining Predictors
We combined informative predictors for in silico prioritization. For a candidate target gene, strategies that combine individual predictors could be either direct or indirect. The direct combinations can be sum (summing up affinity scores), max (taking the maximum) or harmonic (using the harmonic sum), while the idea of indirect combinations is to first transform affinity scores into P-values and then combined via meta-analysis methods such as Fisher’s, logistic or order statistic. Performance for combination strategies was measured by the area under the ROC curve (AUC) separating clinical proof-of-concept targets in endometriosis from simulated controls. Existing proof-of-concept targets in endometriosis were defined as therapeutic target genes of drugs reaching development phase 2 and above [sourced from the ChEMBL database (44)].
Step 4: Benchmarking Prioritization Approaches
We benchmarked competing approaches for performance evaluation. Benchmarking was also based on AUC (that is, separating clinical proof-of-concept targets in endometriosis from simulated controls) to compare the performance of ‘END’ (this study), ‘Naïve’ (an approach prioritizing a gene by how often it has been targeted by existing drugs), and ‘Open Targets’ (using the harmonic sum to aggregate individual evidence except for the information on known drugs) (45).
Target Set Enrichment Analysis
The dnet package (46) was used to conduct target set enrichment analysis quantifying the degree to which a predefined gene list is enriched at the leading prioritization. The leading prioritization, visually defined as the left-most region of the peak in running enrichment plot, is the core subset of the prioritized target genes accounting for the enrichment signal. The analysis was applied to clinical proof-of-concept targets in endometriosis, showing that they tended to be highly ranked. This analysis was also applied to: (i) cell-type-specific gene signatures (47) for exploring the cellular basis of all prioritized target genes in endometriosis; (ii) MSigDB hallmark gene sets (48) for revealing molecular hallmarks based on the top 10% prioritized target genes in endometriosis; and (iii) one fibroblast gene expression signature specific to ectopic endometrium (49) and four gene expression signatures involving two endometriosis stages (I/II and III/IV) at two menstrual cycle phases (proliferative and secretory) (50) for examining the highly prioritized target gene expression.
Pathway Crosstalk-Based Attack Analysis
There were three steps to achieve this. First, the XGR package (51) and a collection of KEGG organismal system pathways (52) were used to identify enriched pathways based on the top 1% prioritized genes. Second, pathway crosstalk was identified by searching for a subset of gene interactions (extracted from enriched pathways identified in the previous step); identified so in a manner that the resulting pathway crosstalk contained highly prioritized and interconnected genes (53). Third, attack analysis was conducted to select optimal targeting combinations that maximized the effect of removing nodes on the crosstalk; done so in a manner to maximize the effect of removing either single nodes or specific node combinations. The combinatorial node removal can be 2-node combinations removed in the context of a fixed node ‘ATK1’, and 3-node combinations removed in the context of two fixed nodes ‘AKT1 + ESR1’.
Construction of Cross-Disease Prioritization Map
Focusing on the top 5% prioritized target genes (n = 689) in endometriosis, the supraHex package (54) was used to construct cross-disease prioritization map. In brief, a supra-hexagonal map, consisting of 91 hexagons, was trained by the prioritization matrix (containing priority ratings) of 689 target genes versus 7 diseases including endometriosis (this study) and 6 immune-mediated diseases (53). These immune diseases include: (i) inflammatory bowel diseases, subdivided into Crohn’s disease (CRO) and ulcerative colitis (UC); and (ii) inflammatory systemic diseases, including multiple sclerosis (MS), rheumatoid arthritis (RA), Sjögren’s syndrome (SJO), and systemic lupus erythematosus (SLE). The trained map was used to illustrate a gene prioritization profile per disease, and together with a consensus neighbour-joining tree built from the prioritization matrix, to further illustrate inter-disease relationships. The trained map was also divided into target gene clusters in a topology-preserving manner. Enrichment analysis for genes within a cluster was based on one-sided Fisher’s exact test to identify enrichments in terms of: (i) approved drug targets obtained from the ChEMBL database (44); (ii) immune system pathways from the Reactome database (55); (iii) Gene Ontology functional annotations from NCBI (56); and (iv) mouse phenotype annotations using Mammalian Phenotype Ontology (57).
Identification of Druggable Pockets
The fpocket software (58) was used to predict druggable pockets of a target gene based on its known protein structure(s). The known protein structures were obtained from the PDB database (59), for example, the access code ‘4N78’ for the WAVE regulatory complex (60). A gene was defined to be tractable if predicted to have drug-like binding sites (that is, druggable pockets). The PDB structure was viewed in 3D as cartoon (secondary structure abstraction), colored by PDB chains and embedded with druggable pockets in blue (61).
Results
Leveraging Genomic Predictors for In Silico Prioritization of Therapeutic Targets in Endometriosis
We performed genomics-led prioritization, taking GWAS summary statistics in endometriosis as inputs and leveraging the informativeness of regulatory genomics in diverse cell types, activation states and tissues. As outlined in Figure 1 (see Materials and Methods for details), our multi-step prioritization process consisted of: (i) the preparation of genomic predictors; (ii) the evaluation of predictor importance to identify informative predictors; (iii) the assessment of how to combine informative predictors; and (iv) performance evaluation to benchmark competing approaches. We found that using the meta-analysis-like combination strategy, particularly based on order statistic to combine genomic predictors, achieved much better performance than using the direct combination strategy (Figure 2A). Benchmarking results showed that our prioritization approach (called ‘END’) was superior to competing approaches (Figure 2B). We considered two competing approaches, namely, Open Targets (also using genetics and genomics for target identification and prioritization) (45) and Naïve prioritization (prioritizing a gene by how often it has been targeted by existing drugs), that were similar in performance. Notably, Naïve prioritization, based on the concept of drug repurposing, was limited in being unable to predict new targets. The precision-recall analysis showed that our prioritization achieved the 91% precision at the 56% recall (prioritization coverage; Figure 2C).
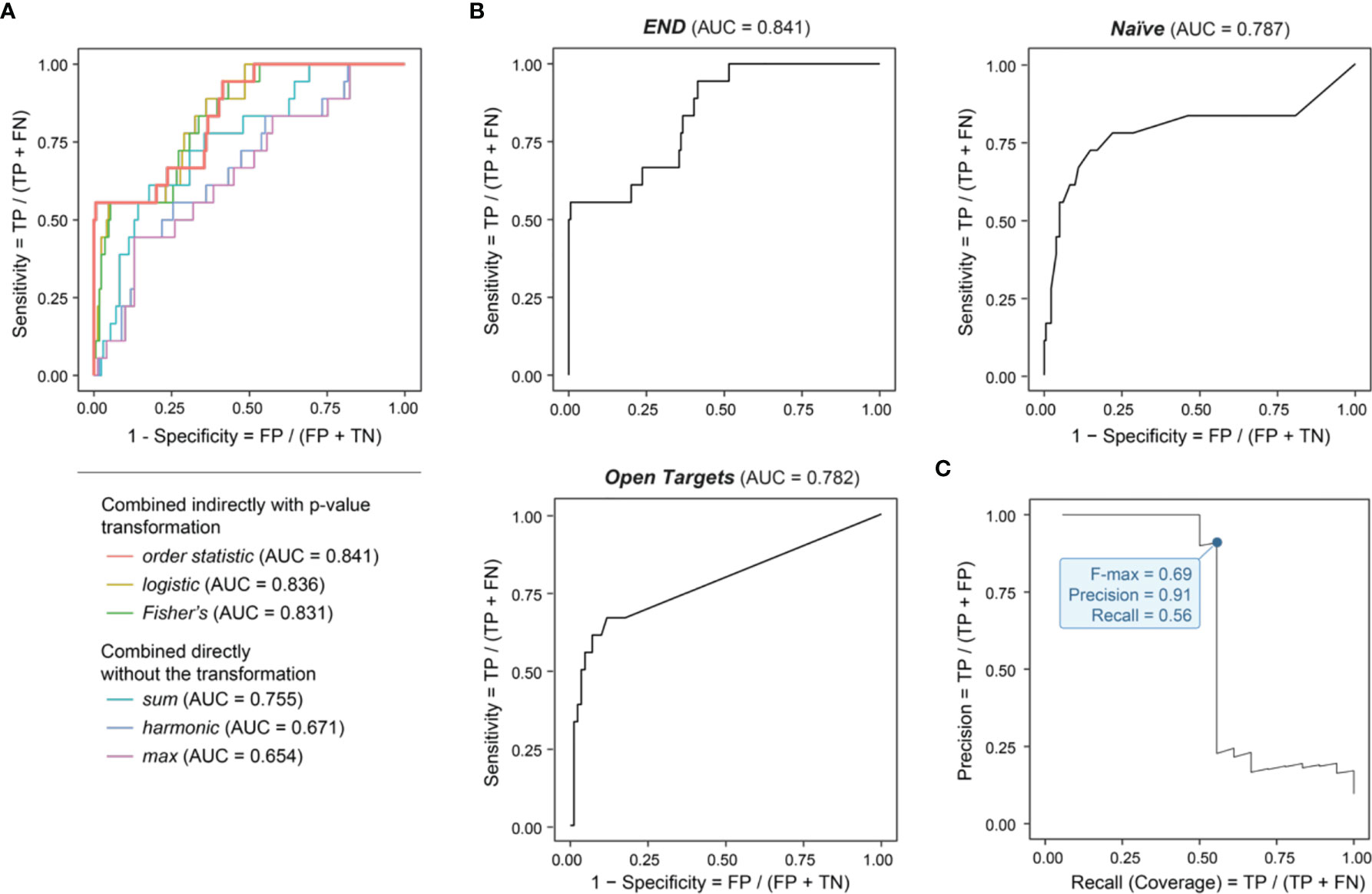
Figure 2 Genomic prioritization in endometriosis. (A) Comparisons of strategies combining predictors. The combination strategy based on order statistic achieves the optimal performance, measured by the area under the ROC curve (AUC) separating clinical proof-of-concept targets from simulated negative controls. FN, false negatives; FP, false positives; TN, true negatives; TP, true positives. (B) Benchmarking prioritization approaches as described in Figure 1, in terms of AUC. (C) Precision-Recall curve for END, with the F-max metric indicated (corresponding to 91% precision and 56% recall).
We prioritized a total of >14,000 candidate target genes [with the knowledge of gene interactions sourced from the STRING database (42)], ranked by priority rating (Table S1). The top prioritized targets included genes essential in inflammation and cellular responses, such as JUN (ranked 1st), NFKB1 (12th), RUNX1 (17th), PTEN (21st), MAPK14 (24th), FOS (20th), SMAD3 (22nd), and RELA (25th) (Figure 3A). Our prioritization recovered existing clinical proof-of-concept target genes in endometriosis (Figures 3B, C). These proof-of-concept targets in endometriosis were all at the top 1% prioritized gene list, including: MAPK8 (9th), MAPK10 (46th), and MAPK9 (47th) targeted by bentamapimod, a JNK inhibitor; ESR1 (19th) targeted by estradiol, an estrogen receptor (ER) alpha agonist; ESR2 (99th) targeted by prinaberel, an ER beta agonist; AR (26th) targeted by danazol, an androgen receptor (AR) agonist, and cyproterone acetate, an AR antagonist; NR3C1 (53th) targeted by cyproterone acetate and mifepristone, two glucocorticoid receptor antagonists; NGF (91th) targeted by tanezumab, a beta-nerve growth factor inhibitor; PGR (116th) targeted by selective progesterone receptor modulators (SPRMs); and TNF (136th) targeted by infliximab, a TNF-alpha inhibitor. According to the latest ESHIR guideline (62), SPRMs such as medroxyprogesterone acetate and levonorgestrel are highly recommended to reduce endometriosis-associated pain, and norethindrone acetate (alongside GnRH) highly recommended to prevent bone loss and hypoestrogenic symptom, but no longer recommended for danazol. Figure 3C also provides a summary of the predictors used and their relative importance (informativeness). We treated the nearby gene predictor as the baseline defining predictive informativeness. We identified informative regulatory genomic predictors that were mostly derived from immune blood cells (with a few from other cell types and tissues). These results necessitated the use of genomic datasets from diverse contexts for target prioritization and identification, which is consistent with endometriosis that is increasingly recognized as a systemic disease (4).
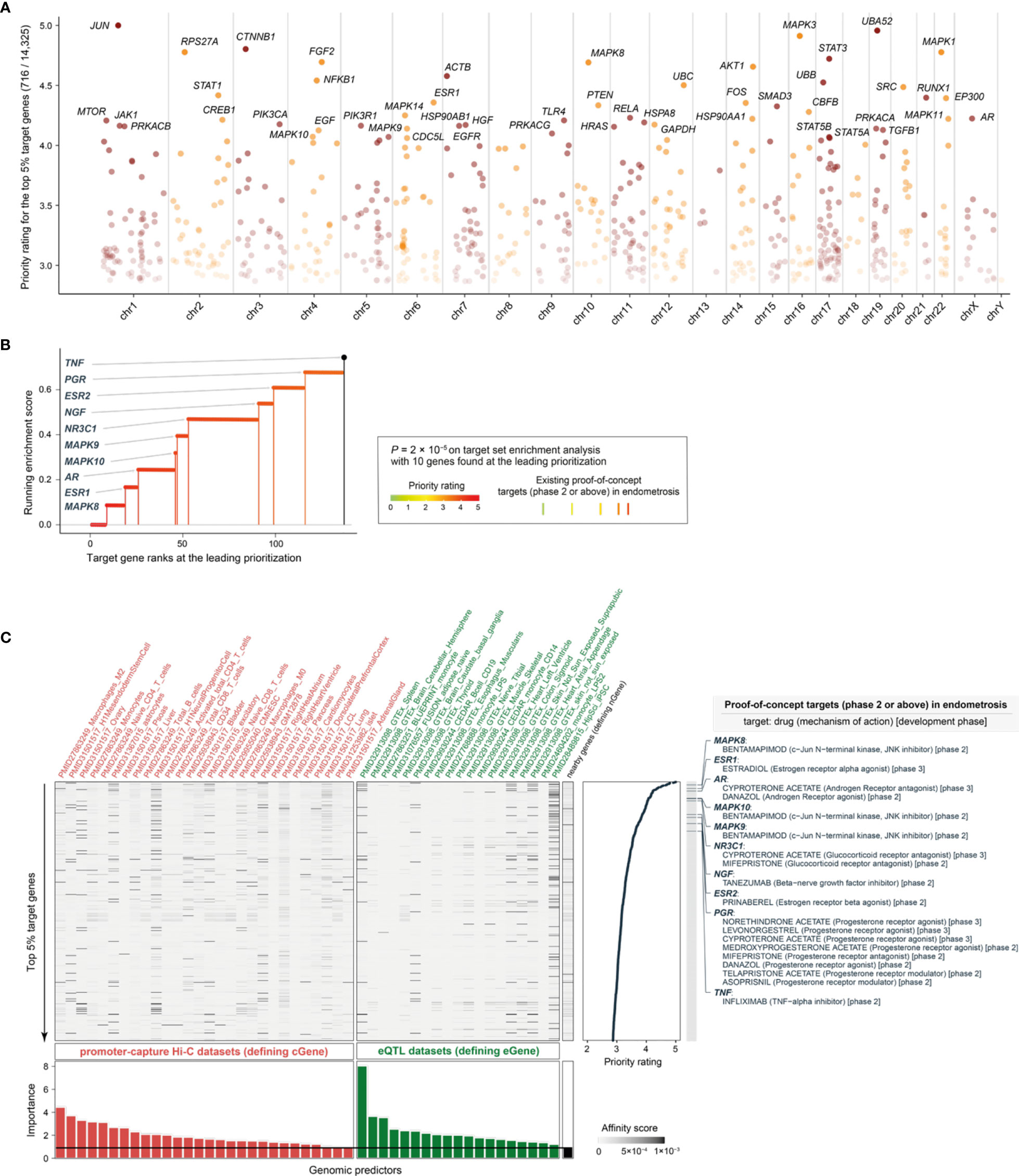
Figure 3 Performance evaluation for prioritization in endometriosis. (A) Manhattan plot illustrating priority rating (y-axis) for top 5% target genes across chromosomes (x-axis), with top 30 named. (B) Leading prioritization plot for clinical proof-of-concept targets in endometriosis, which are indicated in vertical lines (also color-coded by priority rating). (C) Heatmap plot illustrating predictor matrix used in END. Shown beneath is relative importance of predictors (color-coded by dataset types), quantified by the accuracy decrease (disabling a predictor), and the horizontal line indicates the importance for nGene (that is, the nearby gene predictor that is conventionally used and thus here treated as a baseline predictor). Shown on the right is the information of prioritized target genes (only the top 5% shown for simplicity), together with the information on existing proof-of-concept targets in endometriosis (including drugs, mechanisms of action, and development phases). eQTL, expression quantitative trait locus.
Characterizing the Cellular Basis and Molecular Hallmarks of Target Gene Prioritization in Endometriosis
We used rank-based target set enrichment analysis to characterize prioritized target genes using cell-type-specific gene signatures (47). We found the enrichment for immune cells; these included cells in myeloid lineages (such as monocytes, neutrophils, and mast cells) and lymphoid lineages (such as gamma delta T-cells, CD4 + memory T-cells, and CD8+ effector-memory T-cells) but not in B-cell lineages (Figure 4 and Table S2). In addition to immune cells (for example, gamma delta T-cells; Figure S1A), we also found the enrichment for stromal cells (Figure S1B) and epithelial cells (Figure S1C). Taken together, our findings are consistent with evidence of altered immunity and inflammation (local and systemic) in endometriosis (3), and also reveal the cellular basis that might involve multiple lineages underlying the pathogenesis of endometriosis.
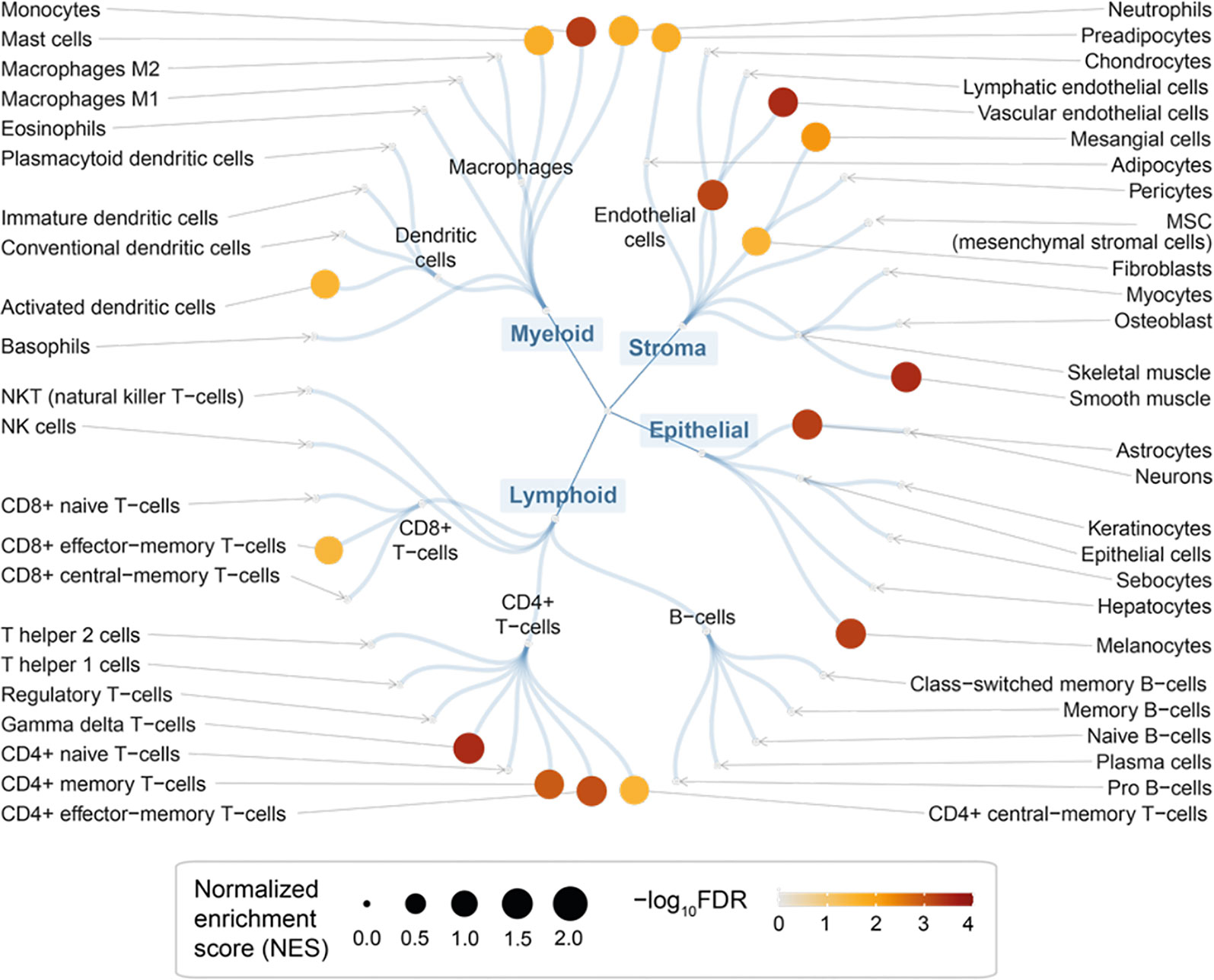
Figure 4 Cellular basis of therapeutic targeting in endometriosis. Circular overview of cell type enrichments, with nodes sized by the normalized enrichment score (NES) and colored by the enrichment significance (FDR) calculated using target set enrichment analysis.
Next, using MSigDB hallmark gene sets (48), we identified 8 molecular hallmarks enriched in highly prioritized genes (Figure 5A and Table S3). These included: cellular responses to low oxygen levels (hypoxia), ultraviolet radiation and estrogen; signaling pathways (NF-kB signaling in response to TNF, and activation of the PI3K/AKT/mTOR pathway); immune processes (allograft/transplant rejection, and the complement system); and the apical junction complex. For each of these hallmarks, genes found at the leading prioritization are shown in Figure 5B. We identified 21 leading prioritized genes that are responsive to hypoxia, with 4 genes (HSPA5, IL6, PDGFB, and SERPINE1) encoding components of the complement system and 7 genes (CDKN1A, DUSP1, FOS, IL6, IRS2, JUN, and VEGFA) regulated by NF-kB in response to TNF (Figure 5C). We identified 26 leading prioritized genes involved in the PI3K/AKT/mTOR pathway (Figure 5B), the pathway relevant to the severity of endometriosis stages (63). Among these 26 genes, 7 (AKT1, EGFR, IL2RG, IL4, LCK, MAP3K7, and PRKCB) are also involved in allograft rejection (Figure 5C), the consequences of which may cause tissue injury and fibrosis. These results revealed diverse but related molecular hallmarks in endometriosis, indictive of being an inflammatory systemic disease.
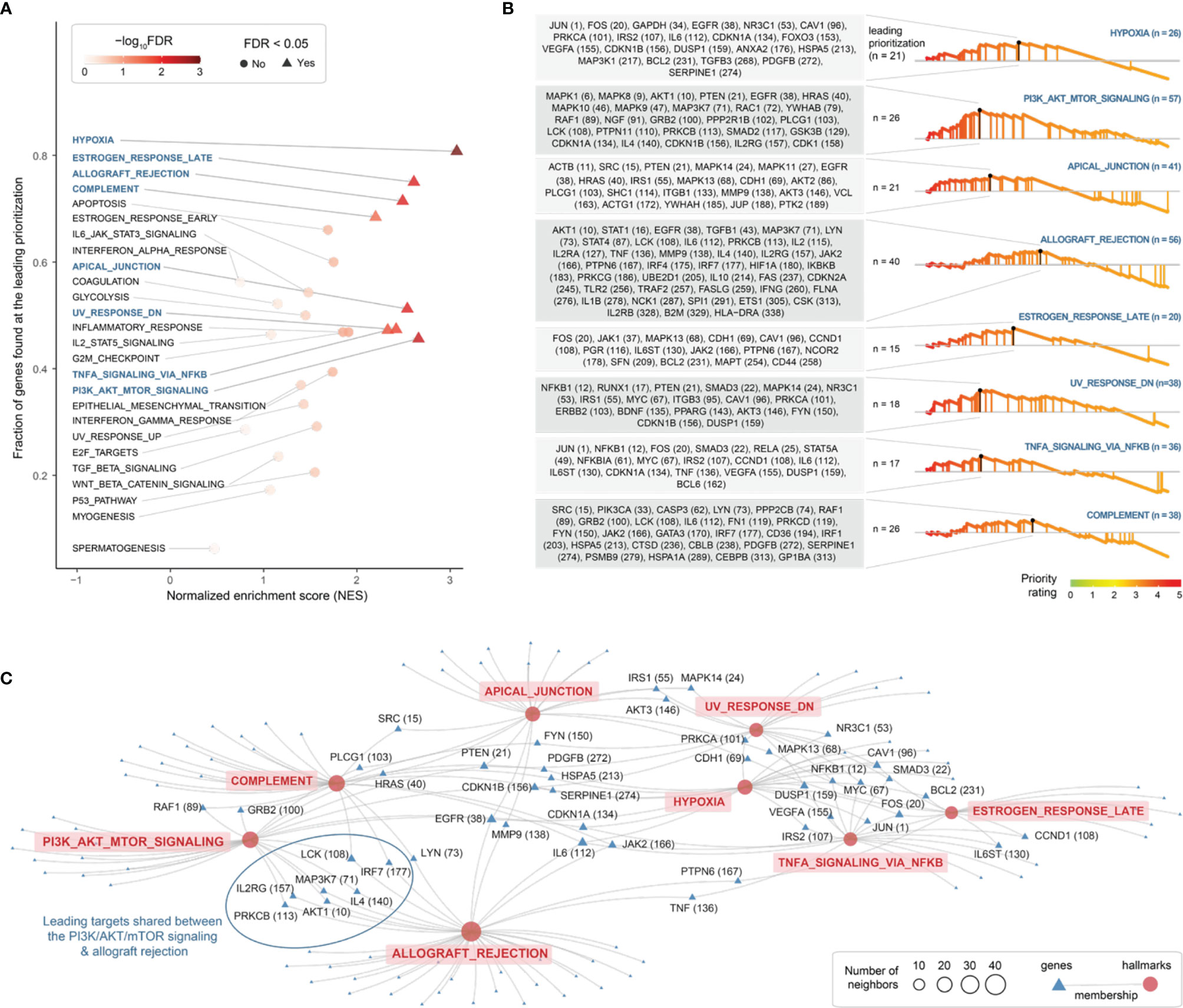
Figure 5 Exploring molecular hallmarks of therapeutic targeting in endometriosis. (A) Scatter plot of molecular hallmark enrichments, including normalized enrichment score (NES; x-axis), fraction of hallmark genes found at the leading prioritization (y-axis), and the enrichment significance (FDR) calculated using target set enrichment analysis. Enriched hallmarks (FDR < 0.05) are shaped in triangle and labelled in blue. (B) Details on enriched hallmarks. Left: the gene list found at the leading prioritization, with the number in the parentheses indicating priority rank. Right: the leading prioritization (visually defined as the left-most region ahead of the peak). (C) Network-like representation of enriched hallmarks and member genes. Inter-hallmark sharing genes are also labelled including priority rank. For example, genes shared between two hallmarks (the PI3K/AKT/mTOR signalling and allograft rejection) are framed in oval shape.
Pathway Crosstalk-Based Attack Analysis Identifies Critical Genes and Repurposed Drugs
Using the KEGG resource (52), we proceeded to perform pathway enrichment analysis for the top 1% prioritized target genes. This identified a wide range of organismal system pathways that can be broadly categorized into immune, endocrine, nervous, and reproductive systems (Figure 6A). It is well-recognized that maladaptation of these organismal systems can occur in chronic inflammatory systemic diseases (64). Integrated analysis of these enriched pathways identified a 31-gene network (P = 3.1 × 10−213 on permutation test), reflective of crosstalk between pathways, that all contained highly prioritized genes in endometriosis (Figure 6B and Table S4). Using the information on approved therapeutics available in the ChEMBL database (44), we found 5 genes that are already targeted by licensed medications (approved drugs) in other diseases (Figure 6B).
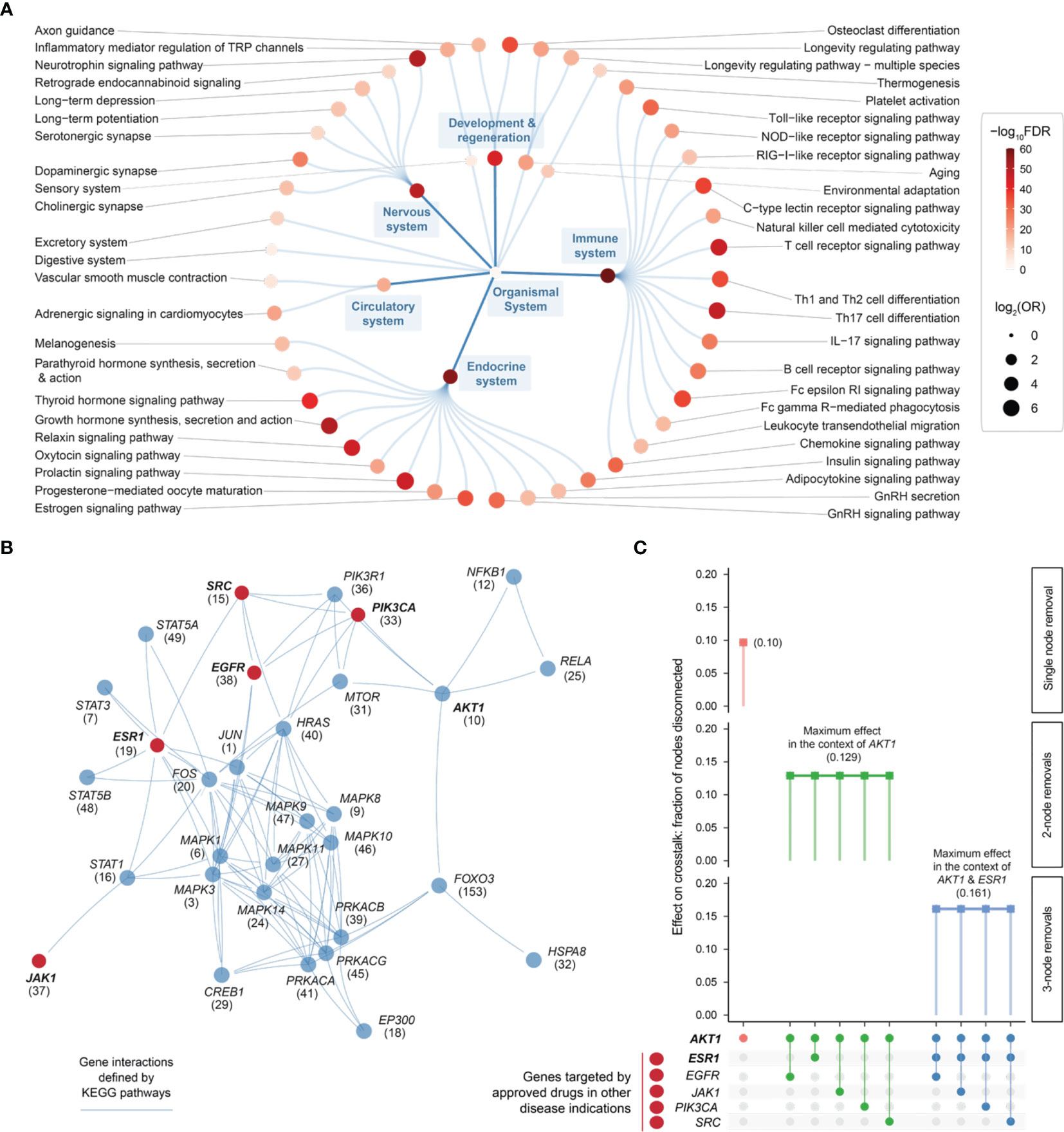
Figure 6 Pathway crosstalk-based attack analysis identifies critical genes and repurposes licensed medications. (A) Pathway enrichments of the top 1% prioritized target genes. Based on KEGG organismal system pathways that are organized circularly. Each dot represents a pathway, sized by the odd ratio (OR) and colored by the significance level (FDR) calculated using one-sided Fisher’s exact test. (B) Pathway crosstalk identified by integrating target prioritization information with gene interactions (extracted from enriched pathways). Nodes are labelled by gene symbols (priority rank) and highlighted in red if currently targeted by approved drugs in other disease indications. (C) Effect of node removals on the crosstalk. Top: the single-node effect maximized by removing ATK1, the maximum effect of removing another approved drug target in the context of AKT1, and the maximum effect of removing another approved drug target in the context of AKT1 and ESR1. Bottom: node removals are indicated by colored circles.
Removing/attacking a node critical for the crosstalk would result in large numbers of disconnected nodes. We found that the crosstalk identified above was very robust to node removal. The maximal effect achieved by any individual node removal was as low as 10% network disconnection (that is, removing AKT1 gene; Figure 6C). The identification of AKT1, or more generally, genes in the PI3K/AKT/mTOR pathway (including MTOR, PIK3CA, and PIK3R1 in addition to ATK1; see also Figure 5), is in line with current interests targeting this pathway in endometriosis (65). This motivated us to further perform combinatorial attack analysis in the context of AKT1. The 2-node maximal effect was observed when removing another approved drug target, ESR1, EGFR, JAK1, PIK3CA or SRC (Figure 6C). Therapeutic agents targeting ESR1 are now under phase 3 clinical trials in endometriosis (66), and thus, we further performed attack analysis in the context of AKT1 + ESR1, with maximal network disconnection (~16%) achieved by 3-node removal (Figure 6C). These findings supported the usefulness of our identified crosstalk genes for drug repurposing (Figure 7). For example, estrogen receptor modulators/agonists targeting ESR1 are now used in the clinic; this includes bazedoxifene acetate and conjugated estrogens for preventing postmenopausal osteoporosis (related to oestrogen deficiency) (67).
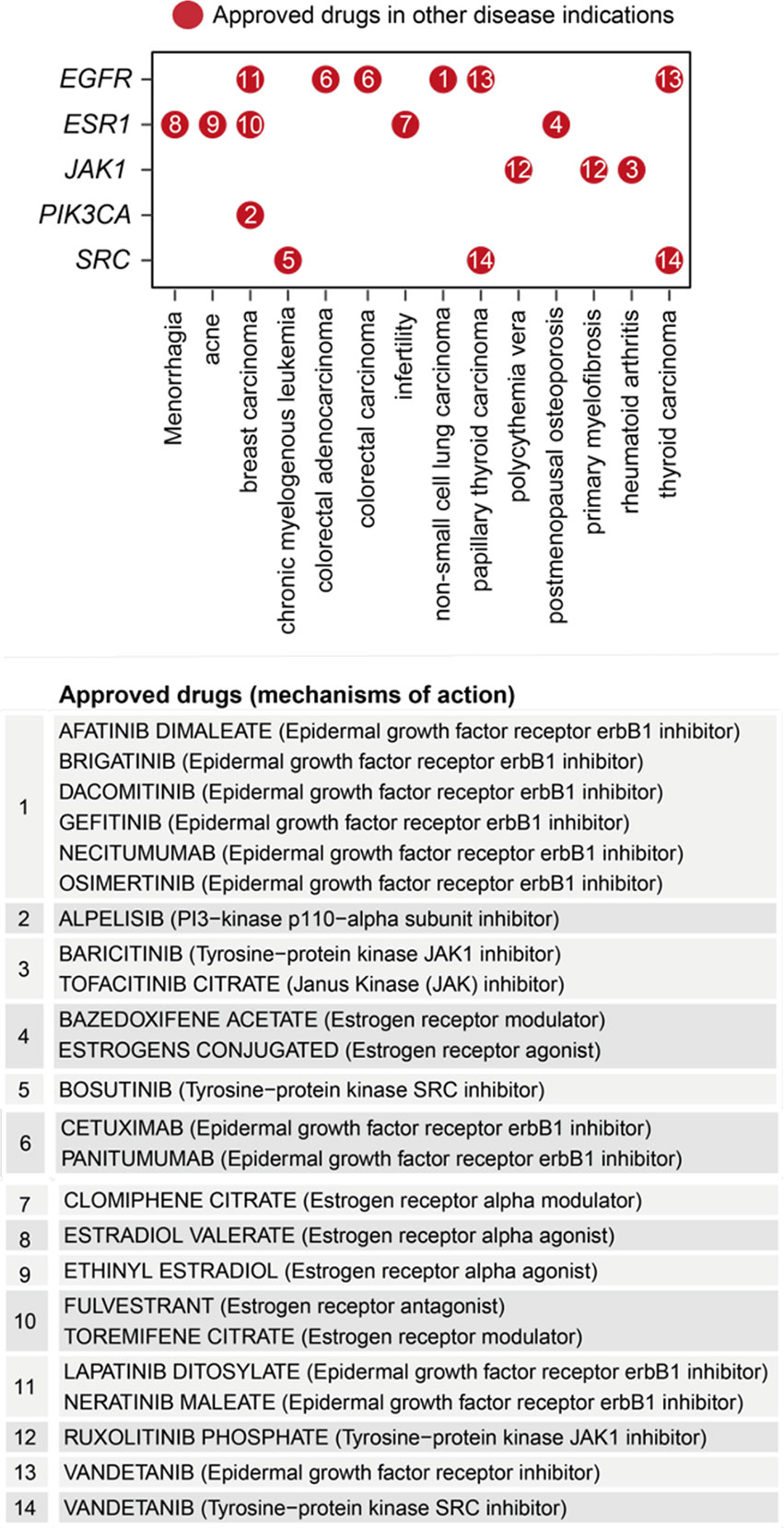
Figure 7 Repurposing analysis of licensed medications based on pathway crosstalk genes. Dot plot shows approved drug target genes (y-axis) and disease indications (x-axis), with dots indexed in number and referenced beneath showing the information on drugs and mechanisms of action.
Cross-Disease Prioritization Map Between Endometriosis and Immune Diseases Identifies Shared and Distinct Target Gene Clusters
A systematic review has confirmed that endometriosis has statistically significant associations with immune-mediated diseases (CRO, MS, RA, SJO, SLE, and UC) (8). Hereinafter, we explored such relationships based on the top 5% prioritized genes in endometriosis. We indeed observed significant correlations of priority ratings between endometriosis and immune diseases (Figure 8A). To further capture such correlations, we next employed a supra-hexagonal map (54) for cross-disease comparisons (Figure 8B, Figure S2 and Table S5). Each presentation illustrates a disease-specific prioritization profile (in which hexagons are color-coded to show priority ratings for genes thereof), while consensus neighbor-joining tree captures the (dis)similarity of prioritization profiles between diseases. The observed relationships are consistent with therapeutic/phenotypic similarity. For example, inflammatory bowel diseases (CRO and UC) are grouped together, and inflammatory systemic diseases (SJO, SLE, and RA) stay closer.
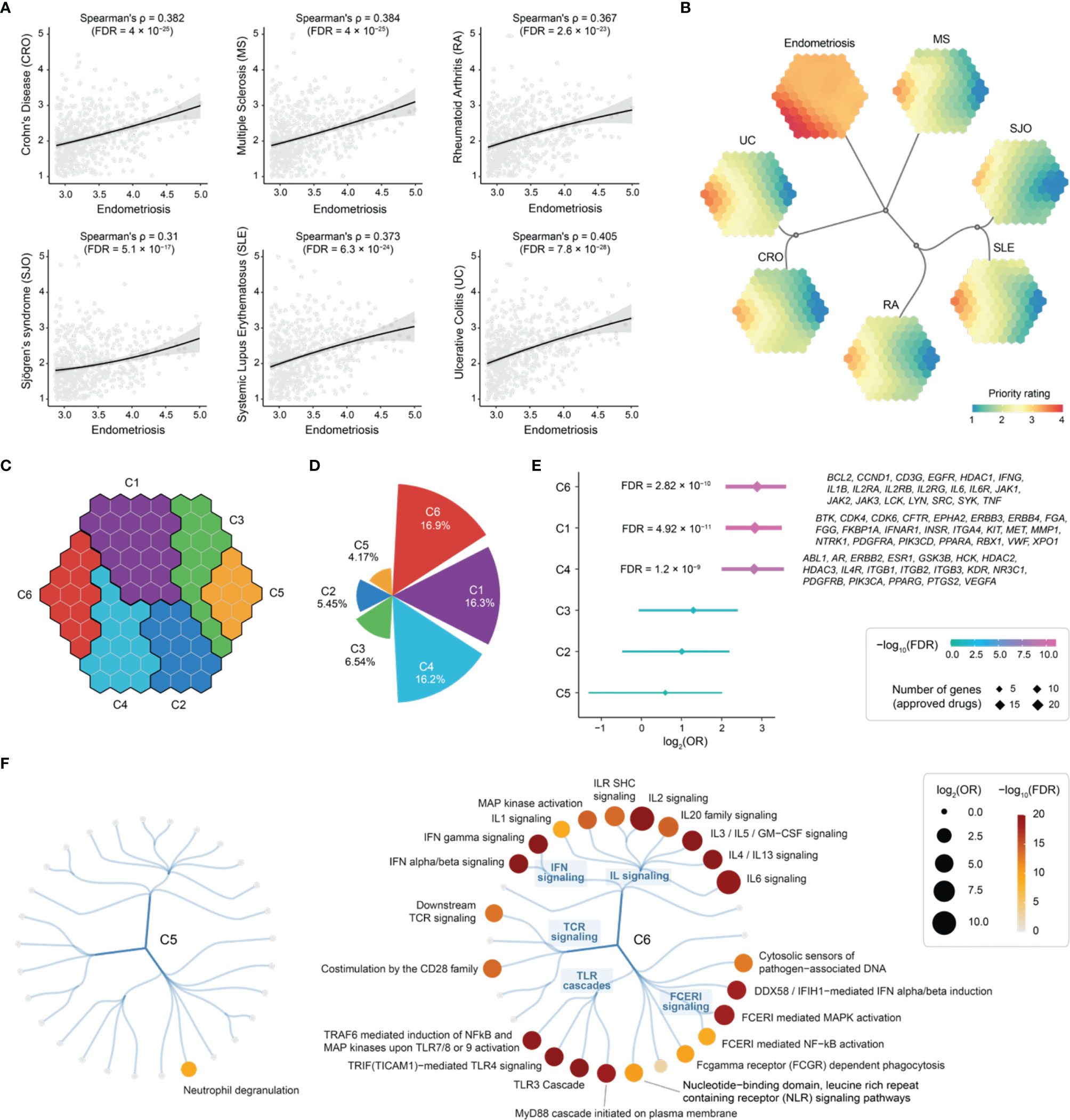
Figure 8 Cross-disease prioritization map between endometriosis and immune-mediated diseases identifies shared and distinct target gene clusters. (A) Scatter plots showing priority rating between endometriosis (x-axis) and immune diseases (each indicated in y-axis). Spearman’s rank correlation calculated, with the significance level (FDR) accounting for multiple tests. Such correlation was calculated based on the top 5% prioritized genes (n = 689) in endometriosis. (B) Prioritization map. Learned using a supra-hexagonal map to compare prioritizations between endometriosis and immune diseases. Each map illustrates a disease-specific gene prioritization profile, while consensus neighbor-joining tree captures the similarity of inter-disease prioritization profiles. (C) Target gene clusters. The prioritization map divided into 6 clusters (C1-C6), each covering continuous hexagons as color-coded. (D) Polar bar summarizing the percentage of approved drug targets per cluster. (E) Forest plot of approved drug target enrichments. (F) Pathway enrichments for C5 (left) and C6 (right). Reactome immune-related pathways are organized circularly, with each dot representing a pathway, sized by odds ratio (OR) and colored by FDR. FCER1, Fc epsilon receptor 1; GM-CSF, granulocyte-macrophage colony-stimulating factor; IFIH1, interferon induced with helicase C domain 1; IFN, interferon; IL, interleukin; ILR, interleukin receptor; SHC, Src homology 2-domain-containing; TCR, T-cell receptor; TLR, toll-like receptor.
To systematically identify target genes that are shared between diseases and are unique to specific diseases, we grouped nearby hexagons and identified 6 target clusters (C1-C6), each containing genes with similar prioritization patterns (Figure 8C and Table S5). Among these, C6 was highly rated in all diseases analyzed, displaying the highest percentage of approved therapeutics (Figure 8D). Notably, C5 was highly rated in endometriosis only and contained a relatively low proportion of approved drug genes (Figure 8D), indicative of being under-explored. In agreement with this, we found the highest degree of support from clinical evidence (approved drugs) for genes in C6, and no enrichment was observed for genes in C5 (Figure 8E).
Next, we characterized C5 and C6 using Reactome pathways (55). We found the enrichment for genes in C6 that are engaged in all major immune system pathways, except for neutrophil degranulation that was specific to C5 (Figure 8F). To confirm this, we performed functional enrichment analysis using Gene Ontology (56). We found that genes in C5 are mostly functionally relevant to neutrophil degranulation and are localized in the secretory granule lumen (Figure S3A). Using Mammalian Phenotype Ontology (57), we also observed phenotypic enrichments for genes in C5; these genes, when knocked out, tended to cause lethality phenotypes (Figure S3B).
Exploring Repurposing Opportunities Based on Shared Target Genes
Genes in C6 were highly prioritized across diseases (Figure 8). Based on these genes, we explored repurposing opportunities via a heatmap-like illustration (Figure 9). In addition to priority ratings, this illustration collectively showed how these genes are particulate in immune system pathways, including cytokine signaling (interferon signaling and interleukin signaling), innate immunity (toll-like receptor and Fc epsilon receptor 1) and adaptive immunity (T-cell receptor). We identified 20 approved drug targets that are mostly immune-related. These genes provide opportunities for licensed immunomodulatory drugs that could be repurposed for the potential use in endometriosis, particularly disease-modifying anti-rheumatic drugs (DMARDs). They include biological DMARDs (such as inhibitors of TNF, IL6, and IL6R) and targeted synthetic DMARDs (such as kinase inhibitors targeting JAK1/2/3, SRC, LYN, SYK, and LCK). Of particular interest, TNF is a well-established therapeutic target for chronic inflammatory diseases (68). We suggest that drugs targeting this gene should be studied in cohort-scale clinical trials for repurposing in endometriosis, such as infliximab (known as anti-TNF biologics).
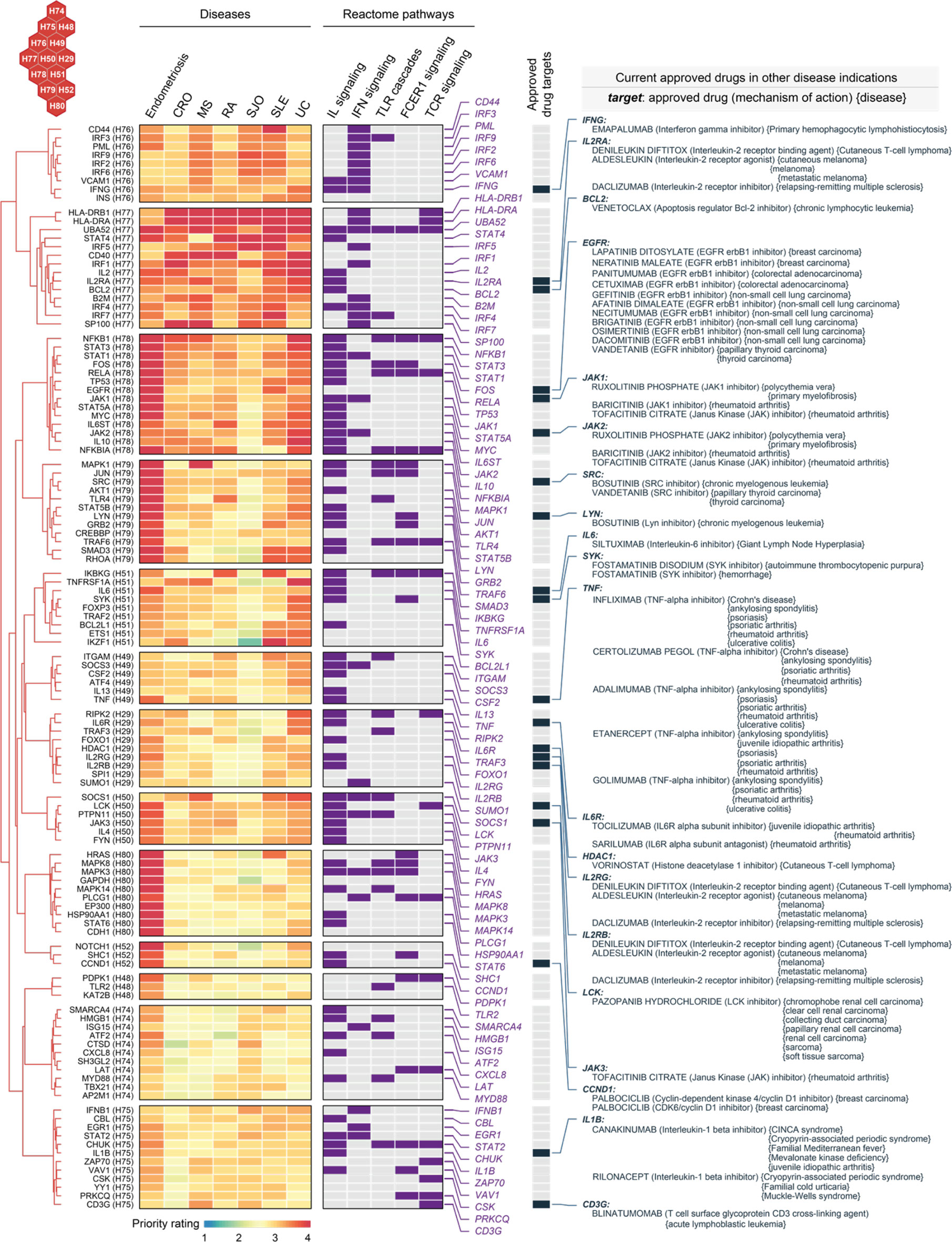
Figure 9 Repurposing evidence for immunomodulatory drugs based on shared target genes in the cluster C6. Heatmap illustrates target genes in C6 prioritized across 7 diseases, with annotations to interleukin (IL), interferon (IFN), toll-like receptor (TLR), Fc epsilon receptor 1 (FCER1), and T-cell receptor (TCR) signalings. Also annotated is the information on approved drug targets, repurposed drugs, mechanisms of action, and disease indications.
Exploring Tractable Evidence for Neutrophil Degranulation Target Genes Specific to Endometriosis
As shown in the heatmap visualization in Figure 10A, genes in C5 were specifically prioritized in endometriosis and characterized by neutrophil degranulation. Neutrophils are highly versatile and plastic. Plastic neutrophils are rendered through degranulation, in which cytoplasmic granules are mobilized and fused with the plasma membrane (69). Accumulating evidence supports the tumour metastatic spread model in which neutrophils mediate the migration of cancer cells to distant sites (70). Interestingly, we found that neutrophil degranulation genes in C5 are almost all linked to cancer; for example, those involved in breast cancer are the genes ATG7 (71), CAP1 (72), CCT8 (73), CD14 (74), CYFIP1 (75), and QSOX1 (76). Notably, neutrophil degranulation genes have been reported to be associated with endometriosis, such as A1BG (a diagnostic marker for stage II, III and IV endometriosis) (77) and ATG7 (an autophagy gene in ovarian endometriosis) (78). Finally, we explored the tractable evidence; a tractable gene was defined if its known protein structures were predicted to contain druggable pockets (Figure 10B). We suggest that the tractable genes involved in neutrophil degranulation are of particular interest to inform future clinical studies.
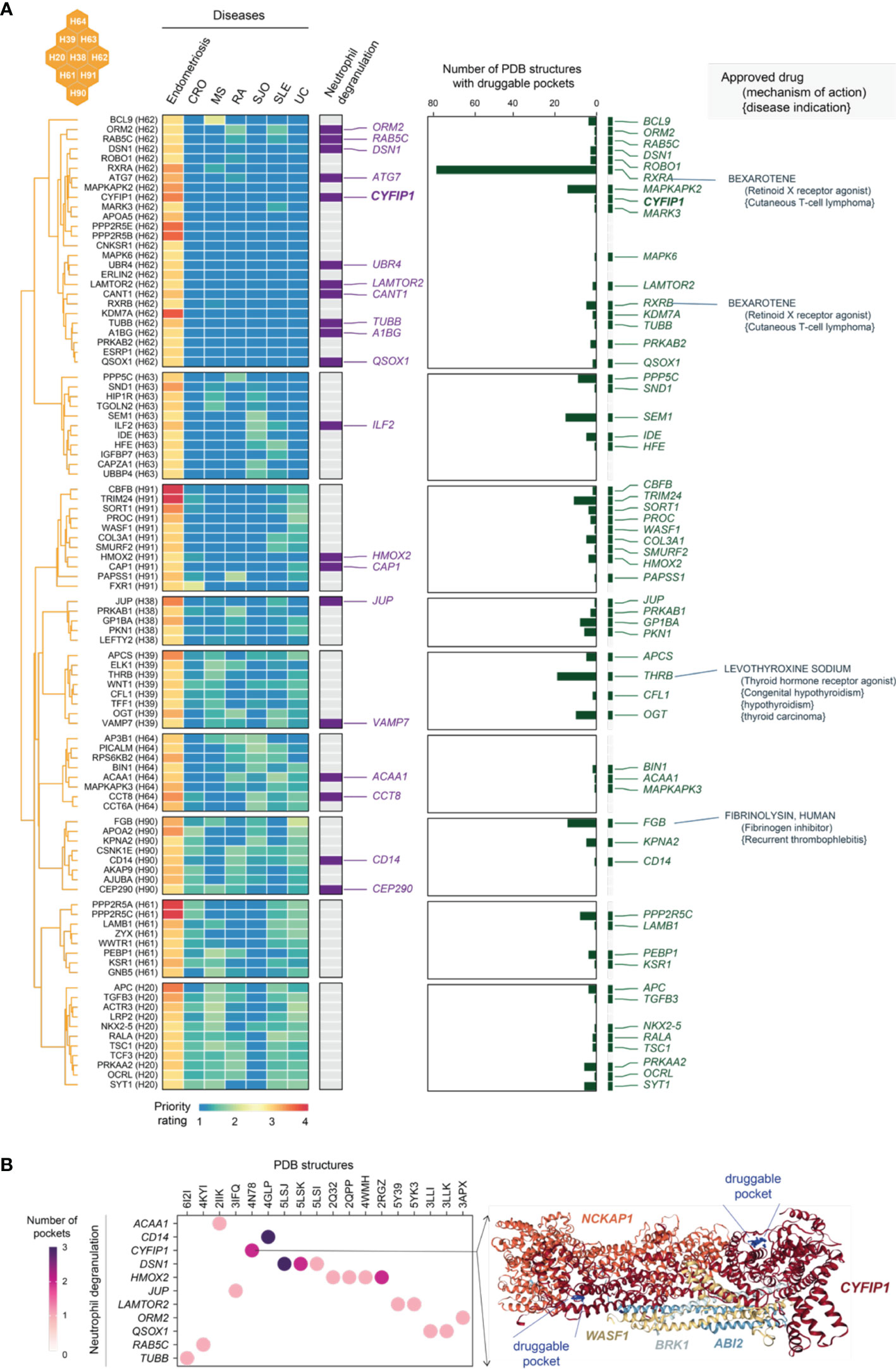
Figure 10 Tractable evidence for targeting neutrophil degranulation based on genes prioritized specific to endometriosis in the cluster C5. (A) Heatmap illustrates target genes in C5 prioritized across 7 diseases, together with the information on functional relevance to neutrophil degranulation, the tractability (the number of druggable pockets), and therapeutic approval (approved drug targets, repurposed drugs, mechanisms of action, and disease indications). (B) Druggable pockets for genes prioritised specific to endometriosis in C5. Dot plot shows 11 tractable genes involved in neutrophil degranulation (y-axis) and their PDB known protein structures (x-axis). Color-coded is the number of druggable pockets predicted based on the PDB structure. On the right is an exemplar illustrating an experimentally resolved WAVE regulatory complex coded as ‘4N78’ in PDB. This structure consists of 5 genes including CYFIP1, where two druggable pockets are predicted (as indicated in blue balls and labelled as well).
Discussion
Our organismal systems (mainly immune, endocrine, nervous, and reproductive) are evolutionarily conserved to maintain homeostasis. If these systems fail to coordinately respond to invaders or ectopic hazards, autoimmunity or other forms of systemic inflammation might occur. If such maladaptation persists, it may eventually lead to chronic inflammatory systemic diseases (64). We have found that genetic target genes prioritized in endometriosis are significantly enriched in a wide range of organismal system pathways (Figure 6) and molecular hallmarks (Figure 5). These findings strongly support the increasing recognition of endometriosis as an inflammatory systemic disease.
Our identification of target genes shared with immune diseases suggests drug repurposing opportunities (Figure 9). Drug repurposing is a strategy that repositions existing effective drugs with acceptable safety from original indications (such as immune diseases) to new indications (here, endometriosis). Immunomodulatory targets and DMARDs are rich in numbers and have been well-studied in therapeutic targeting. In the next steps, DMARDs represent a wish list that should be taken forward for validation, for example, to assess efficacy on perturbation in patient-derived cell assays (53). Given that multiple cell lineages might be involved in endometriosis (Figure 4), we utilized single-cell RNA-seq datasets of endometriosis (49) to examine the expression of shared target genes (listed in Figure 9). We found that these genes were expressed in one or more cell clusters that were assigned to patients with endometriosis (Figure S4A). Using genes specifically expressed in fibroblasts of ectopic endometrium (as compared to fibroblasts of normal endometrium) (49), we showed that these fibroblast-specific genes tended to be highly prioritized (Figure S4B), supporting the importance of fibroblasts in endometriosis.
We know that neutrophils are highly plastic and can mediate the transport of cancer cells into distant sites; plasticity and metastasis-like spread are both enabled by granule mobilization and exocytosis (known as ‘degranulation’) (69, 70). Systemic circulating neutrophils from endometriosis patients display distinct expression profiles when compared to neutrophils from healthy controls, and the lesion microenvironment contains factors such as IL-8 that promote recruitment of neutrophils (79). The identification of endometriosis-specific targets essential for neutrophil degranulation (Figure 10) not only brings forward new ideas for therapies targeting neutrophil degranulation, but also provides important clues about how endometriosis might spread to distant tissues or organs, causing inflammatory-like microenvironments. Our findings expand the existing theories that are proposed to explain the cause of endometriosis (the origin of disease), including: (i) Sampson’s retrograde menstruation (or the implantation theory), stating that at menses, endometrial cells reflux through the fallopian tubes and invade pelvic structures, where coordinated growth of nerves and blood vessels (known as ‘neuroangiogenesis’) may occur and elicit an inflammatory response, fibrotic scarring and pain; (ii) the coelomic metaplasia theory (80), postulating that mesothelial cells may transform into ectopic endometrial tissue through metaplastic transition; and (iii) stem cell trafficking, particularly endometrial mesenchymal stem/stromal cells (81, 82) and bone marrow-derived stem cells (83). We anticipate that consolidating mixed evidence to unify all findings (including ours) might be the most parsimonious explanation for this inflammatory systemic disease.
We have also used gene expression signatures (50) involving two endometriosis stages (I/II and III/IV) at two menstrual cycle phases (proliferative and secretory) to examine the highly prioritized target gene expression (Figure S5A). Target set enrichment analysis revealed the enrichment of gene signatures in stage I/II endometriosis (early disease), independent of menstrual cycle phases. For stage III/IV endometriosis (late disease), the enrichment was observed at the proliferative cycle phase but not at the secretory cycle phase. These results suggested that highly prioritized target genes tended to be expressed in the early stage of disease, which is consistent with the pro-inflammatory profile in stage I/II endometriosis (50). We also found that genes highly prioritized only in endometriosis (listed in Figure 10) were mostly expressed in stage I/II disease (relative to the healthy counterpart; Figure S5B).
In summary, we have shown that multi-layered genomic datasets (including new information on genetic susceptability loci identified in GWAS on endometriosis, regulatory genomics, and protein interactome) can be harnessed with our prioritization approach to generate an atlas of genetic target prioritizations in endometriosis (Figures 1-3). This atlas has enhanced our understanding of endometriosis as an inflammatory systemic disease and has also expanded the existing theories on the origin of disease. More importantly, we have discovered target candidates that are specific to and tractable for disease. Genomic insights and therapeutic candidates arising from this study may change our perspectives on endometriosis and our strategies for coping with disease, envigorating further research and drug repurposing.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. All results, including an atlas of genetic targets and repurposed drugs in endometriosis, are publicly available at https://23verse.github.io/end for download and exploration. Further inquiries can be directed to the corresponding author.
Author Contributions
CB analyzed data, contributed to data curation, wrote and revised the manuscript. HW contributed to data curation and interpretation. HF conceived and supervized the project, curated and analyzed data, wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work is funded by National Natural Science Foundation of China (32170663), Shanghai Pujiang Program (21PJ1409600), Innovative Research Team of High-Level Local Universities in Shanghai, and Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (awarded to HF).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.758440/full#supplementary-material
References
1. Clement PB. The Pathology of Endometriosis: A Survey of the Many Faces of a Common Disease Emphasizing Diagnostic Pitfalls and Unusual and Newly Appreciated Aspects. Adv Anat Pathol (2007) 14:241–60. doi: 10.1097/PAP.0b013e3180ca7d7b
2. Saunders PTK, Horne AW. Endometriosis: Etiology, Pathobiology, and Therapeutic Prospects. Cell (2021) 184:2807–24. doi: 10.1016/j.cell.2021.04.041
3. Zondervan KT, Becker CM, Koga K, Missmer SA, Taylor RN, Viganò P. Endometriosis. Nat Rev Dis Prim (2018) 4:9. doi: 10.1038/s41572-018-0008-5
4. Taylor HS, Kotlyar AM, Flores VA. Endometriosis Is a Chronic Systemic Disease: Clinical Challenges and Novel Innovations. Lancet (2021) 397:839–52. doi: 10.1016/S0140-6736(21)00389-5
5. Goetz LG, Mamillapalli R, Taylor HS. Low Body Mass Index in Endometriosis is Promoted by Hepatic Metabolic Gene Dysregulation in Mice. Biol Reprod (2016) 95:1–8. doi: 10.1095/biolreprod.116.142877
6. Chen LC, Hsu JW, Huang KL, Bai YM, Su TP, Li CT, et al. Risk of Developing Major Depression and Anxiety Disorders Among Women With Endometriosis: A Longitudinal Follow-Up Study. J Affect Disord (2016) 190:282–5. doi: 10.1016/j.jad.2015.10.030
7. Alderman MH, Yoder N, Taylor HS. The Systemic Effects of Endometriosis. Semin Reprod Med (2017) 35:263–70. doi: 10.1055/s-0037-1603582
8. Shigesi N, Kvaskoff M, Kirtley S, Feng Q, Fang H, Knight JC, et al. The Association Between Endometriosis and Autoimmune Diseases: A Systematic Review and Meta-Analysis. Hum Reprod Update (2019) 25:486–503. doi: 10.1093/humupd/dmz012
9. Sapkota Y, Steinthorsdottir V, Morris AP, Fassbender A, Rahmioglu N, De Vivo I, et al. Meta-Analysis Identifies Five Novel Loci Associated With Endometriosis Highlighting Key Genes Involved in Hormone Metabolism. Nat Commun (2017) 8:1–12. doi: 10.1038/ncomms15539
10. Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The Support of Human Genetic Evidence for Approved Drug Indications. Nat Genet (2015) 47:856–60. doi: 10.1038/ng.3314
11. King EA, Wade Davis J, Degner JF. Are Drug Targets With Genetic Support Twice as Likely to be Approved? Revised Estimates of the Impact of Genetic Support for Drug Mechanisms on the Probability of Drug Approval. PloS Genet (2019) 15:1–20. doi: 10.1371/journal.pgen.1008489
12. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of Rheumatoid Arthritis Contributes to Biology and Drug Discovery. Nature (2014) 506:376–81. doi: 10.1038/nature12873
13. Reay WR, Cairns MJ. Advancing the Use of Genome-Wide Association Studies for Drug Repurposing. Nat Rev Genet (2021) 22(10):658-71. doi: 10.1038/s41576-021-00387-z
14. Fang H, Knight JC. Priority Index: Database of Genetic Targets in Immune-Mediated Disease. Nucleic Acids Res (2022) 50:1358–67. doi: 10.1093/nar/gkab994
15. Fang H, Chen L, Knight JC. From Genome-Wide Association Studies to Rational Drug Target Prioritisation in Inflammatory Arthritis. Lancet Rheumatol (2020) 2:e50–62. doi: 10.1016/S2665-9913(19)30134-1
16. Chia R, Saez-Atienzar S, Murphy N, Chiò A, Blauwendraat C, Consortium IMGG, et al. Identification of Genetic Risk Loci and Prioritization of Genes and Pathways for Myasthenia Gravis: A Genome-Wide Association Study. Proc Natl Acad Sci (2022) 119:e2108672119. doi: 10.1073/pnas.2108672119
17. Ghoussaini M, Mountjoy E, Carmona M, Peat G, Schmidt EM, Hercules A, et al. Open Targets Genetics: Systematic Identification of Trait-Associated Genes Using Large-Scale Genetics and Functional Genomics. Nucleic Acids Res (2021) 49:D1311–20. doi: 10.1093/nar/gkaa840
18. Fang H, Jiang L. Genetic Prioritization, Therapeutic Repositioning and Cross-Disease Comparisons Reveal Inflammatory Targets Tractable for Kidney Stone Disease. Front Immunol (2021) 12:687291. doi: 10.3389/fimmu.2021.687291
19. Schwartzentruber J, Cooper S, Liu JZ, Barrio-Hernandez I, Bello E, Kumasaka N, et al. Genome-Wide Meta-Analysis, Fine-Mapping and Integrative Prioritization Implicate New Alzheimer’s Disease Risk Genes. Nat Genet (2021) 53:392–402. doi: 10.1038/s41588-020-00776-w
20. Ochoa D, Hercules A, Carmona M, Suveges D, Gonzalez-Uriarte A, Malangone C, et al. Open Targets Platform: Supporting Systematic Drug-Target Identification and Prioritisation. Nucleic Acids Res (2021) 49:D1302–10. doi: 10.1093/nar/gkaa1027
21. Robertson CC, Inshaw JRJ, Onengut-Gumuscu S, Chen W-M, Santa Cruz DF, Yang H, et al. Fine-Mapping, Trans-Ancestral and Genomic Analyses Identify Causal Variants, Cells, Genes and Drug Targets for Type 1 Diabetes. Nat Genet (2021) 53:962–71. doi: 10.1038/s41588-021-00880-5
22. Orrù V, Steri M, Sidore C, Marongiu M, Serra V, Olla S, et al. Complex Genetic Signatures in Immune Cells Underlie Autoimmunity and Inform Therapy. Nat Genet (2020) 52:1036–45. doi: 10.1038/s41588-020-0684-4
23. Fang H, The ULTRA-DD Consortium, De Wolf H, Knezevic B, Burnham KL, Osgood J, et al. A Genetics-Led Approach Defines the Drug Target Landscape of 30 Immune-Related Traits. Nat Genet (2019) 51:1082–91. doi: 10.1038/s41588-019-0456-1
24. Sobalska-Kwapis M, Smolarz B, Słomka M, Szaflik T, Kępka E, Kulig B, et al. New Variants Near RHOJ and C2, HLA-DRA Region and Susceptibility to Endometriosis in the Polish Population—The Genome-Wide Association Study. Eur J Obstet Gynecol Reprod Biol (2017) 217:106–12. doi: 10.1016/j.ejogrb.2017.08.037
25. Albertsen HM, Chettier R, Farrington P, Ward K. Genome-Wide Association Study Link Novel Loci to Endometriosis. PloS One (2013) 8:e58257. doi: 10.1371/journal.pone.0058257
26. Nyholt DR, Low SK, Anderson CA, Painter JN, Uno S, Morris AP, et al. Genome-Wide Association Meta-Analysis Identifies New Endometriosis Risk Loci. Nat Genet (2012) 44:1355–9. doi: 10.1038/ng.2445
27. Painter JN, Anderson CA, Nyholt DR, MacGregor S, Lin J, Lee SH, et al. Genome-Wide Association Study Identifies a Locus at 7p15.2 Associated With Endometriosis. Nat Genet (2011) 43:51–4. doi: 10.1038/ng.731
28. 1000 Genomes Project Consortium. An Integrated Map of Genetic Variation From 1,092 Human Genomes. Nature (2012) 491:56–65. doi: 10.1038/nature11632
29. Jung I, Schmitt A, Diao Y, Lee AJ, Liu T, Yang D, et al. A Compendium of Promoter-Centered Long-Range Chromatin Interactions in the Human Genome. Nat Genet (2019) 51:1442–9. doi: 10.1038/s41588-019-0494-8
30. Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, et al. Mapping Long-Range Promoter Contacts in Human Cells With High-Resolution Capture Hi-C. Nat Genet (2015) 47:598–606. doi: 10.1038/ng.3286
31. Choy M, Javierre BM, Williams SG, Baross SL, Liu Y, Wingett SW, et al. Promoter Interactome of Human Embryonic Stem Cell-Derived Cardiomyocytes Connects GWAS Regions to Cardiac Gene Networks. Nat Commun (2018) 9:2526. doi: 10.1038/s41467-018-04931-0
32. Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, et al. Lineage-Specific Genome Architecture Links Enhancers and Non-Coding Disease Variants to Target Gene Promoters. Cell (2016) 167:1369–84.e19. doi: 10.1016/j.cell.2016.09.037
33. Miguel-Escalada I, Bonàs-Guarch S, Cebola I, Joan P-C, Mendieta-Esteban J, Rolando DMY, et al. Human Pancreatic Islet 3D Chromatin Architecture Provides Insights Into the Genetics of Type 2 Diabetes. Nat Genet (2019) 51:1137–48. doi: 10.1101/400291
34. Song M, Yang X, Ren X, Maliskova L, Li B, Jones IR, et al. Mapping Cis-Regulatory Chromatin Contacts in Neural Cells Links Neuropsychiatric Disorder Risk Variants to Target Genes. Nat Genet (2019) 51:1252–62. doi: 10.1038/s41588-019-0472-1
35. Leland Taylor D, Jackson AU, Narisu N, Hemani G, Erdos MR, Chines PS, et al. Integrative Analysis of Gene Expression, DNA Methylation, Physiological Traits, and Genetic Variation in Human Skeletal Muscle. Proc Natl Acad Sci USA (2019) 166:10883–8. doi: 10.1073/pnas.1814263116
36. Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, et al. Common Genetic Variation Drives Molecular Heterogeneity in Human iPSCs. Nature (2017) 546:370–5. doi: 10.1038/nature22403
37. Momozawa Y. IBD Risk Loci are Enriched in Multigenic Regulatory Modules Encompassing Putative Causative Genes. Nat Commun (2018) 9:1–18. doi: 10.1038/s41467-018-04365-8
38. Quach H, Rotival M, Pothlichet J, Loh YHE, Dannemann M, Zidane N, et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell (2016) 167:643–56.e17. doi: 10.1016/j.cell.2016.09.024
39. Chen L, Ge B, Casale FP, Downes K, Pastinen T. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells Resource Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell (2016) 167:1398–414. doi: 10.1016/j.cell.2016.10.026
40. Fairfax BP, Humburg P, Makino S, Naranbhai V, Wong D, Lau E, et al. Innate Immune Activity Conditions the Effect of Regulatory Variants Upon Monocyte Gene Expression. Science (2014) 343:1118. doi: 10.1126/science.1246949
41. The GTEx Consortium. The GTEx Consortium Atlas of Genetic Regulatory Effects Across Human Tissues The Genotype Tissue Expression Consortium. Science (2020) 369:1318–30. doi: 10.1101/787903
42. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47:D607–13. doi: 10.1093/nar/gky1131
44. Mendez D, Gaulton A, Bento AP, Chambers J, De Veij M, Félix E, et al. ChEMBL: Towards Direct Deposition of Bioassay Data. Nucleic Acids Res (2019) 47:D930–40. doi: 10.1093/nar/gky1075
45. Carvalho-Silva D, Pierleoni A, Pignatelli M, Ong CK, Fumis L, Karamanis N, et al. Open Targets Platform: New Developments and Updates Two Years on. Nucleic Acids Res (2019) 47:D1056–65. doi: 10.1093/nar/gky1133
46. Fang H, Gough J. The Dnet Approach Promotes Emerging Research on Cancer Patient Survival. Genome Med (2014) 6:64. doi: 10.1186/s13073-014-0064-8
47. Aran D, Hu Z, Butte AJ. Xcell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol (2017) 18:220. doi: 10.1186/s13059-017-1349-1
48. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst (2015) 1:417–25. doi: 10.1016/j.cels.2015.12.004
49. Ma J, Zhang L, Zhan H, Mo Y, Ren Z, Shao A, et al. Single-Cell Transcriptomic Analysis of Endometriosis Provides Insights Into Fibroblast Fates and Immune Cell Heterogeneity. Cell Biosci (2021) 11:1–19. doi: 10.1186/s13578-021-00637-x
50. Poli-Neto OB, Meola J, Rosa-e-Silva JC, Tiezzi D. Transcriptome Meta-Analysis Reveals Differences of Immune Profile Between Eutopic Endometrium From Stage I-II and III-IV Endometriosis Independently of Hormonal Milieu. Sci Rep (2020) 10:1–17. doi: 10.1038/s41598-019-57207-y
51. Fang H, Knezevic B, Burnham KL, Knight JC. XGR Software for Enhanced Interpretation of Genomic Summary Data, Illustrated by Application to Immunological Traits. Genome Med (2016) 8:1–20. doi: 10.1186/s13073-016-0384-y
52. Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res (2021) 49:D545–51. doi: 10.1093/nar/gkaa970
54. Fang H, Gough J. Suprahex: An R/Bioconductor Package for Tabular Omics Data Analysis Using a Supra-Hexagonal Map. Biochem Biophys Res Commun (2014) 443:285–9. doi: 10.1016/j.bbrc.2013.11.103
55. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res (2020) 48:D498–503. doi: 10.1093/nar/gkz1031
56. Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: Gene-Centered Information at NCBI. Nucleic Acids Res (2011) 39:D52–7. doi: 10.1093/nar/gkq1237
57. Smith CL, Eppig JT. The Mammalian Phenotype Ontology: Enabling Robust Annotation and Comparative Analysis. Wiley Interdiscip Rev Syst Biol Med (2009) 1:390–9. doi: 10.1002/wsbm.44
58. Schmidtke P, Barril X. Understanding and Predicting Druggability. A High-Throughput Method for Detection of Drug Binding Sites. J Med Chem (2010) 53:5858–67. doi: 10.1021/jm100574m
59. Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, et al. RCSB Protein Data Bank: Powerful New Tools for Exploring 3D Structures of Biological Macromolecules for Basic and Applied Research and Education in Fundamental Biology, Biomedicine, Biotechnology, Bioengineering and Energy Sciences. Nucleic Acids Res (2021) 49:D437–51. doi: 10.1093/nar/gkaa1038
60. Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, et al. The WAVE Regulatory Complex Links Diverse Receptors to the Actin Cytoskeleton. Cell (2014) 156:195–207. doi: 10.1016/j.cell.2013.11.048
61. Rose AS, Bradley AR, Valasatava Y, Duarte JM, Prlic A, Rose PW. NGL Viewer: Web-Based Molecular Graphics for Large Complexes. Bioinformatics (2018) 34:3755–8. doi: 10.1093/bioinformatics/bty419
62. Group T members of the EGC, Becker CM, Bokor A, Heikinheimo O, Horne A, Jansen F, et al. ESHRE Guideline: Endometriosis. Hum Reprod Open (2022):hoac009. doi: 10.1093/hropen/hoac009
63. Madanes D, Bilotas MA, Bastón JI, Singla JJ, Meresman GF, Barañao RI, et al. PI3K/AKT Pathway is Altered in the Endometriosis Patient’s Endometrium and Presents Differences According to Severity Stage. Gynecol Endocrinol (2020) 36:436–40. doi: 10.1080/09513590.2019.1680627
64. Straub RH, Schradin C. Chronic Inflammatory Systemic Diseases: An Evolutionary Trade-Off Between Acutely Beneficial But Chronically Harmful Programs. Evol Med Public Heal (2016)(1):37-51. doi: 10.1093/emph/eow001
65. Barra F, Ferro Desideri L, Ferrero S. Inhibition of PI3K/AKT/mTOR Pathway for the Treatment of Endometriosis. Br J Pharmacol (2018) 175:3626–7. doi: 10.1111/bph.14391
66. Harada T, Kosaka S, Elliesen J, Yasuda M, Ito M, Momoeda M. Ethinylestradiol 20 μg/Drospirenone 3 Mg in a Flexible Extended Regimen for the Management of Endometriosis-Associated Pelvic Pain: A Randomized Controlled Trial. Fertil Steril (2017) 108:798–805. doi: 10.1016/j.fertnstert.2017.07.1165
67. Yavropoulou MP, Makras P, Anastasilakis AD. Bazedoxifene for the Treatment of Osteoporosis. Expert Opin Pharmacother (2019) 20:1201–10. doi: 10.1080/14656566.2019.1615882
68. Monaco C, Nanchahal J, Taylor P, Feldmann M. Anti-TNF Therapy: Past, Present and Future. Int Immunol (2015) 27:55–62. doi: 10.1093/intimm/dxu102
69. Jaillon S, Ponzetta A, Di Mitri D, Santoni A, Bonecchi R, Mantovani A. Neutrophil Diversity and Plasticity in Tumour Progression and Therapy. Nat Rev Cancer (2020) 20:485–503. doi: 10.1038/s41568-020-0281-y
70. Mollinedo F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol (2019) 40:228–42. doi: 10.1016/j.it.2019.01.006
71. Li M, Liu J, Li S, Feng Y, Yi F, Wang L, et al. Autophagy-Related 7 Modulates Tumor Progression in Triple-Negative Breast Cancer. Lab Investig (2019) 99:1266–74. doi: 10.1038/s41374-019-0249-2
72. Bergqvist M, Elebro K, Elebro K, Sandsveden M, Borgquist S, Borgquist S, et al. Effects of Tumor-Specific CAP1 Expression and Body Constitution on Clinical Outcomes in Patients With Early Breast Cancer. Breast Cancer Res (2020) 22:1–15. doi: 10.1186/s13058-020-01307-5
73. Xu WX, Song W, Jiang MP, Yang SJ, Zhang J, Wang DD, et al. Systematic Characterization of Expression Profiles and Prognostic Values of the Eight Subunits of the Chaperonin TRiC in Breast Cancer. Front Genet (2021) 12:637887. doi: 10.3389/fgene.2021.637887
74. He W, Tong Y, Wang Y, Liu J, Luo G, Wu J, et al. Serum Soluble CD14 Is a Potential Prognostic Indicator of Recurrence of Human Breast Invasive Ductal Carcinoma With Her2-Enriched Subtype. PloS One (2013) 8:1–8. doi: 10.1371/journal.pone.0075366
75. Chang JW, Kuo WH, Lin CM, Chen WL, Chan SH, Chiu MF, et al. Wild-Type P53 Upregulates an Early Onset Breast Cancer-Associated Gene GAS7 to Suppress Metastasis via GAS7–CYFIP1-Mediated Signaling Pathway. Oncogene (2018) 37:4137–50. doi: 10.1038/s41388-018-0253-9
76. Knutsvik G, Collett K, Arnes J, Akslen LA, Stefansson IM. QSOX1 Expression is Associated With Aggressive Tumor Features and Reduced Survival in Breast Carcinomas. Mod Pathol (2016) 29:1485–91. doi: 10.1038/modpathol.2016.148
77. Dutta M, Subramani E, Taunk K, Gajbhiye A, Seal S, Pendharkar N, et al. Investigation of Serum Proteome Alterations in Human Endometriosis. J Proteomics (2015) 114:182–96. doi: 10.1016/j.jprot.2014.10.021
78. Allavena G, Carrarelli P, Del Bello B, Luisi S, Petraglia F, Maellaro E. Autophagy Is Upregulated in Ovarian Endometriosis: A Possible Interplay With P53 and Heme Oxygenase-1. Fertil Steril (2015) 103:1244–51.e1. doi: 10.1016/j.fertnstert.2015.02.007
79. Symons LK, Miller JE, Tyryshkin K, Monsanto SP, Marks RM, Lingegowda H, et al. Neutrophil Recruitment and Function in Endometriosis Patients and a Syngeneic Murine Model. FASEB J (2020) 34:1558–75. doi: 10.1096/fj.201902272R
80. Matsuura K, Ohtake H, Katabuchi H, Okamura H. Coelomic Metaplasia Theory of Endometriosis Evidence From In Vivo Studies and an In Vitro Experimental Model. Gynecol Obstet Invest (1999) 47:18–22. doi: 10.1159/000052855
81. Gargett CE, Gurung S. Endometrial Mesenchymal Stem/Stromal Cells, Their Fibroblast Progeny in Endometriosis, and More. Biol Reprod (2016) 94:1–4. doi: 10.1095/biolreprod.116.141325
82. Gargett CE, Schwab KE, Deane JA. Endometrial Stem/Progenitor Cells: The First 10 Years. Hum Reprod Update (2016) 22:137–63. doi: 10.1093/humupd/dmv051
Keywords: endometriosis, inflammatory systemic disease, therapeutic target prioritization, drug repurposing, neutrophil degranulation
Citation: Bao C, Wang H and Fang H (2022) Genomic Evidence Supports the Recognition of Endometriosis as an Inflammatory Systemic Disease and Reveals Disease-Specific Therapeutic Potentials of Targeting Neutrophil Degranulation. Front. Immunol. 13:758440. doi: 10.3389/fimmu.2022.758440
Received: 14 August 2021; Accepted: 02 March 2022;
Published: 23 March 2022.
Edited by:
Rosa M. Sainz, University of Oviedo, SpainReviewed by:
Hao Shi, St. Jude Children’s Research Hospital, United StatesPhilippa Saunders, University of Edinburgh, United Kingdom
Copyright © 2022 Bao, Wang and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Fang, ZmgxMjM1NUByamguY29tLmNu
 Chaohui Bao
Chaohui Bao Hengru Wang
Hengru Wang Hai Fang
Hai Fang