- Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, Molecular Biology Section, National Institutes of Health, Bethesda, MD, United States
Immune recognition by T lymphocytes and natural killer (NK) cells is in large part dependent on the identification of cell surface MHC molecules bearing peptides generated from either endogenous (MHC I) or exogenous (MHC II) dependent pathways. This review focuses on MHC I molecules that coordinately fold to bind self or foreign peptides for such surface display. Peptide loading occurs in an antigen presentation pathway that includes either the multimolecular peptide loading complex (PLC) or a single chain chaperone/catalyst, TAP binding protein, related, TAPBPR, that mimics a key component of the PLC, tapasin. Recent structural and dynamic studies of TAPBPR reveal details of its function and reflect on mechanisms common to tapasin. Regions of structural conservation among species suggest that TAPBPR and tapasin have evolved to satisfy functional complexities demanded by the enormous polymorphism of MHC I molecules. Recent studies suggest that these two chaperone/catalysts exploit structural flexibility and dynamics to stabilize MHC molecules and facilitate peptide loading.
Introduction
Classical experiments indicate that proteins arrive at their stable three-dimensional conformation at their lowest Gibbs free energy, achieved as a result of their primary amino acid sequence and their interactions with solvent (1, 2). Nevertheless, the potential timescale of searching the myriad possible conformations of a protein as noted by Levinthal (3, 4) raised a conundrum solved only partially by the recognition of the contribution of protein nucleation regions and folding landscapes (5, 6) to the descent along an energy funnel to achieve a final stable structure (7). More recently, the so-called “protein folding problem” has been redefined in terms of the practical utility of predicting a protein’s three-dimensional structure from its primary amino acid sequence. This computational boundary is now being overcome by the concurrence of large and ever increasing structural and sequence databases with innovative artificial intelligence approaches by DeepMind and its implementation of AlphaFold2 (8). However, by contrast to the apparent success of structure prediction of individual proteins in recent years, our understanding of the rules that govern protein interactions remain rudimentary. To paraphrase Donne (9), no protein is an island. During the course of its lifetime, from biogenesis on the ribosome to destruction by the proteasome, a single protein molecule must interact with a multitude of partners. These include chaperones that aid its folding and prevent aggregation, enzymes that add post-translational modifications, transport proteins that escort it to its destinations, the substrate on which it performs its biological function, and the ubiquitinylating enzymes that target it for destruction. The evolutionarily conserved, and crucially important, antigen presentation pathway in vertebrates provides a valuable model system in which to investigate these various events in the life of a protein
Reflected in the pathways that have evolved to permit coassembly of antigenic peptides with their glycoprotein antigen presenting elements, the antigen presentation pathways that govern the biosynthesis, folding, assembly, peptide loading, peptide exchange, and cell surface expression of peptide/protein complexes are crucial to the immune response to tumors, viruses, and a variety of cellular pathogens (10–12). These pathways are based on the major histocompatibility complex (MHC) encoded class I (MHC I) and class II (MHC II) proteins, and their associated molecules. In this speculative review, we will focus on the classical MHC I molecules, HLA-A, -B, and -C in the human and H2-K, -D, and -L in the mouse, obligate cell surface intrinsic membrane proteins, that serve as recognition elements for T cell receptors (TCR) expressed on CD8+ T lymphocytes as well as ligands for various receptors on natural killer (NK) cells and other hematopoietic effector cells.
MHC Molecules, Not All Are the Same
The most remarkable characteristic of classical MHC I molecules is that they are highly polymorphic. That is, the number of allelomorphic variants in the human population, encoded at the three major genetic loci, HLA-A, -B, and -C, is enormous, catalogued by the IMGT database to be greater than 22,000 at current count (13). These are cell surface expressed type I membrane glycoproteins that are complexed with an essentially monomorphic light chain, β2-microglobulin (β2m). In addition, each MHC I molecule of a given cell binds a multitude of peptides derived from an endogenous MHC I pathway, thus generating a large repertoire of surface molecules available for interaction with immune cell receptors. The puzzles of course, are how do all these distinct MHC I molecules fold, how does each one form a stable ternary complex bound to each of thousands of potential peptides, and how does the biological system select for the most thermodynamically stable peptide/MHC I complexes for display at the cell surface.
The Peptide Loading Complex – A Molecular Machine for MHC I Assembly and Peptide Loading
Several decades of experimentation have identified the peptide loading complex (PLC), a multimolecular dynamic machine that sequentially stabilizes the MHC I heavy chain to fold with its light chain β2m, then to access and bind antigenic peptides delivered to the lumen of the endoplasmic reticulum, to exchange and evaluate peptides to identify the best binders, to pass quality control, to access the cis Golgi, and to proceed from there to the cell surface (10–12). Major insights included the identification of the roles of the chaperone/lectins calnexin and calreticulin that monitor the sequential glycosylation of the MHC I heavy chain. Further studies recognized the importance of the transporter associated with antigen processing (TAP) 1 and 2, an ATP-dependent heterodimer that delivers peptides from the proteasome-generated cytoplasmic pool to the ER, and the crucial function of tapasin, an ER protein that bridges TAP to the nascently folding/peptide binding MHC I/β2m complex, and an oxidoreductase, ERp57. Additional steps in the quality control of peptide-loaded MHC I include glycan-dependent interactions (14, 15). These steps of the classical peptide loading pathway are illustrated in Figure 1.
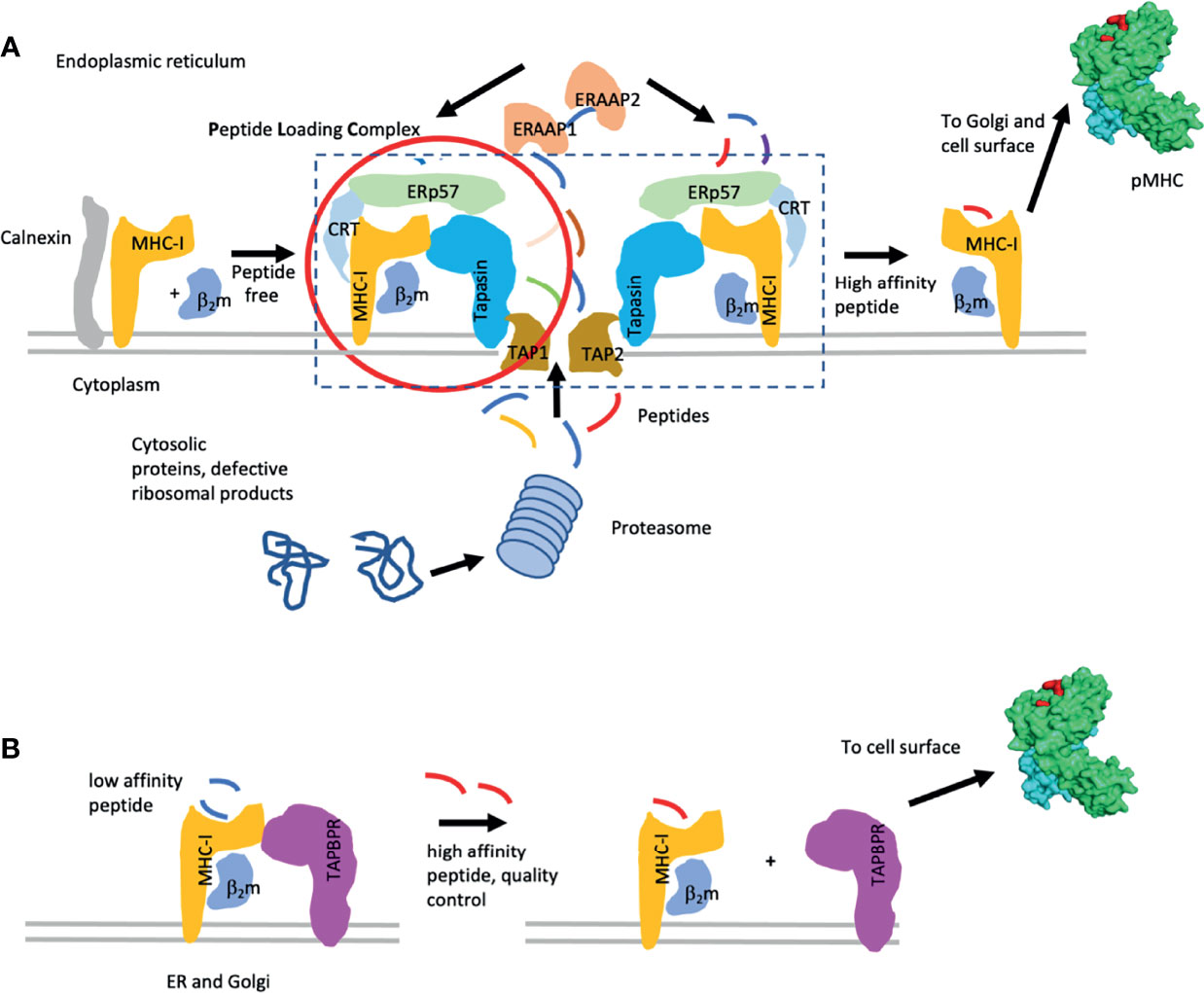
Figure 1 Pathways of MHC-I peptide loading. (A) Classic MHC-I peptide loading pathway is illustrated. Components of the pathway [i.e. the peptide loading complex (PLC)] include the chaperones calnexin, calreticulin (CRT), ERp57, tapasin, and the TAP1/2 peptide transporter. Peptides are indicated. Additional quality control through the UGT pathway is described in the text. (B) Peptide exchange with the chaperone TAPBPR are indicated. Lower affinity peptides are replaced by higher affinity peptides while the MHC I molecules are stabilized by either tapasin or TAPBPR. When high affinity peptide is bound, the MHC I complex dissociates from the chaperone and proceeds to the cell surface. [This figure is a modification of one published elsewhere (16)].
Visualization of structural aspects of tapasin function was first achieved in a classical paper by Dong, Wearsch and colleagues (17), which reported the X-ray crystallographic structure of human tapasin bound to ERp57. This work, complemented by mutational analysis of MHC I molecules and study of MHC I polymorphic variants, provided several molecular models for how tapasin interacts with MHC I, revealing how it might stabilize partially folded MHC I and encourage peptide exchange (18–22).
In the absence of detailed structural information on the nature of the tapasin/MHC I association, a cryo-electron microscopic approach was taken by Blees et al, who established a three-dimensional view, albeit at modest resolution (7.2 Å for the full complex, 5.8 Å for the editing module). This established the relationships between the components of the PLC: β2m, MHC I heavy chain, TAP, tapasin, ERp57, and calreticulin, and confirmed the stoichiometry previously established by pull-down experiments (23, 24) (Figure 2). Thus, the full PLC was visualized as containing one TAP1/2 heterodimer, with each chain flanked by an MHC I/β2m/ERp57/calreticulin complex (see Figure 2). Visualization of peptide was difficult at this resolution.
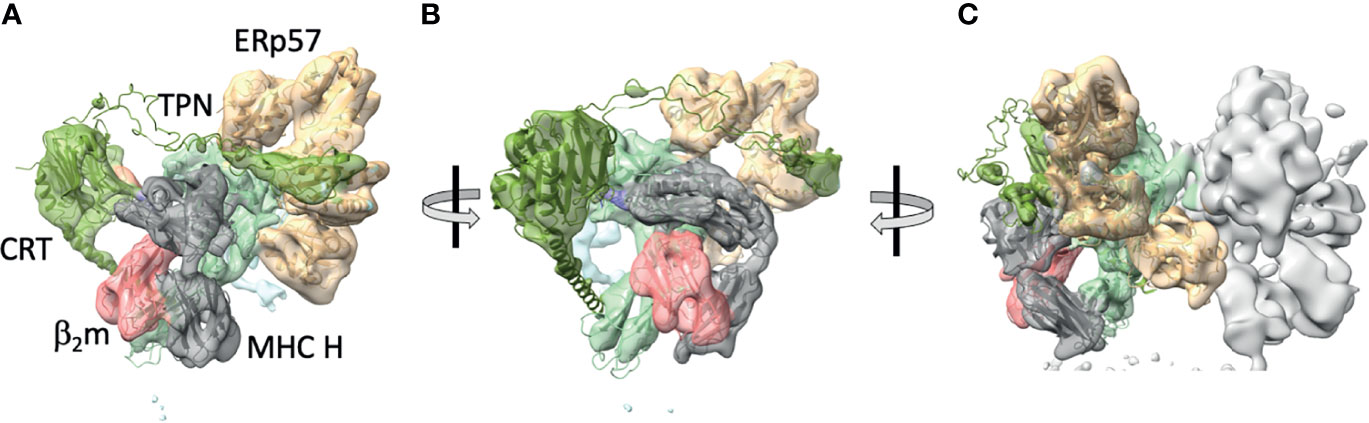
Figure 2 Cryo-EM maps of the complete protein loading complex reveal spatial organization of its components. (A) 5.8 Å cryo-EM map of the PLC editing module (EMD-3906) with superposed X-ray -derived models (PDB-6ENY) as described by (23). (B) PLC rotated approximately 30° to center the β2m/MHC H chain module. (C) 7.2 Å cryo-EM map of the pseudosymmetrical PLC module, with the left side editing module colored and the right side one illustrating by map surface alone. Map and models with generated with ChimeraX. MHC H chain is grey, β2m is pink, calreticulin is green, tapasin is pale blue, and ERp57 is tan.
TAPBPR a Surrogate PLC, Reveals Further Details of CHAPERONE/CATALYST Function
As the complete map of the human genome became available, several groups identified genetic regions paralagous to the extended major histocompatibility complex (25, 26) and Teng et al. identified a gene encoding a tapasin-like molecule (27). Studies of the encoded protein languished until Boyle et al. (28) demonstrated an interaction between TAPBPR and MHC I, independent of other components of the PLC. Further studies not only confirmed the potential for MHC I association, but also established both chaperone and catalytic activities of TAPBPR that mimicked tapasin. Although the precise biological necessity for TAPBPR remains unclear (29), some novel functions, including control of trafficking to the UDP-glucose:glycoprotein glucosyltransferase quality control pathway have been observed (30). Additionally, TAPBPR interactions with MHC I are quantitatively dependent on the glycosylation status of the MHC I molecule (31, 32). In addition, TAPBPR distinguishes different MHC molecules based on their polymorphism (33, 34). Recently, exploitation of the catalytic peptide exchange functions of TAPBPR have given rise to new technologies facilitating the production of recombinant MHC I molecules (35–37).
Structural studies of TAPBPR have offered insight not only into its own function, but also to that of the tapasin homolog (38–40). Initial low resolution small angle X-ray scattering analysis (34) comparing recombinant tapasin with TAPBPR revealed their structural similarity as predicted by their shared amino acid sequences (41, 42).
These results with TAPBPR suggested that higher resolution structure determination of TAPBPR might offer further insight into the mechanism by which TAPBPR, and by inference tapasin, function in their dual roles as chaperones and catalysts. Two reports of X-ray structures of MHC I/TAPBPR complexes were reported at the same time—one of a complex of the mouse MHC I molecule H2-Db complexed with human TAPBPR (39), and another of the mouse H2-Dd with human TAPBPR (38). The models derived in both laboratories are remarkably similar (rmsd for the superposition of the TAPBPR/MHC I/β2m complexes was 1.158 Å for 3385 atoms). The H2-Db complex was generated with H2-Db emptied of a labile peptide by photolysis, and the H2-Dd complex was generated with a covalently-linked truncated peptide. Nevertheless, in both structures, no peptide was visualized. (For the covalent peptide/H2-Dd complex, it is presumed that this is due to structural heterogeneity or mobility of the peptide moiety.) The two structures were in remarkable agreement, with the exception that a peptide loop representing residues K22-D35 of TAPBPR was modeled for the H2-Db complex, while in the absence of reliable electron density in this region, no model was built for the H2-Dd/TAPBPR complex (38). Critical assessment of whether there is solid evidence for such a loop has been presented elsewhere (43, 44). It is also relevant to consider the alignment of a selection of TAPBPR sequences from several species as compared with those of tapasin (Figure 3). Notably, the K22 -D35 loop is significantly longer in all TAPBPR molecules as compared with tapasin (labelled here, TAPBP) molecules.
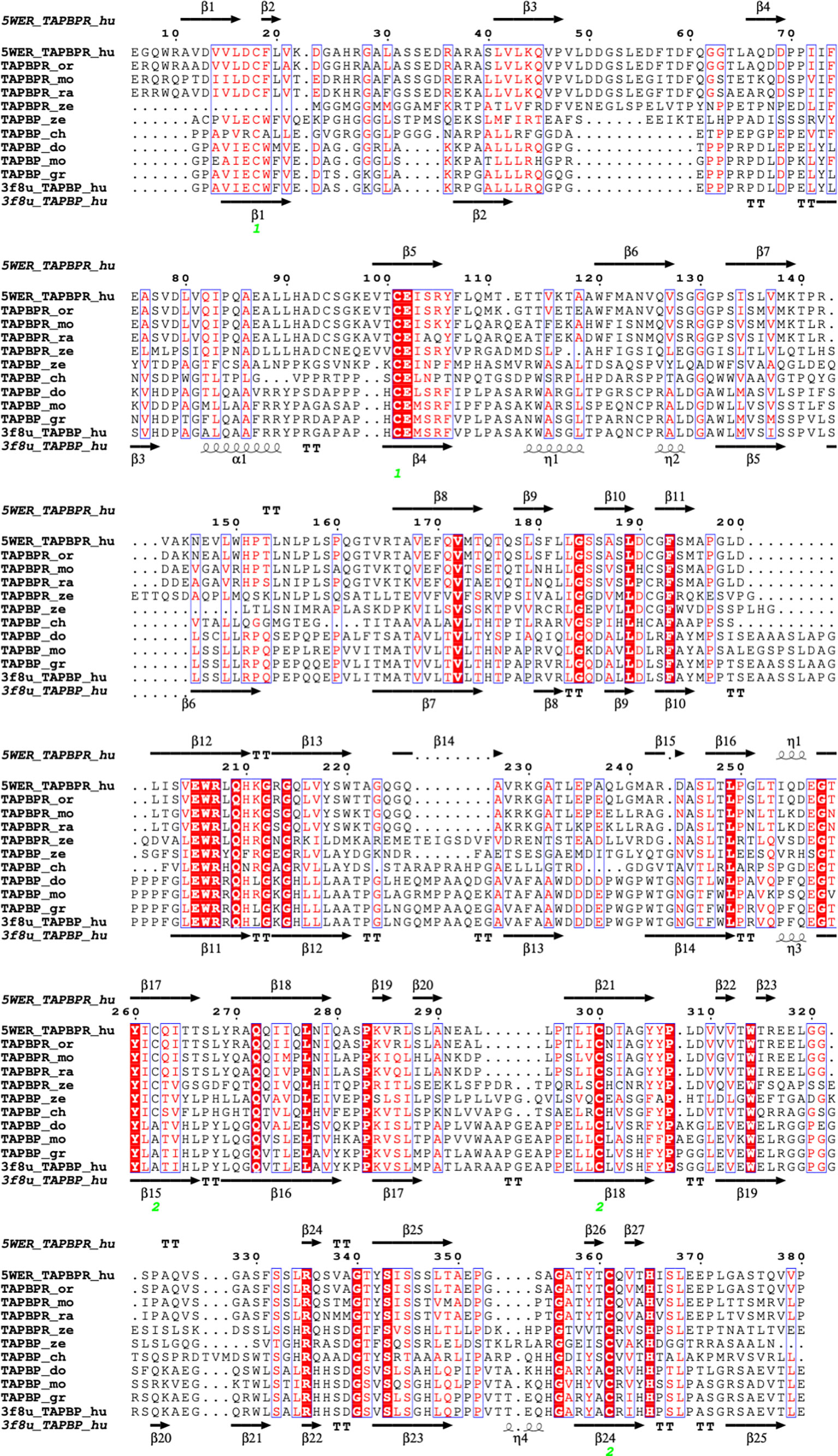
Figure 3 Structure based amino acid sequence alignment of TAPBPR and tapasin (TAPBP) sequences from several different species. Sequences were aligned with ClustalW in MacVector 18.0 and further illustrated with ESPript on the ENDscript server (57). Top and bottom sequences and secondary structure elements are taken from TAPBPR (5WER) and tapasin (TAPBP) (3F8U) respectively. Sequences of TAPBPR_or (orangutan-Pongo abellii-Q5R8H1), TAPBPR_mo (mouse-Mus musculus-Q8VD31), TAPBPR_ra (rat-Rattus novegicus-D4A6L1), and TAPBPR_ze (zebrafish-Danio rerio-X1WBD6), as well as TAPBP_gr (gorilla-Chlorocebus aethiops-Q6PZD2), TAPBP_do (dog-Canis lupus familiaris-Q5TJE4), TAPBP_ch (chicken-Gallus gallus-Q9R233), and TAPBP_ze (zebrafish-Danio rerio-Q1LUU3) are provided.
Indeed, several experimental lines of indirect evidence suggest a competitive role for this loop in protecting the peptide binding groove of the MHC during the process of binding and folding. These include mutational analyses of the loop in TAPBPR (47, 48) and structural studies of truncated peptides complexed with MHC molecules (49). However, more recent nuclear magnetic resonance studies suggest that the TAPBPR loop functions dynamically, forming a lid that modulates the access of peptides to the peptide binding groove (50–52). Other interactions of TAPBPR with the exterior aspects of the peptide binding domains of the MHC I α1 and α2 domains as well as the interaction of the membrane proximal IgC-like domain of TAPBPR with the membrane proximal α3/β2m unit of the MHC I molecule are evidence of a global disruption of the peptide binding groove (16, 38, 43).
These experimental findings on TAPBPR are complemented by molecular dynamics simulations of tapasin (44), MHC I molecules (53–55), and of a model of the entire PLC (56). These studies indicate that the chaperone/catalysts exhibit considerable flexibility to accommodate the structural plasticity of a wide range of peptide/MHC I complexes. As function is embodied in structure, amino acid sequence relationships over evolutionary time may be expected to reveal regions of tapasin or nTAPBPR that are conserved because of conserved function. In Figure 4 we display the surface of X-ray structures of tapasin (Figure 4A) and TAPBPR (Figure 4B) colored according to their evolutionary variability as calculated with Consurf (45, 46). Although considerable variability may be noted, high degrees of conservation are observed in the region of the amino terminal domain of tapasin/TAPBPR that contacts MHC I (Figure 4C), as well as in the membrane proximal IgC domain of these molecules.
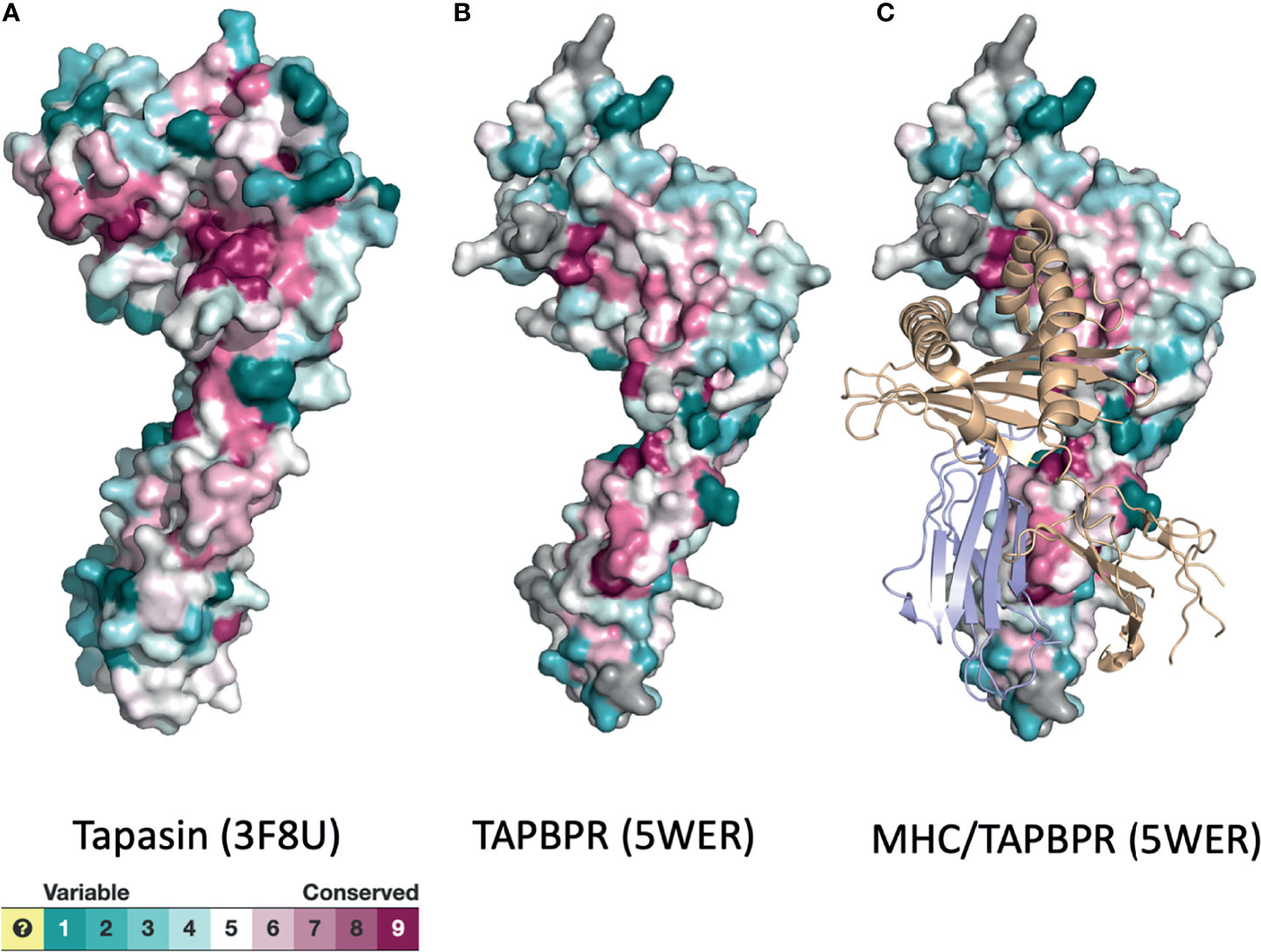
Figure 4 Surface representations of tapasin (A), TAPBPR (B), and MHC/TAPBPR complex (C) reveal regions of conservation and variability. PDB models of tapasin (3F8U) and TAPBPR (5WER) were submitted to the Consurf server (45, 46) to assess the evolutionary conservation of the amino acids of the indicated molecules. Color scheme for the degree of variability is shown.
Structure Predictions Across Species
The recent success of the application of AlphaFold2 to protein structure prediction (8, 58, 59) compels us to exploit this powerful approach to explore likely three-dimensional structures of several additional tapasin and TAPBPR molecules. In Figure 5, we display the experimentally determined models of human TAPBPR (Figure 5A) and human tapasin (Figure 5G) as compared with computationally derived models for h-TAPBPR (Figure 5B) and h-tapasin (Figure 5H) along with examples from other species (TAPBPR - 5C Pongo abelli (Sumatran orangutan); 5D Rattus novegicus; 5E Mus musculus; and 5F Danio rerio). Tapasin comparisons are shown as – 5I Chlorcebus aethiops (green monkey); 5J Rattus novegicus; 5K Mus musculus; and 5L Danio rerio. The overall structures as expected are remarkably similar, revealing the overall N terminal and IgV domain (from the N terminus to TAPBPR residue 278) and the distinctive C terminal IgC domain (TAPBPR residue 279 to C terminus). Structural distinctions are evident in the amino acid sequence alignment of Figure 3. The K22-D35 loop of TAPBPR (as compared to the homologous D12-L18 loop of tapasin) not observed well in the TAPBPR electron density map, is modeled by AlphaFold2 as highlighted by the red dashed oval in Figures 5A–F. Residues T106-K111, another region of poor electron density in h-TAPBPR, is modeled as α-helix by AlphaFold2 (Figures 5B–F).
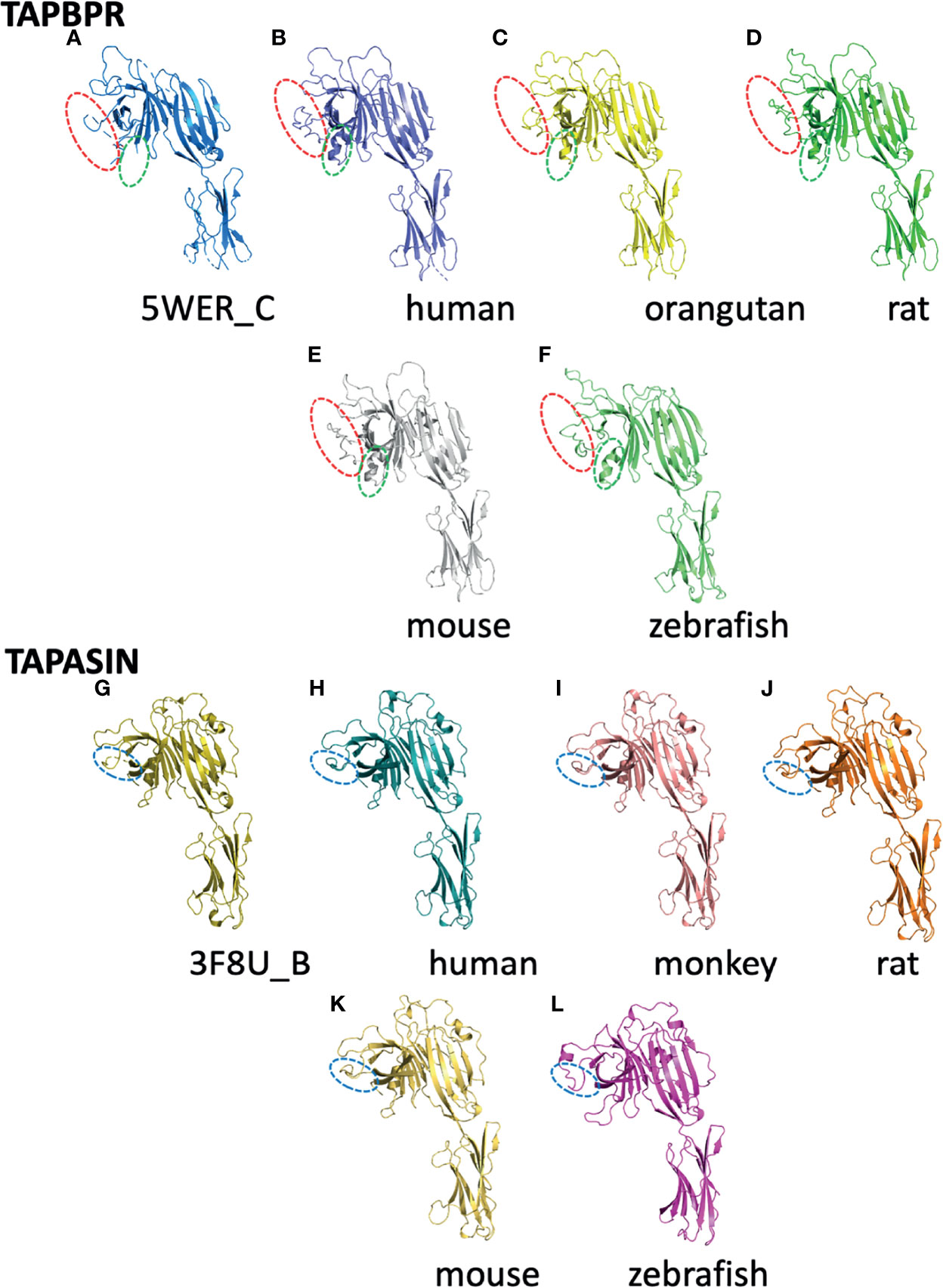
Figure 5 Structures predicted by AlphaFold2 preserve features of experimentally derived structures. The AlphaFold2 Protein Structure Database (https://alphafold.ebi.ac.uk) was queried for TAPBPL (alternate designation for TAPBPR) or TAPBP (for tapasin) and the resulting structural models were superposed on experimentally determined structures for TAPBPR (5WER chain C) (A) and tapasin (3F8U chain B) (G) as indicated. (B) represents the predicted human TAPBPR. (C) Pongo abelii (Sumatran orangutan). (D) Rattus norvegicus. (E) Mus musculus. (F) Danio rerio. Panels (G–L) represent tapasin (TAPBP) structure (3F8U chain B) (G), and models of human (H), Chlorocebus aethiops (I), Rattus novegicus (J), Mus musculus (K), and Danio rerio (L). As described in the text, loops that were not modeled based on X-ray data but were modeled by AlphaFold2 are indicated by dashed ovals.
The Alphafold2 analysis of several tapasin structures reveals modeling for tapasin residues L26-R37 (aligned with L38 to R59 of TAPBPR, Figure 4), which was not built into the original tapasin structure (3F8U) because of poor density. AlphaFold2 also recognizes a conserved α-helix (A83 to T91), unique to tapasin, distinct from the corresponding short loop (Q105-T108) of TAPBPR a region poorly defined in TAPBPR. The conserved loop E72-G101 of tapasin is longer than E98-A114 of TAPBPR (see comparison in Figure 6).
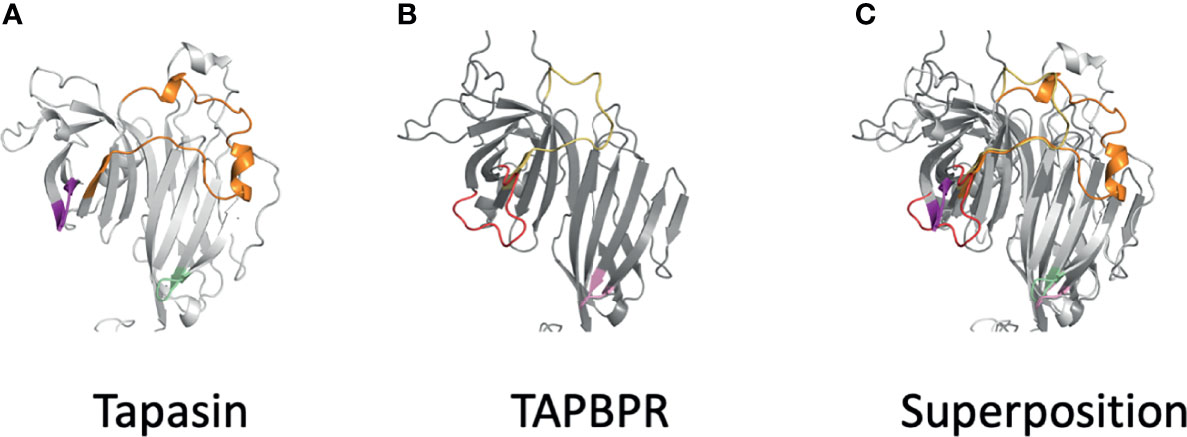
Figure 6 Differences of AlphaFold2 modeled loops of tapasin and TAPBPR suggest distinct details of MHC interactions. (A) Loops of tapasin (E12-L18, magenta), (E72-G101), and (Q189-193, pale green) are shown next to (B) corresponding loops of TAPBPR (D23-D35, red), (E102-A118, yellow), and (Q209-R213). Superposition the two models is shown in (C).
Viewing the Future
The importance of understanding the mechanistic details of peptide loading of MHC I molecules, not only with respect to the obvious applied utility of appreciating the cellular evolution of immunevasion in tumorigenesis (60–62), but also with respect to appreciating the interplay between basic aspects of the protein folding problem, peptide loading, and structure prediction cannot be overemphasized. In this brief review we highlight how structural information—derived both experimentally and computationally—complements our understanding of fundamental aspects of immune function. With improved experimental methods [crystallographic, electron microscopic (both cryogenic and tomographic)], expansion of available sequence and structural databases, and remarkable advances in artificial intelligence and computational approaches, we may anticipate not only a host of solutions to vexing, long-standing questions, but may even look forward to deeper and more exciting questions that result from this enlightenment.
Author Contributions
DM, DT, JJ, LB, JA, MM and KN conceived the concepts and outlined the paper. DM, DT, JJ, LB, and KN designed and prepared figures. All authors wrote and edited the manuscript. All authors contribute to the article and approved the submitted version.
Funding
This work was supported by the Intramural Research Program of the NIH.
Conflict of Interest
The reviewer TJ declared a past co-authorship with one of the authors JJ.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We apologize in advance to colleagues whose work was overlooked because of length limitations or by our own ignorance.
References
1. Anfinsen CB. Principles That Govern the Folding of Protein Chains. Science (1973) 181:223–30. doi: 10.1126/science.181.4096.223
2. Anfinsen CB, Haber E, Sela M, White FH Jr. The Kinetics of Formation of Native Ribonuclease During Oxidation of the Reduced Polypeptide Chain. Proc Natl Acad Sci USA (1961) 47:1309–14. doi: 10.1073/pnas.47.9.1309
3. Levinthal C. Are There Pathways for Protein Folding. J Chim Phys Pcb (1968) 65:44–+. doi: 10.1051/jcp/1968650044
4. Levinthal C. How to Fold Graciously. Univ Illinois Bulletin Mössbaur Spectrosc Biol Syst Proc (1969) 67:22–4.
5. Wetlaufer DB. Nucleation in Protein Folding–Confusion of Structure and Process. Trends Biochem Sci (1990) 15:414–5. doi: 10.1016/0968-0004(90)90275-g
6. Ptitsyn OB. Protein Folding and Protein Evolution: Common Folding Nucleus in Different Subfamilies of C-Type Cytochromes? J Mol Biol (1998) 278:655–66. doi: 10.1006/jmbi.1997.1620
7. Chan HS, Dill KA. Protein Folding in the Landscape Perspective: Chevron Plots and Non-Arrhenius Kinetics. Proteins (1998) 30:2–33. doi: 10.1002/(sici)1097-0134(19980101)30:1<2::aid-prot2>3.0.co;2-r
8. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly Accurate Protein Structure Prediction With AlphaFold. Nature (2021) 596:583–9. doi: 10.1038/s41586-021-03819-2
9. Donne J. In The Works of John Donne Vol. III (ed H. Alford) 574-575 (John W. Parker, 1839, 1635).
10. Blum JS, Wearsch PA, Cresswell P. Pathways of Antigen Processing. Annu Rev Immunol (2013) 31:443–73. doi: 10.1146/annurev-immunol-032712-095910
11. Germain RN, Margulies DH. The Biochemistry and Cell Biology of Antigen Processing and Presentation. Annu Rev Immunol (1993) 11:403–50. doi: 10.1146/annurev.iy.11.040193.002155
12. Rock KL, Reits E, Neefjes J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol (2016) 37:724–37. doi: 10.1016/j.it.2016.08.010
13. Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD-IMGT/HLA Database. Nucleic Acids Res (2020) 48:D948–55. doi: 10.1093/nar/gkz950
14. Wearsch PA, Peaper DR, Cresswell P. Essential Glycan-Dependent Interactions Optimize MHC Class I Peptide Loading. Proc Natl Acad Sci USA (2011) 108:4950–5. doi: 10.1073/pnas.1102524108
15. Zhang W, Wearsch PA, Zhu Y, Leonhardt RM, Cresswell P. A Role for UDP-Glucose Glycoprotein Glucosyltransferase in Expression and Quality Control of MHC Class I Molecules. Proc Natl Acad Sci USA (2011) 108:4956–61. doi: 10.1073/pnas.1102527108
16. Natarajan K, Jiang J, Margulies DH. Structural Aspects of Chaperone-Mediated Peptide Loading in the MHC-I Antigen Presentation Pathway. Crit Rev Biochem Mol Biol (2019) 54:164–73. doi: 10.1080/10409238.2019.1610352
17. Dong G, Wearsch PA, Peaper DR, Cresswell P, Reinisch KM. Insights Into MHC Class I Peptide Loading From the Structure of the Tapasin-ERp57 Thiol Oxidoreductase Heterodimer. Immunity (2009) 30:21–32. doi: 10.1016/j.immuni.2008.10.018
18. Garstka MA, Fritzsche S, Lenart I, Hein Z, Jankevicius G, Boyle LH, et al. Tapasin Dependence of Major Histocompatibility Complex Class I Molecules Correlates With Their Conformational Flexibility. FASEB J (2011) 25:3989–98. doi: 10.1096/fj.11-190249
19. Simone LC, Georgesen CJ, Simone PD, Wang X, Solheim JC. Productive Association Between MHC Class I and Tapasin Requires the Tapasin Transmembrane/Cytosolic Region and the Tapasin C-Terminal Ig-Like Domain. Mol Immunol (2012) 49:628–39. doi: 10.1016/j.molimm.2011.11.002
20. Turnquist HR, Vargas SE, Schenk EL, McIlhaney MM, Reber AJ, Solheim JC. The Interface Between Tapasin and MHC Class I: Identification of Amino Acid Residues in Both Proteins That Influence Their Interaction. Immunol Res (2002) 25:261–9. doi: 10.1385/ir:25:3:261
21. Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC Class I Peptide Cargo Is Dependent on Tapasin. Immunity (2002) 16:509–20. doi: 10.1016/s1074-7613(02)00304-7
22. Yu YY, Turnquist HR, Myers NB, Balendiran GK, Hansen TH, Solheim tJC. An Extensive Region of an MHC Class I Alpha 2 Domain Loop Influences Interaction With the Assembly Complex. J Immunol (1999) 163:4427–33.
23. Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, et al. Structure of the Human MHC-I Peptide-Loading Complex. Nature (2017) 551:525–8. doi: 10.1038/nature24627
24. Panter MS, Jain A, Leonhardt RM, Ha T, Cresswell P. Dynamics of Major Histocompatibility Complex Class I Association With the Human Peptide-Loading Complex. J Biol Chem (2012) 287:31172–84. doi: 10.1074/jbc.M112.387704
25. Kasahara M. The Chromosomal Duplication Model of the Major Histocompatibility Complex. Immunol Rev (1999) 167:17–32. doi: 10.1111/j.1600-065x.1999.tb01379.x
26. Katsanis N, Fitzgibbon J, Fisher EM. Paralogy Mapping: Identification of a Region in the Human MHC Triplicated Onto Human Chromosomes 1 and 9 Allows the Prediction and Isolation of Novel PBX and NOTCH Loci. Genomics (1996) 35:101–8. doi: 10.1006/geno.1996.0328
27. Teng MS, Stephens R, Du Pasquier L, Freeman T, Lindquist JA, Trowsdale J. A Human TAPBP (TAPASIN)-Related Gene, TAPBP-R. Eur J Immunol (2002) 32:1059–68. doi: 10.1002/1521-4141(200204)32:4<1059::AID-IMMU1059>3.0.CO;2-G
28. Boyle LH, Hermann C, Boname JM, Porter KM, Patel PA, Burr ML, et al. Tapasin-Related Protein TAPBPR Is an Additional Component of the MHC Class I Presentation Pathway. Proc Natl Acad Sci USA (2013) 110:3465–70. doi: 10.1073/pnas.1222342110
29. Hafstrand I, Aflalo A, Boyle LH. Why TAPBPR? Implications of an Additional Player in MHC Class I Peptide Presentation. Curr Opin Immunol (2021) 70:90–4. doi: 10.1016/j.coi.2021.04.011
30. Neerincx A, Hermann C, Antrobus R, van Hateren A, Cao H, Trautwein N, et al. TAPBPR Bridges UDP-Glucose:Glycoprotein Glucosyltransferase 1 Onto MHC Class I to Provide Quality Control in the Antigen Presentation Pathway. Elife (2017) 6. doi: 10.7554/eLife.23049
31. Ilca FT, Boyle LH. The Glycosylation Status of MHC Class I Molecules Impacts Their Interactions With TAPBPR. Mol Immunol (2021) 139:168–76. doi: 10.1016/j.molimm.2021.09.007
32. Neerincx A, Boyle LH. Preferential Interaction of MHC Class I With TAPBPR in the Absence of Glycosylation. Mol Immunol (2019) 113:58–66. doi: 10.1016/j.molimm.2018.06.269
33. Ilca FT, Drexhage LZ, Brewin G, Peacock S, Boyle LH. Distinct Polymorphisms in HLA Class I Molecules Govern Their Susceptibility to Peptide Editing by TAPBPR. Cell Rep (2019) 29:1621–32.e1623. doi: 10.1016/j.celrep.2019.09.074
34. Morozov GI, Zhao H, Mage MG, Boyd LF, Jiang J, Dolan MA, et al. Interaction of TAPBPR, a Tapasin Homolog, With MHC-I Molecules Promotes Peptide Editing. Proc Natl Acad Sci USA (2016) 113:E1006–15. doi: 10.1073/pnas.1519894113
35. Ilca T, Boyle LH. The Ins and Outs of TAPBPR. Curr Opin Immunol (2020) 64:146–51. doi: 10.1016/j.coi.2020.06.004
36. O'Rourke SM, Morozov GI, Roberts JT, Barb AW, Sgourakis NG. Production of Soluble pMHC-I Molecules in Mammalian Cells Using the Molecular Chaperone TAPBPR. Protein Eng Des Sel (2019) 32:525–32. doi: 10.1093/protein/gzaa015
37. Overall SA, Toor JS, Hao S, Yarmarkovich M, Sara MOR, Morozov GI, et al. High Throughput pMHC-I Tetramer Library Production Using Chaperone-Mediated Peptide Exchange. Nat Commun (2020) 11:1909. doi: 10.1038/s41467-020-15710-1
38. Jiang J, Natarajan K, Boyd LF, Morozov GI, Mage MG, Margulies DH. Crystal Structure of a TAPBPR-MHC I Complex Reveals the Mechanism of Peptide Editing in Antigen Presentation. Science (2017) 358:1064–8. doi: 10.1126/science.aao5154
39. Thomas C, Tampe R. Structure of the TAPBPR-MHC I Complex Defines the Mechanism of Peptide Loading and Editing. Science (2017) 358:1060–4. doi: 10.1126/science.aao6001
40. Thomas C, Tampe R. MHC I Assembly and Peptide Editing - Chaperones, Clients, and Molecular Plasticity in Immunity. Curr Opin Immunol (2021) 70:48–56. doi: 10.1016/j.coi.2021.02.004
41. Hermann C, van Hateren A, Trautwein N, Neerincx A, Duriez PJ, Stevanovic S, et al. TAPBPR Alters MHC Class I Peptide Presentation by Functioning as a Peptide Exchange Catalyst. Elife (2015) 4. doi: 10.7554/eLife.09617
42. Neerincx A, Boyle LH. Properties of the Tapasin Homologue TAPBPR. Curr Opin Immunol (2017) 46:97–102. doi: 10.1016/j.coi.2017.04.008
43. Margulies DH, Jiang J, Natarajan K. Structural and Dynamic Studies of TAPBPR and Tapasin Reveal the Mechanism of Peptide Loading of MHC-I Molecules. Curr Opin Immunol (2020) 64:71–9. doi: 10.1016/j.coi.2020.04.004
44. van Hateren A, Elliott T. The Role of MHC I Protein Dynamics in Tapasin and TAPBPR-Assisted Immunopeptidome Editing. Curr Opin Immunol (2021) 70:138–43. doi: 10.1016/j.coi.2021.06.016
45. Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, et al. ConSurf 2016: An Improved Methodology to Estimate and Visualize Evolutionary Conservation in Macromolecules. Nucleic Acids Res (2016) 44:W344–50. doi: 10.1093/nar/gkw408
46. Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, et al. ConSurf 2005: The Projection of Evolutionary Conservation Scores of Residues on Protein Structures. Nucleic Acids Res (2005) 33:W299–302. doi: 10.1093/nar/gki370
47. Ilca FT, Neerincx A, Hermann C, Marcu A, Stevanovic S, Deane JE, et al. TAPBPR Mediates Peptide Dissociation From MHC Class I Using a Leucine Lever. Elife (2018) 7. doi: 10.7554/eLife.40126
48. Sagert L, Hennig F, Thomas C, Tampe R. A Loop Structure Allows TAPBPR to Exert Its Dual Function as MHC I Chaperone and Peptide Editor. Elife (2020) 9. doi: 10.7554/eLife.55326
49. Hafstrand I, Sayitoglu EC, Apavaloaei A, Josey BJ, Sun R, Han X, et al. Successive Crystal Structure Snapshots Suggest the Basis for MHC Class I Peptide Loading and Editing by Tapasin. Proc Natl Acad Sci USA (2019) 116:5055–60. doi: 10.1073/pnas.1807656116
50. McShan AC, Devlin CA, Morozov GI, Overall SA, Moschidi D, Akella N, et al. TAPBPR Promotes Antigen Loading on MHC-I Molecules Using a Peptide Trap. Nat Commun (2021) 12:3174. doi: 10.1038/s41467-021-23225-6
51. McShan AC, Devlin CA, Overall SA, Park J, Toor JS, Moschidi D, et al. Molecular Determinants of Chaperone Interactions on MHC-I for Folding and Antigen Repertoire Selection. Proc Natl Acad Sci USA (2019) 116:25602–13. doi: 10.1073/pnas.1915562116
52. McShan AC, Natarajan K, Kumirov VK, Flores-Solis D, Jiang J, Badstubner M, et al. Peptide Exchange on MHC-I by TAPBPR Is Driven by a Negative Allostery Release Cycle. Nat Chem Biol (2018) 14:811–20. doi: 10.1038/s41589-018-0096-2
53. Abualrous ET, Saini SK, Ramnarayan VR, Ilca FT, Zacharias M, Springer S. The Carboxy Terminus of the Ligand Peptide Determines the Stability of the MHC Class I Molecule H-2kb: A Combined Molecular Dynamics and Experimental Study. PloS One (2015) 10:e0135421. doi: 10.1371/journal.pone.0135421
54. Sieker F, Springer S, Zacharias M. Comparative Molecular Dynamics Analysis of Tapasin-Dependent and -Independent MHC Class I Alleles. Protein Sci (2007) 16:299–308. doi: 10.1110/ps.062568407
55. Zacharias M, Springer S. Conformational Flexibility of the MHC Class I Alpha1-Alpha2 Domain in Peptide Bound and Free States: A Molecular Dynamics Simulation Study. Biophys J (2004) 87:2203–14. doi: 10.1529/biophysj.104.044743
56. Fisette O, Schroder GF, Schafer LV. Atomistic Structure and Dynamics of the Human MHC-I Peptide-Loading Complex. Proc Natl Acad Sci USA (2020) 117:20597–606. doi: 10.1073/pnas.2004445117
57. Robert X, Gouet P. Deciphering Key Features in Protein Structures With the New ENDscript Server. Nucleic Acids Res (2014) 42:W320–4. doi: 10.1093/nar/gku316
58. Jones DT, Thornton JM. The Impact of AlphaFold2 One Year on. Nat Methods (2022) 19:15–20. doi: 10.1038/s41592-021-01365-3
59. Castelvecchi D. DeepMind AI Tackles One of Chemistry's Most Valuable Techniques. Nature (2021) 600:371. doi: 10.1038/d41586-021-03697-8
60. Belicha-Villanueva A, McEvoy S, Cycon K, Ferrone S, Gollnick SO, Bangia N. Differential Contribution of TAP and Tapasin to HLA Class I Antigen Expression. Immunology (2008) 124:112–20. doi: 10.1111/j.1365-2567.2007.02746.x
61. Maggs L, Sadagopan A, Moghaddam AS, Ferrone S. HLA Class I Antigen Processing Machinery Defects in Antitumor Immunity and Immunotherapy. Trends Cancer (2021) 7:1089–101. doi: 10.1016/j.trecan.2021.07.006
Keywords: Antigen presentation, peptide loading complex (PLC), tapasin, TAP binding protein, related (TAPBPR), major histocompatibility complex (MHC), protein folding, structural immunology
Citation: Margulies DH, Taylor DK, Jiang J, Boyd LF, Ahmad J, Mage MG and Natarajan K (2022) Chaperones and Catalysts: How Antigen Presentation Pathways Cope With Biological Necessity. Front. Immunol. 13:859782. doi: 10.3389/fimmu.2022.859782
Received: 21 January 2022; Accepted: 11 March 2022;
Published: 07 April 2022.
Edited by:
Qihui Wang, Institute of Microbiology (CAS), ChinaReviewed by:
Malcolm John Wyness Sim, University of Oxford, United KingdomTengchuan Jin, University of Science and Technology of China, China
Copyright © 2022 Margulies, Taylor, Jiang, Boyd, Ahmad, Mage and Natarajan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David H. Margulies, ZG1hcmd1bGllc0BuaWFpZC5uaWguZ292
 David H. Margulies
David H. Margulies Daniel K. Taylor
Daniel K. Taylor Jiansheng Jiang
Jiansheng Jiang Lisa F. Boyd
Lisa F. Boyd Javeed Ahmad
Javeed Ahmad Kannan Natarajan
Kannan Natarajan